Abstract
The pH 6 antigen (pH 6 Ag; PsaA) of Yersinia pestis has been shown to be a virulence factor. In this study, we set out to investigate the possible function of Y. pestis PsaA in a host cell line, RAW264.7 mouse macrophages, in order to better understand the role it might play in virulence. Y. pestis KIM5 derivatives with and without the pCD1 plasmid and their psaA isogenic counterparts and Escherichia coli HB101 and DΗ5α carrying a psaA clone or a vector control were used for macrophage infections. Macrophage-related bacteria and gentamicin-resistant intracellular bacteria generated from plate counting and direct microscopic examinations were used to evaluate these RAW264.7 macrophage infections. Y. pestis psaA isogenic strains did not show any significant difference in their abilities to associate with or bind to mouse macrophage cells. However, expression of psaA appeared to significantly reduce phagocytosis of both Y. pestis and E. coli by mouse macrophages (P < 0.05). Furthermore, we found that complementation of psaA mutant Y. pestis strains could completely restore the ability of the bacteria to resist phagocytosis. Fluorescence microscopy following differential labeling of intracellular and extracellular Y. pestis revealed that significantly lower numbers of psaA-expressing bacteria were located inside the macrophages. Enhanced phagocytosis resistance was specific for bacteria expressing psaA and did not influence the ability of the macrophages to engulf other bacteria. Our data demonstrate that Y. pestis pH 6 Ag does not enhance adhesion to mouse macrophages but rather promotes resistance to phagocytosis.
Yersinia pestis, the causative agent of plague, was originally considered a facultative intracellular parasite that can survive and multiply inside host macrophages (7, 8, 37). The cited studies and others have suggested that the interaction of Y. pestis with host macrophages might be important in the pathogenesis of plague infection. Paradoxically, many of the characterized virulence factors of Y. pestis are involved in maintaining the organism's extracellular location. The best characterized of these virulence determinants is the type III secretion system and the effector proteins, Yops (Yersinia outer proteins). Yops are involved in preventing phagocytosis as well as promoting cytotoxicity and modulation of the immune response (9). Specifically, two of these Yops, YopH and YopE, are involved in the ability of Yersinia to inhibit phagocytosis (30, 32). Grosdent et al. recently reported that YopT and YopO are also involved in resistance to phagocytosis and demonstrated that YopH, YopT, YopO, and YopE act synergistically to increase the resistance of Yersinia enterocolitica to phagocytosis by macrophages (13). Additionally, the Y. pestis-specific capsular antigen F1 also protects the organism from phagocytosis (12). Adhesins such as YadA, Inv, and Ail of Yersinia pseudotuberculosis and Y. enterocolitica have been reported to mediate initial adhesive events between the enteropathogenic Yersinia and eukaryotic cells and thus can manipulate host cell signaling pathways to the advantage of the pathogen (1, 4, 19, 28, 29, 36). However, none of these adhesins appear to be generally functional in Y. pestis (11, 26, 32, 34). Accordingly, the adhesin responsible for Y. pestis attachment and delivery of Yops to host cells remains unidentified.
Two potential adhesins expressed by Y. pestis are Pla and the pH 6 antigen (pH 6 Ag; PsaA). Pla localizes to the outer membrane and has a proteolytic activity that can cleave and activate plasminogen, a property that has been shown to be important for the ability of Y. pestis to infect via the peripheral route (35). Pla could also mediate adherence of Y. pestis to epithelial cells and the mammalian extracellular matrix (10, 22). PsaA of Y. pestis is expressed at 37°C in acidic media, mediates agglutination of erythrocytes of many species, and has been physically recognized as a fimbrial structure (1, 2, 24). Also, psaA mutants of a pigmentation-negative Y. pestis strain have been shown to be attenuated by the intravenous route of infection (23). PsaA has also been suggested to possibly be involved in the binding of the organism to allow effective delivery of Yops to target cells (36). In Y. pseudotuberculosis, PsaA was reported to be a thermoinducible adhesin that allowed binding of the organism to cultured mammalian epithelial cells (18, 20, 39). However, the virulence function of the antigen remains undefined. A recent report demonstrated that purified PsaA selectively binds to apolipoprotein B (apoB)-containing lipoproteins (LDL) in human plasma. LDL at concentrations close to the physiological concentration in human blood (250 μg of human LDL per ml) almost abolished the interaction of purified PsaA with macrophages. This process could prevent recognition of the pathogen by host defense systems (25). It was suggested that immune masking might be important for the ability of the pathogen to cause disease in the susceptible host (25).
