Abstract
Staphylococcus aureus fibronectin-binding proteins (FnBPs) play a critical role in S. aureus pathogenesis. FnBPs mediate adhesion to fibronectin and invasion of mammalian cells, including epithelial, endothelial, and fibroblastic cells, by fibronectin bridging to the host cell fibronectin receptor integrin (α5)β1. Strain Newman is a laboratory strain frequently used for genetic, functional, and in vivo studies. However, despite pronounced production of FnBPs, strain Newman is only weakly adherent to immobilized Fn and weakly invasive. We examined whether these effects are due to a structural difference of FnBPs. Here, we show that both fnbANewman and fnbBNewman contain a centrally located point mutation resulting in a stop codon. This leads to a truncation of both FnBPs at the end of the C domain at identical positions. Most likely, the stop codon occurred first in fnbBNewman and was subsequently transferred to fnbANewman by replacement of the entire region encompassing the C, D, and W domains with the respective sequence of fnbBNewman. Using heterologous expression in Staphylococcus carnosus, we found that truncated FnBPs were completely secreted into the culture medium and not anchored to the cell wall, since they lack the sortase motif (LPETG). Consequently, this led to a loss of FnBP-dependent functions, such as strong adhesion to immobilized fibronectin, binding of fibrinogen, and host cell invasion. This mutation may explain some of the earlier reported conflicting data with strain Newman. Thus, care should be taken when drawing negative conclusions about the role of FnBPs as a virulence factor in a given model.
Staphylococcus aureus fibronectin-binding proteins (FnBPs) appear to play a critical role in S. aureus pathogenesis (35), as has been shown, e.g., for infective endocarditis (29, 49) and osteomyelitis (22). However, so far, often conflicting results have been reported that argue for (3, 22, 29, 38, 49) or against (2, 5, 10, 33, 41) their role as virulence factors in a given model in vivo. In most situations, the adhesive and invasive functions of FnBPs are the likely functional link in pathogenicity. Adhesion to fibronectin and invasion of mammalian cells, including epithelial, endothelial, and fibroblastic cells, depend on fibronectin bridging between FnBPs and the host fibronectin receptor integrin (α5)β1 (54).
During the analysis of the mechanism for cellular invasion of S. aureus, we found that several reference strains are invasion deficient compared to clinical isolates (54). Strain Newman is a laboratory strain frequently used for genetic, functional, and in vivo studies. Despite strong production of FnBPs (57), strain Newman is only weakly adherent to immobilized Fn and weakly invasive (54). Another often-used reference strain, 8325-4, is also only weakly invasive (54); however, this appears to be due to regulatory defects resulting in low FnBP expression rather than structural FnBP modifications. Strain 8325-4 has a regulatory defect (rsbU) (28) and expresses FnBPs at a low level (60). However, upon heterologous expression, both FnBPs are functionally intact (49, 55). Both strains, 8325-4 and Newman, have been widely used for in vivo and in vitro studies. In addition, strain Newman strongly expresses Eap (extracellular adherence protein), which among other ligands binds fibronectin (20), thus rendering the interpretation of data derived from in vivo (4, 5, 8, 23-25, 39, 44, 50, 58, 59) and in vitro (6, 9, 12, 13, 16, 20, 32, 34, 42, 43, 48, 62, 66, 67, 69, 70) studies rather difficult.
Moreover, besides its ability to bind to fibronectin, FnBPA has been shown to bind to fibrinogen (64). This may complicate the interpretation of the resulting data, especially with regard to the roles of FnBPs in several infection models.
This study aimed to determine whether the weak invasiveness and adherence to immobilized fibronectin of strain Newman are due to structurally defective FnBPs rather than altered regulatory pathways. Here, we show that in strain Newman both fnbA and fnbB harbor a central stop codon, leading to FnBPs truncated in the C domain. As expected from the sequence data, using heterologous expression in Staphylococcus carnosus, we found that FnBPs were completely secreted and not anchored to the cell wall. This can be explained by the lack of the LPETG motif for sortase in the truncated protein. Consequently, this led to a loss of FnBP-dependent functions, such as adhesion to immobilized fibronectin, binding of fibrinogen, and host cell invasion. This mutation may explain some of the conflicting data obtained with strain Newman, and care should be taken when drawing negative conclusions about the role of FnBPs as a virulence factor in a given model.
MATERIALS AND METHODS
Reagents and enzymes.
Phosphate-buffered saline (PBS) with and without Ca2+ or Mg2+ was from Gibco-BRL (Karlsruhe, Germany). Recombinant lysostaphin (Ambicin; WAK Chemie/Applied Microbiology, Brooklyn, N.Y.), a murolytic enzyme that specifically degrades the staphylococcal cell wall by cleaving the pentaglycine bridge in peptidoglycan, and human serum albumin (HSA) were from Behring (Marburg, Germany), and the prestained protein ladder (∼10 to 180 kDa) was from MBI Fermentas (St. Leon-Rot, Germany). Lyophilized bacterial culture media were from Merck (Darmstadt, Germany) (M17 broth and brain heart infusion broth), Difco (Augsburg, Germany) (tryptic soy broth), and Mast (Reinfeld, Germany) (Müller-Hinton broth).
Bacterial strains, plasmid DNA, and chromosomal DNA isolation and transformation.
All bacterial strains used are listed in Table 1. S. aureus strain Cowan 1 was used as a reference isolate; strains Newman D2C (ATCC 25904) and 8325-4 were used as donors for cloning fnbA and fnbB. The FnBP-encoding plasmids pFNBA4 and pFNBB4 (15) were isolated from S. carnosus TM300 strains (55) by a modified alkaline lysis (including 10 μg of lysostaphin/ml in the resuspension buffer) and the QIAGEN method (miniprep columns). S. carnosus strain TM300 (52) was transformed by protoplast transformation (14). Briefly, bacteria were grown to an optical density at 578 nm (OD578) of ∼0.5 in B2 broth. After centrifugation, the pellet was resuspended in SMM (sucrose-MgCl2-maleic acid)-Pennassay broth (adjusted for S. aureus and S. carnosus, respectively) with lysostaphin (1.5 μg/ml). The cell suspension was incubated overnight at 30°C and washed thoroughly on ice. The protoplasts were analyzed by microscope and stored at −70°C. The protoplasts were mixed gently with plasmid DNA or ligation products and briefly incubated after addition of the fusogen (40% polyethylene glycol in SMM). After being washed with SMM-Pennassay broth, the protoplasts were plated onto DM-3 (5% agar, 1 m sodium succinate, 5% casein hydrolysate, 10% yeast extract, 50% glucose, 1 M NaCl2, and 5% bovine serum albumin) plates and incubated for 4.5 h. To select transformants, the DM-3 plates were overlayed by soft agar with chloramphenicol (final concentration, 10 μg/ml), as described earlier (14). After 2 days, colonies were picked and tested by restriction analysis and gel electrophoresis of isolated plasmids obtained by the QIAGEN modified alkaline lysis method (miniprep columns).
TABLE 1.
