Preface
Bacteria must divide in order to increase in number and colonize their niche. Binary fission is the most widespread means of bacterial cell division, but even this relatively simple mechanism displays many variations on a theme. In most bacteria, the tubulin homolog FtsZ assembles into a ring structure (Z ring) at the site of cytokinesis and recruits additional proteins to form a large protein machine (divisome) that spans the membrane. Here we discuss current insights into the regulation of Z ring assembly and how the divisome drives membrane invagination and septal cell wall growth while still flexibly responding to various cellular inputs.
Introduction
Most bacterial cells divide by binary fission, which is the splitting of a cell into two roughly equal progeny cells. Fission is orchestrated by the divisome 1, a protein complex whose assembly is directed by the conserved tubulin homolog FtsZ. In several model bacterial species, the divisome has been proposed to assemble in temporally distinct stages (Figure 1A). Initially, FtsZ undergoes GTP-dependent polymerization into filaments that form a defined ring-like structure (Z ring) at the site of cytokinesis. Membrane-associated FtsZ-interacting proteins such as FtsA and ZipA in Escherichia coli tether the Z ring to the cell envelope, resulting in an initial complex called the proto-ring (Figure 1B, top). In the second stage, the proto-ring recruits enzymes involved in cell wall (septum) synthesis such as FtsI in E. coli, and proteins that likely coordinate signaling between the these enzymes and the proto-ring such as FtsN and FtsBQL, thereby forming the mature divisome. (Figure 1A, 1B bottom). The divisome then constricts the cytoplasmic membrane, which in concert with synthesis of new peptidoglycan at the division septum and invagination of the outer membrane (in Gram-negative bacteria), achieves cytokinesis and cell separation.
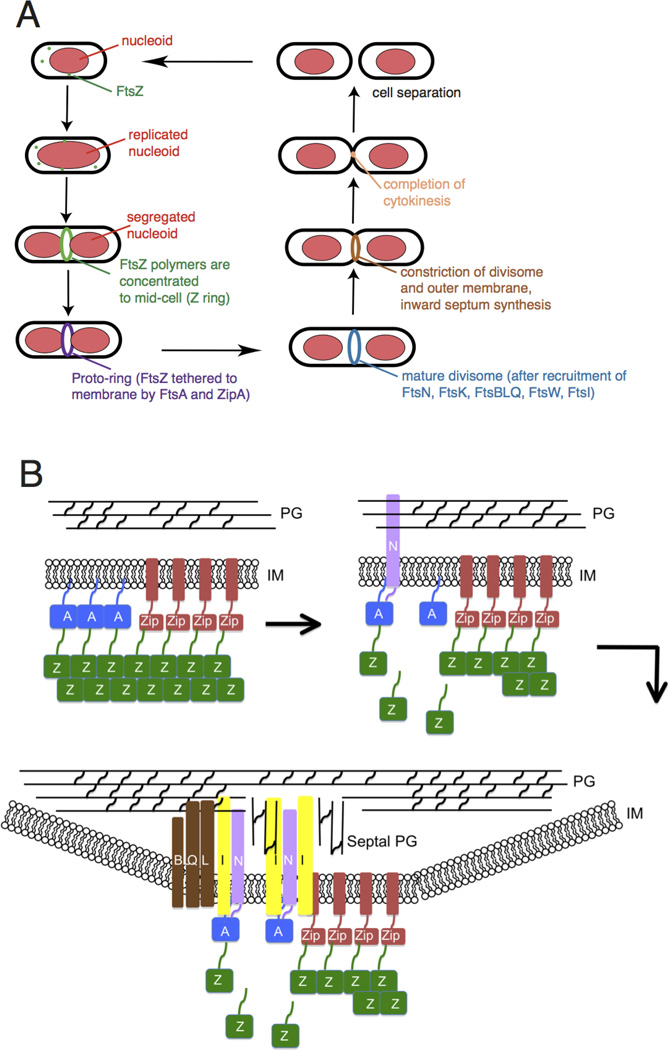
Fig. 1.
Overview of bacterial cytokinesis. (A) After E. coli cells replicate and segregate their chromosomes, organized as nucleoids, FtsZ and its membrane tethers are concentrated at midcell and organize into a Z ring or proto-ring. After recruitment of additional proteins, the proto-ring progresses into a mature divisome, which coordinates constriction of the inner and outer membranes with targeted cell wall hydrolases and ingrowing septal peptidoglycan. The ultimate result is separation into two equal-sized daughter cells. (B) ZipA and FtsA tether FtsZ protofilaments and polymer bundles to the membrane using flexible linkers and either a transmembrane segment (ZipA, red rectangle) or an amphipathic helix (FtsA). ZipA and FtsA are depicted as clustered, but they may be interspersed instead. In a later stage, ZipA bundling of FtsZ protofilaments is offset by FtsA-mediated disassembly. In addition, FtsA recruits FtsN via its cytoplasmic domain, which in turn may disrupt FtsA oligomers. FtsN’s periplasmic domain is targeted to peptidoglycan, reinforcing its localization and potentially stimulating FtsI and septal peptidoglycan synthesis. After septal peptidoglycan synthesis is underway, the other divisome proteins including the FtsBLQ complex transduce signals from the periplasm to the Z ring via FtsA, coordinating Z ring constriction (and FtsZ turnover) with septal peptidoglycan synthesis and targeted cell wall hydrolysis. For simplicity, many of the divisome proteins are not shown here, but they are shown in Fig. 4A.
Model bacteria that divide by binary fission include the Gram-negative E. coli and Caulobacter crescentus, as well as the Gram-positive Bacillus subtilis. When cytokinesis is inactivated in these and similar systems, cells continue to grow as cylindrical rods and chromosomes continue to replicate and segregate, resulting in filamentous cells with multiple nucleoids. Newly available genetic and cytological tools have allowed the study of cytokinesis in additional genera such as streptococci and mycobacteria that are rapidly emerging as additional model systems. Finally, alternatives to binary fission such as polar budding 2, division without FtsZ 3, longitudinal division 4, and other non-canonical mechanisms showcase the vast diversity of bacterial strategies for cell duplication that still await full mechanistic explanations (Box 1). Inactivation of cytokinesis in organisms using such atypical modes of division often gives rise to diverse cell morphologies that are more complex than simple extension of a cylindrical rod.
Box 1 – Variations on the theme of binary fission.
Model bacilli such as E. coli grow by increasing their length and divide by binary fission to produce roughly equal-sized daughter cells, which are a few microns long. This mechanism can be literally stretched to cells up to 120 microns in length. Two newly described ectosymbionts hailing from the sulfur-oxidizing γ-proteobacteria divide using only one Z ring at the center of these long cells 146. Yet C. crescentus, despite being only a few microns long, divides slightly off center to produce cells of different size and fates. A related alpha-proteobacterial group, the rhizobiales, uses the divisome to bud off a new cell at one pole 2. Actinobacteria such as Corynebacterium glutamicum and Mycobacterium tuberculosis also divide asymmetrically 147,148. A more extreme case of asymmetric division occurs in sporulating B. subtilis, where the division septum is placed very near one cell pole. Streptococci divide by binary fission, but often fail to separate, forming long chains. Anabaena, a genus of cyanobacteria, also form chains, but this is because they never finalize the divisome constriction process and end up sharing a continuous outer membrane 149. Recently, proteins important for Anabaena cell septation have been shown to interact directly with proteins important for chain formation 149. More unusual is another γ-proteobacterial ectosymbiont of marine nematodes that grows by increasing its diameter rather than its length, and consequently divides longitudinally using a single ellipsoidal Z ‘ring’ 4. Even the perfectly round coccus S. aureus, which could conceivably divide along an infinite number of equivalently sized planes, divides in alternating three-dimensional planes to create its typical grape-like cell clusters 150. Finally, there are mechanisms other than canonical fission for bacterial reproduction, such as the multiple intracellular offspring produced by giant Epulopiscium spp. 151.
Bacteria have evolved exquisite control over divisome assembly, most ubiquitously through regulation of the timing and placement of Z-ring assembly. The importance of this control for bacterial survival has made FtsZ a frequent target for the discovery and design of novel antibiotics, and other divisome components could be considered as potential targets for specific bacterial families or species. Here we review recent discoveries related to the structure, assembly, and dynamics of the proto-ring, enhanced by new insights from super-resolution microscopy and in vitro reconstitution studies. We then summarize recent insights into the regulation of Z-ring assembly and divisome activity, focusing mainly on the E. coli system, before returning to models to explain how Z rings may exert inward force on membranes and how early and late divisome proteins may signal to each other during the cytokinetic process.
Assembly of the proto-ring
Initiating Z ring assembly
The Z ring needs to be positioned properly at the future cell division site, usually at mid-cell. There are several known spatial regulation systems that act prior to FtsZ, either by forming a negatively acting gradient that keeps Z rings away from cell poles and restricts their assembly at mid-cell, or by localizing directly at midcell and stimulating Z-ring assembly (Figure 2A). Negative spatial regulators include nucleoid occlusion and the Min system, which have been extensively reviewed recently 5,6. Positive spatial regulators include SsgA and SsgB in Streptomyces coelicolor 7, PomZ in Myxococcus xanthus 8, and MapZ (also called LocZ) in Streptoccocus pneumoniae 9,10. Conserved solely in the Actinomycetes, SsgA and SsgB localize to future sites of septum formation between developing spores along elongated S. coelicolor hyphae. In this system, SsgA mediates localization of SsgB, which binds directly to FtsZ, recruits it, and promotes Z-ring assembly. In M. xanthus, cell-cyle regulated PomZ promotes properly localized FtsZ assembly, though unlike SsgA and SsgB it is not essential for septum formation and cytokinesis. So far, there is no evidence that PomZ can interact directly with FtsZ, suggesting that additional proteins are needed to bridge the interaction. As with PomZ, MapZ in S. pneumoniae is not absolutely required for Z ring formation, but is important for proper division site placement, and cells without MapZ are severely deformed. Conserved only in the Streptococcaceae and most other Lactobacillales, MapZ is itself regulated through phosphorylation by the serine-threonine protein kinase StkP, which regulates other divisome functions in cocci 10.
