Abstract
Heme a is an essential metalloporphyrin cofactor of the mitochondrial respiratory enzyme cytochrome c oxidase (CcO). Its synthesis from heme b requires several enzymes, including the evolutionarily conserved heme a synthase (Cox15). Oligomerization of Cox15 appears to be important for the process of heme a biosynthesis and transfer to maturing CcO. However, the details of this process remain elusive, and the roles of any additional CcO assembly factors that may be involved remain unclear. Here we report the systematic analysis of one such uncharacterized assembly factor, Pet117, and demonstrate in Saccharomyces cerevisiae that this evolutionarily conserved protein is necessary for Cox15 oligomerization and function. Pet117 is shown to reside in the mitochondrial matrix, where it is associated with the inner membrane. Pet117 functions at the later maturation stages of the core CcO subunit Cox1 that precede Cox1 hemylation. Pet117 also physically interacts with Cox15 and specifically mediates the stability of Cox15 oligomeric complexes. This Cox15-Pet117 interaction observed by co-immunoprecipitation persists in the absence of heme a synthase activity, is dependent upon Cox1 synthesis and early maturation steps, and is further dependent upon the presence of the matrix-exposed, unstructured linker region of Cox15 needed for Cox15 oligomerization, suggesting that this region mediates the interaction or that the interaction is lost when Cox15 is unable to oligomerize. Based on these findings, it was concluded that Pet117 mediates coupling of heme a synthesis to the CcO assembly process in eukaryotes.
Keywords: cytochrome c oxidase (Complex IV), heme, mitochondria, mitochondrial disease, oligomerization, Cox15, Pet117
Introduction
Both the initial and final steps of heme (heme b; iron protoporphyrin IX) biosynthesis occur in mitochondria, and this process is then followed by the routing and delivery of heme to maturing hemoproteins throughout the cell (1). One major route for newly synthesized heme is its modification to form heme a, which is uniquely used by cytochrome c oxidase (CcO3; Complex IV), the heme-copper terminal enzyme of the mitochondrial electron transport chain (2, 3). Two heme a molecules with different coordination geometries, an isolated heme a moiety and a heterobimetallic cofactor center designated the heme a3-CuB site, reside in the core CcO subunit Cox1, where they are essential for the stability and folding of Cox1 as well as enzymatic activity (3, 4). These heme a cofactors are derived from heme b by the sequential actions of the conserved enzymes heme o synthase (Cox10), which mediates farnesylation of a vinyl group to yield heme o intermediate, and heme a synthase (Cox15), which converts heme o to heme a through oxidation of a methyl group in a reaction dependent upon the ferredoxin-ferredoxin reductase relay (3, 5, 6).
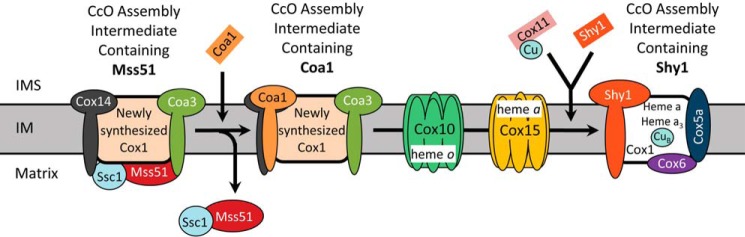
Understanding of how heme a is incorporated into CcO and how these steps are coupled to the complex process of Cox1 maturation during CcO assembly remains limited. After its translation by a mitochondrial ribosome and insertion into the mitochondrial inner membrane (IM), Cox1 undergoes sequential maturation aided by multiple assembly factors and receives its cofactors (2, 3, 7). Before being joined by the other core CcO subunits, Cox1 can be found in at least three distinct assembly intermediate complexes, which can be identified by the characteristic presence of the assembly factors Mss51, Coa1, and Shy1, respectively (Fig. 1). The last of these complexes contains heme a, and thus its formation depends on the activity of Cox10 and Cox15.
FIGURE 1.
Scheme depicting the present understanding of early CcO assembly intermediates formed during Cox1 maturation. Following translation and protein-assisted insertion into the mitochondrial inner membrane, newly synthesized Cox1 is associated with Ssc1, Mss51, Cox14, and Coa3 assembly factors, forming an assembly intermediate that can be identified by the presence of these constituents, particularly the characteristic presence of Mss51. The next assembly intermediate is characterized by the addition of the Coa1 assembly factor and the loss of Mss51 and Ssc1 (which form a distinct binary complex). Subsequently, an intermediate containing the assembly factor Shy1 is formed, which receives cofactors in a process dependent upon Cox11, Cox10, and Cox15 and begins to associate with nucleus-encoded CcO subunits, such as Cox5a and Cox6. IMS, intermembrane space.
Oligomerization of both Cox10 and Cox15 to form independent complexes within the mitochondrial IM appears to be required for heme a biosynthesis and/or transfer to maturing CcO. In the yeast Saccharomyces cerevisiae, Cox10 and Cox15 are large (52 and 55 kDa, respectively) polytopic proteins with 7–9 predicted transmembrane segments (3). They form complexes of ∼300 and ∼250 kDa, respectively, which are probably homooligomeric and may also associate with additional proteins (8–10), and the mammalian orthologs of these heme a biosynthetic enzymes appear to be organized in a similar fashion (8).
We have previously shown that Cox10 oligomerization is directly linked to the sophisticated process of Cox1 synthesis and maturation; multimeric Cox10 is important for coupling heme a synthesis to the incorporation of heme a into maturing Cox1 (9, 11). Cox10 multimerization is promoted by the C-terminal portion of the newly synthesized Cox1 protein and can be uncoupled from the catalytic activity of Cox10 (11). Notably, uncoupling of Cox10 oligomerization and catalytic function is observed in the D336V variant of Cox10 found in the neurological disease Leigh syndrome (11).
In contrast, oligomerization of Cox15 appears to be important for, yet largely independent of, Cox1 maturation (8, 10, 11). We recently determined that the matrix-exposed, unstructured linker region (of 20 amino acid residues) connecting the N- and C-terminal membrane-embedded halves of Cox15 is required for enzymatic oligomerization and activity and that activity is compromised and oligomerization is affected in the yeast analog of the R217W variant of Cox15 observed in patients with mitochondrial disease (8).
Heme a delivery to maturing Cox1 also depends on two additional proteins, Shy1/SURF1 and Coa2 (3, 7). The formation of the heme a and heme a3 centers appears to occur within the Shy1-containing Cox1 assembly intermediate, and Shy1 appears to chaperone maturation of the heme a3 site rather than serve as a direct heme a donor to newly synthesized Cox1 (12). A recent report has suggested that Shy1 may associate with Cox15, but the significance of this association is unclear (10). The small membrane-associated protein Coa2 cooperates with Shy1 and probably functions in Cox1 hemylation; in its absence, hemylation of Cox1 is impaired, and the resulting misfolded protein is rapidly degraded (9, 13, 14). Although Shy1 is well conserved among eukaryotes, it remains to be determined whether Coa2 orthologs exist in metazoans.
To increase our understanding of these stages of CcO maturation, we analyzed the conserved mitochondrial protein Pet117, which has previously been implicated in CcO assembly (15, 16) but whose role was unknown. Here we show that Pet117 is a small, IM-associated protein residing in the mitochondrial matrix that is required for the stable oligomerization of Cox15, suggesting that Pet117 aids in coupling heme a synthase activity to the process of CcO maturation. We demonstrate that Pet117 and Cox15 physically interact and that this interaction persists in the absence of Cox15 catalytic function and in the absence of heme a yet is disrupted by deletion of the 20-amino acid region in Cox15 connecting its two heme-binding domains. We found that Pet117 and Cox15 interact in the absence of the late stage CcO assembly factor Shy1, yet their interaction is severely diminished in the absence of the early CcO assembly factor Mss51. These results suggest that Pet117 is an important assembly factor for linking the vital process of heme a biosynthesis to CcO biogenesis.
