Abstract
Group I introns are inserted into genes of a wide variety of bacteriophages of gram-positive bacteria. However, among the phages of enteric and other gram-negative proteobacteria, introns have been encountered only in phage T4 and several of its close relatives. Here we report the insertion of a self-splicing group I intron in the coding sequence of the DNA polymerase genes of ΦI and W31, phages that are closely related to T7. The introns belong to subgroup IA2 and both contain an open reading frame, inserted into structural element P6a, encoding a protein belonging to the HNH family of homing endonucleases. The introns splice efficiently in vivo and self-splice in vitro under mild conditions of ionic strength and temperature. We conclude that there is no barrier for maintenance of group I introns in phages of proteobacteria.
The first demonstration of an intron in a bacterium or a bacterial virus was the self-splicing group I intron in the thymidylate synthase (td) gene of Escherichia coli phage T4 (1). Group I introns were subsequently discovered in tRNA genes of cyanobacteria (12, 24) and proteobacteria (20), in the rRNA genes of several Thermotoga species (16), and in a variety of phages of gram-positive bacteria and cyanobacteria (9, 15, 18). In view of the intensive investigation of E. coli phages it is surprising that, in the 20 years since their initial discovery in the td gene of T4, the only new examples in phages of E. coli or other proteobacteria are two additional introns in genes of T4 and a small number of its close relatives (the T-even phages) (8, 9, 23).
The apparent limitation of introns to phages of the T-even family might be due to a special property of these phages that would overcome a disadvantage that self-splicing group I introns might confer on other phages of proteobacteria. For example, the large genome of T-even phages (1.7 × 105 bp) might accommodate introns more easily than the genomes of smaller phages, or the extensive modifications of cellular metabolism by T-even phages might overcome some unknown E. coli metabolic process that makes group I intron splicing deleterious to gene expression.
Very early in its history, a group of workers agreed to limit the study of phages of E. coli to seven isolates (phages T1 to T7) (2, 5). Although lambdoid phages and single-stranded DNA and RNA phages eventually became the subjects of intensive study, most of the literature on phages of E. coli concerns a relatively small number of phage isolates. The complete genomic sequences of phages T1, T3, T5, and T7 (GenBank accession numbers NC005833, AJ318471, NC005859, and NC001604) show no evidence of introns (7, 17, 21), supporting a preliminary study that failed to find evidence of introns in these phages (4). Even among the T-even phages, introns are relatively rare (8, 9, 23). If T2, which probably has no introns, had been selected as the model phage for genetics rather than T4, there would have been no reason to conduct a survey of the other T-even phages for introns (8), and the presence of introns in E. coli phages might still be unknown today.
Therefore, the apparent restriction of introns to T4 and its relatives might be due to a lack of comparative data. We screened several isolates of T7-like phages for variability over a 1-kb region by PCR using primers T75.0 (GACCGAAGAGATGCTCGTAGAG) and T75.7 (GTACGGAACAATGCGAGCCTC), which correspond to identically conserved sequences in genes 5 and 5.7 of phages T3 and T7. Eight phage DNAs (ΦI, ΦIIP, ΦIIW, H, and W31, which were obtained from W. F. Studier; and A1122, C21R, and ViIII, obtained from I. Molineux) yielded products in the PCR, with two of them (ΦI and W31) migrating slower than the others (data not shown).
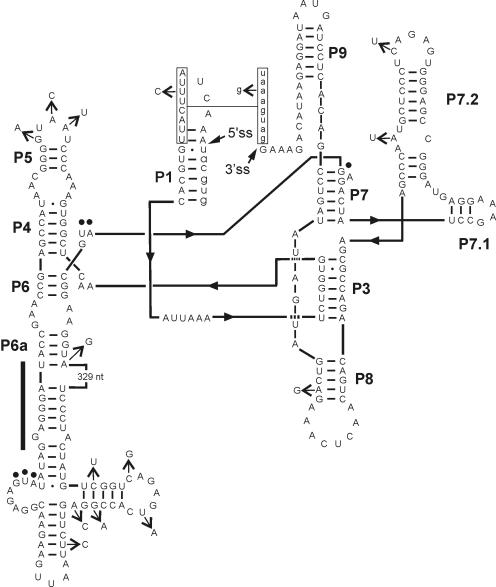
A group I intron in phages ΦI and W31.