Bacterial strains used in this study
| Straina | Relevant genotype or plasmid | Properties | Reference or source |
|---|---|---|---|
| S. aureus | |||
| Cowan 1 | Reference isolate | Isolated from septic arthritis | ATCC 12598; NCTC 8530 |
| 8325-4 | Reference isolate; rsbU | NCTC 8325 cured of prophages and plasmids | 28, 40 |
| DU5883 | 8325-4 fnbA::TcrfnbB::Emr | FnBPA− FnBPB− | 15 |
| DU5883 (pFNBA4) | DU5883(pFNBA4) Cmr | FnBPA8325-4+++ | 15 |
| Newman D2C | Reference isolate | ClfA positive (high level); nonhemolytic; coagulase negative | ATCC 25904; NCTC 10833 |
| Newman | Reference isolate | ClfA positive (high level); strongly β-hemolytic (variable colonies); coagulase positive | NCTC 8178b |
| S. carnosus | |||
| TM300 | Reference isolate; WT | No expression of adhesins | 52 |
| BS100 | TM300(pFNBA4) | FnBPA8325-4+++ | 55 |
| BS101 | TM300(pFNBB4) | FnBPB8325-4+++ | 55 |
| MG8Vec | TM300(pMG8Vec) Cmr | pFNBPA4 without fnbA | This study |
| MG4ExpA | TM300(pMG4ExpA) Cmr | pMG8Vec with fnbANewman | This study |
| MG4ExpB | TM300(pMG4ExpB) Cmr | pMG8Vec with fnbBNewman | This study |
| MG5ExpA | TM300(pMG5ExpA) Cmr | pMG8Vec with fnbA8325-4 | This study |
The fnbA/fnbB isogenic mutant of strain 8325-4 (DU5883) and the expression vectors pFNBPA4 and pFNBPB4 were kindly provided by T. Foster.
The NCTC database refers to strain ATCC 13420, which does not (or no longer) exist. Inversely, the ATCC database refers to strain NCTC 8178 from another isolate, I.J.7 (ATCC 31153), which is not Newman but a variant of Newman D2C. Both entries were verified again, as of 15 July 2004.
Bacterial cultures.
For the invasion assay, staphylococci were cultured in Müller-Hinton broth at 37°C. All transformants were grown in the presence of 10 μg of chloramphenicol/ml. Wild-type (WT) strains were kept on sheep blood agar plates, and transformants were kept on tryptic soy agar plates supplemented with 10 μg of chloramphenicol/ml.
For ligand overlay blotting, the S. aureus and S. carnosus strains were cultured in brain heart infusion broth to an OD578 of ∼1.0.
Construction of plasmids encoding FnBPA and FnBPB of S. aureus strain Newman.
In order to create an expression vector for fnbA or fnbB, the vector part of pFNBA4 (15), including the signal peptide sequence of fnbA8325-4 (Fig. 1), was amplified by PCR. The oligonucleotide primers used were 5′-GAA GAT CTT ATG TCT GAT GAT TGA TAA CGA ACT-3′ (the BglII recognition site is underlined) and 5′-ATA TCT GCA GCT TCT TTG TCT TGT C −3′ (the PstI recognition site is underlined). Deep Vent DNA polymerase (New England Biolabs) was used according to the manufacturer's instructions. The blunt-ended PCR product was purified using the QIAquick PCR purification kit (QIAGEN) and ligated with T4 DNA ligase, resulting in the vector pMG8Vec (Fig. 1). pMG8Vec contains the promoter region as well as the 5′ part of the signal peptide sequence of fnbA8325-4. The chromosomal DNA from S. aureus was isolated with the QIAamp DNA Blood Mini kit (QIAGEN) according to the manufacturer's instructions. The fnbA and fnbB genes from S. aureus strains Newman D2C (ATCC 25904) and 8325-4 were amplified by PCR using Deep Vent DNA polymerase. The oligonucleotide primers used for the amplification of fnbA were 5′-ATA TCT GCA GCA TCA GAA CAA AAG AC-3′ and 5′-GAA GAT CTA ACC AAT GAA GCA ATC AGA A-3′ (the BglII and PstI recognition sites are underlined). The oligonucleotide primers used for the amplification of fnbB were 5′-ACT TTT TAT TAA CTC GCT TTT TTT C-3′ and 5′-GAA GAT CTA CGC CTT CAT AGT GTC ATT GAG T-3′. After purification of the PCR product with the QIAquick PCR purification kit, the vector and the fnb products were digested by BglII and PstI and ligated by T4 DNA ligase.
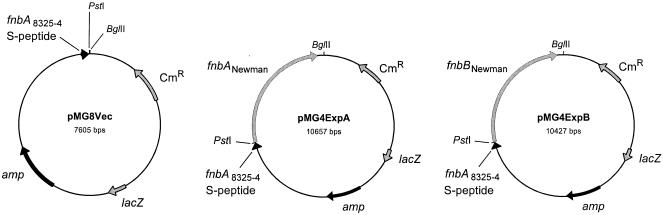
FIG. 1.
Schematic representation of vector construction. pMG8Vec is a modification of pFNBA4 and does not contain most of the fnbA gene but has retained the complete sequence for the signal peptide. pMG8Vec was used as a vector part for the expression of fnbA and fnbB from S. aureus strains Newman and 8325-4. pMG4ExpA is an expression vector for the fnbA gene from S. aureus strain Newman, in which the fnbA gene was ligated into pMG8Vec by PstI and BglII. pMG4ExpB is an expression vector for the fnbB gene from S. aureus strain Newman, constructed by the same procedure as pMG4ExpA. Only restriction sites relevant to the construction of the vector are shown.
Sequence analysis.
For sequence analysis, the fnbA and fnbB genes from the S. aureus strains 8325-4 and Newman were amplified by PCR (see above) and purified with a PCR purification kit (QIAGEN). The DNA sequences of both strands were determined by the dideoxy chain termination method (ABI PRISM BigDye Terminator version 3.0), using the cycle-sequencing protocol with the GeneAmp PCR system 2400 (Perkin-Elmer) on an ABI PRISM 3100-Avant Genetic Analyzer.
Slide agglutination tests.
All transformants of S. aureus and S. carnosus were tested for the functional surface expression of adhesins qualitatively and semiquantitatively by two methods, as described previously (55): a routine slide agglutination test with citrated rabbit plasma (BioMérieux, Marcy L'Etoile, France) and a commercial S. aureus identification latex agglutination kit (Pastorex Staph-plus; Bio-Rad, Marnes La Coquette, France) that recognizes ClfA, SpA, FnBPs (by virtue of their fibrinogen-binding activities) (64), and capsular polysaccharides 5 and 8.
Ligand overlay assays.
FnBPs were identified using soluble fibronectin, as described previously (18, 55), with minor modifications. Briefly, bacteria from 50-ml cultures grown to exponential phase were pelleted (3,500 × g; 20 min; 4°C). Total protein in the supernatants was precipitated by trichloroacetic acid (10% [vol/vol] final concentration), and the pellet was washed with acetone and resuspended in 300 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer. For preparation of cell surface proteins, the bacterial pellet was resuspended in 0.5 ml of hypertonic buffer (30% raffinose-0.05 Tris-HCl, pH 7.5) with phenylmethylsulfonyl fluoride (2 μM) and lysostaphin (10 μg/ml). After incubation for 30 min at 37°C, the protoplasts were pelleted (10,000 × g; 5 min), and the resulting supernatant was used as a cell wall extract. Both bacterial-culture supernatants and cell wall extracts were analyzed by ligand overlay blotting. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto nitrocellulose membrane (Millipore), and blocked with Tris-buffered saline containing 0.05% Tween 20 and 5% bovine serum albumin. The blots were incubated with digoxigenin-3-O-methylcarbonyl-ɛ-aminocaproic acid-N-hydroxysuccinimide ester (DIG; Roche)-labeled human fibronectin for 2 h. Subsequently, the blots were exposed to anti-DIG antibodies (Roche) and developed with a color reaction according to the manufacturer's protocol.
