Nitric oxide may regulate histone acetylation at stress-related genes by inhibiting histone deacetylases.
Abstract
Histone acetylation, which is an important mechanism to regulate gene expression, is controlled by the opposing action of histone acetyltransferases and histone deacetylases (HDACs). In animals, several HDACs are subjected to regulation by nitric oxide (NO); in plants, however, it is unknown whether NO affects histone acetylation. We found that treatment with the physiological NO donor S-nitrosoglutathione (GSNO) increased the abundance of several histone acetylation marks in Arabidopsis (Arabidopsis thaliana), which was strongly diminished in the presence of the NO scavenger 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide. This increase was likely triggered by NO-dependent inhibition of HDAC activity, since GSNO and S-nitroso-N-acetyl-dl-penicillamine significantly and reversibly reduced total HDAC activity in vitro (in nuclear extracts) and in vivo (in protoplasts). Next, genome-wide H3K9/14ac profiles in Arabidopsis seedlings were generated by chromatin immunoprecipitation sequencing, and changes induced by GSNO, GSNO/2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide or trichostatin A (an HDAC inhibitor) were quantified, thereby identifying genes that display putative NO-regulated histone acetylation. Functional classification of these genes revealed that many of them are involved in the plant defense response and the abiotic stress response. Furthermore, salicylic acid, which is the major plant defense hormone against biotrophic pathogens, inhibited HDAC activity and increased histone acetylation by inducing endogenous NO production. These data suggest that NO affects histone acetylation by targeting and inhibiting HDAC complexes, resulting in the hyperacetylation of specific genes. This mechanism might operate in the plant stress response by facilitating the stress-induced transcription of genes.
Nitric oxide (NO) is a key messenger molecule in all kingdoms. In plants, NO participates in the regulation of various physiological processes like flowering, stomatal closure, germination, root development, gravitropism, and the response to abiotic and biotic stresses (Delledonne et al., 1998; Durner et al., 1998; García-Mata and Lamattina, 2002; Pagnussat et al., 2002; He et al., 2004; Hu et al., 2005; De Michele et al., 2009; Šírová et al., 2011). Mutants impaired in NO production or turnover show pleiotropic and severe phenotypes, highlighting the fundamental and multiple roles of NO in Arabidopsis (Arabidopsis thaliana). NO belongs to the group of redox-signaling molecules whose common feature is the ability to covalently modify target residues (mainly Cys) on proteins, thereby altering the function of the protein (Kovacs and Lindermayr, 2013). This redox-signaling mechanism is very important in plant development as well as in the response to abiotic and biotic stresses (Suzuki et al., 2012; Yun et al., 2012). In plants, most of the biological functions of NO are mediated by protein S-nitrosylation. In this process, NO reversibly binds to specific Cys residues of proteins, resulting in the formation of S-nitrosothiols (SNOs), thereby altering the catalytic activity, subcellular localization, or association with binding partners of the protein. In this context, the S-nitrosylated form of the antioxidant tripeptide glutathione (S-nitrosoglutathione [GSNO]) is considered to play a major role as a stable and mobile cellular NO reservoir. On the one hand, GSNO can directly release NO in the presence of reduced glutathione (GSH), copper(II), or hemoglobins (Singh et al., 1996; Spencer et al., 2000). Therefore, GSNO is widely used as an in vivo NO donor (Mur et al., 2013). The direct reaction of NO with Cys thiols is too slow to occur in vivo (Folkes and Wardman, 2004). Instead, it is assumed that NO reacts rapidly with oxygen, resulting in the formation of N2O3, which then directs the de novo formation of SNOs (Ridnour et al., 2004). On the other hand, GSNO can directly donate a nitrosonium ion (NO+), without the intermediate formation of NO, to thiol groups of target proteins, a mechanism known as transnitrosylation (Stamler, 1994; Hogg, 2002). This reaction allows us to analyze the effect of S-nitrosylation on proteins in vitro (Lindermayr et al., 2006; Wang et al., 2009; Yun et al., 2011).
NO treatment leads to substantial transcriptional reprogramming (Huang et al., 2002; Polverari et al., 2003; Palmieri et al., 2008), which might be partially mediated through the S-nitrosylation of transcription factors (Serpa et al., 2007; Tada et al., 2008; Lindermayr et al., 2010; Viola et al., 2013). It remains elusive whether NO also might regulate transcription by affecting the chromatin state, which constitutes the second layer of transcriptional control in eukaryotes.
The basic building blocks of chromatin are nucleosomes, which consist of a histone octamer around which the DNA is wrapped. The nucleosomes are arranged into higher order structures, resulting in a strong compaction of the contour length of the DNA but still allowing the temporally and spatially controlled access of the transcriptional machinery to certain genes. This local accessibility of the DNA is determined by posttranslational modifications of the N termini of histones as well as cytosine methylation, large ATP-dependent remodeler complexes, and histone variants (Pikaard and Mittelsten Scheid, 2014). Among these, histone acetylation plays a major role in the regulation of transcription (Jenuwein and Allis, 2001). First, the acetylation of Lys residues neutralizes positive charges on the histone octamer, thereby weakening the interaction with the negatively charged DNA, resulting in a local opening of the chromatin. Second, acetylated Lys residues may serve as docking platforms for bromo domain-containing proteins, which, in turn, can be regulators of the chromatin state or direct transcriptional regulators (Kurdistani and Grunstein, 2003). Hyperacetylated chromatin often is associated with high transcriptional activity, since the transcriptional machinery has easy access to the DNA. In contrast, hypoacetylated chromatin has a very dense structure, which is usually transcriptionally inactive. Histone acetylation supports the transcription of genes; however, recent reports suggest that histone acetylation alone is not sufficient to activate transcription (Yang et al., 2016).
Histone acetylation is controlled by two enzyme superfamilies. Histone acetyl transferases (HATs) catalyze the formation of the ε-amino bond between Lys residues and acetyl groups using acetyl-CoA as a cofactor, whereas histone deacetylases (HDACs) hydrolyze these bonds. HATs are classified into four families in Arabidopsis, harboring different substrate specificities (Earley et al., 2007). HDACs are grouped into three families. The largest family consists of 12 members, characterized by a highly conserved HDAC domain with a catalytic zinc ion, and shares homology with yeast REDUCED POTASSIUM DEPENDENCY PROTEIN3 (RPD3) or HISTONE DEACETYLASE1 (HDA1; Pandey et al., 2002). Sirtuins (two members in Arabidopsis) are homologs of yeast SILENT INFORMATION REGULATOR2 and display a different catalytic mechanism using NADH as a cofactor. The HD2-like family (HD2A, HD2B, HD2C, and HD2D) is plant specific, since no homologs have been identified in other organisms so far (Dangl et al., 2001).
In animals, NO participates in the regulation of histone acetylation by targeting and modifying several HDACs. In plants, no such link has been reported so far. The aim of this study was to describe the impact of NO on histone acetylation in Arabidopsis. Treatment with the physiological NO donor GSNO increased global histone 3 (H3) and H4 acetylation in Arabidopsis. GSNO-induced hyperacetylation was probably mediated by S-nitrosylation and subsequent inhibition of HDACs, since GSNO and S-nitroso-N-acetyl-dl-penicillamine (SNAP) significantly and reversibly reduced total HDAC activity in vitro (in nuclear extracts) and in vivo (in protoplasts). Chromatin immunoprecipitation sequencing (ChIP-seq) revealed that several hundred genes displayed NO-regulated histone acetylation. Many of these genes were involved in the plant defense response and the abiotic stress response. Furthermore, salicylic acid (SA) and its functional analog 2,6-dichloro-isonicotinic acid (INA) inhibited HDAC activity and increased histone acetylation in protoplasts by stimulating endogenous NO production. Our data suggest that NO affects histone acetylation by targeting and inhibiting HDACs, thereby increasing histone acetylation at specific genes. This mechanism might play a role in the plant stress response by facilitating the stress-induced transcription of genes.
RESULTS
SNO Content in Seedlings after GSNO, GSNO/cPTIO, GSH, and Trichostatin A Treatment
To analyze the effect of NO on histone acetylation, liquid-grown Arabidopsis seedlings were stimulated either with the NO donor GSNO or a combination of GSNO and the NO scavenger 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO). In vivo GSNO treatment of plant tissue results in the transient release of NO (Mur et al., 2013), which can be prevented by coapplication of the specific NO scavenger cPTIO (Akaike et al., 1993; Maeda et al., 1995). Thus, the comparison of GSNO and GSNO/cPTIO treatments allows us to decipher the contribution of NO on a given process. Since reductive decomposition of GSNO results in the formation of GSH and, additionally, NO induces a rapid change in the glutathione state by activating the biosynthesis of GSH (Kovacs et al., 2015), a GSH treatment was included as an additional control to distinguish the direct effects of GSNO-released NO from those mediated by an increase of the GSH concentration, similar to a recently published study (Begara-Morales et al., 2014). As positive control, 5 µm trichostatin A (TSA) was applied, which is an inhibitor of RPD3-like HDACs and possibly HD-tuins (Yoshida et al., 1990; Bourque et al., 2011).
To ensure that the GSNO-mediated increase of cellular NO lies within a physiologically meaningful range, liquid-grown seedlings were treated with different concentrations of GSNO and the total SNO content (which correlates with the NO level) within the tissue was measured subsequently. Treatment with 250 µm GSNO resulted in an increase of the SNO level from 25 to 275 pmol mg−1 protein after 3 h (Supplemental Fig. S1), which was comparable to that observed after the infection of Arabidopsis plants with Pseudomonas syringae avrB (Feechan et al., 2005). At 16 h after GSNO stimulation, SNO levels had decreased to 75 pmol mg−1 protein (Supplemental Fig. S1), which can be explained either by nonenzymatic cleavage of SNOs by GSH, copper(II), and other cellular reductants or by enzymatic degradation mediated by GSNO reductase, an enzyme capable of degrading GSNO and, thereby, also controlling the amount of protein SNOs (Sakamoto et al., 2002). Coapplication of 500 µm cPTIO reduced the SNO level by 27.2% to 200 pmol mg−1 protein after 3 h, indicating that cPTIO can prevent the formation of SNOs originating from GSNO-released NO. Unexpectedly, we also observed slightly increased total SNO levels after treatment with 250 µm GSH (115 pmol mg−1 protein after 3 h). These results question the suitability of GSH as a no-NO control for GSNO in our experimental setup (see below). Interestingly, TSA also enhanced the formation of SNOs to 125 pmol mg−1 protein after 3 h and to 245 pmol mg−1 protein after 16 h, suggesting that TSA might induce NO formation in seedlings. This finding extends recent data demonstrating TSA-induced hydrogen peroxide (H2O2) and superoxide production (Wang et al., 2015). In contrast to the GSNO treatment, the level of SNOs after stimulation with TSA did not decline between 3 and 16 h. This might be due to the fact that TSA is stable in cells and, therefore, imposes continuous stress, which is generally associated with NO production on the plant. Additionally, TSA-induced hyperacetylation might deregulate the expression of oxidant and antioxidant enzymes, resulting in long-lasting alterations of reactive oxygen species and nitrogen species levels.
