Abstract
The genome of the prolate-headed lytic lactococcal bacteriophage c2 is organized into two divergently oriented blocks consisting of the early genes and the late genes. These blocks are separated by the noncoding origin of DNA replication. We examined the functional role of transcription of the origin in a plasmid model system. Deletion of the early promoter PE1 abolished origin function. Introduction of mutations into PE1 which did not eliminate promoter activity or replacement of PE1 with an unrelated but functional promoter did not abolish replication. The A-T-rich region upstream of PE1, which is conserved in prolate phages, was not required for plasmid replication. Replacement of the PE1 transcript template sequence with an unrelated sequence with a similar G+C content abolished replication, showing that the sequence encoding the transcript is essential for origin function. Truncated transcript and internal deletion constructs did not support replication except when the deletion was at the very 3′ end of the DNA sequence coding for the transcript. The PE1 transcript could be detected for all replication-proficient constructs. Recloning in a plasmid vector allowed detection of PE1 transcripts from some fragments that did not support replication, indicating that stability of the transcript alone was not sufficient for replication. The data suggest that production of a transcript of a specific length and with a specific sequence or structure is essential for the function of the phage c2 origin in this model system.
Bacterial strains of the genus Lactococcus are widely used in the manufacture of cultured milk products. Bacteriophage attack of lactococcal starter strains may result in the lysis of susceptible strains and therefore in fermentation failure. Three genetically unrelated groups of lactococcal phages are responsible for most fermentation failures (23). These phages conform to one of two morphotypes: the prolate-headed c2 species and the isometric-headed 936 and P335 species (24). There are no known temperate phages in the c2 and 936 species groups, whereas the P335 species contains both temperate and lytic phages (24). Recently, the genomes of the small isometric 936 group phages sk1 (8) and bIL170 (11), the small isometric P335 group phages r1t (46), TP901-1 (4), Tuc2009 (GenBank accession no. AF109874), bIL285, bIL286, bIL309 (9), and BK5-T (3, 32), and the prolate-headed phages c2 (29) and bIL67 (43) have been sequenced and analyzed. Although there is a wealth of information at the nucleotide sequence level for these lactococcal phages, our current understanding of some aspects of phage biology, such as the replication of the phage genome, is relatively poor.
The 22,163-bp double-stranded linear DNA genome of the prolate-headed phage c2 contains 39 open reading frames (ORFs), which are organized in two divergently oriented blocks consisting of the early genes and the late genes (29). These blocks are separated by a 611-bp noncoding region that contains the origin of replication (ori) in Lactococcus lactis (47). A 521-bp ori fragment, which includes early promoter 1 (PE1) and late promoter 1 (PL1), was shown to support plasmid replication (pVA891-ori) in L. lactis in the absence of phage proteins (47), whereas a 261-bp subfragment (including the conserved sequence upstream of PE1 but not the sequence downstream of PE1) did not support plasmid replication. Furthermore, it has recently been demonstrated that phage c2 replicates via theta replication initiated at the ori region (5).
Following infection of an L. lactis cell by phage c2, three transcripts in the size range from 260 to 360 nucleotides (nt) are made from the PE1 promoter (28). The level of transcripts made increases during the course of c2 infection (28). Sequence analysis has suggested that these transcripts are not translated, and Lubbers et al. (28) speculated that they might be involved in DNA replication, although no supporting evidence was available. The region upstream of PE1 is highly A-T rich (78%) and consists of several inverted and direct repeats. It is also highly conserved in the closely related prolate phages bIL67 and φ197 (47), in contrast to the region downstream of PE1, which is not conserved. It has recently been shown that a large panel of lactococcal prolate phage could be divided into three distinct groups based on the sequence of the PE1 transcript (40). All three ori types supported plasmid replication.
Transcription in the origin is essential for replication of numerous phages and plasmids (reviewed in reference 13), but the mechanisms involved vary widely. In coliphage λ, an ori transcript is formed, which does not act as a primer for replication (18). The RNA polymerase β subunit also serves as a contact site for DnaA to act as a transcription activator at pR and thus stimulates transcription-mediated activation of ori λ (44). In phage T4, the transcript made from the middle-modeorigin promoter forms a persistent DNA-RNA hybrid within ori(uvsY) and is required for initiation of replication from ori(uvsY) (6). Initiation of replication of plasmid ColE1 requires synthesis of a transcript by RNA polymerase (35). This transcript assumes a particular conformation (33, 34) and then forms a persistent hybrid with the template DNA near the replication origin. The hybridized RNA may be cleaved by RNase H to act as a primer. In the absence of RNase H, the unprocessed transcript can act directly as a primer or indirectly by displacing the nontranscribed DNA strand (12).
Little is known about mechanisms of lactococcal prolate phage DNA replication, and only slightly more is known about the mechanisms in other lactococcal phage species. A P335 species DNA replication module was identified in the temperate phage Tuc2009, which encoded putative single-stranded DNA binding proteins, a topoisomerase I, a methylase, and a replisome organizer protein (36). The putative replisome organizer protein (Rep2009) has been shown in gel retardation assays to bind to ori2009 (36). In another temperate phage, TP901-1, a single-stranded DNA binding protein and a putative replication initiation protein, which are essential for in vivo phage replication, have been identified (37). P335 phages require phage-encoded proteins for DNA replication. This was demonstrated by the ability of cloned phage ori fragments to bind and presumably titrate out proteins essential for phage replication, thus conferring the Per phenotype (phage-encoded resistance; φ50, φ31, BK5-T, TP901-1, and Tuc2009) (20, 32, 36, 37, 38). Phage c2 ori does not confer a Per phenotype (39), suggesting that DNA replication of this phage relies on a cis-acting origin-encoded product and/or host-encoded proteins.
