Abstract
Transport of RsaA, the crystalline S-layer subunit protein of Caulobacter crescentus, is mediated by a type I secretion mechanism. Two proteins have been identified that play the role of the outer membrane protein (OMP) component in the RsaA secretion machinery. The genes rsaFa and rsaFb were identified by similarity to the Escherichia coli hemolysin secretion OMP TolC by using the C. crescentus genome sequence. The rsaFa gene is located several kilobases downstream of the other transporter genes, while rsaFb is completely unlinked. An rsaFa knockout had ∼56% secretion compared to wild-type levels, while the rsaFb knockout reduced secretion levels to ∼79%. When expression of both proteins was eliminated, there was no RsaA secretion, but a residual level of ∼9% remained inside the cell, suggesting posttranslational autoregulation. Complementation with either of the individual rsaF genes by use of a multicopy vector, which resulted in 8- to 10-fold overexpression of the proteins, did not restore RsaA secretion to wild-type levels, indicating that both rsaF genes were required for full-level secretion. However, overexpression of rsaFa (with normal rsaFb levels) in concert with overexpression of rsaA resulted in a 28% increase in RsaA secretion, indicating a potential for significantly increasing expression levels of an already highly expressing type I secretion system. This is the only known example of type I secretion requiring two OMPs to assemble a fully functional system.
Crystalline surface protein layers (S-layers) are common in many genera of microorganisms, including gram-negative bacteria, gram-positive bacteria, and archaebacteria. S-layers may function as protective barriers and molecular sieves, promote cell adhesion and surface recognition, and maintain cell shape and envelope rigidity (23). The S-layer of the gram-negative bacterium Caulobacter crescentus has been shown to act as a physical barrier to a Bdellovibrio-like parasitic bacterium (26).
The C. crescentus S-layer is a two-dimensional hexagonal array composed of the 98-kDa protein RsaA (41, 42) that is anchored to the outer membrane via an interaction with smooth lipopolysaccharide (S-LPS) (4, 48). The C. crescentus S-layer has been estimated to be 10 to 12% of the total cell protein produced, with approximately 40,000 RsaA subunits attached to the surface of the cell (5, 8, 41). RsaA synthesis occurs without need for induction, and the protein is produced continuously throughout the cell cycle (17, 39).
While most of the characterized S-layer transport systems for gram-negative bacteria involve type II secretion (in which an N-terminal signal system directs export across the inner membrane by use of the general secretory pathway and secretion from the bacterium then occurs via a protein-specific mechanism), the S-layer protein of C. crescentus is secreted by a type I mechanism (3). This mechanism is also found for the S-layer of Campylobacter fetus (46) and the S-layer-like protein (SlaA) in Serratia marcescens (24). The sheer amount of protein secreted by the rsaA secretion apparatus suggests a well organized transport system, but we wish to determine if there are bottlenecks in the overall secretion process that can be addressed by control of gene expression levels.
Type I secretion is a sec-independent pathway where protein is transported from the cytoplasm across the inner and outer membranes without interaction with the periplasm. Type I secreted proteins utilize an uncleaved C-terminal secretion signal, and, in general, the last 60 amino acids (aa) of the protein are sufficient for secretion (6). A type I secretion apparatus is composed of three components. The ATP-binding cassette (ABC) transporter, which likely forms a dimer, resides in the inner membrane, engaging the C-terminal sequence of the substrate protein and hydrolyzing ATP during transport. The membrane fusion protein (MFP) is anchored in the inner membrane by a single transmembrane domain, as well as by binding to the ABC transporter, and may span the periplasm (45). The MFP is thought to interact with the ABC transporter protein and a trimer of the outer membrane protein (OMP), to form a channel from the cytoplasm to the outside of the cell (25).
For most type I systems, the ABC transporter and MFP genes are immediately 3′ of the gene for the secreted protein, whereas the OMP gene location can vary. In some cases, the genes for all three transport components are immediately adjacent to the substrate gene(s) (13, 27, 46). In other type I systems, only the ABC transporter and MFP genes are located next to the substrate gene (28, 30) and the OMP gene lies far from the others. TolC, the OMP for the Escherichia coli alpha-hemolysin (HlyA) system, is an example (50).
The E. coli HlyA system is the best-characterized type I secretion system (12, 19, 31). The structure of the OMP (TolC) has been determined to 2.1 Å (25), and TolC has been shown to be multifunctional, interacting with numerous efflux systems, including the HlyA, AcrA, and CvaA systems (18, 22, 50). Thus, it appears to interact with inner membrane translocases involved in drug and cation efflux as well as protein secretion. It has been suggested that the reason TolC is multifunctional may be because its gene does not belong to any export operon (2).
The RsaA secretion system has the expected characteristics of a type I secretion system. The secretion signal has been localized to the C-terminal 82 aa of RsaA (11). The ABC transporter (RsaD) and MFP (RsaE) have been characterized (3), and the rsaD and rsaE genes are found immediately downstream of rsaA. However, an OMP gene candidate did not immediately follow the rsaE gene, and despite repeated efforts, no Tn5 mutations were found in any genes encoding OMP-like products. This suggested either that the OMP was essential or that there were multiple functional OMP genes for this type I system.
Later, as part of a study characterizing the S-LPS genes that were adjacent to rsaD and rsaE (and involved with attachment of the S-layer), we identified a putative OMP gene (rsaFa) (4). Another study (35), however, declared that this was not an OMP gene, since disruption of the gene did not abolish RsaA secretion. Those authors apparently had not considered the possibility of multiple functional OMP homologs or made efforts to quantitate the amount of RsaA produced by the mutant strain.
