Abstract
Bacteria are exposed to reactive oxygen species from the environment and from those generated by aerobic metabolism. Catalases are heme proteins that detoxify H2O2, and many bacteria contain more than one catalase enzyme. Also, the nonheme peroxidase alkyl hydroperoxide reductase (Ahp) is the major scavenger of endogenous H2O2 in Escherichia coli. Here, we show that aerobically grown Bradyrhizobium japonicum cells express a single catalase activity. Four genes encoding putative catalases in the B. japonicum genome were identified, including a katG homolog encoding a catalase-peroxidase. Deletion of the katG gene resulted in loss of catalase activity in cell extracts and of exogenous H2O2 consumption by whole cells. The katG strain had a severe aerobic growth phenotype but showed improved growth in the absence of O2. By contrast, a B. japonicum ahpCD mutant grew well aerobically and consumed H2O2 at wild-type rates. A heme-deficient hemA mutant expressed about one-third of the KatG activity as the wild type but grew well aerobically and scavenged low concentrations of exogenous H2O2. However, cells of the hemA strain were deficient in consumption of high concentrations of H2O2 and were very sensitive to killing by short exposure to H2O2. In addition, KatG activity did not decrease as a result of mutation of the gene encoding the transcriptional activator OxyR. We conclude that aerobic metabolism produces toxic levels of H2O2 in B. japonicum, which is detoxified primarily by KatG. Furthermore, the katG level sufficient for detoxification does not require OxyR.
Reactive oxygen species (ROS) are generated during aerobic respiration as a result of the partial reduction of molecular oxygen and the subsequent reactions of those products with metals and other compounds. Furthermore, bacterial membranes are permeable to hydrogen peroxide, and thus the environment can be a source of ROS. ROS damage lipids, proteins, and nucleic acids (reviewed in references 12 and 13). Bacterial oxidative stress responses involve the activities of enzymes and small molecules that directly detoxify or protect against ROS and regulatory proteins that control their expression (20). In many bacteria, the transcriptional regulator OxyR (6, 35) senses H2O2 directly and induces numerous genes whose products aid in response to the stress, including those for peroxide defense (1, 36), redox balance (19, 24, 26), and other factors (2, 44).
Catalases detoxify hydrogen peroxide by catalyzing its decomposition to O2 and H2O. Two types of structurally unrelated catalases are common in bacteria: a bifunctional catalase-peroxidase (HPI) and a monofunctional catalase (HPII). Both of these catalases contain heme as the prosthetic group. In addition, a nonheme manganese-containing catalase is present in some bacteria as well (4). Most bacteria appear to express one or more catalases in response to peroxide stress, and the different types of catalases are regulated independently.
In Escherichia coli, HPII, encoded by katE, is part of the σS regulon and functions in stationary phase (21, 29, 41). The expression of the HPI catalase, KatG, is controlled by the OxyR transcriptional activator (7, 36, 37) and is induced in response to exogenous H2O2. However, E. coli mutants defective in both catalases typically exhibit no adverse phenotypes during growth in aerobic media, and those strains are able to rapidly scavenge H2O2 (18, 29, 32). It was shown recently that alkyl hydroperoxide reductase (Ahp) is responsible for the peroxide detoxifying activity in catalase-deficient mutants (32). Cells that lack both Ahp and catalase are unable to scavenge H2O2 and grow poorly in air. Ahp is a more efficient scavenger of H2O2 at low concentrations, and loss of Ahp is sufficient to activate OxyR. Thus, Ahp is the primary scavenger of endogenous H2O2 in E. coli, and catalase activity becomes essential only when the H2O2 concentration is high.
Exposure to H2O2 induces heme biosynthesis genes in E. coli (46), Salmonella enterica serovar Typhimurium (8), and Bacillus subtilis (5, 20), presumably because catalases are heme proteins. This induction is coordinated with the activation of catalase-encoding genes by OxyR in E. coli and Salmonella serovar Typhimurium and by PerR in Bacillus subtilis. A serovar Typhimurium heme biosynthesis mutant displays increased sensitivity to killing by H2O2, but this finding was attributed to an accumulation of reducing equivalents to drive the Fenton reaction rather than to a catalase deficiency alone (8).
Our laboratory is interested in the regulation of heme-dependent processes in the bacterium Bradyrhizobium japonicum. The rhizobia are soil bacteria that exist as free-living organisms or in symbiosis with leguminous plants. These bacteria must be equipped to respond to oxidative stresses under these diverse environments. Alfalfa roots produce superoxide and H2O2 in response to infection by Sinorhizobium meliloti (28). S. meliloti contains a KatG homolog and two HPII-like catalases, of which one is induced by peroxide stress and is essential for resistance to exogenous H2O2 (11, 14, 33). Symbiosis is affected only after loss of both HPII catalases. In contrast to S. meliloti, Rhizobium etli appears to express only a KatG homolog that is induced by H2O2 but is not required for symbiosis (39). Interestingly, none of the studies on catalase mutants in the rhizobia or in the related organism Agrobacterium tumefaciens report an aerobic growth phenotype (22). This finding suggests that these cells do not produce toxic endogenous levels of H2O2 or that there are compensatory enzymes for H2O2 scavenging when catalase function is lost. In E. coli, AhpCF compensates for loss of catalase activity.
