Abstract
Diarrhea-causing Escherichia coli strains are responsible for numerous cases of gastrointestinal disease and constitute a serious health problem throughout the world. The ability to recognize and attach to host intestinal surfaces is an essential step in the pathogenesis of such strains. AIDA is a potent bacterial adhesin associated with some diarrheagenic E. coli strains. AIDA mediates bacterial attachment to a broad variety of human and other mammalian cells. It is a surface-displayed autotransporter protein and belongs to the selected group of bacterial glycoproteins; only the glycosylated form binds to mammalian cells. Here, we show that AIDA possesses self-association characteristics and can mediate autoaggregation of E. coli cells. We demonstrate that intercellular AIDA-AIDA interaction is responsible for bacterial autoaggregation. Interestingly, AIDA-expressing cells can interact with antigen 43 (Ag43)-expressing cells, which is indicative of an intercellular AIDA-Ag43 interaction. Additionally, AIDA expression dramatically enhances biofilm formation by E. coli on abiotic surfaces in flow chambers.
Diarrhea-causing strains of Escherichia coli are responsible for many cases of gastrointestinal disease throughout the world, notably among infants in third world countries, where these pathogens are a major cause for concern (37). According to World Health Organization sources, more than 2 million humans, mainly infants, die from E. coli diarrhea each year. An essential step in the pathogenesis of diarrheagenic E. coli is the initial recognition of and attachment to intestinal tissue surfaces. Bacterial attachment is provided by specific adhesins protruding from the bacterial surface. There are a multitude of bacterial adhesins, and they can crudely be divided into two groups: long organelle-type adhesins (typified by fimbriae) and short, non-organelle-type adhesins (reviewed in reference 27).
The AIDA protein, a non-organelle-type adhesin, was originally found to be associated with adherence to human cells of an E. coli O126:H27 strain isolated from a case of infantile diarrhea (2, 3). AIDA belongs to the autotransporter protein family. One characteristic of this group of proteins, which encompasses many virulence factors, is that the protein itself contains all of the information required for traversing the bacterial membrane system and routing to the bacterial cell surface (20, 21). AIDA is produced as a precursor consisting of 1,286 amino acids, which subsequently undergoes extended posttranslational modifications. First, it is processed by removal of a 49-amino-acid signal peptide. The resultant protein, located in the periplasm, is further processed by autocatalytic action into a C-terminal translocator domain and an N-terminal passenger domain consisting of 439 and 798 amino acids, respectively. The translocator moiety forms a β-barrel porin in the outer membrane through which the adhesin moiety gains access to the surface (31, 50). Additional modifications also occur since the fully mature AIDA adhesin is a glycoprotein. Immediately upstream of the aidA gene, responsible for AIDA production, is a second gene, aah, which encodes a 45-kDa heptosyltransferase (autotransporter adhesin heptosyltransferase [AAH]) that modifies the AIDA adhesin by addition of an average of 19 heptose residues (5). Without this modification AIDA does not bind to human cells (4, 5).
AIDA has been demonstrated to mediate bacterial binding to a wide range of human and nonhuman cell types, exemplified by cell line types such as HeLa human cervical cancer cells, HT29 human colon cancer cells, monkey fibroblast cells, and Chinese hamster ovary cells (2, 28, 46). This suggests that the molecular motif(s) that AIDA interacts with is widely distributed. The broad-range cell recognition characteristics of AIDA are also reflected in the fact that although AIDA was originally found in an E. coli O126:H27 strain associated with a case of infantile diarrhea, it has also been found in E. coli strains isolated from pigs with edema disease and diarrhea (40). Interestingly, AIDA was found to be associated with the Stx2 Shiga-like toxin in such strains.
Several adhesins, such as type 1 fimbriae and curli, have been shown to confer bacterial autoaggregation and/or enhance biofilm formation on abiotic surfaces in addition to their receptor recognition faculty (17, 43, 45, 47). In this study we investigated whether the AIDA adhesin, in addition to its ability to promote bacterial binding to mammalian cells, possesses additional virulence properties, including autoaggregation of bacterial cells and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and plasmids and growth conditions.
The strains, plasmids, and primers used in this study are listed in Tables 1, 2 and 3, respectively. Cells were grown at 37°C on solid or liquid Luria-Bertani medium supplemented with the appropriate antibiotics unless indicated otherwise.
TABLE 1.
