Abstract
Streptococcus pyogenes (the group A streptococcus [GAS]) is a medically significant pathogen of humans, causing a range of diseases from pharyngitis to necrotizing fasciitis. Several important GAS virulence genes are under the control of a pleiotropic regulator called Mga, or the multiple gene regulator of GAS, including the gene encoding the streptococcal collagen-like protein, or sclA. Analysis of the genome sequence upstream of sclA revealed two potential Mga-binding sites with homology to the published Mga-binding element, which were called PsclA-I (distal) and PsclA-II (proximal) based on their location relative to a predicted start of transcription. Primer extension was used to confirm that the Mga-dependent transcriptional start site for sclA was located adjacent to the proximal PsclA-II binding site. By using overlapping PsclA promoter probes and purified Mga-His fusion protein, it was shown by electrophoretic mobility shift assays that, unlike other Mga-regulated promoters, Mga binds only to a distal DNA-binding site (PsclA-I). Binding of Mga to PsclA-I could be competed with cold probes corresponding to known Mga-regulated promoters (Pemm, PscpA, and Pmga) but not with a nonspecific probe or the proximal PsclA-II fragment. With the use of a plasmid-based green fluorescent protein transcriptional reporter system, the full-length PsclA was not sufficient to reproduce normal Mga-regulated activation. However, studies using a single-copy gusA transcriptional reporter system integrated at the native sclA chromosomal locus clearly demonstrated that the distal PsclA-I binding site is required for Mga regulation. Therefore, PsclA represents a new class of Mga-regulated promoters that requires a single distal binding site for activation.
A hallmark of the group A streptococcus (GAS; Streptococcus pyogenes) is its ability to elicit a wide array of human diseases at varied locations throughout the human body. These syndromes may include relatively minor purulent infections that target either the upper respiratory tract or the skin as well as life-threatening invasive infections of deeper tissues, bloodstream, and organs (6, 30). GAS pathogenesis involves a number of different factors that allow the organism to survive within the human host. The initial stages of infection involve cell-associated factors that contribute to colonization, cell tropism, and immune evasion, including M proteins, capsule, and extracellular matrix binding proteins. Later stages often utilize secreted bacterial factors such as proteases, toxins, and other enzymes that allow the organism to spread and invade deeper tissues (6). In order to coordinate the expression of these different determinants with the changing environments that GAS encounters during an infection, the pathogen has developed multigene networks to regulate virulence.
One such regulator is the multiple virulence gene regulator of GAS known as Mga, which activates the transcription of genes encoding primarily surface-associated proteins involved in GAS pathogenesis. These factors include the M family of proteins (emm, mrp, arp, and enn), C5a peptidase (scpA), fibronectin-binding proteins (sof and fbx), and the secreted inhibitor of complement (sic) (4, 5, 12, 20). Mga is also required for its own positive regulation or autoactivation (19, 23). Environmental growth conditions such as increased CO2 and temperature have been shown to activate transcription of genes in the Mga regulon (3, 16, 26). Furthermore, expression of the Mga regulon is maximal during exponential phase and becomes transcriptionally silent as the bacteria enter stationary phase (18). This temporal regulation correlates with the requirement for Mga-regulated surface proteins during early stages of infection when GAS is actively growing but down regulates its expression during times of nutrient deprivation when the proteins may actually hinder the bacterium from spreading to new tissues. Thus, Mga controls its set of virulence factors in response to both environmental and temporal signals through an unknown mechanism.
Mga functions by binding directly to specific sequences found in the promoters of target genes, as demonstrated for emm (Pemm) and scpA (PscpA) in the serotype M6 GAS strain JRS4 (15). An alignment of these two sites was used as the basis for a published consensus Mga-binding site (15). Binding of Mga in these two promoters occurs at similar locations consisting of a single site overlapping the −35 hexamer and just upstream of the start of transcription (15). Interestingly, autoregulation of mga transcription also involves binding of Mga within its own promoter (Pmga); however, Mga interacts with two separate binding sites located more than 100 bp upstream of the major P2 start site (19). Thus, based upon the variable location of binding sites in these three promoters, the mechanisms for Mga activation (Pemm and PscpA) versus autoactivation (Pmga) may be quite different. Although all four of the identified Mga-binding sites are less than 50% identical at the DNA sequence level, recent studies have shown that the same two helix-turn-helix DNA-binding domains of Mga are responsible for recognizing each of these divergent sites (17). Furthermore, the ability of Mga to bind to these promoters in vivo was shown to be essential for virulence gene activation (17). Exactly which nucleotides within a given binding site are required for Mga recognition and binding at these various promoters has not been established.
Recently, a gene encoding a cell-wall-anchored streptococcal collagen-like protein (sclA or scl1) was identified in all serotypes of GAS tested (13, 27). A mutant of the newly described surface protein was found to be defective for adherence to lung epithelial cells, but not pharyngeal cells, and was attenuated for virulence in a mouse model of soft-tissue pathology (13, 27). Expression of sclA was found to be Mga regulated in a serotype M1 GAS, and maximal expression occurred during exponential growth as expected (13, 14, 27). With the use of the published consensus Mga-binding site (15) to search the promoter region upstream of sclA, a potential Mga-binding site was found overlapping the −35 hexamer of a suspected start of transcription (13, 27) in a location similar to that observed for Pemm and PscpA. In this study, we identify a second distal binding site, and we investigate whether Mga plays a direct role in transcriptional activation at the sclA promoter.
MATERIALS AND METHODS
Bacterial strains and media.
S. pyogenes (GAS) SF370 (31) represents an available serotype M1 GAS genome sequence (8). JRS4 is a streptomycin-resistant derivative of serotype M6 strain D471 (29). JRS519 (mga-10) is an Mga− derivative of JRS4 (18). MGAS166 is a streptomycin-resistant M1 strain (22). Escherichia coli strain DH5α (hsdR17 recA1 gyrA endA1 relA1) was used as the host for plasmid constructions. The T7 polymerase E. coli strain BL21(DE3) [F− ompT hsdSB (rB− mB−) gal dcm met] was used for protein expression and purification.
All E. coli strains were grown in Luria-Bertani broth. GAS was cultured in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY), and growth was assayed by optical density with a Klett-Summerson photoelectric colorimeter with the A filter. Antibiotics were used at the concentrations indicated: ampicillin at 100 μg/ml for E. coli, spectinomycin at 100 μg/ml for both E. coli and GAS, kanamycin at 50 μg/ml for E. coli and 300 μg/ml for GAS, and erythromycin at 500 μg/ml for E. coli and 1.0 μg/ml for GAS.
DNA manipulations.
Plasmid DNA was isolated from E. coli by using either the Wizard Miniprep system (Promega) or Maxi/Midi Prep purification systems (Qiagen). DNA fragments were isolated from agarose gels by using the QIAquick gel extraction kit (Qiagen). GAS chromosomal DNA was isolated using the FastDNA kit and a FastPrep cell disruptor (Bio 101, Inc.). PCR for cloning and promoter probes was performed using Pfx high-fidelity DNA polymerase (Invitrogen), and reaction mixtures were purified using the QIAquick PCR purification system. PCR for diagnostic assays was performed using Vent DNA polymerase (New England Biolabs). DNA sequencing was performed either using the Excel II cycle sequencing kit (Epicentre, Inc.) or through the automated sequencing core facility in the McDermott Center at University of Texas Southwestern Medical Center.
