Arabidopsis EXTRA-LARGE G PROTEINs interact with two PUB proteins in mediating the cytokinin response and multiple developmental processes.
Abstract
Heterotrimeric GTP-binding proteins (G proteins) composed of Gα, Gβ, and Gγ subunits are conserved signal transduction molecules in animals and plants. In Arabidopsis (Arabidopsis thaliana), there are three Gα-like proteins named EXTRA-LARGE G PROTEINs (XLGs) in addition to the canonical Gα protein GPA1. XLGs have been reported to be implicated in multiple pathways, although the underlying mechanisms of their action remain elusive. Here, we report that all three XLGs interact with two closely related plant U-box (PUB) E3 ligases, PUB2 and PUB4. Three XLGs are predominantly localized at the plasma membrane, whereas XLG2 and XLG3 also show nuclear localization. The interactions between PUB2/4 and XLGs suggest that they might function in the same pathways. Indeed, we found that a newly obtained xlg1/2/3 triple knockout mutant, the pub4 mutant, and the pub2/4 double mutant all exhibited defects in cytokinin responses, stamen development, tapetum development, and male fertility. However, the xlg single mutants and the pub2 mutant did not exhibit an obvious defect in these processes, which suggests functional redundancy among the three XLGs and between PUB2 and PUB4. Overexpressing ARR10 to enhance the cytokinin response in pub4 or in the xlg1/2/3 triple mutant partially restored several phenotypes caused by the pub4 and xlg1/2/3 mutations. Our findings reveal that the XLGs and PUB2/4 are components in the complex cytokinin signaling networks regulating many developmental and physiological processes.
Heterotrimeric GTP-binding proteins (G proteins) are highly conserved signaling components in eukaryotes. G proteins consist of Gα, Gβ, and Gγ subunits (Hamm, 1998; Oldham and Hamm, 2008). In animals, G proteins are typically regulated by a G-protein-coupled receptor (GPCR). Following recognition of a ligand, GPCR is activated and recruits a G protein. GDP-bound Gα then becomes GTP-bound and separated from Gβγ to activate downstream effectors (Wettschureck and Offermanns, 2005; Li et al., 2007; Oldham and Hamm, 2008).
Arabidopsis (Arabidopsis thaliana) contains one canonical Gα (GPA1), one Gβ (AGB1), and three Gγ proteins (AGGs; Urano et al., 2013). In addition, the Arabidopsis genome encodes three EXTRA-LARGE G PROTEINs (XLGs), which have a Gα-like C terminus and an N-terminal region that shares homology among the three XLG proteins but has little similarity to other proteins (Ding et al., 2008). Like canonical Gα proteins, the C-terminal Gα-like domain has the ability to bind and hydrolyze GTP in the presence of Ca2+ instead of Mg2+ (Heo et al., 2012). All three XLGs were found predominantly localized in the plasma membrane, whereas XLG2 and XLG3 were also found in nuclei (Chakravorty et al., 2015; Maruta et al., 2015). XLGs interact and form complexes with Gβγ and have been reported to share similar function with Gβ and Gγ in many developmental processes and stress responses (Zhu et al., 2009; Chakravorty et al., 2015; Maruta et al., 2015). Like agb1, xlg3 mutants were impaired in the root waving and skewing response and were hypersensitive to ethylene (Pandey et al., 2008). An xlg triple mutant, agb1, and agg1/2 also showed increased sensitivity to salt, tunicamycin, and D-Glc and had an increased stomatal density (Chakravorty et al., 2015). XLGs, AGB1, and AGG1/2 are known to be positive regulators of defense responses to several pathogens, and their loss-of-function mutants were compromised in the pathogen-associated molecular pattern-triggered oxidative burst, in disease resistance, or in BIR1-mediated defense signaling and programmed cell death (Zhu et al., 2009; Delgado-Cerezo et al., 2012; Lee et al., 2013; Liu et al., 2013; Maruta et al., 2015; Liang et al., 2016). XLG2 also interacts with RELATED TO VERNALIZATION1 (RTV1) and is involved in RTV1-mediated flowering time control (Heo et al., 2012).
Ubiquitin-mediated protein degradation is a key regulatory mechanism to modulate protein activities in a wide variety of fundamental cellular processes. In this process, ubiquitin is first activated by the ubiquitin activating enzyme (E1), then transferred to an ubiquitin conjugating enzyme (E2), and finally added to a specific Lys residue of target proteins by ubiquitin ligase (E3). Polyubiquitinated proteins are generally targeted for degradation by the 26S proteasome. Monoubiquitination, however, often regulates protein activities. E3 proteins are highly diversified and are responsible for selecting specific target proteins for ubiquitination. E3 ligases are classified into four categories: HECT, RING-finger, U-box, and PHD-finger. Plant U-box proteins (PUBs) are classified into several subgroups based on the other domains besides the U-box (Yee and Goring, 2009). One subgroup contains Armadillo (ARM) repeats, which are often involved in protein-protein interactions (Hatzfeld, 1999; Samuel et al., 2006). PUB proteins are known to function in diverse biological pathways. Brassica ARC1 interacts with an S-locus receptor kinase to regulate self-incompatibility (Stone et al., 2003). Arabidopsis PUB12 and PUB13 ubiquitinate FLAGELLIN-SENSITIVE2 (FLS2) and regulate FLS2-mediated immunity (Lu et al., 2011). Arabidopsis pub4 mutation causes aberrant expansion and incomplete degeneration of tapetal cells (Wang et al., 2013). The pub4 mutant also shows increased cell proliferation in both shoot apical meristem and root meristem (Kinoshita et al., 2015a, 2015b). A recent report also shows that PUB4 is involved in individual chloroplast quality control upon oxidative stress (Woodson et al., 2015).
Here, we report the identification of PUB2 and PUB4 as XLG-interacting proteins. A newly generated xlg1/2/3 triple mutant, the pub4 single mutant, and the pub2/4 double mutant were found to exhibit similar morphological phenotypes and were compromised in the cytokinin response. They were also defective in stamen and tapetum development. Overexpression of ARR10 (a positive regulator of cytokinin signaling) partially restored the phenotypes of pub4 and the xlg triple mutant. Our study indicates that PUB2/4 and XLGs play important roles in multiple developmental and physiological responses and function in the cytokinin responses.
RESULTS
XLGs Interact with PUB4 and PUB2 in Yeast
We performed yeast two-hybrid (Y2H) screen to identify XLG2-interacting proteins. Full-length XLG2 was fused to the GAL4 DNA-binding domain (BD) and used as a bait to screen a normalized Arabidopsis Y2H cDNA library in which each cDNA was fused to the activation domain (AD) sequence. About 5 × 106 clones were screened on the synthetic defined (SD) selection medium (−Trp, −Leu, +X-α-Gal, and +aureobasidin A). Positive clones were further grown on the SD medium lacking Trp, Leu, Ade, and His. A total of 37 clones were obtained from the screen. Among them, 12 clones harbored PUB2-encoding sequences and five clones harbored PUB4-encoding sequences. In a similar Y2H screen using the full-length PUB4 as bait, 16 XLG1 clones and seven XLG2 clones were identified among the positive colonies harboring putative PUB4-interacting proteins.
