Abstract
Chromatin modification and cellular metabolism are tightly connected. Chromatin modifiers regulate the expression of genes involved in metabolism and, in turn, the levels of metabolites. The generated metabolites are utilized by chromatin modifiers to affect epigenetic modification. The mechanism for this cross-talk, however, remains incompletely understood. The corepressor SIN3 controls histone acetylation through association with the histone deacetylase RPD3. The SIN3 complex is known to regulate genes involved in a number of metabolic processes. Here, we find that Drosophila SIN3 binds to the promoter region of genes involved in methionine catabolism and that this binding affects histone modification, which in turn influences gene expression. Specifically, we observe that reduced expression of SIN3 leads to an increase in S-adenosylmethionine (SAM), which is the major cellular donor of methyl groups for protein modification. Additionally, Sin3A knockdown results in an increase in global histone H3K4me3 levels. Furthermore, decreased H3K4me3 caused by knockdown of either SAM synthetase (Sam-S) or the histone methyltransferase Set1 is restored to near normal levels when SIN3 is also reduced. Taken together, these results indicate that knockdown of Sin3A directly alters the expression of methionine metabolic genes to increase SAM, which in turn leads to an increase in global H3K4me3. Our study reveals that SIN3 is an important epigenetic regulator directly connecting methionine metabolism and histone modification.
Keywords: Drosophila, gene transcription, histone acetylation, histone methylation, methionine, S-adenosylmethionine (SAM), SIN3
Introduction
Cellular function relies on the ability of the cell to sense and respond to the environment. Cellular response is mediated in part by epigenetic and metabolomic information (1, 2). The expression of metabolic gene is under epigenetic control. Reduction of three histone modifiers, the H3K9 demethylase Jhdm2a, the H3K9/H3K56 deacetylase SIRT6, and the histone deacetylase HDAC1, leads to changes in metabolic gene transcription, as well as metabolites in mouse and rat models (3–5). Because histone-modifying enzymes utilize key metabolites, these metabolites could then feedback and impact epigenetic modifications. Indeed, several groups have demonstrated that histone methylation can be altered through changes in metabolism. For example, histone methylation is regulated by threonine metabolism in mouse embryonic stem cells (6), by folate metabolism in yeast and human cells (7), and by methionine metabolism in yeast, fly, mouse, and human cells (7–10). Histone methylation and phosphorylation can also be modulated by changing glycolysis and serine metabolism in yeast (11). Although these studies collectively indicate that epigenetic control and metabolism are tightly connected, the mechanism for this cross-talk remains to be elucidated.
The SIN3 complex is one of the major histone-modifying complexes present in cells. SIN3 is a conserved transcriptional scaffold protein, which interacts with the histone deacetylase (HDAC)2 RPD3 and other associated proteins (12, 13). In Drosophila and mammals, a histone demethylase is also part of a SIN3/RPD3 HDAC complex (14). We previously reported a genetic interaction between Drosophila Sin3A and the genes encoding the histone demethylases KDM2 and dKDM5/LID (15, 16). These biochemical and genetic data suggest that the SIN3 complex may regulate histone methylation in addition to histone acetylation. Sin3A is essential in Drosophila and mammals (17–21). Deficiency of SIN3 leads to changes in expression of many genes involved in multiple biological processes, including cellular metabolism (16, 18, 22). SIN3 regulates genes involved in several metabolic pathways such as glycolysis, gluconeogenesis, and the citric acid cycle and additionally is associated with regulation of genes encoding proteins that process reactive oxygen species and glutathione (16, 18, 22–24). The mechanism of how SIN3 regulates cellular metabolism, however, is not fully understood.
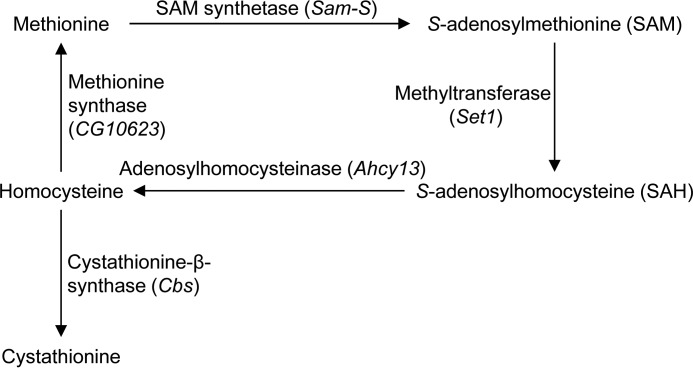
Methionine, an essential amino acid, is converted to the major methyl donor S-adenosylmethionine (SAM) by SAM synthetase (SAM-S) (Fig. 1). SAM is then converted to S-adenosylhomocysteine and methylated substrates by methyltransferases. S-Adenosylhomocysteine is next hydrolyzed by adenosylhomocysteinase (AHCY) to homocysteine, which is in turn either converted to methionine through methionine synthase or to cystathionine by cystathionine-β-synthase (CBS). To date, according to FlyBase (25), Sam-S and Cbs are the only known Drosophila genes encoding SAM synthetase (26) and cystathionine-β-synthase, respectively. Ahcy13 is the major adenosylhomocysteinase gene (27). CG10623 encodes a putative methionine synthase. The metabolites involved in methionine metabolism are critical for multiple pathways and biological processes (28). For example, reduced methionine leads to decreased H3K4me3 in yeast and mammalian cells, at least in part by modulating SAM levels (7–9). Given that SIN3 may regulate histone methylation because of its biochemical or genetic association with the histone demethylases dKDM5/LID and KDM2 (15, 16, 29, 30), we wanted to examine further the relationship between SIN3 and methionine metabolism in Drosophila.