Construction of the Mga− M1 strains KSM165-L and MGAS166.165-L.
To inactivate mga in SF370 and MGAS166, a suicide plasmid was constructed as follows: a 983-bp PCR fragment containing an internal fragment of mga was amplified from M1 SF370 genomic DNA by using primers OYR-4 and DivMgaL6 (Table 1). The resulting fragment was cloned blunt into an SmaI-digested suicide vector, pCIV2 (23), to produce pKSM165-L. Serotype M1 SF370 and MGAS166 were transformed with 200 to 300 μg of pKSM165-L, and chromosomal integrations were selected for by growth on THYA plates containing kanamycin at 37°C. Mutants were verified by PCR analysis of the mga locus and slot blot analysis of mga transcripts (data not shown).
TABLE 1.
PCR primers used in this study
| Target | Primer | Sequence (5′-3′)a | Reference |
|---|---|---|---|
| emm | OM6-16 | GTTTCCTTCATTGGTGCT | 17 |
| OM6-35 | AACAGCAAATTAGCTGCTC | 17 | |
| gusA | gusA-PE | GTTGGGGTTTCTACAGGACG | This study |
| mga | DivMgaL6 | CTTTTAAGAAACTATTTTCC | This study |
| MgaPetI_Nde | ggggcatATGTATGTAAGTAAGTTGTTT | This study | |
| MgaPetII_Xho | aactcgagAGTTGTGGAGGGGG | This study | |
| OYR-4 | GTACCATCAACATTGCG | 19 | |
| Nonspecific | rpsL-1 | GAATGTAGATGCCTACAATTAACCA | 17 |
| rpsL-2 | GTGCGCCACGAACGATATG | 17 | |
| nrdI | nrdI-PE | GGTAATCTGAGGCATAAGTG | This study |
| Pemm6 | Pemm-L1 | GCATGGATCCCATCGCAAAGAGCTTA | 17 |
| Pemm-R1 | GCGGCTCGAGTAGTGTCTATTCGTGTTATT | 17 | |
| Pemm1 | Pemm1-L1 | AAGCGATTATTGACAAGCTC | This study |
| Pemm1-R1 | AGTCAAAGCTACCGCTACTG | This study | |
| Pmga | OYR-25 | GGTTGTACCATAACAGTC | 17 |
| OYL-25 | TACCATAAAATACCTTTC | 17 | |
| PsclA | PsclA-L | TTAAAGAAAGGATCCGGATG | This study |
| PsclA-R | ATGGTGCTTTGATGTCAACA | This study | |
| PsclA-L2 | TGGTAATCGTAATTGTCTGC | This study | |
| PsclA-R2 | GGAGATCAGAGGGCTACTTT | This study | |
| PsclA-L4 | AGCAGTCAAAAAAAGTAACA | This study | |
| PsclA-R4 | TGTACCTTCTTGTCTGGTTG | This study | |
| PsclA-L5 | ACAGCTATGATCATCTATGG | This study | |
| PsclA-R5 | GATTATGGTGCTTTGATGTC | This study | |
| PsclA-L6 | GTGATCCCAAGCCCCAA | This study | |
| PscpA | C5-L1 | AAGAATGAGATTAAGGAGGTCACA | 15 |
| C5-R1 | GCGCAATGGCAAGTTTGTC | 15 | |
| sclA | sclA-F2 | ACGTCGAGTCAAAGAGAGTCAAAAATAAA | This study |
| sclA-R1 | GTTTGGTTAGCTTCTTTGTCA | This study |
Underlined nucleotides are restriction sites, and lowercase nucleotides are anchor sequences introduced into the primer.
Northern blot analysis.
Total RNA was isolated from samples in late exponential phase (65 Klett units) by using the FastRNA kit and a FastPrep cell disruptor as previously described (17). Northern blot assays of total RNA were performed using the NorthernMax kit (Ambion). Briefly, either 1 (emm) or 5 (sclA) μg of total RNA was separated on a 1% agarose denaturing gel (formaldehyde) and transferred to a positively charged nylon membrane. The blots were then UV cross-linked and prehybridized for 30 min at 50°C. Blots were hybridized overnight at 50°C with 5 × 106 cpm of [α32P]dATP-labeled emm or sclA probe (RadPrime labeling system; Invitrogen) followed by two low-stringency washes at room temperature (RT) for 5 min and two high-stringency washes at 50°C for 15 min. Blots were visualized by exposure to a phosphorimaging cassette for 2 h. Probes were PCR amplified from a serotype M1 strain by using the primers listed in Table 1 for emm and sclA.
Primer extension analysis.
Total RNA was extracted from samples in late exponential phase as described previously (17) by using the FastRNA kit and a FastPrep cell disruptor. Primer extensions were performed on 25 μg of total RNA as described previously (19) with the primers PsclA-R and nrdI-PE (Table 1). The primer extension products were run on a 6% denaturing polyacrylamide gel (Amresco), and gels were processed as described above. Sequence was generated using labeled PsclA-R and nrdI-PE primers (Table 1) on a PCR product amplified from SF370 genomic DNA (Table 1; PsclA-L2 and PsclA-R) as described above.
Expression and purification of Mga-His protein from E. coli.
A carboxy-terminal fusion of six histidines to the serotype M6 Mga was constructed as follows: a 1.4-kb PCR fragment containing the mga open reading frame was amplified from GAS JRS4 genomic DNA by using primers MgaPetI_Nde and MgaPetII_Xho (Table 1). The PCR fragment was digested with NdeI/XhoI and cloned into NdeI/SalI-digested pET21a (Novagen) to produce the T7-inducible Mga-His expression vector pKSM170.
The E. coli strain BL21(DE3) contains the T7 polymerase under the control of Plac and allows induction upon addition of IPTG (isopropyl-β-d-thiogalactopyranoside). The pKSM170 mga-his plasmid was introduced into BL21(DE3) for expression and purification. Briefly, cultures of E. coli BL21(DE3) containing pKSM170 (PT7-mga-his) were grown at 30°C in Luria-Bertani broth plus ampicillin (100 μg/ml), and expression of the protein was induced for 1.5 h by addition of 1 mM IPTG. Cells were lysed by two passages through a prechilled French pressure cell, and Mga-His was purified over a nickel-nitrilotriacetic acid resin column (Qiagen) under native conditions. Protein concentrations were determined using the Bio-Rad protein assay kit. DNA-binding activity of purified Mga-His was verified by comparison to the published maltose binding protein (MBP)-Mga fusion protein by electrophoretic mobility shift assay (EMSA) (15, 17, 19). The stability of the purified protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis as described below.
Expression and purification of Mga-His protein from GAS.