In both phages the larger sizes of the PCR products are accounted for by 601-bp insertions 156 bp from the ends of their DNA polymerase genes (gene 5). These insertions can be folded into structures typical of group IA2 introns (3, 14) with a 131-codon open reading frame (ORF) inserted into stem P6a (Fig. 1). The introns are inserted between codons in a conserved region of DNA polymerases (R. P. Bonocora and D. A. Shub, submitted for publication) that specifies a metal ion binding site in the catalytic center of the T7 enzyme (6). Overall, the introns are 86% identical, while the phylogenetically conserved intron structures (without the ORFs) are 97% identical. Most of the sequence divergence occurs between the intron-encoded ORFs, which comprise more than half of the intron and are 81 and 86% identical at the nucleotide and amino acid levels, respectively.
FIG. 1.
Secondary structure of the ΦI group I intron. Exon sequences are in lowercase letters, and intron sequences are in uppercase letters. Filled arrows indicate splice sites (ss). Open arrows indicate sequence differences between the ΦI and W31 introns. Conserved structural elements P1 to P9 are shown. Boxed regions designate the P10 pairing. Filled circles indicate the start and stop codons of the I-TslI coding sequence. The putative RBS in stem P6a is indicated by a thick vertical bar. Additionally, the 329-nucleotide sequence of the intron-encoded ORF is indicated.
Intron secondary structure.
The introns contain helical elements P7.1 and P7.2 separated by a G-U-A sequence, features indicative of group IA2 introns (14), but lack the optional P2 helix. Although they lack a discernible P9.0 interaction at their 3′ ends, this may be compensated by long P10 pairings between the first eight nucleotides of the 3′ exon and a sequence near the 5′ end of the intron (13). All but one of the sequence differences in paired regions either preserve base pairing or are accompanied by a compensatory change (Fig. 1). The lone exception is in the extended P6 stem-loop region that is part of the coding sequence of the ORF.
The ribosome binding sites (RBS) of the intronic ORFs are sequestered in the secondary structure. Similarly, the ORFs in the three introns in phage T4 each have an RBS sequestered in stable base-paired helices with intron sequences. These structures are involved in temporal regulation of ORF expression, preventing translation from the early-transcribed introns. Instead, the ORFs are independently expressed from transcripts under control of T4 late promoters located immediately upstream of the RBS (11). However, no T7-like promoter can be identified upstream of the intronic ORFs of ΦI or W31.
We have shown elsewhere that the intronic ORFs of both phages encode an endonuclease (I-TslI) that cleaves the intronless allele 1 bp from the intron insertion site, activating the introns for homing (Bonocora and Shub, submitted). We suggest that, rather than being involved in temporal regulation of gene expression, sequestering the RBS serves primarily to reduce expression of I-TslI while perhaps also contributing to stability of intron RNA structure. Expression of I-TslI may need to be maintained at levels much lower than those of DNA polymerase (whose transcript it shares), since at high protein concentrations secondary cleavages may occur elsewhere in the genome. In fact, we have observed a secondary cleavage site just 216 bp upstream of the intron insertion site in gene 5 (data not shown). Since this potential cleavage site is not protected by intron insertion, high in vivo concentrations of I-TslI could be very deleterious.
Intron splicing in vivo.