FIGURE 1.
Methionine metabolism in Drosophila. The methionine cycle generates the major methyl donor SAM. The key Drosophila genes encoding SAM synthetase, adenosylhomocysteinase, and cystathionine-β-synthase are listed. Set1 encodes one of methyltransferases. CG10623 encodes a putative methionine synthase.
In this work, we focused on the mechanism through which SIN3 regulates cellular metabolism. We provided evidence that SIN3 binds to the promoters of methionine metabolic genes and affects H3K4me3 and H3K9ac levels at the promoter regions of these genes to control their expression. We observed increased levels of SAM and global H3K4me3 when SIN3 was reduced. Collectively, these results reveal that SIN3 regulates the expression of methionine metabolic genes through controlling histone modification levels at the promoters of these genes, which in turn regulates cellular SAM concentration and global H3K4me3.
Results
SIN3 Regulates Expression of Methionine Metabolic Genes and Histone Modifications at the Promoters of these Genes
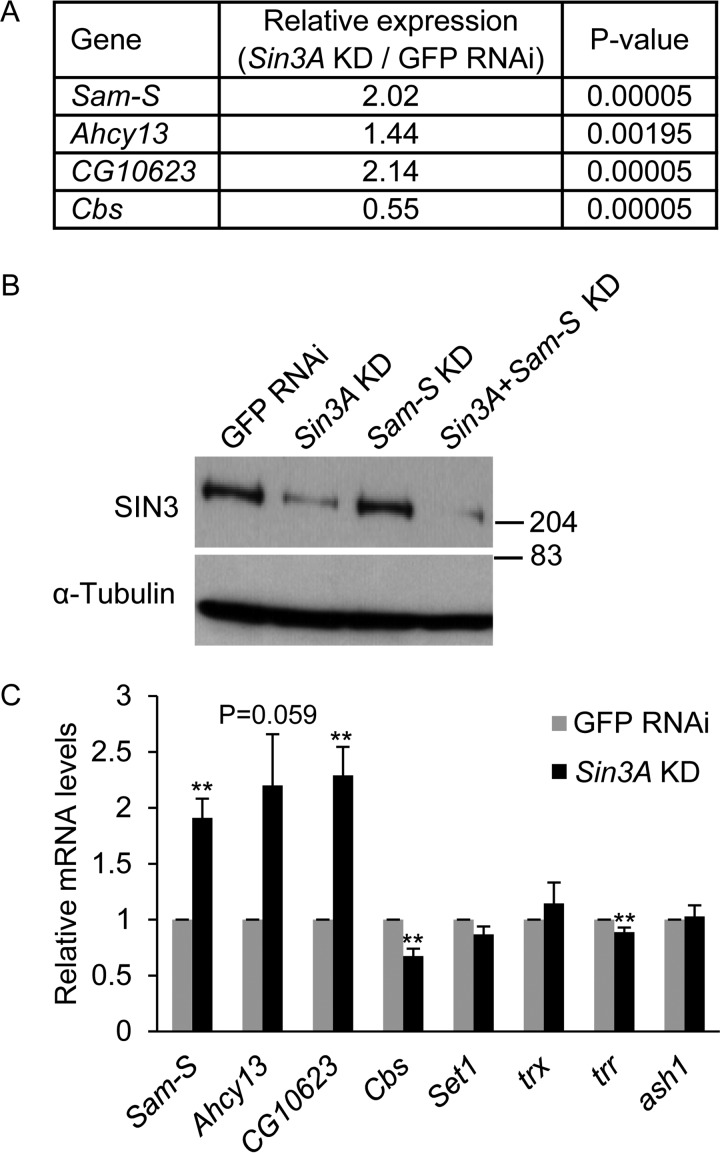
Because SIN3 is a global transcriptional regulator (12, 13) and affects the expression of genes involved in several metabolic pathways such as glycolysis, gluconeogenesis, and the citric acid cycle (16, 18, 22, 23), we sought to determine whether SIN3 regulates the transcription of methionine metabolic genes. Analysis of our recently published RNA-Seq gene expression profiles of S2 and RNAi-mediated Sin3A knockdown cells (16) indicates that reduction of SIN3 alters the expression of Sam-S, Ahcy13, Cbs, and CG10623 (Fig. 2A). To verify the RNA-Seq data, we repeated the Sin3A knockdown experiment and analyzed mRNA levels by real time qRT-PCR. The knockdown of Sin3A was validated by Western blotting analysis (Fig. 2B). Consistent with the RNA-Seq data, transcription of these genes was significantly changed when SIN3 was reduced (Fig. 2C). These results demonstrate that SIN3 regulates the expression of methionine metabolic genes.
