Abstract
A current question in biofilm research is whether biofilm-specific genetic processes can lead to differentiation in physiology and function among biofilm cells. In Pseudomonas aeruginosa, phenotypic variants which exhibit a small-colony phenotype on agar media and a markedly accelerated pattern of biofilm development compared to that of the parental strain are often isolated from biofilms. We grew P. aeruginosa biofilms in glass flow cell reactors and observed that the emergence of small-colony variants (SCVs) in the effluent runoff from the biofilms correlated with the emergence of plaque-forming Pf1-like filamentous phage (designated Pf4) from the biofilm. Because several recent studies have shown that bacteriophage genes are among the most highly upregulated groups of genes during biofilm development, we investigated whether Pf4 plays a role in SCV formation during P. aeruginosa biofilm development. We carried out immunoelectron microscopy using anti-Pf4 antibodies and observed that SCV cells, but not parental-type cells, exhibited high densities of Pf4 filaments on the cell surface and that these filaments were often tightly interwoven into complex latticeworks surrounding the cells. Moreover, infection of P. aeruginosa planktonic cultures with Pf4 caused the emergence of SCVs within the culture. These SCVs exhibited enhanced attachment, accelerated biofilm development, and large regions of dead and lysed cells inside microcolonies in a manner identical to that of SCVs obtained from biofilms. We concluded that Pf4 can mediate phenotypic variation in P. aeruginosa biofilms. We also performed partial sequencing and analysis of the Pf4 replicative form and identified a number of open reading frames not previously recognized in the genome of P. aeruginosa, including a putative postsegregational killing operon.
Bacteria in biofilms often form densely packed, matrix-encased structures (microcolonies) in which steep oxygen and nutrient availability gradients can occur (10, 53). Bacterial adaptation to such highly heterogeneous and changing conditions is thought to include the development of phenotypic variants, which may become established as niche specialists within the biofilms (2, 43, 48). One such example is the significant variation observed among Pseudomonas aeruginosa colonies obtained from laboratory biofilms, as well as from persistent clinical infections caused by P. aeruginosa. The variants include mucoid (11, 37), dwarf (12, 18, 19, 37, 58), lipopolysaccharide-deficient (9), rough (37), hyperpiliated (12, 19), and antibiotic-resistant (14) colonies. Although much remains to be learned about the processes that cause phenotypic variation within biofilms, the variants may reflect inducible mechanisms that generate genetic variability under stress conditions or when there is significant environmental change.
Bacteria possess diverse mechanisms that may lead to an increase in genetic and phenotypic variability under stress conditions. These mechanisms include adaptive mutation (3, 43, 55), phase variation (14), and enhanced gene transfer through conjugation and transformation (17, 20, 41). In addition, the relationship between bacterial stress responses and the mobility of bacteriophages has been extensively documented, and bacteriophage transduction is now increasingly recognized as a process that is important in gene transfer within natural bacterial populations (40). Moreover, bacterial prophages can cause DNA inversions and phenotypic variation (32, 57), and bacteria often acquire phenotypic traits, such as virulence factors (13), from the genome of an infecting bacteriophage.
Recent studies have shown that bacteriophage genes are among the most highly upregulated groups of genes during biofilm development in both gram-positive and gram-negative bacteria (52, 64). In P. aeruginosa, genes of a Pf1-like filamentous bacteriophage, which exists as a prophage in the genome of P. aeruginosa, showed up to 83.5-fold activation during biofilm development compared with expression in planktonic cells (64). Other studies have shown that activation of Pf1 genes in biofilms is regulated by quorum sensing in P. aeruginosa (21, 59). Moreover, the activity of the Pf1-like phage is also linked to the killing and lysis of a subpopulation of P. aeruginosa cells within biofilms (60). Induction of the Pf1-like phage (designated Pf4) in P. aeruginosa may therefore represent an important physiological and developmental event during biofilm development.
Here we show that activity of the Pf4 phage in P. aeruginosa biofilms is linked to the emergence of a subpopulation of cells with a small-colony phenotype in the effluent runoff from the biofilms. These cells have high densities of filamentous phage on the cell surface, exhibit enhanced adhesion and microcolony development, and occur in high numbers in the biofilm runoff. Our data suggest that Pf4 small-colony variants (SCVs) play an important role in biofilm development, as well as in the colonization of new surfaces during biofilm dispersal.
MATERIALS AND METHODS
Strains and culture conditions.
P. aeruginosa strain PAO1 (26) was used in this study. Batch cultures of P. aeruginosa were grown at 37°C with shaking in Luria-Bertani medium. For cultivation of biofilms, M9 medium containing 48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 100 μM CaCl2, and 5 mM glucose was used.
Biofilm experiments.
