Abstract
β-Site amyloid precursor protein (APP) cleaving enzyme 1 (BACE1) is the major neuronal β-secretase for amyloid-β generation and is degraded in lysosomes. The autophagy-lysosomal system plays a key role in the maintenance of cellular homeostasis in neurons. Recent studies established that nascent autophagosomes in distal axons move predominantly in the retrograde direction toward the soma, where mature lysosomes are mainly located. However, it remains unknown whether autophagy plays a critical role in regulation of BACE1 trafficking and degradation. Here, we report that induction of neuronal autophagy enhances BACE1 turnover, which is suppressed by lysosomal inhibition. A significant portion of BACE1 is recruited to the autophagy pathway and co-migrates robustly with autophagic vacuoles along axons. Moreover, we reveal that autophagic vacuole-associated BACE1 is accumulated in the distal axon of Alzheimer's disease-related mutant human APP transgenic neurons and mouse brains. Inducing autophagy in mutant human APP neurons augments autophagic retention of BACE1 in distal axons, leading to enhanced β-cleavage of APP. This phenotype can be reversed by Snapin-enhanced retrograde transport, which facilitates BACE1 trafficking to lysosomes for degradation. Therefore, our study provides new insights into autophagy-mediated regulation of BACE1 turnover and APP processing, thus building a foundation for future development of potential Alzheimer's disease therapeutic strategies.
Keywords: amyloid precursor protein (APP), autophagy, β-secretase 1 (BACE1), protein degradation, trafficking, transport, APP processing, autophagic stress, axonal transport, lysosomal degradation
Introduction
Accumulation of senile plaques is a pathological hallmark of Alzheimer's disease (AD).3 Amyloid-β (Aβ) peptide is the main constituent of senile plaques and is derived from a sequential proteolysis of amyloid precursor protein (APP) by β- and γ-secretases. β-Secretase is the initial and rate-limiting enzyme of this process (1–4). β-Site APP-cleaving enzyme 1 (BACE1) is the major neuronal β-secretase for Aβ generation (1–4). BACE1 levels increase with age (5) and are elevated in AD patient brains (6), thereby making BACE1 a prime target for therapeutic intervention (7–9). BACE1 is synthesized in the endoplasmic reticulum and then delivered to the cell surface from the trans-Golgi network (TGN). Mature BACE1 traffics to endosomes via internalization from the plasma membrane or directly from the TGN (3, 10–12). The endosomal compartments (especially late endosomes (LEs) or multivesicular bodies) provide an acidic environment that is crucial for optimal β-secretase activity (10, 13–17), whereas BACE1 is ultimately degraded within lysosomes (15, 18–20). Given that mature lysosomes are mainly located in the soma of neurons (21–24), proper retrograde transport of LE-loaded BACE1 is critical for the trafficking of BACE1 to lysosomes for turnover (20).
Autophagy is the major cellular degradation pathway for long-lived proteins and damaged organelles (25–28). Altered autophagy has been linked to several major age-related neurodegenerative diseases, including AD (27). Recent studies established that autophagosomes are continuously generated in distal axons and move predominantly in the retrograde direction toward the soma for lysosomal proteolysis (23, 29–33). Such retrograde transport is initiated by fusion of nascent autophagosomes with LEs, forming amphisomes, and is driven by LE-loaded dynein-Snapin (motor adaptor) complexes (32). This raises a fundamental question as to whether autophagosomes also serve as cargoes carrying BACE1 trafficking and thus regulate its turnover and, if so, whether AD-associated autophagic stress and altered trafficking of autophagosomes augment BACE1 accumulation in distal axons.
In this study, we provide evidence that autophagy induction enhances BACE1 turnover in primary cortical neurons. A significant portion of BACE1 is recruited to and co-migrates with autophagic vacuoles (AVs) toward the soma, which facilitates BACE1 trafficking to lysosomes for degradation. Moreover, we provide in vitro and in vivo evidence that AV-associated BACE1 is aberrantly accumulated in distal axons along with impaired retrograde transport in neurons of mutant hAPP J20 transgenic (Tg) mice. Autophagy activation augments BACE1 accumulation in the axon of mutant hAPP Tg neurons, leading to enhanced β-cleavage of APP. Conversely, enhancing retrograde transport by overexpressing the dynein adaptor Snapin reduces accumulation of AV-associated BACE1 and facilitates BACE1 delivery to lysosomes. Therefore, our study provides new insights into how neuronal autophagy regulates BACE1 trafficking and turnover through AV retrograde transport and how impaired AV transport augments BACE1 retention in AD axons, thereby enhancing APP amyloidogenic processing.
Results
Autophagy Activation Enhances BACE1 Turnover
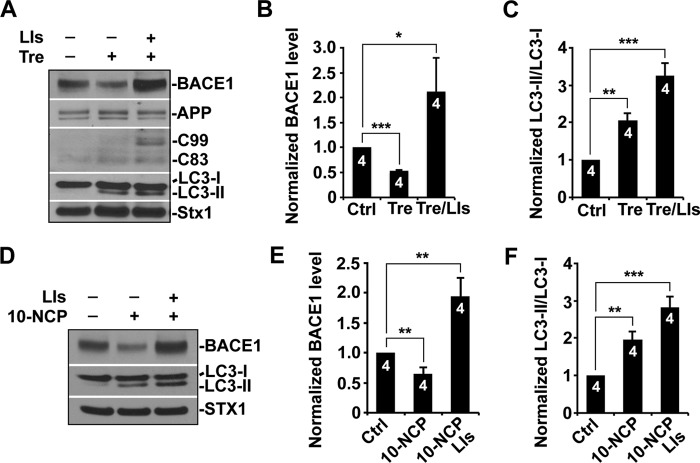
To determine whether BACE1 turnover is regulated by autophagy, cultured primary cortical neurons were treated with trehalose, which induces neuronal autophagy in an mTOR-independent fashion (34). Trehalose treatment led to a significant reduction of BACE1 levels (0.528 ± 0.01; p < 0.001), but a substantial increase of BACE1 was detected in the presence of lysosomal inhibitors (2.12 ± 0.66; p = 0.0365) (Fig. 1, A and B), suggesting that lysosomal inhibition suppressed autophagy-mediated BACE1 turnover. To further evaluate the effect of autophagy induction on BACE1 turnover, we utilized alterative drug 10-NCP, another mTOR-independent autophagy inducer (36). Consistent with trehalose treatment, BACE1 levels were reduced to ∼64% following incubation with 10-NCP for 24 h. Lysosomal inhibition abolished 10-NCP-induced effects on BACE1 turnover, leading to its accumulation (1.94 ± 0.31; p = 0.004) (Fig. 1, D and E). Following a 24-h treatment with trehalose or 10-NCP, we found that LC3-II levels were consistently elevated (trehalose, 2.06 ± 0.195; p < 0.01; 10-NCP, 1.96 ± 0.21; p < 0.01), suggesting an increased autophagic flux. Lysosomal inhibition resulted in an additional increase of LC3-II (trehalose, 3.27 ± 0.33; p < 0.001; 10-NCP, 2.83 ± 0.27; p < 0.001), which is attributed to the blockage of lysosomal proteolysis of autophagic cargoes (Fig. 1, A, C, D, and F). As an internal control, we did not detect any change of syntaxin 1, a neuronal marker (35). Moreover, we tested the effect of activating mTOR-dependent autophagy by treating primary cortical neurons with the mTOR inhibitor rapamycin. We detected similar effects; the levels of BACE1 were decreased in neurons incubated with rapamycin (data not shown). Quantitative analysis indicated ∼25% reduction of BACE1 levels, suggesting that mTOR-dependent autophagy facilitates BACE1 turnover. These observations indicate that the autophagy-lysosomal pathway can effectively regulate BACE1 turnover.
FIGURE 1.
Autophagy activation enhances BACE1 turnover. A–C, representative blots (A) and quantitative analysis (B and C) showing a decrease of BACE1 levels in response to autophagy induction. Primary cortical neurons at DIV 10–14 were treated with trehalose (Tre) (100 mm) for 24 h. The levels of BACE1 were increased when lysosomal inhibitors (LIs) (40 μm pepstatin A and 40 μm E64D) were present. LC3-II, an autophagy marker, was elevated after trehalose treatment and displayed additional increase in the presence of lysosomal inhibitors. Cortical neuron lysates were solubilized, and equal amounts of protein (20 μg) were loaded for sequential detection with antibodies on the same membrane after stripping between applications of each antibody as indicated. Syntaxin 1 (Stx1), a neuronal marker, was detected as a loading control. Ctrl, control. D–F, representative blots (D) and quantitative analysis (E and F) showing reduced BACE1 levels after a 24-h treatment with 10 μm 10-NCP. Incubation with lysosomal inhibitors led to a substantial increase of BACE1. LC3-II levels were increased following a 24-h incubation with 10-NCP or 10-NCP and lysosomal inhibitors. These data were quantified from independent repeats as indicated on the top of bars (B, C, E, and F). Error bars: S.E. Student's t test. (***, p < 0.001; **, p < 0.01; *, p < 0.05.)
