Integration of transcriptomic and proteomic data provides complementary evidence for grape responses to high temperature at transcriptional, posttranscriptional, and translational levels.
Abstract
Heat stress is one of the primary abiotic stresses that limit crop production. Grape (Vitis vinifera) is a cultivated fruit with high economic value throughout the world, with its growth and development often influenced by high temperature. Alternative splicing (AS) is a widespread phenomenon increasing transcriptome and proteome diversity. We conducted high-temperature treatments (35°C, 40°C, and 45°C) on grapevines and assessed transcriptomic (especially AS) and proteomic changes in leaves. We found that nearly 70% of the genes were alternatively spliced under high temperature. Intron retention (IR), exon skipping, and alternative donor/acceptor sites were markedly induced under different high temperatures. Among all differential AS events, IR was the most abundant up- and down-regulated event. Moreover, the occurrence frequency of IR events at 40°C and 45°C was far higher than at 35°C. These results indicated that AS, especially IR, is an important posttranscriptional regulatory event during grape leaf responses to high temperature. Proteomic analysis showed that protein levels of the RNA-binding proteins SR45, SR30, and SR34 and the nuclear ribonucleic protein U1A gradually rose as ambient temperature increased, which revealed a reason why AS events occurred more frequently under high temperature. After integrating transcriptomic and proteomic data, we found that heat shock proteins and some important transcription factors such as MULTIPROTEIN BRIDGING FACTOR1c and HEAT SHOCK TRANSCRIPTION FACTOR A2 were involved mainly in heat tolerance in grape through up-regulating transcriptional (especially modulated by AS) and translational levels. To our knowledge, these results provide the first evidence for grape leaf responses to high temperature at simultaneous transcriptional, posttranscriptional, and translational levels.
Heat stress is one of the main abiotic stresses limiting crop production worldwide. Global warming is predicted to be accompanied by more frequent and powerful extreme temperature events (Lobell et al., 2008). Therefore, a better understanding of the response of plants to heat stress is essential for improving their heat tolerance. In general, heat stress is a complex function of intensity (temperature in degrees), duration, and rate of increase in temperature. At very high temperatures, a catastrophic collapse of cellular organization may occur within minutes, which results in severe cellular injury and even cell death (Wahid et al., 2007). At moderately high temperatures, direct injuries include protein denaturation and aggregation and the increased fluidity of membrane lipids. Indirect or slower heat injuries include inactivation of enzymes, inhibition of protein synthesis, protein degradation, and loss of membrane integrity. These injuries eventually lead to a decline of net photosynthetic rate, reduced ion flux, production of toxic compounds and reactive oxygen species, and inhibition of growth. Actually, plants are not passively damaged by heat stress but respond to temperature changes by reprogramming their composition of certain transcripts, proteins, and metabolites. Such changes are aimed at establishing a new steady-state balance of biological processes that enable the organism to function, survive, and even reproduce at a higher temperature (Bokszczanin, 2013).
In general, most previous studies about heat stress in plants have focused on physiological changes (Bita and Gerats, 2013). With the advancement of biotechnology, more and more studies have investigated transcriptomic or proteomic changes. Lim et al. (2006) found that the expression of 165 genes changed, especially those of heat shock proteins (HSPs), in Arabidopsis (Arabidopsis thaliana) suspension cells during acclimation at a moderate heat. With cDNA microarrays and semiquantitative RT-PCR techniques, Frank et al. (2009) found that HSP70, HSP90, and the heat shock transcription factors (HSF) HSFA2 and HSFA3 were important to tomato (Solanum lycopersicum) microspore resistance to heat stress. González-Schain et al. (2016) conducted genome-wide transcriptomic analyses during anthesis and revealed new insights into the molecular basis of heat stress responses in tolerant and sensitive rice (Oryza sativa) varieties. In addition, proteomic responses to heat stress have been studied in some species, and various organs have been analyzed (Kosová et al., 2011). The accumulation or decrease of specific proteins has been documented by comparison of heat-sensitive and heat-tolerant vegetative tissues, and the significance of certain factors for conferring thermotolerance has been confirmed by genetic engineering (Grover et al., 2013). Although gene expression analyses have offered insights into the mechanisms underlying the high-temperature stress response, there is frequently a poor correlation between transcript and protein levels in data from heat-treated samples. In recent years, a new technique termed iTRAQ (isobaric tags for relative and absolute quantitation) has been applied for proteomic quantification to overcome some of the limitations of two-dimensional gel-based techniques. This technique has a high degree of sensitivity. The amine-specific isobaric reagents of iTRAQ allow the identification and quantification of up to eight different samples simultaneously (Sankaranarayanan et al., 2013). Therefore, it is possible to integrate transcriptomic and proteomic data to reveal the response of plants to heat stress (Wang et al., 2015).