P. aeruginosa PAO1 wild-type and small-colony variant biofilms were grown in continuous-culture flow cells (channel dimensions, 1 by 4 by 40 mm) at room temperature as previously described (42). The channels were inoculated with 0.5 ml of early-stationary-phase cultures containing approximately 1 × 109 cells ml−1 and incubated with no flow for 1 h at room temperature. Flow was then started with a mean velocity in the flow cells of 0.2 mm s−1, corresponding to laminar flow with a Reynolds number of 0.02. Biofilms were stained with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes Inc., Eugene, Oreg.) and were visualized by using a confocal laser scanning microscope (Olympus). The two stock solutions of the stains (SYTO 9 and propidium iodide) were diluted to a concentration of 3 μl ml−1 with biofilm medium and injected into the flow channels. Live SYTO 9-stained cells and dead propidium iodide-stained cells were visualized with a confocal laser scanning microscope (Olympus) by using fluorescein isothiocyanate and tetramethyl rhodamine isocyanate optical filters, respectively. For isolation of colony variants, 1-ml aliquots of the effluent biofilm runoff (spent culture medium emerging from the flow cell reactor and containing detached biofilm cells) were collected after 1, 3, 5, 7, and 9 days of biofilm development. These effluent samples were then serially diluted 10−2 to 10−6 and plated onto Luria-Bertani agar. The plates were observed for colony variants after 48 h of incubation at 37°C.
To obtain statistically based, quantitative measurements during biofilm development, we characterized biofilm morphology by using the COMSTAT program (23). Biofilms were stained with acridine orange (ProSciTech, Kelso, Australia). At each time examined, five image stacks were recorded for three replicate biofilms, which resulted in 15 image stacks for each strain studied. Images were acquired at 2-μm intervals through the biofilm at random positions in the flow cell at three times (1, 3, and 7 days) as previously described (23) by using a confocal laser scanning microscope. The following parameters were assessed: total biovolume (in cubic micrometers per square micrometer), maximum thickness (in micrometers), average thickness (in micrometers), and average microcolony area at the substratum (in square micrometers).
Adhesion assay.
We compared the abilities of wild-type and SCV strains to adhere to wells of polystyrene microtiter plates using an adhesion assay similar to that described previously (45). P. aeruginosa and SCV cultures were grown to an optical density at 600 nm of 0.6. Cells were centrifuged (6,000 × g for 15 min) and resuspended to an optical density at 600 nm of 0.1. Aliquots (200 μl) of cells were then placed into 96-well microtiter plates and incubated for 2 h at 37°C. After this 25 μl of a 1% solution of crystal violet (CV) was added to each well, and the plates were incubated at room temperature for 15 min and rinsed three times with water. Ethanol (200 μl) was then added to each well to extract the CV, and the extent of CV staining was measured with an enzyme-linked immunosorbent assay plate reader (Wallac, Perkin Elmer) at 600 nm.
Bacteriophage experiments.
Isolation of Pf4 PFU from biofilms, determination of phage titers, and large-scale preparation and purification of Pf4 were carried out as described previously (60). Infection of planktonic P. aeruginosa cultures with Pf4 was carried out at a multiplicity of infection of 10. For the preparation of replicative-form (RF) DNA of Pf4, a 1-liter early-log-phase culture of P. aeruginosa PAO1 was infected with Pf4. After 12 h of incubation with shaking (150 rpm), cells were harvested by centrifugation at 7,000 rpm, and the supernatant was discarded. The DNA pellet was washed with 100 mM NaCl-10 mM Tris-1 mM EDTA (pH 8) and then resuspended in 20 ml of 10 mM Tris (pH 8.0). Cell lysis and extraction of RF DNA were performed by using a QIAGEN Maxiprep kit (QIAGEN, Hilden, Germany). The final pellet was resuspended in 150 μl of 10 mM Tris-1 mM EDTA (pH 8) and visualized on an agarose gel, and the 12-kb RF band was excised from the gel and purified with a QIAquick kit (QIAGEN, Hilden Germany).
To confirm production of the Pf4 RF and to accurately delineate Pf4 within the P. aeruginosa PAO1 genome, we amplified a region of the RF predicted to contain the region of recircularization of the RF (i.e., the region containing the direct repeats which flank the predicted phage genome) (28). Thus, we designed primers that amplified a product only from the Pf4 RF and not from the Pf4 prophage in the genome of P. aeruginosa. Primers Pf4F (5′-AGCAGCGCGATGAAGCAAT-3′), corresponding to bp 2756 to 2774 of GenBank accession no. AE004508, and Pf4R (5′-TAGAGGCCATTTGTGACTGGA-3′), targeting bp 1566 to 1546 of GenBank accession no. AE004507, were used for this purpose. The 839-bp PCR product was purified by using a QIAGEN PCR cleanup kit (QIAGEN, Valencia, Calif.) and was sequenced by using the BigDye termination reaction (Applied Biosystems, Melbourne, Australia) and an ABI 3730 sequencer. For analysis of the Pf4 genome (obtained from the P. aeruginosa PAO1 genome sequence) and the Pf1 genome, sequences were compared by using the National Center for Biotechnology Information BLAST and ORF Finder programs.
