GWA studies combined with multiple mutagenesis technologies, including CRISPR/Cas9, were used to functionally validate novel candidate genes contributing to phenotypic variation in symbiosis between legume plants and rhizobial bacteria.
Abstract
Genome-wide association (GWA) studies offer the opportunity to identify genes that contribute to naturally occurring variation in quantitative traits. However, GWA relies exclusively on statistical association, so functional validation is necessary to make strong claims about gene function. We used a combination of gene-disruption platforms (Tnt1 retrotransposons, hairpin RNA-interference constructs, and CRISPR/Cas9 nucleases) together with randomized, well-replicated experiments to evaluate the function of genes that an earlier GWA study in Medicago truncatula had identified as candidates contributing to variation in the symbiosis between legumes and rhizobia. We evaluated ten candidate genes found in six clusters of strongly associated single nucleotide polymorphisms, selected on the basis of their strength of statistical association, proximity to annotated gene models, and root or nodule expression. We found statistically significant effects on nodule production for three candidate genes, each validated in two independent mutants. Annotated functions of these three genes suggest their contributions to quantitative variation in nodule production occur through processes not previously connected to nodulation, including phosphorous supply and salicylic acid-related defense response. These results demonstrate the utility of GWA combined with reverse mutagenesis technologies to discover and validate genes contributing to naturally occurring variation in quantitative traits. The results highlight the potential for GWA to complement forward genetics in identifying the genetic basis of ecologically and economically important traits.
Genome-wide association (GWA) studies offer the promise of identifying genes responsible for quantitative trait variation in plants and animals (Mackay et al., 2009). Relative to forward genetic screens, GWA has the potential to identify genes of much smaller phenotypic effect. Moreover, the genes identified through GWA are expected to contribute to naturally occurring phenotypic variation. By contrast, forward genetic screens identify genes that are functionally necessary but may or may not contribute to natural variation. An important limitation of GWA analyses, however, is that the statistical approach used to identify candidate genes is also expected to identify many false positives. Therefore, functional validation of GWA candidates is essential for confirming the biological importance of the identified genes (Ioannidis et al., 2009).
The symbiosis between legume plants and rhizobial bacteria is responsible for large inputs of nitrogen into both natural and agricultural systems through symbiotic nitrogen fixation (Vance, 2001). The ecological and economic importance of this symbiosis has made identifying its genetic basis an important goal for plant biologists (Oldroyd et al., 2011). More than 30 plant genes that play central roles in the formation and growth of plant nodules, the site of rhizobial symbiosis, and nitrogen fixation have been identified (Popp and Ott, 2011; Pislariu et al., 2012). These genes were identified primarily through forward genetic screens. In standard forward genetic screens, a mutant phenotype is identified based on the effects seen within individual organisms, something that will bias these screens toward detecting genes of large effect. Forward genetic screens are likely to miss genes of small phenotypic effects, such as those that contribute to naturally occurring quantitative variation in nodulation (Stanton-Geddes et al., 2013; Friesen et al., 2014). Identification of these genes of subtle effect will provide a fuller understanding of the symbiosis between legumes and rhizobia, and may provide targets for selection in important agronomic legumes.
Here, we report on the application of three reverse genetics mutagenesis tools: Medicago truncatula Tnt1 retrotransposon mutant collection (Tadege et al., 2008; Pislariu et al., 2012), hairpin RNA interference knockdown constructs (Wesley et al., 2001), and CRISPR/Cas9 site-specific nuclease (SSN; Baltes et al., 2014) to empirically evaluate the functional importance of 10 genes that a previous sequence-based GWA study identified as candidates for naturally occurring phenotypic variation in the legume-rhizobia symbiosis (Stanton-Geddes et al., 2013). We used 17 whole-plant, stable mutants (six Tnt1, three hairpin, and eight CRISPR mutants) in well-replicated phenotypic assays to evaluate the function of each of the 10 GWA candidates. The functional importance of three genes (PHO2-like, PNO1-like, and PEN3-like) was validated by two independent mutations, confirming the importance of genes found in three of the six clusters investigated. These results not only identify genes not previously identified as being involved in legume-rhizobia symbiosis, but they also illustrate the power of combining GWA with newly developed gene disruption technologies to identify genes that contribute to naturally occurring variation in quantitative traits.
RESULTS
Association Mapping of SNF-Associated Candidate Genes in M. truncatula
To generate a list of candidates underlying variation in nodulation, we previously carried out GWA analysis involving over 6 million single nucleotide polymorphisms (SNPs) on a panel of 226 M. truncatula accessions (Stanton-Geddes et al., 2013). In brief, nodulation data were collected on eight replicates of each accession, and GWA was conducted using the GLMM implemented in TASSEL (Bradbury et al., 2007). From this GWA, we selected the 100 SNPs with the strongest association to variation in the number of nodules per plants, producing an initial list of nodulation candidates. This list then was filtered based on several criteria: (1) statistical support and ranking for an association from the GWA, (2) candidate being in linkage-disequilibrium with other SNPs with strong statistical support (i.e. located in a peak), (3) location within or proximate to annotated coding regions, and (4) expression in root and or nodule tissue. GWA was performed twice over the course of the research as a new assembly of the M. truncatula genome (Mt4.0) was released mid experiment (Tang et al., 2014), replacing the earlier assembly (Mt3.5; Young et al., 2011) used in the original analysis.
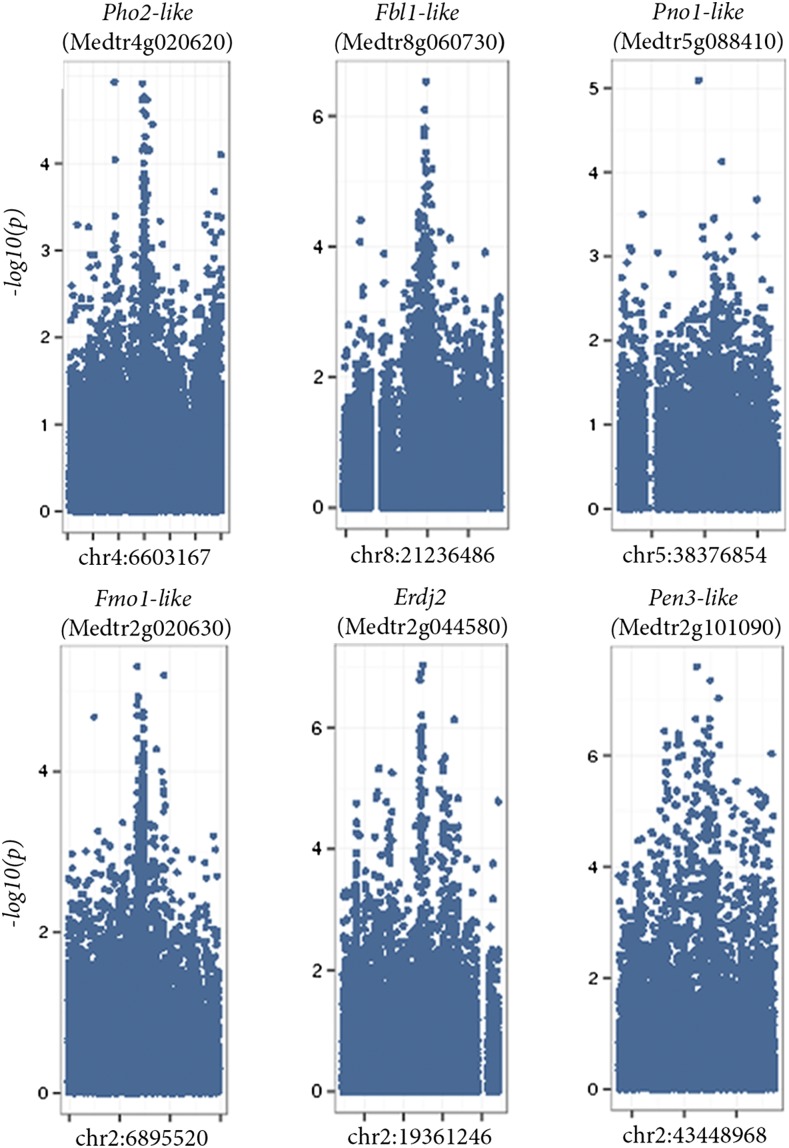
Based on these criteria, we selected 10 candidates (noting that we did not require all candidates to meet all of the filtering criteria) to target for mutagenesis and phenotypic validation (Table I). Six of the candidates were physically proximate to other candidates (two within 15 kb of each other and four within 80 kb of one another). These candidates were chosen to evaluate the potential to identify the causative gene when clusters of statistically associated SNPs identified by GWA spanned multiple genes (Fig. 1).