In the current study, we set out to investigate the possible function of Y. pestis PsaA by analyzing the interaction of bacteria with host cells either expressing or not expressing the pH 6 Ag. Given that the type III secretion system encoded on the large virulence plasmid, pCD1, is known to be a major antiphagocytic virulence determinant, we included in the macrophage infection assay isogenic plasmid-bearing and plasmid-cured derivatives of psaA mutant and wild-type Y. pestis strains. We also included E. coli HB101 and DΗ5α carrying the psaA cloned locus or a vector control in our macrophage infection assay in order to better understand the role PsaA might play in Y. pestis virulence. Our results indicate that PsaA is not necessary for adherence of Y. pestis to these cells and is not involved in delivery of Yops. However, the ability to express PsaA on the surface of the bacteria is antiphagocytic.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Y. pestis isogenic pairs of psaA+ (KIM5-3001) and psaA mutant (KIM5-3001.1) derivatives were described previously (23). We generated pCD1-negative strains of KIM5-3001 and KIM5-3001.1 by plating the organisms on magnesium-oxalate (MgOX) medium at 37°C as previously described (15). These strains were designated Y. pestis KIM5-4001 and KIM5-4002, respectively (Table 1). The psaA clone, pDG5 (24), and its vector pIC20R were transformed into E. coli HB101 and E. coli DH5α, KIM5-3001.1, and KIM5-4002. After transformation, KIM5-3001.1 carrying pDG5 or pIC20R were designated KIM5-4003 and KIM5-4004, respectively, and KIM5-4002 carrying pCD5 or pIC20R were designated KIM5-4005 and KIM5-4006, respectively (Table 1). To grow bacteria for infection of RAW264.7 mouse macrophage cells, Y. pestis was grown in brain heart infusion (BHI; Invitrogen, Rockville, Md.) broth at 26°C overnight and diluted 1/25 in fresh supplemented BHI (SBHI) broth (23). For induction of psaA the medium was adjusted to pH 6 with HCl before sterilization. Some experiments were performed under Yop-inducing conditions by adding MgOX to the medium (15). Different Y. pestis derivatives were grown in broth for 2 h at 26°C followed by 6 h of growth at 37°C with shaking (180 rpm; Innova 4300; New Brunswick Scientific) to allow PsaA or Yop proteins to be synthesized and were used for infection of macrophage monolayers. E. coli harboring pDG5 or pIC20R was grown in BHI broth at 37°C overnight before RAW264.7 infection (23, 24). When necessary, antibiotics were added to the culture medium as follows: ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), kanamycin (40 μg/ml), and streptomycin (25 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant comment(s) | Source or reference |
|---|---|---|
| Strains | ||
| Y. pestis | ||
| KIM5-3001 | Spontaneous Strr mutant of KIM5 (Pgm−), psaA+ | 23 |
| KIM5-4001 | pCD1− from KIM5-3001, Strr | This study |
| KIM5-3001.1 | psaA mutant of KIM5-3001, Chlr | |
| KIM5-4002 | pCD1− from KIM5-3001.1, Chlr | This study |
| KIM5-4003 | KIM5-3001.1 with pDG5, Chlr Ampr | This study |
| KIM5-4004 | KIM5-3001.1 with pIC20R, Chlr Ampr | This study |
| KIM5-4005 | KIM5-4002 with pDG5, Chlr Ampr | This study |
| KIM5-4006 | KIM5-4002 with pIC20R, Chlr Ampr | This study |
| E. coli | ||
| DH5α | Laboratory stock | |
| HB101 | Laboratory stock | |
| DH5α pUT | Kanr | Laboratory stock |
| Plasmids | ||
| pIC20R | pUC-like plasmid with more restriction sites, Ampr, 9.4-kb ClaI fragment with psaA cloned into pIC20R | 24 |
| pDG5 | 24 |
Macrophage infection assay.
RAW264.7 mouse macrophage cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) high-glucose formulation supplemented with 10% fetal bovine serum, 200 mM l-glutamine, and 10 mM HEPES (Invitrogen) at 37°C in 5% CO2. Macrophage cells (5 × 105) were seeded in 24-well tissue culture plates (1 ml/well). The infection assays were performed as previously described (5, 17, 33). Briefly, Y. pestis derivatives and E. coli strains (23) were added to cell monolayers in 24-well tissue culture plates at a multiplicity of infection generally of 10:1 (bacteria to macrophages). After incubation at 37°C for 30 min, duplicate wells of infected cell monolayers were washed three times with Hank's balance salt solution (Invitrogen) and the number of total macrophage cell-associated bacteria was determined. Total cell-associated bacteria were determined by harvesting in 1 ml of 0.1% Triton X-100 in 1× phosphate-buffered saline (PBS). After 10 min, infected cell lysates were collected and serially diluted 10-fold in PBS, and aliquots were inoculated onto BHI agar plates with suitable antibiotics to assess viable bacterial CFU. This number was considered as the total cell-associated bacteria. A second set of duplicate infected monolayer wells were washed twice with Hank's balance salt solution. Dulbecco's modified Eagle's medium containing 50 μg of gentamicin/ml (Invitrogen) was added to these wells for 2 h to kill extracellular bacteria. The infected monolayers then were lysed and treated as above to determine the number of intracellular bacteria. The percentage of cell-associated bacteria was determined by calculating the total number of cell-associated bacteria divided by the total CFU in the inoculum and multiplying by 100. The percent phagocytosis was calculated by dividing the number of intracellular bacteria by the number of cell-associated bacteria and multiplying by 100. Each experiment was repeated two to four times on different days, and each sample of bacteria was used to infect duplicate wells of macrophage monolayers. Since the experiments were performed on different days, the Bonferroni t test was used for statistical analysis (16).
Western blotting.
Protein samples were prepared from bacteria grown under the same conditions as those described above for infection of macrophages. The bacterial suspensions were adjusted to the same optical density at 600 nm (0.5). A 50-μl volume of bacterial suspension was pelleted, washed, and suspended in protein sample loading buffer (Novex, San Diego, Calif.) and boiled for 15 min. A 15-μl volume of material was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 4 to 20% Tris-glycine gel (Novex, San Diego, Calif.) and transferred to polyvinylidene difluoride membranes (Novex). The primary antibody was anti-F1 monoclonal antibody (MabF1-04-AG1; provided by Jeffery Adamovicz and Gerald Andrews, Bacteriology Division, U.S. Army Medical Research Institute of Infectious Disease, Ft. Detrick, Md.) or rabbit anti-pH 6 Ag polyclonal antibody (23) applied for 1 h at room temperature (RT). After being washed (50 mM Tris, 150 mM NaCl [pH 7.4]) the membranes were incubated with alkaline phosphatase-labeled anti-mouse or anti-rabbit antibody (KPL, Gaithersburg, Md.) for 1 h at RT. Blots were developed by the addition of Western Blue Stabilized Substrate (Promega, Madison, Wis.).