NO Enhances Global Histone Acetylation in Arabidopsis
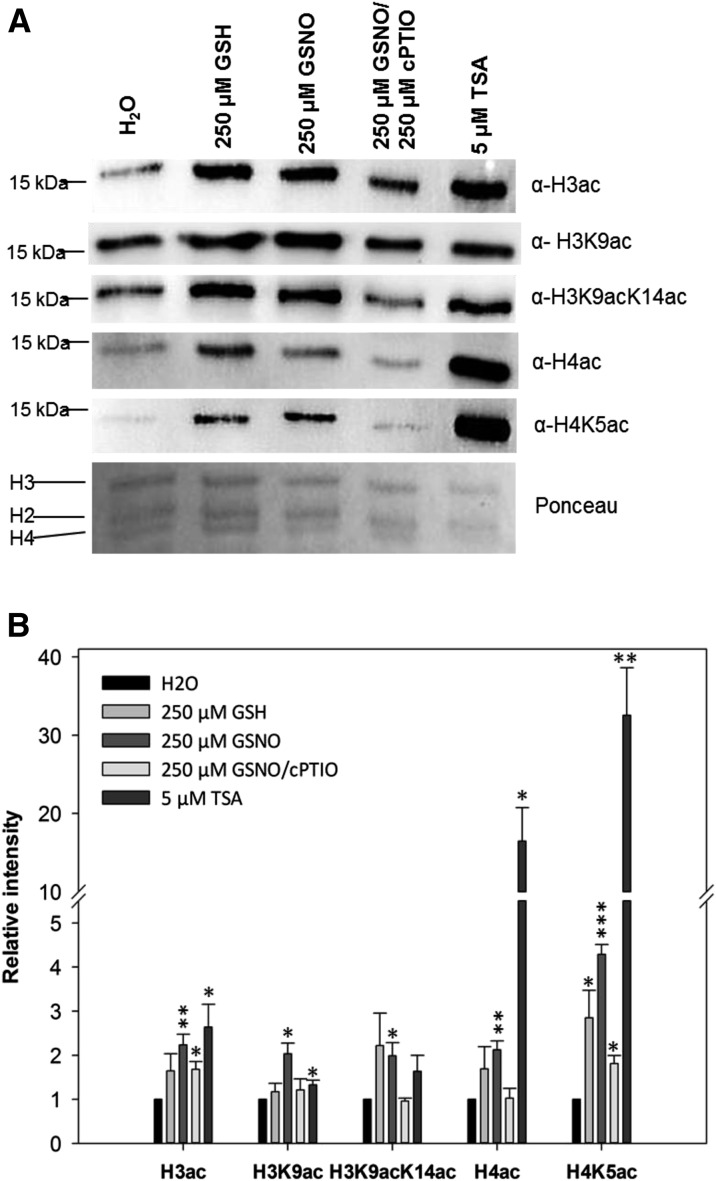
To determine the impact of NO on global histone acetylation levels, liquid-grown Arabidopsis seedlings were treated with 250 µm GSNO, 250 µm GSNO/500 µm cPTIO, 250 µm GSH, or 5 µm TSA. After 16 h, histones were extracted and the acetylation level was determined by western blot. GSNO significantly increased the abundance of all histone acetylation marks tested, namely H3ac (2.3-fold), H4ac (2.2-fold), H3K9ac (2.2-fold), H3K9/14ac (2.1-fold), and H4K5ac (4.5-fold), in comparison with water treatment (Fig. 1). Coapplication of cPTIO largely prevented GSNO-mediated hyperacetylation, suggesting that the increased histone acetylation levels were caused by NO. GSH treatment also enhanced histone acetylation of some of the marks tested (e.g. H4K5ac), although the effect was clearly weaker compared with GSNO. Since GSH increased SNO levels (Supplemental Fig. S1), it seems likely that the effect of GSH on histone acetylation might be indirect, via the formation of NO. As expected, TSA enhanced H3ac (2.5-fold), H3K9ac (1.4-fold), and H3K9/14ac (1.8-fold) as well as H4ac (18-fold) and H4K5ac (32-fold). GSNO-mediated histone hyperacetylation also was detected in Arabidopsis suspension cells, in which 500 µm GSNO induced a significant 1.7-fold increase of total H3ac (Supplemental Fig. S2).
Figure 1.
Histone acetylation in Arabidopsis seedlings after GSNO, GSH, GSNO/cPTIO, and TSA treatments. A, Histones were extracted from liquid-grown seedlings treated with 250 µm GSNO, 250 µm GSH, 250 µm GSNO/500 µm cPTIO, 5 µm TSA, or water (control) and probed against different histone acetylation marks by western blot. As a loading control, the Ponceau S-stained membrane is shown. One representative experiment out of three is shown. B, Quantification of western-blot results. Signal intensities were measured using ImageJ software and normalized to the amount of loaded H3. Values are expressed as fold change over the control treatment. Values shown are means ± se of three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by Student’s t test.
To analyze whether the NO-mediated increase of histone acetylation was caused by transcriptional down-regulation of HDACs, the corresponding mRNA levels were quantified by quantitative PCR (qPCR). For the time of the treatment, GSNO did not change the expression level of HDACs (Supplemental Fig. S3), suggesting that NO might affect histone acetylation by posttranslational mechanisms.
HDAC Activity Is Inhibited by Redox Modifications
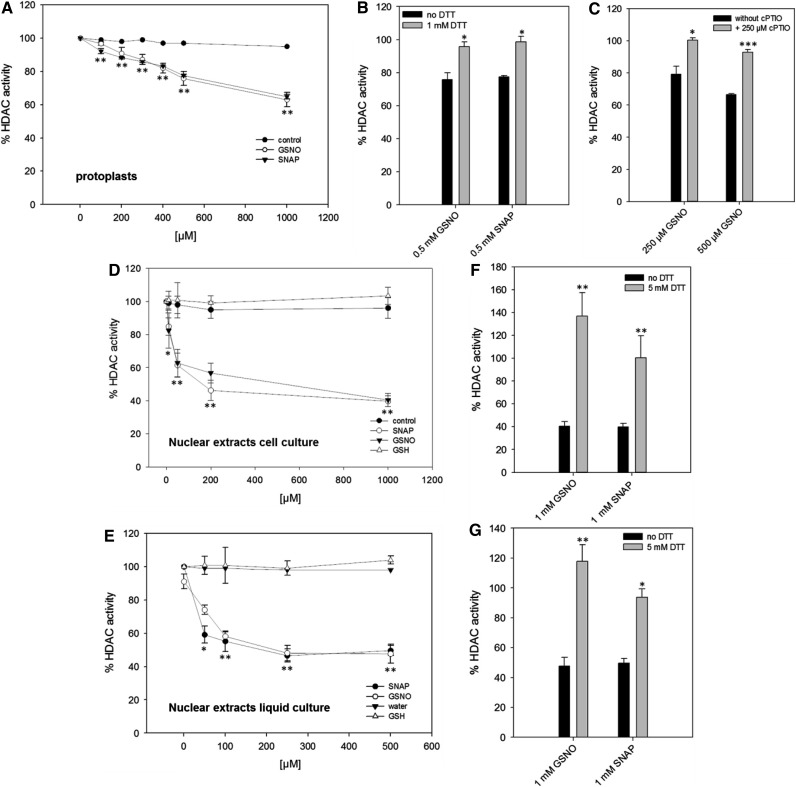
To explain the observed NO-mediated histone hyperacetylation, we hypothesized that NO might directly target HDACs and inhibit their activity, a mechanism that has been described for some mammalian HDACs (Colussi et al., 2008; Nott et al., 2008; Feng et al., 2011; Okuda et al., 2015). To determine the effect of NO on histone deacetylase activity, a commercial fluorescence-based HDAC assay was modified to be used in protoplasts. Protoplasts were chosen since the assay relies on the diffusion of a membrane-permeable substrate into the cells, which was not possible in suspension cells, probably due to cell walls and cell clump formation. Moreover, protoplasts provide a simple model that has proven useful for studying signal transduction mechanisms (Im and Yoo, 2014). GSNO and SNAP, which is another widely used NO donor, inhibited HDAC activity in a concentration-dependent manner (20% inhibition for 500 µm GSNO or SNAP; Fig. 2A). HDAC activity could be restored by the addition of dithiothreitol (DTT), indicating that the inhibition was mediated by oxidative Cys modifications and excluding irreversible chemical side effects of GSNO and SNAP (Fig. 2B). Furthermore, cPTIO prevented the inhibition of HDAC activity by GSNO, demonstrating that NO was responsible for this effect (Fig. 2C). As expected, TSA efficiently blocked HDAC activity in protoplasts, demonstrating the specificity of the assay (Supplemental Fig. S4A). Since GSH increased the total SNO content in seedlings (Supplemental Fig. S1) and also enhanced histone acetylation (Fig. 1), the effect of GSH on HDAC activity in protoplasts also was analyzed. GSH treatment resulted in an inhibition of HDAC activity, but the effect was clearly weaker compared with GSNO (Supplemental Fig. S4A). GSH-induced inhibition of HDACs could be largely prevented by cPTIO (Supplemental Fig. S4B), implying that GSH activates endogenous NO production, thereby affecting HDAC activity. These data support our previous observations (Fig. 1; Supplemental Fig. S1) and reinforce the conclusion that GSH is not an appropriate in vivo control for GSNO in our experimental system. To ensure that the observed reduction of HDAC activity was not caused by dying cells, we analyzed the viability of the protoplasts after the addition of GSNO and SNAP (Supplemental Fig. S5). Neither GSNO nor SNAP affected the viability of the protoplasts within the concentration range used.
Figure 2.
Reversible inhibition of HDAC activity by GSNO and SNAP in protoplasts and nuclear extracts. A, HDAC activity after GSNO and SNAP treatments in protoplasts. Water was used as a solvent control. B, DTT restored HDAC activity after GSNO and SNAP treatments. Protoplasts were first incubated with GSNO and SNAP before the addition of DTT. Subsequently, HDAC activity was measured. C, cPTIO prevented the inhibition of HDACs by GSNO. Protoplasts were preincubated with cPTIO for 5 min before the addition of GSNO, and HDAC activity was recorded. D and E, HDAC activity after GSNO and SNAP treatment in nuclear extracts from suspension cells (D) and liquid-grown seedlings (E). Nuclear extracts were treated with water (solvent control), GSH (control), GSNO, or SNAP for 20 min at room temperature in the dark. After desalting, HDAC activity was measured. F and G, DTT restored HDAC activity after SNAP and GSNO treatments. Nuclear extracts were first incubated with GSNO and SNAP before the addition of DTT. After desalting, HDAC activity was determined. Values are normalized to untreated nuclear extracts or protoplasts. Values shown are means ± se of three independent preparations of nuclear extract or protoplasts. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by Student’s t test.
NO mainly transduces its bioactivity by posttranslational modification of Cys residues (Mengel et al., 2013). To analyze whether NO inhibits HDAC activity through NO-mediated posttranslational modifications, we next studied the effect of GSNO and SNAP on HDAC activity in vitro. In the absence of reduction equivalents, both compounds do not release molecular NO (Singh et al., 1996); instead, they can directly transfer the nitroso group to susceptible protein Cys residues without the intermediate formation of NO (transnitrosylation). Therefore, GSNO and SNAP are widely used to determine the susceptibility of proteins toward S-nitrosylation in vitro (Lindermayr et al., 2005; Chaki et al., 2015). GSH is a commonly used control for in vitro S-nitrosylation studies, since it is structurally similar to GSNO but lacks the ability to modify Cys residues (Lindermayr et al., 2005; Dalle-Donne et al., 2009). Nuclear extracts prepared from Arabidopsis seedlings and suspension cells were incubated with different concentrations of GSNO and SNAP (10–1,000 µm), and HDAC activity was recorded using a modified ELISA-based assay. GSNO and SNAP significantly inhibited nuclear HDAC activity compared with water and GSH controls (Fig. 2, D and E). The dose-response curve dropped sharply for inhibitor concentrations ranging from 0 to 50 µm and flattened for higher concentrations of GSNO and SNAP (60% inhibition for 500 µm GSNO or SNAP). The inhibition of HDAC activity by GSNO and SNAP was fully reversible upon the addition of DTT (Fig. 2, F and G). The specificity of the assay was again demonstrated by incubating nuclear extracts with 1 µm TSA, which strongly inhibited HDAC activity (residual HDAC activity was less than 10%; Supplemental Fig. S6). The importance of reduced thiol groups for the catalytic activity of HDACs was further enforced by the observation that incubation with 0.5 mm N-ethylmaleimide (NEM), a chemical that blocks thiols by alkylation, reduced total nuclear HDAC activity to 35% (seedlings) and 20% (suspension cells) compared with control treatment (Supplemental Fig. S6).
Besides S-nitrosylation, GSNO also can mediate S-glutathionylation (Ji et al., 1999). This redox modification requires similar chemical characteristics of the thiol (high nucleophilicity) but is sterically more demanding and often has comparable effects on the target protein like S-nitrosylation (Dalle-Donne et al., 2009). To analyze the effect of S-glutathionylation on HDAC activity, oxidized glutathione (GSSG) was incubated with nuclear extracts or protoplasts and HDAC activity was monitored. In contrast to GSH, GSSG can S-glutathionylate Cys residues in the absence of oxidizing compounds by a thiol disulfide exchange reaction (Dalle-Donne et al., 2009). GSSG also inhibited total HDAC activity in nuclear extracts (35% inhibition for 500 µm GSSG) and protoplasts (12% inhibition for 500 µm GSSG; Supplemental Fig. S7), suggesting that S-glutathionylation also might participate in the redox regulation of HDACs. In contrast, high concentrations of H2O2, another major player in the concert of redox signaling, only marginally affected HDAC activity (Supplemental Fig. S7). Together, these data suggest that certain plant HDACs might be targets for redox regulation, being particularly sensitive toward S-nitrosylation and S-glutathionylation.