It has been shown that transcription of a divergent region downstream of PE1 occurs in the three groups of lactococcal prolate phages into which our collection was subdivided (40). Here we demonstrate that production of the PE1 transcript is required for phage c2 origin function in a plasmid model system. The sequence encoding the transcript produced from PE1 was essential for the ability to support plasmid replication, and small internal deletions were not tolerated. The data suggest that the transcript probably forms a secondary structure, which is required for its role in replication.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The following bacterial host strains were used in this study. Escherichia coli ER2206 [endA1 thi1 supE44 mcr67 (mcrA) (mcrBC-hsdRMS-mrr)114::IS10 (lac)U169/F′ proAB laqIq Z M15 Tn10] was obtained from New England Biolabs, Beverley, Mass. L. lactis MG1363 is a plasmid-free, prophage-cured derivative of NCDO712 (17). L. lactis NZ9000 is a derivative of strain MG1363 that does not produce nisin and has the nirR-nisK genes integrated in the pepN gene in the chromosome (27). L. lactis NZ9800 is a derivative of strain NZ9700 that does not produce nisin (26). L. lactis was grown at 30°C without aeration in M17 medium supplemented with 0.5% (wt/vol) glucose (45). When required, 5 μg of erythromycin per ml or 5 μg of chloramphenicol per ml was added to the medium. E. coli was grown at 37°C in Luria-Bertani medium (41). E. coli transformants were plated on medium containing 150 μg of erythromycin per ml, 25 μg of chloramphenicol per ml, or 200 μg of ampicillin per ml as appropriate, 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml, and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). For solid media, 15 g of agar per liter was added. Nisin (nisaplin; Aplin and Barrett Ltd., Beaminster, Dorset, England) was added to induce the nisA promoter at a concentration of 150 ng/ml for strains NZ9000 and NZ9800.
DNA preparation and manipulation.
L. lactis was made electrocompetent by growth in the presence of glycine (2.5%) (21). Transformation of L. lactis was performed by electroporation as described previously (25). CaCl2-treated E. coli cells were transformed as described by Sambrook et al. (41). Plasmid DNA was isolated from L. lactis strains by alkaline lysis as described previously (1). Plasmid DNA was isolated from E. coli by alkaline lysis as described previously (41) or with a High Pure plasmid purification kit (Roche, Mannheim, Germany). DNA fragments were isolated from gels with a High Pure PCR product purification kit (Roche). DNA manipulations (restriction enzyme cleavage, ligation, phosphorylation, etc.) were carried out by using conditions specified by the manufacturer or by using standard protocols (41). For all PCRs, Pwo I polymerase (Roche) was used. DNA sequencing was performed by the dideoxy chain termination method (42) by the Allan Wilson Centre DNA Analysis Services, Massey University, Palmerston North, New Zealand.
Construction of plasmids.
The plasmids used or generated in this study are listed in Table S1 in the supplemental material, and the oligonucleotide primers used are listed in Table S2 in the supplemental material. Fragments cloned in recombinant plasmids were verified by DNA sequencing. PL1 was deleted by PCR by using a pLP201 template with oligonucleotides ori22 and ori7. Plasmid pLP204 was created by ligating a PCR product (template pLP201; primers ori24 and ori23) containing the PL1 promoter (coordinates 7056 to 7238 in c2) to a PCR product (template pLP201; primers ori25 and ori22) containing the sequence downstream of PE1 (coordinates 6717 to 7026 in c2). The ligation product was cloned into the EcoRI site of pVA891 (30), resulting in a recombinant ori fragment lacking the −10 and −35 hexamers and the intervening sequence of PE1. Substitutions (T to G) in the consensus −10 region of PE1 were introduced by amplifying the cloned c2 origin in two separate arms by using a primer that contained the two mutations (template pLP201; primers ori26 and ori22 and primers ori27 and ori23) and then ligating the two PCR products and cloning the fragment into the EcoRI site of pVA891 to generate pLP205. To create plasmid pUC-203, the ori fragment of pLP203 was cut out with EcoRI and cloned into the EcoRI site of pUC19. Deletion mutants of the PE1 transcript were created by progressively shortening the PE1 transcript-encoding region from the 3′ end by PCR (template pLP201; primers ori7 and ori1 through ori6) and cloning the fragments into the EcoRI site of pVA891 to generate plasmids pLP206 through pLP211.