Results
Cells Lacking Pet117 Are Respiration-compromised Due to a CcO Defect
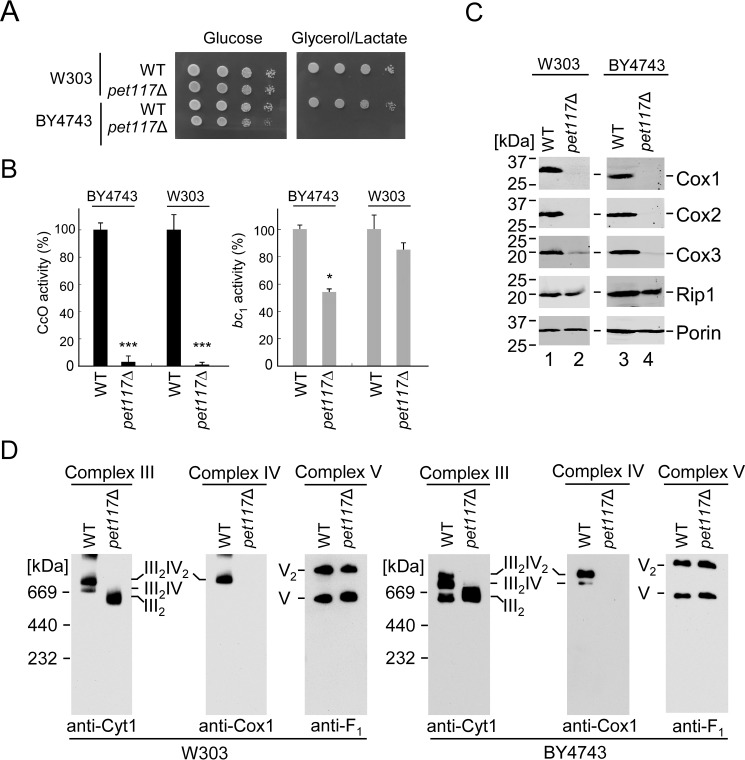
In the attempt to elucidate the role of Pet117 in CcO assembly suggested in the report by McEwen et al. (15), we first examined CcO biogenesis in yeast cells lacking Pet117. To confirm the connection to CcO assembly, PET117 was deleted in two yeast genetic backgrounds, W303 and BY4743. In each case, the pet117Δ strain showed normal growth on fermentable glucose-containing medium but failed to propagate on respiratory medium containing glycerol and lactate (Fig. 2A). Consistent with this respiratory growth defect, mitochondria of Pet117-deficient cells were devoid of CcO-specific activity compared with mitochondria from the isogenic WT cells (Fig. 2B). The activity of ubiquinol-cytochrome c oxidoreductase (cytochrome bc1 complex; Complex III) was ∼60% of WT in the BY4743 background and remained unaffected in W303 cells. Steady-state levels of the core CcO subunits Cox1, Cox2, and Cox3 were dramatically decreased in pet117Δ mitochondria, whereas levels of the cytochrome bc1 complex component Rip1 were unaffected (Fig. 2C).
FIGURE 2.
Pet117 is essential for assembly and function of CcO. A, respiratory growth test at 30 °C of WT and pet117Δ cells in two different genetic backgrounds, BY4743 and W303. Cells were dropped onto yeast extract-peptone medium containing either 2% glucose or 2% glycerol/lactate after normalization and serial dilution of cultures grown in 2% galactose and 0.1% glucose. Results are shown for one experiment, representative of three independent experiments (biological replicates). B, CcO and bc1 enzymatic activity in mitochondria from the above cells determined by spectrophotometric cytochrome c oxidation and reduction assays. Results are presented as a percentage of WT activity. Bars, average and S.D. (error bars) of three technical replicates from one experiment representative of three independent experiments (biological replicates). Asterisks indicate a statistically significant difference compared with WT (*, p < 0.05; ***, p < 0.001). C, steady-state levels of representative subunits of respiratory Complexes IV (Cox1–Cox3) and III (Rip1) with loading control (Porin) in mitochondria of cells described in A analyzed by SDS-PAGE immunoblotting with appropriate antibodies. The positions of molecular mass markers are indicated to the left of the immunoblots. Results are shown for one experiment, representative of three independent experiments (biological replicates). D, BN-PAGE analysis of respiratory complexes in mitochondria of cells described in A. Complexes III, IV, and V were visualized by immunoblotting for Cyt1, Cox1, and Atp2 (F1), respectively, with Complex V serving as the loading control. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots.
Blue native (BN)-PAGE analysis of digitonin-extracted respiratory complexes from these mitochondria revealed that the dimeric and monomeric forms of Complex V as well as Complex II are intact in pet117Δ cells, whereas CcO/bc1 supercomplexes (of the stoichiometries III2IV2 and III2IV found in yeast) were dramatically impaired due to the loss of CcO (Fig. 2D) (data not shown). Pet117-deficient cells accumulated dimeric Complex III in both genetic backgrounds (Fig. 2D), which is the expected result of the loss of Complex IV. Altogether, these results confirm that Pet117 is required for respiration and that its depletion primarily affects CcO.
Pet117 Is a Matrix Protein Peripherally Associated with the Mitochondrial IM
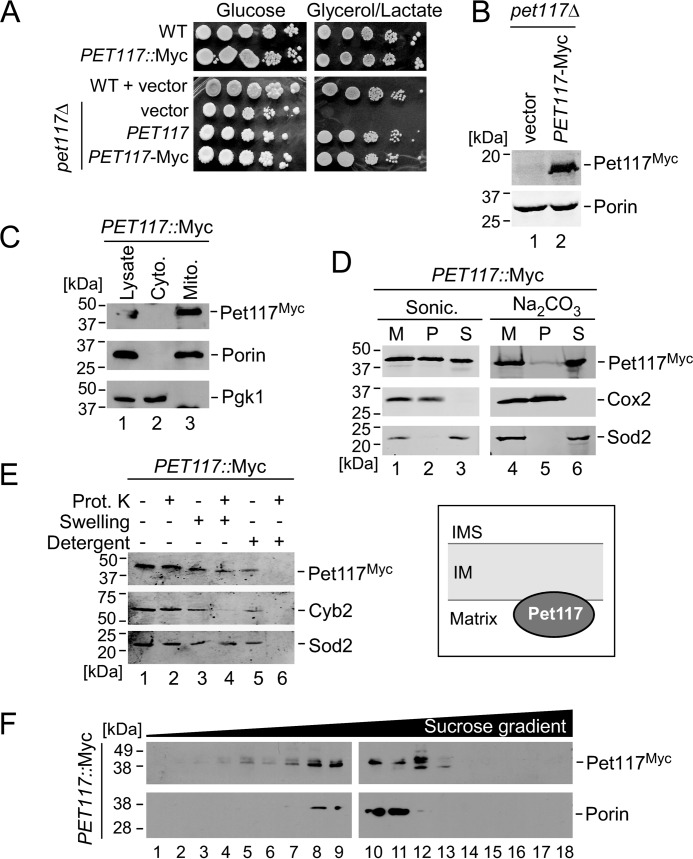
To permit visualization of the Pet117 protein and enable localization studies, chromosomal and plasmid-borne variants of Pet117 were generated bearing the C-terminal Myc13 and Myc1 epitope tags, respectively. As judged by their robust respiratory growth, both variants are expressed and functional (Fig. 3, A and B). Subcellular fractionation analysis confirmed the exclusive localization of Pet117-Myc in purified mitochondria (Fig. 3C). When isolated mitochondria were subjected to sonication, Pet117 was found both in the insoluble fraction containing mitochondrial membranes and in the soluble mitochondrial fraction (Fig. 3D). Incubation of mitochondria with sodium carbonate at high pH resulted in recovery of the majority of the Pet117 protein in the soluble fraction, suggesting that Pet117 is only loosely associated with mitochondrial membranes, probably through a hydrophobic association with either the membrane itself or a membrane-anchored protein (Fig. 3D).
FIGURE 3.