Fluorescence microscopy.
Intra- and extracellular bacteria were identified by a double immunofluorescence assay as previously described (12). Briefly, macrophage cells (∼1 × 105) were seeded in eight-well chamber slides (Lab-Tek II Chamber Slide System154941; Nalge Nunc International, Naperville, Ill.) to make the semiconfluent macrophage monolayers. The infection assay was the same as that described above. After 30 min of infection by KIM5-3001 (psaA+) or KIM5-3001.1 (psaA mutant) the coverslips with the infected monolayers were washed three times with PBS and processed for labeling. To label extracellular bacteria, the infected-monolayer coverslips were incubated with anti-F1 antibody (MabF1-04-AG) for 15 min at RT and rinsed three times with PBS. The coverslips were then fixed in ice-cold methanol for 90 s to permeabilize the macrophage cells and incubated with tetramethyl rhodamine isoctanate-conjugated rabbit anti-mouse immunoglobulin (Jackson ImmunoResearch Laboratories, Inc.) for 30 min at 37°C. To label all macrophage-associated bacteria, the infected-monolayer permeabilized coverslips were again incubated with anti-F1 monoclonal antibody for 30 min at 37°C, washed three times, and finally incubated with fluorescein isothiocyanate-conjugated rabbit anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc.) for 30 min at 37°C. Fluorescent bacteria were viewed with a fluorescence microscope (Nikon E-800) and a confocal system (Bio-Rad Radiance 2100). Under these conditions, intracellular bacteria fluoresce green while extracellular bacteria fluoresce red or orange (12). The overall morphology of macrophage cells was viewed by differential interference contrast microscopy. The infection experiments for fluorescence microscopy were repeated three times. For each sample, five fields from KIM5-derivative-infected monolayers with similar cell densities were observed with the fluorescence microscope. Representative images were photographed with a microscope-mounted camera (Nikon). Extracellular (red-orange) and intracellular (green) bacteria were also counted manually from five fields of one experiment. The Student t test was used for statistical analysis.
RESULTS
Effect of PsaA on phagocytosis and adherence of Y. pestis to RAW264.7 mouse macrophages.
To test whether PsaA of Y. pestis had any influence on adherence to and phagocytosis by macrophages, different Y. pestis KIM5-3001 derivatives were used to infect RAW264.7 mouse macrophage cells. There was no significant difference in the ability of Y. pestis KIM5-3001 (psaA+) and KIM5-3001.1 (psaA mutant) to bind to or associate with RAW264.7 macrophages with or without the virulence plasmid pCD1 (Fig. 1A) as determined by the plate counting method (P > 0.05) under our growth conditions. Approximately 30% of the input bacteria associated with the macrophages regardless of genotype. In contrast, Y. pestis psaA+ strains KIM5-3001 and -4001 showed approximately five- to eightfold lower levels of phagocytosis by the macrophages than did psaA mutant strains KIM5-3001.1 and -4002 (P < 0.05; Fig. 1B). When we compared strain KIM5-3001 (psaA+ pCD1+) with KIM5-4001(psaA+ pCD1−) and KIM5-3001.1 (psaA mutant pCD1+) with KIM5-4002 (psaA mutant pCD1−), we did not detect any significant difference in the phagocytosis of the bacteria, although the trend of uptake of pCD1+ Yersinia was lower than pCD1− Yersinia (Fig. 1B; Table 2). The results of experiments whose results are shown in Fig. 1B were performed with Y. pestis grown in MgOX SBHI at 37°C and pH 6. However, when we grew these Y. pestis derivatives at pH 7 in Yop-inducing MgOX BHI broth instead of pH 6 MgOX SBHI, pCD1-positive strains (KIM5-3001 and -3001.1) showed approximately 24- to 26-fold lower levels of phagocytosis compared to the pCD1− strains (KIM5-4001 and -4002) (Table 2) (P < 0.05). These results suggested that a pH of 7 and a depleted calcium environment are the most efficient conditions for Yop expression and observation of their antiphagocytic effects under our experimental conditions. These results also suggested that the Yops might not be highly expressed under our PsaA-inducing growth conditions. To investigate this possibility, a Western blot experiment was performed. This experiment confirmed that Yops, including YopD, YopN, and YopE, expressed much less or undetectable levels when the KIM5 Y. pestis derivatives were grown in pH 6 MgOX SBHI medium compared to growth in pH 7 MgOX BHI medium (data not shown). Although the pCD1+ psaA+strain KIM5-3001 showed approximately fourfold lower phagocytosis than its psaA mutant counterpart KIM5-3001.1 and the pCD1−psaA+ strain KIM5-4001 showed an approximately fourfold lower rate of uptake than its psaA mutant counterpart, KIM5-4002, after growth at pH 7 in MgOX BHI broth, there was no statistically significant difference (P > 0.05). Taken together, these data suggested that the pH 6 Ag functions at least in part as a newly defined antiphagocytic factor of Y. pestis.
FIG. 1.
RAW264.7 mouse macrophage cell-associated Y. pestis KIM5 derivatives (percent) (A) and phagocytosis (percent) of Y. pestis KIM5 derivatives by the macrophages (B). The infection assay was conducted as described in Materials and Methods. The percent phagocytosis was calculated as the number of intracellular bacteria divided by the macrophage cell-associated bacteria times 100, while the percent cell-associated bacteria was calculated as the number of macrophage cell-associated bacteria divided by the inoculum times 100. Results are shown for KIM5-3001 (psaA+ PCD1+), KIM5-4001 (psaA+ pCD1−), KIM5-3001.1 (psaA mutant pCD1+), and KIM5-4002 (psaA mutant pCD1−). Bars represent means plus standard deviations for four experiments, each with duplicate samples. (A) psaA+ strains KIM5-3001 and KIM5-4001 showed approximately five- to eightfold lower phagocytosis by the macrophages than did psaA mutant strains KIM5-3001.1 and KIM5-4002 (P < 0.05).