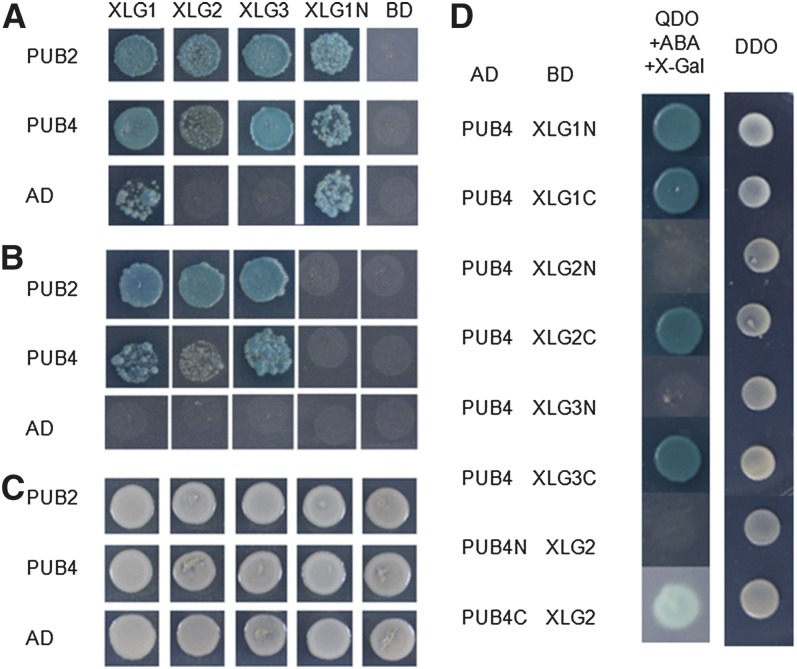
To verify and determine the interactions between XLGs with PUB2 and PUB4 in the Y2H system, all three XLGs (full length) were further tested for their interactions with PUB2 and PUB4. XLG1 and the N terminus of XLG1 were found to cause autoactivation when they were fused to Gal4 DNA-binding domain (Fig. 1A), but the autoactivation could be suppressed by adding 5 mmol 3-amino-1,2,4-triazole (3-AT; Fig. 1B). All of the diploids with AD-PUB2/4 and BD-XLGs could grow on plates with SD/−Ade/−His/−Leu/−Trp/X-α-Gal/Aureobasidin A and activate β-galactosidase activity. 3-AT didn’t impair growth of these diploids or eliminate β-galactosidase activity (Fig. 1B). These data indicate that the three XLGs could interact with both PUB2 and PUB4 in yeast.
Figure 1.
Interactions of XLGs with PUB2/4 in yeast. A, Diploids with BD-XLGs and AD-PUB2/4 were grown on SD plates with SD/−Ade/−His/−Leu/−Trp/X-α-Gal (40 μg/mL)/aureobasidin A (200 ng/mL) for 4 d. B, Diploids with BD-XLGs and AD-PUB2/4 were grown on SD plates with SD/−Ade/−His/−Leu/−Trp/X-α-Gal (40 μg/mL)/aureobasidin A (200 ng/mL) plus 5 mmol 3-AT for 4 d. C, Diploids with BD-XLGs and AD-PUB2/4 were grown on SD plates with SD/−Leu/−Trp for 3 d. D, Diploids with full-length/truncated BD-XLGs and full-length/truncated AD-PUB4 were grown on SD plates with SD/−Ade/−His/−Leu/−Trp/X-α-Gal (40 μg/mL)/aureobasidin A (125 ng/mL) for 4 d (left) and SD plate with SD/−Leu/−Trp for 3 d (right). XLG1N, Amino acids 1 to 481 of XLG1; XLG1C, amino acids 482 to 888 of XLG1; XLG2N, amino acids 1 to 460 of XLG2; XLG2C, amino acids 461 to 861 of XLG2; XLG3N, amino acids 1 to 427 of XLG3; XLG3C, amino acids 428 to 839 of XLG3; PUB4N, amino acids 1 to 525 of PUB4; PUB4C, amino acids 526 to 829 of PUB4.
To further determine which regions of PUB4 and XLGs interact in yeast, the C terminus containing the ARM-repeat domain (526–829) and N terminus (1–525) of PUB4 were tested for their interactions with XLG2. In addition, the N-terminal regions and the C-terminal Gα domain of all three XLGs were tested for their interaction with PUB4. It was found that the interactions between PUB4 and XLGs all occurred between the C-terminal Gα regions of XLGs and the C-terminal ARM-repeat domains of PUB4 (Fig. 1D; Supplemental Fig. S1A).
XLGs Interact with PUB4 and PUB2 in Plants
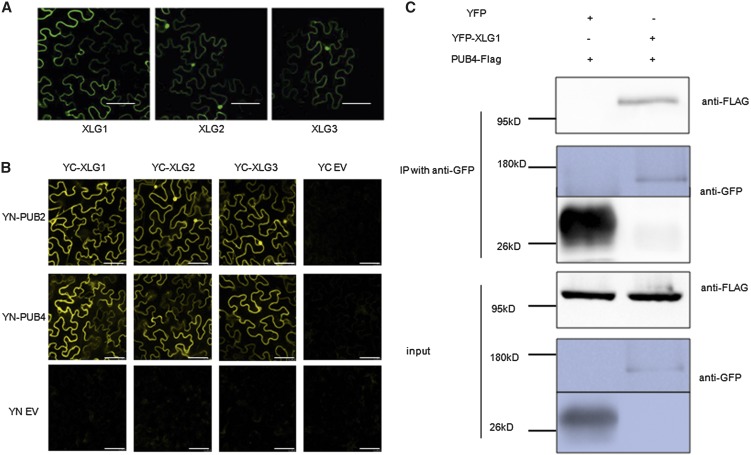
We then tested the interactions of PUB2/4 with XLGs in planta. First, the localization of XLGs was determined by transient expression of the XLG proteins fused to GFP in tobacco (Nicotiana benthamiana). As shown in Figure 2A, the XLG1-GFP protein was mainly localized to the plasma membrane, while XLG2-GFP and XLG3-GFP were localized to both plasma membrane and nuclei in tobacco (Fig. 2A). The subcellular localization patterns of the XLG proteins are consistent with recent reports (Chakravorty et al., 2015; Maruta et al., 2015). PUB4 was previously reported to be localized in cytoplasm, whereas the interactions between PUB2/4 and XLGs would suggest that PUB2 and PUB4 might also be localized in the plasma membrane. We evaluated the possibility that the previous studies might not yield sufficient resolution to distinguish between the cytosol- and plasma membrane-localized fluorescence signals. We extracted membrane fraction and soluble fraction from PUB4-flag transgenic plants. Although most PUB4 proteins were apparently in the cytosol, some PUB4 proteins could be detected as membrane-associated (Supplemental Fig. S2A). Using the plasma membrane marker PIP2A (Nelson et al., 2007) as the control, it was shown that the PUB2-GFP and PUB4-GFP signals were colocalized with PIP2A at the plasma membrane in tobacco cells (Supplemental Fig. S2B). Similarly, colocalization of PUB4-CFP and YFP-XLG3 was also observed in the tobacco cells (Supplemental Fig. S2C).
Figure 2.
XLGs interact with PUB2/4 in planta. A, The fluorescent signal in cells of tobacco following transient expression of XLG1-GFP (left), XLG2-GFP (middle), and XLG3-GFP (right). Localization of the XLG-GFP fusion proteins was analyzed 48 h after agrobacterial infiltration by confocal laser scanning microscopy. Bars = 80 μm. B, XLGs interact with PUB2/4 in tobacco as revealed by BiFC assay. The PUB2/4 proteins were fused to the N terminus of YFP, and the XLG proteins were fused to the C terminus of YFP. YN and YC refer to the empty vectors with just the N terminus or C terminus of YFP, respectively. The signal was observed 48 h after infiltration. Bars = 61.5 μm. C, Co-IP assay showed that XLG1 interacts with PUB4. Total protein was extracted from transgenic plants expressing both YFP-tagged XLG1 and FLAG-tagged PUB4, and plants expressing both YFP and FLAG-tagged PUB4 were used as a negative control. Proteins were immunoprecipitated with anti-GFP magnetic beads, and the precipitated proteins were detected using a FLAG antibody. PUB4-FLAG could be coimmunoprecipitated with YFP-XLG1 but not with YFP.
Interactions between PUB2/4 and XLGs in planta were examined by bimolecular fluorescence complementation (BiFC) assay. PUB2 and PUB4 were fused to the N terminus of eYFP (YN), and XLGs were fused to the C terminus of eYFP (YC). Each pair of YN-PUB and YC-XLG constructs was delivered into tobacco cells via Agrobacterium tumefaciens infiltration. No fluorescence appeared with coinfiltration of empty YN with YC-XLGs or YC with YN-PUB2/4. Reconstituted YFP fluorescence suggested that the interactions between PUB4 and XLGs predominantly occurred in the plasma membrane, whereas PUB2 also interacted with XLG2 and XLG3 in the nuclei (Fig. 2B).
To further confirm the interactions between these two PUBs and XLGs, XLG1 and PUB4 were chosen for coimmunoprecipitation (co-IP) assays using transgenic plants expressing the PUB4-FLAG and YFP-XLG1 fusion proteins. The plants carrying PUB4-FLAG and YFP were used as negative controls. YFP-XLG1 was immunoprecipitated from total leaf protein extract with GFP antibodies, and the co-IPed proteins were then detected using a FLAG antibody (Fig. 2C). PUB4-FLAG was only detected in the precipitate from leaves that expressed both YFP-XLG1 and PUB4-FLAG, but not in that of the negative control, indicating that XLG1 and PUB4 interact in planta.