Table I. Genome location, GWA study statistics, and expression of candidate genes.
| Mt4.0 Designation | Candidate Gene | Rank | P Value | GWA Study | Expression | Confirmed |
|---|---|---|---|---|---|---|
| Medtr2g020630 | Flavin-containing Monooxygenase-like (FMO1-LIKE) | 41 | 8.14E-06 | Mt3.5 | Root and nodule specific | |
| Medtr2g044570 | Ring finger protein1-like (RFP1-like) | 8 | 1.62E-07 | Mt3.5 | All tissues | |
| Medtr2g044580 | Endoplasmic reticulum DNAJ protein (ERDJ2) | 5 | 1.30E-07 | Mt3.5 | Nodule upregulated | |
| Medtr2g101040 | Malic enzyme-like1 (MEL1) | 15 | 2.22E-07 | Mt4.0 | All tissues | |
| Medtr2g101090 | Penetration3-like (PEN3-like) | 1 | 2.56E-08 | Mt4.0 | Root and nodule upregulated | Yes |
| Medtr2g101120 | AVR9/CF-9 rapidly elicited protein 1 (ACRE1) | 23 | 3.55E-07 | Mt4.0 | All tissues | |
| Medtr2g101190 | Homeodomain Leu zipper1-like (HLZ1-like) | 72 | 9.65E-07 | Mt4.0 | All tissues | |
| Medtr4g020620 | Ubiquitin conjugate24 like (PHO2-like) | 12 | 5.58E-06 | Mt3.5 | Nodule upregulated | Yes |
| Medtr5g088410 | Partner of NOB1-like (PNO1-like) | 36 | 7.08E-06 | Mt3.5 | Root and nodule specific | Yes |
| Medtr8g060730 | F-box like 1 (FBL1-like) | 2 | 4.26E-08 | Mt3.5 | Below detection limit |
Figure 1.
GWA study of nodulation candidate regions in M. truncatula. From left to right, Manhattan plots extending 300,000 bp either side of the candidate SNP for each of the six candidate gene regions.
Generation of Whole-Plant Mutants
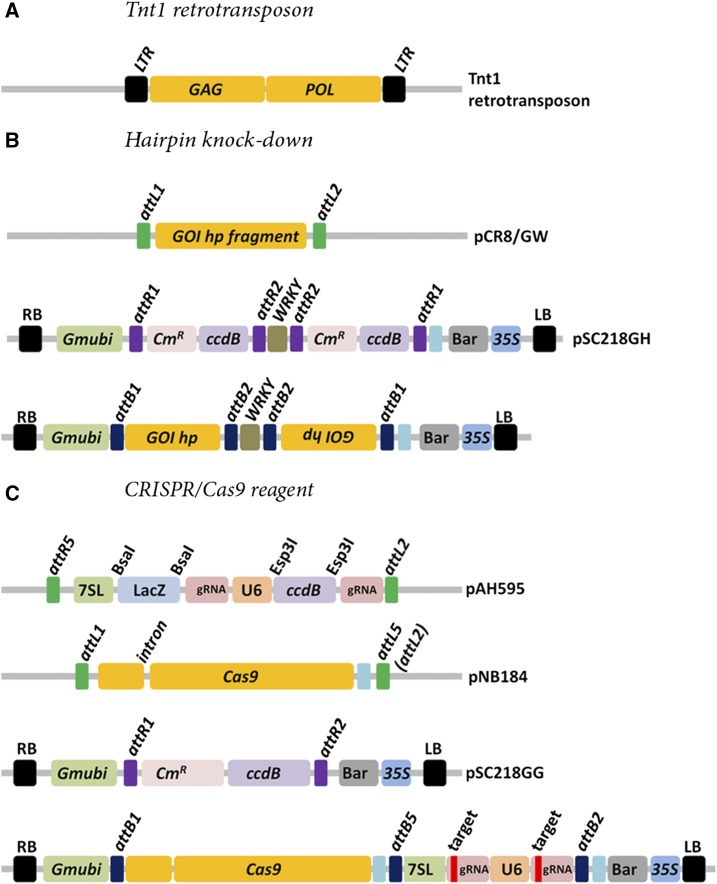
To empirically evaluate the functional importance of GWA-identified candidate genes underlying naturally occurring variation in nodulation, we generated a suite of whole-plant mutants using three mutagenesis platforms: Tnt1 retrotransposons, RNA hairpin knockdown, and CRISPR/Cas9 (SSNs; Fig. 2, A–C). We identified Tnt1 insertions in seven candidates by either querying the Noble Foundation Tnt1 database (Tadege et al., 2008) or through PCR screening of pools of Tnt1 mutant lines (Supplemental Table S1). For each of the seven putative Tnt1 mutants, we used a PCR assay with gene and insertion-specific primers to identify lines that were either homozygous for the Tnt1 insertion or wild type at the respective candidate locus, hereafter referred to as pho2-likeTnt, fbl1-likeTnt, pno1-likeTnt, fmo1-likeTnt, rfp1-likeTnt, hlz1-likeTnt, and pen3-likeTnt (Fig. 3, A–C; Supplemental Figs. S1 and S9). We then used homozygous insertion and wild-type lines identified in this screen as parent plants to generate self-pollinated seeds for use in phenotype assays described below. Preliminary analyses revealed that the fbl1Tnt line as well as the wild-type line derived from the same grandparent plant were also segregating for a supernodulating phenotype (>100 nodules on root systems <5 cm long); therefore, these were excluded from further analyses.