We initiated a study of H2O2 response in B. japonicum. We provide evidence that aerobic metabolism produces toxic levels of H2O2 and that its detoxification is carried out primarily by KatG, the predominant catalase in aerobically grown cells. In addition, the data suggest that KatG levels are sufficiently high to respond to exogenous H2O2 challenge and that this high activity is not the result of activation by OxyR.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth.
B. japonicum I110 is the wild-type strain used in this study. The published genome sequence of strain I110 is fully annotated (http://www.kazusa.or.jp/rhizobase). MLG1 is a hemA deletion mutant containing a Kmr-encoding cassette in place of the deleted portion of the gene (10). Disruptions in strain I110 of katG, oxyR, and ahpCD were constructed in the present study and are described below. Expression of the katG, oxyR, and ahpCD genes in wild-type B. japonicum was examined by reverse transcription (RT)-PCR, and all mutant strains were confirmed by genomic PCR and RT-PCR. Strains were routinely grown in GSY medium as described previously (9) and were harvested at an optical density at 540 nm (OD540) of ∼0.3. As katG mutant cells cannot be aerobically cultured from low density, this strain was routinely grown to an OD540 of ∼0.05 in 1% O2 and then grown aerobically to an OD540 of 0.3. For growth under restricted aeration, flasks containing inoculated media were sealed with rubber stoppers and flushed with N2. O2 was then injected to the desired concentration. Plasmids were maintained and propagated in E. coli DH5α.
The katG locus (blr0778) was cloned with PCR primers (katGa, 5′CAGCAGGAATTTCTCCTCGAACAGG3′; and katGb, 5′CACCGTCGTCAAGGTGATGGAGTTC3′). The katG gene was disrupted by removing a 700-bp BglII fragment, corresponding to codons 198 to 431 of a total of 779, and replacing it with the streptomycin and spectinomycin resistance Ω cassette (BamHI fragment) from pHP45Ω (23). The disruption construct was moved into pLO1 (17) on a SacI fragment and introduced into B. japonicum by conjugation.
The ahpCD locus (bll1777 and bll1776) was cloned with PCR primers (ahp1, 5′GCAAGTGGCGATCGATGTCTTTGC3′; and ahp2, 5′GTCGAAGACAAGCTGTACTACAACC3′). A mutant with a deletion in parts of ahpC and ahpD was constructed by deleting an EcoRI-XhoI fragment that removes sequences corresponding to codons 69 to 184 (of 184) of ahpC and codons 1 to 169 (of 182) of ahpD. These sequences were replaced by a streptomycin and spectinomycin resistance Ω cassette carried on an EcoRI-SalI fragment. The disruption construct was cloned into pBR322 and introduced into I110 by conjugation. Recombinants were selected on streptomycin and spectinomycin and screened for loss of plasmid-borne tetracycline resistance.
The oxyR locus (bll0777) was cloned from B. japonicum strain I110 with PCR primers (oxyR3, 5′CAGATTTGCCAGCAGGAACTCACTC3′; and oxyR4, 5′AAGTGTCATCCATGAATACCTCCTC3′) based on the partial sequence of the strain B. japonicum USDA438. The oxyR gene was disrupted by removing a SacI-XhoI fragment (corresponding to codons 28 to 195 of 309) and replacing it with a streptomycin and spectinomycin resistance Ω cassette (SacI-SalI). The disruption construct was cloned into pBR322 and introduced into I110 by conjugation. Recombinants were selected on streptomycin and spectinomycin and screened for loss of plasmid-borne tetracycline resistance.
The katG locus was cloned into pVK102 (16) for in trans complementation experiments. The gene was cloned downstream of the kanamycin resistance promoter. The constructs were introduced into strains as indicated by conjugation. Plasmid-containing strains were selected based on tetracycline resistance conferred by the plasmid and confirmed by PCR.
In-gel catalase and peroxidase activity assays.
Aerobically grown cells were harvested at log phase (OD540 of 0.3), washed, resuspended in 50 mM NaPO4 buffer (pH 7.0), and lysed by two passages in a French press. Extracts were centrifuged at 12,000 × g at 4°C for 15 min to remove cell debris. Thirty micrograms of total protein was electrophoresed through 7.5% native polyacrylamide gels. Catalase activity was visualized by inhibition of exogenous horseradish peroxidase (HRP)-H2O2-mediated diaminobenzidine (DAB) oxidation as described previously (40). Peroxidase samples were prepared as for the catalase assay except that 50 μg of the extract was used. Peroxidase activity was visualized as dark bands after endogenous peroxidase-H2O2-mediated oxidation of DAB (42).
Measurement of H2O2 in solution.