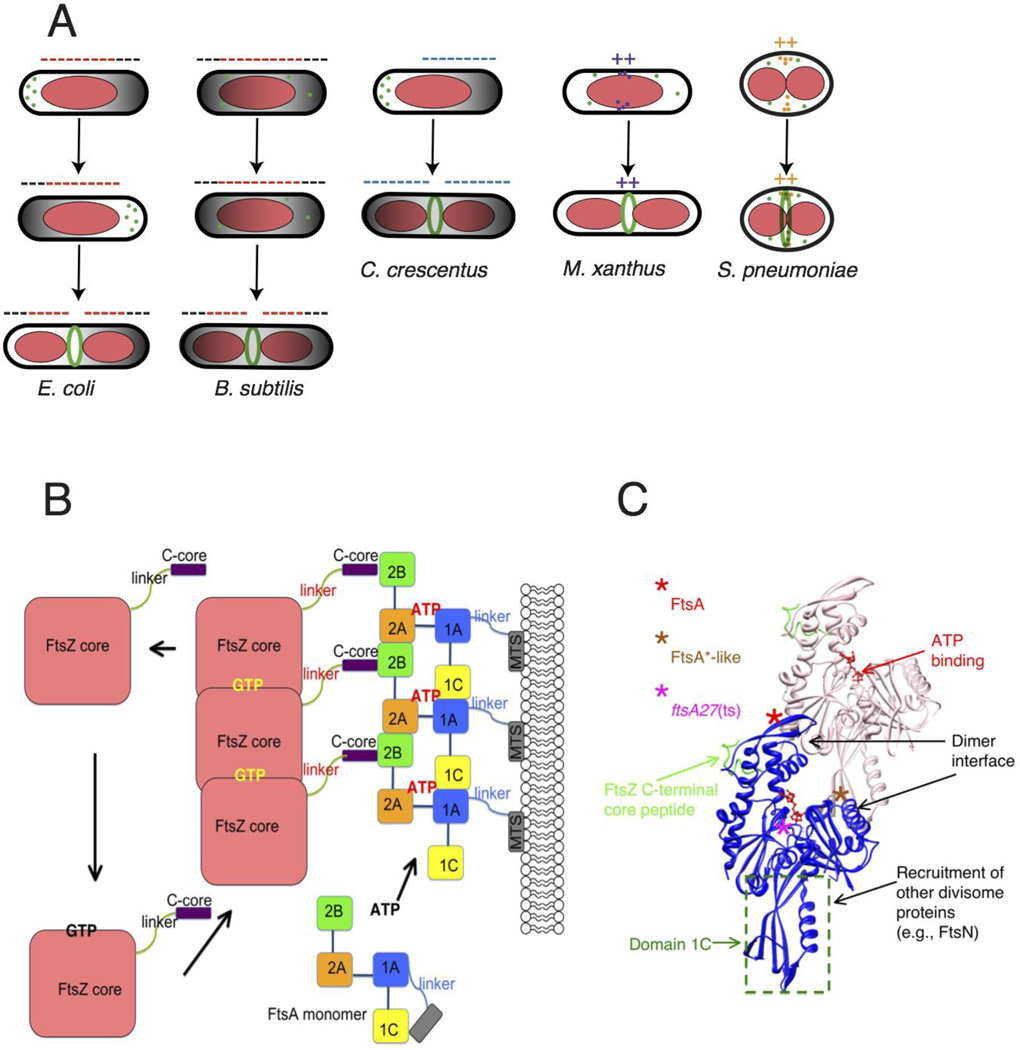
Fig. 2.
Placement and tethering of the Z ring. (A) The Z ring is targeted to the division site by related but distinct mechanisms. In E. coli and B. subtilis, a bipolar gradient formed by the Min system (black; oscillating in E. coli and fixed in B. subtilis) and nucleoid occlusion (red) inhibit assembly of FtsZ (minus signs) so that the Z ring is restricted to the lowest region of negative regulation at midcell. In C. crescentus, MipZ forms a bipolar gradient in response to cell cycle cues and negatively regulates Z ring positioning. In contrast, FtsZ assembly stimulators in M. xanthus and S. pneumoniae localize at midcell via unknown mechanisms, potentiating assembly of the Z ring there (plus signs). (B) GTP promotes assembly of FtsZ into protofilaments, which attach to subdomain 2B of FtsA via FtsZ’s C-terminal core at the terminus of a flexible linker. GTP hydrolysis is coupled to disassembly of FtsZ, as the cycle repeats. FtsA’s four subdomains are schematized, along with the amphipathic C-terminal membrane targeting sequence (MTS) at the end of a short linker. ATP binding promotes oligomerization of S. pneumoniae FtsA, but is not yet confirmed for E. coli FtsA. Reprinted with permission from Ref. 154. (C) Structure of an FtsA dimer and its bound C-terminal core peptide of FtsZ based on the atomic structure from Thermotoga maritima 34. The flexible linker and amphipathic helix are not shown. Loss of function lesions such as the thermosensitive FtsA12 allele map near the ATP binding site, whereas gain of function (FtsA*, FtsA*-like) generally map to the dimer interface. FtsA subdomains have distinct activities, which are outlined.
Some of the differences in activity of these newly discovered proteins result from different growth mechanisms of their hosts. Streptomycetes such as S. coelicolor grow vegetatively as a filamentous mycelium, only occasionally forming crosswalls. Consequently, FtsZ and other divisome proteins are not essential for viability, only for forming the septal crosswalls that lead to spores 11. In the absence of FtsZ, SsgA and SsgB still localize to sites of future spore septa, but form discrete puncta rather than ring-like structures, preventing spore formation. In contrast, after depletion of FtsZ in rod-shaped M. xanthus, PomZ persists at previously formed divisomes in filamenting cells. This suggests that although PomZ may arrive at nascent cell division sites first, it requires FtsZ for stable localization. The effects of FtsZ depletion on MapZ localization are not yet characterized.
Tethering FtsZ to the membrane
FtsZ is a cytoplasmic protein, with no evidence that it can interact with membranes on its own. Therefore, FtsZ polymers require at least one membrane-associated factor to tether them to the inner surface of the cytoplasmic membrane. In E. coli, this task is accomplished by a partnership between FtsA and ZipA proteins, which are both essential for cytokinesis and simultaneously associate with the membrane and bind the conserved carboxyl-terminal core peptide of FtsZ 12. The assembly of these three proteins at the membrane defines the essential part of the proto-ring. The second-best conserved division protein across bacterial species, FtsA, associates with the membrane via a carboxyl-terminal amphipathic helix 13, which forms an independently functioning membrane-targeting sequence (MTS) (Figure 2B). ZipA, on the other hand, is unique to the γ-proteobacteria and contains an amino-terminal transmembrane domain that provides another potentially more permanent membrane tether for FtsZ 14 (Figure 1B). Z rings can still assemble when FtsA or ZipA is inactivated, because only one of the two tethers is needed for ring assembly, but cytokinesis is arrested at the proto-ring stage in these cases 15. In the absence of both membrane tethers, Z rings fail to assemble.
Studies of other model systems have revealed alternative membrane tethers for FtsZ. For example, a protein widely conserved among Gram-positive species called SepF (originally YlmF) serves as a nonessential alternative factor for tethering FtsZ to the membrane in B. subtilis 16. Like FtsA, SepF interacts directly with FtsZ’s C-terminal core peptide 17 and associates with the membrane through an amphipathic MTS, although SepF’s MTS is amino-terminal. Consistent with this, B. subtilis cells can still divide, albeit poorly, in the absence of FtsA, and increased levels of SepF can compensate 18. In Mycobacteria, which naturally lack FtsA, SepF was recently shown to play an essential role in tethering FtsZ to the membrane 19,20, suggesting it is the sole tether.
The EzrA protein is an inhibitor of FtsZ assembly conserved throughout the low-GC Gram-positive bacteria 21. Like ZipA, EzrA contains a single amino-terminal transmembrane domain followed by a cytoplasmic domain. Inactivation of EzrA in B. subtilis leads to Extra Z Rings, resulting in division near the cell poles to form chromosome-free minicells. EzrA not only inhibits FtsZ assembly at the cell poles of rod-shaped species, but also is part of the proto-ring that links it to cell wall synthesis in bacilli and cocci 22,23. The recently solved crystal structure of B. subtilis EzrA without its transmembrane domain places the protein in the eukaryotic spectrin family 24. Spectrin acts as a scaffold to coordinate interaction among membrane proteins 25, and EzrA may have an analogous role in stabilizing membrane-divisome interactions. The carboxyl-terminal coiled coil domains of B. subtilis EzrA are essential for its localization and activity at the midcell divisome 26,27, whereas its amino-terminal coiled coils are involved in inhibiting Z ring assembly at cell poles. In contrast, EzrA in Staphylococcus aureus localizes to the divisome via interactions between its amino-terminal coiled coil domains and FtsZ 28.
FtsA and the dynamics of proto-ring assembly
FtsA is the most conserved member of the bacterial divisome apart from FtsZ, and is a member of the actin family, with structural homology in all but one of four subdomains 29. Early reports of polymerization and ATPase activity of E. coli, B. subtilis and Pseudomonas aeruginosa FtsA proteins in vitro 30–32 were not followed up, presumably due to difficulty in purifying active protein. The first FtsA to exhibit convincing in vitro activity came from Streptococcus pneumoniae, whose FtsA filaments assembled into large helical structures in the presence of ATP but did not exhibit dynamics 33. Several years ago, FtsA from Thermotoga maritima was shown to assemble into actin-like protofilaments that correspond to filaments attached to artificial lipid monolayers 34. Mutations predicted to disrupt subunit interactions and subsequent formation of these filaments 35 were tested in B. subtilis and found to lead to defects in cell division, suggesting that the degree of FtsA oligomerization or polymerization was important for integrity of the proto-ring 36 Interestingly, a recent crystal structure of S. aureus FtsA reveals FtsA oligomers with a 28° inter-subunit twist relative to the T. maritima protein 37. Although this topology has not yet been confirmed on membranes, the evidence underscores the potential species-specific conformational diversity of FtsA, which may complicate generalizations based on a single crystal structure.
What might be the molecular role of ATP binding and oligomerization in FtsA function? The first hint comes from a gain of function mutant of FtsA called FtsA* (Figure 2C), which in addition to performing FtsA-specific functions, can compensate for the loss for ZipA as well as several other divisome defects (see below) 38,39. Upon binding to ATP, purified histidine-tagged FtsA* was shown to destabilize FtsZ polymers by shortening them 40. It was not possible to compare FtsA* with wild-type FtsA in that study because of the difficulty in obtaining active protein. More recently, wild-type FtsA-FtsZ interactions were reconstituted on a supported membrane bilayer 41. Using total internal reflection fluorescence (TIRF) microscopy to visualize fluorescently tagged FtsZ at high resolution, the authors found that GTP-dependent FtsZ protofilaments formed dynamic bundles along the membrane that migrated unidirectionally and at times swirled in vortices. This behavior was dependent on FtsA and adenine nucleotide, and suggests that either ADP- or ATP-bound FtsA oligomers on the membrane promote FtsZ filament treadmilling. During treadmilling, subunits are added to one end of a polymer while removed from the other end, resulting in unidirectional translocation of the filament 41. Together, these data suggest that FtsA plays a role in FtsZ disassembly or turnover (Figure 2B). In support of this idea, purified FtsA from the Gram-positive cocci S. aureus 37 or Deinococcus radiodurans 42 stimulates the GTPase activity of its cognate FtsZ, which would correlate with FtsZ filament disassembly. In contrast, however, D. radiodurans FtsA reduces GTPase activity of E. coli FtsZ. Barring differences resulting from in vitro conditions, this suggests FtsA-FtsZ pairs have closely evolved species-specific regulatory interactions.