In this study, we identify a second OMP gene and characterize the products of rsaFa and rsaFb, showing that both are the only OMP proteins involved in S-layer transport. Eliminating either protein results in decreased RsaA secretion, indicating that either can function as the OMP component but apparently act cooperatively as it was not possible to restore wild-type levels of RsaA secretion by overexpression of individual OMPs. Interestingly, if both were present, overexpression of one led to levels of RsaA secretion significantly above wild-type levels.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli DH5α was grown at 37°C in Luria broth (1% tryptone, 0.5% NaCl, 0.5% yeast extract) with 1.3% agar for plates. C. crescentus strains were grown at 30°C in PYE medium (0.2% peptone, 0.1% yeast extract, 0.01% CaCl2, 0.02% MgSO4) with 1.2% agar for plates. Ampicillin was used at 50 μg/ml, and kanamycin was used at 50 and 25 μg/ml, chloramphenicol was used at at 20 and 2 μg/ml, and streptomycin was used at 50 and 10 μg/ml in E. coli and C. crescentus, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Bacterial strains | ||
| C. crescentus | ||
| NA1000 | Aprsyn-1000; variant of wild-type strain CB15 that synchronizes well | ATCC 19089 |
| JS1001 | S-LPS mutant of NA1000, sheds S-layer into medium | Edwards and Smit (14) |
| JS1003 | NA1000 with rsaA interrupted by KSAC Kmr cassette | Edwards and Smit (14) |
| JS1007 | Smr; NA1000 rsaFa strain | This study |
| JS1008 | Cmr; NA1000 rsaFb strain | This study |
| JS1009 | Cmr Smr; NA1000 rsaFarsaFb strain | This study |
| E. coli DH5α | recA endA | Invitrogen |
| Plasmids | ||
| pHP45Ω | Apr Smr; source of SM cassette removed as-SmaI fragment | Fellay et al. (15) |
| pBSKIIEEH | Apr; modified pBSKSII cloning vector with EcoRI-, EcoRV-, HindIII-modified MCS | This study |
| pBSKIIESH | Apr; modified pBSKSII cloning vector with EcoRI-, StuI-, HindIII-modified MCS | This study |
| pTZ18UCHE | Cmr; cloning vector | This study |
| pTZ18UCHE:rsaFbΔNΔC | Cmr; cloning vector with rsaFb fragment missing N and C termini | This study |
| pBSKIIEEH:rsaFaΩSm | Apr Smr; rsaFa gene fragment with SM cassette inserted at PstI site | This study |
| pBBR4 | Kmr; broad-host-range plasmid derived from pBBR1 | This study |
| pBBR4:rsaFa | rsaFa+ Kmr; rsaFa gene inserted EcoRI/BamHI in pBBR4 | This study |
| pBBR4:rsaFb | rsaF+ Kmr; rsaFb gene inserted EcoRI/BamHI in pBBR4 | This study |
| pWB9:rsaAΔP | Cmr Smr; rsaA gene and rsaA promoter strain | Bingle et al. (9) |
| pWB9Hps12 | Cmr Smr; rsaA containing BamHI site at aa 723 | Bingle et al. (10) |
| pWB9:723/furin | Cmr Smr; rsaA containing furin cleavage site (RKKR) in BamHI site at aa 723 | This study |
| pGEX4T3 | Apr; GST-tagged expression vector | Amersham |
| pGEX4T3:rsaFa | Apr; GST-tagged expression vector with in-frame BamHI-EcoRI rsaFa gene | This study |
| pK18mobsacB | Kmr Sucs; E. coli-based suicide vector | Schafer et al. (37) |
| pK18mobsacB:rsaFaΩSm | Kmr Sucs; E. coli-based suicide vector with rsaFaΩSm fragment | This study |
MCS, multiple cloning site; Sucs, sucrose sensitivity; KSAC, Kmr cassette derived from transposon Tn903.
Plasmid and DNA manipulations.
Standard methods of DNA isolation and manipulation were used (36). Electroporation of C. crescentus was performed as previously described (20). All PCR products were generated using Platinum Pfx DNA polymerase (Invitrogen) following the manufacturer's suggested protocols. Strain NA1000 chromosomal DNA was used as the template for all PCR products. Chromosomal DNA was isolated with standard phenol-chloroform extraction methods (36).
All primers used in this study are listed in Table 2. The PBSKIIEEH vector was constructed from pBSKII (Stratagene). The BssHII fragment containing the multiple cloning site was removed and replaced with annealed oligonucleotides EEH-1 and EEH-2 forming EcoRI, EcoRV, and HindIII sites. pBSKIIESH was created similarly to pBSKIIEEH with annealed oligonucleotides ESH-1 and ESH-2 forming EcoRI, StuI, and HindIII sites.
TABLE 2.
Primers used in this study
| Primer name | Sequence |
|---|---|
| EEH-1 | 5′-CGCGCTGAATTCGGATATCTTAAGCTTGG-3′ |
| EEH-2 | 5′-CGCGCCAAGCTTAAGATATCCGAATTCAG-3′ |
| ESH-1 | 5′-CGCGCTGAATTCGAGGCCTTTAAGCTTGG-3′ |
| ESH-2 | 5′-CGCGCCAAGCTTAAAGGGCTCGAATTCAG-3′ |
| rsaFa-1 | 5′-CGCGGATCCATGCGAGTGCTGTCGAAAGTTCTGTC-3′ |
| rsaFa-2 | 5′-CCGGGAATTCTAGTTGCGGGGCGCGGTCTGGAC-3′ |
| rsaFb-1 | 5′-CGCGGATCCATGTTGATGTCGAACCGTCGACGGG-3′ |
| rsaFb-2 | 5′-CCGGGAATTCTATTTCGAGCCGCTCGGGGGCTT-3′ |
| rsaFbNC-1 | 5′-GAAGCCGACGTGCTGTCT-3′ |
| rsaFbNC-2 | 5′-TGTAGGAGGTTTTCGGGTCA-3′ |
| 1060 | 5′-GAGGCCTAGTACTCTGTCAGACCAAGTTTACTCATA-3′ |
| 1920 | 5′-GAGGCCTACTCTTCCTTTTTCAATATTATTGAA-3′ |
| CHE-1 | 5′-GGAAGATCTGTTAACTTTTCAGGAGCTAAGGAAGCT-3′ |
| CHE-2 | 5′-GGAAGATCTGTTAACACAATAACTGCCTTAAAAAAATTA-3′ |
| RKKR-1 | 5′-TCGAGACCCGATGCGCAAGAAACGGG-3′ |
| RKKR-2 | 5′-CCCGTTTCTTGCGCATCGGGTC-3′ |
| rsaFb-I90 | 5′-GGACGACGCTGACCAGCACCCCCTGCT-3′ |
PCR products containing the rsaFa and rsaFb genes were generated by using primers rsaFa-1 and rsaFa-2 and primers rsaFb-1 and rsaFb-2, respectively. PCR products were ligated into the EcoRV site of the pBSKIIEEH vector and called pBSKIIEEH:rsaFa and pBSKIIEEH:rsaFb, respectively.
pBBR4 was constructed from plasmids pBBR1MCS and pUC4 KISS. The kanamycin resistance (Kmr) fragment from pUC4 KISS was removed with PstI and blunted with T4 polymerase. A 0.3-kbp portion of the chloramphenicol resistance (Cmr)-encoding gene was removed from pBBR1MCS with DraI and replaced with the blunted Kmr fragment, producing a Kmr broad-host-range vector that replicates in C. crescentus. pBBR3 was constructed in a similar manner to that used for pBBR4, using a blunted HindIII-cut streptomycin resistance (Smr) fragment of pHP45Ω.
Plasmids pBBR4:rsaFa and pBBR4:rsaFb and pGEX4T3:rsaFa were made using the BamHI-EcoRI fragment from pBSKIIEEH:rsaFa and pBSKIIEEH:rsaFb, respectively.
pBSKIIEEH:rsaFaΩSm was created by using plasmids pBSKIIEEH:rsaFa and pHP45Ω. The streptomycin resistance insertion cassette was removed as a SmaI fragment and blunt-end ligated into pBSKIIEEH:rsaFa at a blunted PstI site inside rsaFa. The resulting plasmid, pBSKIIEEH:rsaFaΩSm, was then used to make pK18mobsacB:rsaFaΩSm. The EcoRI-HindIII fragment containing the rsaFaΩSm fragment was cloned into EcoRI-HindIII-cut plasmid pK18mobsacB.