Bacterial strains
| E. coli strain | Relevant characteristics | Reference |
|---|---|---|
| MG1655 | E. coli K-12 reference strain | 1 |
| MS427 | MG1655 Δflu | 24 |
| MS528 | MG1655 Δflu Δfim | 24 |
| LH56 | MS427 containing pACYC184 | This study |
| LH73 | MS427 containing pIB264; AAH-AIDA | This study |
| LH75 | MS427 containing pLH44; AAH-AIDA | This study |
| OS45 | SAR19 containing pLH44; Cfp, AAH-AIDA | This study |
| OS46 | SAR20 containing pLH44; Yfp, AAH-AIDA | This study |
| OS47 | MS528 containing pOS32 | This study |
| OS56 | MS427 attB::bla-PA1/04/03-gfpmut3b*-T0; Gfp | This study |
| OS68 | OS56 containing pKKJ128; Gfp, Ag43 | This study |
| OS70 | OS56 containing pACYC184; Gfp | This study |
| OS71 | OS56 containing pLH44; Gfp, AAH-AIDA | This study |
| OS82 | MS427 containing pAR163; DsRed. T3 | This study |
| OS84 | MS427 containing pAR163 and pKKJ128; DsRed.T3, Ag43 | This study |
| OS85 | MS427 containing pOS33; AIDA | This study |
| OS99 | OS56 containing pOS33; AIDA | This study |
| OS101 | MS427 containing pOS37; AAH | This study |
| OS130 | MS427 containing pAR163 and pOS33; DsRed.T3, AIDA | This study |
| PKL1110 | MS427 containing pLH44 and pPKL4; AIDA, fimbriae | This study |
| SAR19 | CSH26 attB::bla-PA1/04/03-cfp*-T0; Cfp | 44 |
| SAR20 | CSH26 attB::bla-PA1/04/03-yfp*-T0; Yfp | 44 |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant genotype or phenotype | Reference |
|---|---|---|
| pAR81 | cat gene from pTP801 ligated into BamHI-digested pTP809; Cmr | This study |
| pAR90 | cfp* inserted between SphI and HindIII sites in pJBA27 | 44 |
| pAR94 | PrrnB, P1 containing SacI-XbaI fragment of pSM1690 inserted into pAR90 digested with SacI and XbaI | This study |
| pAR163 | Amplification of dsred2.T3 with primers Dsred2d and UDsRed2.T3 and ligation into Xbal-HindIII-digested pAR94; results in replacement of cfp* with dsred2.T3, controlled by PrrnB, P1 | This study |
| pAR176 | Amplification of PtetA with primers ar047 and ar048 from pRL27 and ligation into SacI-XbaI-digested pSM1690; results in replacement of PrrnB, P1 with PtetA | This study |
| pAR178 | Amplification of cat gene with primers ar059 and ar060 from pAR81 and ligation with ClaI-SalI-digested p15A ori-containing fragment from pSM1690 (amplified by PCR with primers ar057 and ar058) | This study |
| pAR179 | Insertion of PrrnB, P1-RBSII-dsred2.T3-T0 containing NotI cassette from pAR163 into the NotI site of pAR178 | This study |
| pDsRed.T3 | Encodes a rapidly maturating DsRed.T3 variant | 7 |
| pIB264 | SphI-ClaI aah-aidA-containing fragment from pIB6 | 2 |
| pKKJ128 | flu gene from E. coli MG1655 in pACYC184 | 23 |
| pLH44 | aah-aidA genes from pIB264 ligated into SphI-ClaI-digested pACYC184 | This study |
| pOS32 | gfpmut3b* gene from plasmid pAR176 ligated to XbaI-HindIII-digested pAR179; construct has gfpmut3b* gene under transcriptional control of the E. coli rrnB-P1 promoter | This study |
| pOS33 | aidA gene from pIB264 PCR amplified with primers 482 and 483 and ligated into EcoRV-SalI-digested pACYC184 | This study |
| pOS37 | aah gene from pIB264 PCR amplified with primers 537 and 541 and ligated into BamH1-XmaIII digested pACYC184 | This study |
| pPKL4 | fim gene cluster from E. coli PC31 in pBR322 | 26 |
| pRL27 | mini-Tn5 delivery vector; source of PtetA | 29 |
| pSM1690 | PrrnB, P1-RBSII-gfpmut3b*-T0-T1 NotI fragment in LOW2; Kmr | 49 |
| pTP801 | cat gene in BamHI site of pUC19 | 35 |
| pTP809 | pBR322 derivative with multiple cloning site introduced between EcoRI and NdeI sites | 35 |
TABLE 3.
Primers used in this study
| Primer | Sequence |
|---|---|
| DsRed2d | 5′-CCTCAAGCTTCCCGGGTTACAGGAACAGGTGGTGGCG-3′ |
| UDsRed2.T3 | 5′-GGTCTAGAATTAAAGAGGAGAAATTAAGCATGGCCTCCTCCGAG-3′ |
| ar047 | 5′-GTCGAGCTCAATGATTCTCCGCCAGCA-3′ |
| ar048 | 5′-GCTCTAGAATATGTGGCCTCCGGACC-3′ |
| ar057 | 5′-GCATCGATGACCAAAATCCCTTAACG-3′ |
| ar058 | 5′-GCGGTCGACGGAAATGGCTTACGAA-3′ |
| ar059 | 5′-GCGGTCGACTCCTTACGCATCTGTGC-3′ |
| ar060 | 5′-GCATCGATAGGCGTATCACGAGGC-3′ |
| 482 | 5′-CGCGGATATCATAATAAAGGATCATTTAATGAATAAGGCCTAC-3′ |
| 483 | 5′-CGCGCGGTCGACTCAGGAGCAAGAGTAATCCGCC-3′ |
| 537 | 5′-CGCGGGATCCATAATAAGGAATGACTTTCTTATCACCA-3′ |
| 541 | 5′-CGCGCGGCGGCCGTTAAATGATCCTGTCATGA-3′ |
E. coli strain MS427 was genetically marked by insertion of gfpmut3b* into the chromosomal attachment site of bacteriophage λ (attB) essentially as previously described (14). The gfpmut3b*-containing NotI fragment from pOS32 was ligated to the bla-attP-containing NotI fragment of plasmid pLDR11. The ligation mixture was transformed into MS427(pLDR8) cells expressing λ-Int. Correct chromosomal insertion of gfpmut3b* in an ampicillin-resistant, green fluorescent transformant (OS56) was verified by PCR. E. coli MS427 was labeled with the red fluorescent protein DsRed by transformation with plasmid pAR163 and designated OS82. The differently labeled fluorescent strains were used as hosts for the expression of antigen 43 (Ag43) or AIDA via plasmid-encoded genes.
DNA manipulations and genetic techniques.