Reconstitution of FtsZ-ZipA interactions at the membrane showed different behavior from FtsZ-FtsA interactions 41. FtsZ protofilaments remained dynamic in the presence of ZipA, but large-scale reorganizations of filament bundles did not occur. Moreover, in contrast to FtsA, ZipA could recruit FtsZ monomers as well as polymers to the membrane. A subsequent study found that FtsZ polymerization was necessary for strong interaction with ZipA in solution 43 and argued that the previously observed recruitment of FtsZ monomers by ZipA was an artifact of high magnesium concentrations that can promote FtsZ assembly even in the absence of GTP. This once again highlights differences in vitro activities that may depend on assay conditions.
Although adenine nucleotide is clearly important for FtsA’s ability to stimulate FtsZ dynamics, the role of ATP and FtsA assembly state in this activity remain unclear. For example, FtsA*-like mutants, which may have a self-assembly defect 35, still permitted dynamic FtsZ assembly and streaming 41. Moreover, FtsA ATPase activity was not detected and the FtsZ swirling activity was unchanged when a non-hydrolyzable ATP analog or ADP was added instead of ATP to the reconstituted proteins. However, a recent study of an ftsA thermosensitive mutant (encoding S195P, also known as ftsA27) (Figure 2C) and its intragenic suppressors showed that wild-type FtsA can hydrolyze several ATP molecules per minute, whereas the ATPase activity of equivalently purified thermosensitive FtsA27 protein was significantly lower 44. This suggests that FtsA may have intrinsic ATPase activity, which may be stimulated by as yet unknown factors. Intragenic suppressors of ftsA27 conferring thermoresistance mapped either near the ATP-binding pocket, as expected, or at the FtsA-FtsA subunit interface. As the interface mutations all restored FtsA27 function in vivo and ATP hydrolysis in vitro, it suggests that as with actin, ATP binding and/or hydrolysis may regulate the oligomeric state of E. coli FtsA (Figure 2B, C).
These intragenic suppressors also have shed some light on the little-understood interplay between ZipA and FtsA in the proto-ring. As both membrane-tethered proteins bind the FtsZ C-terminal core peptide, they presumably compete for interaction, with ZipA being the stronger competitor based on in vivo data 45,46. The ftsA27 suppressors that map to the FtsA subunit interface also can bypass the need for ZipA and compete better than native FtsA for binding to the Z ring. This suggests that reducing FtsA oligomerization enhances binding to FtsZ and/or the membrane, perhaps by allowing subdomain 1c, which is normally engaged in FtsA-FtsA interactions, to interact more with integral membrane divisome proteins 35. However, in B. subtilis, which lacks ZipA, reducing FtsA oligomerization seems to inhibit its function in cell division 47, underscoring the idea that much more needs to be learned about the role of FtsA and its interactions with itself and other ring components.
These findings, along with the static behavior of FtsZ-ZipA versus the dynamic behavior of FtsZ-FtsA in vitro, suggest a model in which ZipA is the strong but relatively inflexible membrane tether for FtsZ whereas FtsA, with its potentially reversible membrane tether, provides tethering flexibility. Moreover, there is some evidence that ZipA can enhance bundling of FtsZ polymers in vitro whereas, as mentioned earlier in this section, FtsA seems to destabilize FtsZ polymers and probably polymer bundles (Figure 2B). This evidence for ZipA-mediated FtsZ bundling in vitro is not solid, as bundling seems to depend on electrostatic interactions between the two proteins 43,48, calling into question the relevance of protofilament bundling in vivo (see also below). However, a mutant FtsZ (FtsZL169R) that increases self-bundling in vitro can bypass the need for ZipA in vivo, supporting the idea that ZipA can enhance FtsZ bundling in vivo. In addition, ZipA forms homodimers in vivo via its N terminal domain, which potentially could drive FtsZ polymer bundling 49. It should be pointed out, however, that a mutant FtsZ with increased self-bundling in vitro (E93R) has reduced function in cell division 50,51, perhaps because its bundling is excessive.
It remains to be seen whether the activities of FtsA and ZipA are indeed antagonistic in vivo and whether they are separated spatially or temporally. A clue comes from the effects of excess FtsA on FtsZ. Normally, too much FtsA strongly inhibits Z ring activity, and the only way to reverse this is to concomitantly overproduce FtsZ 52. Strikingly, FtsZL169R rings remain functional in the presence of excess FtsA, suggesting that the negative effects of FtsA on FtsZ polymers can be antagonized by the action of ZipA, potentially via polymer bundling 51. Too much FtsA*, on the other hand, does not inhibit Z ring activity, consistent with its ability to replace ZipA in vivo but at odds with its disassembling effects on FtsZ polymers in vitro. Clearly more experiments with purified active FtsA and FtsA derivatives are needed to integrate their in vitro and in vivo activities. But the fact that FtsA* and other similar FtsA*-like mutants can completely bypass ZipA suggests that the FtsA*-like conformation alone can assume a strong tether role as well as a flexible tether role without much cost, at least under laboratory conditions.
Refining Our View of the Z Ring
Ring architecture illuminated by super-resolution
Upon binding GTP in a pocket conserved among all FtsZ and tubulin homologs, FtsZ polymerizes into single-stranded protofilaments (Figure 2B). The subunit interfaces thus formed can then induce GTP hydrolysis, which in turn leads to filament curving and disassembly in vitro 53,54. Alterations of buffer conditions or addition of certain factors (including other divisome proteins) can promote lateral interactions between FtsZ filaments, leading to FtsZ bundles, tubules, ribbons, and other higher-order structures 55.
Given what is known about these in vitro structures, how do they compare to the ultrastructure of FtsZ in the cell? Conventional imaging of FtsZ by fluorescent tags in live cells or immunofluorescent probes in fixed cells reveals what looks like a continuous ring of FtsZ around the cell periphery at the site of division, visible as a straight band in side view. Recent advances in imaging technology (Box 2) have improved our resolution of this Z ‘ring’ structure 56,57. Studies using electron cryo-tomography (ECT) of C. crescentus proposed that the Z ring is a largely discontinuous structure of loosely-associated, irregularly-spaced protofilaments 58. In contrast, a recent ECT study of C. crescentus and E. coli cells showed multiply-aligned FtsZ filaments along the membrane 47, evidence for a Z ring structure with continuous protofilament segments (Figure 3C). Fluorescence-based super-resolution techniques such as photo-activated localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM) or stimulated emission depletion (STED) microscopy were used to obtain 2D images from labeled FtsZ that support the idea of loosely arranged, patchy Z rings in multiple species 59,60. Another super-resolution method based on a distinct means of detection, 3D-structured illumination microscopy (3D-SIM) (Box 2), was used initially to image the divisome in B. subtilis and S. aureus and again suggested a heterogenous distribution of discontinuous, loosely bundled FtsZ filaments, as the FtsZ localized to patches interspersed with ~100–200 nm gaps 61–63. In B. subtilis, both fixed cells and live cells producing a fluorescent protein fused to FtsZ showed similar patterns, arguing against the possibility that the patches are artifacts of specimen preparation. Moreover, 3D-SIM of other divisome components, EzrA and penicillin-binding protein 2 (PBP2), in S. aureus suggested a similar organization, as did 3D-SIM of FtsZ, FtsA and ZipA in E. coli 61,62. Importantly, gaps between the more intense patches mostly contain some signal above background, suggesting that lower densities of FtsZ polymers are present in these areas (Figure 3A). Refinement of PALM hardware and software to permit high-throughput 3D reconstructions of C. crescentus at specific cell cycle stages further verified the varying, patch-like distribution of FtsZ in rings 59. Given the more continuous Z ring implicated by ECT 47, it will be important to gather more data under different conditions and with different methods to determine the structural model for the Z ring.
Box 2 Recent advances in fine structure imaging of bacteria.
Several new methods have made inroads in our understanding of the structure of the divisome in vivo. Whereas the resolution of two foci by conventional fluorescence microscopy is limited to ~300 nm, two novel fluorescence methods have substantially increased our ability to discern fine structures inside cells 152. One group of technologies imposes patterns on the image, effectively restricting out of focus light, followed by image reconstruction. These include 3D-structured illumination microscopy (3D-SIM) and stimulated emission depletion (STED). This type of imaging harvests all the light emitted from the fluorophore and can thus be used in dynamic studies, and has the advantage of using existing fluorescent proteins or dyes in living or fixed cells. The main disadvantages are the cost of the instruments, and in the case of 3D-SIM, the resolution is still limited to ~100 nm in the x-y plane and ~300 nm in the z plane. The potential resolution of STED and other related methods is much higher as it depends on the size of a mask that can be made around a point source, but this technology is still being optimized. The other group of technologies, called PALM (photoactivated localization microscopy) or STORM (stochastic optical reconstruction microscopy), relies on counting single fluorescent molecules in a ‘pointillist’ approach. By doing so, the exact position of each molecule counted can be determined within several nm, which is a huge increase in resolution. However, PALM and STORM require specialized fluorophores that switch from fluorescent to nonfluorescent in response to the excitation wavelength, allowing single molecules to be switched on or off. This requires specialized instrumentation as well as special photoactivatable dyes or fluorescent proteins. Finally, advances in electron cryo-tomography (ECT) have made important contributions to our understanding of molecular machines at the nanoscale 153, including the divisome. The advantages are that living cells are plunge-frozen, increasing the likelihood of recovering protein machines that were in their native state at the time of freezing without perturbations from fixation and without a requirement of fluorescent protein fusions. However, ECT requires costly instrumentation and time-consuming image processing, images are low in contrast, there are no dynamics, and some data are always lost from the ‘missing wedge’ caused by the tilt series. Moreover, proteins cannot yet be labeled with confidence for ECT, which leaves genetic alteration as the only way to confirm that a protein density visible in the reconstructed images belongs to a specific protein(s).
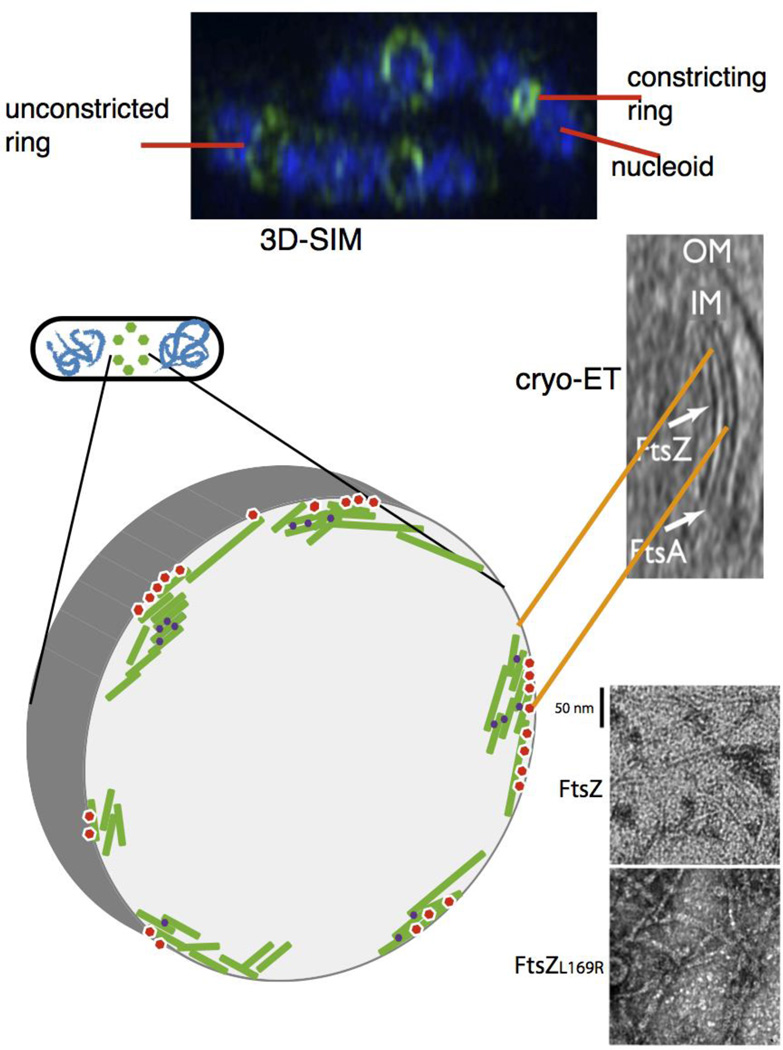
Fig. 3.