Grape (Vitis vinifera) is a popular cultivated fruit throughout the world and represents one of the most important crops with a high economic value. Extreme weather events and temperature shifts create an inimical environment for grape crops, inhibiting physiological activities in plants and resulting in reduced yield and quality. High-temperature events are occurring more often, and the maximum midday air temperature is often 40°C, even exceeding 45°C, in many regions, which causes huge economic losses (Salazar-Parra et al., 2010; Pillet et al., 2012). Although our understanding of the response and adaptation of grape to high temperature is limited and has focused mostly on grape morphological and physiological changes, increasing data on mRNA expression and the proteome during heat stress responses in grape have provided novel insights into the underlying molecular mechanism. Transcriptomic analysis of grape leaves under heat stress and subsequent recovery showed that the effect of heat stress and recovery on grape appears to be associated with multiple processes and mechanisms, including stress-related genes, transcription factors, and metabolism (Liu et al., 2012). Carbonell-Bejerano et al. (2013) revealed that the establishment of a thermotolerance response in berries under high temperatures was marked by the induction of HSPs and the repression of transmembrane transporter-encoding transcripts using the GrapeGen GeneChip. George et al. (2015) reported on proteome changes in cv Cabernet Sauvignon grape cells exposed to sudden high-temperature stresses, with more than 2,000 proteins identified and quantified using spectral counting of the entire data set. Wu et al. (2015) investigated changes of fruit quality and protein expression profiles of grape berries upon hot water treatment during the subsequent 45 d of cold storage, finding immediate increased expression of proteins associated with carbohydrate and energy metabolism. Liu et al. (2014) combined a physiological analysis with iTRAQ-based proteomics of cv Cabernet Sauvignon grape leaves subjected to 43°C stress for 6 h. They proposed that some proteins related to the electron transport chain of photosynthesis, antioxidant enzymes, HSPs, and other stress response proteins, as well as glycolysis, may play key roles in enhancing grapevine adaptation to and recovery capacity from heat stress. However, these transcriptomic and proteomic studies were not conducted on the same grape materials, so it is difficult to compare their results. In addition, the Affymetrix Gene-Chip Vitis vinifera (Grape) Genome Array contains only 15,700 probe sets, which is about half of the grape genome size, and the number of proteins identified in these experiments was lower. Therefore, it is difficult to reveal a complete picture of thermotolerance mechanisms in grape from the previous experiments.
In this study, based on high-throughput sequencing and the iTRAQ labeling technique, we assessed proteomic and transcriptomic changes in grape leaves under four different temperature regimes in order to uncover the cellular responses to sudden temperature changes and characterize the differentially expressed proteins, genes, and pathways. As a result, a new insight into the response of grape leaves to high temperature was revealed after the integration of transcriptomic and proteomic analyses.