FIGURE 2.
Transcription of methionine metabolic genes is regulated by SIN3. A, expression of methionine metabolic genes as determined in an RNA-Seq profile (16). B, verification of Sin3A knockdown. Whole cell extracts from RNAi-treated cells were subjected to Western blotting analysis using the indicated antibodies. α-Tubulin acted as the loading control. Protein size markers are indicated on the right. C, real time qRT-PCR analysis of transcription of methionine metabolic genes and histone methyltransferase genes. The results are the averages of three independent biological replicates. The error bars represent standard error of the mean. Statistically significant results comparing individual knockdown samples to the control are indicated on knockdown samples. GFP RNAi cells are the control cells. **, p < 0.01.
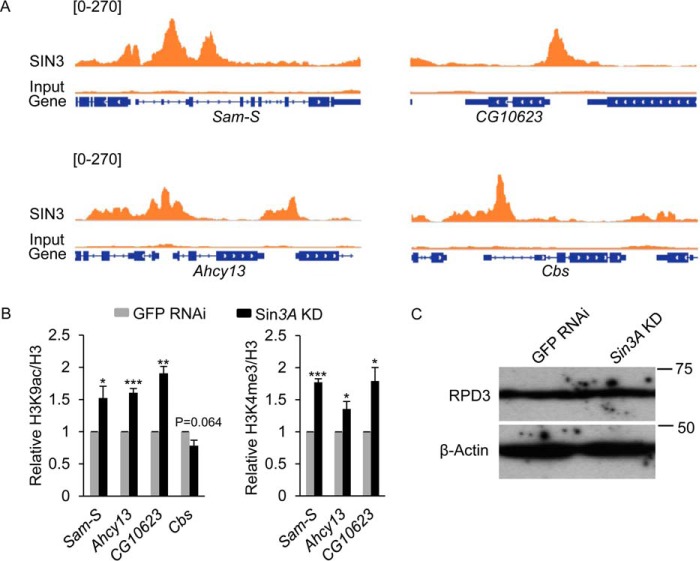
Next, we wanted to investigate how SIN3 affects the expression of these genes. Investigation of our recent ChIP-seq analysis (31), indicates that SIN3 binds to the promoters of Sam-S, Ahcy13, Cbs, and CG10623 (Fig. 3A). Additionally, changing the level of SIN3 alters global and gene specific histone acetylation, especially the H3K9ac mark (16, 29). Proteomic studies indicate that SIN3 copurifies with the H3K4me3 specific histone demethylase dKDM5/LID (29, 30), suggesting that the SIN3 complex may affect histone methylation. Based on these previous findings, we hypothesized that SIN3 directly controls H3K9ac and H3K4me3 levels at the promoters of methionine metabolic genes to regulate their expression. To test this hypothesis, we performed ChIP-qPCR analysis. IgG was used as a nonspecific control. Histone modification levels were normalized to histone H3 signal. Compared with IgG, strong enrichment of signals for tested histone antibodies was observed at all promoter regions sampled (data not shown). Knockdown of Sin3A led to an increase of H3K9ac and H3K4me3 at Sam-S, Ahcy13 and CG10623, whereas little to no change in H3K9ac was observed at Cbs (Fig. 3B). Consistent with published ChIP-seq analysis of H3K4me3 in S2 cells (32), we did not detect H3K4me3 at the promoter of Cbs in either the control or Sin3A knockdown condition (data not shown). Given that H3K9ac and H3K4me3 are associated with active genes (33, 34), their levels at the tested genes are consistent with our gene expression data (Figs. 2, A and C, and 3B).
FIGURE 3.
H3K9ac and H3K4me3 levels at the promoters of methionine metabolic genes are regulated by SIN3. A, SIN3 ChIP-seq signals at methionine metabolic genes. B, ChIP-qPCR analysis of H3K9ac and H3K4me3 levels at the promoters of methionine metabolic genes. The results are the average of three independent biological replicates. The error bars represent standard error of the mean. Statistically significant results comparing individual knockdown samples to the control are indicated on knockdown samples. GFP RNAi cells are the control cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, reduction of SIN3 does not affect the protein level of RPD3. Whole cell extracts from RNAi-treated cells were subjected to Western blotting analysis using the indicated antibodies. β-Actin acted as the loading control. Protein size markers are indicated on the right.