The PE1 promoter was replaced by the inducible nisA promoter (15) by first performing an inverse PCR with the pUC19-ori plasmid, which deleted PE1 (primers ori24 and ori25). The nisA promoter was amplified by PCR (template pNZ8037 [14]; primers ori28 and ori29) to obtain a fragment containing the nisin promoter and a conserved sequence upstream of the position 1 start site (14) but not the nisA ribosome binding site. This fragment was then ligated to the inverse PCR product and cloned into the EcoRI site of pVA891 to create pLP212. An inverse PCR with pUC19-ori and primers ori9 and ori10, each containing 14 bp of the PE1 sequence in an inverted orientation, was performed, and the products were self-ligated. The ori fragment containing the inverted PE1 promoter was cut out with EcoRI and subsequently cloned in pVA891, resulting in pLP213. The PE1 transcript was replaced with a DNA fragment derived from a lactococcal proteinase gene (domain A of the prtP gene) (10) that had a similar G+C content and was a similar length. The proteinase sequence (primers ori11 and ori12; template pHP003) and the PE1 promoter (primers ori13 and ori14; template pLP203) were amplified by overlap extension PCR by using primers that overlapped in the first PCR. The second PCR (primers ori11 and ori14) was performed by using the external primers and the products of the first PCRs as templates in order to join the two sequences together. The product was cloned into the EcoRI site of pVA891 to generate pLP214. Plasmid pLP216 was created by cloning a PCR product (template pLP207; primers ori15 and ori16) into the NcoI and PstI site of plasmid pVAΩ, which contained a transcriptional terminator (Ω terminator) (16, 40). Plasmids pLP215 and pLP217 through pLP223 were made by inverse PCR by using pUC-203 as the template and primers ori30 and ori31 (pLP215), ori44 and ori45 (pLP217), ori42 and ori43 (pLP218), ori40 and ori41 (pLP219), ori38 and ori39 (pLP220), ori36 and ori37 (pLP221), ori34 and ori35 (pLP222), and ori32 and ori33 (pLP223). The ori fragments were recloned into the EcoRI site of pVA891.
Northern blot hybridization.
Total RNA from L. lactis was extracted from frozen cell pellets (−80°C) by the hot phenol method as previously described (28). Formamide-containing gel loading dye (80% deionized formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol) was added to the samples and incubated for 10 min at 75°C before the samples were separated on a 6 M urea-6% polyacrylamide gel by electrophoresis in 1× Tris-borate buffer. RNA was then electrotransferred onto a positively charged nylon membrane (pore size, 0.45 μm; Roche) at 400 mA for 1 h in 1× Tris-borate-EDTA buffer. RNA was fixed to the membrane by UV irradiation. The RNA transfer was visualized by methylene blue staining of blots (19). Hybridizations were performed by using the ECL system according to the manufacturer's instructions (Amersham Pharmacia) and PCR-generated probes.
Primer extension.
The avian myeloblastosis virus reverse transcriptase primer extension system (Promega, Madison Wis.) was used according to the manufacturer's instructions. For the analysis, 20 μg of total RNA isolated from L. lactis MG1363 and primer 6902prex were used. The sequencing reaction was performed by the dideoxy method by using an AmpliCycle sequencing kit (Perkin-Elmer, Foster City, Calif.) according to the manufacturer's instructions and 20 pmol of primer 6902prex, which had been end labeled with [γ-32P]ATP (Amersham Pharmacia Biotech, Little Chalfont, England).
RESULTS
Transcription of the sequence downstream of PE1 is required for origin function.
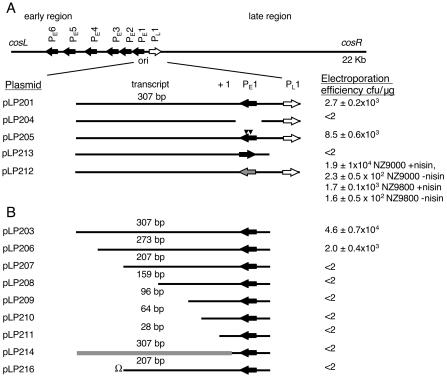
The original ori plasmid pLP201 (pVA891-ori) (30, 47) contained both PE1 and PL1 and also 307 bp of the noncoding region downstream of PE1 (Fig. 1A). This fragment supported replication of the lactococcal origin-screening vector pVA891 (47). To investigate if PL1 and the highly conserved A-T-rich region between PE1 and PL1 played any role in replication, we attempted to introduce plasmid pLP203 into L. lactis cells by electroporation. Plasmid pLP203 contains PE1 and the 307-bp noncoding sequence downstream of PE1 but not PL1 or the A-T-rich region (40) (Fig. 1A). The shortened ori fragment in pLP203 supported plasmid replication in L. lactis (Fig. 1A), with an electroporation frequency that was 1 order of magnitude higher than that for plasmid pLP201. This suggested that PE1 and the sequence downstream of PE1 are sufficient for efficient origin function and that PL1 and the A-T-rich region are not required for replication.
FIG. 1.
Schematic representation of the c2 ori locus. The positions of the origin fragment in the phage c2 genome relative to the early gene promoter (solid arrows) and the late gene promoter (open arrow) are shown at the top. Below this the c2 ori (positions 6717 to 7238 in the c2 sequence) and the derivatives of the origin fragment that were cloned in pVA891 are shown. The abilities of the various constructs (listed on the left) to replicate in L. lactis are indicated on the right as mean transformation frequencies from at least three experiments (in the case of pLP212 in the presence and in the absence of nisin); the values varied because of variations in the L. lactis transformation frequency between experiments. (A) The solid triangles indicate the position of the base pair changes in the pLP205 insert. The gray arrow represents the nisA promoter in the pLP212 insert. In pLP213 PE1 is inverted. (B) Schematic representation of c2 ori fragments cloned in pVA891 carrying shortened DNA sequences coding for the transcript made from PE1 (pLP201 to pLP211). The length of the remaining sequence of each transcript from the transcription start site is indicated above the transcript. The gray line represents the lactococcal prtP sequence replacing the c2 transcript-encoding sequence in the pLP214 insert. Ω in pLP216 represents the transcriptional and translational terminator cloned at the 3′ end of the 207-bp transcript-encoding sequence.