TABLE 2.
Comparison of phagocytosis of Y. pestis KIM5 isogenic strains grown in different media by RAW264.7 mouse macrophages
| Y. pestis strain | Phagocytosis (%)a
|
|
|---|---|---|
| Growth in pH 6 MgOX SBHI | Growth in pH 7 MgOX BHI | |
| KIM5-3001 (pCD1+psaA+) | 2.8 ± 2.8* | 0.08 ± 0.07 |
| KIM5-4001 (pCD1−psaA+) | 5.1 ± 3.1 | 2.1 ± 1.8 |
| KIM5-3001.1 (pCD1+psaA mutant) | 22.1 ± 17.0 | 0.37 ± 0.38 |
| KIM5-4002 (pCD1−psaA mutant) | 24.4 ± 19 | 9 ± 2.9 |
| KIM5-4003 (pDG5 pCD1+psaA mutant) | 0.27 ± 0.15 | NDb |
| KIM5-4005 (pDG5 pCD1−psaA mutant) | 0.33 ± 0.34 | ND |
| KIM5-4004 (pIC20R pCD1+psaA mutant) | 10.3 ± 7.8 | ND |
| KIM5-4006 (pIC20R pCD1−psaA mutant) | 12.4 ± 3.4 | ND |
Values represent means ± standard deviations for three or four experiments each with duplicate samples.
ND, not determined
Complementation of psaA mutants.
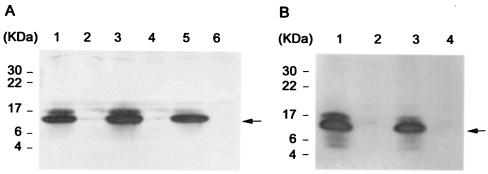
In order to confirm the above results, pDG5 carrying the psaA operon and the vector pIC20R were transformed into psaA mutants KIM5-3001.1 and -4002 (Table 2). Figure 2A shows that psaA was expressed well by pDG5 in those psaA mutants (KIM5-4003 and KIM5-4005). Complementation of Y. pestis psaA mutants resulted in significantly less phagocytosis of the mutant strains (Table 2). The phagocytosis of the psaA mutant strains KIM5-3001.1 and -4002 harboring pDG5 (KIM5-4003 and -4005) by the macrophages was ∼38-fold lower than that of the two psaA mutants carrying the pIC20R vector plasmid (KIM5-4004 and -4006) (Table 2). The influence of pCD1 was not significant in these experiments since the bacteria were not grown under maximum Yop-inducing conditions. As further evidence that PsaA is antiphagocytic, psaA mutant Y. pestis harboring pDG5 (psaA operon) was taken up ∼10-fold less than the isogenic parent strains (compare KIM5-3001 with KIM5-4003 and KIM5-4001 with KIM5-4005 in Table 2). This may have been caused by the higher copy number of the psa operon encoded by pDG5 compared to the chromosomal single copy in the parent strain.
FIG. 2.
Western blot analysis of pH 6 Ag expression by KIM5-3001 derivatives, E. coli HB101, or E. coli DH5α carrying the pH 6 Ag operon clone, pDG5, or the control vector, pIC20R. The arrows indicate pH 6 Ag. (A) Lanes: 1, KIM5-4003 (KIM5-3001.1 carrying pDG5); 2, KIM5-4004 (KIM5-3001.1 carrying pIC20R); 3, KIM5-4005 (KIM5-4002 carrying pDG5); 4, KIM5-4006 (KIM5-4002 carrying pIC20R); 5, KIM5-3001; and 6, KIM5-3001.1. (B) Lanes: 1, E. coli HB101 carrying pDG5; 2, E. coli HB101 carrying pIC20R; 3, E. coli DH5α carrying pDG5; and 4, E. coli DH5α carrying pIC20R.
Levels of F1 antigen expressed by Y. pestis derivatives.
The F1 capsular antigen is well known to be an antiphagocytic factor of Y. pestis (6, 12). In order to investigate if F1 antigen was a factor in the observed antiphagocytosis in these experiments (Fig. 1), the bacteria used for infection of macrophages were analyzed by Western blotting for F1 production. Figure 3 shows that there was no obvious difference in F1 antigen expression by the Y. pestis derivatives. The psaA mutants and the strains with the pDG5 plasmid expressed similar amounts of F1 antigen compared with their parent strains, but the levels of phagocytosis were significantly different (Table 2). These data demonstrated that the observed decrease in phagocytosis of psaA-expressing bacteria was not due to alterations in the level of F1 capsule expression.
FIG. 3.
Western blot analysis of F1 antigen expressed by Y. pestis KIM5-3001 derivatives. The arrowhead indicates the F1 antigen. Lanes:1, KIM5-4003 (KIM5-3001.1 carrying pDG5); 2, KIM5-4004 (KIM5-3001.1 carrying pIC20R); 3, KIM5-4005 (KIM5-4002 carrying pDG5); 4, KIM5-4006 (KIM5-4002 carrying pIC20R); 5, KIM5-3001 (psaA+ pCD1+); 6, KIM5-3001.1 (psaA mutant pCD1+); 7, KIM5-4001 (psaA+ pCD1−); 8, KIM5-4002 (psaA mutant pCD1−). An equal amount of bacteria was loaded in each lane (see Materials and Methods).