Pet117 resides in the mitochondrial matrix, where it is peripherally associated with the IM. A, respiratory growth test of WT and PET117::Myc strains (top) and WT and pet117Δ cells expressing vector control, untagged PET117, and PET117-Myc (bottom). Cells were handled and tested as described in the legend to Fig. 2A, except plates on the bottom contained synthetic medium to maintain plasmid selection. Results are shown for one experiment, representative of three independent experiments (biological replicates). B, expression of Pet117-Myc analyzed by SDS-PAGE immunoblotting with anti-Myc antibody and anti-Porin antibody as a loading control. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. C, Pet117 subcellular localization analysis employing the tagged strain described above. The whole cell lysate, cytosol fraction (cyto.), and isolated mitochondria (mito.) were analyzed by SDS-PAGE immunoblotting for Myc (Pet117-Myc), Porin (mitochondria control), and Pgk1 (cytosol control). Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. D, analysis of Pet117 membrane association using the tagged strain described above. Purified mitochondria were treated with either sonication (sonic.) or sodium carbonate and centrifuged to separate soluble and insoluble fractions. SDS-PAGE immunoblotting shows the untreated mitochondria (M) and the pellets (P) and supernatants (S) following treatment and centrifugation. Cox2 and Sod2 were used as membrane-bound and soluble protein controls, respectively. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. E, analysis of Pet117 submitochondrial compartmentalization using the tagged strain described above. Isolated mitochondria were treated with proteinase K (prot. K) to degrade exposed proteins either alone, after exposure to a hypotonic solution to disrupt the outer mitochondrial membrane (swelling), or after use of 0.5% dodecyl maltoside (detergent) to disrupt the mitochondrial IM. Treated samples and untreated controls were subjected to SDS-PAGE immunoblotting followed by antibody detection of Pet117, the intermembrane space protein Cyb2, and the matrix protein Sod2. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. Localization results are depicted in the graphic to the right of the blots. F, high-velocity sucrose density gradient (12–50%) fractionation of digitonin-solubilized complexes from mitochondria of PET117::Myc cells. Fractions collected following ultracentrifugation were analyzed by SDS-PAGE immunoblotting with anti-Myc (to detect the distribution of Pet117 complexes) and anti-Porin (to detect the 440-kDa Porin complex for size comparison) antibodies. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots.
To determine whether Pet117 resides in the intermembrane space or the matrix, we tested the susceptibility of mitochondria from cells expressing Pet117-Myc to exogenously added proteinase K under different conditions (Fig. 3E). Pet117 was protected from proteinase K-mediated proteolysis in both intact organelles (Fig. 3E, lane 2) and mitochondria with outer membranes disrupted by hypotonic treatment (Fig. 3E, lane 4). This behavior is similar to that of the matrix-localized manganese superoxide dismutase (Sod2) and unlike the behavior of the intermembrane space protein Cyb2, which becomes proteinase K-accessible upon hypotonic swelling of mitochondria. To exclude the possibility that the resistance of Pet117-Myc to proteinase K under these conditions is due to an inherent stability of the protein, mitochondria were also pretreated with the detergent dodecyl maltoside before the addition of proteinase K to disrupt the IM, which resulted in degradation of Pet117-Myc by the proteinase (Fig. 3E, lane 6). These results are consistent with Pet117 residing in the matrix subcompartment of mitochondria, where it is peripherally associated with the matrix side of the IM.
To determine whether Pet117 is part of a larger IM-bound protein complex, mitochondria from cells expressing Pet117-Myc were solubilized with the mild, non-ionic detergent digitonin and subjected to high-velocity density gradient fractionation. Whereas the Myc13-tagged Pet117 migrates at ∼40 kDa, the fractionation analysis revealed that Pet117-Myc migrates in the size range between ∼200 and 450 kDa (Fig. 3F), indicating its association with larger protein complexes.
Pet117 Functions at a Later Stage of Cox1 Maturation before Hemylation and Is Important for Oligomerization of Heme a Synthase
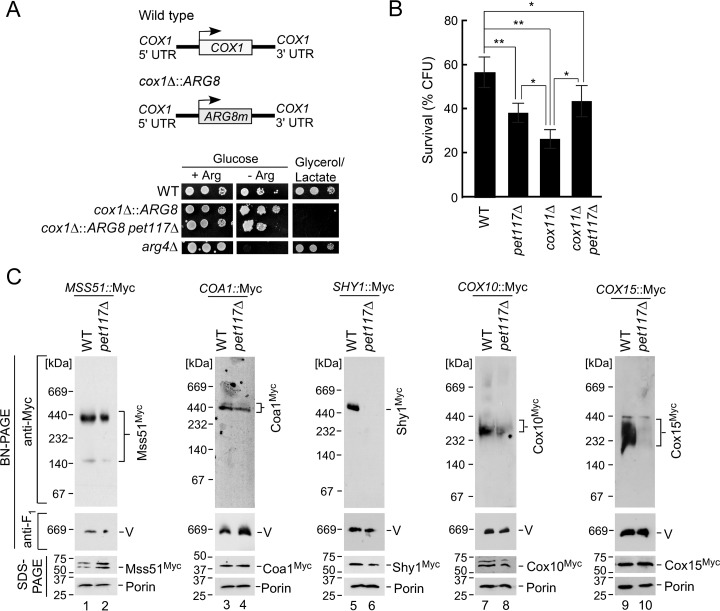
The results described above corroborate previous reports (15, 16) indicating a causal link between Pet117 function and CcO assembly. The study by Barrientos et al. (16) also suggested that Pet117 is likely to be involved in an assembly step pertaining to the core CcO subunit Cox1. To determine which of the many Cox1-centric assembly stages Pet117 engages in, we first tested whether pet117Δ cells have compromised Cox1 synthesis by examining their translational activity at the COX1 locus using a mitochondrial ARG8 genetic reporter strain (17). This strain lacks the authentic nucleus-encoded ARG8 gene encoding the mitochondrial matrix enzyme acetylornithine aminotransferase and instead contains a variant of the ARG8 gene that replaces the endogenous COX1 open reading frame in mitochondrial DNA (cox1Δ::ARG8; Fig. 4A). In these cox1Δ::ARG8 cells, the expression of Arg8 is therefore controlled by all of the regulatory features (such as the 5′- and 3′-untranslated regions) of the COX1 locus, whose translational activity can be directly scored by assessing the growth of the strain in medium lacking arginine. Deletion of PET117 in this strain resulted in a partial growth defect in medium lacking arginine, which is probably the result of translational feedback attenuation due to stalled CcO assembly in pet117Δ cells (16), but the cells were still able to grow in the absence of exogenous arginine, indicating that Pet117 is not essential for Cox1 translation (Fig. 4A). This result is consistent with the study by Barrientos et al. (16), who observed a decrease in Cox1 translation in the absence of Pet117, which was alleviated by a second mutation that deregulates the Cox1 translational feedback system.
FIGURE 4.
Pet117 is not required for translation or early maturation of Cox1 but is essential for CcO maturation before formation of the hemylated Shy1-containing assembly intermediate. A, COX1 translation was tested in the absence of PET117 by deleting the PET117 gene in an arg8Δ strain in which ARG8 replaces the COX1 coding sequence (cox1Δ::ARG8; depicted in the graphic on the top). Cells were cultured in synthetic medium containing 2% glucose and 0.2× arginine (4 mg/liter), normalized, serially diluted, dropped onto synthetic medium containing 2% glucose with (+ Arg) or without (− Arg) 1× arginine and synthetic medium containing 2% glycerol/lactate, and grown at 30 °C. The arg4Δ strain was used as a control. Results are shown for one experiment, representative of three independent experiments (biological replicates). B, hydrogen peroxide sensitivity of WT, pet117Δ, cox11Δ, and cox11Δ pet117Δ cells. Cells were cultured in yeast extract-peptone medium containing 2% glucose at 30 °C to mid-log phase and then normalized and acutely treated with 1 mm hydrogen peroxide for 1 h at 30 °C. Cultures were diluted to 300 cells and plated onto yeast extract-peptone medium containing 2% glucose to test for viable colony-forming units after 48 h of growth. Bars, indicate the average and S.D. (error bars) of 4–7 biological replicates. Asterisks indicate a statistically significant difference by one-way analysis of variance (*, p < 0.05; **, p < 0.01). C, BN-PAGE analysis of digitonin-solubilized complexes containing chromosomally Myc13-tagged CcO assembly factors Mss51, Coa1, Shy1, Cox10, and Cox15 in WT versus the respective pet117Δ strains. Anti-Myc immunoblotting shows complexes containing each assembly factor, and anti-Atp2 (F1) immunoblotting was used to detect Complex V as the loading control. The corresponding steady-state expression of each assembly factor detected by SDS-PAGE immunoblotting with Porin as a loading control is shown below each native gel. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots.