In order to examine in more detail the role of the Pf4 prophage in biofilm development, P. aeruginosa mutant strains with double-crossover knockout insertions in Pf4 were generated in a previous study (60). However, DNA replication in filamentous phages involves the generation of double-stranded circular copies of the genome known as replicative forms, which are stably maintained in generations of bacterial cells. Despite extensive efforts, we have been unable to generate mutations in the prophage and all of its RF copies. PCR amplification of the region encoding the Gmr knockout insertion consistently generated two products corresponding to mutant copies of the phage (containing the Gmr double-crossover insertion) and to the same region of the wild-type phage. We have also tried extensively to cure the P. aeruginosa Pf4 mutant strain of the RF by plate culturing methods without success. The genome of Pf4 contains a putative toxin-antitoxin (plasmid stabilization) operon identified in this study. While we do not yet know if this operon is functionally expressed in P. aeruginosa, it is possible that cured cells may be inhibited or killed by this system, which may explain why inactivation of the prophage has not been possible.
Preparation of antibodies and immunolabeling.
Anti-Pf4 polyclonal antibodies were developed by using a synthetic peptide (Auspep, Parkville, Australia) with the following amino acid sequence: Gly-Val-Ile-Asp-Thr-Ser-Ala-Val-Glu-Ser-Ala-Ile-Thr-Asp-Gly-Cys. This sequence corresponds to residues 1 to 15 of CoaB (PA0723; the major coat protein of the filamentous phage virion) of Pf1 (and of Pf4), which is exposed on the outer surface of the bacteriophage virion (35, 61). An extra cysteine residue was added to the N terminus of this peptide for coupling to the carrier protein, keyhole limpet hemocyanin. A rabbit was immunized by using three subcutaneous injections, each containing 300 μg of the keyhole limpet hemocyanin-conjugated CoaB peptide (Institute of Medical and Veterinary Science, Adelaide, Australia). The serum titer and specificity of the polyclonal antibodies were monitored by an enzyme-linked immunosorbent assay and Western blot analysis.
Electron microscopy and immunogold labeling of Pf4 virions.
Immunogold electron microscopy was carried out with SCV and wild-type cells grown on agar plates for 18 h at 37°C, essentially as described previously (50). Bacteria scraped from an agar surface with a cotton swab were suspended in phosphate-buffered saline (PBS). A drop (50 μl) of this suspension was placed onto a sheet of Parafilm. A carbon- and Formvar-coated nickel grid was placed on the drop, with the coating facing the drop, for 2 min and then sequentially onto drops of the following reagents (at room temperature): PBS containing 0.1% glutaraldehyde (5 min), PBS containing 50 mM NH4Cl (5 min), PBS containing 1% bovine serum albumin (BSA) and 1% normal goat serum (5 min), and rabbit anti-Pf4 antiserum diluted 1/100 in PBS containing 1% BSA and 1% normal goat serum (30 min). After three washes in PBS containing 0.1% BSA (2 min each), the grid was placed on a drop of immunoglobulin-gold-conjugated anti-rabbit immunoglobulin G (heavy and light chains) (12-nm-diameter gold particles; Jackson Immunoresearch) diluted in PBS (30 min). The grid was subjected to three washes in PBS (3 min each), fixed in 1% glutaraldehyde in PBS (5 min), and washed twice in distilled water (5 min each). The grid was then treated with a drop of 1% uranyl acetate for 30 s. The grid was air dried and examined with a Hitachi H7000 transmission electron microscope.
RESULTS AND DISCUSSION
Emergence of SCVs in the biofilm effluent correlates with the release of plaque-forming Pf4 phage variants.
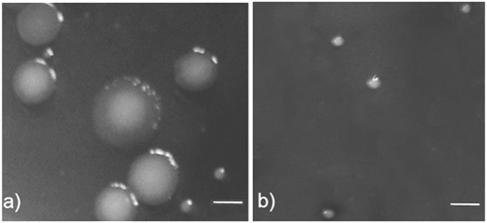
Previously, genes of a Pf1-like filamentous prophage were found to be highly upregulated during P. aeruginosa biofilm development (64), and mature biofilms of P. aeruginosa were found to release a filamentous phage (designated Pf4) capable of forming plaques on the P. aeruginosa host strain (60). Because filamentous phage infection can cause small colonies in Escherichia coli cultures (30), we hypothesized that filamentous phage may also be important in the formation of P. aeruginosa SCVs during biofilm development. We therefore compared the CFU and phage PFU counts on agar plates using the effluent runoff from P. aeruginosa biofilms (Table 1). For the first 5 days of biofilm development we observed between 1.3 × 106 and 3.7 × 106 CFU ml of effluent−1, and all of the colonies resembled those normally formed by the parental P. aeruginosa strain. No phage were detected in the effluent during this period. However, after 7 days, we observed simultaneous emergence of 1 × 105 SCVs ml−1 and 1 × 107 PFU of phage Pf4 ml−1, along with 3.0 × 106 CFU of wild-type large colonies ml−1. Colonies of the SCVs were approximately 0.5 to 1.5 mm in diameter, whereas parental-type colonies were >4 mm in diameter (Fig. 1a).
TABLE 1.
Appearance of SCVs correlates with the emergence of bacteriophage in the fluid runoff from flow cell biofilms
| Biofilm age (days) | Concn of:
|
||
|---|---|---|---|
| Wild type (large colonies) (CFU/ml) | Pf4 (PFU/ml) | SCV (CFU/ml) | |
| 1 | 2.3 × 106 | 0 | 0 |
| 3 | 1.3 × 106 | 0 | 0 |
| 5 | 3.7 × 106 | 0 | 0 |
| 7 | 3.0 × 106 | 1 × 107 | 1 × 105 |
| 9 | 7.0 × 105 | 2 × 107 | 5 × 105 |
FIG. 1.
