The GET protein insertion system inserts the model tail-anchored protein SYP72 into endoplasmic reticulum membranes in Arabidopsis.
Abstract
The Arabidopsis (Arabidopsis thaliana) genome encodes homologs of the Guided Entry of Tail (GET)-anchored protein system for the posttranslational insertion of tail-anchored (TA) proteins into endoplasmic reticulum (ER) membranes. In yeast, TA proteins are loaded onto the cytosolic targeting factor Get3 and are then delivered to the membrane-associated Get1/2 complex for insertion into ER membranes. The role of the GET system in Arabidopsis was investigated by monitoring the membrane insertion of a tail-anchored protein, SYP72, a syntaxin. SYP72 bound to yeast Get3 in vitro, forming a Get3-SYP72 fusion complex that could be inserted into yeast GET1/2-containing proteoliposomes. The Arabidopsis GET system functioned in vivo to insert TA proteins into ER membranes as demonstrated by the fact that the YFP-tagged SYP72 localized to the ER in wild-type plants but accumulated as cytoplasmic inclusions in get1, get3, or get4 mutants. The GET mutants get1 and get3 were less tolerant of ER stress agents and showed symptoms of ER stress even under unstressed conditions. Hence, the GET system is responsible for the insertion of TA proteins into the ER in Arabidopsis, and mutants with GET dysfunctions are more susceptible to ER stress.
Targeting newly made membrane proteins to their appropriate subcellular location is a complex matter because different proteins have different destinations and membrane topologies. There are two general systems for targeting proteins to endoplasmic reticulum (ER) membranes: one is cotranslational and the other is posttranslational. It has been known for over 40 years that many membrane proteins are targeted to the ER membrane by cotranslational mechanisms (Blobel and Dobberstein, 1975). Recently, posttranslational mechanisms have been recognized as a means for the insertion of tail-anchored (TA) proteins with short C-terminal tails into ER membranes after they have been synthesized (Hegde and Keenan, 2011; Mariappan et al., 2011). Three chaperone-assisted pathways have been described in eukaryotes. One involves an unusual posttranslational function of the signal recognition particle and another employs the HSC70/Hsp40 couple (Rabu et al., 2009; Borgese and Fasana, 2011). A third mechanism is referred to as the Guided Entry of Tail (GET) pathway (Schuldiner et al., 2008; Denic, 2012), which is conserved in yeast and mammals and plays a major role in inserting TA proteins into ER membranes (Sherrill et al., 2011).
In the yeast GET system, transmembrane domains (TMDs) on newly synthesized TA proteins are first captured by the cytosolic chaperone Sgt2, a small Gln-rich tetratricopeptide repeat-containing protein (SGTA in humans; Wang et al., 2010; Mariappan et al., 2011). The TA protein complexed with Sgt2 is recruited by Get5 (Ubl4a in humans) into a Get4-Get5 scaffolding complex. Get4 (TRC35 in humans) then recruits Get3, leading to a handover of the TA protein from Sgt2 to Get3 (Chartron et al., 2010). The transfer of the client protein from Sgt2 to Get3 has been demonstrated in vitro and only occurs when Get4 and Get5 are present (Mateja et al., 2015).
Get3, an ATPase, binds to and chaperones the TMDs of TA proteins for their delivery and insertion into the ER (Hegde and Keenan, 2011). Get3 (and its mammalian homolog, TRC40) is a homodimer that assumes a closed conformation in an ATP-bound state (Mateja et al., 2015). In the closed state, the Get3 dimer has a large hydrophobic groove that spans both monomers and constitutes the binding site for a protein with a TMD (Hu et al., 2009; Hegde and Keenan, 2011). In yeast, Get3 chaperones TA proteins to the ER, whereupon the Get3-TA complex interacts with the ER membrane multispanning proteins Get1 and Get2. Following ATP hydrolysis, Get1 is thought to promote substrate release by inserting itself between the Get3 monomers disrupting the closed Get3 conformation (Stefer et al., 2011; Denic, 2012).
TA proteins are defined as proteins that lack other secretory signals and have a predicted TMD within 50 amino acid residues of the C-terminal tail. The number of loci in the human genome encoding proteins predicted to be TA proteins is 411 (Kalbfleisch et al., 2007), while in yeast, the estimate is 55 (Beilharz et al., 2003). In Arabidopsis (Arabidopsis thaliana), bioinformatics analyses has predicted that 454 loci encode TA proteins (Kriechbaumer et al., 2009).
In this study, we characterized the Arabidopsis GET pathway by demonstrating the insertion of a model TA protein, SYP72, into the endoplasmic reticulum. We observed that GET3 interacts with Get4 and Get1, allowing for the recruitment of SYP72 and its subsequent insertion into the ER. Loss-of-function mutants in the GET pathway result in reduced tolerance to ER stress agents and to early flowering phenotypes, indicative of stress.
RESULTS
GET System Gene Homologs in Arabidopsis
The Arabidopsis genome encodes homologs of the main components of the GET pathway (Table I). There are three homologs of Get3 in Arabidopsis (Supplemental Fig. S1); however, we concentrated our efforts on At1g01910 because it is widely expressed throughout the plant at many developmental stages, while the other homologs, such as At3g10350, a plastid stroma protein (Ferro et al., 2010), and At5g60730, a mitochondrial outer membrane factor (Duncan et al., 2011), are expressed during embryo development (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). At1g01910 is also more closely related in sequence to the yeast Get3 and human TRC40; however, it lacks a conserved zinc-binding CXXC motif that in other systems is thought to stabilize the Get3 dimer and serve as a hinge point in the open-to-close transition (Mateja et al., 2009; Supplemental Fig. S1).