In the presence of H2O2, HRP (Sigma, St. Louis, Mo.) oxidizes Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine; Molecular Probes, Eugene, Oreg.) to the resorufin, a chromophore with an absorption at 560 nm. Amplex Red reagent is stable in the presence of H2O2 except when a peroxidase is present. Amplex Red was dissolved in dimethyl sulfoxide to a 20 mM concentration and stored at −20°C in the dark. The HRP stock (2 U/μl) was also stored at −20°C. The working solution of Amplex Red-HRP is 10 mU of HRP/ml and 100 μM Amplex Red in 50 mM NaPO4 (pH 7.0). To measure H2O2, equal volumes of sample in NaPO4 and Amplex Red-HRP working solution were mixed and the absorbance was measured in a DW2000 UV-Vis spectrometer (SLM-Aminco) at OD560. Absorbance was converted to H2O2 concentration by using a curve generated from standard samples and was graphed by GraphPad Prism (GraphPad Software, Inc.).
H2O2 scavenging by cells.
Aerobically grown cells were harvested at log phase (OD540 of 0.3), washed twice in 50 mM NaPO4 buffer (pH 7.0), and resuspended in the same buffer at an OD540 of 0.1. H2O2 was added to a final concentration of 1.5 or 150 μM and cells were incubated at 29°C. At 2-min intervals, aliquots were assayed immediately for remaining H2O2.
Total catalase activity in cell extracts.
Aerobically grown cells were harvested at log phase (OD540 of 0.3), washed in NaPO4 buffer, resuspended in the same buffer, and lysed by two passages in a French press. Extracts were centrifuged at 12,000 × g at 4°C for 15 min to remove cell debris. Catalase activity was measured at 29°C in 1-ml reaction mixtures containing 15 μg of total protein and 1 mM H2O2. At 10-min intervals, aliquots were diluted 1:500 in NaPO4 and assayed for H2O2 by the Amplex Red-HRP method. Any H2O2 associated with the extracts alone was subtracted from the remaining H2O2 calculated.
H2O2 sensitivity.
For the hydrogen peroxide killing assay, cell cultures grown to an OD540 of 0.3 were split and H2O2 was added to aliquots to final concentrations indicated in Fig. 6. Cells were incubated with various concentrations of H2O2 with shaking for 20 min at 29°C, diluted, and spread on GSY plates containing the appropriate antibiotics.
FIG. 6.
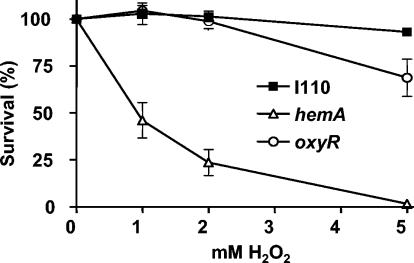
Sensitivity of wild-type and mutant cells to short exposure to H2O2 in liquid culture. Cells grown to mid-log phase were treated with various concentrations of H2O2 for 20 min. Afterwards, cells were plated onto media lacking H2O2 and colonies were counted. The data are presented as averages for two experiments ± standard deviations.
RT-PCR.
Cells were grown to an OD540 of ∼0.3, and RNAprotect (QIAGEN, Valencia, Calif.) was added to 5 × 108 cells. Cells were processed as recommended and frozen at −70°C until RNA isolation. Cells were lysed by using 400 μg of lysozyme/ml for 7 min, and RNA was isolated as recommended with a RNeasy mini kit (QIAGEN). RNA was quantitated and cDNA was synthesized from 1 μg of RNA by using random hexamers and the SuperScript first-strand synthesis system (Invitrogen, Carlsbad, Calif.). RNA was removed by digestion with RNase H, and 1 μl of the cDNA reaction was used in subsequent PCRs with Taq DNA polymerase (Fisher, Pittsburgh, Pa.). Control reactions were carried out by omitting the cDNA synthesis step.
RESULTS
Detection of a single catalase activity in B. japonicum cells.
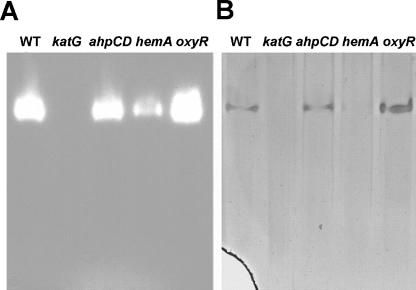
To address whether aerobically grown B. japonicum cells express catalase, an in-gel activity assay was carried out with extracts from parent strain I110 run on nondenaturing polyacrylamide gel electrophoresis (PAGE) gels (Fig. 1). A single activity band was observed, suggesting one protein or protein complex. A peroxidase assay was also carried out on extracts in nondenaturing gels based on H2O2-dependent oxidation of DAB. This assay also yielded a single band, at the same position as the catalase band. These observations suggest that aerobically grown cells contain a single catalase that also possesses peroxidase activity. We note that DAB may not be a substrate for all peroxidases; therefore, we cannot conclude that B. japonicum only has one active peroxidase.
FIG. 1.