Wild-type colonies and SCVs derived from effluent runoff from a 7-day old biofilm (a) and a P. aeruginosa overnight planktonic culture infected with the Pf4 bacteriophage (SCVs only) (b). Bars = 3 mm.
We picked colonies from the 18-h agar plates for culture and found that all of the SCVs, but not wild-type colonies, contained superinfective (able to form plaques on the lysogenic P. aeruginosa strain) Pf4 bacteriophage (approximately 1 × 107 PFU ml−1). To confirm that these SCVs were variants of P. aeruginosa PAO1 and not a contaminant, we sequenced regions of the 16S ribosomal DNA of these variants and found 100% identity with the P. aeruginosa genome sequence (54). We selected one variant, designated SCV7, for subsequent investigation.
Immunoelectron microscopy reveals dense latticeworks of Pf4 filaments surrounding SCV cells.
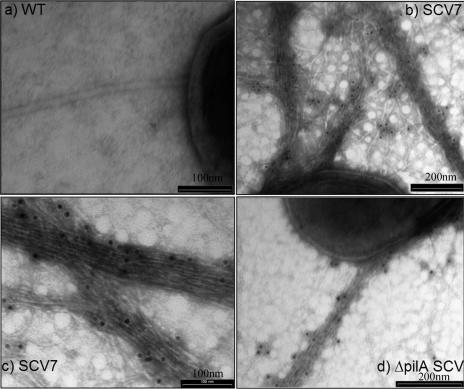
Because SCV colonies contained high numbers of Pf4, we expected that electron microscopic examination would reveal high densities of Pf4 filaments on the surface of SCV cells compared to cells of the wild-type strain. We therefore carried out immunoelectron microscopy of P. aeruginosa cells from normal and SCV7 colonies using antibodies raised against the Pf4 major coat protein. SCV7 colonies, but not wild-type colonies, contained cells that were surrounded by high densities of Pf4 filaments (Fig. 2). The original immunoelectron microscopic descriptions of filamentous phage Pf in P. aeruginosa of Bradley (4) also indicated that there were phage filaments that were often tightly interwoven in skeins, identical to our observations.
FIG. 2.
Transmission electron microscopy and immunogold labeling with anti-Pf4 antibodies, showing high densities of filamentous bacteriophages on the cell surface of SCVs. (a) Wild-type (WT) P. aeruginosa cell, showing a single flagellum. (b) P. aeruginosa SCV7 cell with anti-Pf4 antibodies. (c) Higher magnification of Pf4 filaments tightly woven together. (d) ΔpilA mutant of P. aeruginosa, showing similar Pf4 filament production on the cell surface.
Previously, small-colony variants that emerge from P. aeruginosa biofilms have been reported to overproduce type IV pili on the cell surface (12, 19). Type IV pili are structurally very similar to filamentous phage virions, and they are often indistinguishable by electron microscopy. To obtain further evidence that the hyperfilamentation observed in this study was principally due to Pf4 filaments, we grew biofilms using a P. aeruginosa mutant with a knockout insertion in pilA, which codes for the major structural subunit of the type IV pilus (29). We found that mature ΔpilA biofilms also produced SCVs (ΔpilA-SCVs) that contained Pf4 PFU and exhibited similar high densities of surface Pf4 filaments when they were examined by immunoelectron microscopy (Fig. 2d). These data suggest that Pf4 virions are the principal cause of hyperfilamentation in SCV7.
Addition of Pf4 virions to P. aeruginosa cultures generates SCVs.
To further investigate the role of Pf4 in SCV formation, we infected planktonic cultures of P. aeruginosa with CsCl gradient-purified Pf4. We found that after 12 h of incubation in the presence of the phage, all of the cells in the culture grew with a small-colony phenotype (Fig. 1b), whereas uninfected cultures produced normal-size colonies. The appearance of these SCVs (designated Pf4-SCVs) was identical to the appearance of SCVs that emerged from the biofilm and also exhibited high densities of filamentous phage on the cell surface, just like SCV7 (data not shown).
How might filamentous phage arise and cause an altered colony phenotype in the host bacterium? Wild-type Pf4, like other filamentous phages, establishes a symbiotic state with its host and is continuously released from P. aeruginosa cells under normal culture conditions (60, 64). These wild-type phage do not form visible plaques, have little effect on the growth of the lysogenized P. aeruginosa host strain, and do not generate SCVs. Thus, SCVs are not formed by induction of the wild-type phage. However, filamentous phage that can overcome the lysogenic immunity of the host strain often spontaneously arise in infected cultures (9, 17, 32, 35, 46). Such mutants can cause marked decreases in cellular DNA, RNA, and protein synthesis (25, 26, 35), can kill over 60% of the infected cells (31), and can result in a colony size that is considerably smaller than that of the uninfected cells (31). Phage that emerge from mature P. aeruginosa biofilms may represent such variant forms of Pf4, and P. aeruginosa cells that can propagate variant phage without being killed likely form SCVs. However, the mechanism of lysogenic immunity to Pf4 and the mechanism by which spontaneous phage variants may overcome this immunity are unknown.