A PCR product containing a truncated form of rsaFb missing the N and C termini was generated by using primers rsaFbNC-1 and rsaFbNC-2. This PCR product was blunt-end ligated into the StuI site of pBSKIIESH, creating pBSKIIESH:rsaFbΔNΔC. pTZ18UCHE was constructed by using inverse PCR with primers 1060 and 1920 to create the backbone of pTZ18U without the ampicillin resistance (Apr) cassette. The CHE (containing a chloramphenicol resistance gene) fragment was created as a PCR product by using pMMB206 (32) and primers CHE-1 and CHE-2. The pTZ18U backbone product was cut with StuI and blunt-end ligated with an HpaI-cut CHE fragment, creating pTZ18UCHE. The EcoRI-HindIII fragment from pTZ18UCHE was removed and replaced with the EcoRI-HindIII rsaFbΔNΔC fragment from pBSKIIESH:rsaFbΔNΔC.
pWB9:rsaAΔP was described previously (10). The pWB9:723/furin construct was made using the BamHI site at amino acid (aa) 723 (Hps12). Two oligonucleotides, RKKR-1 and RKKR-2, were annealed and ligated into pUC9CXS as an XhoI-StuI fragment in a manner similar to that for the type IV pilin epitope (9). The RKKR-containing fragment was released by using BamHI and then inserted into the BamHI site at aa 723 in rsaAΔP, and forward orientation of the fragment was confirmed by selection for Cmr. The Cmr cassette was excised using BglII and then ligation of the two complementary ends. The rsaAΔP 723/furin fragment was then removed as an EcoRI-SstI fragment and ligated into EcoRI-SstI-cut pWB9KSAC.
Knockout strain construction.
Knockouts of the two rsaF genes were constructed in the wild-type (S-layer-positive) strain C. crescentus NA1000. The rsaFa gene was destroyed by homologous recombination of an rsaFa gene fragment containing an internal streptomycin resistance insertion cassette by using pK18mobsacB:rsaFaΩSm as a suicide vector. PYE plates containing streptomycin or kanamycin were used to identify single-crossover events, and five rounds of subculturing were used to encourage a second recombination event. Bacteria were grown on 5% sucrose-PYE plates to select the second recombination event. Subsequent replicate plating on PYE-streptomycin and PYE-kanamycin plates confirmed the removal of the Kmr gene. Colonies were then screened by PCR using primers rsaFa-1 and rsaFa-2 to determine if the recombination event resulted in restoration of wild-type rsaFaor incorporation of the rsaFaΩSm gene fragment.
The rsaFb gene was destroyed by inactivation with an N- and C-terminal-deleted form of rsaFb. Selection on PYE-chloramphenicol was used to identify integration of the plasmid into the genome. Colonies were screened by PCR using primers 1060 and rsaFb-I90.
The double rsaF knockout was created by homologous recombination using pK18mobsacB:rsaFaΩSm and JS1008 (rsaFb)-competent cells. Screening for homologous recombination of pK18mobsacB:rsaFaΩSm was done in the same manner as for the single rsaFaΩSm knockout except that chloramphenicol was used in all media to maintain the rsaFb knockout. PCR confirmation of both rsaF gene knockouts was performed with the primers and conditions described above.
Antibody production.
Antibodies to detect RsaA were raised against a form of RsaA containing only the N- and C-terminal portions of the protein, referred to as 188/784. This internal deletion form of the RsaA protein was previously described in reports of linker mutagenesis studies (10). Essentially, the DNA fragment coding for the N-terminal aa 1 to 188 of RsaA was removed as an EcoRI-BamHI fragment, and the portion coding for the C-terminal aa 784 to 1025 was removed as a BamHI-HindIII fragment. The two fragments were ligated together at the BamHI site and then ligated into EcoRI-HindIII-cut pUC8; this maintained the translational frame of the protein. HindIII-cut pUC8:188/784 was ligated to the HindIII-cut pKT215 vector and transformed into C. crescentus, forming aggregated protein which was used to make antibodies against both termini. Aggregated 188/784 protein was collected and washed with distilled water (dH2O) to remove residual C. crescentus cells. Aggregates were solubilized with 4 M urea and dialyzed against dH2O to remove urea. Samples were then injected into a New Zealand White rabbit, and serum was collected and processed by standard protocols (34).
Polyclonal antibodies were produced against RsaFa by using a glutathione S-transferase (GST)-tagged protein. The pGEX4T3:rsaFa plasmid was expressed in E. coli DH5α but produced only inclusion bodies. Inclusion body isolation was performed by growing cells to an optical density at 600 nm of 1.0 and then pelleting and resuspending them in 1× phosphate-buffered saline with lysozyme (100 μg/ml) for 1 h at 25°C; RNase A (50 μg/ml) and DNase I (1 μg/ml) were then added to the suspension, and it was incubated for an additional hour at 25°C. After incubation, 10% (starting concentration) sodium dodecyl sulfate (SDS) was added at a 1:1 ratio with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer and boiled for 5 min, put on ice for 15 min, and centrifuged at 16,000 × g for 10 min. The inclusion body pellet was then recovered and solubilized with 4 M urea. Protein was dialyzed with several changes of dH2O for 2 days to remove the urea. A rabbit was immunized as described above.
Protein analysis methods.
Surface protein from C. crescentus cells was extracted by low-pH extraction as previously described (49). To compare the amounts of S-layer protein extracted from different mutants, cells were normalized and loaded onto SDS-PAGE gels.
Whole-cell protein preparations were made with normalized cultures and were pelleted by centrifugation, and the pellets were washed twice with 10 mM Tris-HCl (pH 8). The cells were resuspended in 10 mM Tris-HCl (pH 8) with lysozyme at 25°C for 15 min, and then RNase A and DNase I were added (as described above) and incubated at 37°C for 30 min. Equal amounts of whole-cell protein preparations were loaded onto protein gels. Note that whole-cell preparation methods were altered for certain strains when required to determine the internal levels of S-layer. RsaA is attached to the cell surfaces of wild-type S-layer-positive strain NA1000, which required removal by low-pH extraction and subsequent washes of the cell pellet before whole-cell preparations could be made to allow assessment of internal levels of RsaA. To remove secreted RsaA from the S-layer-shedding JS1001 strain, culture medium was poured through a fine nylon mesh (350-μm pore size) to remove aggregates so that they did not affect the internal RsaA levels detected in the whole-cell preparation.
Whole-culture preparations were made by using normalized cells as described above. Whole-culture preparations were not centrifuged, ensuring that any aggregated RsaA was small, well dispersed, and included in the preparation. Cultures were incubated with lysozyme, RNase A, and DNase I as described above. Urea was added to the samples to give a final concentration of 2 M to solubilize any microaggregates produced by S-layer-shedding strains. Equal amounts of whole-culture protein preparations were loaded onto SDS-PAGE gels.
SDS-PAGE and Western immunoblotting.
SDS-PAGE was performed with 7.5 or 12% (as indicated) separating gels. Coomassie blue staining of gels and Western immunoblotting were performed by use of standard protocols (36) and 0.2-μm-pore-size BioTrace NT nitrocellulose membranes (Pall Biosciences). Western blots were visualized by colorimetric or electro-chemiluminescence (ECL) developing methods. Colorimetric detection was performed as previously described (40). Chemiluminescent blotting was done with the Amersham Biosciences ECL Western blotting kit in accordance with the manufacturer's protocol. To generate quantitative values and standard deviations, all data reported for ECL Western blots are the result of at least three and as many as seven replicate assays.
Anti-188/784 antibodies were incubated at 1/15,000 for colorimetric Western blotting and 1/30,000 for chemiluminescent Western blotting. Incubation with primary anti-RsaFa was done at a 1/1,000 dilution for colorimetric blotting and at a 1/5,000 dilution for chemiluminescent blotting. Kodak X-OMAT LS film was used for visualization of chemiluminescent blots, while spot densitometry was done with a Bio-Rad VersaDoc 5000 system and the Quantity One (version 4.3.0) program. Within the exposure limits defined by the software, quantitative densitometry using the VersaDoc 5000 allows for a linear response over several orders of magnitude.
Bioinformatic analysis.