Figure 2.
Mutagenesis platforms. A, Tnt1 retrotransposon insertional mutagenesis in M. truncatula was developed in the R108 (HM340) accession. The original transformant harbored five Tnt1 copies. The Tnt1 element can transpose by a copy and paste mechanism during callus regeneration, leading to progeny lines with random stable insertions. B, Schematic representation of the hairpin entry and destination vectors. Typically, a 300- to 500-bp amplicon from the candidate gene of interest is amplified from M. truncatula whole-plant cDNA template and cloned into the pCR8/GW entry vector. The entry vector is then used to clone the hairpin fragment into the pSC218GH binary vector, followed by A. tumefaciens transformation. C, The construction of the CRISPR/Cas9 reagent requires a multisite Gateway cloning reaction utilizing two entry vectors and a destination binary vector. The CRISPR targets are cloned into pAH595 and along with the Cas9 entry vector (pNB184) are cloned into pSC218GG vector, followed by A. tumefaciens transformation. A detail protocol of the CRISPR/Cas9 assembly is provided in the Supplemental Methods.
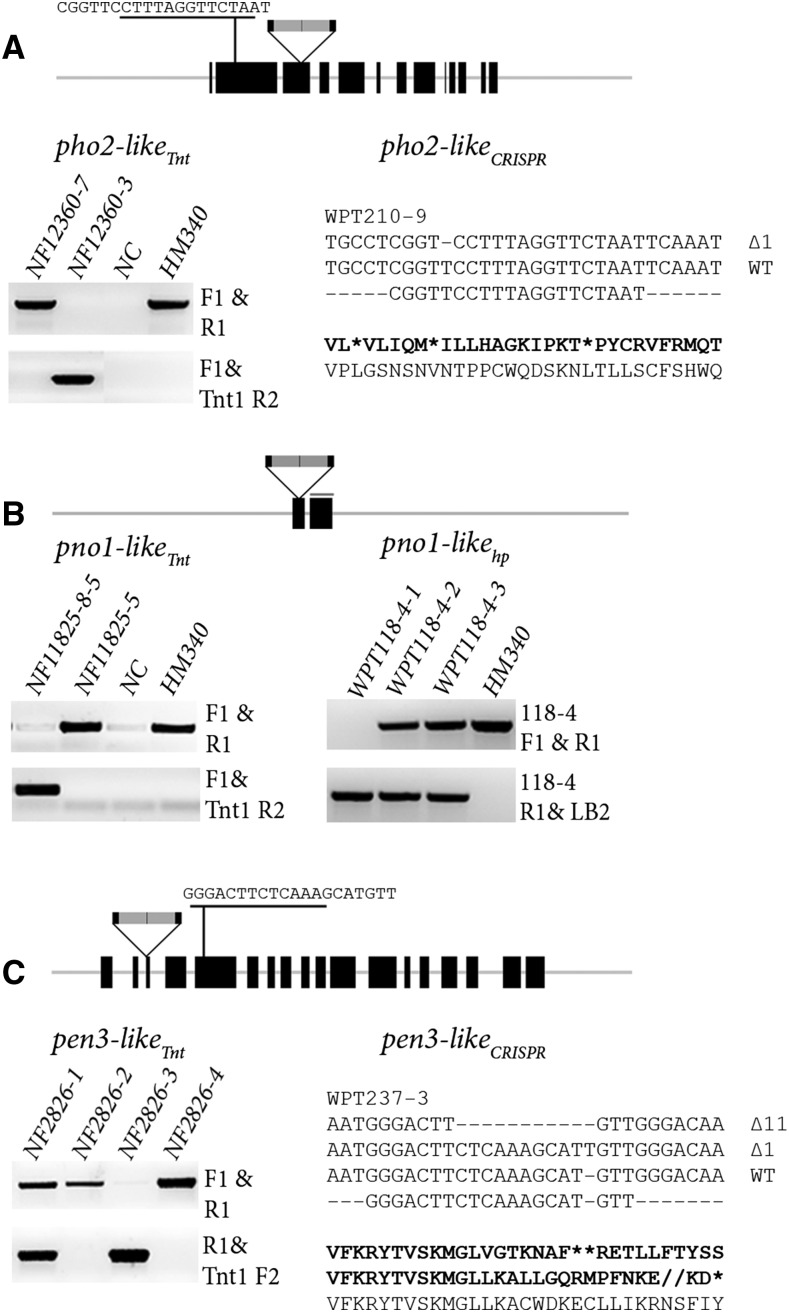
Figure 3.
Screening and identification of candidate mutant alleles. A, Two independent PHO2-like mutant alleles, pho2-likeTnt and pho2-likeCRISPR. Seeds from the Tnt1 line NF12360 were screened by PCR assay to determine homozygous and null insertions at the PHO2-like locus, identifying null insertion NF12360-7 plant and homozygous mutant NF12360-3 (Supplemental Fig. S9). A second mutant allele for PHO2-like was identified using plants transformed with a CRISPR/Cas9 reagent that targeted the open-reading frame of PHO2-like. Several homozygous T0 mutant plants were identified, and the WPT210-9 plant was selected for downstream phenotype analyses. WPT210-9 had a 1-bp deletion in the coding region of both alleles, effectively disrupting the reading frame as indicated by the three stop codons depicted in the amino acid sequence. Heritable transmission of the 1-bp mutation was confirmed by screening T1 plants by a PCR-digestion assay and sequencing. B, Two independent PNO1-like mutant alleles, pno1-likeTnt and pno1-likeHP. Tnt1 mutant alleles were identified for the PNO1-like gene by screening seed from the NF11825 line. Pno1-like mutant and null insertion lines were identified: NF11825-8 and NF11825-5, respectively. Since an insufficient number of seed was recovered from the NF11825-8 plant for phenotype analysis, seed from the next generation (NF11825-8-5) was used as wild-type plant. For the second mutant allele, a hairpin line was identified. TAIL-PCR was used to identify the genomic location of the transgene, and a PCR assay using both the genome location and transgene specific primers was used to identify a plant homozygous for the hairpin transgene (WPT118-4-1). C, Two independent PEN3-like mutant alleles, pen3-likeTnt and pen3-likeCRISPR. Seed from the NF2826 Tnt1 line were used to identify homozygous (NF2826-3) and null (NF2826-2) insertion mutant plants using both PEN3-like and Tnt1 specific primers. For the second mutant allele, we identified several CRISPR/Cas9 homozygous T0 mutant plants such as WPT237-3, which had a segregating a biallelic mutation consisting of a 11-bp deletion and a 1-bp insertion. Screening of WPT237-3 T1 mutant plants confirmed heritable transmission of both mutations.
We also attempted to generate hairpin knockdowns for all 10 candidates and successfully recovered multiple independently transformed whole-plant mutant lines for four: PHO2-like, FBL-like, PNO1-like, and ERDJ2. Using TAIL-PCR, we were able to determine the genomic location of hairpin transgene and identify homozygous hairpin lines for three of these genes, hereafter referred to as fbl-likeHP, pno1-likeHP, and erdj2HP (Fig. 3B; Supplemental Fig. S2, A–C). These lines were grown to produce self-pollinated seed that were used for phenotype assays.