We expected that SCVs would grow more slowly than the wild-type strain in planktonic culture. In fact, SCVs exhibited growth rates similar to those of the wild-type strain (the mean doubling times for the wild-type and SCV7 strains during early logarithmic growth were 47.4 and 43.2 min, respectively); thus, the SCV phenotype is not caused by slower growth of Pf4-infected cells. Small colonies could also be produced if SCV cells adhere more tightly to one another than the wild-type strain cells adhere to one another. Indeed, aggregation of cells in SCV planktonic cultures could normally be observed by eye (data not shown), and SCV cells were found to be more adherent to wells of microtiter plates (see below).
Biofilms formed by SCVs and by Pf4-infected cells show enhanced attachment and microcolony development.
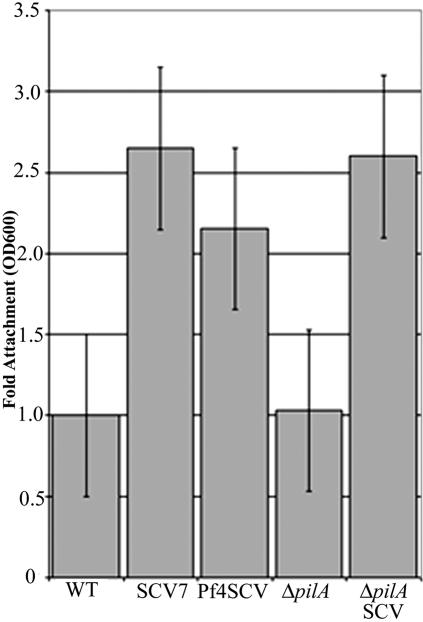
We examined the ability of wild-type, ΔpilA, SCV7, ΔpilA-SCV, and Pf4-SCV strains to attach to an inanimate surface. The SCV strains each showed increased (twofold or more) attachment to the wells of polystyrene microtiter plates (Fig. 3). Enhanced attachment of ΔpilA-SCV compared to the ΔpilA strain suggests that type IV pili are not responsible for the increased attachment observed in this study.
FIG. 3.
Adhesion of wild-type, ΔpilA, and SCVs to wells of tissue culture plates. WT, wild type; OD600, optical density at 600 nm.
The mechanism by which filamentous bacteriophages may lead to increased surface attachment and autoaggregation is unclear. During bacterial attachment, an energy barrier (known as the secondary minimum) is presented to cells, whereby electrostatic repulsion can prevent closer approach of the cell to the surface (7). Because of their extremely small radii, cell surface filaments, such as type IV pili, can extend through this energy barrier and facilitate bridging and permanent attachment of cells to the surface (36). The copious production of interwoven phage filaments could allow a large number of phage filaments to be in contact with the bacterial cell surface at any one time. Possibly, high numbers of Pf4 with low-affinity binding to the substratum or other bacterial cells could result in enhanced adhesion, as described for type IV pili or other cell surface filaments.
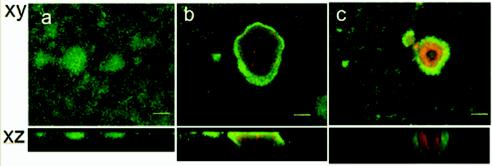
We also grew biofilms of the SCVs in continuous cultures in glass flow cells. Both the biofilm (SCV7) and planktonically derived SCVs were capable of forming much larger attached microcolonies than the wild-type strain formed (Fig. 4). For the wild-type strain, the diameter of microcolonies did not exceed 75 μm at any stage during biofilm development. In contrast, after 5 days of biofilm development, SCVs frequently formed microcolonies that ranged from 200 to 300 μm in diameter (Fig. 4b and c).
FIG. 4.
Biofilms formed by SCVs have enhanced microcolony formation and large regions containing dead cells inside microcolonies. Five-day-old P. aeruginosa biofilms were stained with the BacLight LIVE/DEAD stain. The biofilms were inoculated with wild-type (a), SCV7 (b), and Pf4-infected (c) cells. Bars = 50 μm.
We also compared biofilm development in wild-type and phage-expressing cells using the COMSTAT software (23). The results of this analysis are shown in Table 2. The maximum biofilm thickness and mean microcolony area were significantly greater for the SCV7 strain than for the wild-type strain at each of the times studied. SCVs obtained from Pf4-infected planktonic cultures also showed significantly increased maximum biofilm thickness and microcolony area after 3 days of biofilm development. While not always significantly higher as determined by analysis of variance, the mean biomass and thickness were also consistently greater for SCVs than for the wild type throughout biofilm development (Table 2).
TABLE 2.