Identification of NO-Regulated H3K9/14ac Sites and Associated Genes by ChIP-Seq
Experimental Design
We used ChIP-seq to identify and quantify NO-triggered alterations in the histone acetylation pattern of Arabidopsis seedlings. Chromatin immunoprecipitation (ChIP) was performed with an antibody directed against H3K9/14ac (recognizing H3 acetylated at both Lys-9 and Lys-14), since this modification increased significantly after GSNO treatment (Fig. 1) and, in contrast to H3K9ac and H4K5ac, has not been profiled genome wide in plants (Widiez et al., 2014; Yang et al., 2016). The antibody recognized a single band in nuclear extracts and purified histone extracts from Arabidopsis (Supplemental Fig. S8), thereby fulfilling the criteria for ChIP-seq antibodies of the ENCODE consortia (Landt et al., 2012). An overview of the work flow is presented in Supplemental Figure S9.
Liquid-grown Arabidopsis seedlings treated with water (control), 250 µm GSNO, 250 µm GSH, 250 µm GSNO/500 µm cPTIO, or 5 µm TSA were cross-linked and harvested 3 and 16 h after the onset of the treatment, in order to compare early and later changes of H3K9/14ac. The experiment was performed in two independent biological replicates (Landt et al., 2012). For each sample, a separate input control was kept. Indexed libraries for all ChIP and input samples were pooled and sequenced four times on an Illumina HiSeq2500, resulting in at least 16 × 106 uniquely mapped reads per sample. One replicate of control_16h and one replicate of TSA_16h produced only very low read numbers and, therefore, were excluded from further analysis (Supplemental Fig. S10). All other samples displayed relative strand coefficient values greater than 0.8 (Supplemental Fig. S11), indicating successful enrichment of DNA fragments during the immunoprecipitation (Landt et al., 2012). H3K9/14ac peaks were called for each sample using the corresponding input DNA as a background control. Control experiments on chromatin shearing, library preparation, and peak calling are provided in Supplemental Figures S12 and S13. Finally, differential H3K9/14ac sites were computed for each treatment in comparison with the control using the DiffBind software (Ross-Innes et al., 2012), and NO-affected sites were selected by comparison of the GSNO and GSNO/cPTIO samples (for details, see below). Raw read files, peak files, and DiffBind output files were deposited in the Gene Expression Omnibus database (accession no. GSE82075).
General Features of H3K9/14 Acetylation
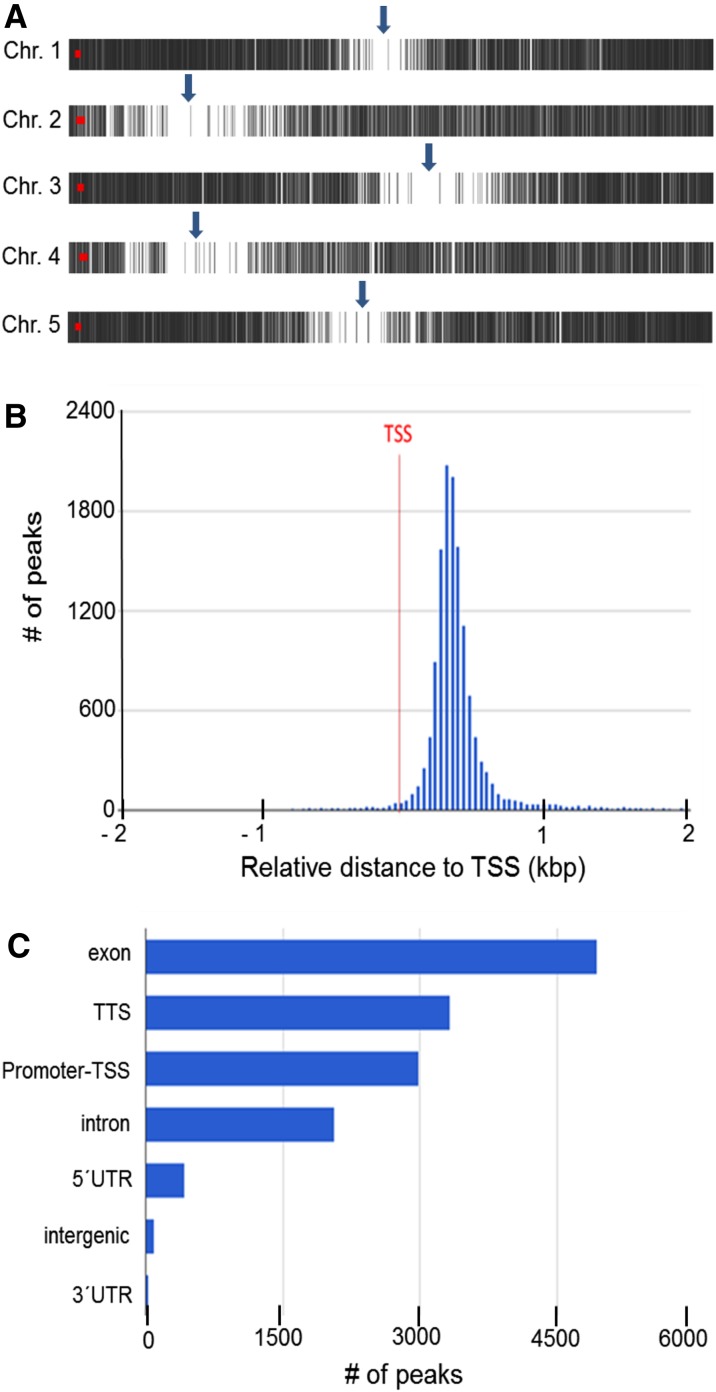
To investigate general features of H3K9/14 acetylation, peaks of the control_3h sample (replicate 1, 13,882 peaks; Supplemental Fig. S11) were annotated (using an annotation window of 2 kb upstream of transcriptional start sites [TSSs] and 1 kb downstream of transcriptional termination sites) and analyzed using the ChIPseek tool (Chen et al., 2014). H3K9/14ac was almost absent from centromeric and pericentromeric regions (Fig. 3A). A histogram of the distance of peak centers to the nearest annotated TSS demonstrates that H3K9/14ac was localized predominantly sharply downstream (around 400 bp) of TSSs (Fig. 3B). Most of the H3K9/14ac sites were located in exons (35%), followed by transcriptional termination sites (23%), proximal promoter regions (22%), and introns (15%), whereas only 5% of all peaks were found in 5′ or 3′ untranslated and intergenic regions (Fig. 3C). Motif enrichment analysis of the peak regions revealed the presence of numerous transcription factor target sites. The most significant one (P = E-168) was found in 49% of all peaks and closely resembled the ARR10 and AGP1 consensus-binding sequences (Supplemental Fig. S14). No differences of these general H3K9/14ac features across the GSNO, GSH, and GSNO/cPTIO treatments could be identified.
Figure 3.
General features of H3K9/14ac. A, H3K9/14ac distribution along Arabidopsis chromosomes. Each vertical gray line represents one H3K9/14ac peak. Arrows indicate the approximate positions of centromers. This image was prepared using PAVIS. B, Histogram of the distance of peak centers to the nearest annotated TSS. C, Location annotation of peaks. Peaks were annotated to functional DNA elements. TTS, Transcription termination site; UTR, untranslated region. B and C were prepared using the ChIPseek software portal (Chen et al., 2014).
Identification of NO-Regulated H3K9/14ac Sites and Associated Genes
Next, we aimed to identify putative NO-regulated histone acetylation sites. First, differential H3K9/14ac sites after GSNO, GSH, GSNO/cPTIO, and TSA treatments in comparison with the control were computed using the program DiffBind. DiffBind allows the quantitative pairwise comparison of ChIP-seq data, considering the appropriate input controls and biological replicates to perform statistical analysis (Ross-Innes et al., 2012). For verification, ChIP-qPCR on six randomly chosen differential H3K9/14ac sites was performed, which confirmed the results predicted by DiffBind (Supplemental Fig. S15). Moreover, visual inspection of representative GSNO-regulated H3K9/14ac sites using the CLC-genome browser also supported the results obtained by DiffBind (Supplemental Fig. S16).
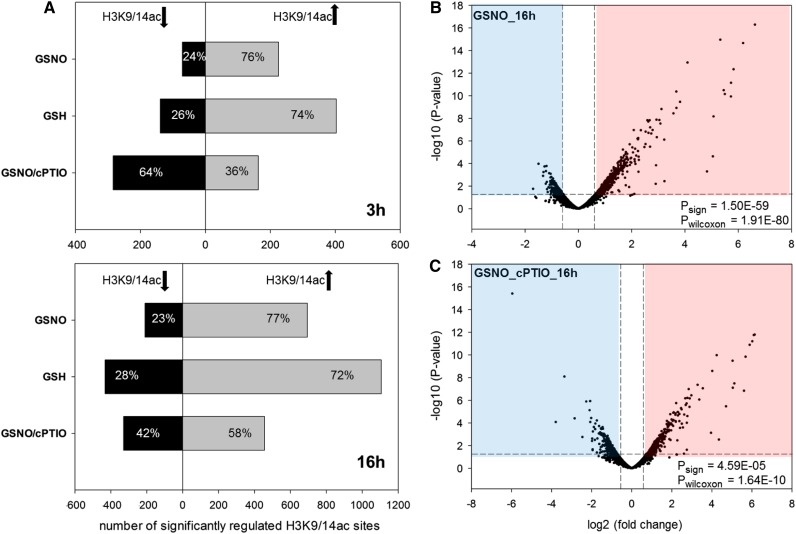
Consistent with our previous results, GSNO (and GSH) enhanced acetylation of the majority of differentially regulated H3K9/14ac sites after both 3 and 16 h of treatment (Fig. 4, A and B). cPTIO clearly reduced the number of peaks displaying increased histone acetylation, reinforcing that NO is responsible for or at least substantially contributes to the GSNO-induced hyperacetylation of H3K9/14ac observed before (Fig. 4C; Supplemental Fig. S17). Interestingly, the 3- and 16-h responses were very distinct from each other; H3K9/14ac sites, which were highly regulated at the 3- or 16-h time point, were not or weakly regulated at the other time point. As a result, the overlap of differentially regulated H3K9/14ac sites between both time points was very low (Supplemental Fig. S18). Although speculative, this might indicate that NO targets different HDACs depending on the duration of the NO exposure.
Figure 4.
Effects of GSNO, GSH, and GSNO/cPTIO treatments on H3K9/14ac. A, For each treatment, significant H3K9/14ac changes (P < 0.05) were determined in comparison with the control treatment using DiffBind software. Shown are the numbers of H3K9/14ac sites that display enhanced (gray bars) or decreased (black bars) acetylation after 3 h (top) and 16 h (bottom) of treatment. B and C, Volcano plots in which the –log10 (P value) of each analyzed H3K9/14ac site is plotted versus the corresponding log2 (fold change [treatment over control]). The horizontal dashed lines correspond to a P value of 0.05, and the vertical dashed lines mark fold changes of ±log2 (1.5). To test for significant differences in the number of peaks with enhanced or decreased H3K9/14ac among the peaks with P < 0.05, sign tests were performed (Psign). To test whether peaks with increased H3K9/14ac show higher absolute log2 (fold change) compared with peaks with decreased H3K9/14ac, Wilcoxon signed-rank tests were performed (Pwilcoxon).
NO-responsive H3K9/14ac sites were then identified by comparison of the GSNO and GSNO/cPTIO treatments. Peaks displaying up-regulation after GSNO (fold change > 1.5 and P < 0.05) but no up-regulation after GSNO/cPTIO treatment (fold change < 1.2 or P > 0.05) and peaks that displayed down-regulation after GSNO (fold change < 0.67 and P < 0.05) but no down-regulation after GSNO/cPTIO treatment (fold change > 0.83 or P > 0.05) were selected and defined as putative NO-regulated H3K9/14ac sites (Supplemental File S1). Totals of 194 and 552 putative NO-regulated H3K9/14ac sites were identified after 3 and 16 h of treatment, respectively (59.5% of all GSNO-regulated sites). Most of the NO-responsive H3K9/14ac sites (3 h) were similarly regulated in the TSA treatment (i.e. enhanced acetylation after GSNO and TSA treatment), whereas very few peaks showed opposite regulation (i.e. enhanced acetylation after GSNO but decreased acetylation after TSA treatment), reinforcing that NO targets and inhibits a subset of TSA-sensitive HDACs (Supplemental Fig. S19).