A GAS-expression plasmid possessing a carboxy-terminal fusion of six histidines to the serotype M6 Mga was constructed as follows: pKSM170 was digested with SpeI and ScaI, and the resulting 2.2-kb fragment containing mga-his was gel purified and cloned into SpeI/SnaBI-digested pJRS525 (18) to form pKSM163. pJRS2050 (1), possessing Pmga, was digested with BamHI/SpeI, and the resulting 2.2-kb fragment was gel purified and cloned into BamHI/SpeI-digested pKSM163 to form pKSM164.
The Pmga-mga-his pKSM164 plasmid was transformed into JRS519, an Mga− M6 JRS4 derivative, for expression and purification. Briefly, cultures of JRS519(pKSM170) were grown to late logarithmic phase at 37°C in THYB plus spectinomycin. Cells (20 50-ml aliquots) were lysed using five 9-s pulses at 4°C on a FastPrep cell disruptor, and Mga-His was purified over a nickel-imino acetic acid resin column (Pharmacia) under native conditions. Protein concentrations were determined using the Bio-Rad protein assay kit. The stability of the purified protein was assessed by SDS-PAGE and Western blot analysis as described below.
Western blot analysis.
Anti-Mga-His immunoblot assays were performed as described previously (17). Briefly, purified Mga-His proteins were separated by SDS-10% PAGE. Proteins were transferred to nitrocellulose membranes and reacted either with the anti-Mga-pep2 antiserum (17) at a 1:250 dilution for 2 h at RT or with the anti-His monoclonal antiserum (Novagen) at a 1:2,000 dilution for 2 h at RT. Blots containing GAS whole-cell extracts were reacted with anti-rat immunoglobulin G horseradish peroxidase-conjugated secondary antibody (Sigma) or anti-mouse immunoglobulin G horseradish peroxidase-conjugated secondary antibody (Chemicon International), respectively; developed using the Renaissance chemiluminescence system (New England Nuclear); and visualized on autoradiography film (Kodak).
EMSAs.
Promoter probes were generated by PCR amplification with either serotype M1 strain SF370 or serotype M6 strain JRS4 chromosomal DNA and the relevant primer pairs (PsclA, Pemm6, Pmga, and PscpA) listed in Table 1. PCR fragments were end labeled with [γ32P]ATP with the use of T4 polynucleotide kinase (New England Biolabs). Labeled fragments were excised from a 5% polyacrylamide gel, extracted by crush and soak elution, and purified using the QIAquick PCR purification system.
EMSA was performed as described previously (15). Briefly, a constant amount of labeled promoter probe DNA (ca. 1 to 5 ng) and increasing amounts of purified Mga-His (1 to 6 μg) were used in each reaction. Competition assays were performed by addition of 750 ng of unlabeled promoter probes to binding reaction mixtures. Gels were dried under vacuum at 80°C for 1 h and exposed overnight either to film or to a phosphorimaging screen. Phosphor screens were scanned using a Storm 820 scanner (Amersham Biosciences), and resulting data were analyzed with the ImageQuant analysis software (version 5.2).
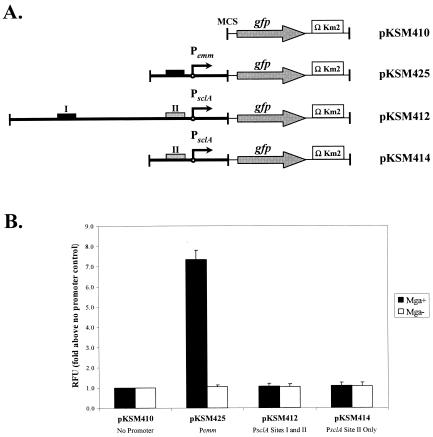
Construction and use of a green fluorescent protein (GFP)-based transcriptional reporter plasmid.
The plasmid pGreenTIR contains a promoterless gfp cassette modified for maximal solubility and fluorescence in bacteria (21). An 854-bp EcoRI fragment containing the promoterless gfp and its ribosomal binding site was cloned into the EcoRI-digested GAS shuttle vector pJRS525 (18) to produce the plasmid pKSM123. A SmaI-digested fragment of pUCΩKm2 (24), possessing ΩKm2 and T4 terminators, was gel purified and cloned at the 3′ end of gfp in the EcoRV-digested pKSM123 to form the transcriptional reporter plasmid pKSM410. Blunt PCR products corresponding to Pemm1 (pKSM425), a 500-bp PsclA-L2-R fragment (pKSM412), and a 253-bp PsclA-L-R fragment (pKSM414) were amplified from the serotype M1 GAS strain SF370 genomic DNA and cloned into XhoI-digested (blunted) pKSM410 (primers shown in Table 1). Plasmids were introduced into the serotype M1 strains MGAS166 (Mga+) and MGAS166.165-L (Mga−) by electroporation as described above.
GAS strains containing GFP reporter plasmids were grown to late logarithmic phase (65 Klett units), pelleted, and resuspended in phosphate-buffered saline. Aliquots (200 μl) in duplicate were transferred to a 96-well black microtiter plate with a clear bottom (Costar) along with a phosphate-buffered saline control. Samples were assayed using a FLUOStar Optima (BMG Lab Technology) fluorometer by using an excitation filter at 485 nm and an emission filter at 510 nm. Relative fluorescence units for each sample were expressed as fold above the level of the promoterless control strain containing pKSM410.
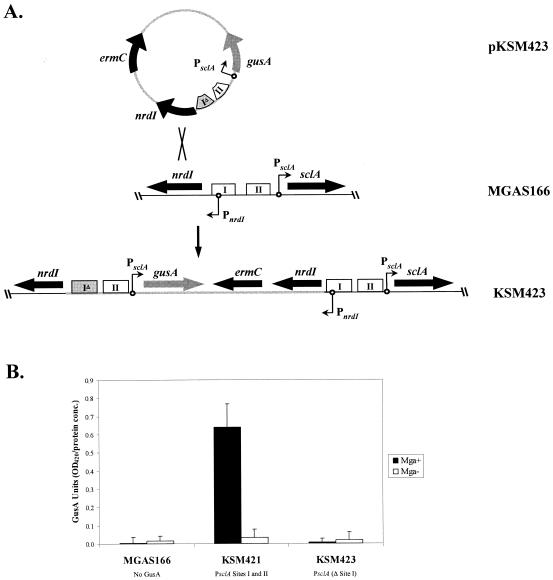
Construction and use of a chromosomal GusA-based transcriptional reporter.
A 257-bp fragment of PsclA containing Mga-binding site II was PCR amplified from the M1 strain SF370 by using the PsclA-L-R5 primer pair (Table 1) and ligated into the EcoRV-digested pBluescript II KS− (Stratagene) to form pKSM417. A 619-bp fragment of PsclA containing sequence upstream of Mga-binding site II was PCR amplified from SF370 by using the PsclA-L5-R4 primer pair (Table 1) and was cloned into the SmaI-digested pKSM417 to form pKSM418. To form the plasmid pKSM422, an 890-bp fragment of pKSM418 was PCR amplified using the PsclA-L5-R primer pair (Table 1) and ligated into the SmaI-digested gusA reporter plasmid pKSM140 (28). The PsclA-Δ site I-gusA allele of pKSM422 was excised by PvuII digestion, gel purified, and cloned into the EcoRV site of the temperature-sensitive pJRS233 (25) to form the PsclA-Δ site I-gusA plasmid pKSM423.