PUB2 and PUB4 Are Partially Functionally Redundant
The PUB E3 ligase family has more than 60 members in Arabidopsis (Yee and Goring, 2009). Among them, PUB2 shares the highest identity with PUB4. Morphological phenotypes caused by loss-of-function mutation of PUB4 have been reported (Wang et al., 2013) and include slightly smaller rosette sizes and low fertility at 22°C. We obtained a pub2 mutant (SALK_086656), which contains a T-DNA insertion at the fourth intron and is apparently a knockout allele since we could not detect any PUB2 transcript by reverse transcription-PCR (Supplemental Fig. S3). The pub2 mutant did not show any obvious morphological phenotype. The PUB2 gene appears to be expressed at a lower level than PUB4 in most tissues according to the public data from Affymetrix Arabidopsis ATH1 Genome Array Data (https://genevisible.com/tissues/AT/Ensembl%20Gene/AT5G67340 and https://genevisible.com/tissues/AT/Ensembl%20Gene/AT2G23140; Supplemental Fig. S4). We also analyzed several sets of microarray and RNA-seq data from EBI-ArrayExpress (http://www.ebi.ac.uk/arrayexpress/). PUB4 appears expressed at a higher level than PUB2 in shoot, seedlings, stems, flower buds, pollen, and siliques, while PUB2 had a much higher expression level in stigma. When treated with either auxin or cytokinin, PUB4 was found to be expressed at a much higher level than PUB2 (Zhao et al., 2010).
When PUB2 was ectopically overexpressed under the control of the 35S promoter in the pub4 mutant, 36 of 43 independent transgenic lines displayed different degrees of fertility restoration (Supplemental Fig. S5). Furthermore, the pub2 pub4 (pub2/4) double mutant is much smaller than pub4 (Fig. 3, A and B). The above results indicate that PUB2 and PUB4 might have overlapping functions.
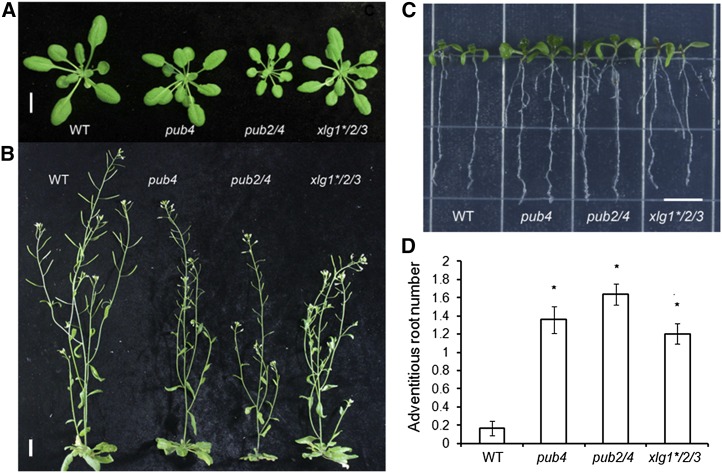
Figure 3.
Morphological phenotypes of pub4, pub2/4, and xlg1*/2/3. A, Four-week-old plants of wild type (WT), pub4, pub2/4, and xlg1*/2/3 grown in soil under the short-day conditions. Bar = 1 cm. B, Six-week-old plants of wild type, pub4, pub2/4, and xlg1*/2/3 grown in soil under the long-day conditions. Bar = 2 cm. C, Nine-day-old seedlings of wild type, pub4, pub2/4, and xlg1*/2/3 grown on Murashige and Skoog plates under long-day conditions. Bar = 1 cm. D, Adventitious root numbers of 9-d-old seedlings of wild type, pub4, pub2/4, and xlg1*/2/3. Vertical bars represent mean ± se (n > 20). Asterisks indicate statistical differences to the wild type (P < 0.05).
XLG1, XLG2, and XLG3 Have Overlapping Functions and Their Triple Mutant Shares Similar Phenotypes to Those of pub4
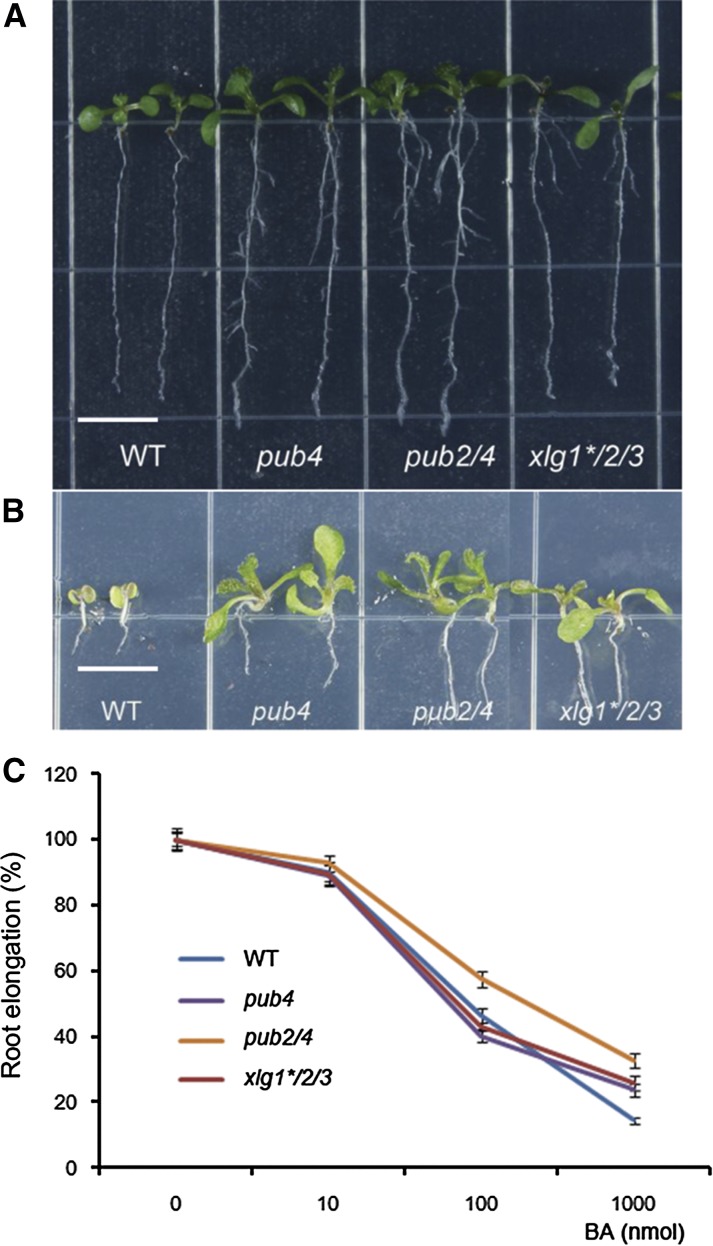
Previous studies revealed that the xlg mutants exhibited multiple developmental phenotypes and showed altered responses to both abiotic and biotic stresses. However, under normal conditions, the xlg1-1 xlg2-1 xlg3-1 triple mutant in the previous reports lacked an obvious morphological phenotype (Ding et al., 2008; Pandey et al., 2008; Zhu et al., 2009; Chakravorty et al., 2015; Maruta et al., 2015). The xlg1-1 allele is considered to be a knockdown as XLG1 expression was still detectable in the mutant, although at a low level (Ding et al., 2008). We obtained another xlg1 T-DNA insertion allele, xlg1-2 (SALK_119657), which contains a T-DNA insertion at the fifth intron. xlg1-2 is apparently a knockout allele since no XLG1 transcript was detected through RT-PCR (Supplemental Fig. S6). The xlg1-2 mutant was used in this study. We generated a new triple mutant of xlg1-2, xlg2-1, and xlg3-1 (in the Columbia-0 [Col-0] background). The triple mutant is named xlg1*/2/3 in this report. Although the xlg single mutants and double mutants did not show easily discernible morphological phenotypes compared with Col-0 wild type, xlg1*/2/3 plants, like pub4, had a smaller stature (Fig. 3A) when grown in soil and had reduced fertility (Fig. 3B). Many siliques in xlg1*/2/3 had few seeds (Supplemental Fig. S7). When grown on the Murashige and Skoog medium, young seedlings of those genotypes were slightly bigger compared with the wild type, and they all showed enhanced root systems (Fig. 3, C and D). The primary roots of pub4 and pub2/4 but not xlg1*/2/3 were also longer than the wild type. More lateral and adventitious roots were observed in all three genotypes (Fig. 3, C and D; Supplemental Fig. S8, A and B).