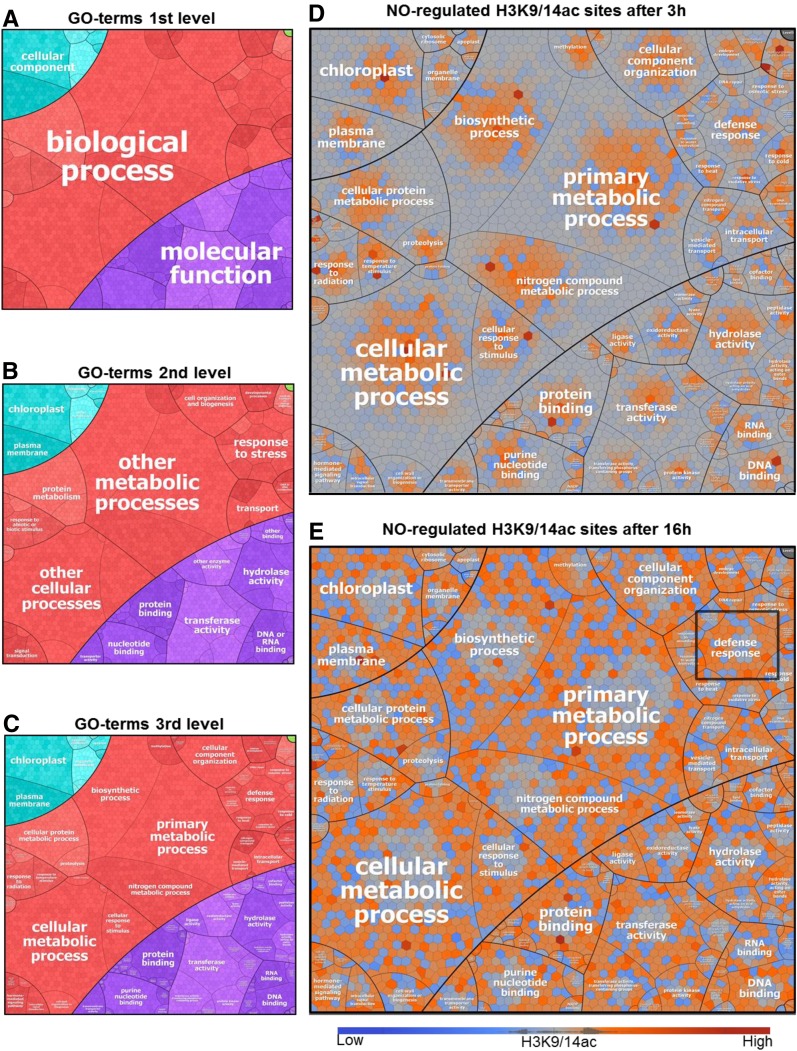
Next, putative NO-regulated H3K9/14ac sites were annotated to the nearest TSS and the corresponding genes were functionally categorized using the Gene Ontology (GO) hierarchy. The results were then visualized using Voronoi treemaps (Fig. 5). Genes with NO-responsive H3K9/14ac were involved in different biological processes (metabolism, transport, and stress response) and displayed diverse molecular functions (protein, DNA, and RNA binding, transferase activity, and hydrolase activity). However, simple assignment of genes to GO categories does not reflect whether certain GO terms are overrepresented. Therefore, separate GO enrichment analyses were conducted for gene subsets with either increased or decreased NO-regulated H3K9/14ac (Supplemental File S2). Enriched GO terms (P < 0.01) could only be identified in the groups of genes displaying increased H3K9/14ac. Among the category biological process, the top-ranking terms were response to cold (3 h) and defense response (16 h; Supplemental Table S1), both being subtypes of the parent term stress response. Defense genes showing NO-responsive H3K9/14ac included intracellular pattern recognition receptors, a mitogen-activated protein kinase (MKK2), PR genes, WRKY, and TGA transcription factors (Table I). Together, these data indicate that NO-mediated histone hyperacetylation might play a role in the plant’s stress response, particularly in the defense response.
Figure 5.
Voronoi treemaps visualizing the functional categorization of genes displaying NO-responsive H3K9/14ac. A to C, Each section in the treemap represents a specific GO term, according to the hierarchical structures depicted, based on GO slim terms and their children (see “Materials and Methods”). D and E, The maps show the log2 fold changes (GSNO versus control) of H3K9/14ac for all genes (Voronoi cells) that display NO-responsive H3K9/14ac (at either time point). Red, Increased H3K9/14ac; blue, decreased H3K9/14ac; gray, no change. The term defense response is highlighted in E.
Table I. Selected defense genes displaying NO-mediated H3K9/14 hyperacetylation.
| GO Term | ATG Number | Gene Name | Description |
|---|---|---|---|
| Defense response (biological process) | AT2G38870 | PR-6 proteinase inhibitor family protein | |
| AT3G25510 | Putative TIR-NBS-LRR class disease resistance protein | ||
| AT4G08450 | TIR-NBS-LRR class disease resistance protein | ||
| AT5G11250 | TIR-NBS-LRR class disease resistance protein | ||
| AT5G15730 | Probable Leu-rich repeat receptor-like Ser/Thr protein kinase | ||
| AT4G29810 | MKK2 | MEK2 | |
| AT5G27520 | PNC2 | Peroxisomal adenine nucleotide carrier2 | |
| AT2G46240 | BAG6 | BCL2-associated athanogene6 | |
| AT3G28930 | AIG2 | avrRpt2-induced protein AIG2 | |
| AT5G42980 | TRX3 | Thioredoxin H3 | |
| AT4G23810 | WRKY53 | Putative WRKY transcription factor 53 | |
| AT5G52830 | WRKY27 | WRKY DNA-binding protein27 | |
| AT5G06950 | TGA2 | Transcription factor TGA2 | |
| AT5G06960 | TGA5 | OCS-element binding factor 5 |
Comparison of GSNO-Induced H3K9/14ac and Gene Expression Changes
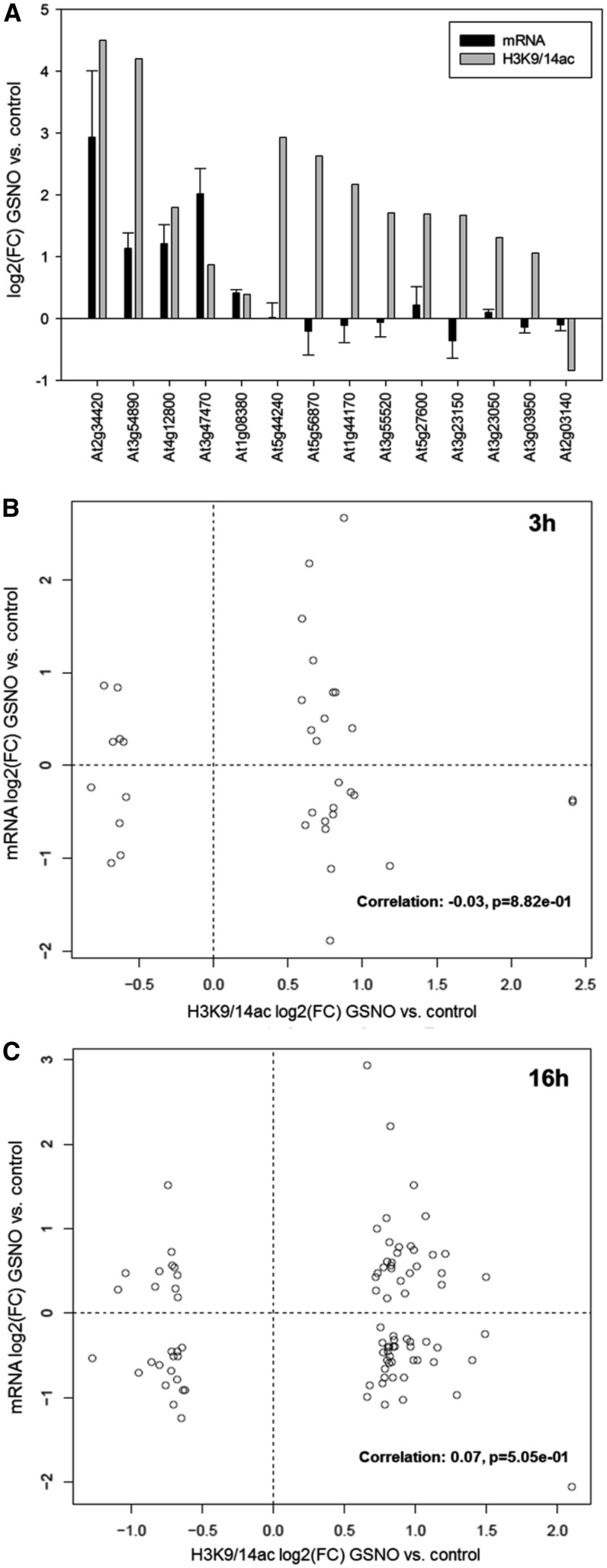
To analyze whether GSNO-induced H3K9/14ac changes also affect the expression of the target genes, the transcript levels of 14 selected genes were quantified by qPCR in GSNO-treated and control seedlings using the same plant material as for the ChIP-seq analysis. Five of these genes showed significantly enhanced H3K9/14ac and also were transcriptionally up-regulated, whereas the mRNA levels of nine genes remained unchanged despite a strong change of histone acetylation (Fig. 6A). To obtain a more complete picture, we also compared our ChIP-seq data with a recently published study, which analyzed GSNO- and GSH-induced transcriptomic changes after 3 h by RNA sequencing (RNA-seq; Begara-Morales et al., 2014). Thirty-five genes showing significant H3K9/14ac changes after 3 h of GSNO treatment also displayed significantly altered transcript levels (Supplemental File S3). Approximately 50% of these genes displayed enhanced H3K9/14ac and increased transcript levels or vice versa (Fig. 6B). In these cases, changes in histone acetylation might be the cause for transcriptional regulation. Since histone acetylation also can be a consequence of transcriptional regulation, acetylation changes might be delayed compared with transcript changes. Therefore, the correlation analysis also was performed for all H3K9/14ac sites regulated after 16 h of GSNO treatment. Again, roughly 50% of the overlapping genes showed the expected correlation between histone acetylation and transcript levels (Fig. 6C). For these genes, changes of H3K9/14ac are probably not the cause but rather the consequence of transcriptional regulation. However, the correlation analysis and our qPCR results clearly demonstrate that histone acetylation changes can be uncoupled from transcriptional regulation. In summary, H3K9/14ac changes mediated by GSNO do partially reflect GSNO-induced transcriptional responses. However, a large proportion seems to be independent of transcriptional activation or repression. Similar results were obtained for the comparison of the GSH data sets (Supplemental Fig. S20).
Figure 6.
Correlation analysis of GSNO-mediated changes of H3K9/14ac and gene expression. A, qPCR analysis of 14 genes displaying significant changes of H3K9/14ac after GSNO treatment in the ChIP-seq experiment. RNA was isolated from the same samples that were used for the ChIP-seq experiment (n = 2). mRNA levels are expressed as means ± se. Since only two replicates per data point were used, a significance test was omitted. B and C, Correlation analysis of genes displaying GSNO-mediated H3K9/14ac changes after 3 h (B) or 16 h (C) and GSNO-induced transcriptional changes (after 3 h), based on a recently published RNA-seq data set (Begara-Morales et al., 2014). FC, Fold change.