Because hairpin technologies are expected to reduce but not eliminate expression of target genes, we also targeted all 10 candidates for whole-plant knockouts using CRISPR/Cas9 and successfully generated whole-plant knockout lines for seven candidate genes: PHO2-like, FMO1-like, ERDJ2, ACRE1, HLZ1-like, MEL1, and PEN3-like (Fig. 3, A–C; Supplemental Figs. S3–S7). We recovered multiple T0 homozygous mutant plants with mutation efficiencies ranging from 50 to 70% for many candidates genes, including PHO2-like, FMO1-like, ERDJ2, and PEN3-like candidates, whereas germ-line mutations in ACRE1, HLZ1-like, MEL1 mutant plants had to be confirmed in later generations (Supplemental Figs. S3–S7). The CRISPR/Cas9 reagent was useful for mutating smaller sized genes (<500 bp) such as Acre1, since the mutagenesis of these genes is typically more challenging using random mutagenesis strategies. Indeed, we could not identify a Tnt1 insertion in this gene by querying the Noble database and were also unsuccessful using the Tnt1 PCR-reverse-screen (Supplemental Fig. S7A). By contrast, using the CRISPR system, we generated several Acre1 candidate mutants and could generate mutations at the ACRE2 paralog located 16 kb upstream of ACRE1 (Supplemental Fig. S7, B and C). For the ERDJ2 candidate, we recovered two T0 homozygous mutant plants, one with a biallelic 3-bp and 85-bp frame-shift deletion and a second plant with two 6-bp mutant alleles (Supplemental Fig. S4) and tested both mutants, since we suspected the plant might be homozygous lethal. We hereafter referred to the CRISPR mutant lines as pho2-likeCRISPR, fmo1-likeCRISPR, erdj2CRISPR-3/85, erdj2CRISPR-6, acre1-likeCRISPR, hlz1-likeCRISPR, mel1CRISPR, and pen3-likeCRISPR. We were unable to confirm homozygous mutant plants for the remaining three candidates (FBL1-like, PNO1-like, and RFP1-like). The apparent poor mutation rate of these three targets might be due to errors in the sequence used to construct the CRISPR reagent. However, we grew all of the stable transformants to produce self-pollinated seeds that were then used for phenotype assays.
Mutation and Knockdown of Gene Candidates Affect Nodulation
We assayed nodule production in 17 mutant and paired control lines in a series of randomized and well-replicated phenotype experiments. The assays were performed with an average of 22 mutant and 22 wild-type plants, including six candidates tested with two independent mutation events to evaluate whether results were robust to the specific mutation and genomic background.
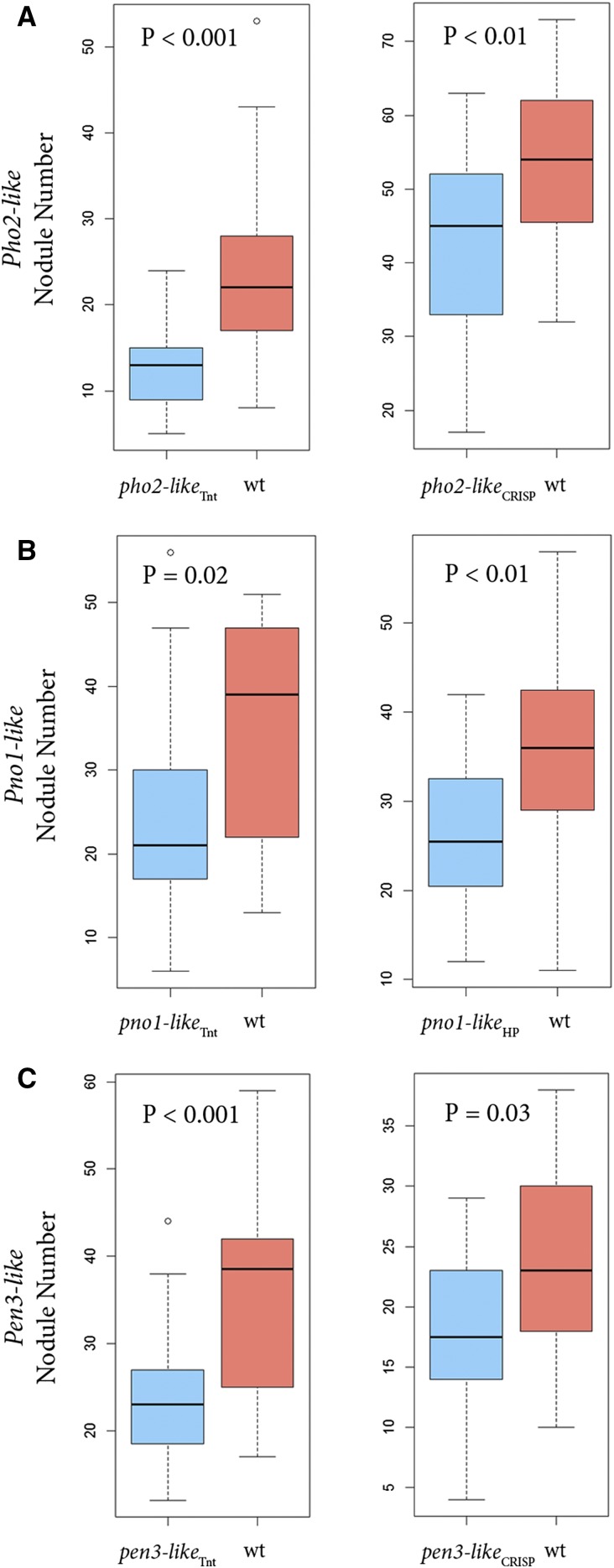
Phenotype assays revealed that lines carrying mutations in three of the candidates, PHO2-like (4g020620), PNO1-like (5g088410), and PEN3-like (2g101090), produced significantly fewer nodules than control plants (P < 0.03 for each mutant by wild-type line comparison; Fig. 4, A–C; Supplemental Table S2). In each of these cases, the phenotypic effects were validated by two independent mutations (Fig. 3). The PHO2-like mutants were generated by the insertion of a Tnt1 element (pho2-likeTnt) in one plant and a CRISPR/Cas9 generated mutant harboring a homozygous 1-bp frame-shift mutation (pho2-likeCRISPR) in the other. PNO1-like mutant plants were generated by insertion of a Tnt1 element (pno1-likeTnt) and a plant homozygous for a hairpin transgene targeting the PNO1-like transcript (pno1-likeHP). Similarly, two PEN3-like mutants were identified by Tnt1 insertion (pen3-likeTnt) and a CRISPR/Cas9 induced 11-bp and 1-bp biallelic frame-shift mutation in each allele (pen3-likeCRISPR; Fig. 3, A–C).
Figure 4.
Nodule number produced by mutant and wild-type plants. Nodules were counted 32 to 42 dpi. Data are shown for two independent mutants for each of three candidate genes. A, PHO2-like; B, PNO1-like; C, PEN3-like. For each figure, the horizontal black bar represents the median value, the box the middle quartiles, and the whiskers the range of data, except for outliers shown as a single data point.