Phagocytosis and adherence of E. coli carrying the psaA gene or a vector control to RAW264.7 mouse macrophages.
In order to further investigate PsaA as a newly defined antiphagocytic factor, we wanted to remove other potential Y. pestis factors that might influence our results. We accomplished this by using E. coli harboring clones of psaA and a vector control in our macrophage infection assay. Figure 2B shows that psaA was expressed well in both E. coli HB101 and DH5α. Figure 4 shows the results of experiments with E. coli HB101. Experiments performed with E. coli DH5α revealed similar results (data not shown). The ability to express psaA had a significant effect on the phagocytosis of E. coli (Fig. 4B, P < 0.05). However, the total number of cell-associated bacteria did not show any significant difference (P > 0.05) as determined by plate counting. This result suggested that the ability to express the antigen had no significant impact on adherence of the organism to cultured macrophages. The presence of pDG5 in both E. coli HB101 and DH5α resulted in an approximately fivefold lower level of phagocytosis by the macrophages compared to that of bacteria containing only the vector. These data strongly suggest that the pH 6 Ag alone is responsible for the reduced phagocytosis of both E. coli strains as well as of Y. pestis.
FIG. 4.
RAW264.7 mouse macrophage cell-associated E. coli HB101 carrying the pH 6 Ag operon clone (pDG5) or the control vector (pIC20R) (percent) (A) and phagocytosis (percent) of E. coli HB101 carrying the pH 6 Ag operon clone (pDG5) or the control vector (pIC20R) by the macrophages (B). The infection assay was conducted as described in Materials and Methods. Macrophage cell-associated bacteria (percent) and phagocytosis (percent) were calculated as for Fig. 1. Shown are results for E. coli HB101 carrying pDG5 and E. coli HB101 carrying vector pIC20R. Bars represent means plus standard deviations for three experiments, each with duplicate samples. The ability to express pH 6 Ag had a significant effect on the phagocytosis of E. coli (P < 0.05).
Coinfection and pH 6 Ag-dependent antiphagocytosis.
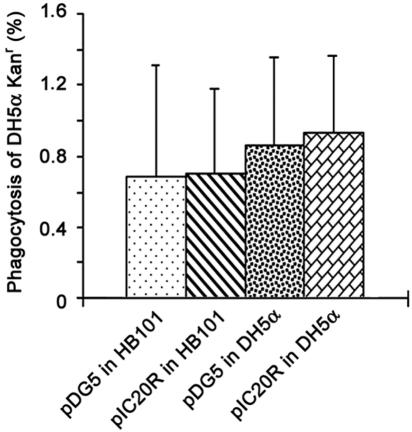
In order to determine whether this antiphagocytosis is specific for PsaA-producing bacteria or a general effect on phagocytosis by the macrophage, we used an E. coli DH5α Kanr strain in coinfection experiments with the cloned locus or vector control in macrophage infection assays. Briefly, E. coli DH5α Kanr was added together with E. coli HB101 carrying pDG5 or pIC20R and E. coli DH5α carrying pDG5 or pIC20R to infect RAW264.7 mouse macrophages. After a 30-min infection, the extracellular bacteria were killed by gentamicin treatment for 2 h and the macrophage monolayers were processed as described in Materials and Methods. Since we did not note any increase in adherence due to PsaA, the efficiency of phagocytosis of E. coli DH5α Kanr was calculated as the number of intracellular bacteria divided by the number of bacteria in the inoculum times 100. There was no significant difference found for phagocytosis of E. coli DH5α Kanr by the macrophages during these coinfections (Fig. 5). These data demonstrate that the antiphagocytosis seen with PsaA-producing bacteria was specifically for cells expressing the antigen and did not affect the general ability of the macrophage to phagocytize E. coli.
FIG. 5.
Phagocytosis (percent) of E. coli DH5α Kanr was calculated as the number of intracellular bacteria divided by the number of bacteria in the inoculum times 100 after coinfection with E. coli HB101 carrying pDG5 or pIC20 and E. coli DH5α carrying pDG5 or pIC20R. Bars represent means plus standard deviations of phagocytosis (percent) of E. coli DH5α Kanr for two experiments, each with duplicate samples. The ratios of the DH5α Kanr bacteria to the competitive bacteria were ∼1:1 to 1:6.
Fluorescence microscopy of Y. pestis KIM5-3001- and KIM5-3001.1-infected RAW264.7 macrophages.