Analysis of biofilm development in wild-type and Pf4-expressing P. aeruginosa strains with the COMSTAT softwarea
| Parameter | Day | Wild type | Wild type + Pf4 | SCV7 |
|---|---|---|---|---|
| Mean biomass (μm−3 μ−2) | 1 | 0.43 ± 0.09 | 0.30 ± 0.02 | 0.47 ± 0.13 |
| 3 | 0.74 ± 0.07 | 0.94 ± 0.13 | 1.19 ± 0.55 | |
| 7 | 1.87 ± 0.07 | 2.48 ± 0.57 | 5.29 ± 0.62 | |
| Mean thickness (μm) | 1 | 0.36 ± 0.09 | 0.32 ± 0.03 | 0.53 ± 0.14 |
| 3 | 0.82 ± 0.06 | 1.01 ± 0.11 | 1.41 ± 0.65 | |
| 7 | 2.02 ± 0.03 | 3.08 ± 0.89 | 6.37 ± 0.93 | |
| Maximum thickness (μm) | 1 | 11.40 ± 1.53 | 11.9 ± 0.33 | 41.60 ± 1.60 |
| 3 | 11.66 ± 0.21 | 15.2 ± 0.02 | 43.20 ± 9.73 | |
| 7 | 12.80 ± 0.32 | 19.2 ± 0.89 | 57.60 ± 7.82 | |
| Mean microcolony area at | 1 | 9.73 ± 2.93 | 5.28 ± 2.36 | 123.49 ± 51.62 |
| substratum (μm2) | 3 | 5.76 ± 5.75 | 8.11 ± 3.61 | 181.31 ± 81.62 |
| 7 | 17.00 ± 2.95 | 26.93 ± 4.64 | 294.70 ± 146.22 |
The values are means ± standard errors for data from 15 image stacks (five image stacks from three replicate biofilms) Boldface type indicates values that are significantly higher than the values for the wild-type strain as determined by analysis of variance.
In biofilms formed with SCVs, we also observed complex heterogeneity within the microcolonies, which contained large regions of dead and lysed cells, as well as hollow voids that did not contain cells. The killing and lysis within microcolonies occurred much earlier in SCV biofilms (4 to 5 days) than in wild-type biofilms (after 7 days, as observed previously) (60). Pf4 has previously been linked with the death and lysis of a subpopulation of cells inside microcolonies in mature (7-day) P. aeruginosa biofilms (60). Filamentous bacteriophages can kill a proportion of host cells (25, 30, 31, 34, 46, 51). The consequences of this cell death for surviving cells within the biofilm and for the propagation and dispersal of the bacteriophage remain to be fully elucidated. For example, it is possible that nutrients released by cell death in this manner are assimilated by other bacteria in the biofilm (49, 56).
Several mechanisms may explain how Pf4 activity leads to the formation of colonies that are larger and more differentiated than the wild-type strain colonies. We observed that SCV7 and phage-producing strains were unable to carry out type IV pilus-mediated twitching motility (data not shown), possibly because filamentous phages use the type IV pilus as the receptor for infection (38) and might therefore interfere with pilus function. Previously, mutant P. aeruginosa strains that were not capable of twitching motility were found to form larger microcolony structures than the wild-type strain (22, 29). However, other studies showed that type IV pilus mutants were unable to form microcolonies (44); thus, the role of type IV pili in microcolony development appears to vary depending on the experimental system used. In this study, biofilms formed by the ΔpilA strain exhibited microcolonies similar to the wild-type strain microcolonies (data not shown); thus, impaired twitching motility is unlikely to be the cause of enhanced microcolony development in SCV strains.
Another possible mechanism for enhanced microcolony formation is that high densities of filamentous phage on the cell surface may play a direct role in the cohesion of biofilm cells. In E. coli, expression of conjugative plasmid-encoded pili can lead to enhanced microcolony and biofilm formation (16, 47). This process is thought to facilitate plasmid maintenance within the population by allowing high rates of infectious transfer (16). Filamentous bacteriophages share striking functional similarities with conjugative plasmids; indeed, several authors have suggested that conjugative pili may have evolutionary links with filamentous bacteriophages (1, 5, 62). Because biofilm formation would similarly enhance the maintenance and infectious transfer of bacteriophages within the host cell population, it is interesting to consider whether the cohesion of biofilm cells by filamentous structures may have its evolutionary origins among the filamentous bacteriophages.
Analysis of the Pf4 genome.