Surprisingly, two Tnt1 insertion lines (fmo1-likeTnt and hlz1-likeTnt) produced significantly more nodules than control lines (Supplemental Table S2). However, second mutations in these candidates, both generated using CRISPR/Cas9, exhibited results inconsistent with the Tnt1 mutants (Pmut_wt x assay < 0.05). Indeed, the number of nodules formed by lines carrying CRISPR mutations in these genes did not differ from controls. Potentially, the greater nodule number in the Tnt1 lines may have been due to Tnt1 insertions in nontarget genes (two of the control Tnt1 plants when assaying the hlz1-likeTnt had no nodules and five others had ≤10 nodules, many fewer than expected). Consequently, we decided to focus on the three candidates with mutants showing reproducibly reduced nodule numbers compared to controls (PHO2-like, PNO1-like, and PEN3-like) for further analysis.
Nitrogen Content and Nodule Morphology of Validated Candidate Lines
Although plants with mutations in the three validated candidate genes (PHO2-like, PNO1-like, and PEN3-like) produced significantly fewer nodules, nitrogen concentration and nodule morphology were not affected. Foliar nitrogen concentrations did not differ significantly between any of the mutants and their control (t tests, all Pdf = 4 > 0.1; Supplemental Table S2). Moreover, longitudinal sections of nodules from all validated mutant lines indicated they were structurally sound, with distinct meristematic, infection, and pink nitrogen fixation zones (Supplemental Figs. S10 and S11). Both pho2-liketnt1 mutants and their corresponding wild-type controls had lighter pink nodules overall; however, pho2-likeCRISPR nodules exhibited similar color intensity compared with the HM340 control (Supplemental Fig. S11). This suggests that differences in the pink color of Pho2-liketnt1 are probably due to the Tnt1 background rather than disruption of the PHO2-like gene itself.
Effects of Forward Genetic Mutants
To better understand the quantitative nature of nodulation mutants, we compared effects of the validated GWA mutants to nodulation mutants in genes originally discovered through forward genetics. Considering only “confirmed” candidates from our GWA study (PHO2-like, PNO1-like, and PEN3-like), GWA mutants produced approximately 60% of the number of nodules on wild-type plants. These results were then compared to phenotype assays on two well-known and previously characterized nodulation mutant lines: the calcium and calmodulin-dependent kinase dmi3 (Catoira et al., 2000) and ipd3-2, which encodes for a protein that interacts with DMI3 (Messinese et al., 2007). Under exactly the same experimental conditions used to test the GWA mutant lines, these forward genetics mutants exhibited drastic reductions in nodule number, an average of just 0.5 nodules on dmi3 lines and 1.5 nodules on ipd3 lines compared with 39 on wild-type plants (P < 0.0005; Supplemental Fig. S8).
DISCUSSION
GWA analyses provide an opportunity for identifying functionally important genes that contribute to naturally occurring variation in quantitative traits, genes that are unlikely to be detected by forward genetic screens. However, the approach is challenged by several statistical issues, including an expected high number of statistical false positives due to the large number of statistical tests and the difficulty of fully eliminating confounding effects of relatedness among assayed individuals. This means that GWA is best viewed as a statistical approach for identifying genomic regions that are likely to contribute to variation. Moving beyond such a candidate status requires empirical validation of gene function (Ioannidis et al., 2009). In this work, we applied a suite of mutagenesis platforms, retrotransposon insertions, hairpin knockdown RNAi constructs, and CRISPR/Cas9, to disrupt the function of 10 genes that GWA had identified as candidates in contributing to variation in the legume-rhizobia symbiosis through nodulation (Stanton-Geddes et al., 2013). Our evaluation experiments revealed that mutations in 3 of the 10 candidate lines produced significantly fewer nodules than wild-type plants, thereby identifying new genes contributing to quantitative variation in nodule production.
Three of 10 candidate genes were empirically validated in this study. However, there was more than one candidate gene for some genomic intervals. Therefore, the validation rate is better represented as three validated genes among the six intervals evaluated. These findings demonstrate that GWA coupled with well-replicated validation experiments is an effective means to identify genes contributing to quantitative variation. The high rate of success in validating the functional importance of GWA candidates is likely due to at least two reasons. First, in creating an initial list of candidates for functional evaluation, we did not limit our efforts to only those variants with the strongest statistical support from the GWA. Rather, we considered the 100 SNPs with the strongest statistical associations with phenotypic variation as potential candidates and then applied genetically informed criteria for identifying which candidates to pursue for validation. These criteria included SNPs in or near annotated genes, genes expressed in roots or nodules, and SNPs that formed clusters of statistical associations (peaks in the Manhattan plots; Fig. 1).
Despite the criterion that chosen SNPs either were in or near a coding region, only four of the GWA variants that we pursued were actually in annotated coding regions, with the remainder anywhere from 168 to 5,819 bp from annotated coding sequence. Our data do not allow us to determine whether the SNPs near, but not in, coding regions cause functionally important variation in gene expression as opposed to linkage-disequalibrium with unassayed SNPs in coding regions. Nevertheless, noncoding SNPs are often associated with phenotypic variation (Hindorff et al., 2009) through effects on gene expression (Cookson et al., 2009; Cubillos et al., 2012). We also prioritized candidates that showed expression in root or nodule tissue, although such a criterion might have biased against lowly expressed genes (like many transcription factors) or gene products expressed elsewhere in the plant affecting nodulation (Krusell et al., 2002). The third criterion we applied, that candidates be physically proximate to other SNPs showing strong associations, is likely to favor alleles that are maintained in contemporary populations through balancing selection where the optimal trait value varies among populations or that have recently increased in frequency (Barton and Turelli, 1989; Yeaman and Whitlock, 2011).
Also central to our evaluation experiments was the use of well-replicated experiments when evaluating phenotypic effects. Each of the phenotypic assays we conducted involved an average of 22 mutant and 22 wild-type (minimum of seven mutant and six wild-type) individuals. This was important because the distributions of mutant and wild-type phenotypes often showed considerable overlap (Fig. 4, A–C). The importance of replication also helps to explain why the genes we identified had not been detected in earlier forward-genetic screens conducted on nodulation in M. truncatula (Catoira et al., 2000; Oldroyd and Long, 2003; Schnabel et al., 2005; Pislariu et al., 2012; Domonkos et al., 2013). If the mutations we evaluated had been tested in only a single individual, the mutant phenotype would likely have fallen into the range of phenotypes exhibited by wild-type plants generally and thus would not have been picked up in a standard genetic screen. The phenotypic effects of genes identified through forward genetic screens relative to those we validated here is highlighted by the comparison of dmi3 and ipd3 to the three GWA-selected candidates: dmi3 and ipd3 plants produced an average of approximately 2% of the number of nodules produced on wild-type plants, whereas the validated genes first identified as candidates through GWA produced roughly 60% of the number of nodules compared with wild-type plants.