Detection of catalase and peroxidase activities in extracts from wild type and mutant cells in nondenaturing PAGE. Extracts from cells grown in GSY media to an OD540 of 0.3 were electrophoresed through native gels. (A) Catalase activity was visualized by the inhibition of exogenous HRP-H2O2-mediated DAB oxidation due to H2O2 consumption by catalase and appeared as a colorless band against a dark background. (B) Peroxidase activity in the same extracts was assayed by DAB oxidation as described in Materials and Methods. Twenty-five micrograms of protein was loaded per lane, and the gels are typical of three independent experiments.
The annotated genome of B. japonicum strain I110 (15), (http://www.kazusa.or.jp/rhizobase/index.html) reveals an open reading frame (ORF) encoding a protein with very high similarity to KatG (blr0778; 63% identity and 73% similarity to E. coli KatG), a bifunctional catalase-peroxidase found in many bacteria (Fig. 2A). In addition, two other ORFs encoding catalase-like proteins were found based on similarities to catalase domains identified by hidden Markov model analysis. These ORFs, blr6251 and bll0292, have some similarity to KatE proteins but are much smaller (308 and 425 amino acids, respectively) than the 753-amino-acid KatE protein from E. coli. These genes may encode catalases, but they are not KatE homologs. Furthermore, ORF bll3758 is homologous to a nonheme catalase characterized for Salmonella serovar Typhimurium (27). We focused on the katG homolog because KatG proteins have both catalase and peroxidase activities, as did the species detected in native gels (Fig. 1).
FIG. 2.
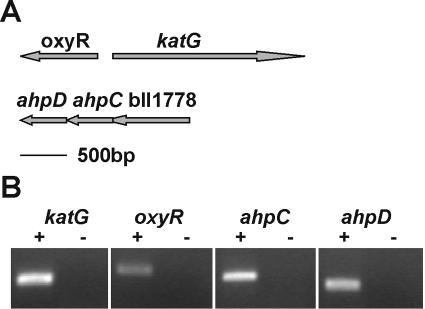
Arrangements of katG, oxyR, ahpC, and ahpD in the B. japonicum genome and detection of transcripts. (A) The katG and oxyR genes are divergently transcribed. The katG translation start site is predicted to be within the oxyR coding sequence, according to the annotation for the published genome. However, sequence alignments with other KatG proteins suggest that the initiation methionine codon is 59 amino acids further downstream. Additionally, a katG construct utilizing the downstream start site fully complements the katG disruption mutation. Thus, the katG gene is drawn to reflect the smaller coding region. An ORF (bll1778) with no known or predicted function is immediately upstream of ahpC. (B) Detection of transcripts by RT-PCR using gene-specific primers and mRNA from wild-type (+) cells or from the respective mutant strains (−). Controls using reaction templates in the absence of reverse transcriptase yielded no PCR products (data not shown). PCR products were run on an agarose gel and stained with ethidium bromide. The gene amplified in each case is indicated above each panel.
The katG gene homolog is a transcribed gene based on a product observed in RT-PCR experiments (Fig. 2B). The katG gene including flanking DNA was isolated by PCR and was used to construct a katG chromosomal mutant derivative of parent strain I110 (see Materials and Methods). The catalase and peroxidase activity bands observed for the parent strain in nondenaturing gels were completely absent in the katG mutant strain (Fig. 1). A quantitative determination of catalase activity was determined in cell extracts of the wild type and the katG strain (Fig. 3); no catalase activity was detected in the mutant, a result which is in good agreement with the gel assay. These findings show that aerobically grown B. japonicum cells have a single detectable catalase activity catalyzed by KatG.
FIG. 3.
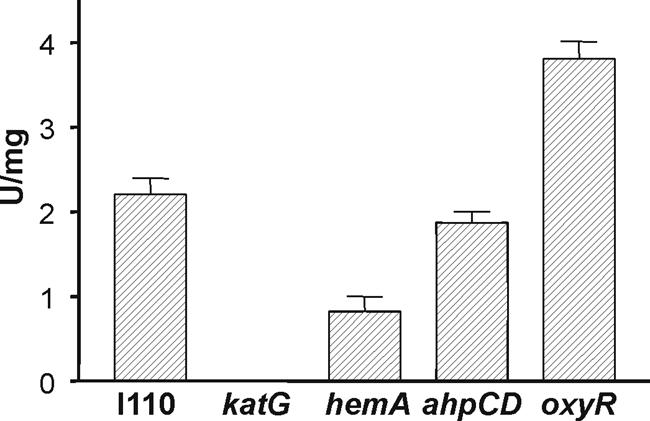
Catalase activity in B. japonicum cell extracts. Catalase activity associated with extracts from wild-type and mutant cells was assessed by incubating 15 μg of extract protein with 1 mM H2O2 at 29°C and measuring remaining H2O2 after 10 min, as described in Materials and Methods. Units are μmol of H2O2 consumed/minute/milligram of protein. Error bars represent the standard deviations for three trials.
A katG mutant displays a severe aerobic growth phenotype.