RESULTS
Heat Injury in Grape Leaves Exposed to Different High Temperatures
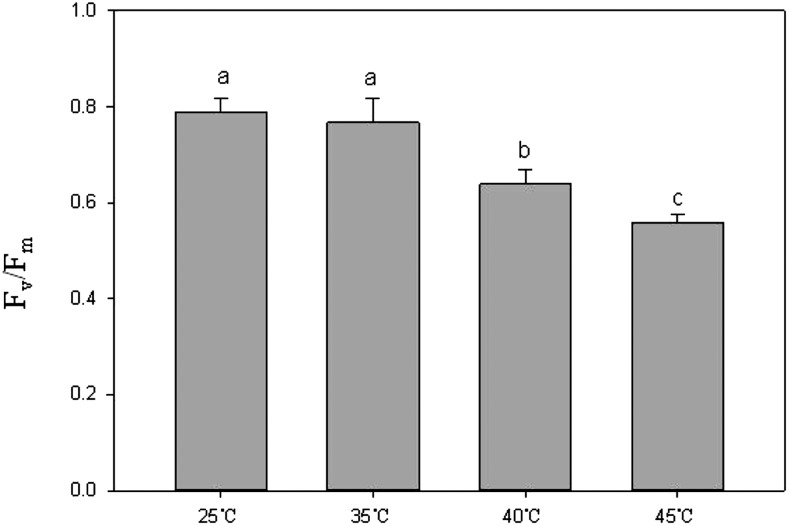
Photosynthesis is a sensitive biological process to high temperature in plants (Ashraf and Harris, 2013). Heat injury may be evaluated rapidly by the chlorophyll a fluorescence transient (O-J-I-P test) through investigating changes in the electron transport chain of PSII. The parameter Fv/Fm represents the potential maximum quantum yield of primary photochemistry and was thought to be suitable for evaluating heat injury in grape leaves (Xu et al., 2014). In this study, before temperature treatments, Fv/Fm values of grape leaves ranged from 0.78 to 0.81 (data not shown), which suggested that these grape leaves were healthy. After 35°C treatment for 2 h, Fv/Fm values of grape leaves did not show significant (P < 0.05, the same as below) sign of change compared with the control group (at 25°C). When heat treatment was 40°C or 45°C for 2 h, the Fv/Fm values declined significantly (P < 0.05). Moreover, the higher the treatment temperature, the lower the Fv/Fm values (Fig. 1). These results suggested that grape leaves were not inhibited or injured by 35°C for 2 h but that they were injured by 40°C or 45°C for 2 h. These results were very similar to previous findings (Sorkel, 2006; Greer and Weedon, 2012).
Figure 1.
Photosynthetic efficiency of grape leaves under different temperatures (25°C, 35°C, 40°C, and 45°C). Each Fv/Fm value represents the mean ± sd of five replicates. Different letters indicate significant differences (P < 0.05) among Fv/Fm values of four temperature treatments.
Transcriptome Sequencing and de Novo Assembly
A cDNA library was constructed using equal amounts of RNA extracted from grape leaves that had been exposed to four different temperatures (25°C, 35°C, 40°C, and 45°C). To characterize the grape transcriptome, the cDNA library was subjected to paired-end read sequencing using the Illumina HiSeq2000 platform. After removing reads of low quality, adaptor sequences, or reads with greater than 5% ambiguous nucleotides, a total of 453,786,102 clean paired-end reads were produced consisting of 68,067,915,300 nucleotides (68.1 Gb), with an average GC content of 46.5%. These high-quality reads were then de novo assembled using the Trinity program, resulting in 397,101 contigs with an average length of 820 nucleotides and an N50 length of 1,340 nucleotides. Based on the paired-end sequence information, the contigs were further assembled to give 314,660 unigenes, which accounted for 205,090,426 nucleotides (205.1 Mb), with an average length of 651 nucleotides. Of these unigenes, 117,055 (37.2%) were longer than 500 nucleotides, 46,687 (14.8%) were longer than 1,000 nucleotides, 25,797 (8.2%) were longer than 1,500 nucleotides, and 15,656 (5%) were longer than 2,000 nucleotides. Each treatment had three replicates, as shown in Supplemental Figure S1, with different replicates having almost unanimous results.