SA-Induced NO Production Inhibits HDAC Activity and Increases Histone Acetylation
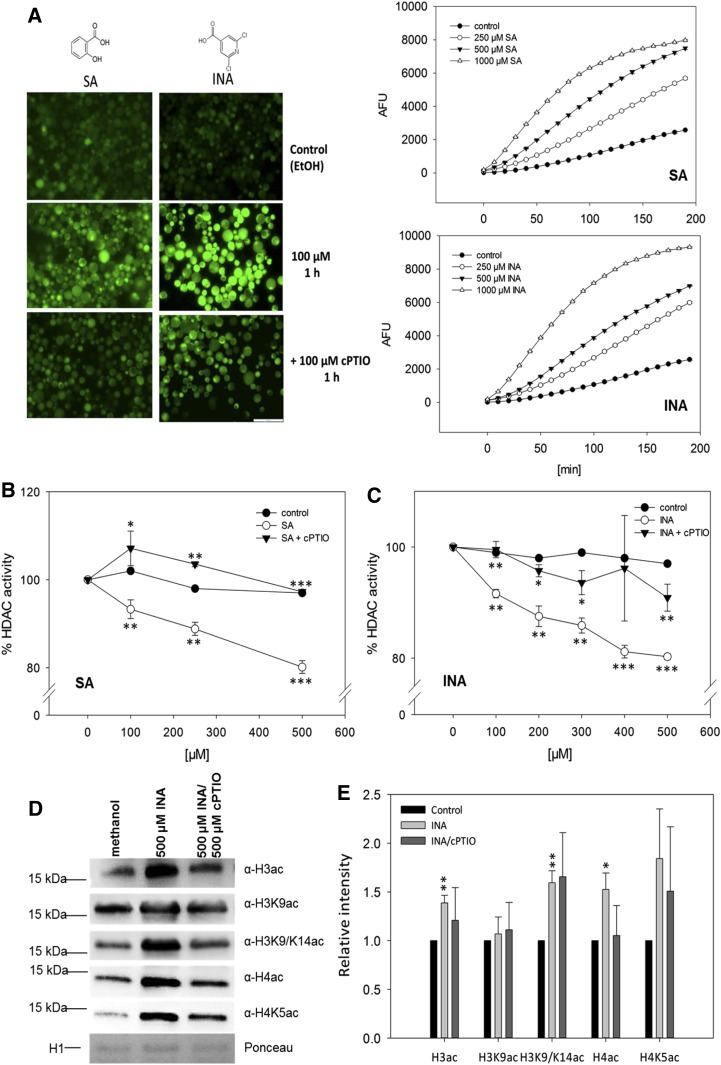
To analyze whether NO-mediated inhibition of HDACs might play a role in the defense response, the effect of SA on HDAC activity was examined in protoplasts. SA is the major plant defense hormone against biotrophic pathogens (Boatwright and Pajerowska-Mukhtar, 2013) but also fulfills important functions during the response to abiotic stresses (Miura and Tada, 2014). Moreover, SA was demonstrated to induce NO production in various model systems (Zottini et al., 2007; Hao et al., 2010; Sun et al., 2010; Gémes et al., 2011). To confirm that SA and its functional analog INA induce endogenous NO production, protoplasts were loaded with 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA), which is a membrane-permeable NO-sensitive fluorescent dye accumulating inside cells. One hour after stimulation with 100 µm SA or INA, DAF-FM fluorescence was analyzed by microscopy. A strong increase in fluorescence indicated the induction of endogenous NO production (Fig. 7A, left). Protoplasts pretreated with cPTIO displayed reduced fluorescence (Fig. 7A, left), demonstrating the specificity of the NO measurement. Moreover, time-course measurements of DAF-FM fluorescence revealed that the amplitude and starting point of the NO burst correlated with the concentration of SA and INA (Fig. 7A, right).
Figure 7.
Effects of SA/INA-induced NO production on HDAC activity and histone acetylation. A, NO production after SA and INA treatments. Fluorescence images show DAF-FM DA-loaded protoplasts after control (ethanol) and SA/INA ± cPTIO treatments. The experiment was repeated twice with similar results. EtOH, Ethanol. Bar = 100 µm. Graphs at right show the quantification of DAF-FM fluorescence in protoplasts stimulated with different concentrations of INA or SA. Fluorescence was detected on a fluorescence microplate reader. Values are means ± se (hidden by symbols) of 12 technical replicates. The experiment was repeated twice with similar results. B and C, NO-dependent inhibition of HDAC activity in protoplasts after SA and INA treatments. Protoplasts were incubated with INA (B) or SA (C) in the presence or absence of 100 µm cPTIO. Ethanol was used as a solvent control. Values are expressed as percentage of HDAC activity in untreated protoplasts. Values are means ± se of three independent protoplast preparations. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by Student’s t test. Asterisks below the white circles mark statistically significant inhibition in comparison with the control treatment, whereas asterisks in proximity to triangles indicate significant differences compared with SA or INA treatment. D, Histone acetylation in Arabidopsis seedlings after control (methanol), INA, or INA/cPTIO treatment. Histones were extracted from liquid-grown seedlings treated with 500 µm INA, 500 µm INA/500 µm cPTIO, or methanol (control) and probed against different histone acetylation marks by western blot. As a loading control, a fraction of the Ponceau-stained membrane is shown (H1). One representative experiment out of three is shown. E, Quantification of western-blot results. Signal intensities were measured using ImageJ software and normalized to the amount of loaded H1. Values are expressed as fold change over control treatment. Values shown are means ± se of three independent experiments. *, P < 0.05 and **, P < 0.01 by Student’s t test.
Next, HDAC activity was measured in protoplasts after stimulation with SA or INA. Both compounds significantly inhibited HDAC activity in a concentration-dependent manner (20% inhibition for 500 µm SA or INA), whereas ethanol (solvent control) and diluted HCl (with the same pH as SA solution; acidic control) had no effect on HDAC activity (Fig. 7, B and C; Supplemental Fig. S21A). Since SA is known to bind directly to some proteins, thereby modulating their activity, the effect of SA on HDAC activity in nuclear extracts was tested. However, no changes of HDAC activity could be detected, excluding a direct binding of SA to HDACs (Supplemental Fig. S21B). Remarkably, preincubation of the protoplasts with cPTIO before SA or INA treatment completely (SA) or largely (INA) prevented the inhibition of HDAC activity, demonstrating that the observed inhibition was due mainly to NO production (Fig. 7, B and C).
To analyze whether INA induces global changes of histone acetylation, Arabidopsis liquid-grown seedlings were treated with methanol (control), 500 µm INA, or 500 µm INA/500 µm cPTIO. After 16 h, histones were extracted and the abundance of several histone acetylation marks was analyzed by western blot. INA treatment significantly increased H3ac, H4ac, and H3K9/14ac, whereas H3K9ac remained unchanged. No significant changes of histone acetylation could be detected after INA/cPTIO treatment, indicating that INA-induced hyperacetylation was largely dependent on the production of NO (Fig. 7, D and E). Together, these results demonstrate that SA and INA affect histone acetylation via the induction of endogenous NO production, resulting in the inhibition of HDACs and subsequent histone hyperacetylation.
DISCUSSION
Histone acetylation is an important mechanism to control the chromatin structure and regulate transcription. Here, the impact of the plant signaling molecule NO on histone acetylation and specifically on H3K9/14ac was analyzed.
Redox Regulation of HDACs
Although HDACs are important transcriptional regulators, information on how this enzyme family is posttranslationally regulated in plants is sparse. Reports on maize (Zea mays) HDACs demonstrated that HD1 and HD2 are phosphoproteins. In the case of HD1, phosphorylation changed the substrate specificity of the enzyme whereas dephosphorylation of HD2 resulted in the inactivation of this protein (Brosch et al., 1992; Lusser et al., 1997; Kölle et al., 1999). Recently, it was shown that NtHD2a and NtHD2b, both belonging to the plant-specific HDAC family, are rapidly phosphorylated after cryptogein treatment of tobacco (Nicotiana tabacum) suspension cells, but the precise function of this modification during the hypersensitive response remained elusive (Bourque et al., 2011).
In vivo and in vitro HDAC activity assays revealed that the physiological NO donors GSNO and SNAP as well as the glutathionylating reagent GSSG reversibly inhibited total and nuclear HDAC activity (Fig. 2; Supplemental Fig. S7), strongly suggesting that certain plant HDACs might be targets for redox regulation, being particularly sensitive toward S-nitrosylation and, to a somewhat lesser extent, S-glutathionylation. Both S-nitrosylation and S-glutathionylation require a high nucleophilicity of the targeted Cys residue, but since the glutathione moiety is larger than the nitroso group, S-glutathionylation is sterically more demanding (Dalle-Donne et al., 2009). It is important to emphasize that both modifications can be triggered by GSNO, since SNOs are highly reactive toward glutathione, resulting in the formation of the thermodynamically more stable glutathionylated Cys, given that the steric constraints allow the attack by glutathione (Ji et al., 1999). Results from our study suggest that certain plant HDACs (or their regulators) contain nucleophilic and surface-localized Cys residues, which are important for the catalytic activity of these proteins and which are targets for redox regulation.
Several mammalian HDACs have been described to be regulated by S-nitrosylation. HDAC2 is nitrosylated at two Cys residues after stimulation of neurons with brain-derived neurotrophic factor, which leads to its dissociation from chromatin and subsequent chromatin remodeling at genes involved in neuronal development (Nott et al., 2008). HDAC6, which mainly deacetylates cytosolic α-tubulin, is S-nitrosylated by cytokine-induced NO production, resulting in the inhibition of this enzyme (Okuda et al., 2015). Finally, HDAC8 was demonstrated to be modified by NO in vitro (Feng et al., 2011). Interestingly, the catalytic activity of SIRTUIN1 is regulated by reversible S-glutathionylation, which guides vascular development in zebrafish (Bräutigam et al., 2013).
HDACs generally operate in large multiprotein complexes, which can affect their activity, subcellular localization, or the genes to which they are targeted (Sengupta and Seto, 2004). Interestingly, in a screen for potential S-nitrosylated nuclear proteins, the plant-specific HDtuins have been identified as putative targets (Chaki et al., 2015). HDtuins interact physically with RPD3-like HDACs (Luo et al., 2012). Hence, these data reinforce that NO directly targets HDAC complexes to inhibit their activity (Fig. 2). The enzymatic activity of HDAC complexes critically depends on the presence of non-HDAC subunits (You et al., 2001). At this stage, we do not know whether the observed inhibition of HDAC activity is mediated by direct S-nitrosylation of HDACs or indirectly by the modification of complex partners (or both), resulting in structural alterations of the complex or even the dissociation of regulatory subunits.
Genome-Wide Distribution of H3K9/14ac
The genome-wide distribution of H3K9/14ac was mapped by ChIP-seq (Fig. 3). Similar to H3K9ac and H3K27ac in Physcomitrella patens (Widiez et al., 2014), H3K9ac in rice (Oryza sativa; He et al., 2010), and H4K5ac in maize (Yang et al., 2016), H3K9/14ac was found predominantly within genes and was almost absent from intergenic and repetitive regions like centromeric and pericentromeric chromatin (Fig. 3A). Genome-wide mapping of H3K9ac and H3K14ac in mouse embryonic stem cells revealed that both marks co-occurred in many regulatory regions around 200 to 400 bp downstream of the TSS (Karmodiya et al., 2012). Similar observations were documented for H3K9ac and H3K27ac in P. patens (Widiez et al., 2014), for H3K36ac in Arabidopsis (Mahrez et al., 2016), and for H4K5ac in maize (Yang et al., 2016). Consistently, it was found in this study that H3K9/14ac is located primarily within 1 kb of the 5′ end of genes, showing a maximum in its distribution at 400 bp downstream of the TSS (Fig. 3B). The majority of H3K9/14ac peaks were located in exons followed by transcription termination sites and promoter regions (Fig. 3C). Similarly, in P. patens, most of the H3K9ac and H3K27ac sites were found to be within promoters, coding sequences, and introns (Widiez et al., 2014). In summary, the global H3K9/14ac distribution closely resembles those of other histone acetylation marks. Therefore, our data extend the concept that different histone acetylation marks display very similar genome-wide patterns.
H3K9/14ac Changes and Gene Expression
To analyze whether H3K9/14ac changes triggered by GSNO are associated with transcriptional regulation, the transcript levels of several selected genes displaying GSNO-regulated H3K9/14ac were quantified by qPCR. Thirty-six percent of these genes showed significantly enhanced H3K9/14ac and also were transcriptionally up-regulated, whereas the mRNA levels of 64% of the genes remained unchanged despite a strong change of histone acetylation (Fig. 6A). Moreover, our ChIP-seq data were compared with a recently published RNA-seq data set (Begara-Morales et al., 2014), which reported on the GSNO-induced transcriptional response (Fig. 6, B and C). Although it was found that several genes showed the expected interdependence of histone acetylation and transcription, we could not detect a clear correlation between both parameters, which supported our qPCR results. These data clearly show that GSNO can induce or repress H3K9/14ac independently of transcriptional changes, excluding the possibility that the histone acetylation differences we detected are solely the consequence of altered transcription. It was shown recently that histone hyperacetylation is not sufficient to trigger gene expression. Loss of HDA101 (a maize HDAC) resulted in increased histone acetylation at more than 2,500 genes, but less than 5% of these genes were transcriptionally up-regulated in the hda101 mutant (Yang et al., 2016). Another study analyzed changes in the histone modification pattern and transcriptional changes triggered by weak salt stress and could not detect a correlation between both parameters, reinforcing that quantitative alterations of histone modifications can be uncoupled from transcriptional activation/repression (Sani et al., 2013). The functional importance of histone modification changes that occur independently of transcriptional regulation remains to be established, but it was proposed that they might prime the chromatin for subsequent stress exposure and allow a faster/stronger transcription of the corresponding genes upon repeated challenge (Jaskiewicz et al., 2011; Sani et al., 2013). Interestingly, NO and the formation of SNOs have been implicated in the priming response of potato (Solanum tuberosum) against the necrotrophic pathogen Phytophtora infestans (Floryszak-Wieczorek et al., 2012). The authors found that different priming agents induced NO synthesis and hypothesized that NO could sensitize epigenetic changes (Floryszak-Wieczorek et al., 2012). Based on our data, it is tempting to speculate that these changes are mediated, at least partially, by NO-dependent inhibition of HDACs.