Table I. Arabidopsis homologs of GET system components in yeast and mammals.
| Yeast Gene | Mammalian Gene | Arabidopsis Gene | Predicted Function |
|---|---|---|---|
| Get 1 | WRB | At4g16444 | Subunit of the membrane insertase complex |
| Get 2 | CAML | – | Subunit of the membrane insertase complex |
| Get 3 | TRC40 | At1g01910 (At3g10350) | TA substrate targeting factor |
| Get 4 | TRC35 | At5g63220 | Subunit of the pretargeting complex |
| Get 5 | Ubl4A | At1g55060 | Subunit of the pretargeting complex |
| Sgt2 | SGTA | At4g08320 | Subunit of the pretargeting complex |
| Bag 6 | Bag6 | – | Subunit of the pretargeting complex |
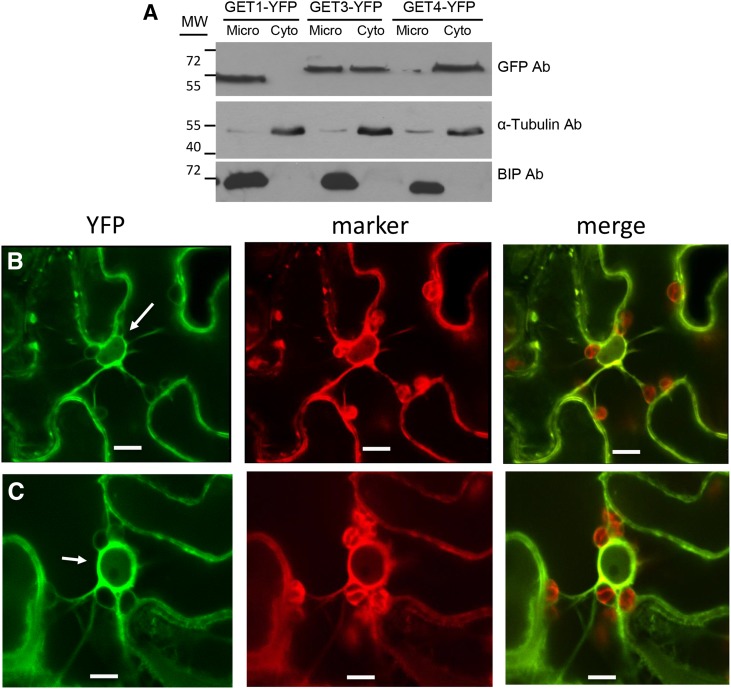
To determine the subcellular localization of the various GET homologs in Arabidopsis, we fused YFP to their C termini and stably expressed them as transgenes. Arabidopsis seedlings expressing the transgenes were cell fractionated and the localization of the tagged proteins in either the microsome or cytosolic fractions was determined (Fig. 1A; Supplemental Fig. S2). Western-blot results showed that Get4-YFP and Get1-YFP were exclusively localized to the cytosolic and microsome fractions, respectively, while GET3-YFP partitioned between both fractions.
Figure 1.
Subcellular localization of GET factors in Arabidopsis. A, Extracts from Arabidopsis seedlings expressing GET1-YFP, GET3-YFP, and GET4-YFP were fractionated into microsomal and cytoplasmic fractions. Immunoblot of proteins obtained from the two fractions was probed with anti-GFP, antitubulin (a cytoplasmic marker), and anti-BiP (an ER marker) antibodies. B and C, Confocal microscopy of N. benthamiana leaf epidermal cells agroinfiltrated with constructs expressing GET1-YFP (B) and GET3-YFP (C), both with the ER marker CDC-960 mCherry. Bar = 10 µm.
To confirm these results, the YFP-tagged constructs were transiently expressed by agroinoculation in Nicotiana benthamiana leaves along with marker proteins and localized by confocal microscopy. In leaf epidermal cells, it was observed that GET1-YFP colocalized with the ER marker CDC-960-mCherry (Figure 1B) and GET3-YFP partly colocalized with the ER marker and also showed diffuse fluorescence, suggesting that GET3-YFP was also in the cytoplasm (Fig. 1C; Supplemental Fig. S2). Fluorescence from GET4-YFP colocalized with the cytoplasmic marker mCherry (Supplemental Fig. S2). The pattern was consistent with the cell fractionation findings in that GET4-YFP was localized in the cytoplasm.