Proto-ring ultrastructure in E. coli. (A) 3D-SIM of several species indicates a patchy distribution of proto-ring proteins; shown is a 3D-SIM image of E. coli cells fixed and stained with DAPI (blue) for nucleoids and anti-FtsZ (green). Image courtesy of V. Rowlett. (B) the modeled structure based on this patchy pattern is depicted, with FtsZ protofilaments (green) attached to membrane tethers (red) and bundled by crosslinking Zap proteins (purple). (C) a cryo-ET image of E. coli producing FtsA and a mutant FtsZ that increases its stability, showing concentric filaments of both against the inner membrane (reproduced with permission from ref. 47). This is also modeled in the schematic in (B). (D) transmission EM images of purified FtsZ or FtsZL169R mutant in the presence of GTP, showing the double protofilaments for FtsZL169R that can compensate for the lack of other FtsZ bundling factors in vivo.
What can be deduced about the nano-structural organization of FtsZ filaments within this overall patchy distribution? Image analysis of optically trapped E. coli cells led to a structural model with a loosely connected, randomly oriented network of FtsZ filaments 64. Consistent with this model, polarized fluorescence microscopy (PFM) of live E. coli cells reveal a disorganized filament organization, although many align in the cell-axis direction 65. In contrast, a strong circumferential orientation bias for C. crescentus FtsZ filaments was observed by ECT 47,58 or PFM 65, suggesting that despite the general agreement on a patchy ring, there is substantial variation in fine-structural organization of FtsZ filaments within that patchy ring among different species. This should not be surprising given the diversity of divisome proteins and modes of cytokinesis—for example, E. coli divides by a combination of septation and constriction, while C. crescentus divides solely by constriction 66 —and thus may require a more aligned set of filaments. These variations clearly indicate that the E. coli system may not always be a universal paradigm of bacterial division organization.
The physiological relevance of FtsZ filament bundling
Despite these advances in imaging, the biological relevance of the various higher order structures formed by FtsZ protofilaments remains unclear. Proteins such as ZipA and SepF, as well as several known ‘Z associated proteins’ including ZapA, ZapC, and ZapD, promote lateral interactions between FtsZ protofilaments in vitro 55. This multitude of proteins suggests that the divisome—at least in E. coli and B. subtilis—is over-built, allowing for the most flexible responses to a variety of inputs including metabolic and stress signals. Therefore, the ability to cluster FtsZ polymers is probably fine-tuned by different physiological conditions. B. subtilis SepF is notable in that it self-assembles into large rings in vitro, which form a scaffold that promotes bundling of FtsZ into long tubules 67. Yet most of these FtsZ bundling proteins are nonessential for cell division and functionally redundant, making their potential bundling roles difficult to dissect in vivo. Moreover, any roles for ZipA and SepF in bundling are difficult to separate from their roles as membrane tethers for FtsZ. If they indeed have separable functions, then it may be possible to obtain mutants that are defective in one but not the other. However, this might be very challenging, as even tethering ZipA to the membrane with a heterologous transmembrane domain abolishes function 68. Other potential advances would be to visualize SepF scaffolds in vivo and reconstitute SepF-FtsZ dynamics in a membrane system as was done with ZipA-FtsA-FtsZ.
Despite their multiple potential roles, it is generally accepted that the net effect of these factors is to stabilize lateral interactions between FtsZ protofilaments in vivo, thus enhancing Z-ring integrity (Figure 3B). Cells that lack ZapA or produce ZapA variants that are defective in interaction with FtsZ assemble abnormal Z rings, reducing cell division efficiency 69. ZapC and ZapD, though functionally redundant, also promote proper Z ring assembly and subsequent cell division 70–72. Strikingly, PALM imaging of E. coli cells lacking ZapA or ZapB (which influences the Z ring by interacting with ZapA) shows broader, more disordered FtsZ filament clusters at the division site, suggesting that the role of these bundling factors may be to orient FtsZ filaments properly 73, perhaps during transition from a spiral-like assembly of FtsZ to the more constrained ring-like structures typical of initial Z ring assembly.
FtsZ protofilaments from different species differ greatly in their intrinsic ability to bundle, because of variations within the extreme carboxyl-terminus of FtsZ. This domain is separated from the conserved amino-terminal conserved do by a species-specific, intrinsically disordered linker of variable length (Figure 2B). The ~50 residue linkers of E. coli or B. subtilis can be replaced by heterologous intrinsically disordered peptides of roughly similar length without significant deleterious effects, although severe truncations of the linker are not functional 74,75. Many alpha-proteobacteria such as C. crescentus have much longer FtsZ linker domains, but surprisingly, replacement of the 250 residue C. crescentus linker with a 14 residue linker still allows function 76. Following the linker and the previously mentioned conserved C-terminal core peptide, which is important for interactions between FtsZ and other proteins, is a highly variable stretch of residues at the extreme carboxyl terminus called the C-terminal variable region, or CTV. The CTV is necessary and sufficient to induce lateral interactions between FtsZ protofilaments in the absence of modulatory proteins 77. For example, a chimera of E. coli FtsZ whose CTV is replaced by the CTV from B. subtilis bundles more readily than the native E. coli FtsZ. This highlights the ability of the B. subtilis CTV to mediate self-bundling, and is consistent with evidence showing that B. subtilis FtsZ protofilaments have a greater intrinsic ability to bundle 77.
The relatively low intrinsic bundling activities of some FtsZs such as E. coli FtsZ may require additional bundling proteins such as ZipA, ZapA, ZapC, or ZapD for optimal Z ring function in vivo. In support of this, the aforementioned FtsZL169R mutant forms protofilament bundles much more readily than wild-type FtsZ (Figure 3D) and can divide E. coli cells even in the absence of the normally essential ZipA 51. In addition to conferring resistance to excess FtsA, FtsZL169R also confers resistance to some endogenous FtsZ assembly inhibitors and suppresses the more subtle division defects associated with loss of zapA and zapC. The L169 residue lies on the lateral face of the FtsZ monomer, so a change to arginine may increase electrostatic interaction with a negatively charged residue on the lateral face of a monomer within another filament, which would promote filament bundling. Although the increased bundling of FtsZL169R results in a gain of function, it also sharply increases the frequency of abnormally shaped division septa, evidence that excess bundling has a fitness cost and the degree of FtsZ bundling in the cell is finely tuned. Regulation of bundling may explain how the axial width of initially assembled Z rings—anywhere from 70 nm (C. crescentus) to 95 nm (S. pneumoniae) to 110 nm (E. coli)—is maintained 59,60,78.
Divisome maturation and cytokinesis
Building the entire divisome
As its name implies, the proto-ring is only an initial stage of divisome assembly, whose major function is to recruit the second group of divisome proteins 79,80. These include the PBPs such as E. coli PBP3 (FtsI), which synthesize peptidoglycan required for formation of the cell division septum, and proteins involved in bridging interactions between these PBPs and the proto-ring (Figure 4A). However, even in the well-studied E. coli system, the precise function of some of these later divisome components remains enigmatic. For example, the conserved essential protein FtsW has a lipid II flippase activity 81 that may not be physiologically relevant 82.
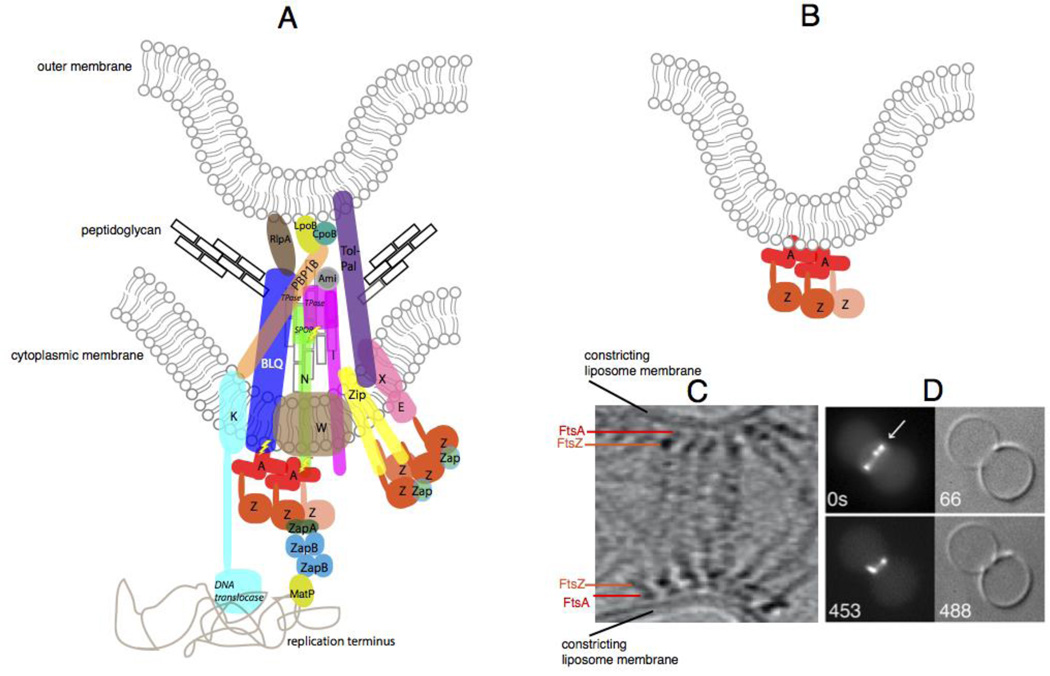
Fig. 4.