Functional Annotation of Unigenes
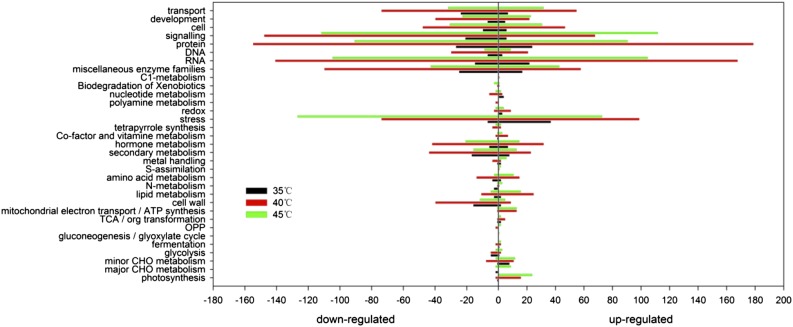
The assembled unigene sequences were used to search public databases, including the NCBI nonredundant protein database, the Swissprot protein database, the Gene Ontology (GO) database, the Clusters of Orthologous Groups database, and the Kyoto Encyclopedia of Genes and Genomes database, using the BLASTX algorithm with an E value threshold of 1e−5. In total, 163,216 (51.9%) unigenes were matched to a sequence in at least one of the above-mentioned databases and more than 9,392 unigenes matched more than 80% of annotated genes in the nonredundant protein and Swissprot nonredundant protein databases, representing almost the whole length. In terms of E value distribution, 37.4% of the homology ranged between 1e−5 and 1e−45, while a majority of the sequences (62.5%) showed a threshold E value of less than 1e−45, indicating strong homology. Among the unigenes annotated using the nonredundant protein database, 74.6% had more than 60% similarity with the corresponding gene sequence. There were more than 10,000 differentially expressed transcripts between different treatments according to the criteria of fold change (≥2), false discovery rate (FDR; ≤0.01), and P < 0.05 (Supplemental Fig. S2). For annotated genes mapped to the grape genome, we used MapMan software to conduct functional categorization. As shown in Figure 2, 35°C treatment of grape leaves resulted in differential expression of only 252 genes compared with 25°C. However, there were 4,551 and 3,490 differentially expressed genes under 40°C and 45°C, respectively. These genes were associated with biological processes that were related mainly to protein, RNA, stress, and signaling. Examples of these genes include HSP90.1, HSP23.6, HSP101, HSFA2, and ribonucleoprotein family proteins U2 and U3.
Figure 2.
Main biology processes of differentially expressed genes under 35°C, 40°C, and 45°C compared with 25°C in grape leaves. The x axis indicates the number of up-regulated (right) and down-regulated (left) genes.
The Influence of Different Temperatures on the Regulation of Genes at the Alternative Splicing Level
Alternative splicing (AS) is an important style of posttranscriptional regulation (Syed et al., 2012). A few studies have explored the relationship between AS and temperature change in plants (Capovilla et al., 2015). Therefore, we further analyzed the patterns of heat-induced AS events from the RNA sequencing data. In this study, AS events including exon skipping (ES), intron retention (IR), mutually exclusive exons (MXEs), and alternative 3′ and 5′ splice sites (A3SS and A5SS) were detected and quantified using rMATS with self-assembled transcript annotation (version 3.0.9; Shen et al., 2014). The inclusion level of each candidate splicing event was calculated using reads mapped to splicing junctions (for detailed information, see “Materials and Methods”). At the normal growth temperature (25°C), about 23.6% of the total assembled genes appeared as AS events in the grape leaves, while the 35°C, 40°C, and 45°C, treatments resulted in AS occurrence of 68.2% genes (data not shown). In the AS events, the number of IR events was largest, followed by ES events, then by A5SS and A3SS, and with the number of MXE events lowest (Supplemental Table S1).