A quantitative functional assay related to the presence of heme a in stalled CcO assembly intermediates was also employed to obtain information on which step of the Cox1 maturation sequence involves Pet117. This assay tested the survival rate of cultures after 1 h of exposure to 1 mm hydrogen peroxide (measured as the number of viable colonies, or colony-forming units, able to grow on a glucose plate following this peroxide stress). We and others have previously used this type of proxy assay to show that deletion of certain CcO assembly factors, including Cox11, Sco1, and Coa2, renders yeast cells sensitive to an acute hydrogen peroxide stress (11, 13, 18, 19), probably due to formation of a pro-oxidant heme a3-containing Cox1 assembly intermediate (18). In this assay, cells lacking Pet117 exhibited a moderate decrease in the ability to form viable colonies upon treatment with hydrogen peroxide compared with similarly treated WT cells, whereas the viability of cells lacking COX11 was more drastically impaired (Fig. 4B). Significantly, the tolerance of the hydrogen peroxide-sensitive cox11Δ strain to peroxide stress was improved to the same level of the pet117Δ cells by the deletion of PET117 in the cox11Δ strain (Fig. 4B). These data suggest that Pet117 functions upstream of Cox11 in the CcO biogenesis process, and that loss of Pet117 may partially prevent the formation of the pro-oxidant assembly intermediate.
As mentioned previously, Cox1 progresses through multiple assembly intermediates during the complicated process of CcO assembly (Fig. 1). To pinpoint the step at which Pet117 functions, the stability of protein complexes pertinent to each stage of Cox1 assembly was monitored by BN gel electrophoresis (Fig. 4C). Formation of both the Mss51-containing, early stage Cox1 assembly intermediate and the downstream assembly complex containing the Coa1 assembly factor was unaffected in mitochondria from pet117Δ cells. In contrast, the late stage, Shy1-containing Cox1 assembly complex was not detectable in the pet117Δ mutant, indicating that Pet117 acts near this later stage of Cox1 maturation. We therefore examined the two protein complexes required for progression of the Coa1-containing intermediate to the hemylated Shy1-containing intermediate: the complexes of heme o synthase (Cox10) and heme a synthase (Cox15) (11, 12). BN-PAGE analysis revealed that whereas the Cox10 oligomer remained largely unaffected in pet117Δ mitochondria, a marked impairment was observed in the Cox15 complex, without any decrease in the steady state levels of Cox15 protein (Fig. 4C). Because Cox15 functions upstream of the Shy1 assembly intermediate, these results suggest that impaired Cox15 oligomerization may be one of the primary consequences of the loss of Pet117.
Pet117 Promotes CcO Assembly by Stabilizing Cox15 Oligomers
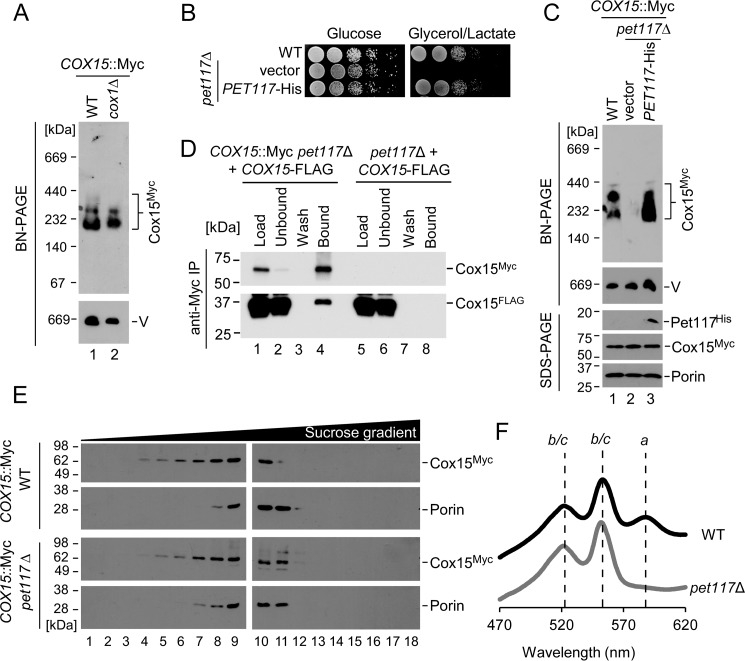
The observed impairment of Cox15 oligomers in pet117Δ cells analyzed by BN-PAGE is unusual because even Cox1-depleted cells retain heme a synthase complexes (Fig. 5A) (10, 11). Therefore, it was hypothesized that Pet117 might be directly involved in the process of Cox15 oligomerization or in maintaining the stability of Cox15 complexes. Expression of Pet117-His in the pet117Δ mutant restored its respiratory competence (Fig. 5B) and reinstated the high-mass Cox15 complexes observed by BN-PAGE (Fig. 5C), reflecting the specificity of the effect of Pet117 on Cox15.
FIGURE 5.
Pet117 is essential for stabilization of Cox15 oligomers and for heme a synthesis. A, BN-PAGE analysis of Cox15 complexes in the COX15::Myc strain in WT and in the absence of Cox1, analyzed as in Fig. 4C. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. B, respiratory growth test of cells expressing the Pet117-His plasmid, as in Fig. 2A, except plates contained synthetic medium. Results are shown for one experiment, representative of three independent experiments (biological replicates). C, BN-PAGE analysis of Cox15 complexes in the COX15::Myc strain in WT and in pet117Δ cells transformed with vector control or Pet117-His plasmid, analyzed as in Fig. 4C with appropriate antibodies. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. D, co-immunoadsorption of differentially tagged variants of Cox15 in the absence of Pet117. Digitonin-solubilized mitochondrial lysates from pet117Δ or COX15::Myc pet117Δ cells expressing Cox15-FLAG were incubated with anti-Myc-agarose beads before SDS-PAGE immunoblotting to show mitochondrial lysates before (Load) and after (Unbound) the incubation with affinity resin, the whole fraction of protein precipitated from the final wash (Wash), and half of the eluate (Bound). Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. E, high-velocity density gradient fractionation of digitonin-solubilized complexes from mitochondria of COX15::Myc cells in the presence and absence of Pet117, analyzed as in Fig. 3F. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. F, heme-pyridine redox difference spectra of mitochondria from WT and pet117Δ cells. Spectra are offset for clarity, and lines are drawn as a guide to the eye through absorption maxima arising from the overlapping heme b and c absorptions and from heme a. Results are shown for one experiment, representative of three independent experiments (biological replicates). IP, immunoprecipitation.
Because heme a synthase oligomers contain multiple copies of Cox15 (8), we tested whether loss of Pet117 disrupted Cox15-Cox15 homotypic interaction. To this end, mitochondria from cells expressing two functional, differentially tagged variants of heme a synthase used previously (8), Cox15-Myc and Cox15-FLAG, were used in co-immunoprecipitation (co-IP) experiments with anti-Myc-agarose beads (Fig. 5D). In this figure, fractions from the mitochondria containing both Cox15-Myc and Cox15-FLAG are shown in lanes 1–4, whereas fractions from the accompanying control IP experiment in which mitochondria contained only Cox15-FLAG are shown in lanes 5–8. The lack of Cox15-FLAG protein in lane 8 demonstrates that Cox15-FLAG does not adsorb nonspecifically to the affinity beads in the absence of Cox15-Myc, suggesting that the Cox15-FLAG present in lane 4 is bound to the beads specifically through its physical interaction with Cox15-Myc.
This result suggests that Cox15-Cox15 interaction remains robust in the absence of Pet117 (Fig. 5D), which is somewhat surprising due to the marked attenuation of Cox15 oligomers observed in the absence of Pet117 by BN-PAGE (Figs. 4C and 5C). BN-PAGE conditions are relatively harsh, and destabilization of labile protein complexes by native electrophoresis is not unprecedented (20–22); therefore, it was hypothesized that Cox15 complexes in the pet117Δ mutant might have reduced stability that is more evident under the conditions of BN-PAGE than under the conditions of the co-IP experiment. To test this, Cox15 oligomers from WT and pet117Δ cells were examined by the more gentle technique of high-velocity density gradient fractionation. Consistent with the hypothesis that Cox15 oligomers are less stable in the absence of Pet117, leading to their disruption under BN-PAGE but not co-IP conditions, Cox15 complexes observed by density gradient fractionation were nearly equally abundant in pet117Δ mitochondria compared with WT, and they fractionated at the expected molecular mass range of ∼200–350 kDa (Fig. 5E) (data not shown). Collectively, these data indicate that Pet117 is required for the stabilization, but not the formation, of heme a synthase oligomers.