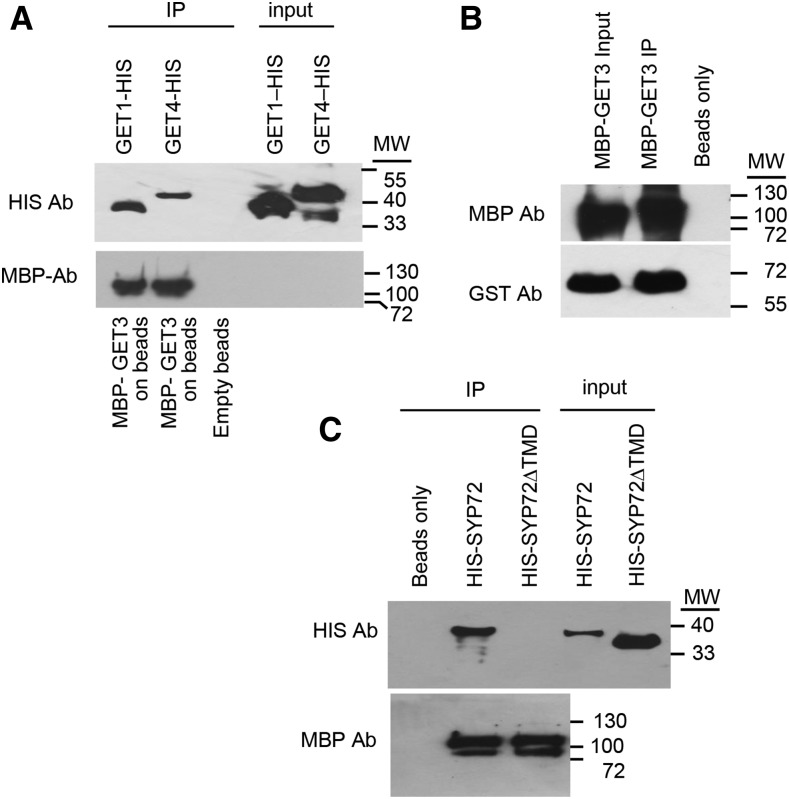
In other systems, GET factors are known to interact with each other in the delivery of client TA proteins to target membranes. We examined the interaction of recombinant forms of Arabidopsis GET factors by tagging them and analyzing their interaction in vitro in coimmunoprecipitation experiments. GET4 is a factor involved in recruiting GET3 to the GET4-GET5 scaffold from which there is a handoff of client TA proteins to GET3. In this regard, we found that Arabidopsis GET4-HIS interacts and coimmunoprecipitates with recombinant MBP-GET3 in vitro (Fig. 2A). From work in other systems, the Get3-TA substrate complex interacts with Get1 and Get2, membrane factors that facilitate the insertion of the C-terminal tail of substrates into the ER membrane. We also observed that that GET1-HIS interacts and coimmunoprecipitates with recombinant MBP-GET3 in vitro (Fig. 2A). In addition, Get3 is thought to capture client proteins as a dimer (Mateja et al., 2015). To find out whether Arabidopsis GET3 dimerizes, we determined whether GST-GET3 interacts and coimmunoprecipitates in vitro with itself in the form of MBP-GET3. We found that, indeed, the two forms of GET3 dimerize and/or oligomerize in vitro (Fig. 2B).
Figure 2.
Interactions with GET factors and candidate substrates. A, Coimmunoprecipitation of MBP-GET3 with Get1-HIS and Get4-HIS. Anti-MBP and anti-HIS antibodies were used to probe the immunoblots. B, GET3 dimerizes as demonstrated by the coimmunoprecipitation of MBP-GET3 with GST-GET3. Anti-MBP and anti-GST were used to probe the blots. C, MBP-GET3 binds the model substrate, SYP72, and the binding depends on the presence of the TMD in SYP72 as demonstrated by the coimmunoprecipitation of MBP-GET3 with HIS-SYP72, but not HIS-SYP72ΔTMD. The immunoblot was probed with anti-MBP and anti-HIS.
SYP72 as a Model TA Protein
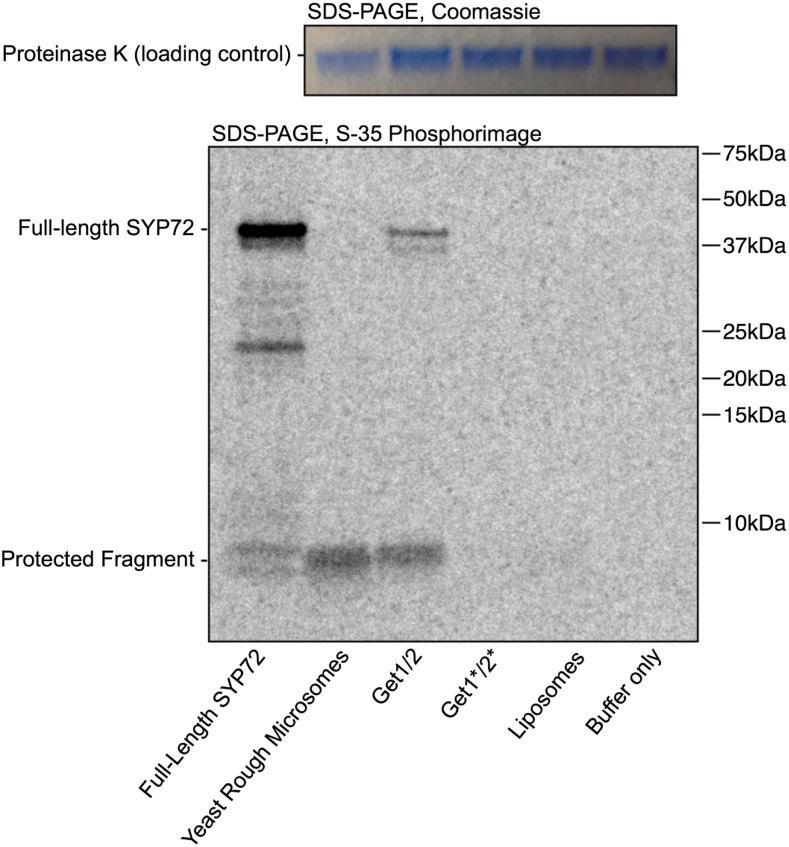
To determine whether the GET system inserts TA proteins into the ER membranes in Arabidopsis, we chose to study the syntaxin SYP72 (At3g45280) as a model substrate. By sequence analysis, SYP72 (At3g45280) is predicted to be a TA protein (Abell and Mullen, 2011), a type II membrane protein with a large cytoplasmic-facing N terminus and with a TMD only four amino acid residues from its C terminus (Supplemental Fig. S3). To demonstrate that, in fact, SYP72 is a conventional TA protein that uses the GET system for membrane insertion, we first tested whether it could be inserted into yeast Get1/2 proteoliposomes. In this assay system, the test protein SYP72 was epitope tagged with a twin-strep-tag (strep II) at its N terminus and 3F4 at its C terminus (Supplemental Fig. S4) and synthesized in a T7-coupled in vitro transcription-translation system supplemented with yeast Get3. After affinity purification, the Get3-SYP72 complex was incubated with proteoliposomes containing yeast Get1/2, and insertion was monitored using a protease protection assay (Brambillasca et al., 2005)
In proteoliposomes containing both Get1/2, the experimental 3F4-tagged SYP72 fragment was protected. In control proteoliposomes containing either a mutated form of Get1/2 that disrupts binding to Get3 or empty liposomes, the 3F4 fragment was poorly protected (Fig. 3). We conclude from the in vitro system that SYP72 interacts with yeast Get3 and is inserted in proteoliposomes in a manner comparable to a conventional TA-protein in yeast.