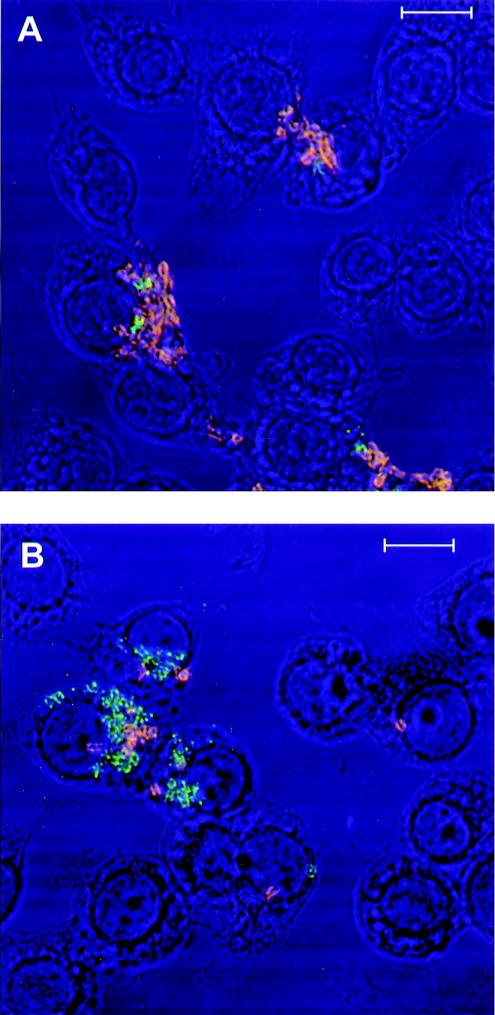
In order to visually confirm the plate counting results presented above fluorescence microscopy was conducted. Figure 6 shows double-fluorescence-labeled bacteria that are located extracellular to (red-orange) or taken up by (green) the macrophages. We examined approximately five fields in each experiment and repeated each experiment three times. In each case it was apparent that the phagocytosis of KIM5-3001.1 (psaA mutant) (Fig. 6B) was significantly more than that of the KIM5-3001 (psaA+) parent strain (Fig. 6A). Fields of Y. pestis producing PsaA always contained a higher proportion of orange-fluorescing bacteria than those of the psaA mutant, visually demonstrating that more psaA-positive bacteria remained outside the macrophages and were not phagocytosed. Table 3 shows that the numbers of phagocytosed (green labeled) KIM5-3001 (psaA+) bacteria are significantly less than those of the KIM5-3001.1 (psaA mutant) (P < 0.01) while the numbers of extracellular (red-orange labeled) KIM5-3001 (psaA+) bacteria are significantly higher than those of the KIM5-3001.1 (psaA mutant) (P < 001). However, there is no significant difference between KIM5-3001 and KIM5-3001.1 when the total numbers of bacteria (extracellular and intracellular) were added together (P > 0.05). This result suggested that the numbers of macrophage-associated bacteria were similar for both KIM5 derivatives. Accordingly, the chance to adhere to the macrophages for both KIM5-3001 (psaA mutant) and KIM5-3001 (psaA+) was similar, but their intracellular or extracellular fate was significantly different.
FIG. 6.
Confocal fluorescence microscopy of RAW264.7 macrophages infected by Y. pestis KIM5-3001(psaA+) (A) and KIM5-3001.1 (psaA mutant) (B). Extracellular bacteria labeled orange, while intracellular bacteria labeled green. Magnification, ×120. Bars, 10 μm.
TABLE 3.
Numbers of Y. pestis KIM5 Isogenic bacteria of phagocytosed by and adherent to RAW264.7 macrophages as determined by fluorescence microscopy
| Strain | No. of extracellular bacteriaa | No. of intracellular bacteriaa | Total no. of extra- and intracellular bacteriab |
|---|---|---|---|
| KIM5-3001 (pCD1+psaA+) | 115.4 ± 27.3 | 65.6 ± 16.5 | 181.0 ± 43.8 |
| KIM5-3001.1 (pCD1+psaA mutant) | 70.0 ± 9.1 | 129.0 ± 22.0 | 199.0 ± 31.1 |
Extracellular bacteria imaged as red-orange (labeled with rhodamine and Fluorescein isothiocyanate [FITO]), while intracellular bacteria imaged as green (labeled with FITC) (see Materials and Methods and Fig. 6). Values represent means ± standard deviations for five fields of bacteria in the semiconfluent mouse macrophage monolayer under a fluorescence microscope.
Values represent means ± standard deviations of total numbers of extra- and intracellular bacteria described in footnote a.
DISCUSSION
Previously, it has been shown that Y. pestis, once grown inside phagocytes, was able to resist subsequent phagocytosis (7). Several other Yersinia factors have been shown to be antiphagocytic. Four of the type III secretion system effector proteins, YopH, YopT, YopO, and YopE, are particularly important in the ability of Y. pestis to inhibit phagocytosis (9). Also, F1 capsular antigen has been reported to block phagocytosis (12). The current study adds to the antiphagocytic armament of Y. pestis by showing that the pH 6 Ag also has the function of inhibiting phagocytosis. In our experiments, Y. pestis with and without the pCD1 plasmid (KIM5-3001 and -4001) always revealed an approximately five- to eightfold lower level of phagocytosis than the psaA-negative isogenic strain when analyzed by plate counting. This difference was not due to differences in the level of F1 antigen expression. Furthermore, E. coli HB101 and DH5α harboring a psaA operon clone (pDG5 plasmid) also showed significantly lower phagocytosis than the bacteria carrying the vector only. The psaA clone, pDG5, completely restored the ability of Y. pestis psaA mutants (KIM5-3001.1 and -4002) to resist phagocytosis. Additionally, expression of some Yops (YopE, YopN, and YopD) had been greatly reduced when growth was at pH 6. These results demonstrate that besides Yops and F1 antigen, PsaA is another factor that contributes to resistance to phagocytosis by Y. pestis and is Yop and F1 antigen independent. In the current study, we did not see a significant difference between the total numbers of cell-associated bacteria (P > 0.05) when psaA isogenic strains were compared. However, there was a significant difference between the extracellular and intracellular bacterial distribution between psaA+ and psaA mutant strains (Table 3 and Fig. 6). Under the fluorescence microscope KIM5-3001 (psaA+) showed an approximately twofold lower level of phagocytosis than the psaA mutant counterpart (Table 3). The lower ratio of internalized mutant bacteria noted by microscopy may have been caused by the bacteria overlapping each other. Superimposed bacteria cannot be recognized by microscopy and would be counted as one bacterium. However, the trend for both methods was the same. Taken together, these results indicated that the pH 6 Ag does not appear to promote adherence of the bacteria to macrophages but rather functions at least partially as an antiphagocytic factor. Furthermore, our data did not show any significant difference between pCD1+ and pCD1− strains in either psaA wild-type or mutant Y. pestis backgrounds and the ability to resist phagocytosis under pH 6 Ag-inducing conditions. Accordingly, pH 6 Ag is unlikely to be the adhesin responsible for the delivery of Yops to host cells in this species, and the adhesin responsible for delivery of Yops to host cells by Y. pestis remains unknown.