The Cytokinin Response Is Impaired in pub4, pub2/4, and xlg1*/2/3
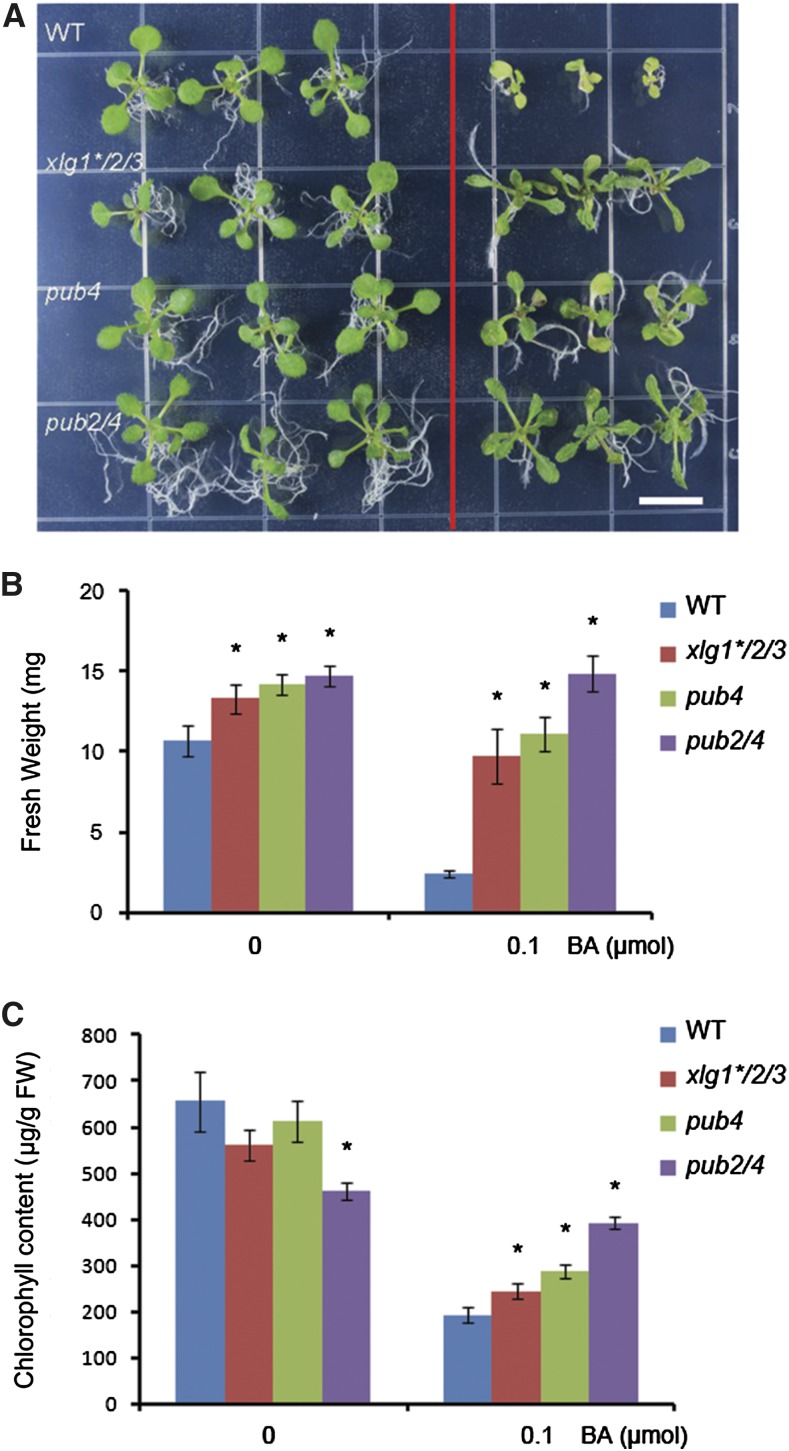
Some of the pub4, pub2/4, and xlg1*/2/3 phenotypes are reminiscent of the mutants defective in the cytokinin response (Hutchison et al., 2006; Riefler et al., 2006). To assess their roles in cytokinin signaling, we treated the pub and xlg mutants with 0.1 μm 6-benzylaminopurine (BA) in Murashige and Skoog plates for 2 weeks. In the control medium, the fresh weight of pub4, pub2/4, and xlg1*/2/3 seedlings was slightly higher than that of wild-type seedlings, likely due to their enhanced root systems (Supplemental Fig. S8, C and D), and the total chlorophyll contents were comparable to the wild type, except a lower chlorophyll level in pub2/4. In the presence of exogenous cytokinin, which is known to inhibit seedling growth and development, growth of the wild-type plants was severely arrested and their rosette leaves were pale with a decreased chlorophyll level. In contrast, the pub4, pub2/4, and xlg1*/2/3 seedlings were much less sensitive to the BA-caused growth inhibition (Fig. 4A). The pub4 and xlg1*/2/3 seedlings had slightly lower fresh weight than the seedlings on the control medium and kept a higher chlorophyll level than the wild type under the BA treatment. The pub2/4 seedlings were the most insensitive toward cytokinin and only slightly lost their chlorophyll contents under the BA treatment compared with the control seedlings (Fig. 4). pub2 and the xlg single and double mutants were not significantly different from the wild type in their sensitivity to exogenous BA treatment (Supplemental Fig. S9, A and B). This BA-insensitivity phenotype of pub4 and xlg1*/2/3 could be genetically complemented by introduction of PUB4 into pub4 or by introducing XLG1, XLG2, or XLG3 into xlg1*/2/3 (Supplemental Figs. S10 and S11). In addition, another pub4 mutant, pub4-3, displayed a similar phenotype as the pub4-1 mutant (Supplemental Fig. S9B). These data indicate that the cytokinin-insensitive phenotype is caused by the xlg and pub mutations.
Figure 4.
Shoot inhibition by cytokinin in wild type (WT), pub4, pub2/4, and xlg1*/2/3. A, Fourteen-day-old seedlings grown on the Murashige and Skoog medium without (left) or with 0.1 μmol BA (right). Bar = 1 cm. B, Fresh weight of seedlings of the different genotypes in the medium without and with BA. Fresh weight was determined based on three replicates of 12 14-d-old seedlings for each genotype shown in A. Vertical bars represent mean ± sd. Asterisks indicate statistical differences to the wild type (P < 0.05). C, Total chlorophyll contents of seedlings of different genotypes under the conditions shown in A were determined based on three replicates of 12 seedlings each. Vertical bars represent mean ± sd. Asterisks indicate statistical differences to the wild type (P < 0.05).