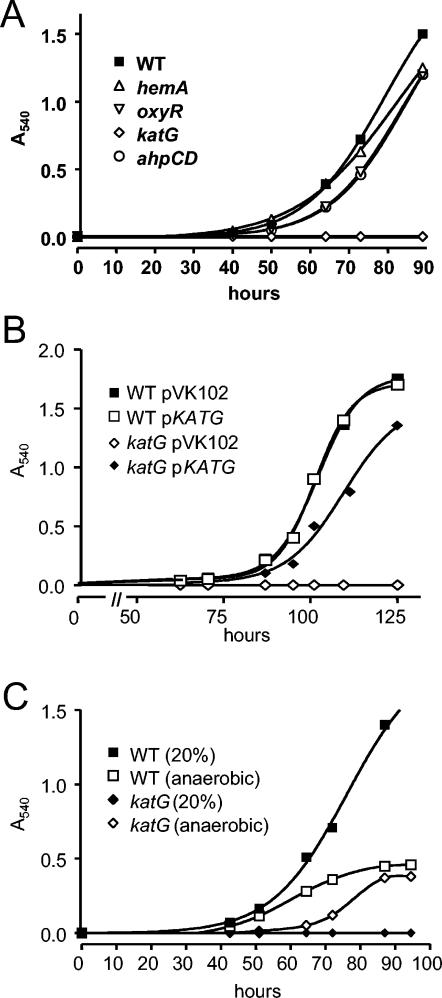
Parent strain I110 grows aerobically, with a doubling time of 8 to 9 h. However, the katG strain inoculated at low cell density under ambient oxygen concentrations was unable to grow (Fig. 4A). The mutant grew aerobically on plates only when streaked as a patch at high density. Normal aerobic growth was restored by the introduction of the katG gene in trans on a broad-host-range plasmid (Fig. 4A), thus verifying that the mutation in the katG gene is responsible for the phenotype. The katG strain was able to grow anaerobically with nitrate as the terminal electron acceptor to an optical density of ca. 0.5 (Fig. 4B) or under 1 to 2% O2 tension (data not shown). Furthermore, the mutant grew if initially cultured in 1% O2 to an optical density (540 nm) of ca. 0.05 before exposure to air. Cells were grown this way for all aerobic experiments. The growth phenotype of strain I110katG suggests that H2O2 produced as a function of aerobic metabolism is sufficient to be toxic to cells and that KatG is responsible for its detoxification in B. japonicum.
FIG. 4.
Growth curves of wild-type (WT) and mutant cells under various conditions. Flasks of GSY media were inoculated with 5 × 105 log phase cells/ml at time zero and grown at 29°C with shaking. Aliquots were taken at the indicated time points for A540 measurements. (A) Cells were grown aerobically. (B) In trans complementation of aerobic growth of the katG strain was assessed by the introduction of pkatG, which harbors the katG gene. pVK102 is the empty vector. The increased lag time compared to the growth in panel A is likely the result of the presence of a plasmid or of the tetracycline (50 μg/ml) in the media for plasmid selection. (C) Cells were grown anaerobically in GSY medium supplemented with 5 mM KNO3 as a terminal electron acceptor.
KatG, not Ahp, is the primary scavenger of H2O2 in B. japonicum.
The growth phenotype of the B. japonicum katG strain was surprising in light of the fact that catalase mutants in other bacteria do not display growth or viability anomalies unless challenged with exogenous H2O2 (32, 33, 39, 43). In E. coli, mutation of the ahpCF genes encoding Ahp and mutation of both catalase genes (katG and katE) are necessary to impair aerobic growth (32). Furthermore, scavenging of low concentrations of H2O2 appears to be carried out exclusively by the Ahp proteins in E. coli (32). The B. japonicum genome contains ahpC and ahpD gene homologs arranged in tandem that are transcribed in cells (Fig. 2A and B). AhpD is found in numerous gram-positive bacteria, and it serves to shuttle electrons to AhpC as does AhpF in organisms that express it (3).
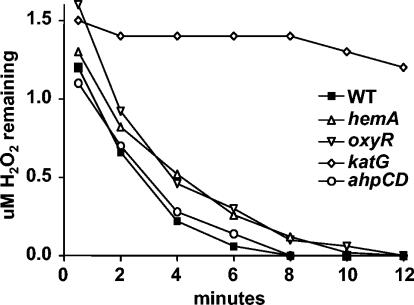
To determine the respective roles of KatG and Ahp in H2O2 detoxification, an ahpCD mutant of B. japonicum was constructed and compared to the katG strain. Unlike katG, the ahpCD mutant grew well in aerobic cultures (Fig. 4). We assessed the contributions of KatG and Ahp in scavenging a low concentration of H2O2 by exposing whole cells to an initial H2O2 concentration of 1.5 μM and measuring its disappearance (Fig. 5). Cells of the wild-type and ahpCD strains rapidly consumed H2O2 at similar rates, with none detectable after 8 min. The sizes and intensities of the catalase activity bands in extracts of the ahpCD mutant in gels were similar to those of the wild type (Fig. 1); therefore, the mutation of ahpCD was not compensated for by an increase in KatG activity or induction of another catalase. This result strongly suggests that H2O2 consumption in whole cells of the ahpCD mutant was catalyzed by KatG. Finally, katG mutant cells had extremely low H2O2 consumption activity (Fig. 5). Collectively, these data indicate that KatG, not Ahp, is the primary scavenger of endogenous H2O2.