Genetics of Nodulation
Although we identified candidates without prior knowledge of their function, the molecular functions of homologs for the validated candidates are known, and all are interesting in the context of nodulation. One of the validated candidates, PHO-like (Medtr4g020620), suggests a potentially important role for phosphorous in quantitative variation in nodulation. PHO2-like is annotated as encoding an E2 ubiquitin conjugating enzyme, and its Arabidopsis (Arabidopsis thaliana) ortholog (35% amino acid identity, E value = 4e-104) is involved in phosphorous accumulation (Park et al., 2014). Not only can phosphorous availability affect nodulation and N2 fixation (Graham and Rosas, 1979; Jakobsen, 1985; Graham, 1992; Sulieman, 2013) but in Arabidopsis, AtPHO2 ortholog interacts directly with the E3 product of Nitrogen Limitation Adaption, which mediates plant responses to N limitation (Park et al., 2014). Moreover, E2 ubiquitin conjugate enzymes are presumed to interact with multiple E3 partners that could lead to the proteolytic degradation of a larger range of uncharacterized substrates (Smalle and Vierstra, 2004). M. truncatula PHO2-like is expressed at modest levels throughout nodule development (and also in uninoculated roots) but reaches a distinct peak in expression at 20 d postinoculation (dpi; Supplemental Fig. S12). This is a timepoint when nodules are fully differentiated, actively fixing nitrogen, and still prior to the initiation of senescence.
A second interesting validated gene is the annotated ABC transporter (PEN3-like; Medtr2g101090). ABC transporters are involved in a wide range of plant activities, including pathogen metabolite detoxification, nonhost pathogen resistance, stomatal function, and Ca2+ sensing associated plant immunity (Rea, 2007; Campe et al., 2016). The Arabidopsis gene with greatest similarity to Medtr2g101090 is PEN3 (66% identical amino acid sequence across 97% of the gene length; E-value = 0), which has been shown to be an essential component of salicylic acid-related defense against biotrophic fungi (Stein et al., 2006; Johansson et al., 2014). The formation of nodules requires direct interaction with bacteria, and there is growing evidence that defense responses can negatively affect nodule formation (Ramu et al., 2002; Lopez-Gomez et al., 2012; Larrainzar et al., 2015). Interestingly, ABC transporters have been shown to be significantly and highly up-regulated in several nodulation mutants relative to wild-type controls (Lang and Long, 2015). PEN3-like is strongly expressed in M. truncatula nodules throughout development, though even more highly expressed in roots, with a slight peak in nodule expression at 14 dpi (Supplemental Fig. S12).
The final validated candidate is annotated as encoding an RNA-binding PNO1-like protein (Medtr5g088410). How this protein might affect nodulation is not clear, although recent work has identified roles for members of this gene family in the regulation of miRNAs (Karlsson et al., 2015), suggesting that Medtr5g088410 could affect the expression of genes involved in nodulation. It is expressed at a comparatively low level in both nodules and roots with no distinct peak in time of expression (Supplemental Fig. S12).
Mutation Platforms for GWA Validation
Efficient means to validate the functional importance of GWA candidate genes are necessary for GWA studies to be effective in discovering genes of subtle effects, fulfilling the promise of linking molecular genetics with quantitative trait variation. Here, we applied multiple genomic technologies (retrotransposon mutagenesis, hairpins, and CRISPR/Cas9) to make this link. Each of these technologies has advantages and disadvantages when being applied as a reverse genetics tool. The M. truncatula Tnt1 collection has the advantage of ease: the Noble Foundation in Ardmore, OK has over 21,000 mutant lines that have largely saturated the genome with random Tnt1 insertion events, and the collection is easily searchable online. Although easy to use for validation experiments, there is the problem of background mutations (e.g. Supplemental Fig. S9). On average, there are 25 insertions in each Tnt1 line and as many as 97 independent Tnt1 loci have been reported (Veerappan et al., 2016). These background mutations can make phenotyping data potentially unreliable, complicating their use as the sole validation tool in reverse genetics phenotyping experiments. In our experiments, one Tnt1 line expressed an (unexpected) supernodulation phenotype, while two Tnt1 lines produced more nodules than control plants, though neither of these phenotypes could be validated with a second (non-Tnt1) mutation.
Both hairpin and CRISPR require more effort to create, involving construction of vectors, infecting plants with Agrobacterium tumefaciens, and regeneration of whole plants from callus. Despite these challenges, we had a high rate of success in creating stable whole-plant transformants using both hairpin and CRISPR/Cas9 at high efficiency and frequency. For example, CRISPR/Cas9 targeting the PHO2-like and PEN3-like genes induced homozygous mutations in multiple independent T0 plants (Supplemental Figs. S3–S7).
Hairpin technologies seem promising in two situations: when genes of interest are members of multigene families where gene redundancy might mask phenotype effects and for genes that are gametophytic or homozygous lethal. This is illustrated by the ERDJ2 candidate in our mutant screening. We identified a CRISPR/Cas9 ERDJ2 mutant with in-frame 3-bp and frame-shift 85-bp mutant alleles and confirmed that the gene is homozygous lethal by segregation analysis. PCR analysis of approximately 30 erdj2CRISPR_3/85 mutants failed to identify plants with both frame-shift 85-bp mutant alleles, suggesting that the combination of frame-shift mutations results in a homozygous lethal phenotype (Supplemental Fig. S4). Nevertheless, we successfully created a hairpin mutant that decreased transcript expression without the lethality of a knockout to use in candidate validation. For some genes, however, “knocking down” rather than “knocking out” expression could also limit the phenotypic effects of the mutations.
CRISPR technologies have advanced rapidly in recent years and hold promise for altering gene sequence and expression in many systems including plants (Baltes and Voytas, 2015; Luo et al., 2016). Although producing stable germ-line whole-plant mutants has proven challenging in some species, including Arabidopsis (Fauser et al., 2014; Feng et al., 2014), we were able to produce stable mutant lines with high efficiency. The mechanism behind this success rate is not clear, although we suspect that the transformation method might be a factor. M. truncatula transformation generates callus tissue that undergoes strong herbicide selection in contrast to floral dip transformation that requires infection of individual embryos. Moreover, we were often able to remove the CRISPR transgene from the mutant lines by genetic segregation to obtain nontransgenic mutant plants that can be used for future complementation studies. Given the rapid development of CRISPR multiplex platforms, it should soon be feasible to mutate and test large numbers of GWA identified candidates. Doing so would allow researchers to move beyond validating small numbers of candidates and allow robust empirical evaluation of the genes that control variation underlying complex quantitative traits.
MATERIALS AND METHODS
Insertional Mutant Screening and Genotyping
To identify mutants in the 10 candidates selected for validation, we first searched for Tnt1 insertional mutants using the Noble Foundation’s Medicago truncatula Mutant Database (http://medicago-mutant.noble.org/mutant/blast/blast.php; Tadege et al., 2008; Pislariu et al., 2012). Searches of this database, which is comprised of more than 21,000 mutants, identified insertions in five of the seven candidates with the remaining two candidates identified by a PCR reverse screen performed at the Noble Foundation (Supplemental Table S1). Seeds for Tnt1 mutant lines were obtained from the M. truncatula Tnt1 collection, planted, and genotyped for homozygous and wild-type (null) insertion alleles at the locus of interest. Mutant plants for Erdj2, Acre1, and Mel1 candidates could not be identified from either the Tnt1 database or the PCR reverse screen, and we therefore relied upon the CRISPR/Cas9 platform to generate these mutants.