In general, a rather low overlap between our ChIP-seq study and the RNA-seq study was observed, which could largely be explained by substantial differences in the experimental setup and data analysis, since NO elicits quite diverse transcriptional responses, depending on the tissue, method of NO application, and developmental stage (Huang et al., 2002; Polverari et al., 2003; Palmieri et al., 2008). We used 7-d-old liquid-grown seedlings to profile H3K9/14ac changes (using 250 µm GSNO or GSH), whereas Begara-Morales et al. (2014) separately analyzed GSNO-induced transcript changes in the roots and leaves of 30-d-old plants grown in a hydroponic system (stimulated with 1 mm GSNO or GSH). Moreover, Begara-Morales et al. (2014) filtered GSNO-regulated transcripts by those that responded to GSH, which was not suitable for our data set since GSH induced NO production in the experimental setup used (Supplemental Figs. S1 and S4). Despite the rather low overlap at the single gene level, we found that GSNO-regulated genes belong to similar functional categories to those displaying GSNO-mediated H3K9/14ac changes. GSNO-regulated genes were involved mainly in the response to stress, metabolism, or transport and mostly showed binding, hydrolase, and transferase activity (Begara-Morales et al., 2014), thereby mirroring our ChIP-seq results (Fig. 5). Furthermore, several reports analyzing transcriptomic changes induced by NO also identified the defense response as one of the top-ranking categories to be influenced by NO (Huang et al., 2002; Polverari et al., 2003; Palmieri et al., 2008; Begara-Morales et al., 2014), again reflecting the results from this study (Table I; Supplemental Table S1).
Possible Role of NO-Mediated H3K9/14ac Changes during the Defense Response
Our ChIP-seq analysis revealed that NO affected histone acetylation of genes involved in several different physiological processes (Fig. 5). Many of the affected genes were assigned to cellular and primary metabolism, indicating that GSNO might influence the chromatin state and thereby the expression of metabolic enzymes. This is supported by other reports that demonstrated transcriptional alterations of metabolic enzymes in response to GSNO (Begara-Morales et al., 2014). Moreover, NO might affect the activity of a variety of enzymes involved in cellular metabolism by posttranslational mechanisms (Holtgrefe et al., 2008; Palmieri et al., 2010; van der Linde et al., 2011). Together, these data suggest that GSNO induces a metabolic shift in the cell, which is the result of transcriptional as well as posttranslational regulation of metabolic enzymes. We found that genes involved in the plant defense response are overrepresented among the genes displaying NO-regulated H3K9/14ac (Table I). Moreover, SA, which is the major plant defense hormone against biotrophic pathogens, inhibited HDAC activity in vivo and increased global H3 and H4 acetylation in a NO-dependent manner (Fig. 7). Interestingly, the effect of INA on global histone acetylation was weaker than for the GSNO treatment (Fig. 1), indicating that endogenously produced NO might selectively target a subset of NO-sensitive HDACs. Together, these data strongly suggest that NO-mediated histone acetylation changes might be important during the expression of plant immunity, thus extending the multiple roles of NO during plant-pathogen interactions (Delledonne et al., 1998; Durner et al., 1998; Feechan et al., 2005). We hypothesize that HDACs get inactivated by NO upon pathogen attack, leading to enhanced acetylation and a supportive chromatin state for the expression of defense genes. Transcription of these genes is then initiated by transcription factors, which are activated by additional defense signaling cascades. Interestingly, hyperacetylation of defense genes after INA treatment has been observed before. Choi et al. (2012) found that spraying with INA enhanced H3K9 and total H3 acetylation at the PR1, PR2, GDG1, and EDS5 genes, resulting in the NPR1-dependent expression of these genes. Furthermore, it was demonstrated that PR1, PR2, and GDG1 are direct target genes of HDA19, which represses their expression under nonchallenged conditions by deacetylation of the corresponding promoters. Therefore, the authors concluded that, in response to pathogen attack, either HDA19 has to be excluded or HATs have to be recruited to mediate the hyperacetylation and expression of these genes (Choi et al., 2012). Our data suggest that the HDA19 complex might be targeted directly by NO, resulting in the inhibition of its activity and subsequent hyperacetylation of HDA19 target genes. This hypothesis is supported by the observation that HDA19 underwent oxidative Cys modifications in response to salicylate treatment, as demonstrated in a proteomic approach aimed to identify early redox-regulated proteins in the defense response (Liu et al., 2015). Furthermore, approximately 12.5% of all NO-regulated H3K9/14ac sites discovered in our work are putative HDA19-binding sites (Zhou et al., 2013), reinforcing that HDA19 might be one of the HDAC isoforms targeted by NO.
CONCLUSION
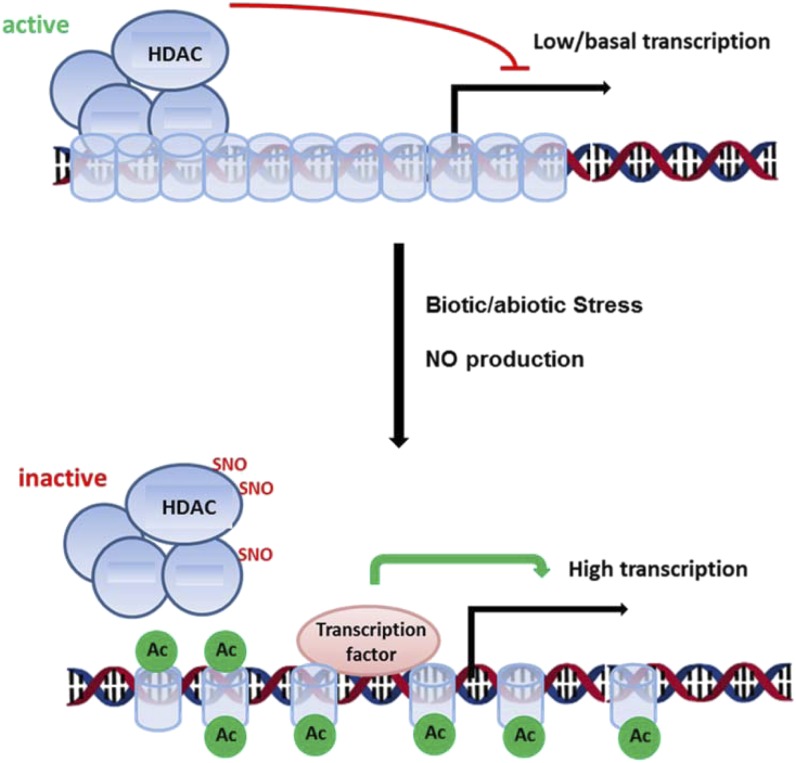
Global histone hyperacetylation has been observed after heat, salt, and cold stress in a variety of plant species (Sokol et al., 2007; Zhao et al., 2014; Wang et al., 2015), and these stresses are generally characterized by the extensive production of NO (Zhao et al., 2007, 2009; Bouchard and Yamasaki, 2008; Cantrel et al., 2011). In general, HDACs are transcriptional repressors of stress responses (Choi et al., 2012; Luo et al., 2012; Zheng et al., 2016). Therefore, proper stress gene induction requires the inactivation or exclusion of HDACs from the corresponding promoter regions (Choi et al., 2012). Our data suggest that the inactivation of HDACs upon stress perception might be mediated by NO. We propose the following model. In nonstressed conditions, HDAC complexes help to establish a repressive chromatin state by deacetylation of histones at stress genes. Stress perception initiates NO production, which targets and inhibits HDAC complexes by redox modifications, thereby enhancing histone acetylation and promoting a supportive chromatin state for the expression of stress genes (Fig. 8). This open-chromatin state might be preserved to allow a more efficient induction of stress genes upon subsequent stresses. Our study suggests a new link between NO signaling and chromatin remodeling and, in a broader perspective, indicates how abiotic and biotic stresses might trigger chromatin changes in plants.
Figure 8.
NO-mediated inhibition of HDACs might contribute to stress gene induction. In nonchallenged conditions, HDAC complexes repress the transcription of stress-responsive genes by deacetylation of the corresponding chromatin regions. Upon exposure to stress, NO is produced, resulting in the inhibition of HDAC complexes. This might activate transcription in tight interplay with transcription factors. Blue cylinders, Nucleosomes; green circles, acetyl groups (Ac); black arrow, TSS.
MATERIALS AND METHODS
Chemicals
All chemicals were purchased from Sigma unless stated otherwise.
Plant Material
Liquid-Grown Seedlings
Approximately 20 µL of surface-sterilized seeds (dry volume) from Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was incubated in 70 mL of Murashige and Skoog medium supplemented with 1% Suc in 250-mL flasks. Plants were grown for 7 d under continuous shaking (120 rpm) in short-day conditions (10 h of light/14 h of dark) at a light intensity of 130 µmol s−1 m−2.
Cell Cultures
Suspension cells were generated from Arabidopsis Columbia-0 wild-type seedling roots, which were cut into small pieces and transferred on CIM medium. After several days, calli were transferred to liquid CIM medium and subcultured every 7 d until homogenous cell suspension cultures were obtained. Cell cultures were grown in the dark at 23°C. In all experiments, 7-d-old cells were used.
Histone Extraction
Histones were extracted using the Histone Purification Kit (Active Motif; catalog no. 40025) according to the manufacturer’s instructions with some modifications. Amounts of 0.5 to 0.6 g of frozen seedlings were ground to a fine powder in liquid nitrogen and incubated for 2 h in 2.5 mL of extraction buffer at 4°C with gentle agitation. After centrifugation at 4°C for 10 min at maximal RCF, the supernatants were transferred to PD10 columns (GE Healthcare; catalog no. 17085101) that had been equilibrated with extraction buffer. The eluates were neutralized with one-quarter volumes of 5× neutralization buffer (0.875 mL) to reach a pH of 8 and loaded onto anion-exchange columns sent with the kit. Eluates were desalted using Zeba-Spin columns (Thermo Fisher; catalog no. 89882).
Western Blot
Western-blot analysis was carried using 5 µg of purified histones following standard protocols for western blotting. The antibodies and dilutions used were as follows: anti-acetylated H3 (1:20,000; Millipore; catalog no. 06599), anti-acetylated H3K9 (1:5,000; Abcam; catalog no. 10812), anti-acetylated H3K9K14 (1:2,000; Diagenode), anti-acetylated H4 (1:20,000; Millipore; catalog no. 06866), and anti-acetylated H4K5 (1:10,000; Abcam). Secondary antibody was anti-rabbit IgG (1:2,500; catalog no. W54011) coupled to horseradish peroxidase, and the signal was developed using Western Lightning Plus-ECL chemiluminescent substrate (Perkin-Elmer; catalog no. NEL103001EA).
Protoplast Isolation
Protoplasts were generated from Arabidopsis cell suspension cultures as described (Chaki et al., 2015). Approximately 5 g of cells was resuspended in 20 mL of enzyme solution (0.4 m mannitol, 20 mm KCl, 20 mm MES, pH 5.7, 10 mm MgCl2, 0.1% BSA, 2% cellulase, and 1% macerozyme) and incubated for 3 to 4 h at room temperature with gentle agitation (60 rpm). The reaction was stopped with 20 mL of W5 solution (2 mm MES, pH 5.7, 154 mm NaCl, 125 mm CaCl2, and 5 mm KCl), and the suspension was filtered through two layers of Miracloth. Protoplasts were washed two times with 20 mL of cold W5 (100g, 3 min, 4°C) and resuspended in cold W5. The concentration of the protoplast suspension was adjusted to 106 cells mL−1.