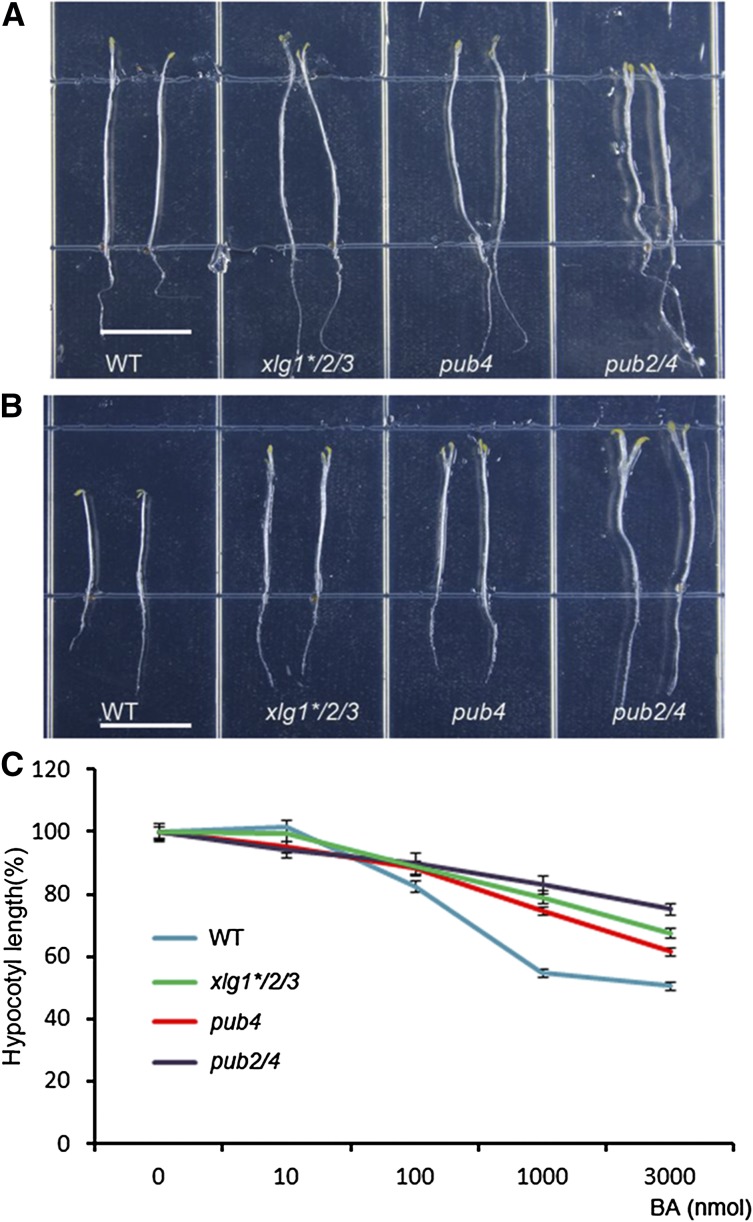
To further examine the possible involvement of XLGs and PUB2/4 in the cytokinin response, the effect of exogenous cytokinin on hypocotyl elongation inhibition was measured in the wild type and the xlg and pub mutants. In the dark, obvious hypocotyl elongation inhibition by cytokinin was observed in wild-type seedlings (Fig. 5, A and B). pub4, pub2/4, and xlg1*/2/3 were all less sensitive than the wild type to cytokinin, with pub2/4 the most insensitive among the three genotypes, particularly under the higher cytokinin concentrations (Fig. 5C). We also noticed that all those three mutant seedlings had elongated petioles and open cotyledons in the presence of 3 μm BA (Fig. 5). Again, the hypocotyl elongation phenotype was not significantly different in the pub2, xlg single, and double mutants from wild type when growing under dark in the presence of cytokinin (Supplemental Fig. S12).
Figure 5.
Hypocotyl elongation inhibition of wild type (WT), pub4, pub2/4, and xlg1*/2/3 seedlings under the treatment of cytokinin. A, Nine-day-old seedlings of wild type, pub4, pub2/4, and xlg1*/2/3 grown on the Murashige and Skoog medium supplemented with DMSO but without BA. Bar = 1 cm. B, Nine-day-old seedlings of wild type, xlg1*/2/3, pub4, and pub2/4 grown on the Murashige and Skoog medium with 3 μm BA. Bar = 1 cm. C, Hypocotyl length of wild type, xlg1*/2/3, pub4, and pub2/4 under different BA concentrations relative to that grown on DMSO control plates. The hypocotyl elongation of each line with different cytokinin treatment was expressed as a percentage of its DMSO control. Each value represents mean ± se (n > 10).
When seedlings were grown in the presence of BA in vertical Murashige and Skoog plates under normal long-day conditions, root sensitivity to cytokinin in pub4 and xlg1/*2/3 was not different from that of wild-type plants at low BA concentrations and only slightly less sensitive to 1 μmol BA in those two genotypes (Fig. 6). Root growth of the pub2/4 mutant, however, showed reduced sensitivity to inhibition by cytokinin at concentrations of 100 nmol and 1 μmol (Fig. 6). These data indicated that those mutations also cause decreased sensitivity to cytokinin for root development, but to a lesser degree compared with the insensitivity to cytokinin for shoot development.
Figure 6.
Root elongation inhibition by cytokinin in wild type (WT), pub4, pub2/4, and xlg1*/2/3. A, Nine-day-old seedlings grown on plates without BA. Bar = 1 cm. B, Nine-day-old seedlings grown on Murashige and Skoog plates with 1 μm BA. Bar = 1 cm. C, Relative root elongation of seedlings grown on Murashige and Skoog plates with different concentrations of BA relative to seedlings in mock. The root elongation of each line with different cytokinin treatment was expressed as a percentage of its DMSO control. Each value represents mean ± se (n ≥ 8).
xlg1*/2/3 Displays the Similar Tapetum and Stamen Developmental Defects to Those of pub4
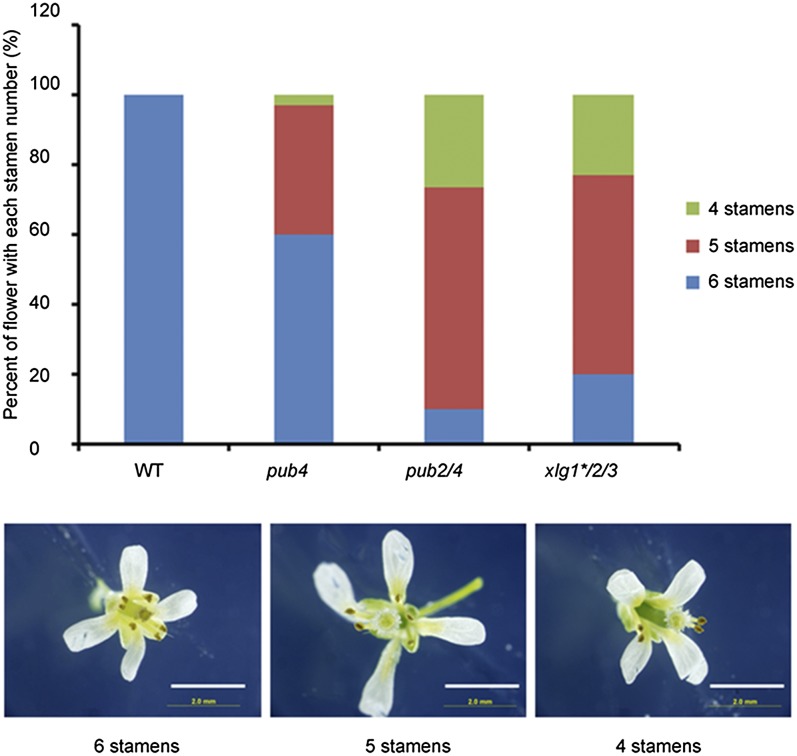
We observed abnormal numbers of stamens in many flowers of the pub4, pub2/4, and xlg1*/2/3 mutants. Normally wild-type plants develop six stamens (four long and two short). However, flowers with four or five stamens frequently appeared in pub4, pub2/4, and xlg1*/2/3 mutants (Fig. 7). The missing stamens were always one or two of the short stamens. In pub4, approximately 40% of flowers had a reduced stamen number. In pub2/4 and xlg1*/2/3, a majority of flowers lacked one or two stamens (Fig. 7).
Figure 7.
Stamen numbers of wild type (WT), pub4, pub2/4, and xlg1*/2/3 mutants. The stamen number was counted from more than 60 flowers for each genotype. Shown below are flowers with normal number (six) of stamens and with reduced numbers of stamens. Bars represent 2 mm.