Solid-phase adherence assay.
The radiometric adherence assay with log-phase cultures was performed as described previously (19, 63). Briefly, polymethylmethacrylate (PMMA) coverslips (8 by 8 mm) were coated by incubation with 50 μg of purified human fibronectin (Chemicon, Temecula, Calif.)/ml in PBS for 60 min at 37°C and then rinsed with PBS. An inoculum containing 4 × 106 CFU (40 μl) of [3H]thymidine-labeled staphylococci was incubated with an Fn-coated coverslip in a tube containing 960 μl of PBS with Ca2+-Mg2+ supplemented with 0.5% HSA and incubated at 37°C for 1 h in a shaking water bath. Thereafter, the PMMA coverslip was removed and washed three times with PBS, and the adherent counts per minute were determined. The results were expressed as percent recovery of the inoculum. In pilot experiments testing different concentrations of Fn (5 to 200 μg/ml), we found a rather flat dose response for adhesion; thus, we used 50 μg of Fn/ml in subsequent experiments.
Preparation of FITC-labeled bacteria.
Bacteria were prepared as described previously (54). The bacteria were grown overnight without shaking, washed in 0.9% NaCl, and then fixed in 0.5% formaldehyde in PBS for at least 1 h and washed. Subsequently, the bacteria were labeled in 3 ml of 0.5 M NaHCO3 buffer (pH ∼9.5) supplemented with 100 μg of fluorescein isothiocyanate (FITC)/ml (isomer I [Molecular Probes, Leiden, The Netherlands] solubilized in 150 μl of dimethyl sulfoxide) for 1 h at 37°C. Finally, the bacteria were resuspended in PBS-1% HSA and used within 24 h after preparation. The suspensions were normalized for OD540 after gentle sonication in a water bath.
Cell culture.
All medium components were from Gibco-BRL. 293 cells (adenovirus type 5 DNA-transformed primary human embryonic kidney cells) were obtained from the American Type Culture Collection (ATCC) (CRL-1573); maintained in Dulbecco's modified Eagle's medium (DMEM)-Nut mix F-12 (containing Glutamax I, a stable glutamine dipeptide) supplemented with 10% fetal calf serum, 50 IU of penicillin/ml, and 50 μg of streptomycin/ml; and split 1:4 twice weekly by trypsinization. They were maintained in humidified air-5% CO2 at 37°C and were used up to passage number 35 after freezing.
Flow cytometric invasion assay.
The flow cytometric invasion assay was performed as described previously (54, 55) with minor modifications (27). A fresh bacterial culture was used for each experiment. Briefly, 293 cells were plated in 24-well plates at 0.4 × 106 cells/well the day before the assay. The cells were washed with DMEM-Nut mix F-12, and then 0.5 ml of 1% HSA-10 mM HEPES (pH 7.4) in DMEM-Nut mix F-12 (invasion medium) was added. The cells were cooled on ice, and 50 μl of fixed, FITC-labeled bacterial suspensions was added, resulting in an estimated multiplicity of infection of ∼25:1. The culture dishes were preincubated for 1 h at 4°C to allow sedimentation of the bacteria and shifted to 37°C for 3 h for invasion. Under the applied conditions, this assay measures only internalized, not adherent, bacteria, as previously shown (54). Furthermore, lysostaphin protection assays show a high correlation with this assay (55). Finally, the cells were harvested, treated with 20 μM monensin (10 min at ambient temperature) to neutralize fluorescence quenching, and analyzed by flow cytometry, after propidium iodide (5 μg/ml) exclusion, as previously described (54). The results were normalized according to the mean fluorescence intensity of the respective bacterial preparation, as determined by flow cytometry (27).
Presentation of results.
For each experiment, a fresh bacterial culture was prepared. The results were expressed as the mean ± standard error of the mean (SEM) of n independent experiments performed in duplicate (the values of n are specified in the figure legends), unless stated otherwise.
Nucleotide sequence accession number.
The complete sequence data were deposited in the EMBL database as follows: fnbANewman, accession number AJ629121; fnbBNewman, accession number AJ629122.
RESULTS
Construction of expression vectors for fnbA and fnbB from strain Newman.
In order to test for potential functional differences between FnBPANewman and FnBPBNewman, the respective genes were heterologously expressed in S. carnosus. For this purpose, fnbANewman and fnbBNewman were cloned into an established staphylococcal expression vector (multicopy plasmid), pFNBA4 (15), by replacing fnbA8325-4 with the respective genes from strain Newman (Fig. 1).
The resulting plasmids all contained the same promoter sequence and signal sequence of fnbA8325-4. Therefore, any functional alterations should be due to structural rather than to regulatory differences between the clones harboring the different constructs. In parallel, fnbA8325-4 was reinserted into the original vector as a control.
S. aureus strain Newman harbors a stop codon in fnbA and fnbB.
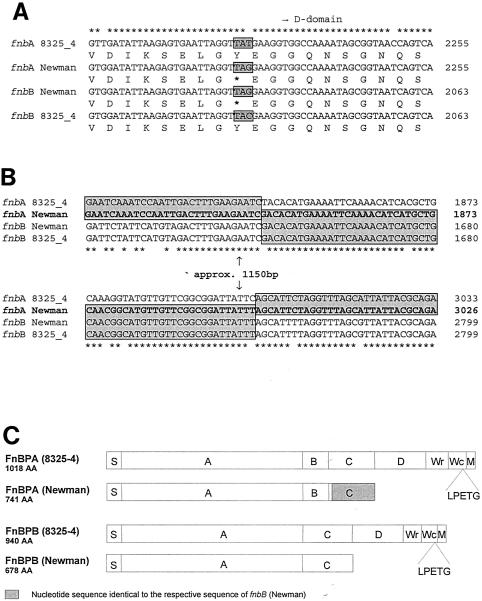
Sequence analysis of fnbBNewman, coding for FnBPB, revealed that strain Newman shows a surprisingly high degree of nucleotide identity (∼99.9%) with the respective prototype sequence of strain 8325-4. One of the few differences between fnbBNewman and fnbB8325-4 is a base pair exchange near the 3′ end of the C domain of fnbBNewman. This exchange (nucleotide position 2224 from the start codon) results in a stop codon near the 3′ end of the C domain of fnbBNewman and leads to a premature termination of FnBPB in strain Newman (Fig. 2A). In addition, in fnbANewman, the C-terminal portion of the gene (from the beginning of the C domain to the beginning of the M domain), including the stop codon, appears to have been replaced by the corresponding region of fnbBNewman (Fig. 2B). Apart from this exchange, there were only 3 nucleotide differences over the rest of the full sequence for fnbA. These two events, the stop codon in the C domain of fnbBNewman in combination with the sequence exchange in fnbANewman, lead to a truncation of both FnBPs in S. aureus strain Newman (Fig. 2C).