Because SIN3 is part of an HDAC complex, which contains both a histone deacetylase RPD3 and a histone demethylase dKDM5/LID (15, 16), we asked whether the increase of both H3K9ac and H3K4me3 at the promoters of methionine metabolism genes in Sin3A knockdown cells is due to instability of the complex and degradation of RPD3 and dKDM5/LID following knockdown of Sin3A. Our published RNA-Seq data indicate that the expression of rpd3 and lid is not changed when SIN3 is decreased (16). Reduction of SIN3 did not affect the protein level of RPD3 (Fig. 3C). Together these findings suggest that SIN3 controls histone modifications at methionine metabolic genes by regulating complex recruitment to their promoters, which in turn affects their transcription.
SIN3 Impacts the Levels of SAM and Global H3K4me3
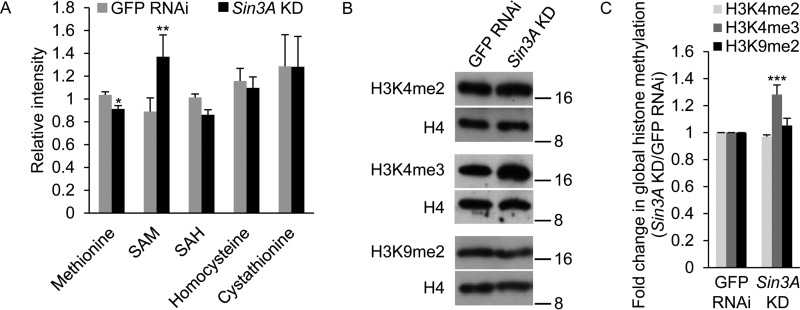
The finding that the expression of methionine metabolic genes is regulated by SIN3 led us to examine whether the levels of metabolites involved in methionine metabolism are also affected by SIN3. Through inspection of the methionine pathway, we noted that homocysteine has two major fates (Fig. 1). Because expression of Sam-S, Ahcy13, and CG10623 was up-regulated, whereas expression of Cbs was down-regulated in Sin3A knockdown cells compared with control cells (Fig. 2, A and C), we hypothesized that homocysteine would be remethylated to methionine to generate SAM rather than be converted to cystathionine when SIN3 is reduced. If this hypothesis is correct, then more SAM should be observed in Sin3A knockdown cells relative to the control. To test our hypothesis, we used liquid chromatography-tandem mass spectroscopy and gas chromatography mass spectroscopy to generate a quantitative metabolomic profile. We found that SAM levels were significantly up-regulated in Sin3A knockdown cells compared with control cells, whereas other metabolites in the pathway showed little change (Fig. 4A). These results indicate that reduced SIN3 alters the expression of methionine metabolic genes to increase the amount of major methyl donor SAM.
FIGURE 4.
Levels of SAM and global H3K4me3 are regulated by SIN3. A, effects of SIN3 on the cellular concentration of the metabolites involved in methionine metabolism. B, whole cell extracts from GFP RNAi control and Sin3A knockdown S2 cells were subjected to Western blotting analysis using the indicated antibodies. Protein size markers are indicated on the right. C, Western blots as shown in B were repeated with protein extracts prepared from at least three independent cultures, and the results were quantified after normalization to histone H4. The error bars represent standard error of the mean. Statistically significant results comparing individual knockdown samples to the control are indicated on knockdown samples. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
A change in the cellular concentration of SAM has been demonstrated to result in altered histone methylation (7–9). To examine whether SIN3 affects global histone methylation, whole cell protein extracts from dsRNA-treated S2 cells were probed with antibodies specific for distinct histone methylation marks, as well as histone H4 as a loading control. Because SIN3 regulates gene specific H3K4me3 levels (Fig. 3B), we first tested this mark. Knockdown of Sin3A resulted in a small, but reproducible, increase in global H3K4me3 levels (Fig. 4, B and C). H3K4me3 is more sensitive to cellular SAM concentration relative to other histone methylation marks (8). We thus hypothesized that the effect of SIN3 on H3K4me3 would be greater as compared with other methylation marks. To test our hypothesis, we examined H3K4me2 and H3K9me2. We did not observe any significant changes in global H3K4me2 and H3K9me2 levels when SIN3 was reduced (Fig. 4, B and C). Collectively, these data suggest that reduction of SIN3 leads to an increase in global H3K4me3 through increasing SAM levels.