The constricting divisome in vivo and reconstituted in vitro. (A) A schematic of a putative individual sub-complex at the constricting divisome of E. coli shows FtsZ polymers being anchored to the membrane by FtsA and ZipA, along with other essential and nonessential divisome proteins of E. coli that cross the inner membrane, some of which contact the peptidoglycan and the outer membrane. Sidewall and septal peptidoglycan are denoted by black or grey rectangles, respectively. All Fts proteins are shown as a single letter, including the FtsBLQ complex. FtsZ dynamics are shown by the lighter colored FtsZ monomer coming off the polymer. Most known direct protein-protein contacts are shown, but some are speculative. Known enzymatic activities are highlighted in italics (TPase = transpeptidase). ZapC and ZapD are denoted as “Zap”, and the several periplasmic amidases of E. coli are denoted as “Ami”. Also shown are contacts made between the divisome and the terminus region of the nucleoid, including the helicase domain of FtsK155 and MatP, which probably interacts with ZapB 156. Regions of putative regulatory signaling are shown with lightning bolts. (B) Schematic of constriction of liposomes in vitro by FtsZ tethered to FtsA in the liposome lumen. (C) Cryo-ET of T. maritima FtsZ and FtsA proteins pinching a liposome from the inside reproduced with permission from Ref 47. and D) and fluorescence/DIC microscopy of E. coli FtsZ-YFP and FtsA* also pinching liposomes from the inside; reproduced with permission from ref 109.
The E. coli second-stage proteins FtsB, FtsL, and FtsQ each possess a short amino-terminal cytoplasmic domain, a single transmembrane domain and a larger periplasmic domain (Figure 4A); their function is also enigmatic. FtsB and FtsL are by themselves unstable in both E. coli and B. subtilis (FtsB is called DivIC in the latter species), but together they associate into a stable complex 83–85 associated with FtsQ (DivIB in B. subtilis and other Gram positives ). Interestingly, the extracellular domain of S. aureus DivIB can bind peptidoglycan 86 despite lacking a binding domain typical of other divisome proteins such as FtsN (see next section). The precise role of this binding, however, is unclear. The essentiality of the FtsBLQ subcomplex varies greatly between species, possibly reflecting their specific roles. Nonetheless, recent findings indicate that this complex likely participates in key signaling pathways important for regulating divisome activity (see below).
Coordinating cell division
As described above, the proto-ring proteins FtsA and ZipA in E. coli are required for the recruitment of subsequent divisome proteins under normal conditions, but little is known about how this recruitment occurs. ZipA can be bypassed by FtsA*-like mutants and others such as FtsZL169R, suggesting that these diverse mutants have gained the recruitment function of ZipA. Therefore, it is likely that ZipA does not directly recruit anything, and we propose that instead it may stimulate divisome protein recruitment indirectly via promotion of a higher-order FtsZ structure such as protofilament bundling 51. Although genetic and cytological assays implicate direct interactions between some proto-ring and later divisome proteins 87,88, the most compelling biochemical evidence to date for interactions between early and late proteins is the direct interaction between subdomain 1c of FtsA and the cytoplasmic tail of FtsN 89. Subdomain 1c, which is unique to FtsA and not present in other actin homologs, is required for recruitment of the late divisome proteins 90 (Figure 2B–C). This interaction likely promotes the initial recruitment of FtsN, which then stimulates septal peptidoglycan synthesis by an unknown mechanism, perhaps by interaction with FtsI. This initial septal synthesis then permits more recruitment of FtsN via its peptidoglycan-binding sporulation related (SPOR) domain in the periplasm (Figure 4A), which binds specifically to septal peptidoglycan 91, resulting in a self-reinforcing accumulation at the septum 92,93. Some hints as to how FtsN might stimulate septal peptidoglycan synthesis have come from studies of mutants that bypass FtsN. One class of such mutants maps to subdomain 1c of FtsA and may allow FtsA to mimic its activation by FtsN 94,95, perhaps by interfering with FtsA oligomerization 96. The other class maps to the periplasmic domains of FtsB or FtsL, which may likewise mimic FtsN-mediated activation on the periplasmic side of the membrane (Figure 4A) 95,97.
There is additional evidence that proto-ring proteins have roles in later divisome functions. The flexible linker domain of C. crescentus FtsZ, which is significantly longer than the analogous domains of most other FtsZ proteins, is important for proper regulation of PBPs, which have a role in sidewall growth that is associated with cytokinesis in that species 76. In E. coli, FtsZ and ZipA, but not FtsA, are required for preseptal peptidoglycan synthesis, a specific stage that occurs prior to divisome-mediated septum synthesis 98. Curiously, the ZipA requirement for preseptal peptidoglycan synthesis can be bypassed by FtsA*, suggesting that assembly state of FtsZ may ultimately govern this activity. In low-GC Gram-positive species such as B. subtilis, EzrA plays an indirect role in recruitment of PBPs 22,99, which is tied to post-translational regulation via the serine-threonine protein kinase family member PrkC 100–102.
Once the divisome is assembled, PBPs for septal growth must couple their enzymatic activity to membrane constriction and localized cell wall hydrolysis (Figure 4A). In Gram-negative bacteria, this regulation needs to extend to the outer membrane. The Tol-Pal complex, which physically connects both cytoplasmic and outer membranes in E. coli, is crucial for coordinating outer membrane constriction and peptidoglycan synthesis 103. At the heart of this system is CpoB (YbgF), which regulates PBP1B activity in response to the outer membrane Tol-Pal complex 104. Following septation and membrane constriction, hydrolases act to separate bacterial daughter cells in non-chaining species. These hydrolases may signal the Z ring via a conserved pair of proteins that form an ABC transporter homolog, FtsEX. In E. coli, FtsEX may signal the status of hydrolases to the proto-ring in order to coordinate ring constriction with septum growth and accompanying wall hydrolysis 105,106 It is possible that the several other proteins with SPOR domains may, like FtsN, be involved in this coordination. For example, the outer membrane lipoprotein RlpA localizes to the divisome but its deletion shows no phenotype in E. coli. However, in Pseudomonas RlpA was recently shown to be a transglycosylase that is needed to regulate cell shape and separation 92,107.
The source of the force?
In vitro reconstitution experiments have made important advances in our understanding of force generation by the Z ring on membranes. In the first of these experiments, a fluorescent FtsZ was artificially fused to an MTS, and mixed with tubular liposomes in a way that some of the protein was internalized within cell-shaped enclosures. Upon addition of GTP, the fluorescent FtsZ chimeras formed multiple ring-like structures, some of which conferred partial constriction of the liposomes 108. A subsequent study used the more physiological combination of wild-type FtsZ plus FtsA* to recapitulate the constriction (Figure 4B), and some potentially complete liposome scission events were observed (Figure 4D) 109. ECT was also used to visualize liposome constrictions in the presence of FtsZ and FtsA (Figure 4C) 47. Moreover, ECT of both C. crescentus and E. coli cells revealed filaments of FtsZ and FtsA at the membrane with overlapping lateral contacts. Together with the previously discussed ECT studies, this observed arrangement is consistent with a model of filament ‘sliding’ causing membrane constriction by pinching of the membrane, followed by reassembly of new sliding filament pairs that then pinch membranes further in subsequent cycles.
Both molecular modeling 110 and FtsZ crystal structure analysis 111 also support a model where a membrane-tethered FtsZ could provide the constrictive force for membrane invagination. However, recent fluorescence recovery after photobleaching (FRAP) studies indicate that FtsZ completely disassembles prior to the completion of constriction 112, suggesting that at least the latter stages of membrane constriction involve additional factors. Importantly, attachment of FtsA to vesicle membranes and subsequent polymerization caused vesicle shrinkage, suggesting that FtsA may also play some role in the constrictive force 113, perhaps similar to actin and myosin in animal cells. However, FtsZ attached to membrane vesicles by ZipA also can induce vesicle shrinkage 114, consistent with the idea that FtsA may not be required for force generation. Among the many questions that remain, one is whether FtsZ and FtsA/ZipA contribute the main driving force of constriction, or instead help guide the cytoplasmic membrane in front of localized cell wall synthesis. As any polymerizing protein can deform a membrane if it is tethered to it, another question is how specifically FtsZ and FtsA/ZipA contribute to constriction and how sensitive are they to membrane curvature. Finally, constriction in reconstitution experiments is not regulated, whereas in vivo it is. Therefore, if FtsZ is the force generator for the ring, it will ultimately be crucial to more fully understand how FtsZ disassembly is coordinated with septum synthesis and cell physiology. As mentioned in the previous section, FtsA is such a potential coordinator, likely by regulating FtsZ assembly dynamics (Figure 4A); recent reviews provide good summaries of how this might occur 115,116.
Metabolic and stress inputs
Some metabolic enzymes moonlight as Z-ring regulators
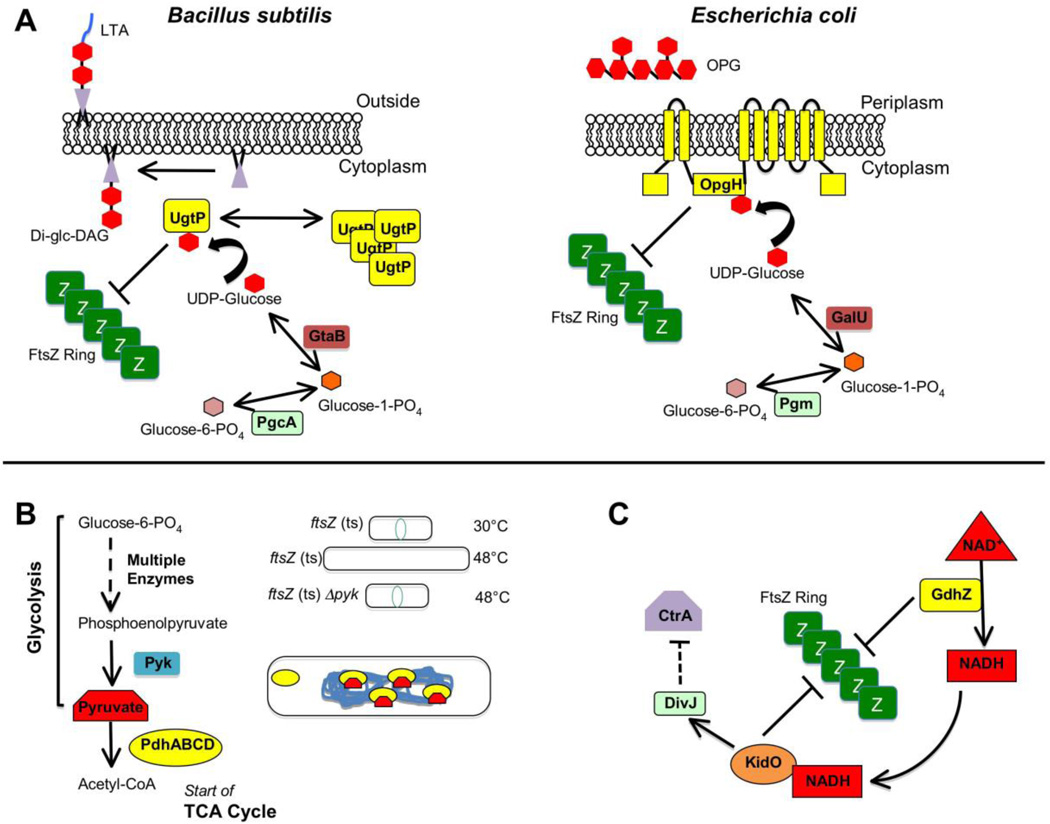
Like all cells, bacteria need to regulate how large they get before they reproduce, and bacilli like E. coli or B. subtilis are longer on average when grown in conditions with high nutrients versus low nutrients. Remarkably, these species increase their cell length simply by delaying formation of the divisome relative to cell growth. In B. subtilis, the glucosyltransferase UgtP achieves this by inhibiting FtsZ assembly specifically in nutrient-rich conditions 117 (Figure 5A, left panel). In nutrient-poor conditions, cells are deficient in UDP glucose, UgtP’s substrate. This induces UgtP to self-assemble into puncta that do not interact with FtsZ, thus permitting normal Z ring activity 118. This means of regulation is conserved in E. coli, but instead of UgtP, the inner-membrane protein OpgH serves as the UDP glucose-associated inhibitor of FtsZ assembly 119 (Figure 5A, right panel).