Figure 3.
GET3 inserts SYP72 into proteoliposomes in vitro. A proteinase K protection assay was used to demonstrate that yeast Get1/2 can insert Arabidopsis SYP72 into proteoliposomes via yeast Get3. The purified Get3-SYP72 complex was incubated with yeast rough microsomes or proteoliposomes containing the Get1/2 insertase complex. Get1*/2* point mutants that disrupt binding to Get3, empty liposomes, and buffer serve as negative controls. After insertion, SYP72 substrate is digested with proteinase K. Digestion of properly inserted SYP72 results in a C-terminal protected fragment, indicating insertion and correct orientation. Full-length SYP72 contains 4 times as many 35S-Met residues as the protected fragment, corresponding to the difference in band intensity.
To determine whether SYP72 interacts with Arabidopsis GET3 (AtGET3) in vitro, we incubated HIS-SYP72 with MBP-AtGET3, both produced and purified from E. coli (Fig. 2C). We found that, indeed, HIS-SYP72 bound to and coimmunoprecipitated with MBP-AtGET3. From work in other systems, Get3 is known to interact with the TMDs of client proteins. We generated a truncated version of HIS-SYP72 lacking the TMD (HIS-SYP72ΔTMD) and observed that the truncated version did not interact and did not coimmunoprecipitate with MBP-GET3, indicating that Arabidopsis recognizes and interacts with the TMD of conventional tail-anchored proteins.
GET System Is Required for TA Protein Insertion
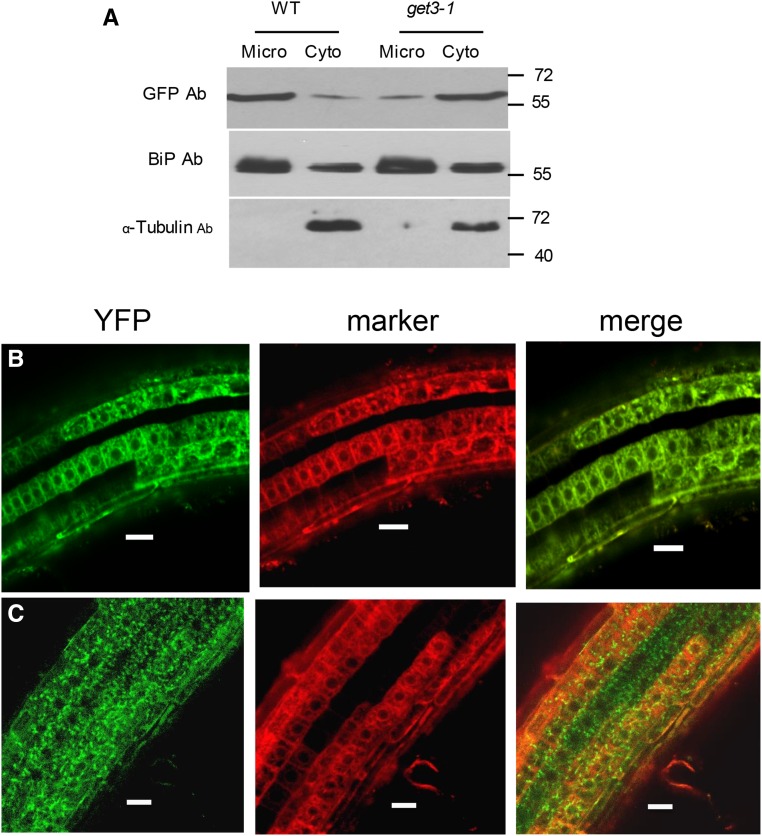
To determine whether the GET system is required for the insertion of SYP72 into Arabidopsis cell membranes in vivo, we analyzed the insertion of YFP-SYP72 into ER membranes in a line with a T-DNA insertion in the GET3 gene (Salk 033189; Supplemental Fig. S5, A and B). Cell fractionation and western-blot analysis revealed that when YFP-SYP72 was expressed as a transgene in a wild-type background, it was largely localized in the crude microsome fraction (Fig. 4A). In contrast, when the YFP-SYP72 was expressed in a get3-1 background, it was mostly recovered in the cytosolic fraction.