Association of BACE1 with Autophagosomes and Robust Movement toward the Soma upon Autophagy Induction
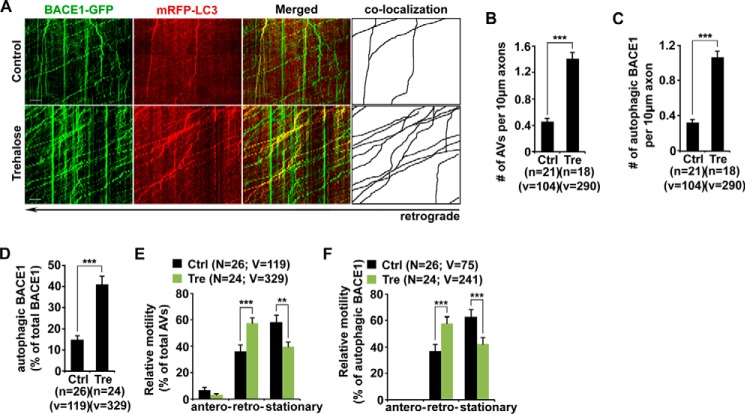
Recent studies demonstrated that autophagosomes are predominantly generated in distal axons and undergo retrograde transport for lysosomal proteolysis in the soma (32, 37). We next asked whether autophagy induction facilitates BACE1 turnover by enhancing their retrograde transport. We examined the distribution and transport of axonal BACE1 and autophagosomes in live neurons transfected with BACE1-GFP and the autophagy marker mRFP-LC3 in the presence and absence of 100 mm trehalose. At the basal condition, a majority of mRFP-LC3 was diffused as the form of cytosolic LC3-I. We detected a few lipidated LC3-II-labeled AVs, which predominantly moved toward the soma (Fig. 2A). Trehalose incubation markedly increased the density of AVs along axons (number of AVs per 10 μm length: basal condition, 0.46 ± 0.04; trehalose, 1.40 ± 0.10; p < 0.001) (Fig. 2, A and B). This result is consistent with our biochemical analysis showing increased LC3-II levels (Fig. 1, A and C), suggesting an increased autophagic flux. Surprisingly, a significant portion of BACE1 was recruited to the autophagy pathway after the treatment with trehalose. The density of AV-associated BACE1 was robustly elevated relative to that in basal conditions (per 10 μm length: basal condition, 0.32 ± 0.04; trehalose, 1.06 ± 0.07; p < 0.001) (Fig. 2, A and C). Under basal conditions, ∼15% of BACE1 was co-localized and co-migrated with AVs along the same axon. However, it was significantly increased following autophagy activation (41.02 ± 3.95%; p < 0.001) (Fig. 2, A and D). These newly formed AVs displayed predominant retrograde transport along axons (57.57 ± 3.73%) (Fig. 2, A and E). More importantly, retrograde transport of AV-associated BACE1 along the same axons was markedly increased (basal condition, 37.02 ± 5.02%; trehalose, 60.72 ± 4.41%; p < 0.001) (Fig. 2, A and F). Therefore, our results suggest that autophagy induction promotes the recruitment of BACE1 to the autophagy pathway and enhances BACE1 retrograde transport from distal axons.
FIGURE 2.
Association of BACE1 with autophagosomes and robust movement in retrograde direction upon autophagy induction. A, representative dual channel kymographs showing enhanced retrograde transport of BACE1 within LC3-marked AVs in live neurons under the condition of autophagy induction. Cultured cortical neurons were transfected with BACE1-GFP and mRFP-LC3, followed by a 24-h incubation with 100 mm trehalose (Tre) prior to time-lapse imaging at DIV 10–12. The axon image was taken ∼100 μm away from the cell body. Whereas BACE1-GFP displayed bi-directional movement, mRFP-LC3 predominantly moved toward the soma of neurons. Note that a significant portion of BACE1 co-localized and co-migrated with LC3-labeled AVs along the same axons toward the soma in the presence of trehalose. B–D, inducing autophagy facilitates BACE1 trafficking along the autophagy pathway. Note that the densities of axonal AVs and BACE1 associated with AVs and the percentage of autophagic BACE1 relative to total BACE1 were increased in response to autophagy activation. The average numbers of AVs and AVs co-labeled with BACE1 per 10-μm axon were quantified in neurons with and without trehalose treatment, respectively (B and C). E and F, autophagy induction enhanced retrograde transport of AV-associated BACE1 in axons. Relative motility of axonal AVs and AV-associated BACE1 in the presence and absence of trehalose was measured. Vertical lines represent stationary organelles; oblique lines or curves to the right (negative slope) represent anterograde movements; and lines to the left (positive slope) indicate retrograde transport. These data were quantified from at least four independent repeats and from a total number of AVs or AV-associated BACE1 (v) from a total number of neurons (n), as indicated in parentheses (B–F). Scale bars, 10 μm. Ctrl, control. Error bars, S.E. Student's t test. (***, p < 0.001; **, p < 0.01.)
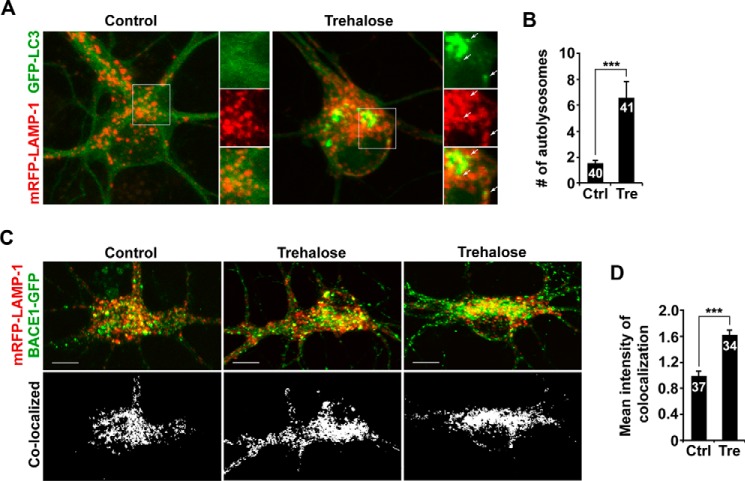
Accumulating evidence demonstrates that mature lysosomes are mainly located in the soma of neurons (21–24). We and others reported that BACE1 relies on lysosomal degradation (15, 18–20). Given that autophagy activation enhanced retrograde transport of AV-associated BACE1 (Fig. 2, A and F), we sought to determine whether BACE1 trafficking to lysosomes could also be enhanced under the same conditions. After a 24-h incubation with trehalose, we found that co-localized intensity of BACE1 with lysosomes, labeled by LAMP-1, was significantly increased in the soma compared with that of control condition (1.62 ± 0.08; p < 0.001) (Fig. 3, C and D). Thus, by enhancing retrograde transport, autophagy activation facilitates the trafficking of BACE1 to somatic lysosomes for degradation, as reflected by reduced BACE1 levels (Fig. 1). Consistently, the density of autolysosomes in the soma was markedly increased upon autophagy induction (basal, 1.55 ± 0.22; trehalose, 6.56 ± 1.27; p < 0.001) (Fig. 3, A and B). Our data are consistent with a recent study that axon-generated autophagosomes enter the soma and then remain in the somatodendritic domain, where mature lysosomes are mainly located (33).
FIGURE 3.
Increased density of autolysosomes and enhanced BACE1 trafficking to somatic lysosomes following autophagy activation. A and B, representative images (A) and quantitative analysis (B) showing increased density of autolysosomes in the soma following a 24-h incubation with trehalose. The average number of autolysosomes (arrows) co-labeled by GFP-LC3 and mRFP-LAMP-1 in the soma of neurons in the presence and absence of trehalose was quantified. C and D, trafficking of BACE1 to lysosomes was increased in the soma of neurons treated with trehalose for 24 h. Co-localized intensity of BACE1 with LAMP-1 in the soma of neurons with and without trehalose treatment was examined. These data were quantified from at least four independent repeats and from a total number of neurons (n), as indicated on the top of bars (B and D). Scale bars, 10 μm. Error bars, S.E. Student's t test. (***, p < 0.001.)