Wild-type PsclA was PCR amplified from M1 SF370 by using the primer pair PsclA-L2-R (Table 1) and digested with HindIII prior to cloning into HindIII/HincII-digested pBluescript II KS− (Stratagene) to form pKSM419. This plasmid was then cut with KpnI/EcoRI and ligated into KpnI/EcoRI-digested gusA plasmid pKSM140 (28) to form pKSM403. The 2.3-kb EcoRI (blunted)/PstI PsclA-gusA fragment from pKSM403 was moved into PstI/SmaI-digested pVIT164 (9) to form pKSM405. The HpaI/PstI PsclA-gusA fragment from pKSM405 was cloned into EcoRV/PstI-digested pJRS233 (25) to form pKSM421.
The pKSM421 (PsclA-gusA) and pKSM423 (PsclA-Δ site I-gusA) plasmids were introduced into MGAS166 (Mga+) and MGAS166.165-L (Mga−) by electroporation. Following growth at the permissive temperature, integrants were selected by passaging the cells in THYB containing erythromycin at the nonpermissive temperature. Integration at the correct position was assessed by PCR amplification with the primer pair PsclA-L6 and gusA-PE (Table 1). The presence of the site I mutation was confirmed by digestion of this PCR product with PstI. A GusA assay on these strains was performed as previously described (7). Briefly, cells were grown to late logarithmic phase (65 Klett units), lysed with a FastPrep cell disruptor, and assayed for GusA activity. Results are reported in GusA units, which are equivalent to the optical density at 420 nm (OD420) of the processed lysate divided by the concentration of total lysate protein (micrograms per microliter) as determined by the Bio-Rad protein assay kit.
RESULTS
Identification of two potential Mga-binding sites upstream of sclA.
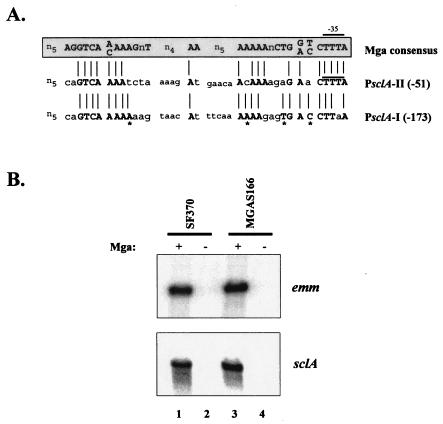
To identify new Mga-regulated virulence genes in GAS, a homology search of the serotype M1 SF370 genome database (AE004092) was performed using the published Mga consensus binding sequence (15). As expected, binding sites were found within promoters of known Mga-regulated genes, including emm, scpA, and sic (data not shown). A putative Mga-binding site was also identified centered 51 bp upstream of the Mga-regulated sclA (Fig. 1A; PsclA-II) and corresponds to a site described previously (13, 27). The site shares 66% identity to the Mga consensus sequence at conserved sites and was located in PsclA overlapping the −35 hexamer of a predicted start of sclA transcription (Fig. 1A; PsclA-II), identical to the established Mga-binding sites for Pemm and PscpA (15). Interestingly, a second site showing 72% identity to the Mga consensus sequence (Fig. 1A; PsclA-I) was identified centered 153 bp upstream from PsclA. Scanning of the 3′ sequence adjacent to PsclA-I did not reveal an obvious second start of transcription, suggesting that Mga bound at this potential element may target the downstream transcriptional start site next to PsclA-II (Fig. 2B). Furthermore, sequences that are 100% identical to the conserved consensus nucleotides for both sites (Fig. 1A) are found upstream of sclA in strains representing a wide array of GAS serotypes, including M2, M3, M6, M12, M18, M28, M49, M52, M56, and M77 (data not shown). Thus, PsclA appears to possess two potential Mga-binding elements arranged in a pattern that is unique to this Mga-regulated promoter and is conserved across GAS serotypes.
FIG. 1.
Mga regulation of sclA in M1 strains. (A) A search of the serotype M1 GAS SF370 genomic sequence upstream of the known Mga-regulated sclA gene with the published Mga consensus binding sequence (15) revealed two potential Mga-binding sites. An alignment of PsclA Mga-binding site I (PsclA-I) and site II (PsclA-II) with published consensus Mga-binding sequence is shown. PsclA-II overlaps the predicted −35 hexamer of this promoter as described previously (13, 27). PsclA-I represents a novel site and is located farther upstream. Bold, capital letters and lines represent nucleotide identity to the Mga consensus binding sequence, while lowercase letters represent nonidentity. Asterisks indicate nucleotides in PsclA-I that are identical to the Mga-binding consensus but are not conserved in PsclA-II. (B) Northern blot analysis of total RNA isolated from M1 GAS strains SF370 (Mga+, lane 1), KSM165-L (Mga−, lane 2), MGAS166 (Mga+, lane 3), and MGAS166.165-L (Mga−, lane 4). Blots were hybridized with probes to emm and sclA amplified from the PCR primers listed in Table 1.
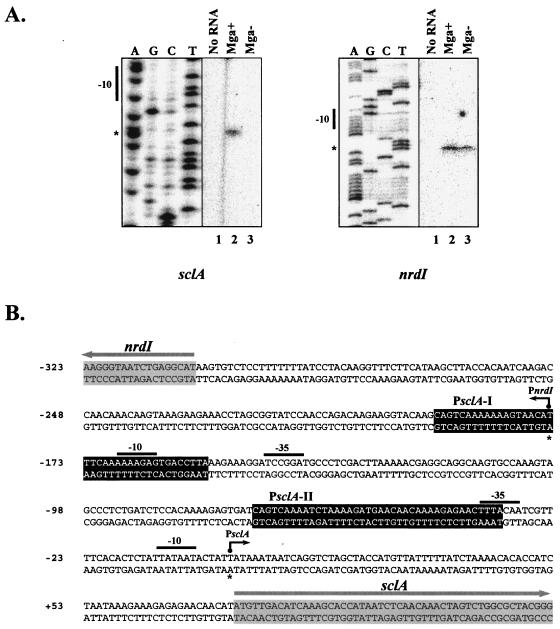
FIG. 2.
Identification of transcriptional start sites for sclA and nrdI. (A) Primer extension analysis was performed on total RNA isolated from serotype M1 SF370 (Mga+) and KSM165-L (Mga−) by using the radiolabeled antisense primers PsclA-R for sclA and nrdI-PE for nrdI (Table 1) as described in Materials and Methods. The start of transcription for sclA and nrdI (asterisks) as well as their corresponding −10 hexamers is shown. (B) Sequence of sclA and nrdI intergenic region from serotype M1 SF370 indicating the identified start sites (asterisk and dark arrow) along with −10 and −35 regions (solid bars). Potential Mga-binding sites (black boxes) and starts of translation (gray arrows) are designated. The sequence is numbered using the PsclA transcriptional start site as +1.