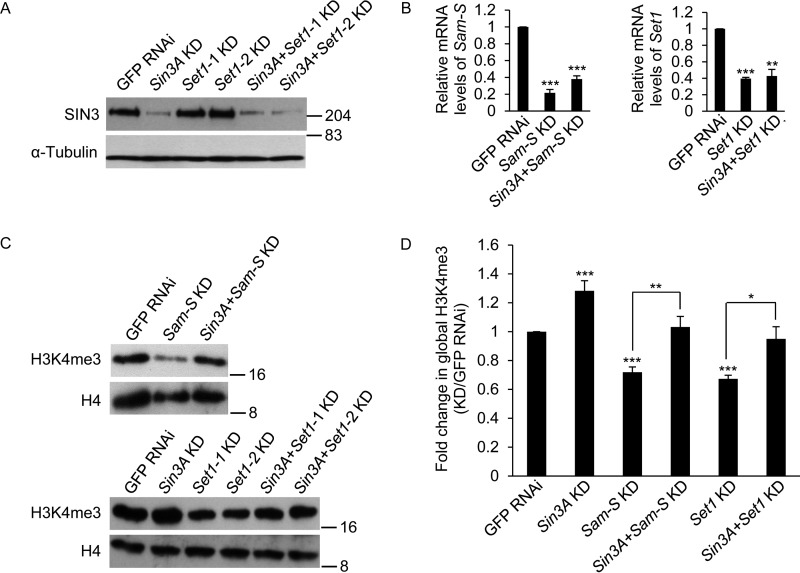
To further confirm the role of SIN3 in regulation of global H3K4me3, we chose two other genes, Sam-S and Set1, for this study. Set1 encodes the main H3K4 di/tri methyltransferase in Drosophila (35–37). We used RNAi to knock down targets, and efficiency was verified by Western blotting and real time qRT-PCR analysis (Figs. 2B and 5, A and B). Consistent with published work (10, 35–37), reduction of SAM-S or SET1 led to reduced global H3K4me3 levels in S2 cells (Fig. 5, C and D). Interestingly, this decrease caused by reduced SAM-S or SET1 was restored to near control levels upon Sin3A knockdown (Fig. 5, C and D), validating the findings that SIN3 affects global H3K4me3. Our previously published RNA-Seq data show that the expression of multiple histone methyltransferase genes, such as Set1, trithorax (trx), trithorax-related (trr), or absent, small, or homeotic discs 1 (ash1), is not affected when SIN3 is reduced (16). To confirm those results, we knocked down Sin3A and analyzed gene expression by real time qRT-PCR. Sin3A knockdown did not influence the transcription of Set1, trx, and ash1 and only very mildly affected trr expression (Fig. 2C). These real time qRT-PCR results thus validate the RNA-Seq data, suggesting that the role of SIN3 in regulating H3K4me3 is not through the control of expression of these histone methyltransferases. Moreover, these data indicate that SIN3 is critical for the global H3K4me3 response to SAM limitation.
FIGURE 5.
Decreased global H3K4me3 levels caused by reduction of SAM-S or SET1 is restored to near control levels upon Sin3A knockdown. A, verification of Sin3A knockdown. Whole cell extracts from RNAi-treated cells were subjected to Western blotting analysis using the indicated antibodies. α-Tubulin acted as the loading control. Protein size markers are indicated on the right. B, real time qRT-PCR analysis of Sam-S and Set1 transcript level. C, whole cell extracts from RNAi-treated cells were subjected to Western blotting analysis using the indicated antibodies. Protein size markers are indicated on the right. D, Western blots as shown in C were repeated with protein extracts prepared from at least three independent cultures, and the results were quantified after normalization to histone H4. Set1-1 KD and Set1-2 KD used two different dsRNA targeting different regions of Set1 mRNA, but these regions overlap. Therefore, we used Set1-1 oligonucleotides for the rest of the study and referred to it as Set1 KD. The error bars represent standard error of the mean. Statistically significant results comparing individual knockdown samples to the control are indicated on knockdown samples. p values were also calculated between the double knockdown samples and each single knockdown sample for the two tested genes, e.g. Sin3A + Sam-S KD to Sin3A KD or Sin3A + Sam-S KD to Sam-S KD. Statistically significant results are indicated with the bars. GFP RNAi cells are the control cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
In this study, we provide mechanistic insight into the relationship between an epigenetic regulator and metabolism. We first confirmed that SIN3 affects the expression of methionine metabolic genes. Next, we found that SIN3 influences histone acetylation and methylation at the promoters of these genes. Importantly, we observed that SIN3 regulates the levels of SAM and global histone methylation. Given that SIN3 is localized to methionine metabolic genes, these findings indicate that SIN3 directly regulates histone modifications at methionine metabolic genes to modulate their expression, which in turn impacts cellular SAM and global H3K4me3 levels. To our knowledge, this is the first demonstration of a regulatory role of SIN3 on methionine metabolism and consequently on global H3K4me3.
It has been reported that changes in the amount of histone modifier enzymes affect metabolism (3–5). In this report, we demonstrate that altering SIN3, the scaffold protein for assembly of one of the two major cellular histone deacetylase complexes, affects the expression of metabolic genes to impact metabolism. The data support a model in which changing a scaffold protein will alter the assembly and/or recruitment of the functional histone-modifying complex, which then impacts gene expression. Correlation analysis of modENCODE development RNA-Seq data provided by FlyBase (25) revealed that the expression of Sin3A was not obviously correlated with the expression of tested methionine metabolic genes during development (data not shown). We hypothesize that control by the complex will occur because of a change in cellular environmental status that will impact SIN3 complex binding or assembly rather than transcriptional changes to complex components. It will be informative to identify complex component gene specific recruitment under different conditions.