The sequence downstream of PE1 is very poorly conserved in prolate phages (28) and is transcribed from PE1 during phage c2 infection. To determine if active transcription was necessary for replication, we constructed several c2 ori fragments with modified PE1 promoters and cloned them into pVA891. The PE1 promoter itself (−10 and −35 promoter hexamers and the intervening sequence) was deleted by a PCR-based strategy in order to generate plasmid pLP204 (Fig. 1A). This plasmid was unable to replicate in L. lactis, as shown by our repeated and reproducible failure to electroporate it into L. lactis strain MG1363 compared with parallel controls (Fig. 1A).
The loss of ori function by deletion of PE1 in pLP204 could have been due to lack of a transcript with a necessary mechanistic role or due to DNA conformational changes in the ori fragment caused by the internal deletion. To help distinguish between these possibilities, PE1 was inverted in plasmid pLP203 to generate pLP213 (Fig. 1A). This construct was a control for conformational changes induced by gross deletion of the promoter, although promoter-inversion-related conformational changes were still possible. In this plasmid, the PE1 promoter was upstream of, and in the same orientation as, the erythromycin resistance gene of the vector. Plasmid pLP213 could not be electroporated into L. lactis cells (Fig. 1A), suggesting that mere binding by RNA polymerase to the ori fragment was not sufficient for replication and that directional promoter activity was required.
Unrelated but functional promoter plus the PE1 transcript-encoding region are sufficient for origin function.
To distinguish whether PE1 promoter activity or the PE1 sequence was required for c2 ori replication, PE1 was replaced by the inducible nisA promoter (14, 15) in order to generate pLP212 (Fig. 1A) and was transformed into L. lactis strains NZ9800 and NZ9000. Neither of these strains produces the nisin peptide (26, 27), but both of them allow regulated gene expression under the control of the inducible nisA promoter upon addition of the nisin peptide itself to the growth medium. Higher levels of expression of genes under the control of the nisin promoter are achieved in NZ9000 (27). This may be because the effective nisin level is lower in NZ9800 because of nisin binding to NisI or other nisin immunity proteins produced in this strain (27). Plasmid pLP212 was successfully introduced into both strains in the presence of nisin (Fig. 1A). The plasmid could also be introduced even without induction by nisin (Fig. 1A), conditions under which the nisA promoter activity was reduced but clearly detectable (see Fig. 4).
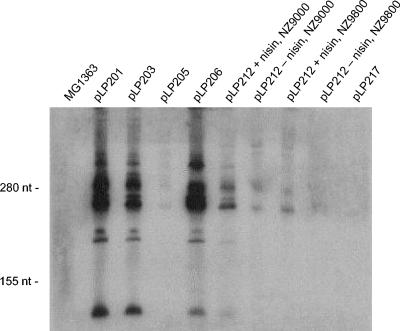
FIG. 4.
Northern hybridization analysis of the c2 ori fragments that support plasmid replication. Twenty micrograms of total RNA from L. lactis cells harboring a plasmid was electrophoresed on an acrylamide gel, blot transferred, and then probed with a PCR product (template pLP206; primers ori1 and ori7) and detected by the ECL system. MG1363, no plasmid; pLP constructs, MG1363 transformed with the plasmids indicated; NZ9000 and NZ9800, two lactococcal host strains for pLP212, in which the ori transcript is placed under control of the nisA promoter; + nisin, growth in the presence of nisaplin; − nisin, growth in the absence of nisaplin.
To further investigate the requirement for a functional promoter for replication, we created two T-to-G substitutions in the consensus −10 region of PE1 (TATAAT was mutated to TAGAAG). The substitutions were designed to eliminate or significantly reduce PE1 promoter activity with minimum impact on the DNA conformation in the origin. The fragment was cloned into pVA891 to produce pLP205 (Fig. 1A), which was assayed for plasmid replication in L. lactis MG1363. Despite the sequence changes in the PE1 promoter, the ori fragment in pLP205 supported plasmid replication.
Functional importance of the PE1 transcript length and sequence.
Waterfield et al. (47) previously showed that a 261-bp subfragment that included PE1, PL1, the conserved A-T-rich region between the two promoters, and a 48-bp PE1 transcript (starting from the position 1 nucleotide) cloned in pVA891 (previously not designated, now designated pLP202) did not support replication in L. lactis. This result suggested that the transcript had to be a particular length to support replication. To identify the minimal DNA fragment that exhibited ori activity, we generated six deletion constructs of the transcript made from PE1 by progressively shortening the DNA sequence coding for the transcript from the 3′ end. The ori fragments containing PE1 and the downstream sequence but not the late promoter (PL1) (pLP206 to pLP211) (Fig. 1B) were cloned into pVA891. The shortened 273-nt transcript (from the position 1 nucleotide) in pLP206 still supported replication, but the next-shortest truncated-transcript construct (207 nt; pLP207) did not replicate. None of the transcripts shorter than 207 nt supported replication (Fig. 1B). To eliminate the possibility of deleterious run-on transcription into the vector in the deletion mutants, we inserted a transcriptional terminator (omega terminator [20]) at the 3′ end of the transcript of pLP207 (to generate pLP216 [Fig. 1B]). The presence of the terminator did not restore replication, indicating that run-on transcription did not make the plasmid genetically unstable.