FIG. 5.
Scavenging of low concentrations of H2O2 by whole cells. Cells in mid-log phase were resuspended in NaPO4 buffer, and H2O2 was added to a concentration of 1.5 μM. Aliquots were taken at the indicated times and assayed immediately for remaining H2O2 as described in Materials and Methods. WT, wild type.
Catalase activity in a heme biosynthesis mutant.
KatG and most other catalases are heme proteins. The B. japonicum hemA gene encodes the heme biosynthetic enzyme δ-aminolevulinic acid synthase. It was reported previously (9) and demonstrated here (Fig. 4) that a hemA deletion mutant grows well aerobically on GSY medium, results which seemed to be at odds with the severe growth phenotype of the katG mutant. Whole cells of the B. japonicum hemA strain were able to scavenge low concentrations of H2O2 almost as well as the parent strain (Fig. 5), which is consistent with the apparent ability of the mutant to detoxify H2O2 produced by aerobic metabolism as judged by its growth phenotype. Extracts of the hemA strain contained about one-third of the catalase activity as the parent strain (Fig. 3). To assess whether a catalase other than KatG was induced in the hemA strain, extracts were assayed for activity in nondenaturing PAGE (Fig. 1A). An activity band with the same mobility as KatG was observed, but with diminished intensity. A faint peroxidase band was also observed, but it is difficult to see in Fig. 1B. Although no cytochrome hemes are spectroscopically detectable in the hemA mutant, evidence suggests that a small amount of heme is made or acquired from the medium and is necessary for viability (9). Expression of katG in trans from a plasmid increased catalase activity in the wild type about fivefold but had no effect on activity in the hemA strain (data not shown). These observations argue against the possibility of a heretofore undescribed heme-independent catalase activity of KatG. The simplest explanation for these data is that the hemA mutant produces or acquires enough heme to synthesize KatG, albeit in lesser quantity than the wild type, and that this amount is sufficient to degrade H2O2 produced by aerobic metabolism.
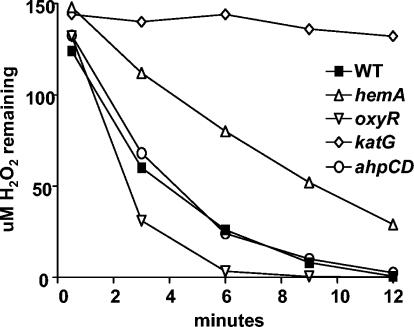
Earlier studies showed that a δ-aminolevulinic acid synthesis mutant of Salmonella is susceptible to oxidative stress (8) and that heme synthesis genes in E. coli are induced by H2O2 (46). In those studies, oxidative stress responses were assessed by exposure to exogenous H2O2. In the present study, the viabilities of cells exposed for 20 min to increasing amounts of H2O2 were evaluated for the parent strain and for the hemA strain MLG1. The wild type was resistant to 5 mM H2O2, whereas the mutant was very sensitive to H2O2 relative to the parent strain, with essentially 100% killing at a concentration of 5 mM (Fig. 6). We note that the viability analysis was not carried out with the katG strain, because it failed to form single colonies on plates (see above). In addition, the ability of the wild-type and hemA strains to decompose exogenous H2O2 at a high initial concentration was measured (Fig. 7). Although the mutant was able to decompose H2O2 much better than the katG strain, the rate was considerably less than was observed with the wild type. We suggest that the KatG level expressed in the hemA strain is sufficient to support aerobic metabolism but is insufficient to detoxify high concentrations of H2O2 supplied exogenously.
FIG. 7.
Scavenging of high concentrations of H2O2 by whole cells. H2O2 (150 μM) was added to cells in NaPO4 buffer. Aliquots were taken at the times indicated and were assayed for remaining H2O2 as described in Materials and Methods. WT, wild type.
KatG activity is not defective in an oxyR mutant.
OxyR is a peroxide sensor and a transcriptional activator of the oxidative stress response (45). In many bacteria, OxyR positively regulates genes involved in the detoxification and protection from peroxide stress. In B. japonicum, an oxyR gene homolog (bll0777) is found adjacent to katG in the opposite orientation, and it was shown to be a transcribed gene in aerobically grown cells (Fig. 2A and B). The known role of OxyR and the position of the oxyR gene with respect to katG led us to ask whether the katG expression observed in this study is dependent on OxyR. We tested this prediction by creating and characterizing an oxyR mutant strain. Interestingly, the mutant grew well aerobically (Fig. 4A), suggesting that it was able to detoxify H2O2 produced by aerobic respiration. Consistent with this result, whole cells of the oxyR strain were able to scavenge low (1.5 μM) and high (150 μM) concentrations of H2O2 as well as or better than wild-type cells (Fig. 5 and 7). Catalase activity in cell extracts was not diminished in the oxyR strain, but rather it was higher in that mutant than in the parent strain (Fig. 3). Accordingly, the KatG activity band was more intense in nondenaturing PAGE (Fig. 1). The higher activity can explain the higher rate of H2O2 consumption in whole cells of the oxyR mutant at a 150 μM concentration of H2O2 (Fig. 7). An increase in catalase expression was also observed with an oxyR mutant of Neisseria gonorrhoeae (38). This finding may be due to a repressor activity of OxyR, or it may be an indirect effect of increased reactive oxygen species in the oxyR strain, resulting in catalase activation. The data show that the high KatG activity in wild-type cells is not the result of OxyR induction.