FIG. 2.
S. aureus strain Newman expresses a regular fusion protein of FnBPA and FnBPB and harbors a stop codon at the ends of the C domains of both fnbA and fnbB. (A) Sequence comparison of fnbA and fnbB of strains Newman and 8325-4 at the beginning of the D domain. The protein translation shows a stop codon in fnbA and fnbB from strain Newman at the end of the C domain (asterisks indicate nucleotide sequence identity of all genes; boldface asterisks indicate stop codons). (B) Sequence comparison of the divergent regions of fnbA and fnbB of strains Newman and 8325-4, respectively. The sequence of fnbANewman was exchanged for that of fnbBNewman from the beginning of the C domain to the end of the M domain. Boxed sequences are 100% identical (asterisks indicate nucleotide sequence identity of all genes). (C) Schematic representation of FnBPA and FnBPB of strains Newman and 8325-4, respectively. S indicates signal peptide; further domain designations (A, B, C, Wr, Wc, and M domains) were used as published previously (26). The sequences shown in the figure were obtained from strain Newman D2C, purchased from the ATCC, by directly sequencing genomic PCR products and fnb genes cloned into the expression plasmids. The regions containing the identified stop codons (in fnbA and fnbB) and crossovers (in fnbA) were verified by sequencing of genomic PCR products in all four strains tested, designated Newman and Newman D2C (including the strain used by Wolz et al. and reference isolates for both strains, freshly purchased from the NCTC), respectively.
Phenotypic characterization of strains.
We tested the transformants of S. carnosus for the surface expression and function of adhesins qualitatively by two methods: a slide agglutination test with rabbit citrate plasma and a commercial S. aureus identification latex agglutination kit. All S. carnosus transformants containing plasmids that encode fnb genes from S. aureus strain 8325-4 tested positive in both agglutination tests (by virtue of the fibrinogen-binding activity of FnBPs [64]). The S. carnosus wild-type strain, as well as the S. carnosus transformants containing expression vectors with fnb genes from S. aureus strain Newman, appeared negative (Table 2).
TABLE 2.
Surface expression of cell wall-anchored adhesins in transformantsa
| Strain | Heterologous expression | Rabbit plasma | NaCl | Latex agglutination | Control |
|---|---|---|---|---|---|
| S. aureus | |||||
| Cowan 1 | − | +++ | − | +++ | − |
| Newman D2C | − | +++ | − | +++ | − |
| S. carnosus | |||||
| WT TM300 | − | − | − | − | − |
| TM300 (pFNBA4) | FnBPA8325-4 | ++ | − | +++ | − |
| TM300 (pFNBB4) | FnBPB8325-4 | +(+) | − | +++ | − |
| TM300 (pMG4ExpA) | FnBPANewman | +/− | − | − | − |
| TM300 (pMG4ExpB) | FnBPBNewman | − | − | − | − |
| TM300 (pMG5ExpA) | FnBPA8325-4 | +++ | − | +++ | − |
Surface expression of adhesins by transformants of S. carnosus (TM300) was tested by induction of clumping in citrated rabbit plasma and a commercial latex agglutination kit (Pastorex Staph-plus), which detects fibrinogen binding by FnBPs, as described in Materials and Methods. Results were expressed semiquantitatively in five categories: −, no clumping; +/−, +, ++, +++ (progressively more efficient and rapid clumping). S. aureus strain Cowan 1 was used as a positive control.
Strain Newman produces truncated forms of FnBPA and FnBPB which are not cell wall anchored.
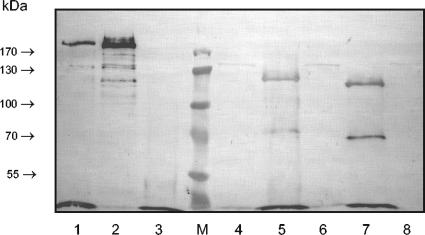
Ligand overlay assays with DIG-labeled fibronectin showed that both FnBPANewman and FnBPBNewman were not found in lysostaphin-solubilized cell wall material and total lysates upon heterologous expression in S. carnosus WT strain TM300 (Fig. 3). Bands corresponding to truncated forms of FnBPANewman (apparent molecular mass, ∼120 kDa) and FnBPBNewman (apparent molecular mass, ∼115 kDa) were visible when bacterial culture supernatants were analyzed. As expected, this was clearly different for both FnBPs of strain 8325-4. Cell wall extracts of S. carnosus transformants heterologously producing FnBPA8325-4 (Fig. 3) and FnBPB8325-4 (data not shown), respectively, contained most of the FnBPs, whereas FnBPs were barely detectable in the culture supernatants. In addition, the FnBPs of strain Newman were detectable at a position corresponding to a smaller molecular mass than the respective FnBPs of strain 8325-4 (Fig. 3). Full-length FnBPs migrate with an apparent molecular mass of ∼200 to 220 kDa (55).
FIG. 3.
FnBPANewman and FnBPBNewman are truncated and secreted, as shown in ligand overlay assays with cell wall-associated proteins of S. carnosus transformants (strain TM300) expressing FnBPs. FnBPs were detected with DIG-labeled fibronectin and peroxidase-coupled anti-DIG antibodies. Supernatant of MG5ExpA with FnBPA8325-4 of ∼200 kDa (lane 1), cell wall preparation of MG5ExpA with FnBPA8325-4 of ∼200 kDa (lane 2), supernatant of S. carnosus (lane 3), prestained protein ladder (∼10 to 180 kDa) (lane M), cell wall preparation of S. carnosus (lane 4), supernatant of MG4ExpA with truncated FnBPANewman of ∼115 kDa (lane 5), cell wall preparation of MG4ExpA (lane 6), supernatant of MG4ExpB with truncated FnBPBNewman of ∼110 kDa (lane 7), and cell wall preparation of MG4ExpB (lane 8). The masses of marker proteins in the separation gel (7.5%) are indicated.
S. carnosus producing either FnBPANewman or FnBPBNewman does not adhere efficiently to immobilized fibronectin.
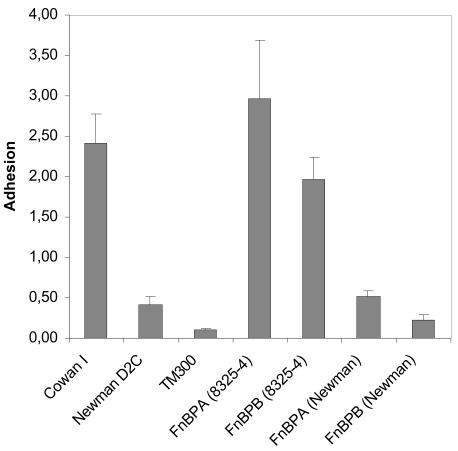
In order to determine whether the mutations observed so far led to a loss of FnBP-mediated functions, we tested S. carnosus strains heterologously expressing FnBPA and FnBPB for adherence to immobilized fibronectin (Fig. 4). As expected, both FnBPs of strain 8325-4 mediated efficient adherence to immobilized fibronectin. Interestingly, the adherence of S. carnosus expressing FnBPANewman did not differ substantially from that of S. aureus strain Newman but was stronger than that of WT S. carnosus. S. carnosus expressing FnBPBNewman was less adherent than S. carnosus expressing FnBPANewman but slightly more than WT S. carnosus.