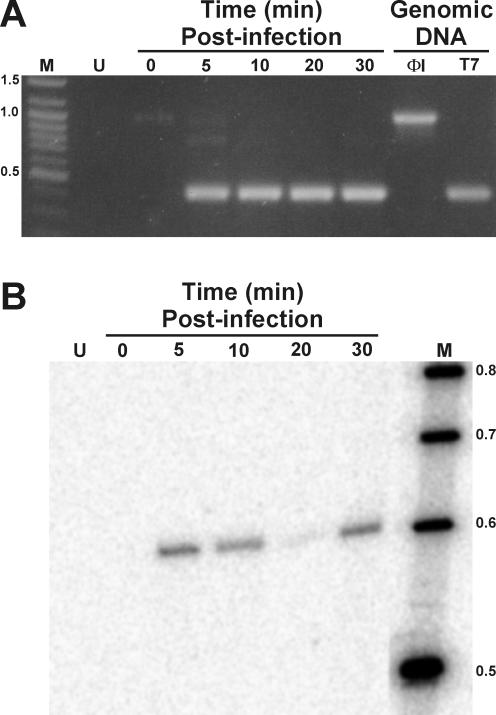
E. coli BE was grown in Luria-Bertani medium (22) at 37°C to an optical density at 590 nm of 0.3 to 0.35 (2 × 108 CFU/ml) and infected with phage ΦI at a multiplicity of 4 per cell. RNA was isolated at various times after infection, using an RNAeasy mini-prep kit and RNase-Free DNase (QIAGEN) and assayed for splicing by reverse transcription. Primer ΦI5.0rev (CAGAAGACACCGGAAGTTCC) was annealed to isolated RNA and incubated at 25°C for 5 min, 37°C for 60 min, and 75°C for 15 min with IMPROMPII reverse transcriptase (Promega) in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 10 mM dithiothreitol, and 3 mM MgCl2. One-tenth of the reverse transcriptase reaction mixture was used as template for PCRs using Taq DNA polymerase with primers ΦI5.0rev and T75.0b (GAAGATTGGNCAGATTGTTGG), and products were analyzed by electrophoresis in a 0.8% agarose gel (Fig. 2A).
FIG. 2.
Splicing of the ΦI group I intron. A. In vivo splicing. RNA isolated from ΦI-infected E. coli was analyzed by reverse transcription-PCR. The times of RNA isolation (in minutes) after infection are indicated. The sizes (in kilobases) of prominent bands of the DNA size standard (lane M) are indicated on the left. Lane U contains RNA isolated from uninfected E. coli. PCR products generated from ΦI and T7 DNA are shown on the right. B. In vitro self-splicing. GTP labeling was performed on the RNA preparations shown in panel A. DNA size standards were 5′ end labeled with 32P.
The size of the amplification product from unspliced transcripts is expected to be approximately 1.1 kb, whereas the size from the spliced transcript is 0.4 kb. At 5 min postinfection and continuing until 30 min the 0.4-kb product predominates, indicating that the intron is spliced from the transcript in vivo. The virtual absence of a product from the unspliced precursor is likely due to the difficulty of reverse transcription through the highly structured intron RNA.
Self-splicing in vitro.
RNA preparations can be screened for self-splicing introns in vitro by exploiting the group I intron splicing mechanism, in which the attacking guanosine nucleophile becomes covalently attached to the 5′ end of the excised intron (10). When the same RNA preparations were incubated with [α-32P] GTP (19) followed by electrophoresis in 4% denaturing polyacrylamide (19:1 ratio of acrylamide to bisacrylamide with 7 M urea) gels, a labeled 0.6-kb band (corresponding to the size of the intron) was present beginning at 5 min postinfection (Fig. 2B), indicating that the ΦI intron is autocatalytically removed by self-splicing.
Apart from phage T4 and some of its close relatives, this is the first description of group I introns in phages of enteric bacteria, with two of the eight phages examined here having an intron at the same genomic site. Although the sample sizes are still small, the proportion of T7-like phages with an intron in their DNA polymerase genes is remarkably similar to the proportion (5 of 22) of independently isolated T-even phages that were previously found to have introns in their td genes (8). Since the genome sizes of phages in the T7 family are only about one-quarter those of the T-even phages, large genome size seems not to be a factor in the ability of a phage to harbor a mobile intron. We suspect that the focused study of only a few E. coli phages, which contributed to their usefulness as model genetic systems, may be responsible for the apparent paucity of introns and that as more genomes of independently isolated phages are investigated, mobile introns will continue to be found.