Isolation of plasmid DNA was carried out by using a QIAprep Spin miniprep kit (QIAGEN). Restriction endonucleases were used according to the manufacturer's specifications (New England Biolabs Inc.). Chromosomal DNA was purified by using a GenomicPrep cell and tissue DNA isolation kit (Amersham Pharmacia Biotech Inc.). All PCRs were performed with the Expand High Fidelity polymerase system (Roche) essentially as previously described (48). The primers used are listed in Table 3. Amplified products were sequenced to ensure fidelity of the PCR (MWG Biotech, Ebersberg, Germany).
Release and purification of AIDA protein.
One-milliliter portions of overnight cultures were standardized to a final optical density at 600 nm (OD600) of ∼1, harvested by centrifugation, washed, and reconstituted in phosphate-buffered saline. The α subunit was released from the surface of the cells by heating the cells at 60°C for 3 min. The cells were immediately removed by centrifugation, and the resultant protein in the supernatant was precipitated with acetone (75%, vol/vol). The pellet was dried, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer, and boiled for 3 min prior to gel electrophoresis.
Autoaggregation assay.
To monitor differences in autoaggregation, we devised an assay to monitor bacterial settling rates over time. Overnight cultures were standardized and mixed vigorously for 15 s prior to the start of the assay. At regular time intervals, a 150-μl sample was taken approximately 0.5 cm from the liquid surface and transferred into a microtiter plate maintained on ice. At the end of the experiment the OD600 values were determined with a microtiter plate reader. When the influence of pH was investigated, the cells were harvested and resuspended in phosphate-buffered saline at different pH values.
Flow chamber experiments.
Flow chamber experiments were performed essentially as previously described (10); the major exception was that cells were grown in morpholinepropanesulfonic acid (MOPS) minimal medium and incubated at 30°C (39). Briefly, biofilms were allowed to form on glass surfaces in a multichannel flow system that permitted online monitoring of community structure. Flow cells were inoculated with OD600-standardized cultures pregrown overnight in MOPS medium containing chloramphenicol. Glucose was used as the sole carbon source at a concentration of 0.2%.
Microscopy and image analysis.
All microscopic observations and image acquisition were performed by using a confocal scanning laser microscope (TCS4D; Leica Lasertechnik, GmbH, Heidelberg, Germany) equipped with detectors and filters for monitoring cyan fluorescent protein (Cfp), yellow fluorescent protein (Yfp), green fluorescent protein (Gfp), and DsRed. Vertical cross sections through the biofilms were generated by using the IMARIS software package (Bitplane AG, Zürich, Switzerland) running on a Silicon Graphics Indigo2 workstation (Silicon Graphics, Mountain View, Calif.). Images were further processed for display by using Photoshop software (Adobe, Mountain View, Calif.).
RESULTS
Cloning and characterization of the AIDA gene system.
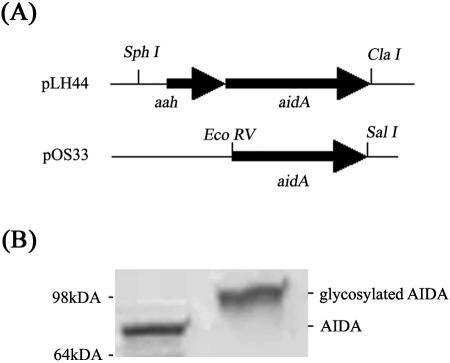
Plasmid pIB264 contains the adjacently linked aah and aidA genes from E. coli O126:H27 strain 2787 (2, 5). In order to perform functional analysis of AIDA with and without glycosylation, we prepared several different plasmid constructs. The aah-aidA fragment from pIB264 was cloned into plasmid pACYC184, resulting in plasmid pLH44. A plasmid containing the aidA gene alone was prepared by PCR amplification and cloning into pACYC184, which resulted in plasmid pOS33. Both constructs were verified by restriction enzyme mapping and DNA sequencing (Fig. 1). The AIDA passenger domain, but not the translocater domain, could be efficiently liberated from cells by brief heating to 60°C. To confirm correct processing, glycosylation (plasmid pLH44), and a surface location for AIDA, the α subunit was released by heat treatment of bacteria harboring plasmids pOS33 and pLH44 and examined by SDS-PAGE (Fig. 1). As expected, proteins with apparent molecular masses of approximately 80 kDa (AIDA) and 100 kDa (glycosylated AIDA) were visible (Fig. 1).
FIG. 1.
(A) Overview of the plasmid constructs used for expression AIDA (pOS33) and glycosylated AIDA (pLH44). The constructs were made in the pACYC184 cloning vector and were constitutively expressed. (B) Coomassie brilliant blue-stained SDS-PAGE gel of AIDA (lane 1) and glycosylated AIDA (lane 2) α-subunit proteins liberated from E. coli MS427 host cells by brief heat treatment.
AIDA mediates autoaggregation, flocculation, and settling of cells.
Cells of E. coli strain MS427 did not aggregate or settle from liquid suspensions (Fig. 2). When plasmid pLH44, containing the aidA and aah genes, was introduced into MS427, colonies of the resultant strain (OS71) were observed to aggregate, and when liquid suspensions of the cells were left standing, they flocculated and settled (Fig. 2). The aggregates formed by AIDA-expressing cells were very compact and rather difficult to separate. This finding was also reflected in the fast settling kinetics of AIDA-expressing cells (Fig. 3). It should also be noted that MS427 hosts that contained other types of plasmids encoding the aidA and aah genes (for example, pIB264 [2]) behaved like OS71 (data not shown). Furthermore, AIDA-induced cell aggregation was not restricted to a particular host strain as it was observed in a range of E. coli K-12 strains.
FIG. 2.