Preparation of Nuclear Extracts
Liquid-Grown Seedlings
One gram of seedlings was ground to a fine powder in liquid nitrogen. The powder was homogenized in 2 mL of LB buffer (20 mm Tris-HCl, pH 7.4, 25% glycerol, 20 mm KCl, 2 mm EDTA, 2.5 mm MgCl2, 250 mm Suc, 1 mm DTT, and cOmplete Protease Inhibitor EDTA-free Cocktail Tablets [Roche]) and filtered first through Miracloth (Millipore) and then through 30-µm nylon mesh (Millipore). After centrifugation (1,500g, 4°C, 10 min), the pellet was washed three times with 3 mL of NRBT buffer (20 mm Tris-HCl, pH 7.4, 25% glycerol, 2.5 mm MgCl2, 0.2% Triton X-100, and cOmplete Protease Inhibitor EDTA-free Cocktail Tablets) and once with 3 mL of NRB buffer (20 mm Tris-HCl, pH 7.4, 25% glycerol, 2.5 mm MgCl2, and cOmplete Protease Inhibitor EDTA-free Cocktail Tablets). To extract nuclear proteins, the nuclear pellet was resuspendend in 340 µL of NLB buffer (10 mm Tris, pH 7.5, 500 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm Na4P2O7, and protease inhibitor cocktail) and subjected to sonification on a Bandelin Sonopuls HD2070 (25 s, 50% duty cycle, 10% power output; repeat seven times with 1 min of rest on ice between each cycle). After centrifugation (12,000g, 4°C, 15 min), the supernatant was recovered and protein concentration was determined by the Bradford assay. Nuclear extracts were stored at –80°C.
Suspension Cells
First, protoplasts were prepared from the suspension cells; then, nuclei were isolated from protoplasts as described (Chaki et al., 2015).
Treatment of Nuclear Extracts
Ten micrograms of nuclear extract was incubated with various concentrations of NEM, GSNO, SNAP, GSSG, and H2O2 for 20 min at room temperature in the dark. In the case of DTT treatment, DTT was added after this incubation. Samples were then desalted using Zeba-Spin (Thermo Fisher) columns taking care to avoid exposure to light. Five micrograms of desalted nuclear extracts was subjected directly to HDAC activity measurement.
HDAC Activity Measurement in Nuclear Extracts
HDAC activity in nuclear extracts was measured using the Epigenase HDAC Activity/Inhibition Direct Assay Kit (Fluorimetric, Epigentek; catalog no. P-4035) according to the manufacturer’s instructions. Briefly, 5 µg of nuclear extract per well was incubated with 50 ng of substrate for 90 min at room temperature. Deacetylated product was immunodetected, and fluorescence at excitation/emission = 530/590 nm was measured on a fluorescence microplate reader (Tecan Infinite 1000).
In Situ HDAC Activity Measurement in Protoplasts
HDAC activity in protoplasts was measured using the InSitu HDAC Activity Fluorometric Assay Kit from BioVision (catalog no. K339-100) with some modifications. A total of 105 protoplasts per well were incubated with 1 µL of substrate at room temperature in the presence or absence of effector chemicals (cPTIO was added 10 min before addition of the substrate). The reaction was stopped by adding fluorescence developer and mixing by pipetting up and down to ensure proper lysis and homogenization of the cells. For each treatment, one well was stopped directly after addition of the substrate (t = 0) and a second well was stopped after 30 min (SA/INA) or 60 min (GSNO, SNAP, GSSG, or H2O2). After lysis, the plate was incubated for 30 min at 37°C to develop the signal. Fluorescence was measured at excitation/emission = 368/442 nm in a fluorescence microplate reader (Tecan Infinite 1000). The difference in fluorescence between t = 0 and 30 or 60 min was used to quantify HDAC activity. All chemicals used (GSNO, SNAP, GSSG, SA, INA, and cPTIO) were tested for interference with the assay by performing standard curves and by monitoring kinetics of the fluorescence development in the presence of this chemical.
Measurement of NO Production in Protoplasts
A total of 2 × 105 protoplasts were incubated with 15 µm DAF-FM DA for 15 min at room temperature in the dark. After centrifugation (100g, 2 min, room temperature), protoplasts were resuspended in 200 µL of W5 buffer. Then, 100 µm cPTIO or water (control) was added. After 10 min at room temperature, cells were centrifuged, resuspended in W5, and subsequently stimulated with 100 µm SA or INA. After 1 h, protoplasts were visualized using an Olympus BX700 microscope with a GFP filter set.
For quantitative measurement of 4-amino-5-methylamino-2′,7′-difluorofluorescein fluorescence, DAF-FM DA-loaded protoplasts were stimulated with different concentrations of SA and INA and split into wells of a black 96-well plate (Greiner; eight technical replicates per treatment, 2 × 105 protoplasts per well). Fluorescence was measured on a Tecan Infinite 1000 microplate reader every 10 min for a total of 20 cycles at excitation/emission = 485/535 nm.
Measurement of SNO Content
SNO content was determined as described (Kuruthukulangarakoola et al., 2017). Briefly, SNOs were reduced with triiodide solution and photochemical detection of the emitted NO after its reaction with ozone. Nitrite was scavenged by sulfanilamide.
RNA Extraction, cDNA Synthesis, and qPCR
A total of 100 mg of 7-d-old liquid-grown Arabidopsis seedlings was ground to a fine powder, followed by RNA extraction using the RNeasy Plant Mini Kit (Qiagen; catalog no. 74904) according to the manufacturer’s instructions. RNA concentration and quality were determined spectrophotometrically (NanoDrop 1000). One microgram of total RNA was used for cDNA synthesis with the QuantiTect Reverse Transcription Kit (Qiagen; catalog no. 205311). Quantitative reverse transcription-PCR consisted of 10 µL of Sybr Green (Bioline; catalog no. QT625-05), 5 µL of deionized water, 0.5 µL of 10 µm specific primers, and 4 µL of 1:20 diluted cDNA template. Cycling conditions were 95°C for 10 min followed by 45 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 45 s. Each sample was run in triplicate and normalized to the mRNA level of the S16 housekeeping gene. Primers are listed in Supplemental Table S2.
ChIP-Seq
ChIP was performed on 7-d-old liquid-grown Arabidopsis seedlings using the Plant ChIP-seq kit from Diagenode (C01010150). Briefly, tissue was cross-linked with 1% methanol-free formaldehyde, chromatin was isolated and sheared to 200-bp fragments using the Bioruptor Pico (Diagenode), and immunoprecipitation was performed with 1 µg of an antibody directed against H3K9/14ac (Diagenode; C15410200). An aliquot of the sheared chromatin was kept as an input control. After reversal of the cross-links, DNA was purified using the IPure Kit from Diagenode (C03010011). For each input and immunoprecipitation sample, an individual indexed library was prepared using the MicroPlex Library Preparation Kit version 2 (Diagenode; C05010014) using 1 ng of DNA and 11 amplification cycles. Ten microliters of each library was pooled, and the combined library was sequenced on an Illumina HiSeq2500. After demultiplexing, reads were filtered (only uniquely mapped reads were kept) and mapped to the Arabidopsis TAIR10 reference genome, and peaks were called using CLC genomics software version 8.5.1 (Qiagen) using a P value cutoff of 0.05.
After removing low-quality samples (rep2_control_16h and rep2_TSA_16h), differential binding sites were computed using the R package DiffBind (Ross-Innes et al., 2012): chromosomic peaks outside the centromeric region that occurred in at least six out of the 18 samples were used in the differential analysis with the edgeR method, and log2 fold changes as well as raw and false discovery rate-adjusted P values were recorded for each treatment-versus-control comparison. Peaks were annotated by genomic region and nearest gene using the ChIPseek tool (Chen et al., 2014). Heat maps were plotted with the gplots R package (Warnes et al., 2013), and other plots and data selections were created with basic R functions (R Core Team, 2014). To compare the number of up- and down-regulated peaks and the log fold changes between the up-regulated and down-regulated sets, the sign test and the Wilcoxon rank-sum test were performed using the R functions binom.test and wilcox.test, respectively (R Core Team, 2014). Enrichment analyses of peak-derived gene lists against the GO, KEGG, and AraCyc databases were done using the package org.At.tair.db (Carlson, 2014) and the R functions fisher.test and p.adjust with the false discovery rate method for multiple testing correction (R Core Team, 2014). Correlations were computed with cor.test (R Core Team, 2014). For the treemaps, GO slim terms below the basic terms molecular function, biological process, and cellular component were taken from TAIR (https://www.arabidopsis.org/download_files/GO_and_PO_Annotations/Gene_Ontology_Annotations/TAIR_GO_slim_categories.txt) on April 26, 2016 (Berardini et al., 2004), and all child terms regarding is_a relationships were determined from the go-basic.obo file (release 2016-04-27) downloaded from http://geneontology.org/page/download-ontology (Gene Ontology Consortium, 2015). Gene annotations were obtained using org.At.tair.db (Carlson, 2014).
ChIP-qPCR
Chromatin immunoprecipitated and input DNA was diluted 1:10 in sterile water, and 4 µL was used for qPCR as described above. Values are expressed as percentage of the input. Primers used are listed in Supplemental Table S3.
Accession Numbers
ChIP-seq raw data as well as peak files and DiffBind output files were deposited in the Gene Expression Omnibus database under accession number GSE82075.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SNO levels in liquid-grown Arabidopsis seedlings after GSNO, GSH, GSNO/cPTIO, TSA, and water (control) treatments.
Supplemental Figure S2. GSNO increases H3ac in Arabidopsis suspension cells.
Supplemental Figure S3. mRNA levels of HDACs after GSNO and GSH treatments.
Supplemental Figure S4. Inhibition of HDAC activity by GSH.
Supplemental Figure S5. Viability of protoplasts after SA, INA, GSNO, and SNAP treatments.
Supplemental Figure S6. Inhibition of nuclear HDAC activity by NEM and TSA.
Supplemental Figure S7. Inhibition of HDAC activity by GSSG and H2O2.
Supplemental Figure S8. Antibody quality control and titration.
Supplemental Figure S9. Work flow to quantitatively determine differences in the H3K9/14ac pattern after GSNO, GSH, GSNO/cPTIO, and TSA treatments.
Supplemental Figure S10. Summary of read mapping.
Supplemental Figure S11. Summary of peak calling parameters.
Supplemental Figure S12. Shearing and library preparation.
Supplemental Figure S13. Verification of peak calling.
Supplemental Figure S14. Motif analysis of H3K9/14ac peak regions.
Supplemental Figure S15. Verification of the quantitative analysis (DiffBind) by ChIP-qPCR.
Supplemental Figure S16. CLC genome browser snapshots of representative GSNO-regulated H3K9/14ac sites.
Supplemental Figure S17. cPTIO reduces the number of hyperacetylated H3K9/14ac sites after 3 h of GSNO treatment.
Supplemental Figure S18. Time-point comparison of GSNO-, GSH-, and GSNO/cPTIO-induced H3K9/14ac changes.
Supplemental Figure S19. Comparison of NO-induced H3K9/14ac changes with TSA treatment.
Supplemental Figure S20. Correlation analysis of GSH-mediated changes of H3K9/14ac and gene expression.
Supplemental Figure S21. HDAC assay control experiments.
Supplemental Table S1. GO enrichment analysis for genes displaying NO-regulated H3K9/14ac.
Supplemental Table S2. Primers used for qPCR.
Supplemental Table S3. Primers used for ChIP-qPCR.
Supplemental File S1. NO-regulated peaks.
Supplemental File S2. GO enrichment analysis.
Supplemental File S3. Correlation analysis (ChIP-seq and RNA-seq).
Supplementary Material
Acknowledgments
We thank Elke Mattes and Lucia Gößl for excellent technical assistance.
Glossary
- NO
nitric oxide
- SNO
S-nitrosothiol
- cPTIO
2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- GSNO
S-nitrosoglutathione
- GSH
reduced glutathione
- SNAP
S-nitroso-N-acetyl-dl-penicillamine
- ChIP-seq
chromatin immunoprecipitation sequencing
- SA
salicylic acid
- INA
2,6-dichloro-isonicotinic acid
- TSA
trichostatin A
- qPCR
quantitative PCR
- DTT
dithiothreitol
- NEM
N-ethylmaleimide
- GSSG
oxidized glutathione
- ChIP
chromatin immunoprecipitation
- TSS
transcriptional start site
- GO
Gene Ontology
- RNA-seq
RNA sequencing
- DAF-FM DA
4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate
Footnotes
This work was supported by the Bundesministerium für Bildung und Forschung. E.G. was supported by the German Plant Phenotyping Network funded by the Bundesministerium für Bildung und Forschung (DPPN, no. 031A053C).
Articles can be viewed without a subscription.