All sequences were obtained from the Institute for Genomic Research (TIGR) C. crescentus genome database. The Biology Workbench website http://workbench.sdsc.edu/ was used for protein sequence alignments with the BLASTP sequence alignment tools (1). ClustalW alignments of the RsaFa (accession no. NC002696, locus tag CC1015), RsaFb (accession no. NC002696, locus tag CC1318) and TolC proteins were done with MacVector 6.0.
Electron microscopy.
Protein A-colloidal gold immunolabeling of C. crescentus by use of the anti-RsaFa antibody and 5-nm-diameter protein A-colloidal gold label was performed as previously described (35). The antibody was preabsorbed with JS1009 cells. Cells (1 ml of cells with an optical density at 600 nm of 1) were centrifuged at 16,000 × g for 5 min. The cell pellet was resuspended in 10 mM Tris-HCl (pH 8) and disrupted by sonication for 5 s with a microprobe at maximum power. Sonicated cells were centrifuged at 16,000 × g for 2 min, and the supernatant was discarded. Anti-RsaFa rabbit serum was added to suspend the pellet; the suspension was incubated on ice for 1 h, followed by centrifugation at 16,000 × g for 3 min. The supernatant was used for labeling. Unstained cells were imaged by whole-mount transmission electron microscopy.
RESULTS
Identification of two OMP genes.
Two possible candidates for the OMP gene with similarity to the gene for the E. coli TolC protein sequence were identified in the C. crescentus genome (33). These were named rsaFa and rsaFb. RsaFa has 23% identity and 45% similarity to TolC, and RsaFb has 25% identity and 47% similarity to E. coli TolC, as determined by local sequence alignment (see Materials and Methods). The rsaFa gene (1,581 bp) is located downstream of the rsaADE genes after a gap of 5,025 bp containing five S-LPS-related genes. The rsaFb gene (1,452 bp) is located 322 kb (303 genes) away from rsaFa and is flanked by genes of unknown function. The two rsaF genes show significant similarity with each other, sharing 39% identity and 60% similarity, and may be the result of gene duplication. A BLAST search of the C. crescentus genome for other candidate OMPs by using RsaFa as a query sequence identified several predicted proteins with moderate E values. However these proteins are likely outer membrane lipoproteins, as percentage identities with lipoproteins are much higher (approximately 40 to 48%), and the TIGRFAM and Pfam protein families group them in the lipoprotein family.
Disruption of the rsaFa and rsaFb genes.
Knockouts of rsaF genes were constructed by homologous recombination of inactivated rsaF genes in the wild-type C. crescentus strain NA1000. Disruption of rsaFa was performed by gene replacement with an rsaFa gene construct containing an antibiotic resistance cassette. A mutant resulting from a double-crossover event was confirmed by PCR and designated JS1007. The rsaFb gene was disrupted via insertional inactivation, using an N- and C-terminal-deleted rsaFb fragment (rsaFbΔNΔC). The rsaFbΔNΔC fragment was inserted into the chromosome via homologous recombination with a nonreplicating plasmid, resulting in two tandem, nonfunctional copies of rsaFb. Confirmation by PCR showed that there were only disrupted forms of rsaFb in the chromosome (data not shown). One confirmed mutant was selected (JS1008). To create the double knockout strain, rsaFa was disrupted and confirmed in JS1008 in the same manner used for JS1007 and was designated JS1009.
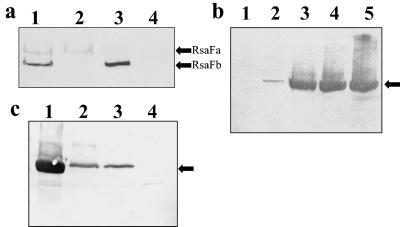
To determine that RsaFa and RsaFb were not produced in the knockout strains, Western blot analysis was performed using polyclonal anti-RsaFa antibodies. Rabbit polyclonal antibodies were generated against a GST-tagged rsaFa gene product. Cross-reactivity between the two RsaF proteins was evident, and thus anti-RsaFb antibodies were not raised since the proteins can be differentiated by size (57.5 kDa for RsaFa and 50.2 kDa for RsaFb). Western immunoblot analysis demonstrated the loss of the RsaF proteins in the rsaF knockout strains (Fig. 1a). Densitometry analysis of the Western blots showed that the levels of the remaining RsaF protein in the single rsaF knockout strains were the same as those observed in the wild-type strains (data not shown).
FIG. 1.
(a) RsaFa and RsaFb levels in wild-type and rsaF knockout strains. Chemiluminescent anti-RsaFa Western blots of whole-culture protein preparation samples are shown. Lanes: 1, NA1000; 2, JS1008; 3, JS1007; 4, JS1009. Arrows indicate RsaFa and RsaFb. (b) Effect of disruption of the rsaF genes on S-layer secretion. Chemiluminescent Western blots of low-pH-extracted protein are shown. Lanes: 1, JS1003; 2, JS1009; 3, JS1007; 4, JS1008; 5, NA1000. RsaA is indicated by an arrow. (c) Determination of internal levels of RsaA in the rsaF double knockout. Whole-cell protein preparations were visualized by colorimetric Western blotting with polyclonal anti-188/784 RsaA antibody. Lanes: 1, NA1000; 2, JS1009; 3, JS1001; 4, JS1003. An arrow indicates RsaA.
Effect of disruption of the rsaF genes on RsaA secretion.
Levels of RsaA secreted by the rsaF knockout strains were analyzed. When low-pH-extracted protein levels (representing secreted protein) of knockout and wild-type strains were compared, there was a progressive decrease in amounts of RsaA secreted as the rsaF genes were lost (Fig. 1b). Disrupting rsaFa decreased S-layer secretion to a greater extent than loss of rsaFb did, but both single rsaF mutants were still capable of secreting RsaA. Elimination of both rsaF genes (JS1009) led to complete loss of RsaA secretion. Levels of RsaA secretion could not be easily determined by SDS-PAGE and Coomassie blue staining, as small amounts of RsaA are not readily detected; therefore, Western immunoblotting was performed.
Quantification of S-layer secretion levels by chemiluminescent Western blotting showed that disruption of rsaFb decreased RsaA secretion to 75.6% ± 1.59% (mean ± standard deviation) of wild-type levels, whereas loss of rsaFa decreased RsaA secretion to 54.5% ± 1.49% of wild-type (NA1000) levels (Table 3). RsaA in the rsaF double knockout (4.44% ± 1.74% of wild-type levels) was detectable only with Western blotting and was thought to be due to release of RsaA from cells that burst during the low-pH extraction.
TABLE 3.
RsaA levels compared to those of the wild type determined by whole-culture preparations or low-pH extractiona
| Strain | % RsaA relative to wild typeb
|
|
|---|---|---|
| Low pH extracted | Whole culture | |
| NA1000 | 100 | 100 |
| JS1008 | 75.6 ± 1.59 | 79.7 ± 3.36 |
| JS1007 | 54.4 ± 1.49 | 56.4 ± 0.47 |
| JS1009 | 4.44 ± 1.74 | 9.56 ± 2.16 |
| JS1003 | 0 ± 0.09 | 0 ± 0.09 |
| JS1009 rsaFa | 78.5 ± 2.18 | 80.0 ± 1.22 |
| JS1009 rsaFb | 56.2 ± 2.08 | 57.3 ± 0.83 |
| JS1001 | NA | 94.8 ± 1.08 |
Spot densitometry of chemiluminescent Western blots was done at least three times with polyclonal anti-188/784 antibodies and used to determine the relative levels of RsaA produced by the cells. Levels determined by densitometry were compared to wild-type NA1000 RsaA levels. All low-pH and whole-culture protein preparations were normalized prior to running samples.