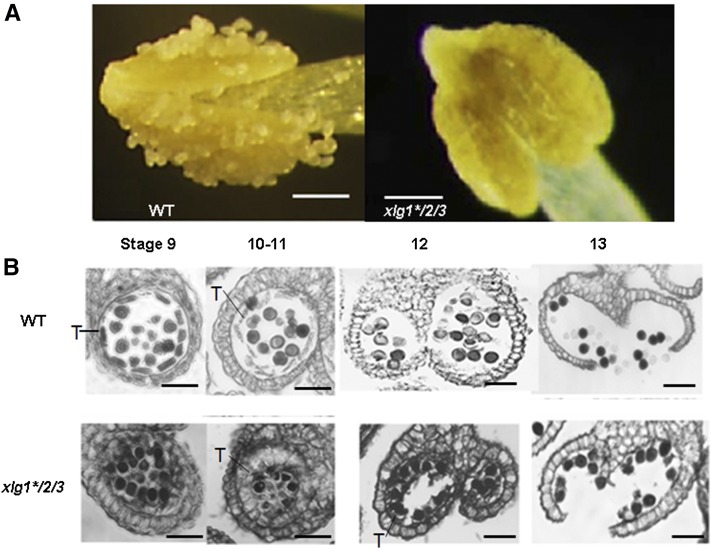
It was previously reported that the pub4 mutant showed aberrant enlargement and incomplete degeneration of the tapetal cells (Wang et al., 2013), leading to failure of pollen release and male sterility. Like pub4 and pub2/4, xlg1*/2/3 displayed reduced fertility under the normal conditions (Fig. 3B). The reduced fertility was not caused by pollen viability because pollen grains from xlg1*/2/3 could be stained purple (an indication of viability) using the Alexander staining solution (Supplemental Fig. S13). We then examined whether the sterility of xlg1*/2/3 is also associated with defect in tapetum development. On dehisced anthers of the wild type, individual pollen grains were visible under light microscopy. However, the pollen grains on the dehisced xlg1*/2/3 mutant anthers were attached to each other (Fig. 8A). The tapetum development was further examined microscopically. At stage 9, the cells in the wild-type tapetum revealed a thin layer and pollen didn’t adhere to each other. However, the tapetum layer in the xlg1*/2/3 plants was much expanded and the tapetal cells were larger than those in the wild type (Fig. 8B). The pollen grains were also packed tightly. The tapetal cells normally start to undergo programmed cell death at stage 10-11 and completely disappear at stage 12-13, leading to release of scattered pollen grains from locules. In the xlg1*/2/3 mutant, the degeneration of tapetal cells was incomplete, causing pollen grains to adhere to the residual tapetal cells and to fail to be released from the locule walls (Fig. 8B).
Figure 8.
xlg1*/2/3 was defective in tapetum development. A, Pollen grains were easily visible on the dehisced anther from the wild type (WT) but not on the anther at the same stages from xlg1*/2/3. Bars = 100 μm. B, Wax sections of anthers at different developmental stage in the wild type and xlg1*/2/3. The mutant anther showed an abnormally expanded tapetum (T) at stage 10-11. The pollen grains were dispersed in the wild-type anther but adhered to the residual tapetal cells and the anther wall in the mutant. Bars = 20 μm.
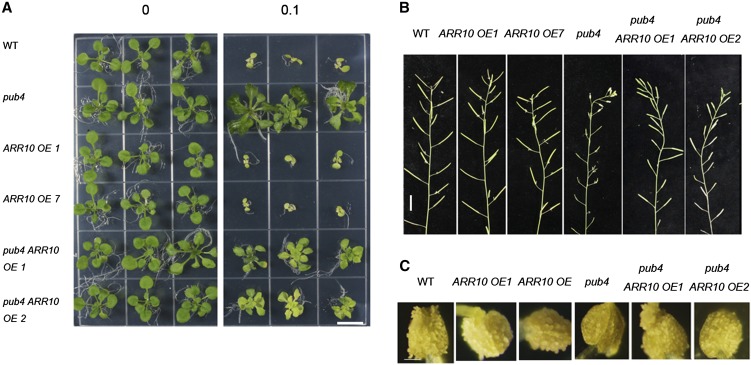
ARR10 Overexpression Can Partially Suppress Several Phenotypes Associated with pub4 and xlg1*/2/3
The data above show that pub4, pub2/4, and xlg1*/2/3 mutants have reduced cytokinin responses. The type-B response regulators, such as ARR1, ARR10, and ARR12, are positive regulators of the cytokinin response, and the triple mutation of those three ARR genes leads to insensitivity to cytokinin and a dwarf phenotype (Argyros et al., 2008). We overexpressed ARR1 and ARR10 in pub4 under the control of the 35S promoter. ARR10 overexpression was found to partially restore some phenotypes caused by pub4, including the cytokinin response in shoot (Fig. 9A) and pollen release and fertility (Fig. 9, B and C). Overexpression of ARR10 in xlg1*/2/3 also partially suppressed, but to a lesser degree, the triple mutant’s phenotype of reduced sensitivity to cytokinin (Supplemental Fig. S14). Overexpression of the ARR1 gene in pub4 was not found to restore the pub4 phenotypes.
Figure 9.
Overexpression of ARR10 suppressed the pub4 phenotypes. A, The 14-d-old wild type (WT), two Pro35:ARR10 lines, pub4, and two Pro-35S:ARR10 lines in the pub4 background were grown on the Murashige and Skoog plates with DMSO (left) or 0.1 μm BA (right). Bar = 1 cm. B, Phenotypes of 7-week-old wild type, two Pro35:ARR10 lines, pub4, and two Pro-35S:ARR10 lines in the pub4 background. Bar = 1 cm. C, Representative anthers of the wild type, two Pro35:ARR10 lines, pub4, and two Pro-35S:ARR10 lines in the pub4 background. Individual pollen grains were visible on the anthers from the other genotypes but not from pub4. Bar = 100 μm.
DISCUSSION
Animals use a large family of plasma membrane GPCRs to perceive various external and internal signals and transduce the signals through a large number of G proteins. Plants have fewer GPCRs, if any, and a much smaller number of G-protein components (Assmann, 2004; Perfus-Barbeoch et al., 2004; Urano et al., 2013; Urano and Jones, 2014), including the single canonical Gα, GPA1. The presence of three XLGs and their known interactions with the multiple Gβγ dimers expand the repertoire of G-protein signaling pathways. Indeed, the XLG mutations have been found to cause various phenotypical changes in developmental and physiological processes (Ding et al., 2008; Pandey et al., 2008; Zhu et al., 2009; Chakravorty et al., 2015; Maruta et al., 2015; Liang et al., 2016). However, the mechanisms by which the XLGs mediate those pathways remain unclear. The lack of a typical GPCR in plants and other evidences have also raised the possibility that G proteins in plants might function downstream of receptor kinases, unlike GPCRs in animals (Urano et al., 2013; Nitta et al., 2015).
A previous study indicated that all XLG proteins are localized in the nucleus (Ding et al., 2008). In more recent reports using transient expression and stable transformation, XLG2 and XLG3 were shown to be localized to both the plasma membrane and nuclei, whereas XLG1 was only associated with the plasma membrane (Chakravorty et al., 2015; Maruta et al., 2015). In this study, we also found plasma membrane localization of all XLGs and additional nuclear location of XLG2 and XLG3. Interactions between XLGs and Gβγ dimers have been reported to occur at the plasma membrane, and overexpression of Gβγ dimers sequestered XLG3 at the plasma membrane (Chakravorty et al., 2015).
The Y2H screen and assay led to the identification of PUB2 and PUB4 as XLGs’ putative interacting proteins. BiFC assay indicated that they interacted in planta. The interaction between XLG1 and PUB4 was further confirmed through the in vivo co-IP assay. A previous observation using PUB4-YFP transgenic plants indicated that the PUB4 protein is predominantly localized in the cytosol (Wang et al., 2013); however, this observation cannot rule out the possibility that some of the eYFP signal came from the plasma membrane due to the limited resolution. Additional experimental data in this study further showed that PUB2 and PUB4 are also localized at the plasma membrane. Nuclear localization of PUB4 was reported in another study, and it was suggested that the nuclear localization was likely due to interaction with other proteins (Kinoshita et al., 2015a). Our BiFC data showed that the interaction between XLGs and PUB4 occurred at the plasma membrane, whereas PUB2 was colocalized with XLG2 and XLG3 in both the plasma membrane and the nucleus. It raises the possibility that XLG proteins might sequester PUB4 and PUB2 to the plasma membrane. The tobacco (Nicotiana tabacum) PUB4 homolog NtPUB4 is associated with a tobacco chitinase-related receptor-like kinase (NtCHRK1) and localized to the plasma membrane (Kim et al., 2003). Several PUB proteins have been reported to undergo translocation between different cellular compartments, often through their interactions with other proteins (Stone et al., 2003; Samuel et al., 2008; Deb et al., 2014).