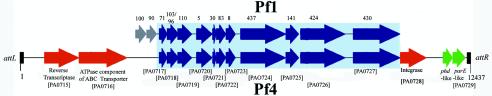
A previous study in our laboratory (60) and the present study confirmed the production of virions encoded by the Pf4 prophage of P. aeruginosa. To provide more information about the genome of Pf4, we extracted the RF plasmid from Pf4-infected cells and used primers Pf4F and Pf4R to amplify the predicted region of recircularization of the phage genome (8, 28) from the RF DNA. Our data confirm that the RF recircularizes by using the repeat sequence TGGAGCGGGCGAAGGGAATCGAACCCT located in the intergenic sequence between PA0714 and PA0715 and within the tRNAGly site of PA0729.1. The sequences predict a 12,437-bp viral genome, based on analysis of the published genome sequence of P. aeruginosa (54). A comparison of the genomes of Pf1 and Pf4 is shown in Fig. 5. Major differences include the presence of genes encoding a putative reverse transcriptase (PA0715) and an ATPase component of an ABC transporter (PA0716) in the Pf4 genome. On the complementary intergenic region between PA0716 and PA0717 (ORF71) is an open reading frame (ORF) with 42% homology to the sequence encoding the repressor C protein of phage P2 (GenBank accession no. WPBPP2). We also found two ORFs, 9 bp apart, encoding proteins with high levels of homology to the prevent host death (phd) antitoxin protein of Pseudomonas syringae (79% identity; GenBank accession no. NP790091.1) (6) and the conserved domain of the ParE plasmid stabilization toxin of broad-host-range plasmid RK2 (84.7% identity; GenBank CD no. COG3668.1) (27). These genes are similar in size and organization to other described toxin-antitoxin modules (15) and suggest that Pf4 may have acquired a host addiction module or programmed cell death operon. These modules can facilitate the maintenance of plasmids (27) and phages (33) by killing cells that have lost the plasmid or phage postsegregation. To our knowledge, a chromosomally located toxin-antitoxin system has not previously been described in P. aeruginosa, and we are currently exploring whether this module is functionally expressed in P. aeruginosa biofilms.
FIG. 5.
Comparison of the Pf4 genome with the genome of Pf1. Genes are indicated as follows: blue, homologous genes found in both Pf1 and Pf4; red, genes occurring only in Pf4; green, new genes or ORFs identified in this study that occur only in Pf4; grey, genes found only in Pf1 and not in Pf4. The numbers above the Pf1 genes are the ORF numbers as presented in the published genome sequence of Pf1 (24). The numbers below the Pf4 genes are gene numbers from the published P. aeruginosa genome sequence (54).
In summary, this study revealed a role for the P. aeruginosa prophage Pf4 in the development of small-colony phenotypic variants during biofilm formation. These variants exhibit high densities of phage filaments on the cell surface, as well as enhanced attachment (two to threefold) and microcolony development. Bacteriophage-mediated SCVs may represent an important dispersal phenotype with enhanced colonization traits, which can originate from established P. aeruginosa biofilms in natural environments. Further studies are also needed to determine the significance of high densities of Pf4 filaments as a structural component within the matrix of both laboratory and clinical P. aeruginosa biofilms. Recently, extracellular DNA has been reported to be an important component of the extracellular matrix in P. aeruginosa (39, 63). Release of high numbers of DNA-containing Pf4 phage within biofilms may provide one mechanism by which DNA can accumulate in the extracellular matrix of P. aeruginosa biofilms.
Acknowledgments
We thank our colleagues at the University of New South Wales, Sydney, Australia, and at Nanyang Polytechnic, Singapore, for their support and Loren Day of the International Committee on Taxonomy of Viruses for advice on bacteriophage nomenclature.
This work was funded by the Australian Research Council, the Centre for Marine Biofouling and Biotechnology at the University of New South Wales, and Nanyang Polytechnic.
REFERENCES
- 1.Agol, V. I. 1976. An aspect on the origin and evolution of viruses. Origins Life 7:119-132. [DOI] [PubMed] [Google Scholar]
- 2.Ali, A., M. H. Rashid, and D. K. Karaolis. 2002. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 68:5773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, D. E. 1973. The adsorption of the Pseudomonas aeruginosa filamentous bacteriophage Pf to its host. Can. J. Microbiol. 19:623-631. [DOI] [PubMed] [Google Scholar]
- 5.Brinton, C. C., Jr. 1971. The properties of sex pili, the viral nature of “conjugal” genetic transfer systems, and some possible approaches to the control of bacterial drug resistance. Crit. Rev. Microbiol. 1:105-160. [DOI] [PubMed] [Google Scholar]
- 6.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busscher, H. J., and A. H. Weerkamp. 1987. Specific and non-specific interactions in bacterial adhesion to solid substrata. FEMS Microbiol. Rev. 46:165-173. [Google Scholar]
- 8.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasgupta, T., T. R. de Kievit, H. Masoud, E. Altman, J. C. Richards, I. Sadovskaya, D. P. Speert, and J. S. Lam. 1994. Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect. Immun. 62:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeBeer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structures on oxygen distribution and mass-transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 11.Deretic, V., M. J. Schurr, J. C. Boucher, and D. W. Martin. 1994. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J. Bacteriol. 176:2773-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobrindt, U., and J. Reidl. 2000. Pathogenicity islands and phage conversion: evolutionary aspects of bacterial pathogenesis. Int. J. Med. Microbiol. 290:519-527. [DOI] [PubMed] [Google Scholar]
- 14.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 15.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 17.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häußler, S., B. Tümmler, H. Weissbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 19.Häußler, S., I. Ziegler, A. Löttel, F. von Götz, M. Rohde, D. Wehmhöhner, S. Saravanamuthu, B. Tümmler, and I. Steinmetz. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295-301. [DOI] [PubMed] [Google Scholar]
- 20.Hendrickx, L., M. Hausner, and S. Wuertz. 2003. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 69:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentzer, M., M. Givskov, and L. Eberl. 2004. Quorum sensing in biofilms: gossip in slime city, p. 118-140. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms. ASM Press, Washington, D.C.