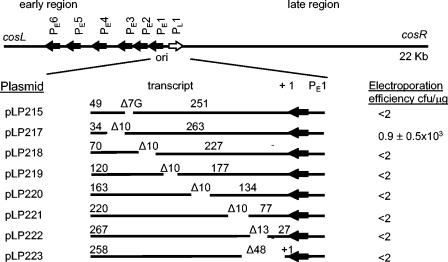
The region downstream of PE1 is not conserved among prolate phages (47). This prompted us to investigate if a particular transcript sequence, and not just a minimum transcript length, was required for replication. Thus, we replaced the PE1 transcript-encoding region with a DNA fragment that had a similar G+C content and was a similar length, which was derived from a lactococcal proteinase gene (domain A of the prtP gene) (10), and cloned it into pVA891 to generate pLP214 (Fig. 1 B). This DNA fragment was designed to encode a transcript that could not be translated, and it was cloned in the antisense direction with respect to the prtP gene upon which it was based. The resulting plasmid, pLP214, did not replicate in L. lactis, suggesting that the specific sequence or conformation of the native ori transcript was critical for its stability or function. To investigate this, scanning mutagenesis was performed with the DNA sequence coding for the transcripts made from PE1. The 10-, 13-, and 48-bp deletions used were designed to disrupt the predicted secondary structures (40) in the transcripts made from PE1 (pLP217 to pLP223). Of the seven deletion plasmids constructed, only pLP217 replicated in L. lactis (Fig. 2).
FIG. 2.
Schematic representation of plasmids used in scanning mutagenesis. The c2 ori fragments carrying deletions in the transcript-encoding sequence were cloned into pVA891 and assayed for the ability to replicate in L. lactis. The numbers above the lines indicate the sizes (in base pairs) of the fragments 5′ and 3′ of the deletion. The spaces between the solid lines indicate the positions of deletions, and the numbers preceded by Δ indicate the lengths of the deletions (in base pairs).
The ColE1 plasmid replication origin contains a G tract consisting of six G residues, which is essential for replication (22). A similar G tract has also been found in the PE1 transcript of the prolate phages c2, biL67, c6A, and 923 (40). The phage c2 G tract consists of seven G residues, whereas the 923 ori has eight G residues and bIL67 ori has seven G residues interrupted by the nucleotides CTA. In order to examine if the G tract played a role in c2 replication, the seven G residues were deleted in the c2 ori fragment by using a PCR-based strategy. The fragment was cloned into pVA891, generating pLP215, which was assayed for its ability to replicate in L. lactis. This plasmid did not replicate in L. lactis (Fig. 2).
Recombinant plasmids from the deletion and modification experiments described above (Fig. 1 and 2) were verified by DNA sequencing of plasmid DNA isolated from L. lactis cells. Thus, we can rule out the possibility that plasmid integration into the chromosome, rather than autonomous replication, was being measured in plasmids harboring recombinant c2 ori fragments.
PE1 promoter analysis in recombinant ori plasmids.
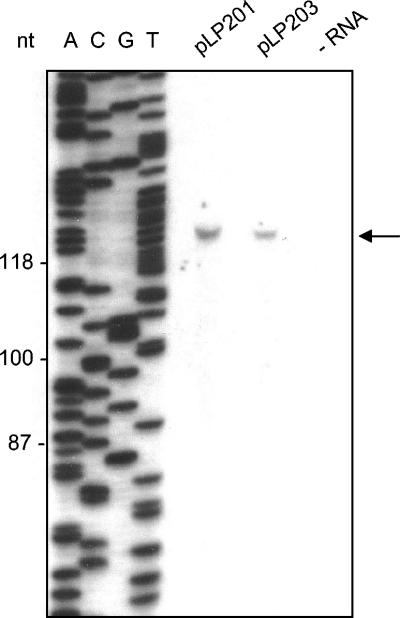
The transcription start site of the early transcripts synthesized from PE1 has been determined for phage c2 (28). To investigate if transcription from PE1 in the plasmid system started from the same position, primer extension analysis was performed. A single cDNA fragment was produced in the primer extension experiments by using the 6902prex primer (Fig. 3). The transcription start site was thus mapped to a nucleotide corresponding to the same A nucleotide identified as the transcription start site in phage c2 during infection (28).
FIG. 3.
Primer extension analysis to identify the transcription start site in the pVA891-ori plasmids. The primer extension reaction was performed with total RNA isolated from L. lactis containing plasmid pLP201 or pLP203 by using primer 6902prex. The primer extension products in both lanes are indicated by the arrow. The same primer was used for the sequencing reaction with pLP201 (lanes A, C, G, and T). Sizes (in nucleotides) are indicated on the left.
To examine if ori transcripts were produced from replicating constructs, we performed Northern blotting using total cellular RNA from the corresponding L. lactis strains harboring replicating plasmids. The probe was generated by amplification of the sequence for the PE1-derived ori transcript in pLP206, including the PE1 promoter. The PE1 transcript was detected in all the replicating plasmids (Fig. 4). The sizes of the major transcripts corresponded to the sizes determined in a previous study for c2-infected cells when RNase protection was used (40), and the major species contained 260, 265, 280, and 295 nt. An additional species consisting of around 150 nt was also present in samples with the largest amounts of transcript (pLP201, pLP203, and pLP206). The substitutions in the −10 region of the PE1 promoter in pLP205 significantly reduced the transcript level compared with the level in plasmid pLP201 but did not abolish promoter activity. The lower level of transcription was obviously sufficient for replication. Plasmid pLP212 (with the nisA promoter) also produced the ori transcript in the absence of nisin in both host genotypes. Plasmid pLP217 produced extremely low levels of the PE1 transcript (Fig. 4). Therefore, the ori transcript was present in all replicating plasmids.
Transcript stability in nonreplicating ori derivatives.