Figure 4.
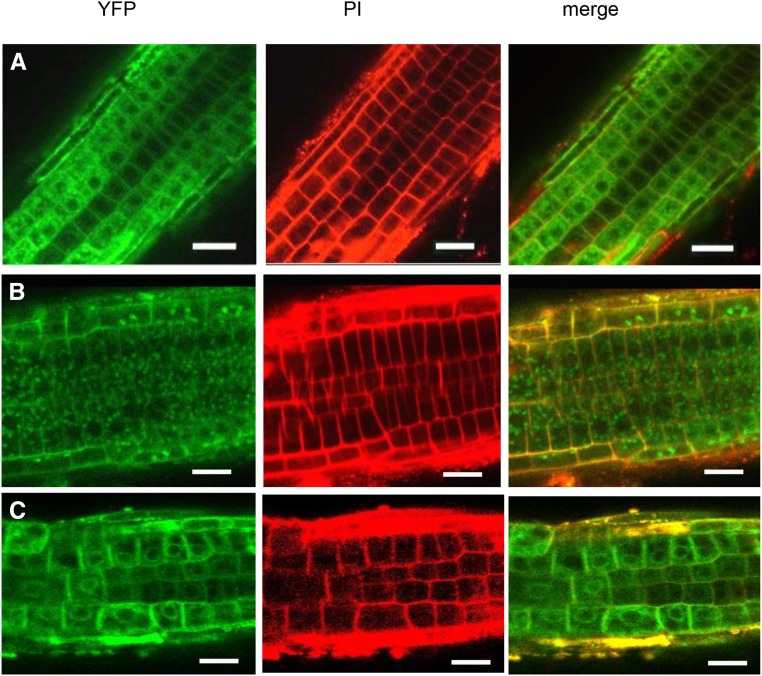
Subcellular localization of YFP-SYP72 in wild-type and get3-1 backgrounds. A, Wild-type and get-3-1 seedlings from lines stably expressing YFP-SYP72 were fractionated into microsomal and cytoplasmic fractions. Immunoblots of fractionated and total extracts were probed with anti-GFP and anti-BiP antibodies. BiP was used as an ER marker. B and C, Confocal microscopy of roots from seedlings coexpressing YFP-SYP72 and the CDC-960 mCherry ER marker in a wild-type background (B) and a get-3 background (C). D, Roots from seedlings expressing YFP-SYP72 in a wild-type background counter stained with propidium iodide. Bar = 50 µm.
We confirmed these cell fractionation observations by confocal microscopy localizing YFP-SYP72 in root cells of transgenic plants with wild-type and mutant backgrounds. In a wild-type background, YFP-SYP72 was observed primarily in perinuclear and cortical ER, coinciding with the ER marker CDC-960-mCherry (Fig. 4B). On the other hand, in a get3-1 background, YFP-SYP72 was mostly found as granular inclusions in the cytoplasm, not associated with the ER marker (Fig. 4C). Thus, functional GET3 is required for proper YFP-SYP72 insertion into ER membranes. To further demonstrate that GET3 is responsible for the failure of the get3-1 line to properly insert YFP-SYP72 into microsome membranes, get3-1 was complemented with a 35S promoter:GET3 construct (Supplemental Fig. S6). It was found by confocal microscopy that transgenic expression of GET3 in the get3-1 mutant partly restored the ER localization of YFP-SYP72 (Fig. 5).
Figure 5.
Subcellular localization of YFP-SYP72 in wild-type and get3-1 backgrounds. Confocal microscopy of roots from seedlings expressing YFP-SYP72 in a wild-type background (A), in a get3-1 background (B), and in get3-1 complemented with 35S:GET3 (C). All were counterstained with propidium iodide (PI). Bar = 50 µm.
The insertion of YFP-SYP72 into ER membranes also required functional GET1 and GET4. In get1-1 or get4-1, T-DNA insertion lines, YFP-SYP72 was not localized in the ER but mainly observed in cytoplasmic granules (Supplemental Fig. S7, B and D). When the mutants were complemented with 35S promoter:GET1 and GET4 constructs, respectively (Supplemental Fig. S6), the ER localization of YFP-SYP72 was mostly restored (Supplemental Fig. S7, C and E). Thus, the Arabidopsis homologs of GET1, 3, and 4 are all required for the proper insertion of YFP-SYP72 in plant cell membranes.
Phenotype of get Mutants
Since the GET system appears to play a role in the insertion of TA proteins in the ER membrane, it was of interest to know whether get mutants might affect Arabidopsis growth and development. Surprisingly, early growth in get mutants appeared similar to the wild type under normal growth conditions (Fig. 6A). However, at later stages of growth, we observed that get1-1 and get3-1 flowered earlier than the wild type, suggesting that the mutants were subject to stress (Fig. 6B).
Figure 6.
Phenotypes of get mutants. A, Wild-type, get3-1, and get1-1 2-week-old plants grown under normal growth conditions. B, The same lines as 4-week-old plants grown under normal growth conditions to show early flowering. C and D, Wild-type, get3-1, and get1-1 4-week-old seedlings on LS medium supplemented with 2.5 mm DTT (C) and on LS medium without DTT (D). E and F, get3-1 (E) and get1-1 (F) 4-week-old seedlings complemented with 35S promoter:GET3 or GET1 cDNA constructs, respectively, and grown on LS medium supplemented with 2.5 mm DTT.