Impaired Retrograde Transport Accumulates AV-associated BACE1 in Axons of Mutant hAPP Neurons
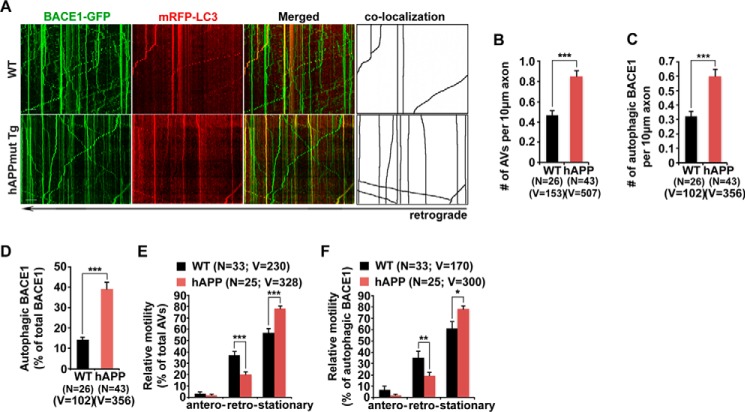
Altered autophagy has been implicated in AD. AD neurons exhibit massive accumulation of AVs within dystrophic neurites (38), highlighting impaired autophagic transport and clearance. We next determined whether such AD-associated autophagic stress augments BACE1 retention in distal axons. Cortical neurons derived from mutant hAPP Tg (J20) mice and their WT littermates were transfected with BACE1-GFP and mRFP-LC3, followed by time-lapse live cell imaging at DIV 17–19. Surprisingly, compared with WT neurons, the density of AVs was increased in mutant hAPP neurons without any treatment (per 10-μm length: WT, 0.47 ± 0.046; hAPP, 0.85 ± 0.056; p < 0.001) (Fig. 4, A and B). A large portion of BACE1 was retained within AVs in the axon of hAPP neurons (per 10-μm length: WT, 0.32 ± 0.036; hAPP, 0.60 ± 0.045; p < 0.001) (Fig. 4, A and C). In WT neurons, ∼14.49% of BACE1 was co-localized with axonal AVs; however, the percentage of BACE1 associated with axonal AVs was significantly increased (39.19 ± 3.16%; p < 0.001) (Fig. 4, A and D). These data indicate that BACE1 was aberrantly accumulated within the autophagy pathway in the axon of mutant hAPP Tg neurons.
FIGURE 4.
Impaired retrograde transport accumulates AV-associated BACE1 in axons of mutant hAPP neurons. A, representative dual channel kymographs showing impaired retrograde transport of AVs and AV-associated BACE1 in the axon of mutant hAPP Tg neurons. Note that LC3-marked AVs and BACE1 co-localized with AVs mainly moved toward the soma of WT neurons, whereas a majority of AVs and BACE1 remained stationary in the axon of mutant hAPP neurons. B–D, BACE1 was retained within AVs in the axon of hAPP neurons. Note that the densities of both axonal AVs and AV-associated BACE1, and BACE1 retention within AVs were increased in mutant hAPP axons. The average numbers of AVs and BACE1 co-labeled with LC3 per 10 μm axon were quantified in the same axon of WT and mutant hAPP neurons, respectively (B and C). The percentage of autophagic BACE1 relative to total BACE1 was examined in the axon of WT and hAPP neurons (D). E and F, impaired retrograde transport of AVs and AV-associated BACE1 in the axon of mutant hAPP neurons. Note that AVs and BACE1 share similarly reduced retrograde motility in the same mutant hAPP axons. Relative motility of axonal AVs and BACE1 co-labeled with LC3 in WT and mutant hAPP neurons was measured. Data were quantified from four independent repeats of four pairs of mice. Data were quantified from the total number of axonal AVs or autophagic BACE1 (v) and from the total number of neurons (n), as indicated in parentheses (B–F). Scale bars, 10 μm. Error bars, S.E. Student's t test. (***, p < 0.001; **, p < 0.01; *, p < 0.05.)
We then assessed the retrograde motility of AVs and AV-associated BACE1 in live mutant hAPP Tg axons. Both AVs and BACE1 associated with AVs displayed similar reduction in retrograde motility in the same distal axon of mutant hAPP neurons (AVs, 20.11 ± 0.02%; p = 0.00012; autophagic BACE1, 19.56 ± 2.75%; p = 0.0076) relative to their retrograde motility in WT neurons (AVs, 36.89 ± 3.56%; autophagic BACE1, 35.48 ± 5.32%) (Fig. 4, A, E, and F). However, such reduction was not found in anterograde transport (AVs, p = 0.2789; AV-associated BACE1, p = 0.1861), suggesting that BACE1 retention in mutant hAPP axons is attributed to impaired retrograde transport of AVs.
Autophagic Retention of BACE1 in the Distal Axon of Mutant hAPP Tg Mouse Brains
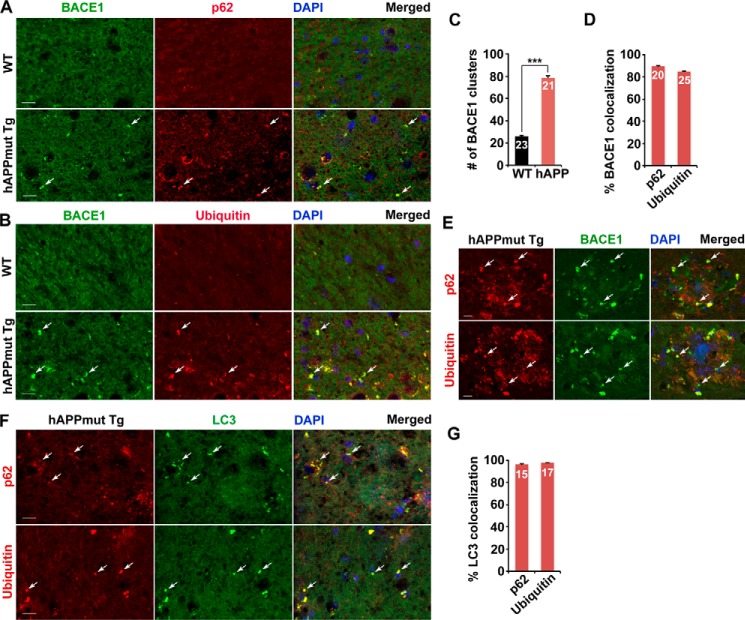
To provide in vivo evidence, we performed co-immunostaining of mutant hAPP Tg (J20) mouse brains with antibodies against BACE1 and autophagy marker p62 or ubiquitin. BACE1 appeared as relatively diffused in the hippocampal mossy fiber processes of WT mouse brains, whereas BACE1 was clustered and accumulated in mutant hAPP Tg (J20) mouse brains (Fig. 5, A and B). The average number of BACE1 clusters per slice section was substantially increased relative to that of WT mouse brains (WT, 26.06 ± 0.91; mutant hAPP Tg, 78.57 ± 1.61; p < 0.001) (Fig. 5C). Moreover, the majority of BACE1 co-localized with p62 or ubiquitin-labeled AVs in the hippocampal regions of mutant hAPP mice (p62, 89.52 ± 0.81%; ubiquitin, 84.85 ± 0.55%) (Fig. 5, A, B, and D). A robust co-localization of BACE1 with p62 or ubiquitin was detected within dystrophic neurites surrounding amyloid plaques of hAPP mouse brains (Fig. 5E). 96.3% of p62 and 97.9% of ubiquitin co-localized with LC3-labeled AVs in mossy fibers enriched with axons and axon terminals of mutant hAPP Tg mice (Fig. 5, F and G). Consistent with cultured mutant hAPP neurons (Fig. 4), these observations in brain slice sections suggest autophagic retention of BACE1 in distal axons of AD neurons in vivo.
FIGURE 5.
Autophagic retention of BACE1 in the distal axon of mutant hAPP Tg mouse brains. A–D, representative images (A and B) and quantitative analysis (C and D) showing that BACE1 was retained within AVs in the distal axon of mutant hAPP Tg mice. Note that BACE1 was accumulated in the hippocampal mossy fibers of mutant hAPP Tg mice. A majority of BACE1 was co-localized with p62 or ubiquitin-labeled AV clusters in mutant hAPP Tg mouse brains. The average number of BACE1 clusters per imaging slice sections (320 × 320 μm) was measured (C). The percentages of BACE1 co-localization with p62 and ubiquitin were quantified, respectively (D). E, representative images showing association of BACE1 with AVs within dystrophic axons surrounding amyloid plaques of mutant hAPP Tg mouse brains. F and G, representative images (F) and quantitative analysis (G) showing aberrant accumulation of AV clusters in the mossy fiber of mutant hAPP mouse brains. The percentages of LC3 co-localization with p62 and ubiquitin were quantified, respectively (G). Data were quantified from a total number of imaging slice sections (320 × 320 μm) indicated on the top of bars (C, D, and G). Scale bars, 10 μm. Error bars, S.E. Student's t test. (***, p < 0.001.)