To investigate the PE1 transcript stability in modified ori fragments that did not support replication, we recloned the c2 fragments from several of the nonreplicating pLP series of plasmids into the vector pFX3, which has a functional replication origin for gram-positive bacteria (48). The three smallest ori fragments could not be recloned in pFX3. The inserts of the replication-competent plasmids pLP203, pLP205, and pLP206 were also recloned into pFX3 as controls. Total RNA was analyzed by Northern blot hybridization with a specific probe. For pFX3-214, this analysis was based upon the cloned prtP fragment in the plasmid. For pFX3-213, the probe was based on a fragment that included the ori transcript, the inverted PE1 promoter, and the pFX3 vector sequence (102, 28, and 126 bp, respectively). For all other plasmids, the probe was based on the PE1-derived ori transcript in pLP206 that included PE1.
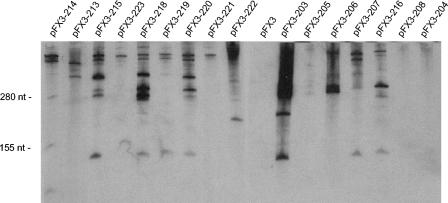
The transcript produced from PE1 was readily detectable in pFX3-203 and pFX3-206 (Fig. 5). The ori fragment of pLP205 cloned into pFX3 produced very low levels of the PE1 transcript, similar to the levels observed with pLP205 (Fig. 4). No transcript was detected in pFX3-204 (complete PE1 promoter deletion). In the deletion mutant pFX3-207, the PE1 transcript was clearly detectable, although this fragment did not support replication. For pFX3-208, no transcript was detected. Addition of the omega terminator to the 3′ end of the pLP207 ori insert significantly increased the stability of the transcript (corresponding to pLP216 and pFX3-216). The presence of the omega terminator did not eliminate the larger hybridizing species produced by this and other plasmids, which probably represented transcript read-through into the vector. Comparison of the transcripts in pFX3-207 and pFX3-216 (Fig. 5) did show that there was an increase in the abundance of the ori transcript of the appropriate size and that there was a significant reduction in the amount of larger transcripts.
FIG. 5.
Northern hybridization analysis of the c2 ori fragments that did not support plasmid replication. The fragments were recloned into a plasmid (pFX3) that contains an origin of replication for gram-positive bacteria. Twenty-microgram portions of total RNA from L. lactis strain MG1363 harboring the plasmids were electrophoresed on acrylamide gels, blot transferred, and then probed with different PCR products and detected by the ECL system. The migration positions of two marker bands (280 and 155 nt) are indicated on the left. Each lane is labeled with the designation of the plasmid construct (see Fig. 1 and 2 for schematic representations of the constructs). The following PCR-generated probes were used: lane pFX3-214, template pLP214 and primers ori11 and ori12; lane pFX3-213, template pFX3-213 and primers ori4 and M13 Rev; lanes pFX3-215, pFX3-223, pFX3-218, pFX3-219, pFX3-220, pFX3-221, pFX3-222, pFX3, pFX3-203, pFX3-205, pFX3-206, pFX3-207, pFX3-216, pFX3-208, and pFX3-204, template pLP206 and primers ori1 and ori7.
The Northern blot analysis of pFX3-213 showed that the inverted PE1 promoter was still active and initiating transcription in the opposite direction. A very low level of transcript could be detected in pFX3-214. Among the internal deletion mutants, a transcript of the expected size could not be detected in pFX3-219, pFX3-221, and pFX3-223, but the transcripts were present in abundance in pFX3-215, pFX3-218, and pFX3-220.
DISCUSSION
In this study we examined the relationship between the structure and organization of the phage c2 origin and its ability to mediate DNA replication. To our knowledge, this is the most detailed investigation to date of lactococcal prolate phage DNA replication, which is characterized in this phage by a lack of a requirement for phage-encoded proteins. We elected to use a model system in which the phage ori supported replication of a plasmid in L. lactis. The ori region was originally inferred from sequence analysis and plasmid cloning (29, 47) and was also recently examined by using the two-dimensional gel technique (5) to analyze replicating c2 phage. Furthermore, the function of the corresponding ori region in prolate phages bIL67 and 923 was recently confirmed (40). In contrast to several isometric-headed phages (φ50, φ31, TP901-1, BK5-T, and Tuc2009) (31, 32, 36, 37, 38), the c2 ori does not confer a Per phenotype (39). The cloned c2 ori is also obviously functional in the absence of phage proteins, in contrast to the ori regions of some isometric-headed phages (BK5-T and φ31) (22, 32). The cloned sk1 origin of replication could support plasmid replication if the N-terminal 179 codons of ORF 47 and the intergenic region between ORFs 47 and 48 were present on a plasmid (8). In contrast, the data for phage c2 suggest that phage-encoded proteins are not required for DNA replication, which is therefore entirely dependent on host replication proteins.
The PE1 promoter gives rise to high levels of early transcript in c2-infected cells (28), and the sequence of the PE1 promoter was strictly conserved in eight diverse prolate phages (40), although the transcript sequence was not conserved. This suggests that there was some selection for the PE1 promoter sequence. However, although ori transcription was essential for supporting replication in the present study, the sequence of the promoter was not critical, as long as the promoter was functional. Introduction of two base pair changes in the −10 region of PE1 (TATAAT to TAGAAG) was tolerated, and the resulting plasmid (pLP205) replicated. Analysis of the ori transcription in pLP205 confirmed that the ability to drive transcription was retained by the mutated promoter, although it was dramatically reduced. This observation is consistent with a previous report that some mutations in the −10 region of L. lactis promoters weaken the promoter strength but do not abolish it completely (25). The PE1 promoter could also be replaced with the inducible nisA promoter (pLP212) without affecting replication. The nisA promoter showed some activity even in the absence of nisin, and the amount of transcript made was clearly sufficient to allow replication in the plasmid system. Leakiness of the nisA promoter in L. lactis has been reported only when cells were grown in lactose or galactose (7), which is not a potential explanation for our findings. However, our construct lacked the nisA gene ribosome binding site and conserved sequence around it, which were present in previous studies on this promoter regulation (14). These sequences were omitted to eliminate a translation signal (which is absent from the PE1 transcript) and to avoid changing the essential 5′ end of the transcript and its position 1 start site. Overall, the 152-bp nisin promoter fragment in our construct lacked 15 nucleotides at the 5′ end and 153 bp at the 3′ end compared with the nisA promoter fragment characterized in pNZ8008 (14). The absence of these regions may have contributed to the nisR-independent transcription which we observed.