SIN3 and methionine metabolism are conserved from yeast to mammals. The data of this report using the Drosophila model system are consistent with the reports on histone modifiers regulating metabolism in yeast and mammals, which strongly suggests that this process is evolutionarily conserved across different species. In addition to histone modifications, in mammals, but not in Drosophila, DNA can also be modified by methylation. The possible role of SIN3 in the control of DNA methylation is largely unexplored. One report indicates that reduction of SIN3 in human netra-2 cells leads to decreased SIN3 binding at the promoter of the gene encoding glial fibrillary acidic protein (GFAP) and increased Gfap gene expression but no change to DNA methylation (38). More work will be necessary to fully examine the link between SIN3 and DNA methylation in those organisms in which DNA methylation is a critical epigenetic mark.
Our work indicates that SIN3 impacts both global and gene specific H3K4me3. These results are consistent with the previous finding that reduction of both mammalian SIN3A and SIN3B leads to increased H3K4me3 at a specific group of genes in differentiated C2C12 myotubes (39). Given that H3K4me3 is associated with active genes (33, 34), the regulatory role of SIN3 on histone methylation may contribute to the mechanism of how SIN3 affects transcription, which in turn regulates biological processes. Previous studies have found that methionine metabolism is sufficient to determine histone methylation at least in part by modulating SAM levels (7–9). Our data indicate that SIN3 impacts H3K4me3 through affecting the expression of the genes encoding enzymes in this pathway, which ultimately controls the levels of key metabolites. Given that the H3K4me3 specific demethylase dKDM5/LID interacts with SIN3 (29, 30), it is possible that dKDM5/LID also contributes to the effect of SIN3 on H3K4me3, which is an interesting area for further research.
Experimental Procedures
Cell Culture
Drosophila Schneider cell line 2 (S2) cells were cultured at 27 °C in Schneider's Drosophila medium (1×) containing l-glutamine (Life Technologies), 10% heat-inactivated fetal bovine serum (Invitrogen), and 50 mg/ml gentamycin.
dsRNA Production
The protocols for generation of constructs containing targeting sequences in pCRII-Topo vector and production of dsRNA are previously described (10, 40).
RNA Interference
The RNAi procedure is previously described (10, 40). Western blotting analysis and RT-PCR assays were routinely carried out for both single- and double-RNAi-treated cells to verify efficient knockdown of Sin3A and methionine metabolic genes, respectively.
Real Time Quantitative RT-PCR Assay (qRT-PCR)
The protocols for RNA extraction, cDNA preparation and qRT-PCR are previously described (10). Taf1 was used as a normalizer. The primers used for Taf1, Sam-S, Ahcy13, CG10623, and Cbs are previously described (10). Primers for Set1 were taken from a previously published report (37). Primers used for other genes are listed in Table 1. The gene expression changes are represented as the mean ± S.E. of the fold changes observed in Sin3A knockdown cells compared with GFP RNAi control cells. The qRT-PCR experiment utilized RNA isolated from three biological replicates for each cell type.
TABLE 1.
Primers used for qRT-PCR analysis
| Gene | Primer orientation | Primer sequence (oriented 5′ to 3′) |
|---|---|---|
| trr | Forward | CAT TGG CGA GGT TAT CCG |
| Reverse | TTC ATC CAG CCG GAA CAT | |
| trx | Forward | CCG AAA TGC CCA ATG AAG |
| Reverse | GTC CCC AGA AGA GGC CAT | |
| ash1 | Forward | CGT TGA AAA AGC GCC ACT |
| Reverse | CCC ATT GGC GAA TGC TAC |
Metabolomics
Five biological replicates of RNAi-treated Drosophila S2 cells were harvested, flash frozen, and sent to Metabolon Inc. Sample preparation and metabolomic analysis were conducted at Metabolon Inc. as previously described (41). The extracted samples were split into equal parts for analysis with UPLC-MS/MS and GC/MS. Statistically significant differences were determined using one-way analysis of variance with post hoc contrasts (t tests).
Western Blotting Analysis
The Western blotting analysis protocol is previously described (10, 40). Primary antibodies included SIN3 (1:2000 (42)), α-tubulin (1:1000; Cell Signaling), H3K4me2 (1:5000; Millipore), H3K4me3 (1:2500; Active Motif), H3K9me2 (1:500; Millipore), and H4 (1:15,000; Abcam). Donkey anti-rabbit HRP-conjugated IgG (1:3000; GE Healthcare) was used as the secondary antibody. The antibody signals were detected using the clarity western ECL substrate (Bio-Rad) for H3K4me2 and H3K4me3 or ECL prime Western blotting detection system (GE Healthcare) for SIN3, α-tubulin, H3K9me2, and H4. A minimum of three biological replicates was performed.