To assess the functional significance of this stabilization in vivo, heme a levels were examined in mitochondria isolated from WT versus pet117Δ cells. Redox difference spectra of lysates from these mitochondria show a dramatic decrease in heme a in pet117Δ cells (Fig. 5F). This result suggests that the stability of Cox15 oligomers in vivo is affected severely enough to result in the loss of heme a synthase activity, which is probably the cause of the observed respiratory growth defect for pet117Δ cells.
Pet117 Physically Interacts with Cox15
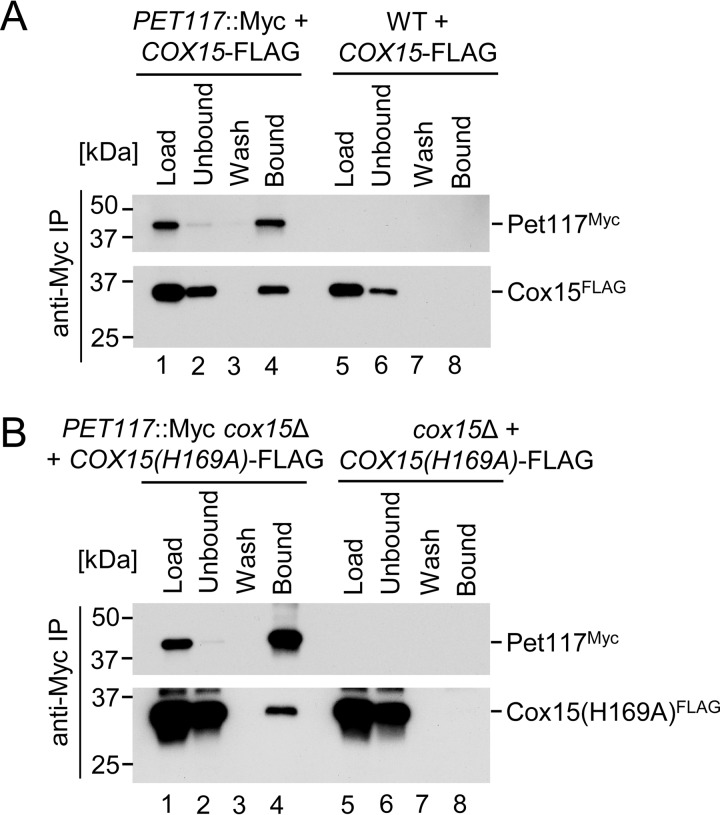
To gain further insight into the Cox15-Pet117 functional interaction and its significance, a series of co-IP experiments was performed. Mitochondrial lysates from cells bearing chromosomally tagged Pet117-Myc (or WT control) expressing vector-borne Cox15-FLAG were exposed to anti-Myc-agarose beads to determine whether Cox15 and Pet117 physically interact. Immunoadsorption of Pet117-Myc to the beads led to the specific co-IP of Cox15-FLAG, suggesting physical interaction between the two proteins (Fig. 6A).
FIGURE 6.
Pet117 and Cox15 physically interact, and their interaction is not strictly dependent upon Cox15 catalytic activity. A, co-immunoadsorption of differentially tagged Pet117 and Cox15. Digitonin-solubilized mitochondrial lysates from WT or PET117::Myc cells expressing Cox15-FLAG were analyzed as in Fig. 5D. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. B, co-immunoadsorption of differentially tagged Pet117 and a catalytically inactive variant of Cox15. Digitonin-solubilized mitochondrial lysates from cox15Δ or PET117::Myc cox15Δ cells expressing Cox15(H169A)-FLAG were analyzed as in Fig. 5D. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. IP, immunoprecipitation.
To assess whether this observed Cox15-Pet117 interaction is dependent upon active heme a biosynthesis, the interaction of Pet117-Myc with the H169A catalytically impaired variant of Cox15-FLAG (8) was examined in the absence of WT Cox15. This experiment yielded successful co-IP of the catalytically inactive form of Cox15 upon Pet117-Myc immunoadsorption (Fig. 6B). The amount of total protein that co-purified with Pet117-Myc was reduced for Cox15(H169A)-FLAG (Fig. 6B) compared with that of the WT Cox15-FLAG (Fig. 6A), suggesting that the impairment in Cox15 catalytic activity may induce some destabilization of the interaction with Pet117, but neither catalytic activity nor the presence of heme a is a strict requirement for the interaction.
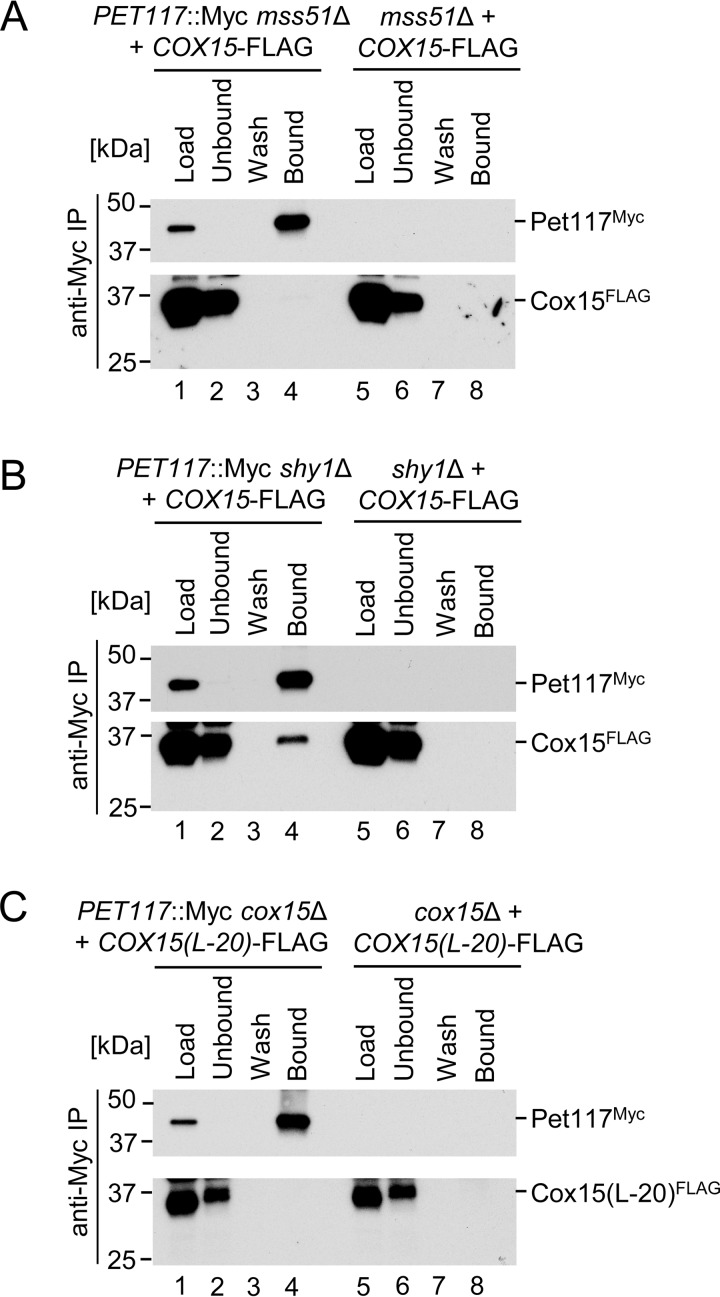
To assess whether the Pet117-Cox15 interaction is direct or instead mediated by other proteins, such as the multiprotein Cox1-containing CcO assembly intermediates, co-IP experiments were performed in the absence of the Mss51 and Shy1 assembly factors, which were chosen because the mss51Δ and shy1Δ deletions block Cox1 synthesis/assembly at early and late stages, respectively (2, 7, 12). The Cox15-Pet117 interaction was drastically attenuated in mitochondria from mss51Δ cells (Fig. 7A) with only marginal amounts of co-purified Cox15-FLAG observed upon extended exposures of the immunoblot to film (data not shown). In contrast, loss of Shy1 had no effect on co-IP of Cox15-FLAG with the anti-Myc-agarose-bound Pet117-Myc (Fig. 7B).