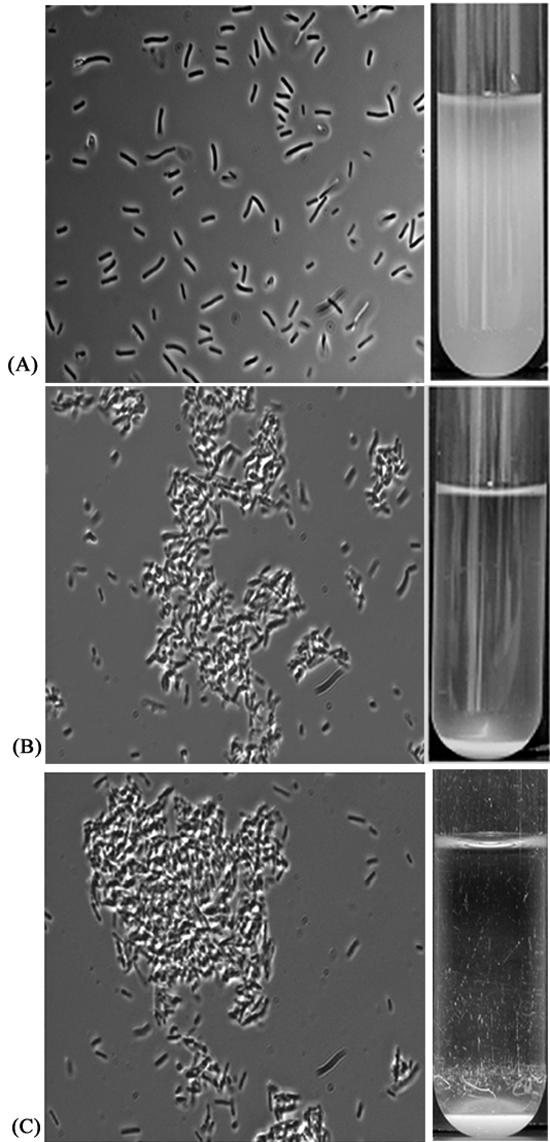
Cell-cell aggregation characteristics (left panels) and settling from static liquid suspensions (right panels) of E. coli MS427 containing either the vector control plasmid pACYC184 (A), the aah-aidA-encoding plasmid pLH44 (B), or the aidA-encoding plasmid pOS33 (C).
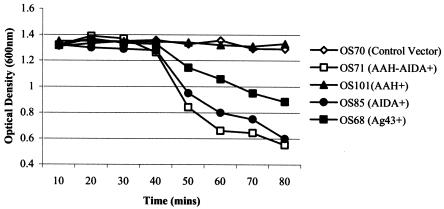
FIG. 3.
Autoaggregation assay demonstrating the settling profiles of liquid suspensions of E. coli strains OS70 (vector control), OS71 (AAH-AIDA+), OS101 (AAH+), OS85 (AIDA+), and OS68 (Ag43+).
AIDA-mediated cell-cell aggregation is independent of glycosylation.
AIDA-mediated attachment to human cells has been reported to be dependent on the glycosylation status of the protein; i.e., in the absence of the glycosylation conferred by the aah heptosyltransferase gene product AIDA does not bind human cells (4, 5). It was therefore conceivable that glycosylation played a role in AIDA-mediated bacterial autoaggregation. To test this, plasmid pOS33, which harbored the aidA gene but not the aah gene, was used. pOS37 harboring the aah gene but not the aidA gene was constructed as a control plasmid. When plasmid pOS33 was introduced into MS427, the transformant (OS85) was observed to aggregate (Fig. 2) and settle from liquid suspensions in the same way that cells expressing the glycosylated version of AIDA aggregated and settled (Fig. 3). When plasmid pOS37 was introduced into MS427 (OS101), this strain did not aggregate or settle from liquid suspensions (Fig. 3), suggesting that the aidA gene and not the aah gene is responsible for bacterial autoaggregation. Taken together, these results indicate that AIDA mediates autoaggregation and that glycosylation of AIDA does not play a role in this phenotype.
AIDA-AIDA interaction is responsible for bacterial autoaggregation.
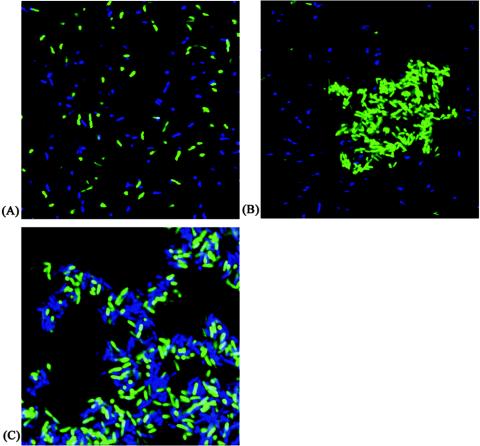
The observed AIDA-mediated cell aggregation could be due to either (i) an intercellular AIDA-AIDA interaction or (ii) an intercellular interaction between AIDA and some other cell surface component. To distinguish between these possibilities, plasmid pLH44, containing the aidA and aah genes, was transformed into E. coli strain CSH26 expressing either Cfp or Yfp (SAR19 and SAR20, respectively). The resulting strains were designated OS45 and OS46, respectively. First, cultures of SAR19 and SAR20 were mixed and examined. No interaction between these E. coli strains was evident, as expected (Fig. 4A). When OS46 cells (expressing glycosylated AIDA and Yfp) were mixed with SAR19 cells, aggregates composed of only Yfp-tagged OS46 cells were evident, and no interaction between these cells and Cfp-tagged SAR19 cells was evident (Fig. 4B). Finally, when OS45 cells (expressing glycosylated AIDA and Cfp) and OS46 cells (expressing glycosylated AIDA and Yfp) were mixed, aggregates consisting of both Cfp- and Yfp-tagged cells were produced (Fig. 4C). This result suggests that an intercellular AIDA-AIDA interaction was responsible for the observed cell aggregation. AIDA-mediated cell aggregation was found to be sensitive to pH extremes. For example, when glycosylated AIDA-expressing cells were subjected to pH 11, no aggregation took place, while a range of pH 10 to 4 demonstrated that aggregation occurred progressively faster as the pH decreased and was most efficient at pH 4. This suggests that charged amino acid side chains could participate in the aggregation process. In this context it should also be mentioned that alkaline conditions ranging from pH 9 to 11 do not release the passenger domain from the cells (data not shown). The AIDA passenger domain, but not the translocater domain, can be efficiently liberated from cells by brief heating to 60°C. Cells depleted in the passenger domain do not interact with AIDA-expressing cells. This suggests that an intercellular interaction between AIDA passenger domains is required for AIDA-assisted cell aggregation.