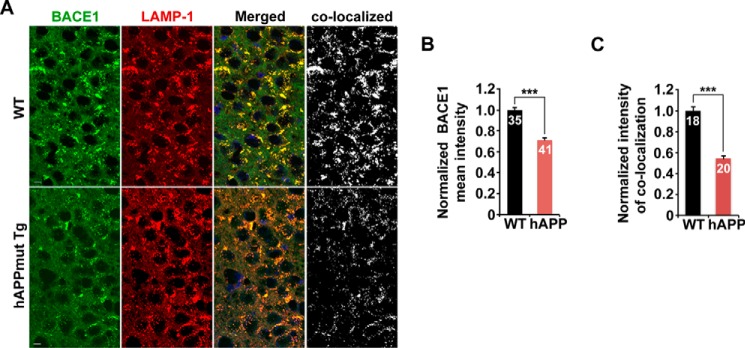
To determine whether aberrant distal retention of BACE1 impairs its delivery to the somatic lysosomes, we examined lysosomal targeting of BACE1 in the hippocampal regions of WT and hAPP mouse brains. We found that the fluorescence intensity of BACE1 in the soma was reduced to 71.28% in the hippocampal CA3 regions of mutant hAPP Tg mice compared with that of WT mice (p < 0.001) (Fig. 6, A and B). Furthermore, the co-localized mean intensity of BACE1 with LAMP-1, a lysosomal marker, was significantly reduced to 54.3% in hAPP mouse brains (p < 0.001) (Fig. 6, A and C). Thus, our data support the notion that AD-linked autophagic stress exacerbates BACE1 accumulation in distal axons, which may augment axonal pathology.
FIGURE 6.
Reduced delivery of BACE1 to somatic lysosomes in mutant hAPP Tg mouse brains. A–C, representative images (A) and quantitative analysis (B and C) showing reduced lysosomal delivery of BACE1 in the soma of the hippocampal CA3 regions in mutant hAPP Tg mice. The mean intensities of BACE1 fluorescence and BACE1 co-localization with LAMP-1 in the soma were quantified, respectively. Data were quantified from a total number of imaging slice sections (320 μm × 320 μm) indicated on the top of bars (B and C). Scale bars, 10 μm. Error bars, S.E. Student's t test. (***, p < 0.001.)
Autophagy-induced BACE1 Turnover Is Impaired in Mutant hAPP Tg Neurons
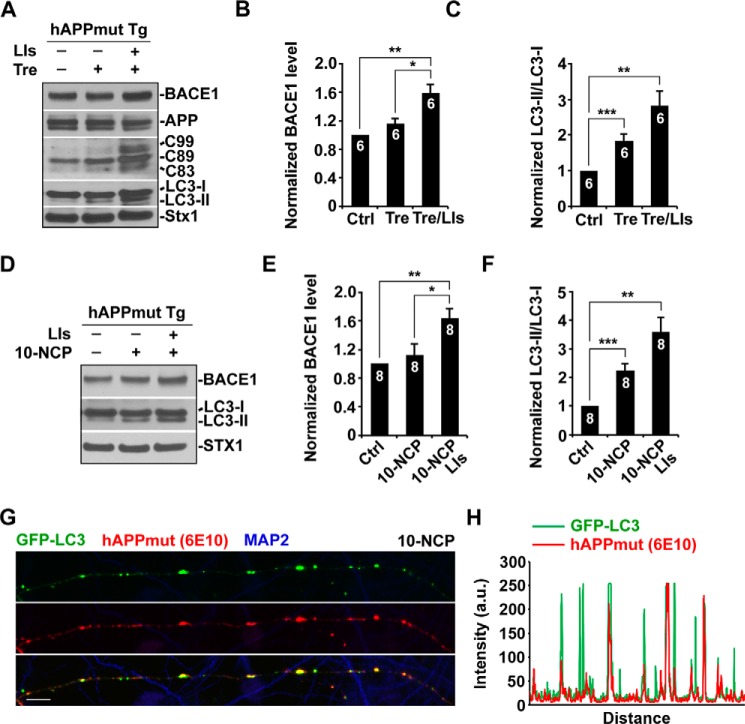
We next examined whether inducing autophagy in mutant hAPP Tg neurons alters BACE1 levels. Whereas autophagy induction enhances BACE1 turnover in WT neurons (Fig. 1), BACE1 levels in mutant hAPP neurons were maintained after a 24-h incubation with trehalose (1.16 ± 0.08; p > 0.05). Substantial increase of BACE1 was detected when lysosomal function was suppressed by lysosomal inhibitors (1.59 ± 0.13; p < 0.01) (Fig. 7, A and B), which suggests that lysosomal activity is unlikely impaired in mutant hAPP neurons. Consistently, trehalose treatment led to increased autophagic flux in mutant hAPP neurons, as evidenced by elevated levels of LC3-II (1.84 ± 0.18; p = 0.001). Further increase of LC3-II was detected in the presence of lysosomal inhibitors (2.81 ± 0.41; p < 0.01) (Fig. 7, A and C). Treatment with the second mTOR-independent autophagy inducer 10-NCP showed similar effects; autophagy-induced BACE1 turnover was abolished in mutant hAPP neurons (1.13 ± 0.15; p > 0.05), but BACE1 levels were further increased in the presence of lysosomal inhibitors (1.63 ± 0.14; p < 0.01) (Fig. 7, D and E). The levels of LC3-II were consistently elevated following a 24-h incubation with 10-NCP (2.25 ± 0.24; p < 0.001; with lysosomal inhibitors, 3.58 ± 0.53; p < 0.01) (Fig. 7, D and F). This result suggests that AV-associated reduction of AV retrograde transport disrupts autophagy-induced BACE1 turnover in mutant hAPP neurons. Consistently, elevated levels of C99 and C89 were also observed in mutant hAPP neurons following autophagy induction (Fig. 7A), suggesting an enhanced BACE1 processing of APP. To determine autophagic APP processing, we carried out co-immunostaining with MAP2 and 6E10 antibodies in mutant hAPP neurons following incubation with 10-NCP. GFP-LC3-labeled AVs were co-stained with 6E10-marked APP or its cleaved products (β-C-terminal fragments (CTFs) or Aβ) along MAP2-negative axons (Fig. 7, G and H). Thus, our data indicate that increasing autophagic induction in AD neurons triggers autophagic retention of BACE1 and enhances BACE1 processing of APP.
FIGURE 7.
Autophagy-induced BACE1 turnover is impaired in mutant hAPP Tg neurons. A–C, representative blots (A) and quantitative analysis (B and C) showing that BACE1 levels were maintained in mutant hAPP neurons upon autophagy activation. Cultured cortical neurons derived from mutant hAPP Tg mice were incubated with trehalose (Tre) (100 mm) or trehalose and lysosomal inhibitors (LIs) (40 μm pepstatin A and 40 μm E64D) for 24 h. Cell lysates were solubilized, and equal amounts of protein (20 μg) were loaded for sequential detection with antibodies on the same membrane after stripping between applications of each antibody as indicated. Note enhanced BACE1 processing of APP in mutant hAPP neurons following a 24-h trehalose treatment. The levels of LC3-II were determined by measuring the ratio of LC3-II/LC3-I, which were elevated after incubation with trehalose or trehalose and lysosomal inhibitors. D–F, autophagy activation did not reduce BACE1 levels in mutant hAPP neurons treated with 10 μm 10-NCP for 24 h. Note that lysosomal inhibition induced BACE1 accumulation. 10-NCP treatment-induced autophagy elevated LC3-II levels in mutant hAPP neurons. Further increase of LC3-II was detected in the presence of lysosomal inhibitors. G and H, representative image showing β-cleavage of APP within AVs along the axon of mutant hAPP neurons following incubation with 10-NCP. The corresponding profile reflects the fluorescence intensities of GFP-LC3-labeled AVs and 6E10 antibody-marked APP or its cleaved products (β-CTFs or Aβ) along MAP2-negative axons (H). Data were quantified from independent repeats as indicated on the top of bars (B, C, E, and F). Scale bars, 10 μm. Error bars, S.E. Student's t test. (***, p < 0.001; **, p < 0.01; *, p < 0.05.)