Hairpin and CRISPR/Cas9 Transgenes and Whole-Plant Transformation
We attempted to create hairpin constructs to reduce gene expression in all 10 candidates and successfully identified homozygous hairpin transgenic plant lines for three: FBL1-like, PNO1-like, and ERDJ2. To create hairpins, targeted coding sequences were amplified by PCR using primer sequences (Supplemental Table S3) from cDNA derived from M. truncatula accession R108. These amplicons were cloned into the pCR8/GW/TOPO entry vector (Life Technologies) and incorporated into the hairpin destination vectors pSC218GH (see below) by a Gateway LR clonase reaction.
Finally, we generated CRISPR/Cas9 SSNs to introduce frame-shift mutations in all 10 candidates and recovered homozygous mutants for seven. Target sequences were identified by BLAST searches of the M. truncatula R108 (HM340) genome assembly (www.medicagohapmap.org/downloads/r108) using the Mt 4.0 reference sequence as a query. Resulting sequences were then used to query the sgRNA designer v.1 for selection of target guide RNAs (www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design-v1; Doench et al., 2014). The targets were created by a primer annealing assay and cloned into the Esp3I and BsaI sites of the U6/At7SL guide RNA entry vector (pAH595; Supplemental Table S3). The completed guide RNA entry vector sequence was confirmed and combined with a second entry vector that harbored the Arabidopsis (Arabidopsis thaliana) optimized Cas9 cassette (pNJB184) by a multisite Gateway LR clonase reaction into the pSC218GG destination vector (see below) for Agrobacterium tumefaciens transformation (Li et al., 2013; Baltes et al., 2014). A detailed protocol can be found in the Supplemental Methods.
Whole-plant transformation of R108 plants, the same genetic background used in the M. truncatula Tnt1 collection, with hpRNA and CRISPR/Cas9 constructs was carried out using a slightly modified version of an established protocol (Cosson et al., 2006). In brief, transformation was carried out by inoculating leaf tissue explants with A. tumefaciens strain EAH105 transformed with either a hairpin or CRISPR SSN. The transformations were conducted using a modified version of the binary vector backbone pNB96 (pSC218; Curtin et al., 2011) with an introduced inverted-repeat cassette to facilitate cloning of hairpin RNA fragments (pSC218GH) and a reading-frame cassette to incorporate both the Cas9 and target guide RNA entry vectors (pSC218GG). The inverted-repeat and reading-frame cassettes were derived from previously reported pTDT-DC-RNAi and pTDTO vectors (Valdés-López et al., 2008) and were driven by the constitutively expressed Glycine max Ubiquitin promoter (Hernandez-Garcia et al., 2010).
After cocultivating the leaf tissue with A. tumefaciens for 3 d, explants were washed in an antibiotic liquid media and transferred to a media for callus induction with biweekly transfers. After approximately 6 weeks, callus tissue was transferred to a shoot induction media and incubated in 16-h-light/8-h-dark photoperiod. Developing shoots were transferred to a rooting media to encourage root development before planting to soil. Both binary vectors pSC218GH and pSC218GG contain the 35S::BAR cassette for phosphinothricin herbicide selection of transgenic plants. T0 plants were rapidly screened using a previously reported DNA extraction method and PCR-CAPS assay to identify target mutations (Curtin et al., 2011).
Nodulation Assays
To evaluate the effects of candidate gene mutations on nodulation, we grew mutant and wild-type plants in well-replicated (an average of 22 mutant and 22 wild-type plants per assay; see Supplemental Table S2 for details) and completely randomized experiments. Prior to planting, seeds were treated in concentrated sulfuric acid for 5 min, washed with sterile deionized water, and then kept in the dark on wet filter paper at 4°C for 7to 10 d before planting into 65-mL containers with 4:1 parts autoclaved sand:perlite mixture. Five days after planting, seedlings were inoculation with a 30-mL suspension (OD600 ∼0.015) of Sinorhizobium meliloti strain Sm2011 carrying the hemA::LacZ reporter (Ardourel et al., 1994). Plants were grown at 22°C to 25°C, 75% humidity, and 200 to 350 μmol m−2s−1 on a 16-h-light/8-h-dark photoperiod, fertilized 15 and 21 d postinoculation with Fähreus nutrient solution, and otherwise watered as needed. Thirty-five to 41 d after inoculation, plants were harvested and nodule number was determined. Each mutant was compared against a separate set of wild-type control plants. To reduce the effects of background insertions, controls for the Tnt1 mutants were grown from seeds from a full-sib of the mutant plants. R108 seeds were used for the control treatment for hairpin and CRISPR lines. For each assay experiment, we tested for statistically significant differences between the phenotype of the mutant line and the phenotype of the wild-type control line using a t test using R (R Development Core Team, 2013). For the six candidates (PHO2-like, PNO1-like, FMO1-like, ERDJ2, HLZ1-like, and PEN3-like) where we evaluated two independent mutation lines, we also used ANOVA to test for between-line differences in nodulation. Similarly, for the two mutant lines (pho2CRISPR and mel1CRISPR) for which the phenotypic assays were repeated, we used ANOVA to test for differences between mutant and wild-type lines between assays. All data are available in the DRYAD repository at http://dx.doi.org/10.5061/dryad.3t4g7.
Nodule Morphology and Nitrogen Content
To complement phenotypic data collected during initial validation experiments, we also grew the three mutants exhibiting validated nodulation phenotypes (PHO2-like, PNO1-like, and PEN3-like) to test for differences in foliar nitrogen concentration and examine nodule morphology. These plants were grown under the same conditions as those for initial phenotype assays. At 14 dpi, three plants per treatment were harvested to make visual observations of nodules. Plants were washed to remove surrounding substrate, and 10 cm of roots with nodules was isolated to obtain close-up pictures of nodules (Supplemental Fig. S10). One nodule (second or third from the top) was then harvested from each plant, sliced in half longitudinally by hand, and then observed under a stereo microscope (Supplemental Fig. S11). At 28 dpi, four plants grown in parallel were harvested for nitrogen concentration assays, which were conducted by the University of Minnesota Research Analytical Laboratory using the Dumais method (Matejovic, 1995).
Accession Numbers
HM340 (R108) genomic sequence data for Pho2-like, Pno1-like, and Pen3-like genes can be found in the GenBank/EMBL data libraries under accession numbers KY458051, KY458052, and KY458053, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The characterization of three Tnt1 mutant lines.
Supplemental Figure S2. The characterization of two whole-plant hairpin lines.
Supplemental Figure S3. The characterization of the Pho2-like CRISPR/Cas9 mutants.
Supplemental Figure S4. The characterization of the Erdj2 CRISPR/Cas9 mutants.
Supplemental Figure S5. The characterization of the Acre-1 CRISPR/Cas9 mutants.