Previous studies found that expression of sclA is Mga regulated in the M1 strains AP1 (27) and JRS301 (14). In order to confirm that sclA is also Mga regulated in the M1 SF370 and MGAS166 strains used in this study, a Northern analysis was performed on wild-type and mga-inactivated versions of each strain on RNA isolated at the exponential phase of growth (Fig. 1B). As a control, Northern analysis was also performed on the established Mga-regulated gene emm (Fig. 1B). As expected, transcript levels for both emm and sclA are dramatically lower in the Mga− versions of SF370 and MGAS166 than in their wild-type counterparts, confirming Mga regulation of sclA in these M1 strains.
Identification of the Mga-regulated start of transcription for sclA.
Since the predicted location of the Mga-regulated start site for sclA expression has not been experimentally verified, primer extension analysis was performed on total RNA isolated from both wild-type (SF370) and mga-inactivated (KSM165-L) serotype M1 GAS strains by using an antisense primer, PsclA-R (Table 1), located in the 5′ end of sclA. An extension product was clearly observed in a background producing Mga (Fig. 2A) that corresponded exactly to the start of transcription predicted in the literature (13, 27). However, the product was clearly absent in the mga-inactivated KSM165-L (Fig. 2A). As a control for the primer extension analysis, the transcriptional start for the divergently transcribed nrdI gene found directly upstream of sclA was mapped in both strains tested (Fig. 2A). Interestingly, although the nrdI start site is located within the putative distal PsclA-I Mga-binding element, there was no alteration in nrdI transcription either in the presence or in the absence of Mga. These data indicate that sclA contains a single Mga-regulated start of transcription that is located directly downstream of the potential PsclA-II Mga-binding site (Fig. 2B).
Mga-His purified from E. coli binds to the distal PsclA-I site but not the promoter-proximal PsclA-II site.
To determine whether Mga directly interacts with the two potential binding elements within PsclA, EMSAs were performed on overlapping PCR-generated promoter probes encompassing the PsclA region (Fig. 3A). Previous EMSA experiments have successfully utilized an MBP-Mga protein generated from a serotype M6 GAS strain (15, 17, 19); however, this chimera is much larger in size than Mga alone (105 versus 62 kDa) and exhibits only 60% of wild-type Mga activity when expressed in vivo (18). In order to bypass these limitations, a carboxy-terminal fusion of six histidines to the full-length Mga from the M6 GAS strain JRS4 (Mga-His; 63 kDa) was constructed and purified from E. coli lysates as described in Materials and Methods. Purified Mga-His showed DNA-binding activity to known binding sites in vitro comparable to that of MBP-Mga and was capable of wild-type transcriptional activation of Mga-regulated genes in vivo (data not shown).
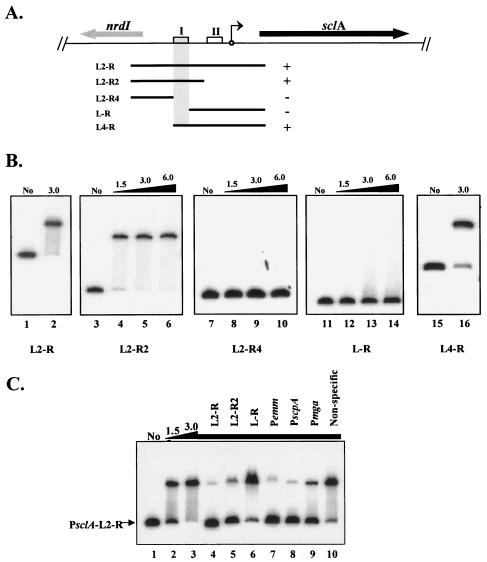
FIG. 3.
Identification of a specific Mga-binding site in the sclA promoter region. The ability of purified serotype M6 Mga-His to bind different PCR-generated PsclA promoter probes was determined by EMSA. (A) Schematic representation of the M1 GAS genomic region surrounding sclA, including nrdI encoding a putative ribonucleotide reductase (thick arrows). The start of transcription for sclA (circle with arrow) and potential Mga-binding sites (boxes) are shown. The ability of purified Mga-His to bind to overlapping PCR-amplified promoter probes (thin lines) in vitro is represented by either plus (+) or minus (−). The defined binding region is shaded. (B) EMSAs of PsclA promoter probes shown above: PsclA-L2-R (lanes 1 and 2), PsclA-L2-R2 (lanes 3 to 6), PsclA-L2-R4 (lanes 7 to 10), PsclA-L-R (lanes 11 to 14), and PsclA-L4-R (lanes 15 and 16). Constant amounts (1 to 2 ng) of labeled promoter probes were incubated with increasing amounts (1.5 to 6.0 μg) of Mga-His for 15 min at 16°C prior to separation on a 5% polyacrylamide gel. Shown are results representative of at least two separate experiments performed with independently purified protein preparations. (C) The specificity of Mga-His binding to PsclA was assayed by addition of unlabeled competitor promoter probes (Table 1). Radiolabeled PsclA-L2-R probe was analyzed by EMSA as described above following incubation with 0, 1.5, and 3.0 μg of Mga-His (lanes 1 to 3). To the remaining binding reaction mixtures (lanes 4 to 10, 3.0 μg of Mga-His), a constant amount of unlabeled competitor probe (750 ng) corresponding to PsclA, Pemm, PscpA, Pmga, and a nonspecific rpsL fragment was added using the PCR primer pairs listed in Table 1.
All three promoter probes that contained the distal PsclA-I binding site demonstrated a slower migration consistent with binding when incubated in increasing amounts of the purified Mga-His protein (Fig. 3B; L2-R, L2-R2, and L4-R). In contrast, no detectable binding of Mga-His was observed to a probe that lacked the distal site I but possessed the proximal PsclA-II binding element and the sclA start of transcription (see above) at any of the concentrations tested (Fig. 3B; L-R). Furthermore, no additional binding sites were found in the region located upstream of PsclA-I (Fig. 3B; L2-R4). Thus, Mga appears to show in vitro DNA binding only to the distal PsclA-I binding site situated more than 100 bp upstream of the Mga-regulated start of transcription.
Specific binding of Mga-His to PsclA-I in vitro.
The specificity of Mga-His binding to the PsclA-I element was investigated by EMSA with the large PsclA-L2-R probe (Fig. 3A) in the presence of different unlabeled competitor probes (Fig. 3C). The bound species was chased back to unbound only in the presence of cold probes containing PsclA-I (L2-R and L2-R2) and not when unlabeled PsclA-II probe (L-R) was added (Fig. 3C, lanes 3 to 6). Mga binding at PsclA-I was also competed by addition of other established Mga-binding site probes such as Pemm, PscpA, and Pmga but not a nonspecific probe internal to rpsL (Fig. 3C, lanes 7 to 10). In fact, an Mga mutated in the major DNA-binding domain (HTH-4), necessary for interaction at all known Mga-binding sites (17), was also incapable of binding to PsclA at any concentration tested (data not shown). These results verify that Mga binds in a highly specific fashion to PsclA and further demonstrate that this interaction occurs only at the upstream PsclA-I site and not at the promoter-proximal PsclA-II site.