The interactions between XLGs and PUB2/PUB4 would suggest that they could function in the same pathways. However, although many phenotypes caused by mutations in these XLG and PUB genes have been described previously, these phenotypes did not suggest that the XLG and PUB proteins are involved in the same biological processes. The xlg1 allele in the xlg1/2/3 triple mutant used in the previous studies for characterizing the XLG biological functions carried a T-DNA insertion at the promoter region. This allele is a weak knockdown allele as the mutant still accumulates about 20% XLG1 transcripts (Ding et al., 2008). In this study, we used another xlg1 allele (xlg1-2), which is likely to be a knockout allele, to generate a new xlg1/2/3 triple mutant (xlg1*/2/3). The new xlg1*/2/3 mutation indeed causes similar pleiotropic effects on many developmental processes to those caused by pub4 and pub2/4. Those similar phenotypes include male sterility likely caused by aberrant expansion and incomplete degeneration of tapetal cells and a smaller rosette size that were previously reported for the pub4 mutant. We also found that both the xlg and pub mutations lead to insensitivity to cytokinin and reduced numbers of stamens. The multiple similar phenotypes shared by the xlg and pub mutations, together with their physical interactions, provide convincing evidence that these proteins play roles in the same biological pathways, although the XLG and PUB proteins might also function in other different pathways.
Many PUB proteins have been identified as protein kinase-interacting proteins. In addition to NtPUB4 that interacted with NtCHRK1 in yeast (Kim et al., 2003), the Arabidopsis S-Domain 1 receptor kinases ARK1 and ARK2 phosphorylated PUB9 and PUB13 in vitro (Samuel et al., 2008). PUB13 was also identified as a protein that interacts with BAK1 in the Y2H assay (Lu et al., 2011). BAK1 phosphorylates PUB12 and PUB13, which then form complexes with FLS2 to ubiquitinate FLS2 for degradation, thereby attenuating flagellin-triggered defense signaling (Lu et al., 2011). PUB1 of Medicago truncatula interacts with and is phosphorylated by the LYK3 symbiotic receptor and negatively regulates the LKY3 signaling pathway (Mbengue et al., 2010). Our previous study showed that PUB4-FLAG purified from plants appeared to catalyze monoubiquitination of itself (Wang et al., 2013), suggesting that it might also catalyze monoubiquitination of its targets. It is unlikely that PUB2/4 target XLGs for polyubiquitination-mediated degradation since the xlg triple mutant and pub2/4 showed very similar phenotypes, instead of opposite phenotypes as would be expected if PUB2/4 negatively regulate XLGs. One possibility is that PUB2/4 could be direct effectors of XLGs and activation of PUB2/4 by XLGs leads to ubiquitination of their target proteins, which remain to be identified. Alternatively, PUB2/4 might activate XLGs by monoubiquitination.
Although knockout of PUB2 didn’t lead to any obvious phenotype, ectopic overexpression of PUB2 could complement the low fertility phenotype of pub4. The pub2/4 had a smaller size than pub4. Besides, pub2 also enhanced the cytokinin-insensitivity phenotype of pub4. All these data indicate that PUB2 and PUB4 have partially redundant functions in multiple developmental processes.
Recently, pub4 was isolated as a mutant for its reduced sensitivity to root growth inhibition by the CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) peptides, and the mutation alters cell proliferation in root meristem in a cytokinin-independent manner (Kinoshita et al., 2015a). The cytokinin level in the pub4 mutant was not changed, but its auxin (indole-3-acetic acid) level was found to be 30% higher than in the wild type (Kinoshita et al., 2015a). We found that the pub4, pub2/4, and xlg1*/2/3 mutants all displayed obvious insensitivity to the cytokinin-caused shoot and hypocotyl elongation inhibition. The root elongation inhibition by BA was only slightly reduced in these mutants. The reduced numbers of stamens in the xlg1*/2/3, pub4, and pub2/4 mutants are also reminiscent of the rice (Oryza sativa) mutant lonely guy (log; Kurakawa et al., 2007). LOG encodes a cytokinin-activating enzyme and functions in the final step in cytokinin biosynthesis in rice. These results indicate that XLGs and PUB2/4 are positive regulators of the cytokinin signaling pathways. The partial complementation of the pub4 phenotypes by overexpression of ARR10, a positive regulator of the cytokinin response, further supports such a notion. In contrast with some well-studied cytokinin signaling mutants, such as ahk2/3/4, ahp2/3/5, and arr1/10/12 that show a severe dwarf phenotype (Nishimura et al., 2004; Hutchison et al., 2006; Riefler et al., 2006; Argyros et al., 2008), the xlg1*/2/3 and pub2/4 mutants show a moderate reduction in their sizes, suggesting that XLGs and PUB2/4 might function in more specific developmental and physiological processes. Alternatively, the mild phenotype could be due to functional/pathway redundancy.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
pub4, xlg2, xlg3, xlg2/3, PUB4-Flag, and the pub4 complementation line (gPUB4) have been described previously (Ding et al., 2008; Zhu et al., 2009; Wang et al., 2013). pub2 (SALK_086656) and xlg1-2 (SALK_119657) were obtained from the Arabidopsis Biological Resource Center at The Ohio State University and crossed to pub4 or xlg1-1 xlg2-1 xlg3-1 for generating the pub2/4 double or xlg1*/2/3 triple mutant. The mutant alleles are listed in Supplemental Table S1. All lines used in this study are in the Col-0 background, and the primers for genotyping of the mutants are listed in Supplemental Table S2.
Surface-sterilized seeds were plated on the Murashige and Skoog medium with 1% Suc, 0.05% (w/v) MES (pH 5.7), and 0.7% Phytoagar (Sigma-Aldrich). The plates were kept on 4°C for 48 h and transferred to a growth room with either 16 h light/8 h dark (long day) or 9 h light/15 h dark (short day) condition with a light intensity of 100 mol m−2 s−1.
Generation of Transgenic Lines
The full-length coding sequences of XLG1, XLG2, XLG3, PUB4, PUB2, ARR10, and the XLG2 genomic DNA fragment were amplified and cloned into pDONR207 and then transferred into corresponding destination vectors (pEarleyGate 102, pEarleyGate 104, pEarleyGate 201, pMDC83, pEarleyGate 301, modified pEarleyGate 101without YFP) using LR Clonase II (Invitrogen). The primers used for constructing those constructs are listed in Supplemental Table S2. All the constructs were introduced to Agrobacterium tumefaciens strain GV3101, which was then used for transformation of Arabidopsis (Arabidopsis thaliana) plants by floral dipping methods (Clough and Bent, 1998).
Y2H Assay
Full-length coding sequences of the XLGs, PUB2/4, and truncated XLGs or PUB4 were amplified and cloned into pGADT7 or pGBKT7 (Clontech), and the corresponding vectors were introduced into Saccharomyces cerevisiae strain Y187 or Y2H Gold yeast cells for mating. The mated strains were grown on the SD medium, the SD medium lacking Trp/Leu, or the SD medium lacking Trp/Leu/His/Ade plus X-α-Gal and aureobasidin A. For library screening, Y2H Gold yeast strains with XLG2-pGBKT7/PUB4-pGBKT7 plasmids were mated to the Y187 Mate and Plate Library (Clontech) and plated on the SD medium lacking Trp/Leu plus X-α-Gal and aureobasidin A plates. The blue clones were picked after 5 d and cultured on the SD medium lacking Trp/Leu and the SD medium lacking Trp/Leu/His/Ade. DNA sequences amplified using T7 and 3AD primers from the positive clones were sequenced and analyzed by BLASTing to TAIR10 transcripts.
BiFC Assay
For BiFC assay, the 35S-YN-pCambia1300 and 35S-YC-pCambia1300 constructs, modified from pSY736 and pSY735 (Bracha-Drori et al., 2004) carrying destination genes, were coexpressed in tobacco (Nicotiana benthamiana) cells by agroinfiltration (Chang et al., 2013). Briefly, overnight cultures of agrobacteria were harvested and resuspended to OD600 = 0.1 in infiltration buffer containing half-strength Murashige and Skoog liquid medium with 2% (w/v) Suc, 10 mm MES, and 100 μm acetosyringone. Equal volumes of bacterial suspensions were mixed together and infiltrated to 3- to 4-week-old tobacco leaves. Signals were detected 24 to 48 h after infiltration.
Microscopy Analyses
Confocal imaging was carried out with Olympus fluoview 1000 or Leica TCS SP5II confocal microscopes. GFP signal was excited with an argon laser at 488 nm for transient expression of GFP-tagged XLGs. The YFP signal was excited at 514 nm for BiFC assays. CFP was excited at 458 nm, and mCherry was excited at 594 nm. Images were acquired and analyzed with the corresponding software FV10-ASW for Olympus and LAS-AF for Leica.