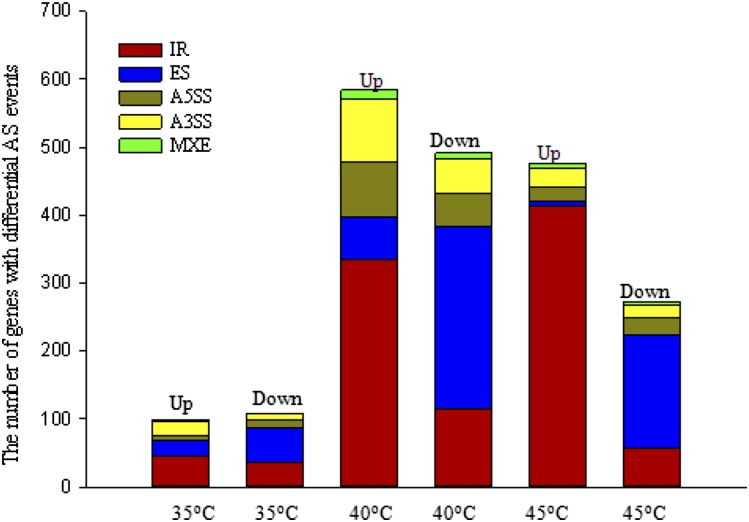
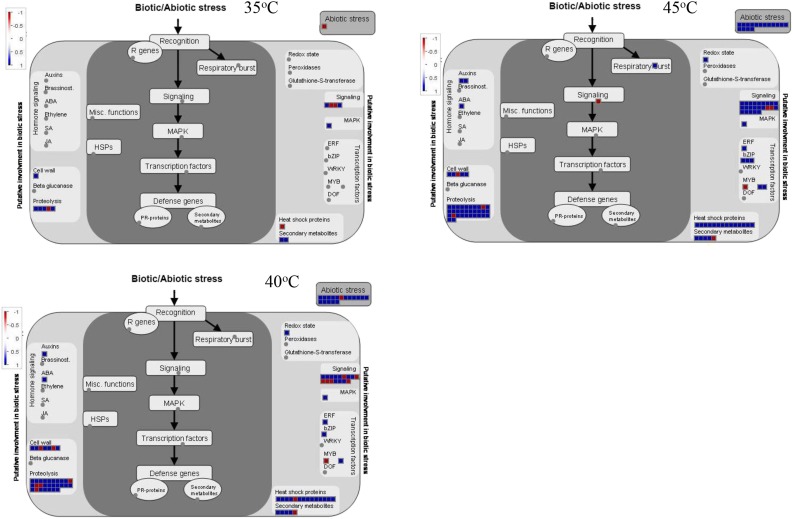
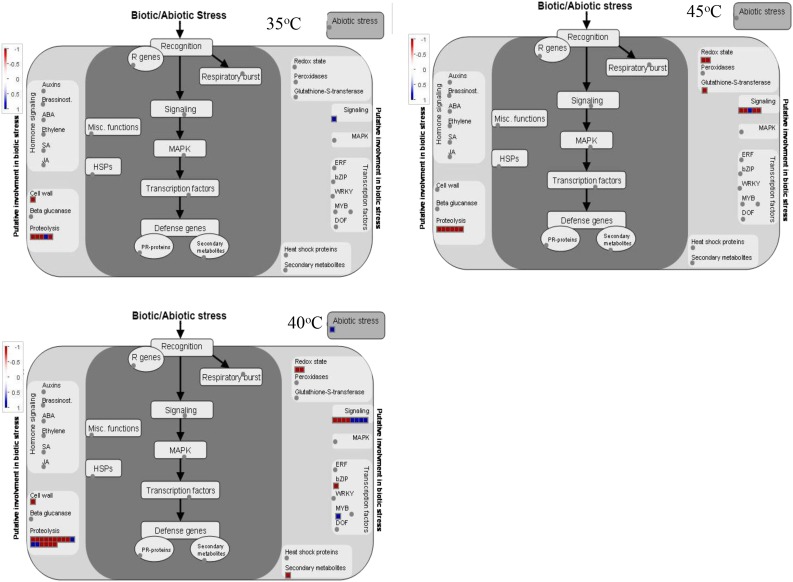
Based on the data described above, differential splicing events were selected with FDR ≤ 0.05. As shown in Figure 3 and Supplemental Table S2, a high temperature of 35°C resulted in the occurrence of 206 significant AS events compared with 25°C. Ninety-eight AS events were up-regulated, while 108 AS events were down-regulated. At 40°C, significant AS events increased to 1,075. Of these, 584 AS events were up-regulated, in which IR events (334) dominated, while 491 AS events were down-regulated, in which IR and ES events were most common, reaching 115 and 269, respectively. When grape leaves were held at 45°C for 2 h, 748 AS events occurred significantly, in which 476 AS events were up-regulated (413 IR events) and 272 AS events were down-regulated. These results indicated that IR is the most abundant up-regulated AS event in the response of grape leaves to high temperature, with IR events at 40 and 45°C far more abundant than those at 35°C. With increased temperatures, up-regulated IR events increased gradually, and up- or down-regulated ES, A5SS, and A3SS events rose from 35°C to 40°C but then declined at 45°C. We identified overrepresented GO terms (biological processes) among functionally annotated genes with IR events (Supplemental Data Sets S1 and S2) and ES events (Supplemental Data Sets S3 and S4). The results revealed that these differentially spliced genes were involved in several biological processes, including responses to abiotic stimuli and RNA processing, suggesting that high temperature may impact biological processes through changing pre-mRNA splicing. In particular, the response-to-heat or stress-stimulus functional category was markedly increased among the differentially spliced genes at 40°C and 45°C. Indeed, further analysis using MapMan software suggested that genes with aberrant splicing in grape leaves with high temperatures of 40°C and 45°C were involved in various stress response pathways, including heat shock protein pathways, proteolytic pathways, transcription regulation of stress responses, and secondary metabolites (Figs. 4 and 5). Interestingly, there were almost no IR events for HSP genes at 35°C. However, those genes with up-regulation of IR events at 40°C or 45°C were involved mainly in protein folding, such as HSP21, HSC70.2, HSC70-5, HSC70.7, HSP90.1 (HSP81.1 and HSP83), HSP101, HSFB1, HSFA2, HSF4, and HSFA6B. In addition, those down-regulated genes in ES events at 40°C or 45°C were involved mostly in proteolysis, such as ATMC5 (Cys-type endopeptidase), DegP protease, FtsH protease9 (metallopeptidase), FtsH protease1, FtsH protease11, UPL7 (ubiquitin-protein ligase), and UBP5 (ubiquitin-specific protease).