FIG. 4.
Confocal scanning laser microscopy showing cell autoaggregation mediated by specific AIDA-AIDA interactions. (A) SAR19 (Cfp+) and SAR20 (Yfp+). (B) SAR19 (Cfp+) and OS46 (Yfp+ AAH-AIDA+). (C) OS45 (Cfp+ AAH-AIDA+) and OS46 (Yfp+ AAH-AIDA+).
AIDA- and Ag43-expressing cells interact and aggregate.
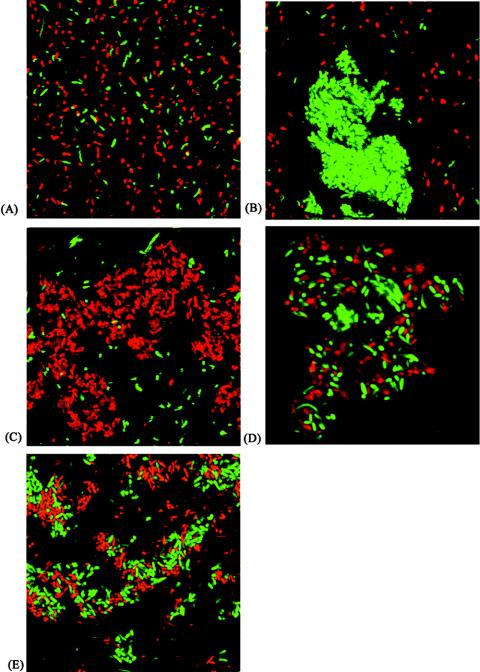
AIDA, TibA, and Ag43 belong to an autotransporter subfamily whose members contain repetitive amino acid sequence motifs (20). One member of this subfamily, Ag43, has been shown to be a self-recognizing protein and to cause cell-cell aggregation and settling of bacteria from suspensions (13, 18, 19). A comparison of the settling kinetics of AIDA and those of Ag43 showed that AIDA consistently settled at a higher rate than Ag43 (Fig. 3). These similarities prompted us to study possible interactions between the two self-recognizing proteins. To this end, E. coli MS427 was tagged with either Gfp or DsRed, resulting in OS56 and OS82 (see Materials and Methods). The fluorescence-tagged strains were transformed with either plasmid pLH44 (glycosylated AIDA expression), pKKJ128 (Ag43 expression), or pOS33 (AIDA expression), resulting in strains OS71, OS84, and OS130, respectively. These organisms were mixed in various combinations and examined (Fig. 5). It is clear that control strains OS56 and OS82 did not interact (Fig. 5A). The combination of OS71 (glycosylated AIDA+ Gfp+) mixed with OS82 (DsRed+) produced only one type of aggregate, consisting uniquely of Gfp-tagged cells (Fig. 5B). Likewise, the combination of OS56 (Gfp+) plus OS84 (DsRed+ Ag43+) produced only aggregates composed of DsRed-tagged cells (Fig. 5C). When OS71 (glycosylated AIDA+ Gfp+) and OS84 (DsRed+ Ag43+) cells were mixed, aggregates composed of both green and red fluorescent cells resulted (Fig. 5D). Likewise, the combination of OS84 (DsRed+ Ag43+) and OS130 (AIDA+ Gfp+) produced aggregates composed of both green and red fluorescent cells (Fig. 5E). These results indicate that Ag43 and AIDA are capable of interacting with each other and that the interaction is not dependent on glycosylation of AIDA. Also, when liquid suspensions containing equivalent quantities of OS71 and OS84 cells were mixed and left standing, the entire population settled, indicating that there was an efficient AIDA-Ag43 interaction (data not shown).
FIG. 5.
Confocal scanning laser microscopy showing cell aggregation mediated by glycosylated AIDA-Ag43 and AIDA-Ag43 interactions. (A) MS427 (Gfp+) and MS427 (DsRed+). (B) MS427 (Gfp+ AAH-AIDA+) and MS427 (DsRed+). (C) MS427 (DsRed+ Ag43+) and MS427 (Gfp+). (D) MS427 (Gfp+ AAH-AIDA+) and MS427 (DsRed+ Ag43+). (E) MS427 (Gfp+ Ag43+) and MS427 (DsRed+ AIDA+).
AIDA-mediated cell aggregation is blocked by fimbriae.