Since the get mutants fail to insert TA proteins into the ER, the earlier flowering phenotype suggests that these mutants might have ER dysfunctions. Therefore, we tested whether the get mutants could tolerate ER stress. When the mutants were grown in the presence of an ER stress agent, 2.5 mm DTT, it was seen that get1-1 and get3-1 were more sensitive to DTT compared to the wild type (Fig. 6, C and D). At 4 weeks on plates containing 2.5 mm DTT, the mutant seedlings were stressed compared to the wild type. The mutant seedlings were underdeveloped, had senesced, and lost chlorophyll content. The complemented lines recovered tolerance to DTT treatment (Fig. 6, E and F). Furthermore, we also observed that an indicator of ER stress, BIP3, was constitutively expressed in get3-1 and get1-1 plants (Supplemental Fig. S8). The early flowering phenotype and BIP3 expression indicate that the get mutants are ER stressed, a condition that correlates with the failure in the insertion of tail-anchored proteins into the ER.
DISCUSSION
The GET system for insertion of TA proteins has been well characterized in mammalian and yeast systems (Sherrill et al., 2011). This study has identified components of the GET system in Arabidopsis and demonstrated using the ER resident tail-anchored protein SYP72 that the GET system functions in Arabidopsis. We identified homologs of components of the GET pathway in Arabidopsis with the exception of GET2 and Bag6 (Table I). Knockout mutant lines get1-1, get3-1, and get4-1 led to the mislocalization of the ER localized tail-anchored protein SYP72, which could be restored by complementation using 35S promoter-driven constructs of the respective genes. Get1 and Get2 work in concert in yeast to insert TA proteins into ER membranes. The absence of a readily identified GET2 homolog in Arabidopsis likely reflects sequence divergence from yeast. For example, the functional homolog of Get2 in mammalian cells, CAML (Yamamoto and Sakisaka, 2012), has no significant sequence similarity to the yeast Get2 or to any protein in Arabidopsis. Alternatively, Arabidopsis GET1 may serve the role of both yeast Get1 and Get2 in inserting TA proteins into the ER membrane in Arabidopsis.
There are a many TA proteins that populate the outer membranes of different organelles, and an interesting question is how different TA proteins are targeted to specific organelles (Abell and Mullen, 2011). The GET system provides some answers about the specificity of targeting TA proteins to the ER, but raises other questions. The specific targeting to the ER is brought about by the binding of GET3 to a client protein and delivering the GET3/client protein complex to the GET1/GET2-containing complex on the ER membrane. However, that mechanism begs the question as to how GET3 recognizes ER targeted proteins. In yeast, the cytosolic chaperone Sgt2 (SGTA in humans) first recognizes and captures the newly synthesized TA proteins (Wang et al., 2010; Mariappan et al., 2011). Then the TA protein/Sgt2 (TRC35 in humans) complex is handed over to the Get4-Get5 scaffolding complex. Thus, some of the specificity for ER targeting must lie in the hands of Sgt2 and/or GET4/GET5 in Arabidopsis. It will be of interest to determine whether these components interact specifically with ER-targeted TA proteins in Arabidopsis.
If there is competition for the targeting of TA proteins to different organelles, then one might expect that in the loss-of-function get mutants in our study, that YFP-SYP72 might have been inserted into another organelle. However, we did not observe that. Rather it appeared that the misdirected YFP-SYP72 aggregated. Thus, it is not clear that YFP-SYP72 was picked up by another organelle TA protein insertion system in the absence of GET system function. Okreglak and Walter (2014) described the mislocalization in yeast of Pex15, a TA protein that is normally inserted by the GET pathway into the ER and transported to peroxisomes. Under normal conditions and especially in get mutants, Pex15 can be mislocalized to the outer mitochondrial membrane whereupon it is extracted by Msp1, a AAA-ATPase. Thus, yeast have a quality control system to prevent the misdirection of ER or peroxisomal TA proteins to the mitochondrial outer membrane.
The loss-of-function get mutants in our study did not show obvious morphological phenotypes at early vegetative stages of growth under normal conditions. However, the get-3 and get-1 mutant lines flowered earlier than the wild type. Early flowering is a sign of stress, and these mutants showed other features characteristic of stress. For example, BIP3 expression (a marker for ER stress) was elevated in the get1-1 and get3-1 lines even under normal conditions of growth. The mutant plants were also highly sensitive to the ER stress agent DTT. These signs of ER stress in the get mutant lines may be due to the loss in proper insertion of a several TA proteins that play a role in protection from ER stress. For example, the stress-induced membrane-associated transcription factor NAC089 is a TA protein that resides in the ER membrane until it relocates to the nucleus in response to stress (Yang et al., 2014). However, the fact that get mutants are viable is likely due to alternative mechanisms for the insertion of TA proteins into ER membranes. These mechanisms may provide alternative routes for some TA proteins better than others. The failure of SYP72 to properly locate in get mutants suggests that alternative routes are less available for this TA protein.
In summary, we show in this study an operational GET system in plants and its direct role in TA protein insertion and indirect role in ER stress.
MATERIALS AND METHODS
Plant Material and Stress Treatments
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used in this study. Seeds were stratified at 4°C for 3 d prior to germination. Plants were grown under continuous white light at 23 to 25°C in soil or on Linsmaier/Skoog medium (1× LS salts, 1% Suc, and 0.8% agar). Agrobacterium tumefaciens-mediated transformation was performed using the floral dip method (Bechtold et al., 1993). A. tumefaciens strain GV3101 was used in all transformation experiments.