Enhancing Retrograde Transport Reverses AD-associated Axonal Retention of Autophagic BACE1
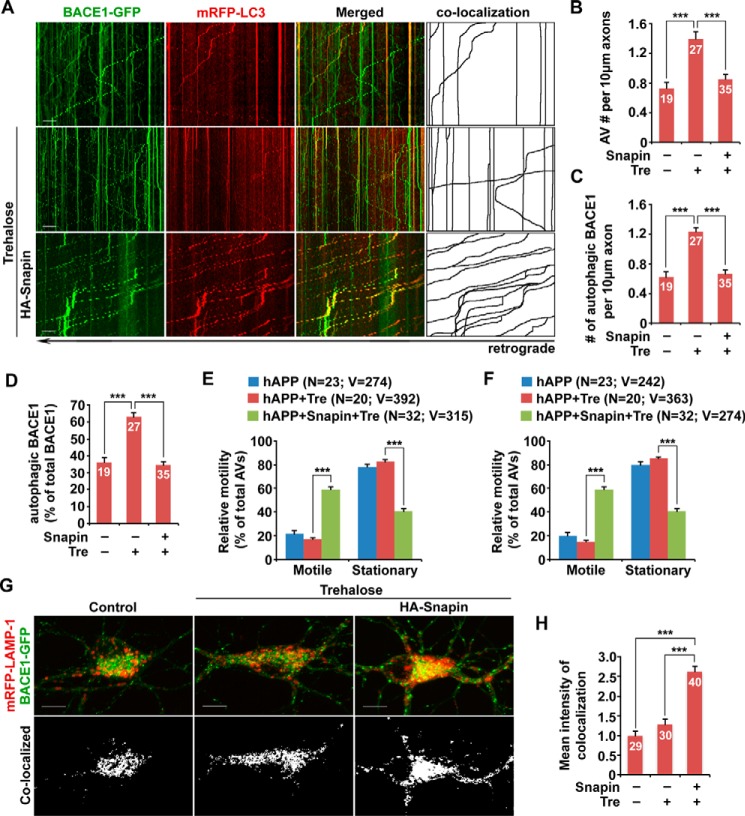
We further determined whether enhanced autophagic retention BACE1 is attributed to impaired retrograde transport in mutant hAPP neurons. We found that autophagy induction augmented autophagic stress in mutant hAPP neurons, as reflected by the additional increase in axonal AVs (per 10-μm length: basal condition, 0.73 ± 0.08; trehalose, 1.4 ± 0.09; p < 0.001) (Fig. 8, A and B). Similarly, accumulation of AV-associated BACE1 was also exacerbated following trehalose treatment (per 10-μm length: basal condition, 0.63 ± 0.06; trehalose, 1.23 ± 0.06; p < 0.001) (Fig. 8, A and C). Moreover, the portion of BACE1 associated with axonal AVs was significantly increased (basal condition: 36.17 ± 2.77%; trehalose, 62.95 ± 2.7%; p < 0.001), suggesting that inducing autophagy triggers more recruitment of BACE1 to AVs in mutant hAPP neurons (Fig. 8, A and D). In contrast to WT neurons, trehalose treatment did not enhance motility of AVs and AV-associated BACE1 in mutant hAPP axons (AVs: basal condition, 21.84 ± 2.44%; trehalose, 17.17 ± 1.45%; p > 0.05; AV-associated BACE1: basal condition, 20.16 ± 2.47%; trehalose, 14.82 ± 1.38%; p > 0.05) (Fig. 8, A, E, and F). It was reported that overexpressing Snapin, a dynein motor adaptor, enhances retrograde transport of axonal AVs (32). We asked whether Snapin-enhanced retrograde transport could reduce autophagy-induced BACE1 retention in AD neurons. Strikingly, the motility of axonal AVs and AV-associated BACE1 was markedly increased in mutant hAPP Tg neurons overexpressing Snapin in response to autophagy induction (AVs, 61.79 ± 2.57%; p < 0.001; AV-associated BACE1, 59.23 ± 2.31%; p < 0.001) (Fig. 8, A, E, and F). Consistently, we also detected reduced densities of AVs and AV-associated BACE1 in trehalose-treated mutant hAPP axons (per 10-μm length: AVs, 0.85 ± 0.07; p < 0.001; AV-associated BACE1, 0.67 ± 0.05; p < 0.001) (Fig. 8, A–C). The percentage of BACE1 association with AVs was significantly decreased to 35% (p < 0.001) (Fig. 8, A and D). Furthermore, BACE1 targeting to lysosomes was increased in mutant hAPP neurons expressing Snapin (2.62 ± 0.14; p < 0.001), compared with untreated mutant hAPP neurons (Fig. 8, G and H). These observations support our view that impeded retrograde transport augments autophagic stress and autophagic retention of BACE1 in distal axons of mutant hAPP neurons following autophagy activation. Enhancing retrograde transport by overexpressing Snapin ameliorates autophagy-induced BACE1 accumulation and facilitates BACE1 trafficking to somatic lysosomes for turnover.
FIGURE 8.
Enhancing retrograde transport reverses AD-associated axonal retention of autophagic BACE1. A–D, representative dual channel kymographs (A) and quantitative analysis (B–D) showing that inducing autophagy augmented accumulation of AV-associated BACE1 in the axon of mutant hAPP neurons, which was reversed by Snapin-enhanced retrograde transport. Cultured cortical neurons were transfected with BACE1-GFP and mRFP-LC3 and HA-Snapin or HA vector, followed by a 24-h incubation with 100 mm trehalose (Tre) prior to time-lapse imaging at DIV 10–12. Note that the axonal densities of both AVs and AV-associated BACE1 and the percentage of BACE1 associated with AVs were increased in mutant hAPP neurons following autophagy induction. The average numbers of AVs and BACE1 co-labeled with LC3 per 10-μm axon were quantified in mutant hAPP neurons in the presence and absence of trehalose, respectively (B and C). The percentage of AV-associated BACE1 relative to total BACE1 was examined in the axon of hAPP neurons (D). E and F, quantitative analysis showing enhanced motility of AVs and AV-associated BACE1 in mutant hAPP axons expressing Snapin but not vector control upon autophagy induction. G and H, overexpressing Snapin facilitates BACE1 trafficking to lysosomes in the soma of mutant hAPP neurons. Co-localized mean intensity of BACE1 with LAMP-1 with and without trehalose treatment was examined. Data were quantified from at least four independent repeats and from a total number of AVs or AV-associated BACE1 (v) from a total number of neurons (n), as indicated in parentheses (E and F) or on the top of the bars (B–D, and H). Scale bars, 10 μm. Error bars, S.E. Student's t test. (***, p < 0.001.)
Discussion
Autophagy is the major cellular degradation pathway, which consists of the following two key steps: 1) formation of double membrane autophagosomes engulfing misfolded proteins and damaged organelles to be degraded; 2) the fusion of the autophagosome with the lysosome into autolysosome, where autophagic cargoes are degraded (39–41). Microtubule-based long-distance axonal transport is essential for autophagic clearance because autophagosomes are predominantly generated in distal axons and mainly rely on retrograde transport to somatic lysosomes for degradation (23, 29, 31–33). Defective autophagy has been implicated in AD pathogenesis (27, 42, 43). The presence of massively accumulated AVs in dystrophic (swollen) neurites is a unique feature linked to AD pathology (38, 44). These pathological features raise the following fundamental question. Does AD-associated autophagic stress interfere with BACE1 trafficking and thus augment BACE1 cleavage of APP in AD neurons?
Although many studies have been focused on BACE1 trafficking and turnover in non-neuronal cell types (16, 45, 46) or neuron-derived cell lines (12, 15, 47, 48), its relevance to BACE1 long distance axonal transport and its retention and cleavage of APP in distal axons remains unknown. This study examined, for the first time, the autophagy-mediated regulation of BACE1 trafficking to lysosomes for degradation in WT and AD-related neurons. We show that BACE1 turnover is enhanced in primary WT cortical neurons upon autophagy induction but is suppressed when lysosomal function is inhibited (Fig. 1). Inducing autophagy triggers robust recruitment of BACE1 to the autophagy pathway; a significant portion of BACE1 co-localizes and co-migrates with AVs from distal axons toward the soma, thereby enhancing BACE1 trafficking to lysosomes (Figs. 2 and 3). To our surprise, this autophagy-induced BACE1 turnover is largely abolished in mutant hAPP neurons. Impaired retrograde transport results in aberrant accumulation of AV-associated BACE1 within the distal axons of neurons from AD-related mutant hAPP J20 Tg mice (Fig. 4). Consistently, mutant hAPP Tg mouse brains recapitulate the phenotype of autophagic BACE1 retention in distal axons and reduced lysosomal targeting in the soma of the hippocampal regions (Figs. 5 and 6). Inducing autophagy in mutant hAPP neurons results in further accumulation of AV-associated BACE1, thereby augmenting β-cleavage of APP within AVs in distal axons (Figs. 7 and 8). Importantly, rescuing retrograde transport by overexpressing Snapin in mutant hAPP neurons reduces axonal retention of autophagic BACE1 and facilitates its targeting to lysosomes (Fig. 8). Therefore, we proposed that autophagy is one of the primary pathways regulating BACE1 trafficking and turnover in WT neurons. This pathway, however, is impaired under AD-linked autophagic stress, thus providing a new mechanistic insight as to why BACE1 is largely retained in distal axons, where β-cleavage of APP mainly occurs.