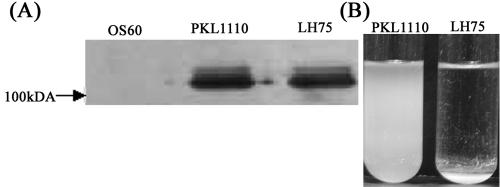
AIDA belongs to the class of short non-organelle-type adhesins and has been predicted to protrude about 10 nm from the bacterial surface (22). Thus, AIDA-mediated cell aggregation requires close cell-cell contact. Expression of type 1 fimbriae (stiff organelles that are ∼1 μm long) is thought to physically shield and block the close cell-cell contact required for non-fimbria-mediated cell aggregation. To determine if AIDA-mediated cell aggregation is blocked by type 1 fimbria expression, we introduced plasmid pPKL4 (conferring expression of type 1 fimbriae) and plasmid pLH44 (conferring glycosylated AIDA expression) into MS427 cells, which resulted in strain PKL1110.
To confirm that the surface location of AIDA was not affected by type 1 fimbria expression, the α subunit was released by brief heat treatment of host cells expressing AIDA (viz., strain LH75) or both AIDA and type 1 fimbriae (viz., strain PKL1110), and the supernatants were examined by SDS-PAGE (Fig. 6A). Clearly similar quantities of α subunit were released from the surfaces of both LH75 and PKL110, indicating that type 1 fimbria expression did not affect surface presentation of AIDA. However, PKL1110 cells were unable to aggregate or settle from static liquid solutions (Fig. 6B). Arguably, fimbria expression physically shields AIDA-AIDA interactions. A similar phenomenon with fimbrial abolishment of Ag43-mediated cell aggregation has been observed previously (18).
FIG. 6.
AIDA-mediated cell aggregation is blocked by fimbriae. (A) Coomassie brilliant blue-stained SDS-PAGE gel of glycosylated AIDA α-subunit proteins liberated from E. coli MS427 and E. coli MS427 (Fim+) host cells, showing that overexpression of type 1 fimbriae does not affect surface presentation of AIDA. (B) Fimbria production prevents AIDA settling in static liquid suspensions.
AIDA display enhances biofilm formation under continuous-flow growth conditions.
Given the excellent cell aggregation characteristics of AIDA, we speculated whether this facility could influence biofilm formation. To evaluate the role of AIDA in biofilm formation, we compared strain OS71 (AAH-AIDA+ Gfp+) with the vector control strain OS70 (Gfp+). Biofilm formation was initially assessed in polystyrene microtiter plates by quantitative crystal violet staining of adhering cells, and the results indicated that AIDA expression enhanced biofilm formation (data not shown).
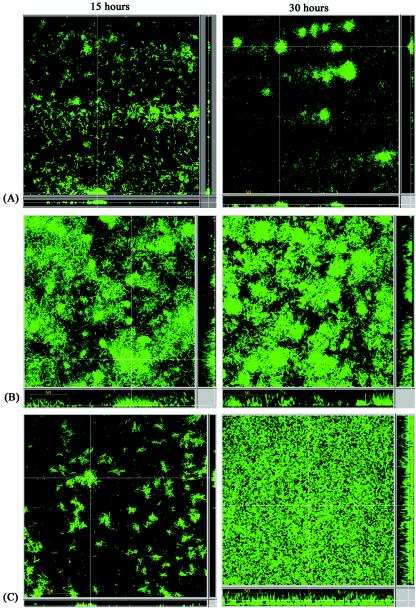
To obtain a more detailed picture of this phenomenon, biofilm experiments were performed in flow chambers with strains OS70 (Gfp+), OS71 (AAH-AIDA+ Gfp+), and OS68 (Ag43+ Gfp+). The Ag43-expressing strain was included in these experiments to permit comparison with AIDA. The experimental design enabled us to monitor bacterial distribution within an evolving biofilm under continuous-flow conditions. Furthermore, the spatial distribution of individual bacterial cells could be assessed due to the combination of fluorescence-tagged cells and scanning confocal laser microscopy.
Biofilms were established on glass surfaces in separate flow cells and analyzed. Image analysis of optical sections was performed to examine the distribution and thickness of the established biofilm. The control strain, OS70, produced small, flat microcolonies (Fig. 7A). A distinct change in the surface colonization pattern was observed when Ag43 was expressed (OS68). Initially, the OS68 microcolonies were both larger and more elevated than those of OS70 (Fig. 7B), and by the end of the experiment the biofilm consisted of fluffy aggregates of cells. In the case of OS71 (AAH-AIDA+) the effect was particularly striking (Fig. 7C). Initially, tight and compact microcolonies were produced. These colonies developed further into macrocolonies protruding from the thin-layer sessile community, and eventually after 30 h the biofilm was dense and uniform and covered the entire surface of the flow cell. The biofilm formed by AIDA+ cells was interspersed with void spaces devoid of any bacteria. The average depth of this biofilm was 40 μm. Furthermore, the AIDA biofilm was extremely stable and resistant to sheer flow forces, in contrast to the Ag43 or control biofilm. Even after exposure of the AIDA biofilm to a 10-fold increase in the flow rate followed by treatment with SDS, a substantial portion of the OS71 biofilm remained attached to the glass surface of the flow cell. Neither the negative control nor the Ag43 biofilm remained after this treatment. In fact, these biofilms were removed after the first 5 min of flushing of the flow cell. There were no differences between the biofilms formed by glycosylated AIDA (OS71) and the biofilms formed by the nonglycosylated protein (OS130) (data not shown), indicating that glycosylation of AIDA is not required for induction of biofilm formation.
FIG. 7.