Fig. 5.
Regulation of divisome activity by stress and nutritional status. (A) The evolutionarily divergent B. subtilis and E. coli each use unrelated glucosyltransferases to coordinate growth rate with cell division through a metabolic determinant: levels of UDP-glucose 109,111,110. For B. subtilis (left), high levels of UDP-glucose during nutrient-rich growth permits UgtP synthesize the diglucosyl-diacylglycerol (Di-glc-DAG) anchor for lipoteichoic acid (LTA), an anionic polymer that is a major component of the Gram-positive cell wall. Additionally some UDP-glucose-bound UgtP interacts with FtsZ and inhibits FtsZ assembly through an uncertain mechanism, delaying division until cells have reached appropriate size for their rapid growth rate. In the absence of UDP-glucose substrate, UgtP is sequestered from FtsZ in cytoplasmic puncta. For E. coli (right), OpgH is a sugar transferase involved in synthesis of osmoregulated periplasmic glucans (OPGs). As with its functional counterpart in Bacillus, OpgH inhibits FtsZ ring formation in a UDP-glucose-dependent manner, likely through sequestration of FtsZ monomers. Although OpgH also only seems to interact with FtsZ during nutrient-rich growth conditions, it does so independently of UDP-glucose, suggesting the existence of other metabolic determinants for this coordination in E. coli. (Modified with permission from ref. 119). (B) Enzymes and intermediates involved in glycolysis and the TCA cycle (left) also play a role in B. subtilis cell division 121. Loss of the pyk gene encoding pyruvate kinase suppresses division defects of a temperature sensitive ftsZ allele under non-permissive conditions (upper & mid, right). In the cell (lower right), binding of pyruvate substrate by the E1alpha subunit (encoded by pdhA) of pyruvate dehydrogenase permits localization of the enzyme/substrate complex to the bacterial nucleoid. E1alpha subunits unable to bind pyruvate substrate (such as in a pyk-null mutant) no longer associate with the nucleoid. (C) A simplification of the highly complex integrated genetic circuit in Caulobacter crescenus linking cell division with metabolism and DNA replication 122,123. Active GdhZ promotes FtsZ disassembly through stimulation of GTP hydrolysis by FtsZ and through its catalysis of glutamate catabolism into alpha-ketoglutarate provides NADH substrate for KidO, which in turn also can inhibit FtsZ assembly through an unknown mechanism. KidO stimulates the kinase activity of DivJ, which leads to a signal transduction pathway leading through multiple mechanisms to inhibition of the activity of CtrA, a master regulator of both cell division and DNA replication timing in C. crescentus. Activated DivJ additionally feeds back on this circuit to inhibit KidO and GdhZ through proteolytic control governed by ClpXP (not shown).
Another metabolic regulatory gene, yvcL, was isolated in a screen for genes synthetically lethal with a zapA-deficient B. subtilis mutant 120. Cells lacking both zapA and yvcL exhibit inefficient Z ring assembly and thus divide poorly. YvcL shares sequence similarity with WhiA, a S. coelicolor transcription factor involved in ftsZ expression. However, although B. subtilis YvcL localizes to the nucleoid, it does not seem to regulate ftsZ gene expression. Interestingly, suppressors of zapA yvcL double mutants mapped to genes involved in the UDP glucose pathway, and inactivation of ugtP suppressed zapA yvcL division defects. This hints at new links among metabolism, cell division, and the nucleoid 120.
Metabolic regulation of FtsZ assembly is not limited to pathways involving UDP glucose. In B. subtilis, deletion of pyk, encoding pyruvate kinase, suppresses a thermosensitive ftsZ mutant and leads to formation of extraneous Z rings in otherwise normal cells 121, much like the effects of an ezrA deletion 21. Addition of exogenous pyruvate reverses these pyk phenotypes. The proposed mechanism involves the E1α subunit of pyruvate dehydrogenase, which localizes over the nucleoid in a pyruvate-dependent manner 121 (Figure 5B). Although the exact mechanism is not yet clear, it suggests a way that the status of glycolysis can regulate cytokinesis and perhaps nucleoid organization.
Central metabolism also influences Z ring assembly in C. crescentus (Figure 5C). This is accomplished via two genes, kidO 122 and gdhZ 123, which are regulated by the global transcription factor CtrA, and whose gene products are under proteolytic regulation by ClpXP, all in conjunction with the C. crescentus developmental cycle. KidO is an oxidoreductase that stimulates the kinase activity of DivJ, a protein involved in CtrA turnover 122. KidO inhibits FtsZ polymer bundling when bound to NADH 123. GdhZ is an NAD-dependent glutamate dehydrogenase that links the tricarboxylic acid cycle cycle to nitrogen metabolism and directly promotes FtsZ GTPase activity 123. A further link between cell growth, replication, and division comes from B. subtilis PlsX, an acyl–acyl carrier protein phosphate acyltransferase that serves as a key enzyme in phospholipid and fatty acid catabolism. PlsX localizes mainly to cell division sites prior to FtsZ and FtsA in response to the progression of chromosome replication and interacts with FtsA to stabilize the proto-ring 124. Finally, although not a metabolic enzyme itself, ClpP protease can moonlight as an FtsZ disassembly factor in B. subtilis 125,126, proteolytically degrade FtsZ in E. coli 125,126 or both FtsZ and FtsA in C. crescentus 127.
Stressing out the divisome
Cellular stress also plays an important role in regulating cell division, by influencing assembly of the Z ring or later divisome proteins. The SOS response to DNA damage is the archetypal pathway linking stress and cytokinesis. In E. coli and related bacteria, the SulA protein is induced by the SOS response and transiently inhibits cell division to allow time for DNA repair. SulA directly binds to FtsZ, sequestering FtsZ monomers away from the pool available for Z ring assembly 128. Interestingly, in C. crescentus the unrelated SOS-induced factor SidA targets a later divisome protein, FtsW, to inhibit division 129. A second factor, DidA, responds to C. crescentus stress independently of the SOS pathway to inhibit division via interaction with FtsN 130. Similarly, an as-yet-unidentified component of the B. subtilis late divisome serves as a target for SOS-induced YneA 131. SulA acts transiently because it is proteolytically unstable, whereas instability of yneA mRNA ensures transient activity of its gene product 132. Oxygen and heat stress also modulate FtsZ assembly. ZapE, an ATPase present in E. coli and the related Shigella flexneri and conserved in other Gram-negative bacteria, is nonessential but becomes important to maintain normal cell division in low oxygen or high temperature conditions 133. It localizes to the divisome and, like FtsA, seems to disassemble FtsZ polymers in an ATP-dependent manner.
Mechanical stress may also influence bacterial cell division, which would not be surprising given that cytokinesis exerts a substantial inward force against turgor pressure. One of the small mechanosensitive channel proteins in E. coli, MscS, interacts with FtsZ through its cytoplasmic C-terminal domain 134. This domain changes conformation in response to channel opening or inactivation, which can be induced by cell envelope stresses such as membrane bulging from exposure to β-lactam antibiotics. Although there is no evidence for a direct effect of MscS on FtsZ activity, it is possible that MscS is enriched at invaginating membranes at the divisome and may modulate Z ring activity in some way. Interestingly, an E. coli mutant lacking all three mechanosensitive channels (MscS, MscL, and MscK), while able to grow and divide normally, assembles excess Z rings compared with wild-type cells when a later stage of septum synthesis is inhibited 135. This is consistent with potential inhibition of FtsZ assembly by one or more of these channels under some conditions.
Outlook
Understanding how bacteria divide at the molecular level is not only of fundamental importance for cell biology, but also provides new candidates for antimicrobial therapies. Although some bacteria without cell walls, called L-forms, can divide without FtsZ or other divisome components 136, the diverse proteins of the divisome provide intriguing targets for novel antibiotic development 137–139. The most progress has been made with chemical inhibitors of FtsZ assembly, including molecules that compete with GTP 140. In addition, there has been a recent resurgence in understanding how bacteriophage factors alter host cell shape and division. For example, the E. coli phages T7 and λ produce peptides that inhibit host cell division during lytic growth by targeting FtsZ assembly 141–143. In contrast, the B. subtilis phage φ29 binds to FtsZ but does not inhibit its function 144. Peptide inhibitors of FtsZ are not restricted to phages: B. subtilis produces a peptide that caps FtsZ protofilaments to prevent Z ring formation in terminally differentiated mother cells during sporulation 145. Alongside the revisiting of phage therapy as an alternative to antibiotics, the mining of phage as well as host genomes for small peptides that target growth or division of specific bacterial species could be an attractive resource for understanding antibacterial activity or identifying novel narrow-range antibiotic targets and compounds. Overall, the expansion of our knowledge beyond the E. coli model system has begun to illustrate the enormous diversity of bacterial cell division mechanisms, which can help us understand the flexibility of these mechanisms but also creates a challenge to dissect out the key players and their relevant activities. However, the E. coli model system is far from solved. The main near-term challenges for that system include understanding how divisome proteins signal each other to coordinate divisome assembly, constriction and disassembly with cell wall synthesis, and the role of connector proteins such as FtsA in this process. This will require more complete understanding of mutant phenotypes in vivo and how they reflect different activities in various in vitro assays, including reconstitution experiments.
KEY POINTS.
All cells must divide in order to proliferate, and most bacteria divide by splitting themselves into two, called cytokinesis. Many bacteria divide by splitting into roughly equal halves, called binary fission. Cytokinesis in bacteria is accomplished by a dedicated protein machine located at the site of cell division called the divisome. Recent advances in ultrastructural imaging, biochemistry and genetics of Escherichia coli and other model bacterial species have helped to refine models for divisome function and regulation.
FtsZ, the bacterial homolog of tubulin, is the principal driver of bacterial cytokinesis. In vitro, FtsZ assembles into single protofilaments in the presence of GTP. In vivo, these protofilaments loosely assemble to encircle the cell at the site of division—called the Z ring—and are positioned there by species-specific spatial positioning proteins.
As FtsZ is a soluble protein, FtsZ protofilaments must be tethered to the inner surface of the cytoplasmic membrane by additional proteins, including FtsA and ZipA in E. coli. This complex of FtsZ and membrane tethers is called the proto-ring and displays highly dynamic behavior.