Autophagosomes are predominantly generated in distal axons and then undergo predominant retrograde transport toward the soma (23, 29, 30, 32, 33). By recruiting LE-loaded dynein-Snapin (motor adaptor) transport complexes, nascent autophagosomes gain retrograde motility through fusion with LEs into amphisomes (32), which delivers autophagic cargoes to somatic lysosomes for clearance. LEs are enriched with amyloidogenic machinery and provide an optimal acidic environment necessary for β-secretase activity (10, 13–17, 20). BACE1 ultimately undergoes degradation within lysosomes (15, 18, 19). Thus, the traffic route for AV-associated BACE1 to lysosomes is critical for controlling its level and activity. Through autophagosome fusion with LEs into amphisomes, we showed that a significant portion of LE-loaded BACE1 is recruited to the autophagy pathway and co-migrates with AVs in the retrograde direction within axons upon autophagy induction. As a result, BACE1 utilizes AVs as a ride for its trafficking toward somatic lysosomes through dynein-Snapin-driven retrograde transport, which facilitates its turnover within lysosomes. Therefore, we proposed that autophagy serves as an alternative trafficking route to regulate BACE1 degradation in neurons.
Our current findings provide a mechanistic interpretation of many reported observations. Altered autophagy has been implicated in the pathogenesis of AD by its involvement in the formation of amyloid plaques (49, 50). AVs are massively accumulated and clustered within dystrophic neurites of AD brains, and these AVs have been proposed as an alternative source for Aβ generation (38, 49, 51). Purified AVs from AD mice are enriched with Aβ peptide and its immediate precursor, βCTF, suggesting that BACE1 cleavage of APP occurs on the membrane of AVs (49, 51). In this study, we show that a significant portion of BACE1 is recruited to the autophagy pathway in response to autophagy induction in healthy neurons (Figs. 1–3). However, under AD-linked autophagic stress with defective retrograde transport, we provide in vitro and in vivo evidence that AV-associated BACE1 is accumulated in distal axons along with impaired retrograde transport (Figs. 4 and 5). Under such pathological conditions, inducing autophagy exacerbated autophagic retention of BACE1 in distal axons of mutant hAPP neurons (Figs. 7 and 8). Thus, our study supports the notion that AD-associated autophagic stress retains BACE1 within the autophagy pathway and thus impairs its trafficking to lysosomes for degradation, thereby enhancing BACE1 processing of APP in AD axons.
Autophagy induction may represent a promising therapeutic strategy to prevent or treat AD and other neurodegenerative diseases. Studies have shown the beneficial effects of autophagy activation on reducing the accumulation of polyglutamine expansions and α-synuclein and on ameliorating disease-associated pathology (52–54). However, the role of autophagy in AD is controversial (55–61). Several studies reported that increasing autophagy showed the rescue effects when AD mice were administered rapamycin prior to the development of AD pathology (58, 60, 61) but had no effect on mice with already established AD-like pathology and cognitive deficits (60). Autophagy activation has also been shown to trigger an increase in Aβ levels and is involved in amyloidogenesis (49, 50). Consistent with these findings, we proposed that impaired retrograde transport in AD axons compromises beneficial effects of increasing autophagy activation on AD pathology by exacerbating autophagic accumulation of BACE1 and augmenting β-cleavage of APP and axonal pathology. Therefore, our study establishes a foundation for future investigation into the potential therapeutic strategy of increasing autophagy induction at the early disease stages.
Experimental Procedures
Mice
hAPP mice (C57BL/6J) from line J20 were purchased from The Jackson Laboratory. The platelet-derived growth factor (PDGF) β chain promoter was used to direct neuronal expression of the familial AD-linked human APP Swedish and Indiana mutants in this J20 mouse line (62).
Materials
The constructs encoding Snapin and Snapin-L99K were prepared as described previously (20, 22, 63). Sources of other antibodies and reagents are as follows: polyclonal anti-syntaxin 1 antibody (Santa Cruz Biotechnology); monoclonal anti-ubiquitin and polyclonal anti-LC3 antibodies (Cell Signaling Technology); monoclonal anti-p62/SQSTM1 antibody (Abnova); polyclonal anti-LC3 and anti-APP C-terminal antibodies (Sigma); monoclonal anti-β-amyloid (6E10) antibody (Covance); Alexa Fluor 546- and 633-conjugated secondary antibodies (Invitrogen). Monoclonal anti-LAMP1 antibody was developed by D. Messner and was obtained from Developmental Studies Hybridoma Bank.
Transfection and Immunocytochemistry of Cultured Cortical Neurons
Cortices were dissected from E18 to E19 mouse embryos as described (22, 64, 65). Cortical neurons were dissociated by papain (Worthington) and plated at a density of 100,000 cells per cm2 on polyornithine- and fibronectin-coated coverslips. Neurons were grown overnight in plating medium (5% FBS, insulin, glutamate, G5, and 1× B27) supplemented with 100× l-glutamine in Neurobasal medium (Invitrogen). Starting at DIV 2, cultures were maintained in conditioned medium with half-feed changes of neuronal feed (1× B27 in Neurobasal medium) every 3 days. Primary hAPP Tg neurons were cultured from breeding mice of hemizygous mutant hAPPSwe/Ind Tg (J20 line) with WT animals (62). Genotyping assays were performed following culture plating to verify mouse genotypes. In our study, we examined both transgenic neurons and non-transgenic neurons derived from their littermates. Neurons were transfected with various constructs using Lipofectamine 2000 (Invitrogen) followed by time-lapse imaging at DIV 10–12 transfection prior to qualification analysis.
For immunostaining, cultured neurons were fixed with 100% ice-cold methanol at −20 °C for 10 min, washed three times with 1× PBS for 5 min each, and then incubated in 0.4% saponin, 5% normal goat serum, and 2% bovine serum albumin (BSA) in PBS for 1 h. Fixed cultures were incubated with primary antibodies in PBS with 2% BSA and 0.4% saponin at 4 °C overnight. Cells were washed four times with PBS at room temperature for 5 min each, incubated with secondary fluorescent antibodies at 1:400 dilution in PBS with 2% BSA and 0.4% saponin for 30 min, re-washed with PBS, and then mounted with Fluor-Gel anti-fade mounting medium (EMS) for imaging.
For semi-quantitative analysis, protein bands detected by ECL were scanned into Adobe Photoshop CS6 and analyzed using ImageJ (National Institutes of Health). Care was taken during exposure of the ECL film to ensure that intensity readouts were in a linear range of standard curve blot detected by the same antibody. Paired Student's t tests were carried out, and results are expressed as mean ± S.E.
Tissue Preparation and Immunohistochemistry
Animals were anesthetized with 2.5% avertin (0.5 ml per mouse) and transcardially perfused with fixation buffer (4% paraformaldehyde in PBS, pH 7.4). Brains were dissected out, postfixed in fixation buffer overnight, and then placed in 30% sucrose at 4 °C. The 10-μm-thick coronal sections were collected consecutively to the level of the hippocampus and used to study co-localization of various markers. After incubation with blocking buffer (2.5% goat serum, 0.15% Triton X-100, 1.5% BSA, 0.5% glycine in H2O) at room temperature for 1 h, the sections were incubated with primary antibodies at 4 °C overnight, followed by incubating with secondary fluorescence antibodies at 1:400 dilution at room temperature for 1 h. After fluorescence immunolabeling, the sections were stained with DAPI and washed three times in PBS. The sections were then mounted with anti-fading medium (Vector Laboratories, H-5000) for imaging. Confocal images were obtained using an Olympus FV1000 oil immersion ×40 objective with sequential acquisition setting. Eight to 10 sections were taken from top-to-bottom of the specimen, and the brightest point projections were made.
Quantification of Co-localization
A threshold intensity was determined in the thresholding function of ImageJ, which was preset for the fluorescent signals as described previously (66). The co-localized pixels above the threshold intensity were automatically quantified and scored by ImageJ based on the fluorescence intensity profile, which was expressed as co-localized mean intensity positive for both channels. The co-localization was presented as the percentage of the co-localized intensity relative to total fluorescence intensity.