Spatial distribution of biofilm formation for Gfp-labeled E. coli strains OS70 (vector control) (A), OS68 (Ag43+), (B), and OS71 (AAH-AIDA+) (C). Biofilm development was monitored by confocal scanning laser microscopy at 15 h (left panels) and 30 h (right panels) after inoculation. The images are representative horizontal sections collected within each biofilm and vertical sections (to the right and below of each larger panel, representing the yz plane and the xz plane, respectively) at the positions indicated by the white lines.
DISCUSSION
Adherence and colonization of host tissues are generally considered key events in bacterial pathogenesis. Moreover, it is also apparent that the ability to form aggregates is a common theme in many bacterial pathogens. A diverse range of both gram-positive and gram-negative bacteria, such as Staphylococcus aureus (32), Streptococcus pyogenes (9, 16), Bordetella pertussis (33), and Mycobacterium tuberculosis (34), are able to form aggregates. Such aggregates are known to be able to resist various host defenses (e.g., complement attack and phagocytosis) more efficiently than planktonic bacteria (6, 41). For example, mice infected with aggregated S. pyogenes cells were reported to develop abscesses more frequently than mice infected with planktonic bacteria (41). These observations support the notion that aggregation is an important virulence mechanism. Formation of aggregates usually takes place through autoaggregation of cells.
Many E. coli strains are capable of autoaggregation and employ a range of different systems to do this. Curli, which are thin surface fibers formed on the cell surface by the precipitation of secreted soluble subunit proteins, confer autoaggregation (17). Likewise, bundle-forming pili mediate autoaggregation (8). In enteroaggregative E. coli strains, two thin 2- to 3-nm-wide fimbrial types designated aggregative adherence fimbriae I and II have been identified (12, 36, 37). Expression causes prominent autoagglutination of bacterial cells (38).
In contrast to these aggregating systems, the self-recognizing AIDA adhesin is anchored directly to the outer membrane. Thus, AIDA-mediated aggregation results in more intimate cell-cell contact than that seen with systems in which the intercellular interactions are based on polymeric structures that reach far out from the bacterial surface (i.e., fimbriae and curli). AIDA, together with Ag43 and TibA, belongs to a subfamily of E. coli autotransporters that is defined by criteria such as sequence homology and the presence of repetitive sequence motifs (3, 25, 30). Furthermore, the passenger domains of both AIDA and Ag43 have been predicted to fold into extended β-helices (22, 25).
These characteristics shared by AIDA and Ag43 prompted us to investigate if AIDA, like Ag43, is a self-recognizing molecule capable of conferring bacterial autoaggregation. Our observations indicate that AIDA does possess this property. In fact, AIDA-expressing cells were found to aggregate and settle more efficiently than Ag43-expressing cells and formed very tightly packed cell aggregates. Interestingly, we observed that AIDA readily interacted with Ag43, and the resultant intercellular AIDA-Ag43 interaction caused cell aggregation. The biological implications of this phenomenon could have interesting consequences. In contrast to Ag43-expressing cells, AIDA-expressing cells can efficiently attach to human cells via a specific interaction mediated by the glycosylated AIDA adhesin. However, glycosylation does not play a role in AIDA-mediated cell aggregation, such as that investigated here. Our data support the notion that Ag43-expressing cells could bind to human cells indirectly by docking onto bound AIDA-expressing cells via specific AIDA-Ag43 interactions. This concept becomes even more interesting when one considers the locations of the corresponding genes. The AIDA-encoding genes are plasmid located, and expression seems to be limited to small number of diarrheagenic strains, while the gene encoding Ag43 (flu) is chromosomally located and is widespread in E. coli. Indeed, a survey of enteropathogenic and uropathogenic strains showed that 77 and 60% of these strains, respectively, could express Ag43 (42). It is not clear which molecular motifs are involved in the AIDA-Ag43 interaction. The passenger domains of Ag43 and AIDA are not very similar, consisting of 499 and 798 amino acids, respectively. Furthermore AIDA is normally extensively glycosylated, while Ag43 is not.
AIDA appears to be a two-in-one protein. In addition to being a bona fide adhesin (with human cells), it also plays a role as an autoaggregator, as demonstrated in this study. Specific attachment to a host tissue is of paramount importance in bacterial pathogenesis. The self-association function of AIDA may also have implications in bacterial pathogenesis. Bacteria expressing the AIDA aggregating phenotype may exist as tight communities of cells having all of the beneficial aspects of this type of existence. In this respect it is interesting to speculate that the autoaggregative function of AIDA may aid survival of the organism on route to a mammalian host. Furthermore, AIDA expression enhanced biofilm formation and gave rise to very compact sessile communities that resisted shear flow forces and detergent treatment very efficiently. The ability to form biofilms is a trait that is closely associated with bacterial persistence and virulence, and many persistent and chronic bacterial infections, including periodontitis, otitis media, biliary tract infections, and endocarditis, are now believed to be linked to the formation of biofilms (11, 15).
A novel picture of the AIDA autotransporter is emerging. This molecule seems to be a highly versatile virulence factor that plays multiple potential roles in bacterial pathogenesis, as follows: (i) it is a potent adhesin with affinity for different human cell types; (ii) it is capable of mediating bacterial aggregation via intercellular self-recognition; (iii) it is able to mediate aggregation with other bacteria that express the distantly related protein Ag43 due to an intercellular AIDA-Ag43 interaction; and (iv) it is a highly efficient initiator of biofilm formation.
Acknowledgments
We thank Birthe Jul Jorgensen for expert technical assistance and Alexander Schmidt (Westfälische Wilhelms-Universität) for providing plasmid pIB264. W. W. Metcalf, K. C. Murphy, and B. Glick are acknowledged for providing plasmids, and Mikkel Klausen and Janus Haagensen are acknowledged for biofilm expertise.