Values are means ± standard deviations. NA, not applicable.
To confirm that apparent RsaA secretion in JS1009 was due to burst cells during low-pH extraction, whole-cell preparations were analyzed to determine the RsaA levels inside the cells. To ensure that only internal RsaA was analyzed, surface protein was extracted from the cell surface (as outlined in Materials and Methods) before whole-cell protein preparations were examined. Colorimetric Western blots show levels of internal RsaA (Fig. 1c). Levels of RsaA observed with a whole-cell preparation of the double rsaF knockout strain appeared to be higher than those seen where the S-layer was removed by low-pH extraction. RsaA levels in JS1009 were similar to levels seen in the JS1001 strain, an S-layer-shedding strain, which was filtered and washed by centrifugation to remove secreted RsaA aggregates. Both the JS1009 and the JS1001 internal RsaA levels were less than those seen in the NA1000 parent strain. The levels of RsaA in the JS1001 strain were only internal levels of S-layer, as transported RsaA was shed and removed, suggesting that the RsaA detected in the double rsaF knockout was located internally. Furthermore, immunofluorescence studies of whole JS1009 cells with an anti-RsaA antibody did not detect the presence of any RsaA on the surface (data not shown).
Complementation of the secretion-deficient JS1009 strain.
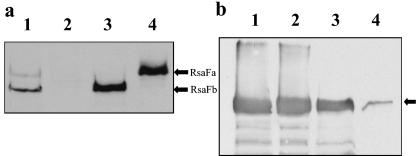
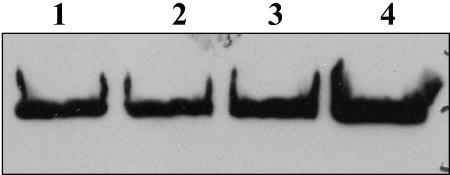
To demonstrate that the rsaF knockouts were responsible for reduction or loss of the S-layer secretion, a multiple-copy broad-host-range plasmid containing either rsaFa (pBBR4:rsaFa) or rsaFb (pBBR4:rsaFb) was used to complement the double knockout strain. To determine if the plasmid-borne rsaF genes were expressed, whole-culture preparations were made using polyclonal antibodies against RsaFa. Western blotting showed that the plasmid-derived RsaF proteins were in fact overexpressed, at levels that were 9.72 ± 0.71 (RsaFa)- and 8.01 ± 0.66 (RsaFb)-fold higher than wild-type strain levels (Fig. 2a).
FIG. 2.
(a) Complementation of JS1009 with the rsaF genes. Whole-culture preparations were compared by Western blotting and visualized by chemiluminescence using the anti-RsaFa antibodies. Lanes: 1, NA1000; 2, JS1009; 3, JS1009 rsaFb; 4, JS1009 rsaFa. (b) Complementation of the rsaF genes in trans restores S-layer secretion. Chemiluminescent Western blotting of low-pH-extracted protein using anti-188/784 RsaA antibodies is shown. Lanes: 1, NA1000; 2, JS1009 rsaFa; 3, JS1009 rsaFb; 4, JS1009. The arrow refers to RsaA.
Despite overexpression mediated by multicopy vectors, in both cases complementation of the individual rsaF genes only restored S-layer secretion to levels comparable to that seen with corresponding single rsaF knockouts (Fig. 2b). The levels of RsaA secretion in the JS1009 rsaFa and JS1009 rsaFb strains were restored to levels that were only about 2% greater than those seen in the single rsaF knockouts (Table 3). Due to the lack of additional antibiotic selection markers, complementation of both rsaF genes in the double knockout was not done.
Production of RsaA appears to be regulated when secretion is impeded.
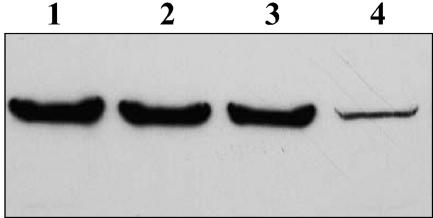
The production of RsaA in whole-culture protein preparations was analyzed when secretion was inhibited in the RsaF mutants. Densitometry performed with Western blots showed that the levels of RsaA in the whole-culture preparations were almost identical to those observed by low-pH extraction (Fig. 3.), with the rsaFb knockout expression of RsaA at 79.7% ± 3.36% of wild-type levels and the rsaFa knockout expressing 56.4% ± 0.47% of wild-type levels (Table 3). However, the levels of RsaA seen with the double knockout strain were higher (9.56% ± 2.16% of wild-type NA1000 levels) than those seen by low-pH extraction (4.44% ± 1.74% of wild-type levels).
FIG. 3.
RsaA production and RsaA secretion levels are comparable. Whole-culture preparations run on SDS-12% PAGE gels were transferred to a nitrocellulose membrane and analyzed with anti-188/784 RsaA antibodies. Lanes: 1, NA1000; 2, JS1008; 3, JS1007; 4, JS1009.
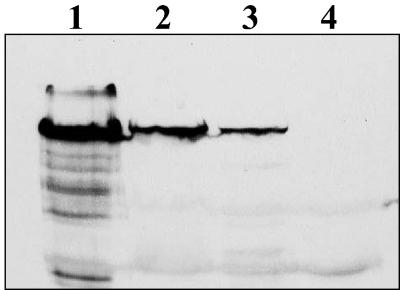
These results suggested that there was regulation of RsaA production in the cell when transport was impeded. To examine this further, the secretion of a recombinant RsaA with the charged RKKR furin cleavage sequence inserted at amino acid 723 (723/furin) was expressed in the various mutants. When this recombinant protein was expressed in JS1003, an S-layer-negative strain, no recombinant protein was secreted, apparently due to the inserted positive-charge cluster. Internal RsaA levels in the transporter mutants and JS1003::pWB9:723/furin were similar to those seen in the RsaF double knockout strain (Fig. 4): 5% of wild-type levels in the strain expressing this modified RsaA compared to 9% for the RsaF double knockout strain. There were no apparent breakdown products seen in any of the transporter or modified RsaA mutants. It is interesting, however, that both the JS1009 rsaF mutant strain and the JS1003::pWB9:723/furin strains grew much slower, with generation times of 114 and 108 min, respectively, compared to a generation time of 78 min for JS1003 and the wild-type NA1000, suggesting that retention of residual RsaA inside the cell is deleterious.
FIG. 4.
Impeded RsaA transport in rsaF mutant (JS1009) and RsaA mutant (JS1003 Hps12furin). Whole-cell preparations were loaded on SDS-12% PAGE gels and compared by Western blotting with anti-188/784 RsaA antibodies. The NA1000 strain was subjected to low-pH extraction before whole-cell preparations were made to minimize the levels of S-layer. No RsaA breakdown products are present in the mutants. Lanes: 1, NA1000; 2, JS1009; 3, JS1003::pWB9:723/furin; 4, JS1003.
Overexpression of RsaA and RsaF.
Because RsaF expression could be significantly increased in trans complementation experiments (see above), we wished to determine if the number of transporter units or the amount of RsaF was a limiting factor in the amount of RsaA that was secreted. Vector-borne copies of a single rsaF gene were inserted into S-layer-positive C. crescentus cells to determine if levels of RsaA secretion could be increased. An S-LPS-negative strain, JS1001, was used instead of the wild-type NA1000 to ensure that there was no possibility of RsaA regulation by surface crystallization, i.e., the possibility that RsaA secretion was impeded by full coverage of the bacterial surface. Levels of RsaA were determined by Western blot analysis of whole-culture preparations to ensure that any secreted RsaA was included in the sample.