PsaA of Y. pestis has been shown to be induced inside macrophages (24, 25). It may be one of the reasons why Y. pestis, once grown inside phagocytes, is able to resist uptake (7). Previously it was shown that Y. pestis bacteria strongly producing PsaA were more rapidly fatal to mice than bacteria not expressing this antigen (1). Furthermore, animal studies have found an ∼200-fold increase in the 50% lethal dose of the psaA mutant compared with that of the parent Y. pestis (23). These previous results suggest that PsaA is involved in the pathogenesis of plague. Our current results suggest that PsaA contributes to virulence by at least preventing phagocytosis of Y. pestis. Our demonstration that the antiphagocytic Yops may be reduced in expression under conditions that induce psaA expression suggest a potential role for the antigen during the early stages of Y. pestis infection. Given that PsaA is produced inside macrophages under acidic conditions (24), the antigen may be the first antiphagocytic factor produced by the organism after release from infected macrophages. Further experimentation will be required to define the relationship of PsaA to other previously defined antiphagocytic factors and determine if pH 6 Ag performs other functions that contribute to virulence of the organism.
Our results are consistent with the findings and hypothesis of Makoveichuk et al. (25), who showed that purified pH 6 Ag selectively binds to apoB-containing lipoproteins in human plasma. These researchers suggest that pH 6 Ag after binding to the lipoprotein could prevent recognition of the pathogen by the host defenses. The lower efficiency of uptake of pH 6 Ag-positive organisms, both Y. pestis and E. coli, by mouse macrophages compared with that of their pH 6 Ag-negative counterparts might be caused by the pH 6 Ag binding to the apoB-containing lipoprotein in fetal calf serum in cell culture media and preventing the bacteria from being phagocytosed by the macrophages. Previously, Y. pestis pH 6 Ag was shown to bind to β1-linked galactosyl residues in glycosphingolipids, which are likely found on a range of host cells (27). Our observation that the pH 6 Ag did not appear to promote significant binding of Y. pestis to macrophages may be due to prebinding of lipoprotein and thus competition with receptors on the host cell surface. Our data is in contrast with the results of Yang et al. (39), who found that the psa locus is responsible for thermoinducible binding of Y. pseudotuberculosis to cultured cells. This may be due to the use of a different host cell type and/or culture conditions. Also Y. pseudotuberculosis encodes different adhesins than Y. pestis, as has been shown for the inv, ail, and yadA genes (11, 26, 32, 34). Accordingly, these other adhesin/invasin proteins may play an interactive role in the ability of pH 6 Ag to promote adherence of Y. pseudotuberculosis to host cells.
PsaA-mediated antiphagocytosis seemed specific to the pH 6 Ag-expressing organisms, as shown in our coinfection assays with differentially marked E. coli bacteria (Fig. 5). These results suggested that the mechanism of PsaA-mediated antiphagocytosis is different from that of Yops. YopE has been demonstrated to function as a GTPase-activating protein to downregulate multiple Rho GTPases (3, 38), which leads to disruption of actin microfilaments in the target cell (30, 31). YopH is homologous to eukaryotic protein tyrosine phosphatases (14), which induce overall dephosphorylation of host proteins and are able to interfere with early tyrosine phosphorylation signals that occur in the cell during phagocytosis. YopT and YopO both are involved in preventing actin rearrangement in host cells and inhibition of phagocytosis (13, 21, 40). The effect of these Yop proteins results in a general failure of macrophages to phagocytose extracellular bacteria. F1 antigen has been reported to block phagocytosis by a mechanism different from that of the type III secretion system, presumably by preventing bacterium-host cell receptor interaction that potentially could result in the uptake of the pathogen (12). The mechanism of pH 6 Ag-mediated phagocytosis resistance may be similar to that of the F1 antigen since it also did not have any influence on the general phagocytic ability of the macrophages. Additionally, our results indicate that there may be other Y. pestis-encoded factors independent of Yops (e.g., pCD1) that promote antiphagocytosis by expression of pH 6 Ag. This is suggested by the ∼38-fold decrease in uptake of pDG5-complemented Y. pestis psaA mutant strains compared to the ∼5-fold decrease seen when comparing E. coli clones to vector controls, given that the copy number of these plasmids is similar in the two different genera.
In summary, PsaA of Y. pestis provides antiphagocytic protection in addition to that provided by Yops and the F1 capsule antigen, which help the bacteria escape host defense mechanisms. PsaA mediates antiphagocytosis probably by preventing adhesin-receptor interaction similar to the F1 antigen or possibly by selectively binding to apoB-containing lipoproteins to avoid recognition by host macrophages. The fact that PsaA is antiphagocytic and has been shown to be induced inside macrophages suggests a possible role for the protein in the early stages of Y. pestis infection. Experiments to address this question are currently under way.
Acknowledgments
We thank Sara Cohn for helpful discussions. Jeffery Adamovicz and Gerald Andrews provided anti-F1 and anti-Yops monoclonal antibodies. Morrissette Craig performed statistical analysis. Michael Zidanic is gratefully acknowledged for performing the fluorescence microscopy. We thank Lee Collins for artwork.
This work was supported by the U.S. Army Medical Research and Materiel Command.
The views reported in this paper are those of the authors and do not reflect those of the U.S. Army or of the Department of Defense.