FIGURE 7.
Pet117-Cox15 physical interaction is dependent upon the presence of Mss51 but not Shy1 and is lost in an oligomerization-deficient Cox15 mutant. A, co-immunoadsorption of differentially tagged Pet117 and Cox15 in the absence of Mss51. Digitonin-solubilized mitochondrial lysates from mss51Δ or PET117::Myc mss51Δ cells expressing Cox15-FLAG were analyzed as in Fig. 5D. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. B, co-immunoadsorption of differentially tagged Pet117 and Cox15 in the absence of Shy1. Digitonin-solubilized mitochondrial lysates from shy1Δ or PET117::Myc shy1Δ cells expressing Cox15-FLAG were analyzed as in Fig. 5D. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots. C, co-immunoadsorption of differentially tagged Pet117 and a variant of Cox15 lacking the matrix-exposed 20-amino acid region linking the two heme-binding domains of Cox15. Digitonin-solubilized mitochondrial lysates from cox15Δ or PET117::Myc cox15Δ cells expressing Cox15(L-20)-FLAG were analyzed as in Fig. 5D. Results are shown for one experiment, representative of three independent experiments (biological replicates). The positions of molecular mass markers are indicated to the left of the immunoblots.
The final co-IP experiment tested whether the matrix-exposed, unstructured linker region of Cox15 might serve as the Cox15-Pet117 binding interface, in light of the data shown in Fig. 3 indicating that Pet117 is a mitochondrial matrix protein loosely associated with the IM. Moreover, loss of this 20-amino acid-long region results in impaired Cox15 oligomerization and heme a synthase activity (8), which mimics the consequences of PET117 deletion. The Cox15-Pet117 interaction was undetectable in mitochondria from cells expressing the Cox15(L-20)-FLAG variant of heme a synthase lacking this unstructured linker region (Fig. 7C), underscoring the importance of this region for physical association between Pet117 and heme a synthase.
Discussion
CcO assembly is a complex process about which much has been discovered, yet many details remain unclear, including the mechanism of heme a biosynthesis and transfer to Cox1 as well as the roles of assembly factors in that process. In this study, we have characterized the previously unexplored CcO assembly factor Pet117 and provide evidence for its role in coupling heme a biosynthesis to CcO maturation.
The present results support the mitochondrial residency of Pet117 that has been previously established through the observation of the respiration-deficient phenotype of pet117Δ cells (15) and by high-throughput proteomic studies (23) and further identify Pet117 as a resident of the mitochondrial matrix that is peripherally associated with the IM. The size distribution of Pet117-containing complexes identified by density gradient fractionation (∼200–450 kDa) overlaps with that of Cox15 complexes (∼200–350 kDa), implying that Pet117 may be both a component of Cox15 complexes, as suggested by co-IP results, and a member of larger complexes lacking Cox15.
Because Pet117 is not required for Cox1 translation and can be placed at a later Cox1 maturation stage before hemylation, with its absence severely compromising Cox15 oligomerization, Pet117 bears some resemblance to the CcO assembly factor Coa2. Both proteins are small IM-associated matrix proteins required for the stability and/or oligomerization of Cox15 (9, 11, 13). However, Coa2 is also important for Cox10 stability and/or oligomerization, and although mammalian cells contain a Pet117 ortholog (NM_001164811), the evolutionary conservation of Coa2 remains unclear. The Cox15-specific effect of Pet117 is in line with previous reports indicating that the oligomerization processes of heme o and heme a synthases are not controlled in the same manner (9, 11).
The dramatic impairment in heme a levels observed in pet117Δ cells in the present study suggests that the functional consequence of the destabilization of Cox15 oligomers is the loss of heme a biosynthetic activity (24). This heme a biosynthetic defect as well as the defects observed in CcO assembly and activity resulting from loss of Cox15 complex stability in the absence of Pet117 are consistent with our previous study of Cox15 that linked its multimerization to heme a biosynthesis and/or transfer of heme a to Cox1 during CcO assembly (8). The phenotype of pet117Δ cells thus underscores the importance of Cox15 multimerization for Cox15 function.
This phenotype and the observed physical interaction between Pet117 and Cox15 together suggest that Pet117 serves as a stabilizing member of the Cox15 complex required to synthesize heme a and/or transfer heme a from Cox15 to the maturing Cox1-containing CcO assembly intermediate. It is also possible that Pet117 has a stabilizing association with a CcO assembly intermediate. However, because Cox15 oligomers stably form in the absence of successful CcO maturation (Fig. 5A) (10, 11) yet are destabilized in the absence of Pet117 (Figs. 4C and 5C), it is unlikely that the sole function of Pet117 is to stabilize a newly synthesized Cox1-containing assembly intermediate.
Some additional insight into the process of CcO assembly can be gained from the analysis of the Pet117-Cox15 association. Consistent with the interaction occurring upstream of the Shy1-containing CcO assembly intermediate, loss of Shy1 does not affect Pet117-Cox15 association. However, loss of Cox1 translation due to MSS51 deletion dramatically attenuates Cox15-Pet117 physical interaction, a somewhat unexpected finding because Cox15 oligomerization is not affected by either loss of Cox1 or attenuation of Cox1 translation (Fig. 5A) (10, 11). These data suggest that Cox1-containing early assembly intermediates are probably required to either nucleate or stabilize the Pet117-Cox15 association. Additionally, whereas the catalytically inactive but oligomerization-competent H169A variant of Cox15 was able to interact with Pet117, the Cox15(L-20) variant defective in forming Cox15 oligomers (8) is compromised in its ability to associate with Pet117. This latter result implicates the linker region of Cox15 as a potential association interface between Cox15 and Pet117, although its lack of sequence conservation (8) suggests that the salient feature of the Cox15(L-20) variant that disrupts its association with Pet117 may be its inability to oligomerize.
Structural modeling predicts that Pet117 is composed of several interrupted α helices, one of which is predicted to serve as a transmembrane helix. However, the results of the present study suggest that this region is unlikely to be a transmembrane segment, because Pet117 is not integral to the IM. This helix might instead comprise a hydrophobic interaction site for Cox15. An earlier study by Broadley et al. (25) predicted that Pet117 might contain a degenerate variant of the tetratricopeptide repeats motif known to be involved in protein-protein interactions. However, mutations changing conserved residues in this putative motif or deletion of the motif resulted in no appreciable defect in Pet117 function (data not shown), indicating that this element is unlikely to play a significant role in Pet117-Cox15 functional association.
The present study of the evolutionarily conserved yet underexplored CcO assembly factor Pet117 reveals its critical role in coupling heme a biosynthesis to CcO maturation and further underscores the importance of Cox15 oligomerization for enzymatic function. The results suggest two potential models for the function of Pet117: 1) Pet117 enables heme a synthase activity by conferring stability to the Cox15 oligomer, and/or 2) Pet117 is involved in facilitating subsequent heme a transfer to maturing Cox1. Although the results of the present study suggest the likelihood of the former scenario, it is possible that Pet117 could perform both of these roles. Further studies are warranted to discern between these possibilities.