Levels of RsaA production were not significantly increased when additional copies of rsaFa or rsaFb were expressed in JS1001 by addition of pBBR4:rsaFa or pBBR4:rsaFb. Both JS1001 rsaFa and JS1001 rsaFb produced levels of RsaA only slightly greater than those produced by JS1001 and the wild-type strain NA1000 (Table 4). Levels of RsaFa and RsaFb were determined by immunoblotting and densitometry analysis and were similar to those seen in the trans complementation strains described above (data not shown).
TABLE 4.
RsaA levels compared to those of JS1001 determined by whole-culture preparationsa
| Strain | % RsaA relative to JS1001b |
|---|---|
| JS1001 | 100 |
| JS1001 rsaFa | 103.3 ± 2.95 |
| JS1001 rsaFb | 102.8 ± 2.38 |
| JS1001::pWB9rsaAΔP | 100.8 ± 1.47 |
| JS1001 rsaFa::pWB9rsaAΔP | 128.3 ± 3.69 |
Spot densitometry of chemiluminescent Western blots was done in triplicate using polyclonal anti-188/784 antibodies and used to determine the relative levels of RsaA produced by the cells. Levels determined by densitometry were compared to JS1001 RsaA levels. All whole-culture protein preparations were normalized prior to running samples.
Values are means ± standard deviations.
To be assured that additional RsaFa was properly targeted to the outer membrane, protein A-colloidal gold labeling with anti-RsaF antibody was used to assess the levels of RsaF on the outer membrane surface. A uniform low-level labeling was noted with JS1001 (Fig. 5), indicating that some portion of the RsaF OMPs were surface exposed when the oligosaccharide chains of the S-LPS fraction of total LPS were eliminated (see below). Labeling of JS1001 rsaFa and JS1001 rsaFb (data not shown) showed two major classes of cells: those labeled at levels similar or slightly greater than that seen with JS1001 and a fraction (approximately 20% of the total) where dense labeling was noted (Fig. 5.). We interpreted that finding as an indication that plasmid copy numbers for the moderate-copy-number plasmid pBBR4 vary significantly from cell to cell, suggesting that it is not a stably maintained plasmid. Nevertheless, it appeared that some cells were expressing much higher levels of RsaFa and that the protein was targeted correctly to the outer membrane.
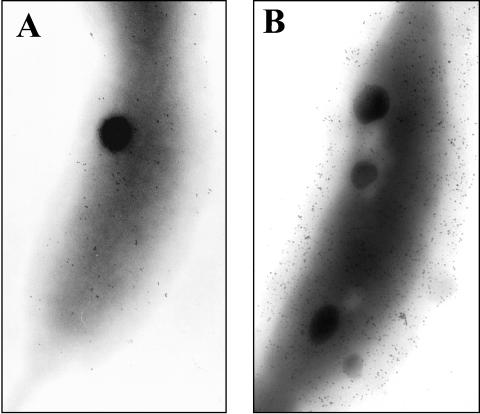
FIG. 5.
Colloidal gold labeling of surface-displayed RsaF. Surface-displayed RsaF (A and B) was determined for the JS1001 strains overexpressing rsaFa or rsaFb (data not shown) by electron microscopy with anti-RsaFa and colloidal gold labeling. (A) JS1001 shows wild-type levels of RsaF with moderate labeling. (B) The JS1001 rsaFa strain shows a significant increase in surface display of RsaF. Note that only about 20% of the JS1001 rsaFa cells showed a significant increase in RsaF display.
Interestingly, in contrast to the findings for JS1001 (which has no S-LPS), there was no detectable label with strain JS1003, which has no S-layer but does have a normal complement of S-LPS (data not shown). Presumably, this smooth form of LPS effectively blocks antibody access to the OMPs, perhaps indicating that their exposure on the surface is minimal.
Since JS1001 has only the single chromosome resident copy of rsaA, we considered whether, in the presence of elevated OMP levels, RsaA transcription or translation might then determine the maximum levels of RsaA secretion. To address this possibility, a multicopy plasmid-borne rsaA gene was introduced into the JS1001 rsaFa strain, and whole-culture protein levels were compared (Fig. 6). The resulting strain, JS1001 rsaFa::pWBrsaAΔP, produced 28.3% ± 3.69% more S-layer protein than the wild-type strain JS1001 or strain JS1001::pWBrsaAΔP. Thus, elevated secretion of RsaA is possible but requires both overexpression of RsaA and at least the RsaFa OMP.
FIG. 6.
Effect of RsaF overexpression in the JS1001 strain. Whole-culture preparations were run on SDS-12% PAGE gels and compared by Western blotting with anti-188/784 antibodies. Lanes: 1, JS1001; 2, JS1001 rsaA; 3, JS1001 rsaFa; 4, JS1001 rsaFa rsaA.
DISCUSSION
Here we have described the discovery of two OMPs that function as part of the type I secretion machinery for RsaA in C. crescentus. This arrangement of the OMP components, where there are two similar but independent proteins functioning as the OMP component, is different from any other known type I secretion system and is the first report of a type I secretion system which can utilize either of two OMPs for the secretion of the same protein. Of particular note is that while both proteins were capable of acting alone as the OMP component of the transport system, neither alone could enable wild-type secretion levels of RsaA. The amount of RsaA produced and secreted was consistently lower in the single OMP knockout strains as well as the complemented strains. Indeed, when overexpressed, the individual OMPs could restore secretion only to the level of their apparent contribution in the normal, single-gene-copy situation. This finding suggests that the level of secretion may not be limited solely by the number of active transporter complexes (when only one OMP is expressed) but also by the transporter composition. We suggest that the presumed trimeric OMP complex may function most effectively when assembled as a heterotrimer of RsaFa and RsaFb and that this may be the native situation. This possibility is tentative, and we are presently devising approaches to study this possibility more directly. In this scenario, heterotrimers are technically not required for RsaA secretion, although survival in a natural environment may require maximal secretion of RsaA (and thereby heterotrimer formation) to maintain complete surface coverage of this protective device. Although an attractive hypothesis, heterotrimer formation is not proven, and other explanations for the inability of a single RsaF to accomplish wild-type expression levels, even when overexpressed, are possible.
A related finding is that if one rsaF gene is destroyed, the level of the remaining rsaF gene product did not increase to compensate for the loss of the other rsaF gene (Fig. 1a). This finding is perhaps predictable given the remote location of rsaFb but does suggest that although the roles of RsaFa and RsaFb are functionally additive, their expression is not tightly coordinated. This is a significant departure from other type I systems, where (except for E. coli TolC and S. marcescens HasF) the OMP may be cotranscribed with the other transporter components (7).
We had previously assigned a tentative transporter function for RsaFa (then called RsaF) (4) and its proximity to other S-layer related genes, but had not provided experimental data to confirm a role in RsaA transport. In contrast to other opinions (21), it was our view that no predicted gene products in the C. crescentus genome sequence other than RsaFa and RsaFb had sufficiently high homology to TolC to be seriously considered OMP candidates. The direct genetic data presented here confirm that assessment, dismissing the notion of a third OMP (21). Although both RsaF proteins are required to secrete wild-type levels of S-layer in C. crescentus, RsaFa plays a more significant role as its loss reduced RsaA secretion to 54.4% ± 1.49% of wild-type levels whereas loss of the RsaFb protein led to only 75.6% ± 1.59% of wild-type secretion (Fig. 1b). For this reason, we chose RsaFa for overexpression in these initial efforts to increase RsaA secretion.