Image Acquisition and Quantification
Confocal images were obtained using an Olympus FV1000 oil immersion ×60 objective (1.3 numerical aperture) with sequential acquisition setting. For fluorescent quantification, images were acquired using the same settings below saturation at a resolution of 1024 × 1024 pixels (8-bit). Eight to 10 sections were taken from top-to-bottom of the specimen, and the brightest point projections were made. Morphometric measurements were performed using ImageJ. Measured data were imported into Excel software for analysis. The thresholds in all images were set to similar levels. Data were obtained from at least three independent experiments, and the number of cells or imaging sections used for quantification is indicated in the figures. All statistical analyses were performed using the Student's t test and are presented as mean ± S.E.
For live cell imaging, cells were transferred to Tyrode's solution containing 10 mm Hepes, 10 mm glucose, 1.2 mm CaCl2, 1.2 mm MgCl2, 3 mm KCl, and 145 mm NaCl, pH 7.4. Temperature was maintained at 37 °C with an air stream incubator. Cells were visualized with a ×60 oil immersion lens (1.3 numerical aperture) on an Olympus FV1000 confocal microscope, using 488 nm excitation for GFP and 543 nm for mRFP. Time-lapse sequences of 1024 × 1024 pixels (8-bit) were collected at 1–2-s intervals with 1% intensity of the argon laser and maximum pinhole opening to minimize laser-induced bleaching and damage to cells. Dual color time-lapse images were captured by a total of 100 frames. All recordings started 6 min after the coverslip was placed in the chamber. The stacks of representative images were imported into ImageJ. A membranous organelle was considered stopped if it remained stationary for the entire recording period; a motile one was counted only if the displacement was at least 5 μm.
Criteria for Axon Selection in Cultured Neurons
For analyzing the motility of AVs or late endosomes in live neurons, we selected axons for time-lapse imaging and measuring motility because axons, but not dendrites, have a uniform microtubule organization and polarity. Axonal processes were selected as we reported previously (22, 64, 67). Briefly, axons in live images were distinguished from dendrites based on the following known morphological characteristics: greater length, thin and uniform diameter, and sparse branching (68). Only those that appeared to be single axons and separate from other processes in the field were chosen for recording axonal transport of AVs or BACE1 vesicles. Regions where crossing or fasciculation occurred were excluded from analysis.
Kymographs were used to trace axonal anterograde or retrograde movement of membranous organelles and to count stationary ones as described previously (67, 69) with extra plug-ins for ImageJ. Briefly, we used the “Straighten” plugin to straighten curved axons and the “Grouped ZProjector” to z-axially project re-sliced time-lapse images. The height of the kymographs represents recording time (100 s unless otherwise noted), whereas the width represents the length (μm) of the axon imaged. Counts were averaged from 100 frames for each time-lapse image to ensure accuracy of stationary and motile events. Measurements are presented as mean ± S.E. Statistical analyses were performed using unpaired Student's t tests.
Author Contributions
T. F., P. T., and Q. C. designed the project; T. F., P. T., C. A., Y. Y. J., and Q. C. performed experiments and analyzed data; Q. C. wrote the manuscript.
Acknowledgments
We thank H. Cai for anti-BACE1 antibody; W. Song for BACE1-GFP; T. Yoshimori for mRFP-LC3; D. Sabatini for LAMP-1-mRFP; C. Agrawal and S. McEwan for editing; E. Gavin, S. Zhu, J. Lam, D. Aikal, and other members of the Cai laboratory for helpful discussions and technical assistance.
This work was supported by the National Institutes of Health Grants R00AG033658 and R01NS089737 (to Q. C.) and Alzheimer's Association Grant NIRG-14-321833 (to Q. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- AD
- Alzheimer's disease
- APP
- amyloid precursor protein
- BACE1
- β-site APP-cleaving enzyme 1
- CTF
- C-terminal fragment
- Aβ
- amyloid-β
- ER
- endoplasmic reticulum
- TGN
- trans-Golgi network
- LE
- late endosome
- AV
- autophagic vacuole
- Tg
- transgenic
- LAMP
- lysosome-associated membrane protein
- DIV
- days in vitro
- h
- human
- Tre
- trehalose
- mTOR
- mammalian target of rapamycin
- 10-NCP
- 10-[4′-(N-diethylamino)butyl]-2-chlorophenoxazine
- mRFP
- monomeric red fluorescent protein.
References
- 1.Cai H., Wang Y., McCarthy D., Wen H., Borchelt D. R., Price D. L., and Wong P. C. (2001) BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nat. Neurosci. 4, 233–234 [DOI] [PubMed] [Google Scholar]
- 2.Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., et al. (1999) Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 3.Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., et al. (1999) β-Secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 4.Yan R., Bienkowski M. J., Shuck M. E., Miao H., Tory M. C., Pauley A. M., Brashier J. R., Stratman N. C., Mathews W. R., Buhl A. E., Carter D. B., Tomasselli A. G., Parodi L. A., Heinrikson R. L., and Gurney M. E. (1999) Membrane-anchored aspartyl protease with Alzheimer's disease β-secretase activity. Nature 402, 533–537 [DOI] [PubMed] [Google Scholar]
- 5.Fukumoto H., Rosene D. L., Moss M. B., Raju S., Hyman B. T., and Irizarry M. C. (2004) β-Secretase activity increases with aging in human, monkey, and mouse brain. Am. J. Pathol. 164, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., and Shen Y. (2003) Elevated β-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 9, 3–4 [DOI] [PubMed] [Google Scholar]
- 7.Vassar R., Kovacs D. M., Yan R., and Wong P. C. (2009) The β-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willem M., Lammich S., and Haass C. (2009) Function, regulation and therapeutic properties of β-secretase (BACE1). Semin. Cell Dev. Biol. 20, 175–182 [DOI] [PubMed] [Google Scholar]
- 9.Evin G., Barakat A., and Masters C. L. (2010) BACE: therapeutic target and potential biomarker for Alzheimer's disease. Int. J. Biochem. Cell Biol. 42, 1923–1926 [DOI] [PubMed] [Google Scholar]
- 10.Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., and Doms R. W. (2000) Maturation and endosomal targeting of β-site amyloid precursor protein-cleaving enzyme. The Alzheimer's disease β-secretase. J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 11.He X., Li F., Chang W. P., and Tang J. (2005) GGA proteins mediate the recycling pathway of memapsin 2 (BACE). J. Biol. Chem. 280, 11696–11703 [DOI] [PubMed] [Google Scholar]
- 12.Kang E. L., Biscaro B., Piazza F., and Tesco G. (2012) BACE1 protein endocytosis and trafficking are differentially regulated by ubiquitination at lysine 501 and the di-leucine motif in the carboxyl terminus. J. Biol. Chem. 287, 42867–42880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter J., Fluhrer R., Hartung B., Willem M., Kaether C., Capell A., Lammich S., Multhaup G., and Haass C. (2001) Phosphorylation regulates intracellular trafficking of β-secretase. J. Biol. Chem. 276, 14634–14641 [DOI] [PubMed] [Google Scholar]
- 14.Fukumoto H., Cheung B. S., Hyman B. T., and Irizarry M. C. (2002) β-Secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol. 59, 1381–1389 [DOI] [PubMed] [Google Scholar]
- 15.Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., and Tanzi R. E. (2007) Depletion of GGA3 stabilizes BACE and enhances β-secretase activity. Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sannerud R., Declerck I., Peric A., Raemaekers T., Menendez G., Zhou L., Veerle B., Coen K., Munck S., De Strooper B., Schiavo G., and Annaert W. (2011) ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc. Natl. Acad. Sci. U.S.A. 108, E559–E568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J., Petralia R. S., Kurushima H., Patel H., Jung M. Y., Volk L., Chowdhury S., Shepherd J. D., Dehoff M., Li Y., Kuhl D., Huganir R. L., Price D. L., Scannevin R., Troncoso J. C., et al. (2011) Arc/Arg3.1 regulates an endosomal pathway essential for activity-dependent β-amyloid generation. Cell 147, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh Y. H., von Arnim C. A., Hyman B. T., Tanzi R. E., and Tesco G. (2005) BACE is degraded via the lysosomal pathway. J. Biol. Chem. 280, 32499–32504 [DOI] [PubMed] [Google Scholar]
- 19.Lefort R., Pozueta J., and Shelanski M. (2012) Cross-linking of cell surface amyloid precursor protein leads to increased β-amyloid peptide production in hippocampal neurons: implications for Alzheimer's disease. J. Neurosci. 32, 10674–10685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye X., and Cai Q. (2014) Snapin-mediated BACE1 retrograde transport is essential for its degradation in lysosomes and regulation of APP processing in neurons. Cell Rep. 6, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overly C. C., and Hollenbeck P. J. (1996) Dynamic organization of endocytic pathways in axons of cultured sympathetic neurons. J. Neurosci. 16, 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Q., Lu L., Tian J. H., Zhu Y. B., Qiao H., and Sheng Z. H. (2010) Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 68, 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S., Sato Y., and Nixon R. A. (2011) Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J. Neurosci. 31, 7817–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y., Zhou B., Lin M. Y., Wang S., Foust K. D., and Sheng Z. H. (2015) Endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron 87, 355–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue Z., Friedman L., Komatsu M., and Tanaka K. (2009) The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim. Biophys. Acta 1793, 1496–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinsztein D. C., Mariño G., and Kroemer G. (2011) Autophagy and aging. Cell 146, 682–695 [DOI] [PubMed] [Google Scholar]