Experimental Procedures
Yeast Strains, Plasmids, Media, and Cell Culture
Yeast strains used in this study (listed in Table 1) are derivatives of the genetic background W303 or BY4743. The pet117Δ, cox11Δ pet117Δ, MSS51::Myc pet117Δ, COA1::Myc pet117Δ, SHY1::Myc pet117Δ, COX10::Myc pet117Δ, COX15::Myc pet117Δ, PET117::Myc cox15Δ, PET117::Myc mss51Δ, and PET117::Myc shy1Δ strains were made by in vivo homologous recombination using PCR-amplified gene-specific knock-out cassettes containing KanMX4 or URA3MX selection markers surrounded by sequences with homology to the chromosomal regions upstream and downstream of the coding sequence for the deleted gene. Chromosomally tagged strains contain the Myc13 epitope.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference/Source |
|---|---|---|
| W303-1B | MATα ade2-1 can1-100 his3-11,15 leu2-3, 112 trp1-1 ura3-1 | ATCC |
| BY4743 | Diploid cross of BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) | Open Biosystems |
| W303–1B pet117Δ | MATa, ade2-1 can1-100 his3-11, 15 leu2-3, 112 trp1-1, ura3-1 pet117Δ::URA3MX | This study |
| BY4743 pet117Δ | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 LYS2/lys2Δ0 pet117::KanMX4/pet117::KanMX4 | Invitrogen |
| PET117::Myc | MATa ade2 can1 his3 leu2 ura3 trp1Δ PET117-Myc13::TRP1 | D. Winge |
| cox1Δ::ARG8 | MATα lys2 leu2-3, 112 arg8::hisG, ura3-52 [cox1Δ::ARG8m] | Ref. 17 |
| cox1Δ::ARG8 pet117Δ | MATα lys2 leu2-3, 112 arg8::hisG, ura3-52 [cox1Δ::ARG8m] pet117Δ::URA3MX4 | D. Winge |
| DTY 833 (arg4Δ) | MATa ade1Δ arg4Δ aro2Δ his7Δ lys5Δ ura2Δ | D. Winge |
| cox11Δ | MATa ade2-1 his3-11, 15 leu2-3, 112 ura3-1 canr galr cox11::HIS3 | Ref. 26 |
| cox11Δ pet117Δ | MATa ade2-1 his3-11, 15 leu2-3, 112 ura3-1 canr galr cox11::HIS3 pet117Δ::KanMX4 | This study |
| MSS51::Myc | MATa ade2-1 his3-1,15 leu2-3, 112 trp1Δ ura3-1 COA1-HA3::TRP1 MSS51-Myc13::HIS3MX6 | Ref. 27 |
| MSS51::Myc pet117Δ | MATa ade2-1 his3-1, 15 leu2-3, 112 trp1Δ ura3–1 COA1-HA3::TRP1 MSS51-Myc13::HIS3MX6 pet117Δ::URA3MX4 | This study |
| COA1::Myc | MATa ade2-1 his3-1, 15 leu2-3, 112 trp1Δ ura3-1 COA1-Myc13::HIS3MX6 | Ref. 27 |
| COA1::Myc pet117Δ | MATa ade2-1 his3-1, 15 leu2-3, 112 trp1Δ ura3-1 COA1-Myc13::HIS3MX6 pet117Δ::URA3MX4 | This study |
| SHY1::Myc | MATa ade2-1 his3-1, 15 leu2–3, 112 trp1Δ ura3-1 SHY1-Myc13::TRP1 | Ref. 27 |
| SHY1::Myc pet117Δ | MATa ade2-1 his3-1, 15 leu2-3, 112 trp1Δ ura3-1 SHY1-Myc13::TRP1 pet117Δ::URA3MX4 | This study |
| COX10::Myc | MATa ade2-1 his3-1, 15 leu2-3, 112 trp1Δ ura3-1 COX10-Myc13::TRP1 | Ref. 9 |
| COX10::Myc pet117Δ | MATa ade2-1 his3-1, 15 leu2-3, 112 trp1Δura3-1 COX10-Myc13::TRP1 pet117Δ::URA3MX4 | This study |
| COX15::Myc | MATa ade2-1 his3-11, 15 leu2-3, 112 trp1Δ ura3-1 COX15-Myc13::TRP1 | Ref. 9 |
| COX15::Myc pet117Δ | MATa ade2-1 his3-11, 15 leu2–3, 112 trp1Δ ura3-1 COX15-Myc13::TRP1 pet117Δ::URA3MX | This study |
| COX15::Myc cox1Δ | MATα lys2 leu2-3, 112 arg8::hisG, ura3-52 [cox1Δ::ARG8m] COX15-Myc13::KanMX4 | E. Hegg |
| PET117::Myc cox15Δ | MATa ade2 can1 his3 leu2 ura3 trp1Δ PET117-Myc13::TRP1 cox15Δ::KanMX4 | This study |
| PET117::Myc mss51Δ | MATa ade2 can1 his3 leu2 ura3 trp1Δ PET117-Myc13::TRP1 mss51Δ::KanMX4 | This study |
| PET117::Myc shy1Δ | MATa ade2 can1 his3 leu2 ura3 trp1Δ PET117-Myc13::TRP1 shy1Δ::KanMX4 | This study |
The plasmid-borne Pet117-Myc (containing the Myc1 epitope), Pet117-His (containing the His6 epitope), Pet117-FLAG (containing the FLAG1 epitope), and untagged Pet117 constructs were made by PCR amplification of PET117 from genomic DNA followed by restriction enzyme cloning into pRS415 or pRS426 vectors containing the MET25 promoter and CYC1 terminator. The pRS426 Cox15(L-20)-FLAG and pRS426 Cox15(H169A)-FLAG plasmids under control of the MET25 promoter and CYC1 terminator were made from the Cox15-FLAG plasmid (8) with use of the Q5 site-directed mutagenesis kit (New England BioLabs). The former construct lacks the 20 amino acid residues (ECKWIKNPVQAISLFKKLDN) that are predicted to connect helices IV and V of Cox15 (8). All constructs were confirmed by DNA sequencing.
Cells were cultured and handled in accordance with published protocols (28), and cells were transformed with plasmids via the lithium acetate method (29). Cells were cultured in either yeast extract-peptone-dextrose (Amresco) medium containing 2% glucose (dextrose) as the carbon source or yeast extract-peptone with 2% galactose and 0.1% glucose as the carbon source. To maintain plasmid selection, cells transformed with plasmids were cultured in Brent Supplement Mixture synthetic selective medium (Sunrise Science Products) containing 2% galactose and 0.1% glucose. Cell cultures were used for growth tests (described below), lysed for mitochondrial isolation and further analysis (described below), or lysed to generate whole-cell protein lysates as described previously (14).
Growth Tests
Respiratory growth tests were performed at 30 °C as described previously (8). Quantitative hydrogen peroxide sensitivity tests were also performed at 30 °C in accordance with the previously published protocol (22).
Mitochondrial Isolation and Assays
Whole mitochondria were isolated from yeast by established protocols (30). Total mitochondrial protein concentration was determined by the Bradford method using the Coomassie Plus assay kit (Thermo Scientific). CcO and cytochrome bc1 complex activity of isolated mitochondria was assessed as described previously (31, 32). Kaleidagraph version 4.1 (Synergy Software) was used to determine the statistical significance of activity assay results via unpaired t tests. Heme-pyridine redox difference spectra were determined from SDS-solubilized mitochondrial lysates according to the method of Berry and Trumpower (33). Blue native (BN)-PAGE separation of mitochondrial protein complexes solubilized with 1% digitonin was performed as described (8, 12) using 5–13% gradient polyacrylamide gels (self-made) or 3–12% gradient polyacrylamide gels (Life Technologies, Inc.). Pet117 localization experiments were performed by treating mitochondria that were purified by nycodenz gradient ultracentrifugation (34) with subsequent sonication, sodium carbonate, and proteinase K as described previously (35). Pet117 and Cox15 complex formation was analyzed by high-velocity density gradient fractionation as described previously (8, 12) using a continuous 12–50% sucrose gradient.
Co-immunoprecipitation
Immunoprecipitation of Cox15-Myc from COX15::Myc pet117Δ cells (or pet117Δ cells as a control) expressing Cox15-FLAG and of Pet117-Myc from PET117::Myc (or WT control), PET117::Myc cox15Δ (or cox15Δ), PET117::Myc mss51Δ (or mss51Δ), and PET117::Myc shy1Δ (or shy1Δ) cells expressing either Cox15-FLAG, Cox15(L-20)-FLAG, or Cox15(H169A)-FLAG was performed with mouse anti-c-Myc-agarose beads (sc-40 AC, Santa Cruz Biotechnology, Inc.). These immunoprecipitations followed previously described procedures (8, 36), except all buffers were made in TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.4) and beads were preincubated with 3 mg/ml BSA (Sigma-Aldrich) for ∼22 h before use. The mitochondrial lysis buffer and bead washing buffer additionally contained 1 mm PMSF and 0.2% (lysis) or 0.1% (wash) digitonin.