FIG. 4.
FnBPANewman and FnBPBNewman do not mediate adherence of staphylococci to immobilized fibronectin. Adhesins were expressed in strain TM300 by multicopy plasmids. PMMA coverslips were incubated with [3H]thymidine-labeled S. aureus strains Newman and Cowan I and S. carnosus strains TM300, MG4ExpA, MG4ExpB, MG5ExpA, and BS101. The results are means plus SEM of two independent experiments run in quadruplicate. The data are shown as percentages of adherent bacteria compared to the inoculum.
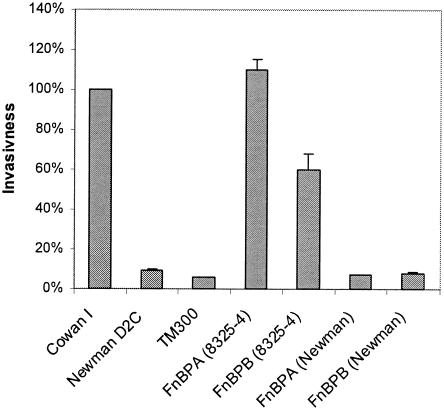
Neither FnBPANewman nor FnBPBNewman confers invasiveness on S. carnosus.
Neither FnBPANewman nor FnBPBNewman renders S. carnosus invasive, as opposed to the respective very closely related FnBP prototype sequences of strain 8325-4 (Fig. 5). As observed previously, FnBPA8325-4 appears to be slightly more effective for invasion than FnBPB8325-4 (54, 55). Competition experiments with bacterial culture supernatants of TM300 expressing FnBPANewman or FnBPBNewman demonstrated that the truncated forms of FnBPs can act as inhibitors of FnBP-dependent functions (data not shown), supposedly by occupying binding sites on fibronectin by the residual Fn-binding sites in the B-C domain of the secreted FnBPs.
FIG. 5.
FnBPANewman and FnBPBNewman do not confer invasiveness on noninvasive S. carnosus. Adhesins were expressed in strain TM300 by multicopy plasmids. The total amount of internalized bacteria after invasion of 293 cells was measured by flow cytometry with fixed bacteria. The results are means plus SEM of four independent experiments run in duplicate and are expressed as relative invasiveness compared to strain Cowan 1.
DISCUSSION
The importance of FnBPs for S. aureus is underlined by the high prevalence of FnBPs in clinical isolates: all of 25 strains (56) and all but one of 30 strains (36) tested carried at least one fnb gene, as detected by PCR analysis of the entire region encompassing the adjacent fnbA and fnbB genes and subsequent Southern blotting. More recent studies found a prevalence of 100% for at least one fnb gene in clinical isolates (38, 45, 47, 51).
S. aureus critically depends on either of the two known forms of FnBPs, FnBPA or FnBPB, to mediate efficient adherence to and invasion of host cells. The lack of FnBPs largely abolishes efficient internalization of the bacteria, with up to a 500-fold reduction (7, 11, 30, 46, 54), whereas it can be conferred by expression of either form of FnBP on noninvasive strains of three different species (i.e., S. aureus, S. carnosus, and Lactococcus lactis) (55). In the absence of functional FnPBs, as observed in strain Newman, Eap, which is highly expressed in this strain (18), appears to play a role in adherence to fibroblasts (20). Eap may partially compensate for the loss of Fn binding and in consequence mediate residual invasiveness (16).
The major high-affinity binding sites of S. aureus FnBPs have been located in 3.5 tandem repeats of ∼38 amino acids close to the C-terminal wall-spanning region and an additional repeat ∼100 amino acids upstream of these (21). However, full-length FnBPA8325-4 was >100 times more effective on an equimolar basis than either of the two fragments encompassing the A and B domains and the C to W domains, respectively (11), suggesting a synergistic effect of the two adhesin domains. Subsequently, it was shown that only FnBPA8325-4 constructs with a combined deletion of the B, C, and D domains lose their adhesive and invasive functions (31). Using phage display, a fragment of the B domain has been identified as a potential additional fibronectin-binding region of FnBPs involved in binding of bone tissue (65). The concept of multiple substituting binding sites on FnBPs has been extended recently, suggesting a “beads on a string” model, where each interacting domain would correspond to a bead (53). Besides binding fibronectin, FnBPA8325-4 has been shown to bind fibrinogen via a region of the A domain that has not yet been further characterized (64). This appears to be functionally relevant, since FnBP-producing S. carnosus transformants can be agglutinated by latex beads sensitized with fibrinogen (this study and reference 55). Additionally, it has been shown recently that FnBPs are also involved in platelet binding (17).
Here, we found that strain Newman produces only truncated FnBPs, which are secreted, leading to a loss of FnBP-dependent functions. In both FnBPs, the truncation is due to a stop codon in the C-terminal end of the C domain, which deprives the FnBPs of the sortase motif (LPETG) required to anchor them covalently to the peptidoglycan layer. We have observed moderate discrepancies for FnBPANewman in adherence to immobilized fibronectin and invasiveness. Whereas transformants expressing FnBPANewman showed slightly higher adherence to immobilized fibronectin than WT S. carnosus, i.e., in a range similar to that of WT S. aureus strain Newman (Fig. 4), their invasiveness was similar to the background, i.e., not higher than of WT S. carnosus (Fig. 5). This could be explained by a partial readthrough at the stop codon, resulting in the production of a low proportion of full-length protein sufficient to mediate moderate adherence. By contrast, the amount of FnBPs required for invasion may not have been reached. This difference was much less pronounced with FnBPBNewman.
Together with S. aureus strain 8325-4, strain Newman is a reference strain that is frequently used for genetic, functional, and in vivo studies. The described mutation in strain Newman should allow us to better interpret apparently divergent results of in vitro and in vivo studies (Table 3). Our data provide some explanations for these discrepancies with regard to known FnBP-dependent functions. A recent study found strain Newman to be only weakly adherent and not invasive when tested on confluent human umbilical vein endothelial cells (HUVEC) despite strong fnbA transcription and FnBP production (57). FnBPs have been shown to substantially contribute to the virulence of S. aureus: the fnb-deficient mutant of strain 8325-4 was clearly less virulent in a murine foreign-body osteomyelitis model (22). However, a mutant of strain Newman (DU5886; ΔclfA ΔfnbA ΔfnbB) did not differ in virulence from WT strain Newman, which had been complemented genomically with cna (clfA+ fnbA+ fnbB+ cna+) in a rabbit foreign body osteomyelitis model (5). In light of our data, it appears that these results are strain dependent rather than model dependent.
TABLE 3.