Although they do not form microtubules, FtsZ protofilaments self-associate to form bundles, either via interactions with other FtsZ subunits or with multiple FtsZ-binding proteins that enhance bundling, including ZipA and the Zap proteins. These lateral interactions among FtsZ protofilaments may play a significant role in FtsZ’s ability to divide a cell.
FtsA, a bacterial homolog of actin, is a key connector between the Z ring and other divisome proteins, all of which span the membrane and some of which bind to the peptidoglycan layer. Once the divisome is completely assembled, it coordinates the inward constriction of the Z ring and cytoplasmic membrane with the synthesis of the cell division septum, which is composed of peptidoglycan. FtsA is a key player in this coordination, which likely involves feedback signaling between the peptidoglycan-binding divisome proteins and the Z ring. Biochemical characterization of FtsA remains a major challenge.
In addition to signaling within the divisome during the process of cytokinesis, the divisome is also regulated by mechanical, metabolic, and stress inputs. FtsZ is a major target for these regulators, but other divisome proteins are also targets. Understanding how divisome proteins are inhibited or stimulated will be valuable in the future design of divisome-specific antimicrobial compounds.
Acknowledgments
The authors gratefully acknowledge support from the National Institute of General Medical Sciences (R01-GM61074, to W.M.).
Glossary
- Cytokinesis
The splitting of a cell’s contents to make two cells
- Nucleoid
the nucleus-like organized structure of bacterial chromosomes
- Mycelium
a filamentous, branched network of multinucleate cells growing on a surface
- Thermosensitive ftsZ mutant
A point mutation in the ftsZ gene that permits normal growth and division at 30°C but ceases division at 42°C despite continued growth, resulting in filamentous, multinucleate cells (thus the term fts for filamentous temperature sensitive mutants)
- Central metabolism
metabolic pathways, such as the tricarboxylic acid cycle, that provide precursor metabolites for all other pathways needed for growth
- ClpXP
A protein chaperone-protease machine that targets certain proteins for unfolding and degradation
- Lipid II flippase
A membrane protein that transfers lipid-linked peptidoglycan precursors from the inner to the outer leaflet of the cytoplasmic membrane, so they can be incorporated into the cell wall
- Sidewall
the peptidoglycan layer in many rod-shaped bacteria that is active in elongating the cell and comprises most of the straight wall of the cell with the exception of cell poles and the division septum
- SOS response
Induced by DNA damage, SOS is a stress response that results in the expression of many genes important for protection of the chromosomal DNA
Biographies
Daniel Haeusser first studied bacterial cell division during his Ph.D. in the laboratory of Petra Levin at Washington University in St. Louis, and continued his work on cell division and bacteriophage inhibitors during his postdoctoral studies with William Margolin at the University of Texas Medical School at Houston. He recently started a faculty position at Canisius College in Buffalo, NY.
William Margolin is a Professor of Microbiology and Molecular Genetics at the University of Texas Medical School (now called the McGovern Medical School) in Houston, Texas. Over the last 25 years he has made many contributions towards the molecular understanding of bacterial cell division and cellular organization of protein machines, focusing mainly on Escherichia coli.
REFERENCES
- 1.Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann NY Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown PJ, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2011;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 4.Leisch N, et al. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr Biol. 2012;22:R831–R832. doi: 10.1016/j.cub.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 5.Monahan LG, Liew ATF, Bottomley AL, Harry EJ. Division site positioning in bacteria: one size does not fit all. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowlett VW, Margolin W. The Min system and other nucleoid-independent regulators of Z ring positioning. Front. Microbiol. 2015;6:478. doi: 10.3389/fmicb.2015.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. doi: 10.1101/gad.600211. The first example of positive spatial regulation of Z ring positioning.
- 8.Treuner-Lange A, et al. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol Microbiol. 2013;87:235–253. doi: 10.1111/mmi.12094. [DOI] [PubMed] [Google Scholar]
- 9.Holeckova N, et al. LocZ Is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. mBio. 2014;6:e01700–e01714. doi: 10.1128/mBio.01700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleurie A, et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormick JR, Su EP, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 12.Rico AI, Krupka M, Vicente M. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J Biol Chem. 2013;288:20830–20836. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 14.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 15.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duman R, et al. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc. Natl. Acad. Sci. 2013;110:E4601–E4610. doi: 10.1073/pnas.1313978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krol E, et al. Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS One. 2013;7:e43293. doi: 10.1371/journal.pone.0043293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 2006;60:1364–1380. doi: 10.1111/j.1365-2958.2006.05184.x. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S, et al. The essential protein SepF of mycobacteria interacts with FtsZ and MurG to regulate cell growth and division. Microbiology. 2015 doi: 10.1099/mic.0.000108. [DOI] [PubMed] [Google Scholar]
- 20.Gola S, Munder T, Casonato S, Manganelli R, Vicente M. The essential role of SepF in mycobacterial division. Mol. Microbiol. 2015;97:560–576. doi: 10.1111/mmi.13050. [DOI] [PubMed] [Google Scholar]
- 21.Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steele VR, Bottomley AL, Garcia-Lara J, Kasturiarachchi J, Foster SJ. Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Mol Microbiol. 2011;80:542–555. doi: 10.1111/j.1365-2958.2011.07591.x. [DOI] [PubMed] [Google Scholar]
- 23.Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol. 2004;52:801–814. doi: 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleverley RM, et al. Structure and function of a spectrin-like regulator of bacterial cytokinesis. Nat. Commun. 2014;5:5421. doi: 10.1038/ncomms6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machnicka B, et al. Spectrins: a structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim. Biophys. Acta. 2014;1838:620–634. doi: 10.1016/j.bbamem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Haeusser DP, Garza AC, Buscher AZ, Levin PA. The division inhibitor EzrA contains a seven-residue patch required for maintaining the dynamic nature of the medial FtsZ ring. J. Bacteriol. 2007;189:9001–9010. doi: 10.1128/JB.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Land AD, Luo Q, Levin PA. Functional domain analysis of the cell division inhibitor EzrA. PloS One. 2014;9:e102616. doi: 10.1371/journal.pone.0102616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Son SH, Lee HH. The N-terminal domain of EzrA binds to the C terminus of FtsZ to inhibit Staphylococcus aureus FtsZ polymerization. Biochem Biophys Res Commun. 2013;433:108–114. doi: 10.1016/j.bbrc.2013.02.055. [DOI] [PubMed] [Google Scholar]
- 29.Van Den Ent F, Löwe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez M, Valencia A, Ferrandiz M-J, Sandler C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- 32.Paradis-Bleau C, Sanschagrin F, Levesque RC. Peptide inhibitors of the essential cell division protein FtsA. Protein Eng Sel. 2005;18:85–91. doi: 10.1093/protein/gzi008. [DOI] [PubMed] [Google Scholar]
- 33. Lara B, et al. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol. 2005;55:699–711. doi: 10.1111/j.1365-2958.2004.04432.x. The first demonstration that FtsA has ATP-dependent polymerization activity.
- 34. Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. This study structurally defined the FtsA-FtsA and FtsA-FtsZ interfaces.
- 35. Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. This study showed that many lesions that interefere with FtsA oligomerization render ZipA nonessential.
- 36.Shiomi D, Margolin W. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol. 2007;66:1396–1415. doi: 10.1111/j.1365-2958.2007.05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita J, et al. Crystal structure of FtsA from Staphylococcus aureus. FEBS Lett. 2014;588:1879–1885. doi: 10.1016/j.febslet.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Geissler B, Elraheb D, Margolin W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beuria TK, et al. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J Biol Chem. 2009;284:14079–14086. doi: 10.1074/jbc.M808872200. The first evidence that FtsA can affect the assembly state of FtsZ.
- 41. Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. This study showed that FtsA can regulate FtsZ assembly dynamics on a physiological membrane surface.
- 42.Modi K, Misra HS. Dr-FtsA, an actin homologue in Deinococcus radiodurans differentially affects Dr-FtsZ and Ec-FtsZ functions In vitro. PloS One. 2014;9:e115918. doi: 10.1371/journal.pone.0115918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du S, Park K-T, Lutkenhaus J. Oligomerization of FtsZ converts the FtsZ tail motif (CCTP) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol. Microbiol. 2014 doi: 10.1111/mmi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herricks JR, Nguyen D, Margolin W. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol. Microbiol. 2014;94:713–727. doi: 10.1111/mmi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen B, Lutkenhaus J. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol. 2009;72:410–424. doi: 10.1111/j.1365-2958.2009.06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowlett VW, Margolin W. Asymmetric constriction of dividing Escherichia coli cells induced by expression of a fusion between two min proteins. J Bacteriol. 2014;196:2089–2100. doi: 10.1128/JB.01425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez-Rocamora VM, et al. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J Biol Chem. 2012;287:30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skoog K, Daley DO. The Escherichia coli cell division protein ZipA forms homodimers prior to association with FtsZ. Biochemistry (Mosc.) 2012;51:1407–1415. doi: 10.1021/bi2015647. [DOI] [PubMed] [Google Scholar]
- 50.Jaiswal R, et al. E93R substitution of Escherichia coli FtsZ induces bundling of protofilaments, reduces GTPase activity, and impairs bacterial cytokinesis. J Biol Chem. 2010;285:31796–31805. doi: 10.1074/jbc.M110.138719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haeusser DP, Rowlett VW, Margolin W. A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol. Microbiol. 2015;97:988–1005. doi: 10.1111/mmi.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJ. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry (Mosc.) 2002;41:521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- 55.Huang KH, Durand-Heredia J, Janakiraman A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol. 2013;195:1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier EL, Goley ED. Form and function of the bacterial cytokinetic ring. Curr. Opin. Cell Biol. 2014;26:147. doi: 10.1016/j.ceb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 57.Rowlett VW, Margolin W. The bacterial divisome: ready for its close-up. Philos Trans R Soc Lond B Biol Sci. 2015;370:20150028. doi: 10.1098/rstb.2015.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. The first glimpse of the Z ring at high resolution by electron tomography.
- 59.Holden SJ, et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacq M, et al. Remodeling of the Z-ring nanostructure during the Streptococcus pneumoniae cell cycle revealed by photoactivated localization microscopy. mBio. 2015;6:e01108–e01115. doi: 10.1128/mBio.01108-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Strauss MP, et al. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. The first direct visual evidence that the Z ring may be a non-uniform structure.