- 27.Nixon R. A. (2013) The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997 [DOI] [PubMed] [Google Scholar]
- 28.Schneider J. L., and Cuervo A. M. (2014) Autophagy and human disease: emerging themes. Curr. Opin. Genet. Dev. 26, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maday S., Wallace K. E., and Holzbaur E. L. (2012) Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maday S., and Holzbaur E. L. (2014) Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell 30, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng X. T., Zhou B., Lin M. Y., Cai Q., and Sheng Z. H. (2015) Axonal autophagosomes use the ride-on service for retrograde transport toward the soma. Autophagy 11, 1434–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng X. T., Zhou B., Lin M. Y., Cai Q., and Sheng Z. H. (2015) Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maday S., and Holzbaur E. L. (2016) Compartment-specific regulation of autophagy in primary neurons. J. Neurosci. 36, 5933–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krüger U., Wang Y., Kumar S., and Mandelkow E. M. (2012) Autophagic degradation of τ in primary neurons and its enhancement by trehalose. Neurobiol. Aging 33, 2291–2305 [DOI] [PubMed] [Google Scholar]
- 35.Su Q., Cai Q., Gerwin C., Smith C. L., and Sheng Z. H. (2004) Syntabulin is a microtubule-associated protein implicated in syntaxin transport in neurons. Nat. Cell Biol. 6, 941–953 [DOI] [PubMed] [Google Scholar]
- 36.Tsvetkov A. S., Miller J., Arrasate M., Wong J. S., Pleiss M. A., and Finkbeiner S. (2010) A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc. Natl. Acad. Sci. U.S.A. 107, 16982–16987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong Y. C., and Holzbaur E. L. (2015) Autophagosome dynamics in neurodegeneration at a glance. J. Cell Sci. 128, 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nixon R. A., Wegiel J., Kumar A., Yu W. H., Peterhoff C., Cataldo A., and Cuervo A. M. (2005) Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 64, 113–122 [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., and Ohsumi Y. (1998) A protein conjugation system essential for autophagy. Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 40.Ohsumi Y. (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216 [DOI] [PubMed] [Google Scholar]
- 41.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., and Ohsumi Y. (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funderburk S. F., Marcellino B. K., and Yue Z. (2010) Cell “self-eating” (autophagy) mechanism in Alzheimer's disease. Mt. Sinai J. Med. 77, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nixon R. A., and Yang D. S. (2011) Autophagy failure in Alzheimer's disease–locating the primary defect. Neurobiol. Dis. 43, 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki K., and Terry R. D. (1967) Fine structural localization of acid phosphatase in senile plaques in Alzheimer's presenile dementia. Acta Neuropathol. 8, 276–284 [DOI] [PubMed] [Google Scholar]
- 45.Qing H., Zhou W., Christensen M. A., Sun X., Tong Y., and Song W. (2004) Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 18, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 46.Prabhu Y., Burgos P. V., Schindler C., Farías G. G., Magadán J. G., and Bonifacino J. S. (2012) Adaptor protein 2-mediated endocytosis of the β-secretase BACE1 is dispensable for amyloid precursor protein processing. Mol. Biol. Cell 23, 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang E. L., Cameron A. N., Piazza F., Walker K. R., and Tesco G. (2010) Ubiquitin regulates GGA3-mediated degradation of BACE1. J. Biol. Chem. 285, 24108–24119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeates E. F., and Tesco G. (2016) The endosome-associated deubiquitinating enzyme USP8 regulates BACE1 enzyme ubiquitination and degradation. J. Biol. Chem. 291, 15753–15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu W. H., Cuervo A. M., Kumar A., Peterhoff C. M., Schmidt S. D., Lee J. H., Mohan P. S., Mercken M., Farmery M. R., Tjernberg L. O., Jiang Y., Duff K., Uchiyama Y., Näslund J., Mathews P. M., et al. (2005) Macroautophagy–a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 171, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson P., Loganathan K., Sekiguchi M., Matsuba Y., Hui K., Tsubuki S., Tanaka M., Iwata N., Saito T., and Saido T. C. (2013) Aβ secretion and plaque formation depend on autophagy. Cell Rep. 5, 61–69 [DOI] [PubMed] [Google Scholar]
- 51.Yu W. H., Kumar A., Peterhoff C., Shapiro Kulnane L., Uchiyama Y., Lamb B. T., Cuervo A. M., and Nixon R. A. (2004) Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for β-amyloid peptide over-production and localization in Alzheimer's disease. Int. J. Biochem. Cell Biol. 36, 2531–2540 [DOI] [PubMed] [Google Scholar]
- 52.Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., and Rubinsztein D. C. (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 53.Sarkar S., Perlstein E. O., Imarisio S., Pineau S., Cordenier A., Maglathlin R. L., Webster J. A., Lewis T. A., O'Kane C. J., Schreiber S. L., and Rubinsztein D. C. (2007) Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 3, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crews L., Spencer B., Desplats P., Patrick C., Paulino A., Rockenstein E., Hansen L., Adame A., Galasko D., and Masliah E. (2010) Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of α-synucleinopathy. PLoS ONE 5, e9313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Lafay-Chebassier C., Pérault-Pochat M. C., Page G., Rioux Bilan A., Damjanac M., Pain S., Houeto J. L., Gil R., and Hugon J. (2006) The immunosuppressant rapamycin exacerbates neurotoxicity of Aβ peptide. J. Neurosci. Res. 84, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 56.Hung S. Y., Huang W. P., Liou H. C., and Fu W. M. (2009) Autophagy protects neuron from Aβ-induced cytotoxicity. Autophagy 5, 502–510 [DOI] [PubMed] [Google Scholar]
- 57.Ling D., Song H. J., Garza D., Neufeld T. P., and Salvaterra P. M. (2009) Aβ42-induced neurodegeneration via an age-dependent autophagic-lysosomal injury in Drosophila. PLoS ONE 4, e4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spilman P., Podlutskaya N., Hart M. J., Debnath J., Gorostiza O., Bredesen D., Richardson A., Strong R., and Galvan V. (2010) Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-β levels in a mouse model of Alzheimer's disease. PLoS ONE 5, e9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caccamo A., Majumder S., Richardson A., Strong R., and Oddo S. (2010) Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-β, and τ: effects on cognitive impairments. J. Biol. Chem. 285, 13107–13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Majumder S., Richardson A., Strong R., and Oddo S. (2011) Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS ONE 6, e25416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caccamo A., De Pinto V., Messina A., Branca C., and Oddo S. (2014) Genetic reduction of mammalian target of rapamycin ameliorates Alzheimer's disease-like cognitive and pathological deficits by restoring hippocampal gene expression signature. J. Neurosci. 34, 7988–7998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mucke L., Masliah E., Yu G. Q., Mallory M., Rockenstein E. M., Tatsuno G., Hu K., Kholodenko D., Johnson-Wood K., and McConlogue L. (2000) High-level neuronal expression of Aβ1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J. Neurosci. 20, 4050–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou B., Cai Q., Xie Y., and Sheng Z. H. (2012) Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Rep. 2, 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai Q., Zakaria H. M., Simone A., and Sheng Z. H. (2012) Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 22, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goslin K., and Banker G. (1998) Rat hippocampal neurons in low-density culture. Culturing Nerve Cells 2, 339–370 [Google Scholar]
- 66.Nagahara A. H., Mateling M., Kovacs I., Wang L., Eggert S., Rockenstein E., Koo E. H., Masliah E., and Tuszynski M. H. (2013) Early BDNF treatment ameliorates cell loss in the entorhinal cortex of APP transgenic mice. J. Neurosci. 33, 15596–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang J. S., Tian J. H., Pan P. Y., Zald P., Li C., Deng C., and Sheng Z. H. (2008) Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banker G. A., and Cowan W. M. (1979) Further observations on hippocampal neurons in dispersed cell culture. J. Comp. Neurol. 187, 469–493 [DOI] [PubMed] [Google Scholar]
- 69.Miller K. E., and Sheetz M. P. (2004) Axonal mitochondrial transport and potential are correlated. J. Cell Sci. 117, 2791–2804 [DOI] [PubMed] [Google Scholar]