Compilation of selected results for FnBP-dependent functions in strain Newman
| Function | Result | Reference | Comments |
|---|---|---|---|
| Adherence to immobilized Fn | Higher than ISP479Cb | 67c | Regulation of adhesive mechanism different from FnBPs examined? |
| Adherence to immobilized Fn | Very low | 61 | Can be explained by fnbNewman mutation |
| Adherence to immobilized Fn | Weak | 46 | Can be explained by fnbNewman mutation |
| Adherence to confluent HUVEC | Weak | ||
| Adherence to confluent HUVEC | Weak | 57d | Can be explained by fnbNewman mutation |
| Invasion of confluent HUVEC | Background | ||
| Invasion of subconfluent 293 cells | Weak | 54 | Can be explained by fnbNewman mutation |
| Invasion of human lung fibroblasts and HACATa | Newman Δeap reduced compared to WT | 16 | Role of strongly expressed Eap most likely detected due to lack of functional FnBPs |
| Virulence in rabbit osteomyelitis (foreign body) | No difference for fnb-negative mutant of Newman | 5 | FnBP dependence in fact not tested; results are in contrast to reference 22, which uses 8325-4 |
HACAT, human keratinocyte cell line.
ISP479C, 8325-4 derivative.
fnbA mRNA detected, as determined by Northern blotting, strongly upregulated in Newman Δagr; FnBPs detected, as determined by fibronectin overlay assays (resulting band, ∼ 200 kDa); cell wall extracts, upregulated in Newman Δagr; culture supernatants, strongly upregulated in Newman Δagr.
fnbA mRNA detected, as determined by Northern blotting; FnBPs detected, as determined by fibronectin overlay assays of cell wall extracts (resulting band, ∼180 kDa, additional very faint ∼200 kDa).
The roles of ClfA and FnBPs in the pathogenesis of infective endocarditis have been a longstanding controversy. Using an approach of adoptive pathogenicity (heterologous expression of adhesins in the large apathogenic species Lactococcus lactis subspecies cremoris), the respective contributions of ClfA and FnBPs to virulence have been clearly dissected in a rat model of traumatic endocarditis (49). FnBPA and ClfA are equally effective for the initiation of the disease, reducing the 80% infectious dose of L. lactis subspecies cremoris transformants ∼100-fold to the level of wild-type S. aureus or streptococci in this model. However, FnBPA-expressing but not ClfA-expressing lactococci are able to reproduce the progression of pathology, marked clinical illness, and invasion of the vegetations and surrounding endothelium. In light of these data, and the results presented in this study, earlier work can be interpreted more clearly. A study using strain 8325-4 (ClfA+) and its fnb-deficient mutant in a rat model of traumatic endocarditis found no difference in virulence (as determined by culture-positive vegetations and recovered CFU from vegetations) at 24 h postinfection (10). This is most likely due to the redundant functions of ClfA and FnBPs in the initiation of infection. Another study using strain Newman and isogenic coa-, clfA-, and double-deficient mutants found ∼50%-reduced virulence (as determined by the number of positive cultures of blood and vegetations, using an inoculum 80% infectious dose) of the ClfA- and double-deficient mutants (but no role for coagulase) (39). Most likely, this effect could be detected only because strain Newman does not possess cell wall-anchored FnBPs, as shown in this study. Thus, functionally, the four strains compared were ClfA+ Coa+ FnBPA− FnBPB−, ClfA+ Coa− FnBPA− FnBPB−, ClfA− Coa+ FnBPA− FnBPB−, and ClfA− Coa− FnBPA− FnBPB−. In addition, fibronectin binding could have been antagonized by secreted FnBPs.
By extension, our findings should be rather important for studies which directly involve FnBP-dependent or partly FnBP-mediated functions, e.g., adherence to fibronectin (61), binding of fibrinogen (61, 62), binding to platelets (17), invasion of host cells (7, 11, 30, 54, 55), and T-cell activation (37). Adhesion to fibronectin for strain Newman has been examined by several studies (32, 67). This may be relevant for the activation of platelets (42). In addition, in experiments where culture supernatant is used, and when live bacteria are allowed to produce FnBPs during the course of the experiment, this may lead to competition for FnBP binding, as has been shown in vitro (11, 54). Based on this, one may even expect results apparently suggesting no role for FnBPs in the model investigated.
Even with our findings of truncated FnBPs, some discrepancies still remain unexplained. A 200-kDa protein, probably full-length FnBPA, was detected in lysostaphin cell wall extracts of strain Newman by fibronectin overlay assays (67). The reason for this is not clear, since the fnb sequences of this strain contain the stop codon. However, it is conceivable that full-length FnBPs may be expressed in very small amounts by readthrough of the stop codon. This would explain the detected 200-kDa protein (67) and the slightly increased adhesion of S. carnosus heterologously expressing fnbANewman (Fig. 4). For the interpretation of data focusing on fnbA promoter activity and global regulation in strain Newman (1, 57, 67, 68), we would not expect to see an influence of the FnBP truncation. However, some of these studies use adherence to immobilized fibronectin (fibronectin-binding capacity) to functionally monitor FnBP expression, which probably would reflect mostly adhesion by a mechanism different from FnBPs. This could include the strongly expressed anchorless adhesin Eap (and potentially Emp), as discussed above. An inverse correlation of the fibronectin-binding capacity and protease production has been observed, except for strain Newman, where fibronectin binding was low despite low protease production (1). This is in line with the lack of surface-anchored FnBPs in strain Newman.
Another reason for divergent results obtained with strain Newman may be the evolution of different subclones after decades of in vitro culturing in different laboratories. A further level of confusion is added because two different isolates have been deposited in the National Collection of Type Cultures (NCTC) (Newman and Newman D2C) (Table 1). Furthermore, apparently identical isolates were deposited in different strain collections with different reference numbers, and referral to other strain collections is sometimes made using invalid reference numbers (Table 1, footnote a).
In summary, this argues for an extremely cautious use and interpretation of data, albeit for different reasons, with strains Newman and 8325-4 in studies where FnBP-dependent functions, including binding of fibrinogen, are investigated or may play a role. Additionally, control of surface expression of FnBPs may be warranted. On the other hand, if used deliberately, strain Newman may allow us to examine the role of adhesins in an FnBP-negative background. However, the redundant fibronectin-binding capacity of strain Newman (67) has to be considered.
Addendum in Proof
Since acceptance of our paper, a paper reporting a major role of fibronectin-binding proteins for adhesion to elastin, which is mediated by the A domain, has been published (F. M. Roche, R. Downer, F. Keane, P. Speziale, P. W. Park, and T. J. Foster, J. Biol. Chem. 279:28433-28440, 2004). Thus, our findings most likely extend to processes that depend on elastin binding by Staphylococcus aureus strain Newman.
Acknowledgments
We thank Katrin Strangfeld and Susanne Weber for excellent experimental assistance and Christiane Wolz for discussions and for kindly providing the strain Newman used in her laboratory. Alexander Mellmann and Alexander Friedrich kindly verified spa-type identities of all isolates of strain Newman analyzed in this study.
This work was supported by the Interdisciplinary Center for Clinical Research (IZKF Münster; grant C20-Si2/048/04 to B.S. and G.P.) and in part by the Deutsche Forschungsgemeinschaft (Collaborative Research Center 492, project B9, to B.S. and G.P.).