Because type I secretion relies on a C-terminal secretion signal, the target protein (here RsaA) must be completely synthesized before the export process begins. Therefore, a highly expressed type I secretion system leads to problems in biochemical logistics for the affected bacterium. As there is little primary sequence homology among various type I secretion signals yet cross-expression can often be demonstrated, it has been suggested by us and others that a degree of folding must occur to generate a structural motif that is recognized by the secretion machinery (10, 29). Thus, if secretion is somehow impaired, rapid accumulation of large (98-kDa) monomers could be deleterious. One solution to this problem could be rapid degradation of protein that cannot be exported. We have suggested that the S-layer-associated protease (the product of sapA), which appears poised to recognize defects in the nascent RsaA monomer, might play such a role (47). The data here, however, gave no indication that detectable degradation occurs. Instead, we simply noted a much reduced level (9.56% of wild-type RsaA) of full-length monomer, retained inside the cells in both the double RsaF knockout and in cells attempting to export the recombinant RsaA displaying the furin cleavage site (RKKR) through a fully functional transporter complex. On this latter point, a preliminary threading of the RsaFa and RsaFb proteins onto the homologous TolC protein (for which the structure has been determined) predicted exposed charge-dense regions in the interior of the transporter channel (data not shown). We estimate that this may be the reason why the addition of the furin cleavage site (RKKR) leads to a blockage of RsaA transport, while a variety of other recombinant RsaA proteins with 100 to 200 amino acid insertions have been readily secreted (34, 47).
These data are consistent with a feedback mechanism involving down-regulation of RsaA production, though we cannot completely exclude the possibility that rapid and complete degradation of unsecreted protein occurs. In such situations, the cells grew much slower, there were few motile cells, and most cells were misshapen (data not shown). It may be that if autoregulation was triggered it may not be specific, affecting the synthesis of other proteins as well. If autoregulation of internal RsaA occurs, we suggest that the overall level of RsaA produced in the cell may in effect be determined by the amount of protein that can be secreted.
The rsaA gene, like other S-layer genes, is transcribed continuously throughout the cell cycle (17, 39). In Lactobacillus brevis, the slpA gene is transcribed during stationary phase even when the S-layer protein is not produced (23). Therefore, it is likely that any sort of regulation would occur at the translational level. The S-layer of Thermus thermophilus HB8 has been shown to autoregulate the translation of the slpA gene by binding a C-terminal SlpA fragment to the 5′ mRNA (16). It may be that a similar situation occurs in C. crescentus, and this possibility is currently being investigated.
Our results suggest that RsaA expression levels may be increased only by increasing both the number of transporters and the expression of RsaA. Overexpression of an individual rsaF gene in the absence of the other rsaF gene was not an appropriate course, since it did not even lead to normal levels of RsaA secretion, so we instead began overexpressing individual transporter components in the presence of a normal complement of remaining transporter proteins. By using the S-layer-shedding strain JS1001, which produces the same amount of RsaA as the parental strain NA1000, any RsaA feedback regulation due to the presence of a “full” S-layer on the bacterial surface would not affect the results. When both rsaF genes were present and additional copies of rsaFa were introduced, the secretion of RsaA was not significantly increased (Fig. 6.). Similarly, a strain containing additional vector-borne copies of the rsaA gene with a wild-type complement of rsaFa and rsaFb genes did not have RsaA levels exceeding those of the wild type. However, the strain overexpressing both RsaA and RsaFa produced 28.3% ± 3.69% more protein than the wild type (Table 4). This is a remarkable increase in RsaA secretion as wild-type levels of RsaA already represent 10 to 12% of total cell protein. Furthermore, the vectors used for overexpression are seemingly unstable, which may mean that only 20% of the cells were truly overexpressing RsaFa. Stable expression of RsaFa by all cells may lead to still higher levels of RsaA secretion.
These results can be compared to those of experiments on hemolysin secretion where additional plasmid-borne copies of tolC were expressed in gram-negative bacteria containing the HlyA secretion apparatus (43). Despite the presence of additional copies of TolC, neither enhancing nor deteriorating effects were noted. The authors suggested that the HlyB and HlyD protein levels might be the limiting factor when only the OMP levels were increased. We suggest instead that translation levels of the transported protein, HlyA, may have been the limiting factor.
It is likely that RsaA secretion is determined by the type and level of both RsaF proteins and the level of rsaA translation. The type I secretion apparatus is able to transport normal levels of RsaA produced by the cell but cannot accommodate increased RsaA levels unless OMP levels are increased. The ability to increase RsaA secretion may be a function of increasing the number of transporter “holes” in the bacteria. It has been shown that the HlyB and HylD proteins exist in a preformed complex in the inner membrane and that when HlyA is engaged, the complex recruits the TolC protein (44). Presuming that the RsaA transport complex is analogous, increasing the number of OMPs in the outer membrane may make it easier for the secretion apparatus components to engage and transport RsaA. Additional studies are under way to determine if coordinate up-regulated expression of various combinations of the transporter elements will lead to still greater levels of RsaA expression or whether other factors, such as membrane stability or available ATP pools, will set limits.
Acknowledgments
We thank Andrea Pusic for technical assistance at various stages of this work. We thank Wade H. Bingle for pWB:rsaAΔP.