The ori deletion studies reported here and the ability of the PE1 transcript template sequences of other prolate phage to support replication in the absence of the upstream A-T-rich region and PL1 (40) show that these two conserved regions are not required for replication in the plasmid system. However, during lactococcal infection, the phage c2 genome replicates via theta forms emanating from the ori region (5) and another replication mechanism that could not be clearly resolved by the two-dimensional gel analysis but that gave rise to replication forks throughout the circularized genome. A role for the A-T-rich region in this mechanism cannot be excluded. Notwithstanding this, the plasmid model system facilitated a range of manipulations that would be difficult or prohibitively laborious in whole phage. However, it does have certain limitations. We used the ability of constructs to be electroporated into L. lactis as a qualitative assay for replication, but we could not distinguish between replication efficiencies once the plasmid had been established. The growth rate under erythromycin resistance selection could not be used as an indication of the copy number of the constructs, because one copy of the gene is sufficient to protect the cells. Future experiments with reporter genes or quantitative PCR may be used to explore the plasmid copy number in order to establish the replication efficiency of derivative ori constructs.
The essential role of transcription in the ability of the phage c2 ori region to support plasmid replication is consistent with observations for other phages, plasmids, and bacteria (2, 6, 18, 35). In phage c2, this promoter activity might be essential for localized strand melting to allow binding of replication proteins or might be required for producing a transcript with a mechanistic role. The former option now seems unlikely for two reasons. First, the promoter inversion abolished replication; hence, RNA polymerase-mediated melting of the transcript template region was not sufficient for replication. Second, the integrity of the nucleotide sequence of the PE1 transcript was essential, suggesting a role for the RNA product. This template sequence could not be replaced by an unrelated fragment with a similar G+C content, and progressive deletion from the 3′ end abolished replication. Deletion of a G tract abolished replication, and the resulting ΔG tract transcript carried the 3′-most internal deletion in a series of internal deletions that did not support replication. Disruption of the transcript, beginning somewhere between positions 263 and 251, and anywhere further upstream abolished replication. These internal deletions were designed to coincide with regions that were predicted to form the stem-loop structure elements of the PE1 transcript. The transcripts produced from three of these scanning deletion constructs were detectable, ruling out the possibility that RNA instability or enhanced degradation interfered with the function of transcripts capable of supporting replication. It was reported recently that chimeric fragments from the PE1 transcript template sequences of phages c2 and 923 did not support replication (40) and that the modeled RNA structures of the PE1 transcripts of these two phages, as well as the third type identified, were very different. Based on these findings and the data obtained with the progressive and internal deletions in this study, the most likely function of the PE1 transcript is that it assumes a specific secondary structure and anneals to the origin region to initiate or facilitate replication. This annealing event could expose the noncoding strand or provide a primer. For example, in ColE1 plasmid replication, DNA polymerase I catalyzes leading strand formation after cleavage of an RNA-DNA hybrid by RNase H (22), and DNA polymerase I is therefore required for replication of ColE1 in the presence of RNase H. It is therefore noteworthy that phage c2 plates with 100% efficiency on a lactococcal strain deficient for DNA polymerase I (40), ruling out the possibility that the function of the PE1 transcript is as a primer for initiation of replication by this polymerase. The primary transcript might be processed by other enzymes, and such a processing event might even depend on a specific transcript conformation. With or without processing, the c2 ori transcript might have a mechanism of action similar to that involved in R-loop formation during initiation of chromosomal replication at oriC of E. coli. This transcript does not form a primer (2) and functions even when it lacks a 3′-hydroxyl group. The continuing production of the phage c2 PE1 transcript while progeny phage genomes accumulate within the cell suggests that there is an ongoing requirement for substantial amounts of the transcript. Further work is required to determine the mechanistic role of the c2 ori transcript in the replication of phage DNA.
Supplementary Material
Acknowledgments
This work was supported by the Marsden Fund of the Royal Society of New Zealand and by the New Zealand Foundation for Research, Science and Technology. A. Schiemann was supported by a Massey University scholarship.