This work was supported by grants from the Danish Technical Research Council (grant 26-02-0183), the Australian National Health and Medical Research Council (grant 301163), and the University of Queensland.
REFERENCES
- 1.Bachmann, B. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 2.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 1992. Isolation and serologic characterization of AIDA, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787(O126:H27). Infect. Immun. 60:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787(O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 6.Berge, A., B.-M. Kihlberg, A. G. Sjöholm, and L. Björck. 1997. Streptococcal protein H forms soluble complement activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J. Biol. Chem. 272:20774-20781. [DOI] [PubMed] [Google Scholar]
- 7.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. [DOI] [PubMed] [Google Scholar]
- 8.Bieber, D., S. W. Ramer, C.-Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 9.Caparon, M. G., D. S. Stephens, A. Olsen, and J. R. Scott. 1991. Role of M protein in adherence of group A streptococci. Infect. Immun. 59:1811-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Møller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Biofilms: a common cause of persistent infections. Science 284:2137-2142. [DOI] [PubMed] [Google Scholar]
- 12.Czeczulin, J. R., S. Balepur, S. Hicks, A. Phillips, R. Hall, M. H. Mahendra, F. Navarro-Garcia, and J. P. Nataro. 1997. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 65:4135-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for the integration into the λ attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 15.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frick, I.-M., M. Mörgelin, and L. Björck. 2000. Virulent aggregates of Streptococcus pyogenes are generated by homophilic protein-protein interactions. Mol. Microbiol. 37:1232-1247. [DOI] [PubMed] [Google Scholar]
- 17.Hammar, M., Z. Bian, and S. Normark. 1996. Nucleator dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:6562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasman, H., M. A. Schembri, and P. Klemm. 2000. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J. Bacteriol. 182:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 22.Kajava, A. V., N. Cheng, R. Cleaver, M. Kessel, M. N. Simon, E. Willery, F. Jacob-Dubuisson, C. Locht, and A. C. Steven. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42:279-292. [DOI] [PubMed] [Google Scholar]
- 23.Kjærgaard, K., M. A. Schembri, H. Hasman, and P. Klemm. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182:4789-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjærgaard, K., M. A. Schembri, C. Ramos, M. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 25.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283-296. [DOI] [PubMed] [Google Scholar]
- 26.Klemm, P., B. J. Jorgensen, I. van Die, H. de Ree, and H. Bergmans. 1985. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol. Gen. Genet. 199:410-414. [DOI] [PubMed] [Google Scholar]
- 27.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: structure and function. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 28.Laarmann, S., and M. A. Schmidt. 2003. The Escherichia coli autotransporter adhesin recognizes an integral membrane glycoprotein as receptor. Microbiology 149:1871-1882. [DOI] [PubMed] [Google Scholar]
- 29.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 30.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibronectin receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 33.Menozzi, F. D., P. E. Boucher, G. Riveau, C. Gantiez, and C. Locht. 1994. Surface associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect. Immun. 62:4261-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menozzi, F. D., J. H. Rouse, M. Alawi, M. Laude-Sharp, J. Muller, R. Bischoff, M. J. Brennan, and C. Locht. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Immunol. 184:993-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 36.Nataro, J. P., Y. Deng, D. R. Maneval, A. L. German, W. C. Martin, and M. M. Levine. 1992. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60:2297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nataro, J. P., J. B. Kaper, R. Robins-Browne, V. Prado, P. Vial, and M. M. Levine. 1987. Patterns of adherence of diarrheagenic Escherichia coli. Pediatr. Infect. Dis. J. 6:829-831. [DOI] [PubMed] [Google Scholar]
- 39.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niewerth, U., A. Frey, T. Voss, C. Le Bouguénec, G. Baljer, S. Franke, and M. A. Schmidt. 2001. The AIDA autotransporter system is associated with F18 and Stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin. Diagn. Lab. Immunol. 8:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochiai, K., T. Kurita-Ochiai, Y. Kamino, and T. Ikeda. 1993. Effect of co-aggregation on the pathogenicity of oral bacteria. J. Med. Microbiol. 39:183-190. [DOI] [PubMed] [Google Scholar]
- 42.Owen, P., M. Meehan, H. de Loughry-Doherty, and I. Henderson. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16:63-76. [DOI] [PubMed] [Google Scholar]
- 43.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Eschericha coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 44.Reisner, A., S. Molin, and E. L. Zechner. 2002. Recombinogenic engineering of conjugative plasmids with fluorescent marker cassettes. FEMS Microbiol. Ecol. 42:251-259. [DOI] [PubMed] [Google Scholar]
- 45.Schembri, M. A., G. Christiansen, and P. Klemm. 2001. FimH-mediated auto-aggregation of Escherichia coli. Mol. Microbiol. 41:1419-1430. [DOI] [PubMed] [Google Scholar]
- 46.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186:1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schembri, M. A., and P. Klemm. 2001. Biofilm formation in a hydrodynamic environment by novel FimH variants and ramifications for virulence. Infect. Immun. 69:1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stentebjerg-Olesen, B., L. Pallesen, L. B. Jensen, G. Christiansen, and P. Klemm. 1997. Authentic display of a cholera toxin epitope by chimeric type 1 fimbriae: effects of insert position and host background. Microbiology 143:2027-2038. [DOI] [PubMed] [Google Scholar]
- 49.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suhr, M., I. Benz, and M. A. Schmidt. 1996. Processing of the AIDA precursor: removal of AIDAc and evidence for the outer membrane anchoring as a β-barrel structure. Mol. Microbiol. 22:31-42. [DOI] [PubMed] [Google Scholar]