Co-IP Assay
Leaves from 4-week-old plants were harvested and used for co-IP assays. Proteins were extracted with the buffer containing 50 mm Tris (pH 7.5), 150 mm NaCl, and 0.1% IGEPAL CA-630 and Proteinase Inhibitor Cocktail (Sigma-Aldrich). After a brief sonication, the samples were centrifuged at 12,000g for 15 min at 4°C. The supernatant was incubated with anti-GFP magnetic beads (catalog no. D153-11; MBL) for 2 h to overnight at 4°C with gentle rotation. The beads were then washed four times with PBS. The immunoprecipitated proteins were eluted in the SDS-PAGE loading buffer with heating at 70°C for 10 min. The presence of the corresponding proteins was detected using a FLAG antibody (catalog no. a8592; Sigma-Aldrich) and a GFP antibody (catalog no. 11814460001; Roche), respectively.
Cytokinin Response Assay
For shoot inhibition, Arabidopsis seedlings were grown on horizontal Murashige and Skoog plates containing BA or 0.1% dimethyl sulfoxide (DMSO; as a control). The seedlings were photographed, and the fresh weight and chlorophyll level were determined at day 14 (Argyros et al., 2008). The experiment was repeated twice.
For root elongation assay, Arabidopsis seedlings were grown on vertical Murashige and Skoog plates containing BA or 0.1% DMSO (as a control). The photos were taken and the main root lengths were measured at day 9. The experiment was repeated twice.
For hypocotyl elongation, seedlings of different genotypes were grown on vertical Murashige and Skoog plates in dark. The photos were taken and the hypocotyl lengths were measured at day 9. The experiment was repeated twice.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource Web site (www.arabidopsis.org) under the following accession numbers: XLG1 (AT2G23460), XLG2 (AT4G34390), XLG3 (AT1G31930), PUB4 (AT2G23140), PUB2 (AT5G67340), and ARR10 (AT4G31920).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. No autoactivation of BD-XLG1C, XLG2C, XLG3C, and AD-PUB4C was detected.
Supplemental Figure S2. PUB2 and PUB4 were partially localized to the plasma membrane.
Supplemental Figure S3. pub2-1 is most likely a knockout allele.
Supplemental Figure S4. Relative PUB4 and PUB2 expression levels according to Affymetrix Arabidopsis ATH1 Genome Array Data.
Supplemental Figure S5. Overexpression of PUB2 in pub4 restored the pub4 phenotype.
Supplemental Figure S6. xlg1-2 is most likely a knockout allele.
Supplemental Figure S7. xlg1*/2/3 produced fewer seeds than the wild type.
Supplemental Figure S8. Primary root length, lateral root numbers, root weight, and root/whole plant fresh weight ratio in seedlings of the wild type, pub4, pub2/4, and xlg1*/2/3.
Supplemental Figure S9. Shoot growth inhibition by cytokinin in the xlg and pub mutants.
Supplemental Figure S10. Complementation of the xlg1*/2/3 cytokinin-insensitive shoot growth phenotype by transgenes of XLG1, XLG2, and XLG3.
Supplemental Figure S11. Complementation of the pub4 cytokinin-insensitive phenotype in shoot growth inhibition by the genomic PUB4 transgene.
Supplemental Figure S12. Hypocotyl elongation inhibition by cytokinin in pub2 and xlg single and double mutants.
Supplemental Figure S13. The fertility of xlg1*/2/3 is temperature-dependent.
Supplemental Figure S14. Cytokinin response of the wild type, xlg1*/2/3, and two xlg1*/2/3 Pro-35S:ARR10 lines.
Supplemental Table S1. Mutant alleles used in this article.
Supplemental Table S2. List of primer sequences used in this article.
Supplementary Material
Glossary
- GPCR
G-protein-coupled receptor
- 3-AT
3-amino-1,2,4-triazole
- BiFC
bimolecular fluorescence complementation
- co-IP
coimmunoprecipitation
- BA
6-benzylaminopurine
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the Research Grants Council of Hong Kong (grant nos. HKBU262111, HKBU262212, HKBU262213, and HKBU12159916, and AoE Grant HKBU/CU/AOE/1314/01 to Y.X.) and by the Hong Kong Baptist University’s Strategic Development Fund (grant no. SDF 15-1012-P04 to Y.X.).
Articles can be viewed without a subscription.
References
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM. (2004) Plant G proteins, phytohormones, and plasticity: three questions and a speculation. Sci STKE 2004: re20. [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Gookin TE, Milner MJ, Yu Y, Assmann SM (2015) Extra-large G proteins expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiol 169: 512–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH (2013) Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25: 1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deb S, Sankaranarayanan S, Wewala G, Widdup E, Samuel MA (2014) The S-domain receptor kinase Arabidopsis Receptor Kinase2 and the U box/armadillo repeat-containing E3 Ubiquitin Ligase9 module mediates lateral root development under phosphate starvation in Arabidopsis. Plant Physiol 165: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cerezo M, Sánchez-Rodríguez C, Escudero V, Miedes E, Fernández PV, Jordá L, Hernández-Blanco C, Sánchez-Vallet A, Bednarek P, Schulze-Lefert P, et al. (2012) Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol Plant 5: 98–114 [DOI] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53: 248–263 [DOI] [PubMed] [Google Scholar]
- Hamm HE. (1998) The many faces of G protein signaling. J Biol Chem 273: 669–672 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. (1999) The armadillo family of structural proteins. Int Rev Cytol 186: 179–224 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S, Assmann SM (2012) Ca2+-dependent GTPase, extra-large G protein 2 (XLG2), promotes activation of DNA-binding protein related to vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. J Biol Chem 287: 8242–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Cho HS, Kim DM, Lee JH, Pai HS (2003) CHRK1, a chitinase-related receptor-like kinase, interacts with NtPUB4, an armadillo repeat protein, in tobacco. Biochim Biophys Acta 1651: 50–59 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Seo M, Kamiya Y, Sawa S (2015b) Mystery in genetics: PUB4 gives a clue to the complex mechanism of CLV signaling pathway in the shoot apical meristem. Plant Signal Behav 10: e1028707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, ten Hove CA, Tabata R, Yamada M, Shimizu N, Ishida T, Yamaguchi K, Shigenobu S, Takebayashi Y, Iuchi S, et al. (2015a) A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 142: 444–453 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Lee S, Rojas CM, Ishiga Y, Pandey S, Mysore KS (2013) Arabidopsis heterotrimeric G-proteins play a critical role in host and nonhost resistance against Pseudomonas syringae pathogens. PLoS One 8: e82445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wright SJ, Krystofova S, Park G, Borkovich KA (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol 61: 423–452 [DOI] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5: e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR (2015) Membrane-localized extra-large G proteins and Gbg of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol 167: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al. (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta Y, Ding P, Zhang Y (2015) Heterotrimeric G proteins in plant defense against pathogens and ABA signaling. Environ Exp Bot 114: 153–158 [Google Scholar]
- Oldham WM, Hamm HE (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol 9: 60–71 [DOI] [PubMed] [Google Scholar]
- Pandey S, Monshausen GB, Ding L, Assmann SM (2008) Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J 55: 311–322 [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol 7: 719–731 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR (2008) Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Salt JN, Shiu SH, Goring DR (2006) Multifunctional arm repeat domains in plants. Int Rev Cytol 253: 1–26 [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Chen JG, Botella JR, Jones AM (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol 3: 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones AM (2014) Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol 65: 365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lu Y, Jiang T, Berg H, Li C, Xia Y (2013) The Arabidopsis U-box/ARM repeat E3 ligase AtPUB4 influences growth and degeneration of tapetal cells, and its mutation leads to conditional male sterility. Plant J 74: 511–523 [DOI] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S (2005) Mammalian G proteins and their cell type specific functions. Physiol Rev 85: 1159–1204 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salomé PA, Weigel D, Fitzpatrick JA, Chory J (2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
- Zhu H, Li GJ, Ding L, Cui X, Berg H, Assmann SM, Xia Y (2009) Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gbeta subunit of heterotrimeric G protein and functions in disease resistance. Mol Plant 2: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.