DISCUSSION
In the present study, we show that KatG is the predominant catalase in aerobically grown B. japonicum cells and provide evidence that it is the primary scavenger of H2O2 under those conditions. The severe growth phenotype of the katG strain indicates that B. japonicum produces sufficient H2O2 under aerobic conditions to be toxic and that KatG is primarily responsible for its detoxification. This aerobic growth defect was unexpected for two reasons.
(i) E. coli and numerous other bacteria have more than one catalase, and therefore mutation of a single catalase-encoding gene does not have a growth phenotype (32, 33, 39, 43). The B. japonicum genome contains several possible catalase-encoding genes, but only KatG was detectable. Interestingly, in R. etli, KatG appears to be the only active catalase in aerobically grown cells, but no growth defect was reported in that study (39). It is plausible that R. etli does not produce toxic levels of endogenous H2O2 or else it consumes it via a peroxidase that was not detectable in the assays used.
(ii) E. coli mutants lacking all catalase activity grow well aerobically, nevertheless, because endogenous H2O2 is detoxified by the peroxidase Ahp in that organism (32). Seaver and Imlay showed convincingly that Ahp is the primary scavenger of endogenous H2O2 in E. coli even though it has a much lower activity (32). This result is possible because Ahp has a higher affinity for H2O2 than does catalase. Catalase is predominant when the H2O2 concentration is high. B. japonicum has genes encoding Ahp, but an ahpCD mutant had no aerobic growth phenotype (Fig. 4), and it consumes both low and high concentrations of H2O2 as well as the wild type (Fig. 5 and 7). Thus, Ahp may be involved in H2O2 detoxification, but it is not essential, and it cannot compensate for a defect in katG in aerobically cultured cells.
A heme biosynthesis mutant defective in the hemA gene had about one-third of the KatG activity as does the parent strain (Fig. 1 and 3), but the mutant grew well aerobically despite this reduced activity (Fig. 1, 3, and 4). It was shown previously that endogenous respiration in the hemA strain is comparable to that in the wild type (9); therefore, it is likely that the mutant produces H2O2 at levels similar to those of the wild type. Thus, these observations suggest that the KatG level in wild-type cells is higher than that needed to consume endogenously produced H2O2. However, the KatG level necessary for endogenous H2O2 detoxification is apparently lower than that needed to respond to an exogenous H2O2 challenge, since the hemA strain was sensitive to killing by H2O2 (Fig. 6). The katG gene may be expressed more or less constitutively at a high level, or else the endogenous H2O2 in B. japonicum is sufficiently high to induce katG but not high enough to be toxic to the hemA mutant. The transcriptional regulator OxyR activates catalase gene expression in numerous bacteria (34). However, an oxyR mutant of B. japonicum grew well aerobically, consumed H2O2, and expressed KatG activity. Thus, catalase activity observed with aerobically grown cells is not OxyR dependent. It is possible that OxyR is involved in catalase expression under conditions that we have not examined. KatG levels were actually modestly higher in the oxyR strain than in the wild type (Fig. 1), and the oxyR strain consumed H2O2 at a slightly higher rate (Fig. 7). This finding could suggest a negative regulatory role for OxyR. Alternatively, an oxyR mutant may have elevated reactive oxygen species, which in turn activate the katG gene.
Evidence suggests that there is very little free heme in wild-type B. japonicum cells (25), and the total heme content in a hemA strain is spectroscopically indiscernible (9). Therefore, it is noteworthy that the hemA mutant has detectable catalase activity, and this finding suggests a mechanism other than free diffusion for the incorporation of heme into the apoprotein. This issue is dealt with by specific chaperones for c-type cytochromes (30, 31), and it will be interesting to learn whether catalase and other heme proteins have similar systems for heme incorporation.
Acknowledgments
This work was supported by National Institutes of Health grant GM067966 to M.R.O'B. H.R.P. was supported by National Institutes of Health training grant AI07614.