For confocal microscopy and cell fractionation experiments, single and double transgenic plants were generated by successive floral dips. The double transgenics were as follows: YFP-SYP72 in the wild type, get-1, get-3, and get-4 background with the CDC-960 ER-mCherry marker. Complementation lines were generated by expressing 35S promoter GET1, 2, or 3 constructs in their cognate mutant backgrounds. Single transgenic lines were generated for GET1-YFP, GET3-YFP, GET4-YFP, GET1Stop, GET3Stop, and GET4Stop.
T-DNA mutant lines were obtained from ABRC. An insertion in the first exon of the gene At4G16444 designated as get1-1 (SAIL line CS861559) was screened and used as a knockout line. The get3-1 mutant line (SALK_033189) was an insertion in the first intron of At1g01910. An insertion in the fifth exon of At5g63220 designated get4-1 (Salk_121195) was used as a knockout line for GET4. The T-DNA lines were genotyped by PCR using gene-specific primers and a left border T-DNA primer. The mutant lines were confirmed for loss of the transcript using cDNA-specific primers as described in Supplemental Table S1.
Plasmid Construction
The open reading frames of Get1 (At4G16444), Get3 (At1g01910), and Get4 At5g63220 were amplified from 10-d-old Arabidopsis seedlings. The products were cloned in pSKM36 and pSKY (Srivastava et al., 2013) at the AscI and SpeI sites for GET1 and 3 and AscI and NheI sites for Get4 to generate Get1pSK, GET3pSK, and Get4pSK. The C-myc tag in pSKM36 and YFP tag in pSKY were designed in frame with the genes. The cloned templates of GET1, 3, and 4 were used for amplifying and cloning Get3 and Get4 in pMALCH using MBPGET3 and MBPGET4 primers at the BamHI-SalI and BamHI, and HindIII sites, respectively. GET3 was cloned in pET42a using GET3GST primer at the BamHI-NotI sites, and GET1 and GET4 were cloned in pET28a using HISGET1 and HIS GET4 primers at the BamHI-NotI sites.
The open reading frame of SYP72 (At3G45280) was amplified from 10-d-old seedlings by RT-PCR using the primer YFP-SYP to generate pMSYP72. The product was cloned into pSKM36 at the AscI and SpeI sites, resulting in pMSYP72. pMSYP72 was tagged with YFP by amplifying YFP from a YFP vector using YFPAsc primers and inserting it into the AscI site to generate N-terminal YFP-tagged SYP72. pMSYP72 was used as template to amplify and clone SYP72 in pET28a using the primers HIS-SYP with the His tag at the N terminus using BamHI and NotI sites. A TMD minus truncation of SYP72 was generated using HIS-SYP-TMD. The various primers used in the study are shown in Supplemental Table S2.
Transient Transformation in Tobacco
Transient expression in tobacco (Nicotiana benthamiana) leaf epidermal cells was performed as described previously (Batoko et al., 2000) using A. tumefaciens (OD600 = 0.05) containing the genes in the binary vector pSKY.
Immunoprecipitations Using Recombinant Proteins from Escherichia coli
The proteins were tagged with MBP, GST, or His tags based on the requirements of the experiment. The cells were grown at 37°C for 8 h followed by protein induction for 20 h at 16°C. MBP (NEB) or Glutathione beads from Sigma-Aldrich containing 2 μg each of MBP, GST, MBP-Get3, or GST-GET3 were incubated in 20 μL (1 to 2 μg) of the potential interactor in pull-down buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% NP-40, and 1 mm DTT) plus 1 mg/mL BSA. The mixture was rotated in a 4°C cold room for 2 h and the beads were washed five times with pull-down buffer. The proteins were stripped off the beads by boiling with 12 μL 2× SDS buffer and loaded onto a PAGE gel. The anti-HIS antibody (27-4710-01) used was from GE Healthcare. The Anti-MBP antibody was a kind gift from Dr. Yanhai Yin at Iowa State University.
Cell Fractionation
Three grams of 10-d-old seedlings was collected on ice. The seedlings were ground in a mortar and pestle until completely homogeneous. Two milliliters of ice-cold buffer containing 0.3 m Suc, 0.1 m Tris-HCl (pH 7.5), 1 mm EDTA, and 0.1 mm PMSF (CF buffer) was added and ground again. The extract was strained through two layers of cheesecloth. The extract was centrifuged for 5 min at 10,000g at 4°C. The pellet was discarded, and the supernatant was centrifuged at 40,000 rpm in a 70Ti rotor for 45 min. The crude microsomal pellet obtained was washed and resuspended in the CF buffer for further use. The protein was quantified by Bradford assay.
Immunoblot Analysis
Immunoblots were performed as described by Srivastava et al. (2013). To examine the processing of bZIP28, plants were grown vertically on petri plates containing agar medium. Ten-day-old seedlings were treated with 2 mm DTT in LS medium, and 300 mg of root material was harvested from the treated plants. Roots were homogenized in liquid nitrogen, and 30 μg of protein was loaded per lane on gels. T8203, monoclonal antitubulin antibody from Sigma-Aldrich was used to detect tubulin as a loading control. Anti-GFP antibodies (11814460001) were obtained from Roche, and BiP antibody, ADI-SPA-818 D, was procured from Enzo Life Sciences.