Figure 3.
Summary of all genes with significantly differential AS events in grape leaves under each high-temperature (35°C, 40°C, and 45°C) treatment compared with the control (25°C). Up indicates up-regulation, and Down indicates down-regulation.
Figure 4.
A network generated by MapMan indicates that genes with up- and down-regulated IR splicing events are involved in various stress response pathways in grape leaves under different high temperatures (35°C, 40°C, and 45°C) compared with 25°C.
Figure 5.
A network generated by MapMan indicates that genes with up- and down-regulated ES splicing events are involved in various stress response pathways in grape leaves under different high temperatures (35°C, 40°C, and 45°C) compared with 25°C.
Protein Responses to High Temperature in Grape Leaves Revealed by iTRAQ Analysis
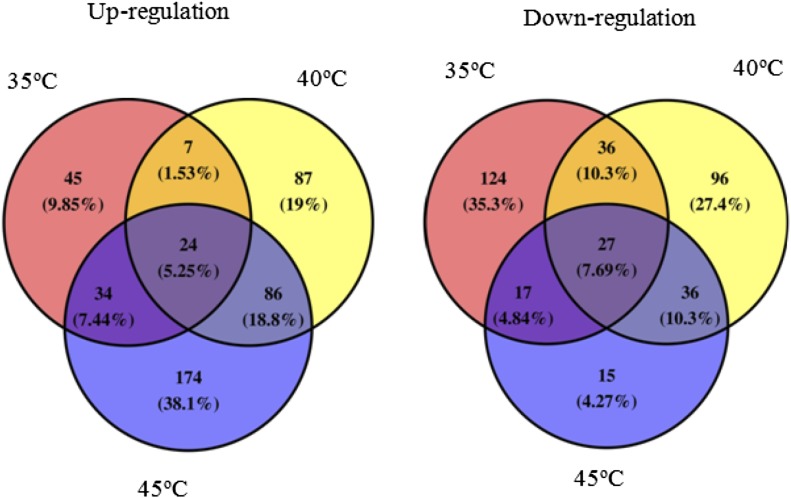
To reveal whether different temperatures had distinct effects on protein expression, we performed an iTRAQ analysis of grape leaves treated with 25°C, 35°C, 40°C, and 45°C. Proteins extracted from these leaves were labeled with iTRAQ (114 for 25°C treatment, 115 for 35°C treatment, 116 for 40°C treatment, and 117 for 45°C treatment) after being digested with trypsin. To improve proteome coverage, high-pH reverse-phase HPLC was used for peptide mixture fractionation. The selected fractions were analyzed with nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a rapid-separation liquid chromatography system (interfaced with a Q Exactive device [Thermo Fisher Scientific]; Supplemental Fig. S3). A total of 524,912 high-resolution tandem mass spectra acquired from 20 × 3 LC-MS/MS runs were searched using Andromeda integrated in MaxQuant (version 1.3.5.8; Cox and Mann, 2008; Cox et al., 2011) against the UniProt grape protein database (UP000009183; 29,907 sequences). In total, 6,482 nonredundant UniProt-annotated proteins were identified with at least two unique peptides (protein and peptide FDR < 0.01), and among them, 5,108 (78.80%) proteins were commonly identified in three biological repeats (Supplemental Fig. S4). The iTRAQ reporter ion intensity of protein in three biological repeats was further integrated, with the filter of minimum reporter precursor intensity fraction = 0.75, total intensity of certain protein > 10,000, and unique peptide ≥ 2, and the 5,651 proteins were eventually quantified. When setting a quantification ratio > 1.3 as the up-regulated threshold and a ratio < 0.7 as the down-regulated threshold and P < 0.05, 808 differentially expressed proteins were obtained under the different high temperatures compared with the control (25°C), as shown in Figure 6. Under 35°C, 40°C, and 45°C, 110, 204, and 318 proteins were up-regulated, respectively, in which 24 proteins were co-up-regulated. There were 31 common up-regulated proteins between 35°C and 40°C, 58 between 35°C and 45°C, and 110 between 40°C and 45°C. Under 35°C, 40°C, and 45°C treatments, 204, 195, and 95 proteins were down-regulated, respectively, in which 27 proteins were co-down-regulated. In addition, there were 63 common down-regulated proteins between 35°C and 40°C, 44 between 35°C and 45°C, and 63 between 40°C and 45°C.