Comparison of E. coli- and GAS-purified Mga.
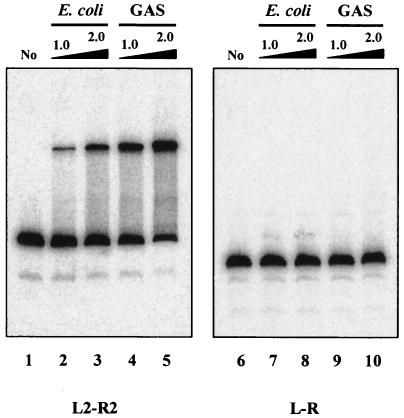
It is possible that Mga-His purified from a heterologous E. coli host lacks a modification that allows it to bind PsclA-II in vitro. To investigate whether Mga obtained from its native background possesses a different in vitro binding activity towards PsclA, Mga-His was purified directly from GAS lysates as described in Materials and Methods. EMSA was then performed on PsclA-I and PsclA-II by using Mga-His purified from both sources (Fig. 4). Interestingly, Mga purified from GAS does appear to bind PsclA-I more efficiently (Fig. 4, lanes 4 and 5) than does an equal quantity of Mga purified from E. coli (Fig. 4, lanes 2 and 3). Binding to the proximal PsclA-II with equivalent amounts of either protein was not observed (Fig. 4, lanes 7 to 10). However, upon significant overexposure of the image, barely detectable binding of GAS-purified Mga-His to PsclA-II was seen (data not shown). Thus, Mga may bind to the proximal PsclA-II site but with markedly lower affinity than that observed for the distal PsclA-I site.
FIG. 4.
Binding of native and recombinant Mga to sclA promoter region. M6 Mga-His proteins were purified either from E. coli (lanes 2, 3, 7, and 8) or directly from GAS (lanes 4, 5, 9, and 10) and compared in their abilities to bind both PsclA site I (L2-R2, lanes 1 to 5) and PsclA site II (L-R, lanes 6 to 10) in an EMSA. Radiolabeled promoter probes (1 ng) were incubated with 1.0 or 2.0 μg of each Mga-His protein for 15 min at 16°C followed by separation on a 5% polyacrylamide gel. Probes were amplified from M1 SF370 by using the primers listed in Table 1.
Mga-dependent regulation of PsclA is not observed if located on a multicopy plasmid.
To investigate the role of Mga binding to the upstream PsclA-I site in regulation of sclA expression, a transcriptional reporter system based on GFP was developed. Use of GFP as a reporter allows direct quantification of promoter activity via levels of fluorescence without the need to lyse the bacteria or add reagents. Unfortunately, attempts to generate single-copy promoter fusions of the strong Pemm promoter to gfp in the chromosome of GAS did not allow detection over background levels (data not shown). Therefore, a multicopy GFP transcriptional reporter plasmid called pKSM410 was constructed that is capable of replicating in GAS as well as E. coli (see Materials and Methods).
Promoter fragments corresponding to Pemm (pKSM425), the entire sclA-nrdI intergenic region (pKSM412), and PsclA lacking the distal PsclA-I Mga-binding site (pKSM414) were cloned in front of the promoterless gfp in pKSM410 (Fig. 5A) and introduced into both wild-type MGAS166 and mga-inactivated MGAS166.165-L serotype M1 GAS. Strains were grown to late logarithmic phase, and the levels of fluorescence were quantified using a fluorometer as described in Materials and Methods. As expected for an Mga-regulated promoter, the control Pemm construct pKSM425 showed extremely high expression in MGAS166 but was reduced 7.5-fold in MGAS166.165-L (Fig. 5B; pKSM425). However, the full-length PsclA-L2-R construct pKSM412, which has both the PsclA-I binding site and the Mga-regulated start of transcription, demonstrated very little promoter activity over background and was not affected by the presence or absence of Mga (Fig. 5B; pKSM412). Likewise, the transcriptional start site without the distal PsclA-I binding site showed little change in activity in the presence or absence of Mga (Fig. 5B; pKSM414). Therefore, the presence of the upstream Mga-binding site PsclA-I in the sclA promoter was not sufficient to show Mga-regulated gfp expression in vivo and suggests that additional cis sequences not included in this construct are required.
FIG. 5.
Analysis of PsclA activity in vivo using a GFP reporter plasmid. (A) Schematic representation of different promoter fusions in the GFP transcriptional reporter plasmid pKSM410 as described in Materials and Methods. Plasmids pKSM425 (Pemm1), pKSM412 (PsclA-L2-R), and pKSM414 (PsclA-L-R) are shown. The multiple cloning site (MCS), starts of transcription (circles with arrows), modified promoterless gfp (gray arrows), kanamycin resistance cassettes containing T4 transcriptional terminators (open boxes), and functional (dark boxes) or nonfunctional (gray boxes) Mga-binding sites are indicated. (B) GFP fluorescence assay on M1 GAS strain MGAS166 (Mga+, dark bars) or MGAS166.165-L (Mga−, white bars) containing the reporter plasmids described above (shown on bottom). Equal numbers of cells taken from late-logarithmic growth were used for each assay. Results are presented as fold increase above the promoterless control strain containing pKSM410 (shown as 1.0). Data represent at least two independent experiments, and standard error bars are provided. RFU, relative fluorescence units.
PsclA-I Mga-binding site is essential for Mga regulation of PsclA at its native chromosomal location.
Due to the lack of Mga-dependent regulation of PsclA in a plasmid system, analysis of the promoter at its native chromosomal location was undertaken. Initial attempts to introduce a deletion of the PsclA-I binding site into the MGAS166 chromosome via allelic exchange were unsuccessful, possibly due to the concomitant deletion of the divergently transcribed nrdI promoter (Fig. 2B; data not shown). Therefore, a merodiploid approach was used to allow the temperature-sensitive integration of single-copy PsclA-gusA reporter fusions into the native chromosomal location while retaining a second wild-type promoter expressing both nrdI and sclA (Fig. 6A). Merodiploid insertion-duplication mutants containing either wild-type PsclA (KSM421) or the PsclA-I deletion allele (KSM423) fused to a promoterless gusA were constructed in MGAS166 (Mga+) as well as MGAS166.165-L (Mga−), and GusA liquid assays were performed on exponential-phase cultures as described in Materials and Methods. The wild-type KSM421 exhibited strong Mga-regulated GusA activity, whereas the PsclA-I deletion strain KSM423 showed GusA activity at background levels regardless of the presence or absence of Mga (Fig. 6B). These results support in vivo the results of the in vitro EMSAs and indicate an essential role for the distal PsclA-I binding site in the Mga-dependent expression of sclA.
FIG. 6.