Confocal Microscopy
Subcellular localization experiments with fluorescent-tagged proteins were performed using a NikonC1si confocal scanning system attached to a 90i microscope (Nikon Instruments). The 3.90.869.4 version of Nikon EZ-C1 FreeViewer was used to analyze the images obtained from the confocal microscope. Roots from Arabidopsis and leaves from tobacco were used for microscopy. The roots and tobacco leaves were observed under 20× and 60× water lenses. Some roots were counterstained with 50 mg mL−1 propidium iodide. The emission signals for YFP, propidium iodide, and mCherry were acquired using sequential scanning mode to eliminate crosstalk and emission signal bleed-through. Fluorescence emission was obtained by laser excitation of YFP at 488 nm and for mCherry and propidium iodide at 591 nm. Emission was in the range 500 to 575 nm and 590 to 700 nm, respectively. Propidium iodide (P4864) was obtained from Sigma-Aldrich.
Proteinase K Protection Assays
35S-SYP72 substrates were synthesized using the T7-coupled PURExpress In Vitro Protein Synthesis Kit (New England Biolabs) in the presence of 35S-Met and 25 μm Get3. The Get3-SYP72 complex was purified on Streptactin sepharose and eluted with buffer containing 5 mm biotin.
The protease protection assay, as well as purification and reconstitution of recombinant Get1, Get2, and Get3, was performed as described previously (Mariappan et al., 2011) Assays were performed in 10-μL reactions at roughly 20 nm of both Get3-SYP72 and Get1/2. Buffer contained 50 mm HEPES, pH 7.5, 150 mm KAc, 7 mm MgAc2, 2 mm ATP, and 1 mm DTT. After 30 min at 32°C, 1 μL of 5 mg/mL Proteinase K was added. After 2 h on ice, PMSF was added and the reaction was reverse quenched into 10 μL boiling 2× LDS sample buffer to quickly destroy the Proteinase K. Tris-Tricine SDS-PAGE gels (12%) were used, dried, and imaged using a phosphor imager.
Gene Expression Analysis
Total RNA was isolated from ground plant tissues using an RNeasy kit, treated with RNase-free DNase I, according to the manufacturer’s instructions (Qiagen), and quantified by 260/280-nm UV light absorption. A 1-μg portion of total RNA was reverse transcribed using the Supertranscript III RT kit (Invitrogen). A 2-μL volume of cDNA was used for RT-PCR. All primers are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: GET1 (At4g16444), GET3 (At1g01910), GET4 (At5g63220), GET5 (At1g55060), Sgt2 (At4g08320), SYP72 (At3g45280), and NAC089 (At5g22290).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment by Clustal omega (EMBL-EBI) of protein sequences for yeast (Saccharomyces cerevisiae) GET3 and homologs in other organisms.
Supplemental Figure S2. Subcellular localization of components of the GET pathway.
Supplemental Figure S3. Analysis of syntaxin 72 (At3g45280) protein sequence.
Supplemental Figure S4. Synthesis of the components used in the proteinase K protection assays.
Supplemental Figure S5. Gene models for Arabidopsis GET gene homologs as indicated.
Supplemental Figure S6. Expression of the GET transgenes in complemented mutant lines.
Supplemental Figure S7. Localization of YFP-tagged SYP72 in various get mutant lines.
Supplemental Figure S8. Expression of an ER stress indicator in get mutant lines.
Supplemental Table S1. Primers used in the study.
Supplementary Material
Glossary
- ER
endoplasmic reticulum
- TA
tail-anchored
- TMD
transmembrane domain
Footnotes
Articles can be viewed without a subscription.
This work was supported in part by the Iowa State University Plant Sciences Institute and by a National Science Foundation grant to S.H.H. (IOS90917).
References
- Abell BM, Mullen RT (2011) Tail-anchored membrane proteins: exploring the complex diversity of tail-anchored-protein targeting in plant cells. Plant Cell Rep 30: 137–151 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T (2003) Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem 278: 8219–8223 [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B (1975) Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol 67: 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Fasana E (2011) Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta 1808: 937–946 [DOI] [PubMed] [Google Scholar]
- Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, Hegde RS, Borgese N (2005) Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J 24: 2533–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Suloway CJ, Zaslaver M, Clemons WM Jr (2010) Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA 107: 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V. (2012) A portrait of the GET pathway as a surprisingly complicated young man. Trends Biochem Sci 37: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan O, Taylor NL, Carrie C, Eubel H, Kubiszewski-Jakubiak S, Zhang B, Narsai R, Millar AH, Whelan J (2011) Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol 157: 1093–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Keenan RJ (2011) Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol 12: 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Li J, Qian X, Denic V, Sha B (2009) The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One 4: e8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch T, Cambon A, Wattenberg BW (2007) A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic 8: 1687–1694 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Shaw R, Mukherjee J, Bowsher CG, Harrison AM, Abell BM (2009) Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic 10: 1753–1764 [DOI] [PubMed] [Google Scholar]
- Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, Keenan RJ (2011) The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ (2015) Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347: 1152–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ (2009) The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461: 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Walter P (2014) The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci USA 111: 8019–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C, Schmid V, Schwappach B, High S (2009) Biogenesis of tail-anchored proteins: the beginning for the end? J Cell Sci 122: 3605–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS (2008) The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134: 634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill J, Mariappan M, Dominik P, Hegde RS, Keenan RJ (2011) A conserved archaeal pathway for tail-anchored membrane protein insertion. Traffic 12: 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH (2013) BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25: 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Löhr F, Bernhard F, et al. (2011) Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science 333: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V (2010) A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40: 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sakisaka T (2012) Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell 48: 387–397 [DOI] [PubMed] [Google Scholar]
- Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX (2014) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10: e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.