References
- Akaike T, Yoshida M, Miyamoto Y, Sato K, Kohno M, Sasamoto K, Miyazaki K, Ueda S, Maeda H (1993) Antagonistic action of imidazolineoxyl N-oxides against endothelium-derived relaxing factor/.NO through a radical reaction. Biochemistry 32: 827–832 [DOI] [PubMed] [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Luque F, Leyva-Pérez MO, Leterrier M, Corpas FJ, Barroso JB (2014) Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol 55: 1080–1095 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatwright JL, Pajerowska-Mukhtar K (2013) Salicylic acid: an old hormone up to new tricks. Mol Plant Pathol 14: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard JN, Yamasaki H (2008) Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: a possible linkage between nitric oxide and the coral bleaching phenomenon. Plant Cell Physiol 49: 641–652 [DOI] [PubMed] [Google Scholar]
- Bourque S, Dutartre A, Hammoudi V, Blanc S, Dahan J, Jeandroz S, Pichereaux C, Rossignol M, Wendehenne D (2011) Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol 192: 127–139 [DOI] [PubMed] [Google Scholar]
- Bräutigam L, Jensen LD, Poschmann G, Nyström S, Bannenberg S, Dreij K, Lepka K, Prozorovski T, Montano SJ, Aktas O, et al. (2013) Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1. Proc Natl Acad Sci USA 110: 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch G, Georgieva EI, López-Rodas G, Lindner H, Loidl P (1992) Specificity of Zea mays histone deacetylase is regulated by phosphorylation. J Biol Chem 267: 20561–20564 [PubMed] [Google Scholar]
- Cantrel C, Vazquez T, Puyaubert J, Rezé N, Lesch M, Kaiser WM, Dutilleul C, Guillas I, Zachowski A, Baudouin E (2011) Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol 189: 415–427 [DOI] [PubMed] [Google Scholar]
- Carlson M. (2014) org.At.tair.db: Genome wide annotation for Arabidopsis. R package version 2.10.1, http://bioconductor.org/packages/release/data/annotation/html/org.At.tair.db.html
- Chaki M, Shekariesfahlan A, Ageeva A, Mengel A, von Toerne C, Durner J, Lindermayr C (2015) Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci 238: 115–126 [DOI] [PubMed] [Google Scholar]
- Chen TW, Li HP, Lee CC, Gan RC, Huang PJ, Wu TH, Lee CY, Chang YF, Tang P (2014) ChIPseek, a web-based analysis tool for ChIP data. BMC Genomics 15: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SM, Song HR, Han SK, Han M, Kim CY, Park J, Lee YH, Jeon JS, Noh YS, Noh B (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J 71: 135–146 [DOI] [PubMed] [Google Scholar]
- Colussi C, Mozzetta C, Gurtner A, Illi B, Rosati J, Straino S, Ragone G, Pescatori M, Zaccagnini G, Antonini A, et al. (2008) HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc Natl Acad Sci USA 105: 19183–19187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A (2009) Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci 34: 85–96 [DOI] [PubMed] [Google Scholar]
- Dangl M, Brosch G, Haas H, Loidl P, Lusser A (2001) Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213: 280–285 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, Careri M, Zottini M, Sanità di Toppi L, Lo Schiavo F (2009) Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol 150: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102: 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JH, Jing FB, Fang H, Gu LC, Xu WF (2011) Expression, purification, and S-nitrosylation of recombinant histone deacetylase 8 in Escherichia coli. Biosci Trends 5: 17–22 [DOI] [PubMed] [Google Scholar]
- Floryszak-Wieczorek J, Arasimowicz-Jelonek M, Milczarek G, Janus L, Pawlak-Sprada S, Abramowski D, Deckert J, Billert H (2012) Nitric oxide-mediated stress imprint in potato as an effect of exposure to a priming agent. Mol Plant Microbe Interact 25: 1469–1477 [DOI] [PubMed] [Google Scholar]
- Folkes LK, Wardman P (2004) Kinetics of the reaction between nitric oxide and glutathione: implications for thiol depletion in cells. Free Radic Biol Med 37: 549–556 [DOI] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2002) Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol 128: 790–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gémes K, Poór P, Horváth E, Kolbert Z, Szopkó D, Szepesi A, Tari I (2011) Cross-talk between salicylic acid and NaCl-generated reactive oxygen species and nitric oxide in tomato during acclimation to high salinity. Physiol Plant 142: 179–192 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2015) Going forward. Nucl Acids Res 43: D1049–D1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao F, Zhao S, Dong H, Zhang H, Sun L, Miao C (2010) Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J Integr Plant Biol 52: 298–307 [DOI] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Hogg N. (2002) The biochemistry and physiology of S-nitrosothiols. Annu Rev Pharmacol Toxicol 42: 585–600 [DOI] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant 133: 211–228 [DOI] [PubMed] [Google Scholar]
- Hu X, Neill SJ, Tang Z, Cai W (2005) Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol 137: 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, von Rad U, Durner J (2002) Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Im JH, Yoo SD (2014) Transient expression in Arabidopsis leaf mesophyll protoplast system for cell-based functional analysis of MAPK cascades signaling. Methods Mol Biol 1171: 3–12 [DOI] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhänsel C (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Ji Y, Akerboom TP, Sies H, Thomas JA (1999) S-Nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch Biochem Biophys 362: 67–78 [DOI] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L (2012) H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölle D, Brosch G, Lechner T, Pipal A, Helliger W, Taplick J, Loidl P (1999) Different types of maize histone deacetylases are distinguished by a highly complex substrate and site specificity. Biochemistry 38: 6769–6773 [DOI] [PubMed] [Google Scholar]
- Kovacs I, Durner J, Lindermayr C (2015) Crosstalk between nitric oxide and glutathione is required for NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1)-dependent defense signaling in Arabidopsis thaliana. New Phytol 208: 860–872 [DOI] [PubMed] [Google Scholar]
- Kovacs I, Lindermayr C (2013) Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Front Plant Sci 4: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M (2003) Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 4: 276–284 [DOI] [PubMed] [Google Scholar]
- Kuruthukulangarakoola GT, Zhang J, Albert A, Winkler B, Lang H, Buegger F, Gaupels F, Heller W, Michalke B, Sarioglu H, et al. (2017) Nitric oxide-fixation by non-symbiotic haemoglobin proteins in Arabidopsis thaliana under N-limited conditions. Plant Cell Environ 40: 36–50 [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012) ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 281: 4285–4291 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zhang H, Yu B, Xiong L, Xia Y (2015) Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci Rep 5: 8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63: 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A, Brosch G, Loidl A, Haas H, Loidl P (1997) Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277: 88–91 [DOI] [PubMed] [Google Scholar]
- Maeda H, Akaike T, Yoshida M, Sato K, Noguchi Y (1995) A new nitric oxide scavenger, imidazolineoxyl N-oxide derivative, and its effects in pathophysiology and microbiology. Curr Top Microbiol Immunol 196: 37–50 [DOI] [PubMed] [Google Scholar]
- Mahrez W, Arellano MS, Moreno-Romero J, Nakamura M, Shu H, Nanni P, Köhler C, Gruissem W, Hennig L (2016) H3K36ac is an evolutionary conserved plant histone modification that marks active genes. Plant Physiol 170: 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel A, Chaki M, Shekariesfahlan A, Lindermayr C (2013) Effect of nitric oxide on gene transcription: S-nitrosylation of nuclear proteins. Front Plant Sci 4: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Tada Y (2014) Regulation of water, salinity, and cold stress responses by salicylic acid. Front Plant Sci 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, Hall MA, Harren FJ, Hebelstrup KH, Gupta KJ (2013) Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants 5: pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A (2008) S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature 455: 411–415 [DOI] [PubMed] [Google Scholar]
- Okuda K, Ito A, Uehara T (2015) Regulation of histone deacetylase 6 activity via S-nitrosylation. Biol Pharm Bull 38: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L (2002) Nitric oxide is required for root organogenesis. Plant Physiol 129: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri MC, Lindermayr C, Bauwe H, Steinhauser C, Durner J (2010) Regulation of plant glycine decarboxylase by s-nitrosylation and glutathionylation. Plant Physiol 152: 1514–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri MC, Sell S, Huang X, Scherf M, Werner T, Durner J, Lindermayr C (2008) Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J Exp Bot 59: 177–186 [DOI] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Mittelsten Scheid O (2014) Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 6: a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverari A, Molesini B, Pezzotti M, Buonaurio R, Marte M, Delledonne M (2003) Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol Plant Microbe Interact 16: 1094–1105 [DOI] [PubMed] [Google Scholar]
- R Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Ridnour LA, Thomas DD, Mancardi D, Espey MG, Miranda KM, Paolocci N, Feelisch M, Fukuto J, Wink DA (2004) The chemistry of nitrosative stress induced by nitric oxide and reactive nitrogen oxide species: putting perspective on stressful biological situations. Biol Chem 385: 1–10 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Ueda M, Morikawa H (2002) Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett 515: 20–24 [DOI] [PubMed] [Google Scholar]
- Sani E, Herzyk P, Perrella G, Colot V, Amtmann A (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E (2004) Regulation of histone deacetylase activities. J Cell Biochem 93: 57–67 [DOI] [PubMed] [Google Scholar]
- Serpa V, Vernal J, Lamattina L, Grotewold E, Cassia R, Terenzi H (2007) Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem Biophys Res Commun 361: 1048–1053 [DOI] [PubMed] [Google Scholar]
- Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR (1996) The chemistry of the S-nitrosoglutathione/glutathione system. Proc Natl Acad Sci USA 93: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šírová J, Sedlářová M, Piterková J, Luhová L, Petřivalský M (2011) The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci 181: 560–572 [DOI] [PubMed] [Google Scholar]
- Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M (2007) Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227: 245–254 [DOI] [PubMed] [Google Scholar]
- Spencer NY, Zeng H, Patel RP, Hogg N (2000) Reaction of S-nitrosoglutathione with the heme group of deoxyhemoglobin. J Biol Chem 275: 36562–36567 [DOI] [PubMed] [Google Scholar]
- Stamler JS. (1994) Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell 78: 931–936 [DOI] [PubMed] [Google Scholar]
- Sun LR, Hao FS, Lu BS, Ma LY (2010) AtNOA1 modulates nitric oxide accumulation and stomatal closure induced by salicylic acid in Arabidopsis. Plant Signal Behav 5: 1022–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K, Gutsche N, Leffers HM, Lindermayr C, Muller B, Holtgrefe S, Scheibe R (2011) Regulation of plant cytosolic aldolase functions by redox-modifications. Plant Physiol Biochem 49: 946–957 [DOI] [PubMed] [Google Scholar]
- Viola IL, Güttlein LN, Gonzalez DH (2013) Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol 162: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao L, Hou H, Zhang H, Huang Y, Wang Y, Li H, Gao F, Yan S, Li L (2015) Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol 56: 965–976 [DOI] [PubMed] [Google Scholar]
- Wang YQ, Feechan A, Yun BW, Shafiei R, Hofmann A, Taylor P, Xue P, Yang FQ, Xie ZS, Pallas JA, et al. (2009) S-Nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem 284: 2131–2137 [DOI] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al. (2013) gplots: Various R programming tools for plotting data. R package version 2.12.1, http://CRAN.R-project.org/package=gplots [Google Scholar]
- Widiez T, Symeonidi A, Luo C, Lam E, Lawton M, Rensing SA (2014) The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. Plant J 79: 67–81 [DOI] [PubMed] [Google Scholar]
- Yang H, Liu X, Xin M, Du J, Hu Z, Peng H, Rossi V, Sun Q, Ni Z, Yao Y (2016) Genome-wide mapping of targets of maize histone deacetylase HDA101 reveals its function and regulatory mechanism during seed development. Plant Cell 28: 629–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265: 17174–17179 [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL (2001) CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci USA 98: 1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, et al. (2011) S-Nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268 [DOI] [PubMed] [Google Scholar]
- Yun BW, Spoel SH, Loake GJ (2012) Synthesis of and signalling by small, redox active molecules in the plant immune response. Biochim Biophys Acta 1820: 770–776 [DOI] [PubMed] [Google Scholar]
- Zhao L, Wang P, Hou H, Zhang H, Wang Y, Yan S, Huang Y, Li H, Tan J, Hu A, et al. (2014) Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PLoS ONE 9: e106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Chen L, Zhang LL, Zhang WH (2009) Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol 151: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Tian QY, Zhang WH (2007) Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol 144: 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ding Y, Sun X, Xie S, Wang D, Liu X, Su L, Wei W, Pan L, Zhou DX (2016) Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J Exp Bot 67: 1703–1713 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu CW, Yang S, Dong S, Ruan J, et al. (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zottini M, Costa A, De Michele R, Ruzzene M, Carimi F, Lo Schiavo F (2007) Salicylic acid activates nitric oxide synthesis in Arabidopsis. J Exp Bot 58: 1397–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.