Editor: V. J. DiRita
REFERENCES
- 1.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouillette, E., G. Grondin, L. Shkreta, P. Lacasse, and B. G. Talbot. 2003. In vivo and in vitro demonstration that Staphylococcus aureus is an intracellular pathogen in the presence or absence of fibronectin-binding proteins. Microb. Pathog. 35:159-168. [DOI] [PubMed] [Google Scholar]
- 3.Brouillette, E., B. G. Talbot, and F. Malouin. 2003. The fibronectin-binding proteins of Staphylococcus aureus may promote mammary gland colonization in a lactating mouse model of mastitis. Infect. Immun. 71:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dajcs, J. J., M. S. Austin, G. D. Sloop, J. M. Moreau, E. B. Hume, H. W. Thompson, F. M. McAleese, T. J. Foster, and R. J. O'Callaghan. 2002. Corneal pathogenesis of Staphylococcus aureus strain Newman. Investig. Ophthalmol. Vis. Sci. 43:1109-1115. [PubMed] [Google Scholar]
- 5.Darouiche, R. O., G. C. Landon, J. M. Patti, L. L. Nguyen, R. C. Fernau, D. McDevitt, C. Greene, T. Foster, and M. Klima. 1997. Role of Staphylococcus aureus surface adhesins in orthopaedic device infections: are results model-dependent? J. Med. Microbiol. 46:75-79. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson, R. B., J. A. Nagel, D. McDevitt, T. J. Foster, R. A. Proctor, and S. L. Cooper. 1995. Quantitative comparison of clumping factor- and coagulase-mediated Staphylococcus aureus adhesion to surface-bound fibrinogen under flow. Infect. Immun. 63:3143-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dziewanowska, K., J. M. Patti, C. F. Deobald, K. W. Bayles, W. R. Trumble, and G. A. Bohach. 1999. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect. Immun. 67:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Entenza, J. M., T. J. Foster, E. D. Ni, P. Vaudaux, P. Francioli, and P. Moreillon. 2000. Contribution of clumping factor B to pathogenesis of experimental endocarditis due to Staphylococcus aureus. Infect. Immun. 68:5443-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flock, J. I., G. Fröman, K. Jönsson, B. Guss, C. Signäs, B. Nilsson, G. Raucci, M. Höök, T. Wadström, and M. Lindberg. 1987. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 6:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flock, J. I., S. A. Hienz, A. Heimdahl, and T. Schennings. 1996. Reconsideration of the role of fibronectin binding in endocarditis caused by Staphylococcus aureus. Infect. Immun. 64:1876-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler, T., E. R. Wann, D. Joh, S. Johansson, T. J. Foster, and M. Höök. 2000. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79:672-679. [DOI] [PubMed] [Google Scholar]
- 12.François, P., J. Schrenzel, C. Stoerman-Chopard, H. Favre, M. Herrmann, T. J. Foster, D. P. Lew, and P. Vaudaux. 2000. Identification of plasma proteins adsorbed on hemodialysis tubing that promote Staphylococcus aureus adhesion. J. Lab. Clin. Med. 135:32-42. [DOI] [PubMed] [Google Scholar]
- 13.Fröman, G., L. M. Switalski, P. Speziale, and M. Höök. 1987. Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J. Biol. Chem. 262:6564-6571. [PubMed] [Google Scholar]
- 14.Götz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 15.Greene, C., D. McDevitt, P. François, P. E. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 16.Haggar, A., M. Hussain, H. Lonnies, M. Herrmann, A. Norrby-Teglund, and J. I. Flock. 2003. Extracellular adherence protein from Staphylococcus aureus enhances internalization into eukaryotic cells. Infect. Immun. 71:2310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmann, C., S. Niemann, B. Sinha, M. Herrmann, B. E. Kehrel, and G. Peters. 2004. Staphylococcus aureus fibronectin-binding protein (FnBP)-mediated adherence to platelets, and aggregation of platelets induced by FnBPA, but not by FnBPB. J. Infect. Dis. 190:321-329. [DOI] [PubMed] [Google Scholar]
- 18.Hussain, M., K. Becker, C. von Eiff, G. Peters, and M. Herrmann. 2001. Analogs of Eap protein are conserved and prevalent in clinical Staphylococcus aureus isolates. Clin. Diagn. Lab. Immunol. 8:1271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183:6778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain, M., A. Haggar, C. Heilmann, G. Peters, J. I. Flock, and M. Herrmann. 2002. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 70:2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Höök. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18:211-223. [DOI] [PubMed] [Google Scholar]
- 22.Johansson, A., J. I. Flock, and O. Svensson. 2001. Collagen and fibronectin binding in experimental staphylococcal osteomyelitis. Clin. Orthop. 382:241-246. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson, I. M., S. K. Mazmanian, O. Schneewind, T. Bremell, and A. Tarkowski. 2003. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 5:775-780. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson, I. M., S. K. Mazmanian, O. Schneewind, M. Verdrengh, T. Bremell, and A. Tarkowski. 2002. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J. Infect. Dis. 185:1417-1424. [DOI] [PubMed] [Google Scholar]
- 25.Jonsson, I. M., C. von Eiff, R. A. Proctor, G. Peters, C. Rydén, and A. Tarkowski. 2003. Virulence of a hemB mutant displaying the phenotype of a Staphylococcus aureus small colony variant in a murine model of septic arthritis. Microb. Pathog. 34:73-79. [DOI] [PubMed] [Google Scholar]
- 26.Jönsson, K., C. Signäs, H. P. Müller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 27.Juuti, K. M., B. Sinha, C. Werbick, G. Peters, and P. I. Kuusela. 2004. Reduced adherence and host cell invasion by methicillin-resistant Staphylococcus aureus expressing the surface protein Pls. J. Infect. Dis. 189:1574-1584. [DOI] [PubMed] [Google Scholar]
- 28.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative σ factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuypers, J. M., and R. A. Proctor. 1989. Reduced adherence to traumatized rat heart valves by a low-fibronectin-binding mutant of Staphylococcus aureus. Infect. Immun. 57:2306-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammers, A., P. J. Nuijten, and H. E. Smith. 1999. The fibronectin binding proteins of Staphylococcus aureus are required for adhesion to and invasion of bovine mammary gland cells. FEMS Microbiol. Lett. 180:103-109. [DOI] [PubMed] [Google Scholar]
- 31.Massey, R. C., M. N. Kantzanou, T. Fowler, N. P. Day, K. Schofield, E. R. Wann, A. R. Berendt, M. Höök, and S. J. Peacock. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell Microbiol. 3:839-851. [DOI] [PubMed] [Google Scholar]
- 32.Maxe, I., C. Rydén, T. Wadström, and K. Rubin. 1986. Specific attachment of Staphylococcus aureus to immobilized fibronectin. Infect. Immun. 54:695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McElroy, M. C., D. J. Cain, C. Tyrrell, T. J. Foster, and C. Haslett. 2002. Increased virulence of a fibronectin-binding protein mutant of Staphylococcus aureus in a rat model of pneumonia. Infect. Immun. 70:3865-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mempel, M., T. Schmidt, S. Weidinger, C. Schnopp, T. Foster, J. Ring, and D. Abeck. 1998. Role of Staphylococcus aureus surface-associated proteins in the attachment to cultured HaCaT keratinocytes in a new adhesion assay. J. Investig. Dermatol. 111:452-456. [DOI] [PubMed] [Google Scholar]