Chromatin Immunoprecipitation and Real Time Quantitative PCR (ChIP-qPCR)
ChIP-qPCR procedure and antibodies are previously described (10, 16). Primers used for qPCR are listed in Table 2. Three biological replicates were performed.
TABLE 2.
Primers used for ChIP-qPCR analysis
| Gene | Primer orientation | Primer sequence (oriented 5′ to 3′) |
|---|---|---|
| Sam-S | Forward | CCA CAC CTC CAC CGT CTA CT |
| Reverse | CCT CTG TTC AAG TCG TGC AA | |
| Ahcy13 | Forward | CGA AGC CCA GCT ACA AAG TC |
| Reverse | AAT AGA TGC AAT TCA CCC GC | |
| CG10623 | Forward | CGG AAA ACG TAC AGC AGT GA |
| Reverse | GCA TTT GAC CAG AAT TGG CT | |
| Cbs | Forward | CCC TTC CTG TTT CCA TCT GA |
| Reverse | TGC GAA ATT GCG TGA GAT TA |
Statistical Analyses
All significance values, except metabolomics experiment, were calculated by the unpaired two sample Student's t test from GraphPad Software.
Author Contributions
M. L. and L. A. P. designed the study. M. L. performed experiments. M. L. and L. A. P. wrote the paper.
Acknowledgments
We thank members of the Pile laboratory and Drs. Joy Alcedo and Smiti Gupta for critical reading of the manuscript. We thank Dr. Russell L. Finley, Jr., for providing the template DNA to make dsRNA against GFP.
This work was supported by a Grants Plus Award from Wayne State University and NIGMS, National Institutes of Health Grant R01GM088886-01A2 (to L. A. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HDAC
- histone deacetylase
- SAM
- S-adenosylmethionine
- SAM-S
- SAM synthetase
- AHCY
- adenosylhomocysteinase
- CBS
- cystathionine-β-synthase
- KD
- knockdown
- qRT-PCR
- real time quantitative RT-PCR
- ChIP-qPCR
- chromatin immunoprecipitation and real time quantitative PCR.
References
- 1.Kaelin W. G. Jr., and McKnight S. L. (2013) Influence of metabolism on epigenetics and disease. Cell 153, 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sassone-Corsi P. (2013) Physiology: when metabolism and epigenetics converge. Science 339, 148–150 [DOI] [PubMed] [Google Scholar]
- 3.Tateishi K., Okada Y., Kallin E. M., and Zhang Y. (2009) Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., Clish C. B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y., et al. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonneaud A., Turgeon N., Boisvert F. M., Boudreau F., and Asselin C. (2015) Loss of histone deacetylase Hdac1 disrupts metabolic processes in intestinal epithelial cells. FEBS Lett. 589, 2776–2783 [DOI] [PubMed] [Google Scholar]
- 6.Shyh-Chang N., Locasale J. W., Lyssiotis C. A., Zheng Y., Teo R. Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J. J., Zhu H., Asara J. M., Daley G. Q., and Cantley L. C. (2013) Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadhu M. J., Guan Q., Li F., Sales-Lee J., Iavarone A. T., Hammond M. C., Cande W. Z., and Rine J. (2013) Nutritional control of epigenetic processes in yeast and human cells. Genetics 195, 831–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mentch S. J., Mehrmohamadi M., Huang L., Liu X., Gupta D., Mattocks D., Gómez Padilla P., Ables G., Bamman M. M., Thalacker-Mercer A. E., Nichenametla S. N., and Locasale J. W. (2015) Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F., and Kume S. (2014) Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19, 780–794 [DOI] [PubMed] [Google Scholar]
- 10.Liu M., Barnes V. L., and Pile L. A. (2015) Disruption of methionine metabolism in Drosophila melanogaster impacts histone methylation and results in loss of viability. G3 6, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., Swanson S. K., Gogol M., Florens L., Washburn M. P., Workman J. L., and Suganuma T. (2015) Serine and SAM responsive complexSESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell 60, 408–421 [DOI] [PubMed] [Google Scholar]
- 12.Grzenda A., Lomberk G., Zhang J. S., and Urrutia R. (2009) Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverstein R. A., and Ekwall K. (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 14.Hayakawa T., and Nakayama J. (2011) Physiological roles of class I HDAC complex and histone demethylase. J. Biomed. Biotechnol. 2011, 129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaminathan A., Barnes V. L., Fox S., Gammouh S., and Pile L. A. (2012) Identification of genetic suppressors of the Sin3A knockdown wing phenotype. PLoS One 7, e49563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gajan A., Barnes V. L., Liu M., Saha N., and Pile L. A. (2016) The histone demethylase dKDM5/LID interacts with the SIN3 histone deacetylase complex and shares functional similarities with SIN3. Epigenetics Chromatin 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowley S. M., Iritani B. M., Mendrysa S. M., Xu T., Cheng P. F., Yada J., Liggitt H. D., and Eisenman R. N. (2005) The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol. Cell. Biol. 25, 6990–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dannenberg J. H., David G., Zhong S., van der Torre J., Wong W. H., and Depinho R. A. (2005) mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 19, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David G., Grandinetti K. B., Finnerty P. M., Simpson N., Chu G. C., and Depinho R. A. (2008) Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 4168–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neufeld T. P., Tang A. H., and Rubin G. M. (1998) A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pennetta G., and Pauli D. (1998) The Drosophila Sin3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev. Genes Evol. 208, 531–536 [DOI] [PubMed] [Google Scholar]