This study was funded by grants to J.S. from the Canadian Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, C. 2003. Channel-tunnels: outer membrane components of type I secretion systems and multidrug efflux pumps of Gram-negative bacteria. Rev. Physiol. Biochem. Pharmacol. 147:122-165. [DOI] [PubMed] [Google Scholar]
- 3.Awram, P., and J. Smit. 1998. The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J. Bacteriol. 180:3062-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awram, P., and J. Smit. 2001. Identification of lipopolysaccharide O-antigen synthesis genes required for attachment of the S-layer of Caulobacter crescentus. Microbiology 147:1451-1460. [DOI] [PubMed] [Google Scholar]
- 5.Beveridge, T. J., P. H. Pouwels, M. Sara, A. Kotiranta, K. Lounatmaa, K. Kari, E. Kerosuo, M. Haapasalo, E. M. Egelseer, I. Schocher, U. B. Sleytr, L. Morelli, M. L. Callegari, J. F. Nomellini, W. H. Bingle, J. Smit, E. Leibovitz, M. Lemaire, I. Miras, S. Salamitou, P. Beguin, H. Ohayon, P. Gounon, M. Matuschek, K. Sahm, H. Bahl, R. Grogono-Thomas, J. Dworkin, M. J. Blaser, R. M. Woodland, D. G. Newell, M. Kessel, and S. F. Koval. 1997. Functions of S-layers. FEMS Microbiol. Rev. 20:99-149. [DOI] [PubMed] [Google Scholar]
- 6.Binet, R., S. Letoffe, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by gram-negative bacterial ABC transporters—a review. Gene 192:7-11. [DOI] [PubMed] [Google Scholar]
- 7.Binet, R., and C. Wandersman. 1996. Cloning of the Serratia marcescens hasF gene encoding the Has ABC exporter outer membrane component: a TolC analogue. Mol. Microbiol. 22:265-273. [DOI] [PubMed] [Google Scholar]
- 8.Bingle, W. H., H. D. J. Kurtz, and J. Smit. 1993. An “all-purpose” cellulase reporter for gene fusion studies and application to the paracrystalline surface (S)-layer protein of Caulobacter crescentus. Can. J. Microbiol. 39:70-80. [DOI] [PubMed] [Google Scholar]
- 9.Bingle, W. H., J. F. Nomellini, and J. Smit. 1997. Cell-surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus. Mol. Microbiol. 26:277-288. [DOI] [PubMed] [Google Scholar]
- 10.Bingle, W. H., J. F. Nomellini, and J. Smit. 1997. Linker mutagenesis of the Caulobacter crescentus S-layer protein: toward a definition of an N-terminal anchoring region and a C-terminal secretion signal and the potential for heterologous protein secretion. J. Bacteriol. 179:601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingle, W. H., J. F. Nomellini, and J. Smit. 2000. Secretion of the Caulobacter crescentus S-layer protein: further localization of the C-terminal secretion signal and its use for secretion of recombinant proteins. J. Bacteriol. 182:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blight, M. A., and I. B. Holland. 1994. Heterologous protein secretion and the versatile Escherichia coli haemolysin translocator. Trends Biochem. Sci. 12:450-455. [DOI] [PubMed] [Google Scholar]
- 13.Duong, F., E. Bonnet, V. Geli, A. Lazdunski, M. Murgier, and A. Filloux. 2001. The AprX protein of. Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene 262:147-153. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, P., and J. Smit. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J. Bacteriol. 173:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for invitro insertional mutagenesis of gram-negative bacteria. Gene 2: 147-154. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Herrero, L. A., G. Olabarria, and J. Berenguer. 1997. Surface proteins and a novel transcription factor regulate the expression of the S-layer gene in Thermus thermophilus HB8. Mol. Microbiol. 24:61-72. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, J. A., J. Smit, and N. Agabian. 1988. Transcriptional analysis of the major surface array gene of Caulobacter crescentus. J. Bacteriol. 170:4706-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentschev, I., G. Dietrich, and W. Goebel. 2002. The E. coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 10:39-45. [DOI] [PubMed] [Google Scholar]
- 20.Gilchrist, A., and J. Smit. 1991. Transformation of freshwater and marine caulobacters by electroporation. J. Bacteriol. 173:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn, H. P., and B.-U. von Specht. 2003. Secretory delivery of recombinant proteins in attenuated Salmonella strains: potential and limitations of type I protein transporters. FEMS Immunol. Med. Microbiol. 37:87-98. [DOI] [PubMed] [Google Scholar]
- 22.Hwang, J. W., X. T. Zhong, and P. C. Tai. 1997. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and TolC. J. Bacteriol. 179:6264-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahala, M., K. Savijoki, and A. Palva. 1997. In vivo expression of the Lactobacillus S-layer gene. J. Bacteriol. 179:284-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai, E., H. Akatsuka, A. Idei, T. Shibatani, and K. Omori. 1998. Serratia marcescens S-layer protein is secreted extracellularly via an ATP-binding cassette exporter, the Lip system. Mol. Microbiol. 27:941-952. [DOI] [PubMed] [Google Scholar]
- 25.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 26.Koval, S. F., and S. H. Hynes. 1991. Effect of paracrystalline protein surface layers on predation by Bdellovibrio bacteriovorus. J. Bacteriol. 173:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letoffe, S., P. Delepelaire, and C. Wandersman. 1990. Protease secretion by Erwinia chrysanthemi: the specific secretion functions are analogous to those of. Escherichia coli alpha-haemolysin. EMBO J. 9:1375-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letoffe, S., J. M. Ghigo, and C. Wandersman. 1994. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc. Natl. Acad. Sci. USA 91:9876-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letoffe, S., and C. Wandersman. 1992. Secretion of CyA-PrtB and HylA-PrtB fusion proteins in Escherichia coli: involvement of the glycine-rich repeat domain of Erwinia chrysanthemi protease B. J. Bacteriol. 174:4920-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackman, N., J. M. Nicaud, L. Gray, and I. B. Holland. 1985. Identification of polypeptides required for export of haemolysin 2001 from E. coli. Mol. Gen. Genet. 201:529-536. [DOI] [PubMed] [Google Scholar]
- 31.Mollenkopf, H. J., I. Gentschev, and W. Goebel. 1996. Conversion of bacterial gene products to secretion-competent fusion proteins. BioTechniques 21:856-860. [DOI] [PubMed] [Google Scholar]
- 32.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 33.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. Debroy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus strain CB15. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomellini, J. F., M. C. Toporowski, and J. Smit. 2004. Secretion or presentation of recombinant proteins and peptides mediated by the S-layer of Caulobacter crescentus, p. 477-524. In F. Banexy (ed.), Expression technologies: current status and future trends. Horizon Scientific Press, Wymondham, United Kingdom.
- 35.Reichelt, M., B.-U. von Specht, and H. P. Hahn. 2001. The Caulobacter crescentus outer membrane protein Omp58 (RsaF) is not required for paracrystalline S-layer secretion. FEMS Microbiol. Lett. 201:277-283. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 45: 69-73. [DOI] [PubMed] [Google Scholar]
- 38.Smit, J. 1987. Localizing the subunit pool for the temporally regulated polar pili of Caulobacter crescentus. J. Cell Biol. 105:1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit, J., and N. Agabian. 1982. Cell surface patterning and morphogenesis of a periodic surface array during Caulobacter development. J. Cell Biol. 95:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smit, J., and N. Agabian. 1984. Cloning of the major protein of the Caulobacter crecentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J. Bacteriol. 160:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit, J., H. Engelhardt, S. Volker, S. H. Smith, and W. Baumeister. 1992. The S-layer of Caulobacter crescentus: three-dimensional image reconstruction and structure analysis by electron microscopy. J. Bacteriol. 174:6527-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smit, J., D. A. Grano, R. M. Glaeser, and N. Agabian. 1981. Periodic surface array in Caulobacter crescentus: fine structure and chemical analysis. J. Bacteriol. 146:1135-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spreng, S., G. Dietrich, S. Niewiesk, V. Meulen, I. Gentschev, and W. Goebel. 2000. Novel bacterial systems for the delivery of recombinant protein or DNA. FEMS Immunol. Med. Microbiol. 27:299-304. [DOI] [PubMed] [Google Scholar]
- 44.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from. E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, S. A., O. I. Shedd, K. C. Ray, M. H. Beins, J. P. Jorgensen, and M. J. Blaser. 1998. Campylobacter fetus surface layer proteins are transported by a type I secretion system. J. Bacteriol. 180:6450-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umelo-Njaka, E., W. H. Bingle, F. Borchani, K. D. Le, P. Awram, T. Blake, J. F. Nomellini, and J. Smit. 2002. Caulobacter crescentus synthesizes an S-layer-editing metalloprotease possessing a domain sharing sequence similarity with its paracrystalline S-layer protein. J. Bacteriol. 184:2709-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker, S., D. N. Kurunaratne, N. Ravenscroft, and J. Smit. 1994. Characterization of mutants of Caulobacter crescentus defective in surface attachment of the paracrystalline surface layer. J. Bacteriol. 176:6312-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, S. G, S. H. Smith, and J. Smit. 1992. Isolation and comparison of the paracrystalline surface layer proteins of freshwater caulobacters. J. Bacteriol. 174:1783-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]