- 35.Menzies, B. E. 2003. The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr. Opin. Infect. Dis. 16:225-229. [DOI] [PubMed] [Google Scholar]
- 36.Minhas, T., H. A. Ludlam, M. Wilks, and S. Tabaqchali. 1995. Detection by PCR and analysis of the distribution of a fibronectin-binding protein gene (fbn) among staphylococcal isolates. J. Med. Microbiol. 42:96-101. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto, Y. J., E. R. Wann, T. Fowler, E. Duffield, M. Höök, and B. W. McIntyre. 2001. Fibronectin-binding protein A of Staphylococcus aureus can mediate human T lymphocyte adhesion and coactivation. J. Immunol. 166:5129-5138. [DOI] [PubMed] [Google Scholar]
- 38.Mongodin, E., O. Bajolet, J. Cutrona, N. Bonnet, F. Dupuit, E. Puchelle, and S. de Bentzmann. 2002. Fibronectin-binding proteins of Staphylococcus aureus are involved in adherence to human airway epithelium. Infect. Immun. 70:620-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreillon, P., J. M. Entenza, P. Francioli, D. McDevitt, T. J. Foster, P. François, and P. Vaudaux. 1995. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect. Immun. 63:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 41.Nozohoor, S., A. Heimdahl, P. Colque-Navarro, I. Julander, B. Soderquist, and R. Mollby. 1998. Virulence factors of Staphylococcus aureus in the pathogenesis of endocarditis. A comparative study of clinical isolates. Zentbl. Bakteriol. 287:433-447. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien, L., S. W. Kerrigan, G. Kaw, M. Hogan, J. Penades, D. Litt, D. J. Fitzgerald, T. J. Foster, and D. Cox. 2002. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol. Microbiol. 44:1033-1044. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien, L. M., E. J. Walsh, R. C. Massey, S. J. Peacock, and T. J. Foster. 2002. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol. 4:759-770. [DOI] [PubMed] [Google Scholar]
- 44.Palmqvist, N., T. Foster, A. Tarkowski, and E. Josefsson. 2002. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 33:239-249. [DOI] [PubMed] [Google Scholar]
- 45.Peacock, S. J., N. P. Day, M. G. Thomas, A. R. Berendt, and T. J. Foster. 2000. Clinical isolates of Staphylococcus aureus exhibit diversity in fnb genes and adhesion to human fibronectin. J. Infect. 41:23-31. [DOI] [PubMed] [Google Scholar]
- 46.Peacock, S. J., T. J. Foster, B. J. Cameron, and A. R. Berendt. 1999. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145:3477-3486. [DOI] [PubMed] [Google Scholar]
- 47.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70:4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pöhlmann-Dietze, P., M. Ulrich, K. B. Kiser, G. Döring, J. C. Lee, J. M. Fournier, K. Botzenhart, and C. Wolz. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect. Immun. 68:4865-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Que, Y. A., P. François, J. A. Haefliger, J. M. Entenza, P. Vaudaux, and P. Moreillon. 2001. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 69:6296-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rennermalm, A., Y. H. Li, L. Bohaufs, C. Jarstrand, A. Brauner, F. R. Brennan, and J. I. Flock. 2001. Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine 19:3376-3383. [DOI] [PubMed] [Google Scholar]
- 51.Rice, K., M. Huesca, D. Vaz, and M. J. McGavin. 2001. Variance in fibronectin binding and fnb locus polymorphisms in Staphylococcus aureus: identification of antigenic variation in a fibronectin binding protein adhesin of the epidemic CMRSA-1 strain of methicillin-resistant S. aureus. Infect. Immun. 69:3791-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schleifer, K. H., and U. Fischer. 1982. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153-156. [Google Scholar]
- 53.Schwarz-Linek, U., J. M. Werner, A. R. Pickford, S. Gurusiddappa, J. H. Kim, E. S. Pilka, J. A. Briggs, T. S. Gough, M. Höök, I. D. Campbell, and J. R. Potts. 2003. Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature 423:177-181. [DOI] [PubMed] [Google Scholar]
- 54.Sinha, B., P. P. François, O. Nüβe, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell. Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 55.Sinha, B., P. P. François, Y.-A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smeltzer, M. S., A. F. Gillaspy, F. L. Pratt, Jr., M. D. Thames, and J. J. Iandolo. 1997. Prevalence and chromosomal map location of Staphylococcus aureus adhesin genes. Gene 196:249-259. [DOI] [PubMed] [Google Scholar]
- 57.Steinhuber, A., C. Goerke, M. G. Bayer, G. Döring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullam, P. M., A. S. Bayer, W. M. Foss, and A. L. Cheung. 1996. Diminished platelet binding in vitro by Staphylococcus aureus is associated with reduced virulence in a rabbit model of infective endocarditis. Infect. Immun. 64:4915-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Wamel, W., Y. Q. Xiong, A. S. Bayer, M. R. Yeaman, C. C. Nast, and A. L. Cheung. 2002. Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb. Pathog. 33:73-79. [DOI] [PubMed] [Google Scholar]
- 60.Vaudaux, P., P. François, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaudaux, P. E., P. François, R. A. Proctor, D. McDevitt, T. J. Foster, R. M. Albrecht, D. P. Lew, H. Wabers, and S. L. Cooper. 1995. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect. Immun. 63:585-590. (Erratum, 63:3239.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaudaux, P. E., V. Monzillo, P. François, D. P. Lew, T. J. Foster, and B. Berger-Bachi. 1998. Introduction of the mec element (methicillin resistance) into Staphylococcus aureus alters in vitro functional activities of fibrinogen and fibronectin adhesins. Antimicrob. Agents Chemother. 42:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaudaux, P. E., F. A. Waldvogel, J. J. Morgenthaler, and U. E. Nydegger. 1984. Adsorption of fibronectin onto polymethylmethacrylate and promotion of Staphylococcus aureus adherence. Infect. Immun. 45:768-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wann, E. R., S. Gurusiddappa, and M. Höök. 2000. The fibronectin-binding MSCRAMM FnBPA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 65.Williams, R. J., B. Henderson, and S. P. Nair. 2002. Staphylococcus aureus fibronectin binding proteins A and B possess a second fibronectin binding region that may have biological relevance to bone tissues. Calcif. Tissue Int. 70:416-421. [DOI] [PubMed] [Google Scholar]
- 66.Wolz, C., D. McDevitt, T. J. Foster, and A. L. Cheung. 1996. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 64:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 68.Xiong, Y. Q., A. S. Bayer, M. R. Yeaman, W. van Wamel, A. C. Manna, and A. L. Cheung. 2004. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect. Immun. 72:1832-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yonemasu, K., T. Sasaki, H. Hashimoto, and S. Kashiba. 1988. Opsonic effect of fibronectin on staphylococcal phagocytosis by human polymorphonuclear leukocytes: its relative inefficiency in post-phagocytic metabolic activities and in intracellular killing. Microbiol. Immunol. 32:795-805. [DOI] [PubMed] [Google Scholar]
- 70.Yonemasu, K., T. Sasaki, R. Ohmae, and S. Kashiba. 1991. Expression of clumping and fibrinogen-binding activities of Staphylococcus aureus at various growth stages. Microbiol. Immunol. 35:405-409. [DOI] [PubMed] [Google Scholar]