We thank Q. Deng for excellent technical help. We are grateful to NIZO Food Research, Ede, Holland, for the nisin promoter plasmids and host strains.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/
REFERENCES
- 1.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, T. A., and A. Kornberg. 1988. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell 55:113-123. [DOI] [PubMed] [Google Scholar]
- 3.Boyce, J. D., B. E. Davidson, and A. J. Hillier. 1995. Sequence analysis of the Lactococcus lactis temperate bacteriophage BK5-T and demonstration that the phage DNA has cohesive ends. Appl. Environ. Microbiol. 61:4089-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 5.Callanan, M. J., P. W. O'Toole, M. W. Lubbers, and K. M. Polzin. 2001. Examination of lactococcal bacteriophage c2 DNA replication using two-dimensional agarose gel electrophoresis. Gene 278:101-106. [DOI] [PubMed] [Google Scholar]
- 6.Carles-Kinch, K., and K. N. Kreuzer. 1997. RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J. Mol. Biol. 266:915-926. [DOI] [PubMed] [Google Scholar]
- 7.Chandrapati, S., and D. J. O'Sullivan. 1999. Nisin independent induction of the nisA promoter in Lactococcus lactis during growth in lactose or galactose. FEMS Microbiol. Lett. 170:191-198. [DOI] [PubMed] [Google Scholar]
- 8.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 9.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensson, C., C. J. Pillidge, L. J. H. Ward, and P. W. O'Toole. 2001. Nucleotide sequence and characterization of the cell envelope proteinase plasmid in Lactococcus lactis subsp. cremoris HP. J. Appl. Microbiol. 91:334-343. [DOI] [PubMed] [Google Scholar]
- 11.Crutz-Le Coq, A., M. B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 12.Dasgupta, S., H. Masukata, and J. Tomizawa. 1987. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell 51:1113-1122. [DOI] [PubMed] [Google Scholar]
- 13.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodd, H. M., N. Horn, and M. J. Gasson. 1990. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J. Gen. Microbiol. 136:555-566. [DOI] [PubMed] [Google Scholar]
- 16.Frey, J., and H. M. Krisch. 1985. Ω mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene 36:143-150. [DOI] [PubMed] [Google Scholar]
- 17.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hase, T., M. Nakai, and Y. Masamune. 1989. Transcription of a region downstream from lambda ori is required for replication of plasmids derived from coliphage lambda. Mol. Gen. Genet. 216:120-125. [DOI] [PubMed] [Google Scholar]
- 19.Herrin, D. L., and G. W. Schmidt. 1988. Rapid, reversible staining of Northern blots prior to hybridization. BioTechniques 6:196-197. [PubMed] [Google Scholar]
- 20.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1990. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J. Bacteriol. 172:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh, T., and J. Tomizawa. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA 77:2450-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis, A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, P. R., and K. Hammer. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 27.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 28.Lubbers, M. W., K. Schofield, N. R. Waterfield, and K. M. Polzin. 1998. Transcription analysis of the prolate-headed lactococcal bacteriophage c2. J. Bacteriol. 180:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubbers, M. W., N. R. Waterfield, T. P. Beresford, R. W. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 31.Madsen, S. M., D. Mills, G. Djordjevic, H. Israelsen, and T. R. Klaenhammer. 2001. Analysis of the genetic switch and replication region of a P335-type bacteriophage with an obligate lytic lifestyle on Lactococcus lactis. Appl. Environ. Microbiol. 67:1128-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahanivong, C., J. D. Boyce, B. E. Davidson, and A. J. Hillier. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masukata, H., and J. Tomizawa. 1986. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell 44:125-136. [DOI] [PubMed] [Google Scholar]
- 34.Masukata, H., S. Dasgupta, and J. Tomizawa. 1987. Transcriptional activation of ColE1 DNA synthesis by displacement of the nontranscribed strand. Cell 51:1123-1130. [DOI] [PubMed] [Google Scholar]
- 35.Masukata, H., and J. Tomizawa. 1990. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62:331-338. [DOI] [PubMed] [Google Scholar]
- 36.McGrath, S., J. F. M. Seegers, G. F. Fitzgerald, and D. van Sinderen. 1999. Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl. Environ. Microbiol. 65:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Østergaard, S., L. Brøndsted, and F. K. Vogensen. 2001. Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan, D. J., C. Hill, and T. R. Klaenhammer. 1993. Effect of increasing the copy number of bacteriophage origins of replication, in trans, on incoming-phage proliferation. Appl. Environ. Microbiol. 59:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polzin, K. M., L. J. Collins, and M. W. Lubbers. 1999. Protecting dairy starters from bacteriophage infection: development of novel resistance mechanisms against lactococcal prolate bacteriophage c2. Recent Res. Dev. Microbiol. 3:125-133. [Google Scholar]
- 40.Rakonjac, J., L. J. H. Ward, A. H. Schiemann, P. P. Gardner, M. W. Lubbers, and P. W. O'Toole. 2003. Sequence diversity and functional conservation of the origin of replication in lactococcal prolate phages. App. Environ. Microbiol. 69:5104-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sanger, F., J. E. Donelson, A. R. Coulson, H. Kossel, and D. Fischer. 1974. Determination of a nucleotide sequence in bacteriophage f1 DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 90:315-333. [DOI] [PubMed] [Google Scholar]
- 43.Schouler, C., S. D. Ehrlich, and M. C. Chopin. 1994. Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140:3061-3069. [DOI] [PubMed] [Google Scholar]
- 44.Szalewska-Palasz, A., A. Wegrzyn, A. Blaszczak, K. Taylor, and G. Wegrzyn. 1998. DnaA-stimulated transcriptional activation of ori λ: Escherichia coli RNA polymerase β subunit as a transcriptional activator contact site. Proc. Natl. Acad. Sci. USA 95:4241-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. J. Ruiters, G. Venema, and A. Nauta. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 47.Waterfield, N. R., M. W. Lubbers, K. M. Polzin, R. W. Le Page, and A. W. Jarvis. 1996. An origin of DNA replication from Lactococcus lactis bacteriophage c2. Appl. Environ. Microbiol. 62:1452-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, F. F., L. E. Pearce, and P. L. Yu. 1991. Construction of a family of lactococcal vectors for gene cloning and translational fusion. FEMS Microbiol. Lett. 61:55-60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.