Analysis of PsclA activity in vivo with a chromosomal GusA reporter. (A) Construction of an insertion-duplication allele that produces a chromosomal PsclA fused to a gusA transcriptional reporter while allowing the preservation of the nrdI promoter. Briefly, the PsclA-gusA transcriptional fusion was cloned into a temperature-sensitive plasmid (pKSM423). Integrants into the chromosome of MGAS166 were isolated after growth at the nonpermissive temperature, which created the strain KSM423. Genes (thick black arrows), gusA (thick gray arrows), starts of transcription (circles with arrows), and wild-type (open boxes) and mutated (gray boxes) Mga-binding sites are indicated. (B) GusA reporter assay on M1 GAS strains MGAS166 (Mga+), MGAS166.165-L (Mga−), KSM421 (Mga+, PsclA-gusA), KSM421.165-L (Mga−, PsclA-gusA), KSM423 (Mga+, PsclA-Δ site I-gusA), and KSM423.165-L (Mga−, PsclA-Δ site I-gusA). Data are reported in GusA units (OD420/concentration of total protein [micrograms per microliter]) and represent an average of the results of three independent experiments. The error bars express the standard deviation for each strain measured.
DISCUSSION
What constitutes an Mga-binding site?
Since every Mga-regulated gene described to date encodes a protein involved in GAS pathogenesis, the ability to identify potential Mga-regulated genes through an Mga-binding site in their promoter would be highly advantageous. The published consensus Mga-binding element was constructed using known binding sites (Pemm and PscpA from serotype M6) as well as predicted sites from sequenced promoters corresponding to emm, mrp, arp, enn, and scpA from various other GAS serotypes (15). The resulting 45-bp consensus element contains three regions of conserved nucleotides totaling 29 nucleotides separated by several stretches of irrelevant nucleotides (Fig. 1A). However, this consensus was not able to predict the location of the two Mga-binding sites found in Pmga (19), indicating that Mga can interact with diverse sequences and that any true consensus must reflect these various sites. Since Mga appears to use the same two helix-turn-helix domains of the protein for binding to all of the known promoter sites (17), there likely exists a core consensus binding element that is shared by all sites.
Although the two sites identified within PsclA shared significant identity (72 and 66%, respectively) to the nucleotides in the published consensus, we have shown here that Mga was able to bind only the distal PsclA-I site in vitro (Fig. 2 to 4). In fact, PsclA-I contains only four nucleotides identical to conserved consensus sequences that are lacking in PsclA-II (Fig. 1A, asterisks). Thus, it is likely that one or more of these nucleotides are critical for the in vitro interaction of Mga and provide excellent targets for mutational analysis of Mga-binding sites. Attempts to introduce both Pmga and the PsclA-I binding site into the Mga-binding consensus resulted in a very nonspecific sequence consisting of two short stretches of primarily adenine nucleotides (data not shown). Therefore, future experiments will focus on changing the nonbinding PsclA-II sequence to resemble PsclA-I to allow for a gain-of-function phenotype and identification of those nucleotides that are essential for binding. These types of directed biochemical and mutational analyses of Mga-binding sites should provide a more reliable way than sequence alignment to establish the essential core Mga-binding sequence.
Interestingly, when Mga is purified from GAS compared to being purified from E. coli, binding to PsclA-I is noticeably enhanced (Fig. 4). In addition, Mga purified from E. coli is still sufficient to activate transcription of Mga-regulated promoters in an in vitro transcription assay (A. C. Almengor and K. S. McIver, unpublished results), but not as effectively as Mga purified from GAS. This difference may indicate several possibilities: that an additional factor present only in GAS copurifies with Mga and augments its activity, that a specific modification of Mga occurs in GAS and not E. coli, or simply that the recombinant expression of Mga in a heterologous host is not optimal for its activity (e.g., improper folding). Regardless of the mechanisms involved, these results support the importance of using Mga purified from GAS in future experiments dealing with its activity.
PsclA represents a new category of Mga-regulated promoters.
Mga is a DNA-binding protein that functions through direct interaction at binding sites within its target promoters. Based upon the biochemical analysis of three Mga-regulated promoters from the serotype M6 strain JRS4 (15, 19), the location of Mga binding at these promoters falls into two types. In the case of Pemm and PscpA, Mga binds to a single site centered at −51 bp from the start of transcription and juxtaposed with the −35 hexamer. This places Mga in a position similar to class I transcriptional activators that stabilize RNA polymerase through direct contact with the carboxy-terminal domain of the α subunit and increase initiation (11, 15). In contrast, Pmga contains two Mga-binding sites situated further upstream (−100 bp and −181 bp) from the P2 start of transcription, and both of these sites are required for the autoactivation of the downstream start site (19). Currently, it is not clear by what mechanism Mga is able to activate transcription initiation at such a distance.
In this study, we have shown that Mga binds specifically within the promoter of the Mga-regulated sclA, which encodes a streptococcal collagen-like protein involved in GAS virulence. Although the sclA promoter (PsclA) contains two potential binding sites based upon homology to the published consensus sequence, Mga interacts only with a single binding site (PsclA-I) located upstream (−168 bp) of the Mga-regulated start of transcription (Fig. 2 to 4). Additionally, the distal PsclA-I was found to be necessary for activation of sclA transcription in vivo (Fig. 6). The location of PsclA-I is somewhat similar to the most distal Pmga Mga-binding site (−181 bp) and may suggest some commonality of activation between these two promoters. Activators that bind at a distance will often use DNA looping or bending to bring the regulator into physical contact with the transcriptional machinery (2, 10). On the other hand, the fact that PsclA uses only a single bound Mga, while autoactivation at Pmga requires two binding sites, suggests that PsclA represents a unique type of Mga-regulated promoter. Unpublished work from our laboratory indicates that an Mga-regulated promoter (Psof) from a class II strain of GAS also possesses a single distal Mga-binding site of this type.
Based on the differences observed in the location of Mga-binding sites with respect to the start of transcription in Mga-regulated promoters, we propose that the promoters be grouped into three categories: a single proximal binding site (category A, Pemm and PscpA), a single distal binding site (category B, PsclA and Psof), and two distal binding sites (category C, Pmga). At some time in the past, the second putative PsclA binding site (PsclA-II) may have been fully functional but is no longer utilized to regulate sclA expression. Alternatively, PsclA-II may be functional for binding only under very specific temporal or environmental conditions in vivo. Evidence of the possible functionality of PsclA-II comes from our observation of barely detectable Mga binding to this site upon significant overexposure of the EMSA, several orders of magnitude below that seen at PsclA-I (data not shown). Therefore, PsclA may have once represented an Mga-regulated promoter containing both a proximal (category A) and a distal (category B) binding site. However, PsclA has evolved such that it no longer requires the proximal site for Mga-specific activation.
Mga regulation of PsclA occurs at its native locus but is not observed on a multicopy plasmid.