- 22.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 24.Hill, D. F., N. J. Short, R. N. Perham, and G. B. Petersen. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349-364. [DOI] [PubMed] [Google Scholar]
- 25.Hohn, B., H. von Schutz, and D. A. Marvin. 1971. Filamentous bacterial viruses (II): killing of bacteria by abortive infection with fd. J. Mol. Biol. 56:155-165. [DOI] [PubMed] [Google Scholar]
- 26.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, E. P., A. R. Strom, and D. R. Helinski. 1996. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J. Bacteriol. 178:1420-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. H., K. B. Lee, J. S. Lee, and Y. H. Cho. 2003. Genome diversification by phage-derived genomic islands in Pseudomonas aeruginosa. J. Microbiol. Biotechnol. 13:783-788. [Google Scholar]
- 29.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella, and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, M. Y., M. K. Yang, W. P. Chen, and T. T. Kuo. 2000. High-frequency interconversion of turbid and clear plaque strains of bacteriophage f1 and associated host cell death. Can. J. Microbiol. 46:841-847. [PubMed] [Google Scholar]
- 31.Kuo, T. T., C. C. Chiang, S. Y. Chen, J. H. Lin, and J. L. Kuo. 1994. A long lytic cycle in filamentous phage Cf1tv infecting Xanthomonas campestris pv. citri. Arch. Virol. 135:253-264. [DOI] [PubMed] [Google Scholar]
- 32.Kutsukake, K., and T. Iino. 1980. Inversions of specific DNA segments in flagellar phase variation of Salmonella and inversion systems of bacteriophages P1 and Mu. Proc. Natl. Acad. Sci. USA 77:7338-7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233:414-428. [DOI] [PubMed] [Google Scholar]
- 34.Lin, S. H., W. P. Chen, and T. T. Kuo. 2001. Mechanism of host cell death induced by infection of Escherichia coli with the c2 clear-plaque mutant of phage f1. Bot. Bull. Acad. Sin. 42:45-52. [Google Scholar]
- 35.Liu, D. J., and L. A. Day. 1994. Pf1 virus structure: helical coat protein and DNA with paraxial phosphates. Science 265:671-674. [DOI] [PubMed] [Google Scholar]
- 36.Marshall, K. C. 1985. Mechanisms of bacterial adhesion at solid-water interfaces, p. 133-161. In D. C. Savage and M. Fletcher (ed.), Bacterial adhesion—mechanisms and physiological significance. Plenum Press, New York, N.Y.
- 37.Martin, C., M. A. Ichou, P. Massicot, A. Goudeau, and R. Quentin. 1995. Genetic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis revealed by restriction fragment length polymorphism of the rRNA gene region. J. Clin. Microbiol. 33:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marvin, D. A. 1998. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8:150-158. [DOI] [PubMed] [Google Scholar]
- 39.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, R. V. 2001. Environmental bacteriophage-host interactions: factors contribution to natural transduction. Antonie Leeuwenhoek 79:141-147. [DOI] [PubMed] [Google Scholar]
- 41.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 42.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 45.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 46.Pratt, D., H. Tzagoloff, and W. S. Erdahl. 1966. Conditional lethal mutants of the small filamentous coliphage (I): isolation, complementation, cell killing, time of cistron action. Virology 30:397-410. [DOI] [PubMed] [Google Scholar]
- 47.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 48.Sadowska, B., A. Bonar, C. von Eiff, R. A. Proctor, M. Chmiela, W. Rudnicka, and B. Rozalska. 2002. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol. Med. Microbiol. 32:191-197. [DOI] [PubMed] [Google Scholar]
- 49.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauvonnet, N., P. Gounon, and A. P. Pugsley. 2000. PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J. Bacteriol. 182:848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shieh, G. J., Y. C. Charng, B. C. Yang, T. Jenn, H. J. Bau, and T. T. Kuo. 1991. Identification and nucleotide sequence analysis of an open reading frame involved in high-frequency conversion of turbid to clear plaque mutants of filamentous phage Cf1t. Virology 185:316-322. [DOI] [PubMed] [Google Scholar]
- 52.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 55.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tominaga, A. 1997. The site-specific recombinase encoded by pinD in Shigella dysenteriae is due to the presence of a defective Mu prophage. Microbiology 143:2057-2063. [DOI] [PubMed] [Google Scholar]
- 58.von Götz, F., S. Haussler, D. Jordan, S. S. Saravanamuthu, D. Wehmhoner,A. Strussmann, J. Lauber, I. Attree, J. Buer, B. Tummler, and I. Steinmetz. 2004. Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. J. Bacteriol. 186:3837-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welsh, L. C., M. F. Symmons, and D. A. Marvin. 2000. The molecular structure and structural transition of the alpha-helical capsid in filamentous bacteriophage Pf1. Acta Crystallogr. D Biol. Crystallogr. 56:137-150. [DOI] [PubMed] [Google Scholar]
- 62.Whitchurch, C. B., and J. S. Mattick. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13:1079-1091. [DOI] [PubMed] [Google Scholar]
- 63.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 64.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]