- 62.Rowlett VW, Margolin W. 3D-SIM super-resolution of FtsZ and Its membrane tethers in Escherichia coli cells. Biophys. J. 2014;107:L17–L20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsui H-CT, et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol. Microbiol. 2014;94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piro O, Carmon G, Feingold M, Fishov I. Three-dimensional structure of the Z-ring as a random network of FtsZ filaments. Env. Microbiol. 2013;15:3252–3258. doi: 10.1111/1462-2920.12197. [DOI] [PubMed] [Google Scholar]
- 65.Si F, Busiek K, Margolin W, Sun SX. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys J. 2013;105:1976–1986. doi: 10.1016/j.bpj.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Judd EM, et al. Distinct constrictive processes, separated in time and space, divide caulobacter inner and outer membranes. J. Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gundogdu ME, et al. Large ring polymers align FtsZ polymers for normal septum formation. Embo J. 2011;30:617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roach EJ, Kimber MS, Khursigara CM. Crystal structure and site-directed mutational analysis reveals key residues involved in Escherichia coli ZapA function. J. Biol. Chem. 2014;289:23276–23286. doi: 10.1074/jbc.M114.561928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol. 2011;193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hale CA, et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol. 2011;193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buss J, et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gardner KA, Moore DA, Erickson HP. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol. 2013;89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buske PJ, Levin PA. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol. 2013;89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sundararajan K, et al. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 2015;6:7281. doi: 10.1038/ncomms8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buske PJ, Levin PA. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem. 2012;287:10945–10957. doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu G, et al. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aarsman ME, et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 80.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohammadi T, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sham L-T, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van den van Saparoea Berg HB, et al. Fine-mapping the contact sites of the Escherichia coli cell division proteins FtsB and FtsL on the FtsQ protein. J. Biol. Chem. 2013;288:24340–24350. doi: 10.1074/jbc.M113.485888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khadria AS, Senes A. The transmembrane domains of the bacterial cell division proteins FtsB and FtsL form a stable high-order oligomer. Biochemistry (Mosc.) 2013;52:7542–7550. doi: 10.1021/bi4009837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Glas M, et al. The soluble periplasmic domains of E. coli cell division proteins FtsQ/FtsB/FtsL form a trimeric complex with sub-micromolar affinity. J. Biol. Chem. 2015;290:21498–21509. doi: 10.1074/jbc.M115.654756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bottomley AL, et al. Staphylococcus aureus DivIB is a peptidoglycan-binding protein that is required for a morphological checkpoint in cell division. Mol. Microbiol. 2014;94:1041–1064. doi: 10.1111/mmi.12813. [DOI] [PubMed] [Google Scholar]
- 87.Grenga L, Rizzo A, Paolozzi L, Ghelardini P. Essential and non-essential interactions in interactome networks: the Escherichia coli division proteins FtsQ-FtsN interaction. Env. Microbiol. 2013;15:3210–3217. doi: 10.1111/1462-2920.12157. [DOI] [PubMed] [Google Scholar]
- 88.Alexeeva S, Gadella JTW, Verheul J, Verhoeven GS, den Blaauwen T. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol. 2010;77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. [DOI] [PubMed] [Google Scholar]
- 89.Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through Its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol Microbiol. 2004;53:1359–1371. doi: 10.1111/j.1365-2958.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- 91.Yahashiri A, Jorgenson MA, Weiss DS. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11347–11352. doi: 10.1073/pnas.1508536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gerding MA, et al. Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Busiek KK, Margolin W. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol Microbiol. 2014;92:1212–1226. doi: 10.1111/mmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu B, Persons L, Lee L, DeBoer P. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 2015;95:945–970. doi: 10.1111/mmi.12906. Along with ref. 89, 93 and 97, this study provides additional evidence for multiple signaling pathways within the divisome to activate cytokinesis.
- 96.Pichoff S, Du S, Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol. 2015;95:971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsang M-J, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 2015;95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Potluri LP, Kannan S, Young KD. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol. 2012;194:5334–5342. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Claessen D, et al. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- 100.Land AD, et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol. Microbiol. 2013;90:939–955. doi: 10.1111/mmi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fleurie A, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pompeo F, Foulquier E, Serrano B, Grangeasse C, Galinier A. Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis. Mol. Microbiol. 2015;97:139–150. doi: 10.1111/mmi.13015. [DOI] [PubMed] [Google Scholar]
- 103.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gray AN, et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife. 2015;4 doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol. 2007;189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang DC, et al. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jorgenson MA, Chen Y, Yahashiri A, Popham DL, Weiss DS. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol. Microbiol. 2014;93:113–128. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. The first demonstration that Z rings can be reconstituted in vitro.
- 109.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dow CE, van den Berg HA, Roper DI, Rodger A. Biological insights from a simulation model of the critical FtsZ accumulation required for prokaryotic cell division. Biochemistry (Mosc.) 2015;54:3803–3813. doi: 10.1021/acs.biochem.5b00261. [DOI] [PubMed] [Google Scholar]
- 111.Li Y, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Söderström B, et al. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol Microbiol. 2014;92:1–9. doi: 10.1111/mmi.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krupka M, et al. Role of the FtsA C terminus as a switch for polymerization and membrane association. mBio. 2014;5:e02221. doi: 10.1128/mBio.02221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cabre EJ, et al. Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J Biol Chem. 2013;288:26625–26634. doi: 10.1074/jbc.M113.491688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weiss DS. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Mol. Microbiol. 2015:903–909. doi: 10.1111/mmi.12925. [DOI] [PubMed] [Google Scholar]
- 116.Tsang M-J, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weart RB, et al. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chien AC, Zareh SK, Wang YM, Levin PA. Changes in the oligomerization potential of the division inhibitor UgtP co-ordinate Bacillus subtilis cell size with nutrient availability. Mol Microbiol. 2012;86:594–610. doi: 10.1111/mmi.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hill NS, Buske PJ, Shi Y, Levin PA. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 2013;9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Surdova K, et al. The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis. J. Bacteriol. 2013;195:5450–5460. doi: 10.1128/JB.00507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Monahan LG, Hajduk IV, Blaber SP, Charles IG, Harry EJ. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio. 2014;5:e00935–e00914. doi: 10.1128/mBio.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 123.Beaufay F, et al. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J. 2015;34:1786–1800. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takada H, et al. An essential enzyme for phospholipid synthesis associates with the Bacillus subtilis divisome. Mol. Microbiol. 2014;91:242–255. doi: 10.1111/mmi.12457. [DOI] [PubMed] [Google Scholar]
- 125.Weart RB, Nakano S, Lane BE, Zuber P, Levin PA. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol. 2005;57:238–249. doi: 10.1111/j.1365-2958.2005.04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Camberg JL, Hoskins JR, Wickner S. The interplay of ClpXP with the cell division machinery in Escherichia coli. J Bacteriol. 2011;193:1911–1918. doi: 10.1128/JB.01317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol. Microbiol. 2014;93:853–866. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen Y, Milam SL, Erickson HP. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry (Mosc.) 2012;51:3100–3109. doi: 10.1021/bi201669d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Modell JW, Kambara TK, Perchuk BS, Laub MT. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mo AH, Burkholder WF. YneA, an SOS-induced inhibitor of cell division in Bacillus subtilis, is regulated posttranslationally and requires the transmembrane region for activity. J Bacteriol. 2010;192:3159–3173. doi: 10.1128/JB.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Buchholz M, Nahrstedt H, Pillukat MH, Deppe V, Meinhardt F. yneA mRNA instability is involved in temporary inhibition of cell division during the SOS response of Bacillus megaterium. Microbiology. 2013;159:1564–1574. doi: 10.1099/mic.0.064766-0. [DOI] [PubMed] [Google Scholar]
- 133.Marteyn BS, et al. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. MBio. 2014;5:e00022–e00014. doi: 10.1128/mBio.00022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koprowski P, et al. Cytoplasmic domain of MscS interacts with cell division protein FtsZ: A possible non-channel function of the mechanosensitive channel in Escherichia coli. PloS One. 2015;10:e0127029. doi: 10.1371/journal.pone.0127029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wilson ME, Jensen GS, Haswell ES. Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell. 2011;23:2939–2949. doi: 10.1105/tpc.111.088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mercier R, Kawai Y, Errington J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife. 2014;3 doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lock RL, Harry EJ. Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 2008;7:324–338. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 138.Ojima I, Kumar K, Awasthi D, Vineberg JG. Drug discovery targeting cell division proteins, microtubules and FtsZ. Bioorg Med Chem. 2014;22:5060–5077. doi: 10.1016/j.bmc.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sass P, Brotz-Oesterhelt H. Bacterial cell division as a target for new antibiotics. Curr Opin Microbiol. 2013;16:522–530. doi: 10.1016/j.mib.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 140.Artola M, et al. Effective GTP-replacing FtsZ Inhibitors and Antibacterial Mechanism of Action. ACS Chem. Biol. 2014 doi: 10.1021/cb500974d. [DOI] [PubMed] [Google Scholar]
- 141.Haeusser DP, et al. The Kil peptide of bacteriophage lambda blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 2014;10:e1004217. doi: 10.1371/journal.pgen.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kiro R, et al. Gene product 0.4 increases bacteriophage T7 competitiveness by inhibiting host cell division. Proc. Natl. Acad. Sci. 2013;110:19549–19554. doi: 10.1073/pnas.1314096110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hernandez-Rocamora VM, Alfonso C, Margolin W, Zorrilla S, Rivas G. Evidence that bacteriophage lambda Kil peptide inhibits bacterial cell division by disrupting FtsZ protofilaments and sequestering protein subunits. J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.653329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ballesteros-Plaza D, Holguera I, Scheffers DJ, Salas M, Munoz-Espin D. Phage 29 phi protein p1 promotes replication by associating with the FtsZ ring of the divisome in Bacillus subtilis. Proc Natl Acad Sci U A. 2013;110:12313–12318. doi: 10.1073/pnas.1311524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bisson-Filho AW, et al. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E2130–E2138. doi: 10.1073/pnas.1414242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pende N, et al. Size-independent symmetric division in extraordinarily long cells. Nat. Commun. 2014;5:4803. doi: 10.1038/ncomms5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Donovan C, Bramkamp M. Cell division in Corynebacterineae. Front. Microbiol. 2014;5:132. doi: 10.3389/fmicb.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kieser KJ, Rubin EJ. How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 2014;12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ramos-Leon F, Mariscal V, Frias JE, Flores E, Herrero A. Divisome-dependent subcellular localization of cell-cell joining protein SepJ in the filamentous cyanobacterium Anabaena. Mol. Microbiol. 2015 doi: 10.1111/mmi.12956. [DOI] [PubMed] [Google Scholar]
- 150.Pinho MG, Kjos M, Veening J-W. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 2013;11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 151.Angert ER. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- 152.Tuson HH, Biteen JS. Unveiling the inner workings of live bacteria using super-resolution microscopy. Anal. Chem. 2015;87:42–63. doi: 10.1021/ac5041346. [DOI] [PubMed] [Google Scholar]
- 153.Asano S, Engel BD, Baumeister W. In situ cryo-electron tomography: A post-reductionist approach to structural biology. J. Mol. Biol. 2015 doi: 10.1016/j.jmb.2015.09.030. in press. [DOI] [PubMed] [Google Scholar]
- 154.Busiek KK, Margolin W. Bacterial actin and tubulin homologs in cell growth and division. Curr. Biol. 2015;25:245–254. doi: 10.1016/j.cub.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grainge I. FtsK-a bacterial cell division checkpoint? Mol Microbiol. 2010;78:1055–1057. doi: 10.1111/j.1365-2958.2010.07411.x. [DOI] [PubMed] [Google Scholar]
- 156.Buss J, et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]