Nucleotide sequence accession numbers. The sequences reported in this paper have been deposited in GenBank under accession numbers AY769989 (ΦI) and AY769990 (W31).
ADDENDUM IN PROOF
We have recently learned (D. Savalia, I. Molineux, and K. Severinov, unpublished results) that FPI, a 17-like podophage, has an insertion in its DNA polymerase gene in the same position as, and highly similar to, the intron and homing endonuclease gene reported here.
REFERENCES
- 1.Belfort, M., J. Pedersen-Lane, D. West, K. Ehrenman, G. Maley, F. Chu, and F. Maley. 1985. Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level. Cell 41:375-382. [DOI] [PubMed] [Google Scholar]
- 2.Brock, T. D. 1990. The emergence of bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Cech, T. R., S. H. Damberger, and R. R. Gutell. 1994. Representation of the secondary and tertiary structure of group I introns. Nat. Struct. Biol. 1:273-280. [DOI] [PubMed] [Google Scholar]
- 4.Chu, F. K., F. Maley, J. Martinez, and G. F. Maley. 1987. Interrupted thymidylate synthase gene of bacteriophages T2 and T6 and other potential self-splicing introns in the T-even bacteriophages. J. Bacteriol. 169:4368-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demerec, M., and U. Fano. 1945. Bacteriophage-resistant mutants in Escherichia coli. Genetics 30:119-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391:251-258. [DOI] [PubMed] [Google Scholar]
- 7.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 8.Eddy, S. R. 1992. Introns in the T-even bacteriophages. Ph.D. dissertation. University of Colorado, Boulder.
- 9.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garriga, G., and A. M. Lambowitz. 1984. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell 39:631-641. [DOI] [PubMed] [Google Scholar]
- 11.Gott, J. M., A. Zeeh, D. Bell-Pedersen, K. Ehrenman, M. Belfort, and D. A. Shub. 1988. Genes within genes: independent expression of phage T4 intron open reading frames and the genes in which they reside. Genes Dev. 2:1791-1799. [DOI] [PubMed] [Google Scholar]
- 12.Kuhsel, M. G., R. Strickland, and J. D. Palmer. 1990. An ancient group I intron shared by eubacteria and chloroplasts. Science 250:1570-1573. [DOI] [PubMed] [Google Scholar]
- 13.Michel, F., M. Hanna, R. Green, D. P. Bartel, and J. W. Szostak. 1989. The guanosine binding site of the Tetrahymena ribozyme. Nature 342:391-395. [DOI] [PubMed] [Google Scholar]
- 14.Michel, F., and E. Westhof. 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216:585-610. [DOI] [PubMed] [Google Scholar]
- 15.Millard, A., M. R. Clokie, D. A. Shub, and N. H. Mann. 2004. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. USA 101:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nesbø, C. L., and W. F. Doolittle. 2003. Active self-splicing group I introns in 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles. Proc. Natl. Acad. Sci. USA 100:10806-10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pajunen, M. I., M. R. Elizondo, M. Skurnik, J. Kieleczawa, and I. J. Molineux. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 319:1115-1132. [DOI] [PubMed] [Google Scholar]
- 18.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 19.Reinhold-Hurek, B., and D. A. Shub. 1993. Experimental approaches for detecting self-splicing group I introns. Methods Enzymol. 224:491-502. [DOI] [PubMed] [Google Scholar]
- 20.Reinhold-Hurek, B., and D. A. Shub. 1992. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357:173-176. [DOI] [PubMed] [Google Scholar]
- 21.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318:245-266. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sandegren, L., and B. M. Sjöberg. 2004. Distribution, sequence homology, and homing of group I introns among T-even-like bacteriophages: evidence for recent transfer of old introns. J. Biol. Chem. 279:22218-22227. [DOI] [PubMed] [Google Scholar]
- 24.Xu, M. Q., S. D. Kathe, H. Goodrich-Blair, S. A. Nierzwicki-Bauer, and D. A. Shub. 1990. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science 250:1566-1570. [DOI] [PubMed] [Google Scholar]