Figure 6.
Venn diagrams of differentially expressed proteins in grape leaves under 35°C, 40°C, and 45°C compared with 25°C at fold change greater than 1.3 (up-regulation) and less than 0.7 (down-regulation).
Functional Classification and Enrichment Analysis of Differentially Expressed Proteins under Different High Temperatures
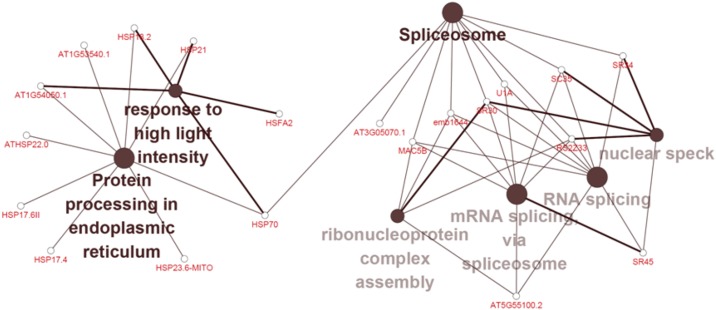
We further conducted functional classification and enrichment analyses of these differentially expressed proteins. The up-regulated biological pathways at 35°C included sugar-mediated signaling pathway, photosynthesis, protein targeting to mitochondria, and purine ribonucleoside monophoshate biosynthetic processes. The down-regulated biological pathways at 35°C included aromatic amino acid family metabolism, sterol biosynthesis, phenylpropanoid metabolism, response to light intensity/UV, establishment of protein localization to membrane, and flavonoid biosynthesis (Supplemental Fig. S5). Different from the 35°C treatment, the up-regulated biological pathways at 40°C were primarily responses to temperature stimulus/heat/high light intensity/reactive oxygen species/hydrogen peroxide, heat acclimation, and α-amino acid catabolic process. The down-regulated biological pathways at 40°C were mainly photosynthesis, aromatic amino acid family metabolism, sterol biosynthesis, phenylpropanoid metabolism, responses to UV light, pigment biosynthesis, formation of translation preinitiation complexes, hyperosmotic salinity responses, and starch biosynthesis (Supplemental Fig. S6). The up-regulated biological pathways at 45°C were mostly involved in mRNA splicing via spliceosomes, sesquiterpenoid metabolism, chlorophyll biosynthesis, cellular responses to heat, cellular protein complex disassembly, sugar-mediated signaling pathways, and aromatic amino acid family metabolism (Supplemental Fig. S7A). The down-regulated biological pathways at 45°C were involved in responses to UV light, Cys-type peptidase activity, phosphopantetheine attachment site binding involved in fatty acid biosynthesis, chloroplast thylakoid lumen, cytoplasmic vesicle membrane, and protein phosphorylated amino acid binding (Supplemental Fig. S7B). As we knew from these results, biological processes of co-up-regulated proteins at 40°C and 45°C compared with the control involved responses to temperature stimuli or heat, mRNA splicing, splicesomal complex assembly, and photosynthesis. Moreover, those proteins that were up-regulated with increasing temperature accounted for ∼16% of total differentially expressed proteins. They were involved in responses to high light intensity, protein processing in the endoplasmic reticulum (HSP17.4, HSP17.6II, HSP23.6, HSP70, HAFA2, HSP21, HSP18.2, and ATHSP22.0), and RNA splicing (U1A, SR33, SR45, SC35, SR34, and SR30; Fig. 7).