REFERENCES
- 1.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 2.Altuvia, S., D. Weinstein-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 3.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 4.Chelikani, P., I. Fita, and P. C. Loewen. 2004. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61:192-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christman, M. F., G. Storz, and B. N. Ames. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA 86:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claiborne, A., and I. Fridovich. 1979. Purification of the o-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J. Biol. Chem. 254:4245-4252. [PubMed] [Google Scholar]
- 8.Elgrably-Weiss, M., S. Park, E. Schlosser-Silverman, I. Rosenshine, J. Imlay, and S. Altuvia. 2002. A Salmonella enterica serovar Typhimurium hemA mutant is highly susceptible to oxidative DNA damage. J. Bacteriol. 184:3774-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frustaci, J. M., I. Sangwan, and M. R. O'Brian. 1991. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J. Bacteriol. 173:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerinot, M. L., and B. K. Chelm. 1986. Bacterial δ-aminolevulinic acid synthase is not essential for leghemoglobin formation in the soybean/Bradyrhizobium japonicum symbiosis. Proc. Natl. Acad. Sci. USA 83:1837-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herouart, D., S. Sigaud, S. Moreau, P. Frendo, D. Touati, and A. Puppo. 1996. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J. Bacteriol. 178:6802-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46:111-153. [DOI] [PubMed] [Google Scholar]
- 13.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 14.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Herouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 16.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 17.Lenz, O., E. Schwartz, J. Dernedde, M. Eitinger, and B. Friedrich. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 176:4385-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loewen, P. C. 1984. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J. Bacteriol. 157:622-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michan, C., M. Manchado, G. Dorado, and C. Pueyo. 1999. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 181:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 21.Mulvey, M. R., J. Switala, A. Borys, and P. C. Loewen. 1990. Regulation of transcription of katE and katF in Escherichia coli. J. Bacteriol. 172:6713-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prapagdee, B., P. Vattanaviboon, and S. Mongkolsuk. 2004. The role of a bifunctional catalase-peroxidase KatA in protection of Agrobacterium tumefaciens from menadione toxicity. FEMS Microbiol. Lett. 232:217-223. [DOI] [PubMed] [Google Scholar]
- 23.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 24.Prieto-Alamo, M. J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 25.Qi, Z., and M. R. O'Brian. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9:155-162. [DOI] [PubMed] [Google Scholar]
- 26.Ritz, D., H. Patel, B. Doan, M. Zheng, F. Aslund, G. Storz, and J. Beckwith. 2000. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 275:2505-2512. [DOI] [PubMed] [Google Scholar]
- 27.Robbe-Saule, V., C. Coynault, M. Ibanez-Ruiz, D. Hermant, and F. Norel. 2001. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σS). Mol. Microbiol. 39:1533-1545. [DOI] [PubMed] [Google Scholar]
- 28.Santos, R., D. Herouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 29.Schellhorn, H. E., and H. M. Hassan. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 170:4286-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz, H., R. A. Fabianek, E. C. Pellicioli, H. Hennecke, and L. Thöny-Meyer. 1999. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc. Natl. Acad. Sci. USA 96:6462-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulz, H., H. Hennecke, and L. Thony-Meyer. 1998. Prototype of a heme chaperone essential for cytochrome c maturation. Science 281:1197-1200. [DOI] [PubMed] [Google Scholar]
- 32.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigaud, S., V. Becquet, P. Frendo, A. Puppo, and D. Herouart. 1999. Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-like catalases during free-living growth and protective role of both catalases during symbiosis. J. Bacteriol. 181:2634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 35.Tao, K., K. Makino, S. Yonei, A. Nakata, and H. Shinagawa. 1989. Molecular cloning and nucleotide sequencing of oxyR, the positive regulatory gene of a regulon for an adaptive response to oxidative stress in Escherichia coli: homologies between OxyR protein and a family of bacterial activator proteins. Mol. Gen. Genet. 218:371-376. [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia, L. A., G. Storz, and B. N. Ames. 1989. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 210:709-719. [DOI] [PubMed] [Google Scholar]
- 37.Triggs-Raine, B. L., B. W. Doble, M. R. Mulvey, P. A. Sorby, and P. C. Loewen. 1988. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J. Bacteriol. 170:4415-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng, H. J., A. G. McEwan, M. A. Apicella, and M. P. Jennings. 2003. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vargas Mdel, C., S. Encarnacion, A. Davalos, A. Reyes-Perez, Y. Mora, A. Garcia-de los Santos, S. Brom, and J. Mora. 2003. Only one catalase, katG, is detectable in Rhizobium etli, and is encoded along with the regulator OxyR on a plasmid replicon. Microbiology 149:1165-1176. [DOI] [PubMed] [Google Scholar]
- 40.Vattanaviboon, P., and S. Mongkolsuk. 2000. Expression analysis and characterization of the mutant of a growth-phase- and starvation-regulated monofunctional catalase gene from Xanthomonas campestris pv. phaseoli. Gene 241:259-265. [DOI] [PubMed] [Google Scholar]
- 41.von Ossowski, I., M. R. Mulvey, P. A. Leco, A. Borys, and P. C. Loewen. 1991. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J. Bacteriol. 173:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wayne, L. G., and G. A. Diaz. 1986. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal. Biochem. 157:89-92. [DOI] [PubMed] [Google Scholar]
- 43.Xu, X. Q., and S. Q. Pan. 2000. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35:407-414. [DOI] [PubMed] [Google Scholar]
- 44.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, M., and G. Storz. 2000. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 59:1-6. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]