Expression of sclA has been shown previously to be strongly activated at the level of transcription in wild-type M1 GAS strains but not in isogenic strains lacking Mga (Fig. 1B) (14, 27). Our primer extension data confirm these results, showing a strong start of transcription in total RNA isolated from wild-type M1 strain SF370 but not in the mga-inactivated KSM165-L (Fig. 2A). Thus, Mga appears to control sclA by activating transcriptional initiation at PsclA. The ability of Mga to bind to the promoter-distal PsclA-I suggests strongly that interaction at this site is directly involved in the observed activation of the downstream transcriptional start site. Further, the presence of PsclA-I at its native position in the GAS chromosome is essential to the activation of sclA transcription in vivo with a GusA reporter system (Fig. 6). However, when a large (500-bp) region of PsclA, including the PsclA-I binding site and the downstream start site, was inserted in a transcriptional reporter plasmid based on a promoterless gfp, it did not exhibit detectable Mga-specific activation, nor did it demonstrate significant promoter activity over background levels of the system (Fig. 5). In contrast, a control plasmid containing the category A Pemm-GFP produced very strong Mga-specific activation in the GFP reporter system. There are several possible explanations for why Mga regulation of sclA occurs only in a single copy at its native locus and not on a multicopy plasmid. First, one or more cis-acting elements located either further upstream or within sclA itself may be required for Mga regulation. Alternatively, there may be a gene dosage effect observed in the plasmid system, resulting in the titration of a factor necessary for Mga regulation to occur. Finally, DNA looping may be required to bring Mga in contact with the RNA polymerase, and this event may not be occurring properly in our plasmid system.
Upstream of sclA in the SF370 M1 genome is the divergently transcribed nrdI gene, which is predicted to encode a putative ribonucleotide reductase (Fig. 2B). As an internal control for primer extension of PsclA, we mapped the start of transcription for nrdI and found that it was located in the center of the PsclA-I Mga-binding site on the antisense strand. Although this might be expected to result in the Mga-specific repression of nrdI transcription, a primer extension product was clearly observed in SF370, an Mga+ strain (Fig. 2A). In fact, the presence of the nrdI promoter in the middle of the distal Mga-binding site may have hindered our attempts to delete PsclA-I at its native site in the chromosome (data not shown), suggesting the importance of nrdI in growth under the conditions used in this study. This suggests an even more complex interplay among Mga, PsclA, and the overlapping PnrdI regions that must be further explored.
In order to assess the importance of the distal Mga-binding site of PsclA while avoiding the deletion of PnrdI, single-copy PsclA-gusA reporter fusions were integrated into the native chromosomal location such that a second wild-type promoter expressing both nrdI and sclA was preserved in each case. These integrations clearly showed that Mga-regulated activity requires the distal Mga-binding site (Fig. 6). As stated above, several reasons for Mga regulation of sclA only at its native locus can be proposed, including DNA topology and gene dosage from a multicopy plasmid. These possibilities might explain why a category A Mga-regulated promoter (Pemm), which presumably does not require DNA looping, shows significant Mga-specific activity in the plasmid-based GFP reporter whereas a category B PsclA does not function in the same way. However, it is clear that the ability of Mga to regulate PsclA from a distal binding site is more complex than activation from a proximal site and may require additional factors.
In conclusion, we have demonstrated that Mga binds to a single distal binding site within the sclA promoter but not to a second putative site located proximal to the Mga-regulated start of transcription. Further, PsclA-I is required for in vivo transcriptional activation of sclA. Notably, an alignment of sequences for PsclA-I found in multiple serotypes of GAS showed no differences in key conserved nucleotides. Therefore, PsclA and the role of Mga in its regulation appear to be conserved among most serotypes of GAS. Analysis of this novel Mga-regulated promoter should help build our overall knowledge of the molecular mechanisms by which Mga controls virulence in GAS.
Acknowledgments
We thank Deborah Ribardo, Cheryl Vahling, and Temekka Leday for critical reading of the manuscript; Steve Lindow for providing the pGreenTIR plasmid; Slawomir Lukomski for unpublished PsclA promoter sequences; and Rhonda Myles for technical assistance.
This work was supported by grants from the American Heart Association, Texas Affiliate (beginning-grant-in-aid no. 0060073Y to K.S.M.), and the National Institutes of Health (NIH AI-47928 to K.S.M.) A.C.A. is supported in part by an NIH/NIAID Molecular Microbiology training grant (5T32 AI07520) and an NIH/NIAID research supplement for underrepresented minorities (RSUM AI-47928-S).
REFERENCES
- 1.Andersson, G., K. McIver, L. O. Heden, and J. R. Scott. 1996. Complementation of divergent mga genes in group A streptococcus. Gene 175:77-81. [DOI] [PubMed] [Google Scholar]
- 2.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 3.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caparon, M. G., and J. R. Scott. 1987. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc. Natl. Acad. Sci. USA 84:8677-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C., N. Bormann, and P. P. Cleary. 1993. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol. Gen. Genet. 241:685-693. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geist, R. T., N. Okada, and M. G. Caparon. 1993. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J. Bacteriol. 175:7561-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gralla, J. D. 1989. Bacterial gene regulation from distant sites. Cell 57:193-195. [DOI] [PubMed] [Google Scholar]
- 11.Ishihama, A. 1993. Protein-protein communication within the transcription apparatus. J. Bacteriol. 175:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kihlberg, B. M., J. Cooney, M. G. Caparon, A. Olsen, and L. Bjorck. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19:299-315. [DOI] [PubMed] [Google Scholar]
- 13.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, R. M. Ireland, S. D. Reid, G. G. Adams, and J. M. Musser. 2000. Identification and characterization of the scl gene encoding a group A streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68:6542-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, B. J. Shelvin, E. A. Graviss, and J. M. Musser. 2001. Identification and characterization of a second extracellular collagen-like protein made by group A streptococcus: control of production at the level of translation. Infect. Immun. 69:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver, K. S., A. S. Heath, B. D. Green, and J. R. Scott. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1602. [DOI] [PubMed] [Google Scholar]
- 18.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIver, K. S., A. S. Thurman, and J. R. Scott. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLandsborough, L. A., and P. P. Cleary. 1995. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of ScpA, FcrA, OF, and M protein. FEMS Microbiol. Lett. 128:45-51. [DOI] [PubMed] [Google Scholar]
- 21.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 22.Musser, J. M., V. Kapur, S. Kanjilal, U. Shah, D. M. Musher, N. L. Barg, K. H. Johnston, P. M. Schlievert, J. Henrichsen, and D. Gerlach. 1993. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin). J. Infect. Dis. 167:337-346. [DOI] [PubMed] [Google Scholar]
- 23.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 26.Podbielski, A., J. A. Peterson, and P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 6:2253-2265. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen, R., A. Eden, and L. Bjorck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribardo, D. A., and K. S. McIver. 2003. amrA encodes a putative membrane protein necessary for maximal exponential phase expression of the Mga virulence regulon in Streptococcus pyogenes. Mol. Microbiol. 50:673-685. [DOI] [PubMed] [Google Scholar]
- 29.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, D. L. 1999. The flesh-eating bacterium: what's next? J. Infect. Dis. 179(Suppl. 2):S366-S374. [DOI] [PubMed] [Google Scholar]
- 31.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]