- 22.Pile L. A., Spellman P. T., Katzenberger R. J., and Wassarman D. A. (2003) The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: implications for the regulation of energy metabolism. J. Biol. Chem. 278, 37840–37848 [DOI] [PubMed] [Google Scholar]
- 23.Barnes V. L., Strunk B. S., Lee I., Hüttemann M., and Pile L. A. (2010) Loss of the SIN3 transcriptional corepressor results in aberrant mitochondrial function. BMC Biochem. 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes V. L., Bhat A., Unnikrishnan A., Heydari A. R., Arking R., and Pile L. A. (2014) SIN3 is critical for stress resistance and modulates adult lifespan. Aging 6, 645–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St. Pierre S. E., Ponting L., Stefancsik R., McQuilton P., and FlyBase Consortium (2014) FlyBase 102: advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42, D780–D788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsson J., and Rasmuson-Lestander A. (1994) Molecular cloning of the S-adenosylmethionine synthetase gene in Drosophila melanogaster. FEBS Lett. 342, 329–333 [DOI] [PubMed] [Google Scholar]
- 27.Caggese C., Ragone G., Barsanti P., Moschetti R., Messina A., Massari S., and Caizzi R. (1997) The S-adenosyl-l-homocysteine hydrolase of Drosophila melanogaster: identification, deduced amino acid sequence and cytological localization of the structural gene. Mol. Gen. Genet. 253, 492–498 [DOI] [PubMed] [Google Scholar]
- 28.Locasale J. W. (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spain M. M., Caruso J. A., Swaminathan A., and Pile L. A. (2010) Drosophila SIN3 isoforms interact with distinct proteins and have unique biological functions. J. Biol. Chem. 285, 27457–27467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moshkin Y. M., Kan T. W., Goodfellow H., Bezstarosti K., Maeda R. K., Pilyugin M., Karch F., Bray S. J., Demmers J. A., and Verrijzer C. P. (2009) Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell 35, 782–793 [DOI] [PubMed] [Google Scholar]
- 31.Saha N., Liu M., Gajan A., and Pile L. A. (2016) Genome-wide studies reveal novel and distinct biological pathways regulated by SIN3 isoforms. BMC Genomics 17, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan Q., Schones D. E., Ho Eun S., Wei G., Cui K., Zhao K., and Chen X. (2010) Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 11, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black J. C., and Whetstine J. R. (2011) Chromatin landscape: methylation beyond transcription. Epigenetics 6, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Black J. C., Van Rechem C., and Whetstine J. R. (2012) Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohan M., Herz H. M., Smith E. R., Zhang Y., Jackson J., Washburn M. P., Florens L., Eissenberg J. C., and Shilatifard A. (2011) TheCOMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31, 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallson G., Hollebakken R. E., Li T., Syrzycka M., Kim I., Cotsworth S., Fitzpatrick K. A., Sinclair D. A., and Honda B. M. (2012) dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 190, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ardehali M. B., Mei A., Zobeck K. L., Caron M., Lis J. T., and Kusch T. (2011) Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 30, 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng P. Y., Lin Y. P., Chen Y. L., Lee Y. C., Tai C. C., Wang Y. T., Chen Y. J., Kao C. F., and Yu J. (2011) Interplay between SIN3A and STAT3 mediates chromatin conformational changes and GFAP expression during cellular differentiation. PLoS One 6, e22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Oevelen C., Wang J., Asp P., Yan Q., Kaelin W. G. Jr, Kluger Y., and Dynlacht B. D. (2008) A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 32, 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pile L. A., Schlag E. M., and Wassarman D. A. (2002) The SIN3/RPD3 deacetylase complex is essential for G(2) phase cell cycle progression and regulation of SMRTER corepressor levels. Mol. Cell. Biol. 22, 4965–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin S. Y., Fauman E. B., Petersen A. K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., Yang T. P., Walter K., Menni C., Chen L., Vasquez L., Valdes A. M., et al. (2014) An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pile L. A., and Wassarman D. A. (2000) Chromosomal localization links the SIN3-RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 19, 6131–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]