Immunoblotting
Following SDS-PAGE or BN-PAGE, proteins or protein complexes were transferred to nitrocellulose or PVDF membranes and blocked in 5% nonfat milk in PBS (with or without Tween 20). Membranes were incubated in the indicated primary antibodies and goat anti-mouse and anti-rabbit HRP-coupled secondary antibodies (sc-2005 and sc-2030, Santa Cruz Biotechnology). Protein bands were visualized by incubation of membranes with chemiluminescence reagents (Millipore and Thermo Scientific) and exposure to X-ray film (BioExpress and Phenix Research Products). The following primary antibodies were used in this study: mouse anti-Cox1 (ab110270, Abcam); mouse anti-Cox2 (ab110271, Abcam); mouse anti-Cox3 (ab110259, Abcam); mouse anti-Porin (459500, Invitrogen); mouse anti-c-Myc (11667149001, Roche Diagnostics); rabbit anti-c-Myc (sc-789, Santa Cruz Biotechnology); mouse anti-Pgk1 (PA5-28612, Invitrogen); mouse anti-His6 (MA1-21315, Thermo Scientific); rabbit anti-FLAG (F7425, Sigma-Aldrich); rabbit anti-F1 (reactive toward the Atp2 β subunit of F1F0 ATPase), kindly provided by Dr. Alexander Tzagoloff; rabbit anti-Sod2, kindly provided by Dr. Valeria Culotta; and rabbit anti-Cyb2, rabbit anti-Cyt1, and mouse anti-Rip1, kindly provided by Dr. Dennis Winge. Antibodies were all tested for reliability to ensure specificity of detection at the expected migration distances in gel electrophoresis.
Bioinformatic Analysis
The Phyre2 protein fold recognition server (37) was used for secondary structural prediction of the Pet117 protein in S. cerevisiae.
Author Contributions
J. L. F. and O. K. designed and coordinated the study, performed experiments, analyzed results, and wrote the manuscript. N. G. T., S. S., N. J. H., and E. M. G. performed experiments and analyzed results. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Drs. Dennis Winge (University of Utah), Valeria Culotta (Johns Hopkins University), Thomas Fox (Cornell University), Eric Hegg (Michigan State University), and Alexander Tzagoloff (Columbia University) for reagents. We thank Dr. Iryna Bohovych for help with the hydrogen peroxide sensitivity tests and acknowledge the expert technical assistance of Nataliya Zahayko.
This work was supported by National Institutes of Health Grants P30GM103335 (to the Nebraska Redox Biology Center), R01GM108975 (to O. K.), T32 GM107001-01A1, 5 P20 RR016461, and 8 P20 GM103499; Cottrell College Science Award 22643 (to J. L. F.) from the Research Corporation for Science Advancement; NSF-Major Research Instrumentation Grant CHE-1229559 (to J. L. F. as co-principal investigator); College of Charleston Summer Undergraduate Research with Faculty grants (to J. L. F., N. G. T., and N. J. H.); and Howard Hughes Medical Institute Grant 52007537 through the Pre-college and Undergraduate Science Education Program (to the College of Charleston). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CcO
- cytochrome c oxidase
- BN
- blue native
- IP
- immunoprecipitation
- IM
- inner membrane.
References
- 1.Hamza I., and Dailey H. A. (2012) One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta 1823, 1617–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soto I. C., Fontanesi F., Liu J., and Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 1817, 883–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H. J., Khalimonchuk O., Smith P. M., and Winge D. R. (2012) Structure, function and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalimonchuk O., and Rödel G. (2005) Biogenesis of cytochrome c oxidase. Mitochondrion 5, 363–388 [DOI] [PubMed] [Google Scholar]
- 5.Barros M. H., Carlson C. G., Glerum D. M., and Tzagoloff A. (2001) Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 492, 133–138 [DOI] [PubMed] [Google Scholar]
- 6.Barros M. H., Nobrega F. G., and Tzagoloff A. (2002) Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 277, 9997–10002 [DOI] [PubMed] [Google Scholar]
- 7.Mick D. U., Fox T. D., and Rehling P. (2011) Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swenson S., Cannon A., Harris N. J., Taylor N. G., Fox J. L., and Khalimonchuk O. (2016) Analysis of oligomerization properties of heme a synthase provides insights into its function in eukaryotes. J. Biol. Chem. 291, 10411–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bestwick M., Khalimonchuk O., Pierrel F., and Winge D. R. (2010) The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bareth B., Dennerlein S., Mick D. U., Nikolov M., Urlaub H., and Rehling P. (2013) The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol. Cell. Biol. 33, 4128–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalimonchuk O., Kim H., Watts T., Perez-Martinez X., and Winge D. R. (2012) Oligomerization of heme o synthase in cytochrome oxidase biogenesis is mediated by cytochrome oxidase assembly factor Coa2. J. Biol. Chem. 287, 26715–26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalimonchuk O., Bestwick M., Meunier B., Watts T. C., and Winge D. R. (2010) Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 30, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., and Winge D. R. (2008) Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Mol. Cell. Biol. 28, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalimonchuk O., Jeong M. Y., Watts T., Ferris E., and Winge D. R. (2012) Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem. 287, 7289–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McEwen J. E., Hong K. H., Park S., and Preciado G. T. (1993) Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 23, 9–14 [DOI] [PubMed] [Google Scholar]
- 16.Barrientos A., Zambrano A., and Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Martinez X., Broadley S. A., and Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalimonchuk O., Bird A., and Winge D. R. (2007) Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282, 17442–17449 [DOI] [PubMed] [Google Scholar]
- 19.Veniamin S., Sawatzky L. G., Banting G. S., and Glerum D. M. (2011) Characterization of the peroxide sensitivity of COX-deficient yeast strains reveals unexpected relationships between COX assembly proteins. Free Radic. Biol. Med. 51, 1589–1600 [DOI] [PubMed] [Google Scholar]
- 20.Dienhart M. K., and Stuart R. A. (2008) The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19, 3934–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claypool S. M., Whited K., Srijumnong S., Han X., and Koehler C. M. (2011) Barth syndrome mutations that cause tafazzin complex lability. J. Cell Biol. 192, 447–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohovych I., Donaldson G., Christianson S., Zahayko N., and Khalimonchuk O. (2014) Stress-triggered activation of the metalloprotease Oma1 involves its C-terminal region and is important for mitochondrial stress protection in yeast. J. Biol. Chem. 289, 13259–13272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schönfisch B., Perschil I., Chacinska A., Guiard B., Rehling P., Pfanner N., and Meisinger C. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U.S.A. 100, 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barros M. H., and Tzagoloff A. (2002) Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 516, 119–123 [DOI] [PubMed] [Google Scholar]
- 25.Broadley S. A., Demlow C. M., and Fox T. D. (2001) Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 7663–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carr H. S., George G. N., and Winge D. R. (2002) Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 277, 31237–31242 [DOI] [PubMed] [Google Scholar]
- 27.Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., and Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 29.Gietz R. D., and Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 30.Diekert K., de Kroon A. I., Kispal G., and Lill R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 31.Capaldi R. A., Marusich M. F., and Taanman J. W. (1995) Mammalian cytochrome-c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol. 260, 117–132 [DOI] [PubMed] [Google Scholar]
- 32.Barrientos A. (2002) In vivo and in organello assessment of OXPHOS activities. Methods 26, 307–316 [DOI] [PubMed] [Google Scholar]
- 33.Berry E. A., and Trumpower B. L. (1987) Simultaneous determination of hemes a, b and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 34.Atkinson A., Khalimonchuk O., Smith P., Sabic H., Eide D., and Winge D. R. (2010) Mzm1 influences a labile pool of mitochondrial zinc important for respiratory function. J. Biol. Chem. 285, 19450–19459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalimonchuk O., Ott M., Funes S., Ostermann K., Rödel G., and Herrmann J. M. (2006) Sequential processing of a mitochondrial tandem protein: insights into protein import in Schizosaccharomyces pombe. Eukaryot. Cell 5, 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui T. Z., Smith P. M., Fox J. L., Khalimonchuk O., and Winge D. R. (2012) Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell. Biol. 32, 4400–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]