Editor: J. B. Bliska
REFERENCES
- 1.Ben-Efraim, S., M. Aronson, and L. Bichowsky-Slomnicki. 1961. New antigenic component of Pasteurella pestis formed under specific conditions of pH and temperature. J. Bacteriol. 81:704-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bichowsky-Slomnicki, L., and S. Ben-Efraim 1963. Biological activities in the extracts of Pasteurella pestis and their relationship to the “pH 6 antigen.” J. Bacteriol. 86:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. S., and J. B. Bliska. 2000. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol. 37:515-527. [DOI] [PubMed] [Google Scholar]
- 4.Bliska, J. B., M. C. Copass, and S. Falkow. 1993. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect. Immun. 61:3914-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland, A., and G. R. Cornelis. 1998. Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect. Immun. 66:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker, R. R. 1991. Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanaugh, D. C., and R. Randall. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J. Immunol. 83:348-363. [PubMed] [Google Scholar]
- 8.Charnetzky, W. T., and W. W. Shuford. 1985. Survival and growth of Yersinia pestis within macrophages and an effect of the loss of the 47-megadalton plasmid on growth in macrophages. Infect. Immun. 47:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 68:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, D. C. Schwartz, S. Zhou, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosdent, N., I. Maridonneau-Parini, M. P. Sory, and G. R. Cornelis. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect Immun. 70:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, K. L., and J. E. Dixon. 1990. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science 249:553-556. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi, K., and J. L. Smith. 1961. Studies on the nutrition and physiology of Pasteurella pestis. VI. A differential plating medium for the estimation of the mutation rate to avirulence. J. Bacteriol. 81:605-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochberg, Y., and A. C. Tamhane 1987. Mutiple comparison procedures. John Wiley & Sons, Inc., New York, N.Y.
- 17.Huang, X. Z., B. Tall, W. Schwan, and D. J. Kopecko. 1998. Salmonella typhi entry into intestinal epithelial cells. Jpn. J. Med. Sci. Biol. 51:S90. [DOI] [PubMed] [Google Scholar]
- 18.Isberg, R. R. 1989. Determinants for thermoinducible cell binding and plasmid-encoded cellular penetration detected in the absence of the Yersinia pseudotuberculosis invasin protein. Infect. Immun. 57:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isberg, R. R. 1991. Discrimination between intracellular uptake and surface adhesion of bacterial pathogens. Science 252:934-938. [DOI] [PubMed] [Google Scholar]
- 20.Isberg, R. R. 1989. Mammalian cell adhesion functions and cellular penetration of enteropathogenic Yersinia species. Mol. Microbiol. 3:1449-1453. [DOI] [PubMed] [Google Scholar]
- 21.Juris, S. J., A. E. Rudolph, D. Huddler, K. Orth, and J. E. Dixon. 2000. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc. Natl. Acad. Sci. USA 97:9431-9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahteenmaki, K., R. Virkola, A. Saren, L. Emody, and T. K. Korhonen. 1998. Expression of plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect. Immun. 66:5755-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindler, L. E., M. S. Klempner, and S. C. Straley. 1990. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect. Immun. 58:2569-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindler, L. E., and B. D. Tall. 1993. Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol. Microbiol. 8:311-324. [DOI] [PubMed] [Google Scholar]
- 25.Makoveichuk, E., P. Cherepanov, S. Lundberg, A. Forsberg, and G. Olivecrona. 2003. pH6 antigen of Yersinia pestis interacts with plasma lipoproteins and cell membranes. J. Lipid Res. 44:320-330. [DOI] [PubMed] [Google Scholar]
- 26.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 27.Payne, D., D. Tatham, E. D. Williamson, and R. W. Titball. 1998. The pH 6 antigen of Yersinia pestis binds to β1-linked galactosyl residues in glycosphingolipids. Infect. Immun. 66:4545-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierson, D. E., and S. Falkow. 1993. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect. Immun. 61:1846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roggenkamp, A., H. R. Neuberger, A. Flugel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol. 16:1207-1219. [DOI] [PubMed] [Google Scholar]
- 30.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 31.Rosqvist, R., A. Forsberg, and H. Wolf-Watz. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334:522-524. [DOI] [PubMed] [Google Scholar]
- 33.Schwan, W. R., X. Z. Huang, L. Hu, and D. J. Kopecko. 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect. Immun. 68:1005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonet, M., B. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sodeinde, O. A., Y. V. Subrahmanyam, K. Stark, T. Quan, Y. Bao, and J. D. Goguen. 1992. A surface protease and the invasive character of plague. Science 258:1004-1007. [DOI] [PubMed] [Google Scholar]
- 36.Straley, S. C. 1993. Adhesins in Yersinia pestis. Trends Microbiol. 1:285-286. [DOI] [PubMed] [Google Scholar]
- 37.Straley, S. C., and P. A. Harmon. 1984. Yersinia pestis grows within phagolysosomes in mouse peritoneal macrophages. Infect. Immun. 45:655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Von Pawel-Rammingen, U., M. V. Telepnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]
- 39.Yang, Y., J. J. Merriam, J. P. Mueller, and R. R. Isberg. 1996. The psa locus is responsible for thermoinducible binding of Yersinia pseudotuberculosis to cultured cells. Infect Immun. 64:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zumbihl, R., M. Aepfelbacher, A. Andor, C. A. Jacobi, K. Ruckdeschel, B. Rouot, and J. Heesemann. 1999. The cytotoxin YopT of Yersinia enterocolitica induces modification and cellular redistribution of the small GTP-binding protein RhoA. J. Biol. Chem. 274:29289-29293. [DOI] [PubMed] [Google Scholar]