Supplemental Figure S6. The characterization of the Pen3-like CRISPR/Cas9 mutants.
Supplemental Figure S7. The characterization of the Fmo1-like, Hlz-1, and Mel1-like CRISPR/Cas9 mutants.
Supplemental Figure S8. Nodulation phenotype of dmi3 and ipd3-2.
Supplemental Figure S9. The Tnt1 insertion background of the NF12360 Tnt1 mutant line.
Supplemental Figure S10. Wild-type and Pho2-like, Pen3-like, and Pno1-like mutant nodules at 14 dpi.
Supplemental Figure S11. Wild-type and Pho2-like, Pen3-like, and Pno1-like mutant dissected nodules at 14 dpi.
Supplemental Figure S12. The expression analysis of validated genes, derived from the Medicago truncatula Gene Expression Atlas (http://mtgea.noble.org/v3/index.php).
Supplemental Table S1. List of Tnt1 mutant lines obtained from Noble Foundation Tnt1 mutant database.
Supplemental Table S2. Statistical tests comparing phenotype between mutant and wild-type control plants.
Supplemental Table S3. Primer sequences for PCR assays and vector construction.
Supplemental Methods S1. CRISPR reagent design and construction.
Supplementary Material
Acknowledgments
We thank Catalina Pislariu, Jianqi Wen, and Sarah Hoerth for assistance with mutant characterization; Nathan Henning for plant care assistance; Aaron Hummel for the pAH595 vector; Ryan Morrow for technical assistance with plant transformation and media preparation; and Jean-Michel Ané for dmi3 and ipd3 mutant seeds.
Glossary
- dpi
days postinoculation
- GWA
genome wide association
- SNP
single nucleotide polymorphism
- SSN
site-specific nuclease
Footnotes
This work was supported by National Science Foundation Award IOS-1237993.
Articles can be viewed without a subscription.
References
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Voytas DF (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33: 120–131 [DOI] [PubMed] [Google Scholar]
- Barton NH, Turelli M (1989) Evolutionary quantitative genetics: how little do we know? Annu Rev Genet 23: 337–370 [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Campe R, Langenbach C, Leissing F, Popescu GV, Popescu SC, Goellner K, Beckers GJM, Conrath U (2016) ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytol 209: 294–306 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V, Durand P, d’Erfurth I, Kondorosi A, Ratet P (2006) Medicago truncatula transformation using leaf explants. Methods Mol Biol 343: 115–127 [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Coustham V, Loudet O (2012) Lessons from eQTL mapping studies: non-coding regions and their role behind natural phenotypic variation in plants. Curr Opin Plant Biol 15: 192–198 [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Zhang F, Sander JD, Haun WJ, Starker C, Baltes NJ, Reyon D, Dahlborg EJ, Goodwin MJ, Coffman AP, et al. (2011) Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol 156: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32: 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos A, Horvath B, Marsh JF, Halasz G, Ayaydin F, Oldroyd GED, Kalo P (2013) The identification of novel loci required for appropriate nodule development in Medicago truncatula. BMC Plant Biol 13: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359 [DOI] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang D-L, Wang Z, Zhang Z, Zheng R, Yang L, et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen ML, von Wettberg EJ, Badri M, Moriuchi KS, Barhoumi F, Chang PL, Cuellar-Ortiz S, Cordeiro MA, Vu WT, Arraouadi S, et al. (2014) The ecological genomic basis of salinity adaptation in Tunisian Medicago truncatula. BMC Genomics 15: 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH. (1992) Stress tolerance in Rhizobiaum and Bradyrhizobium, and nodulation under adverse soild conditions. Can J Microbiol 38: 475–484 [Google Scholar]
- Graham PH, Rosas JC (1979) Phosphorus fertilization and symbiotic nitrogen fixation in common bean. Agron J 71: 925–926 [Google Scholar]
- Hernandez-Garcia CM, Bouchard RA, Rushton PJ, Jones ML, Chen X, Timko MP, Finer JJ (2010) High level transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biol 10: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JPA, Thomas G, Daly MJ (2009) Validating, augmenting and refining genome-wide association signals. Nat Rev Genet 10: 318–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen I. (1985) The role of phosphorus in nitrogen fixation by young pea plants (Pisum sativum). Physiol Plant 64: 190–196 [Google Scholar]
- Johansson ON, Fantozzi E, Fahlberg P, Nilsson AK, Buhot N, Tör M, Andersson MX (2014) Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J 79: 466–476 [DOI] [PubMed] [Google Scholar]
- Karlsson P, Christie MD, Seymour DK, Wang H, Wang X, Hagmann J, Kulcheski F, Manavella PA (2015) KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc Natl Acad Sci USA 112: 14096–14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Lang C, Long SR (2015) Transcriptomic analysis of Sinorhizobium meliloti and Medicago truncatula symbiosis using nitrogen fixation-deficient nodules. Mol Plant Microbe Interact 28: 856–868 [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla-Garcia N, Yu H-J, Hwang H-J, Oh M, Kim GB, Surendrarao AK, Chasman D, et al. (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between Nod factor and ethylene signals. Plant Physiol 169: 233–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gomez M, Sandal N, Stougaard J, Boller T (2012) Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J Exp Bot 63: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Gilbert B, Ayliffe M (2016) Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants. Plant Cell Rep 35: 1439–1450 [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10: 565–577 [DOI] [PubMed] [Google Scholar]
- Matejovic I. (1995) Total nitrogen in plant material determined by means of dry combustion: a possible alternative to determination by Kjedahl digestion. Commun Soil Sci Plant Anal 26: 2217–2229 [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua N-H (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J, Cosson V, Muni RRD, Wang M, Benedito VA, Andriankaja A, Cheng X, Jerez IT, et al. (2012) A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol 159: 1686–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Ott T (2011) Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol 14: 458–467 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
- Ramu SK, Peng H-M, Cook DR (2002) Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol Plant Microbe Interact 15: 522–528 [DOI] [PubMed] [Google Scholar]
- Rea PA. (2007) Plant ATP-binding cassette transporters. Annu Rev Plant Biol 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stanton-Geddes J, Paape T, Epstein B, Briskine R, Yoder J, Mudge J, Bharti AK, Farmer AD, Zhou P, Denny R, et al. (2013) Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PLoS One 8: e65688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou B-H, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman S, Ha CV, Schulze J, Tran LS (2013) Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J Exp Bot 64: 2701–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, et al. (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ 31: 1834–1843 [DOI] [PubMed] [Google Scholar]
- Vance CP. (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127: 390–397 [PMC free article] [PubMed] [Google Scholar]
- Veerappan V, Jani M, Kadel K, Troiani T, Gale R, Mayes T, Shulaev E, Wen J, Mysore KS, Azad RK, et al. (2016) Rapid identification of causative insertions underlying Medicago truncatula Tnt1 mutants defective in symbiotic nitrogen fixation from a forward genetic screen by whole genome sequencing. BMC Genomics 17: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC (2011) The genetic architecture of adaptation under migration-selection balance. Evolution 65: 1897–1911 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.