Figure 7.
Functional classification and enrichment analysis of up-regulated proteins with the increasing of temperature from 25°C to 45°C in grape leaves.
Integration of Transcriptomic and Proteomic Data
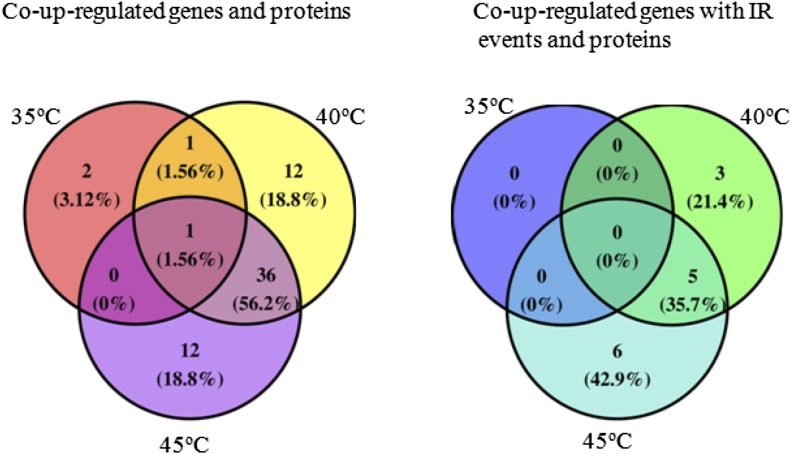
It is not certain that changes in gene expression indicate corresponding protein changes. Integrative analysis of the proteome combined with a comprehensive transcriptome database provides an important verification tool for the expression of key genes. We constructed splicing transcriptomic data, integrated transcriptomic and proteomic data, and then analyzed the regulation of the changes in gene expression with changes in protein expression. Specifically, those genes and proteins whose change trends were similar were examined. As shown in Figure 8, compared with 25°C, four, 50, and 49 co-up-regulated genes and proteins at 35°C, 40°C, and 45°C, respectively, were found. Only one co-up-regulated gene and protein (i.e. MULTIPROTEIN-BRIDGING FACTOR1c [MBF1c]) was identified under the three temperatures. There were 36 co-up-regulated genes and proteins between 40°C and 45°C, which included HSPs (HSP40, HSP70.4, HSP22, HSP18.1, HSP17.6C, HSP17.4, HSP23.6, HSP90.1, HSP101, HSP17.6, and HSP70-1), Ser/Arg-rich splicing factor SR45a, GATA transcription factor, PDX1.2, ABC transporter, Gal-binding protein, 5′-adenylylsulfate reductase1, F-box protein, translation initiation factor SUI1 family protein, Ser acetyltransferase1, and T-complex protein11 (Table I). For these genes, RNA sequencing results were validated using qRT-PCR (Supplemental Fig. S8). However, no co-down-regulated gene and protein was found under the three temperature treatments. Only one down-regulated gene and protein (VIT_14s0036g00070.1) was identified under 40°C and 45°C, which was not named and may be associated with defense. Overall, proteins that exhibited notable changes in their expression levels did not always have a corresponding alteration in transcript levels, indicating that posttranscriptional and posttranslational regulation may play important roles in the grape response to heat.
Figure 8.
Venn diagram analysis of co-up-regulated genes and proteins (left) and co-up-regulated genes with IR events and proteins (right) in grape leaves under 35°C, 40°C, and 45°C compared with 25°C.
Table I. Co-up-regulated genes and proteins with differential expression in grape leaves only under 40°C and 45°C compared with 25°C.
| Gene Identifier | Description |
|---|---|
| VIT_01s0011g04820.1 | Chaperone DnaJ domain-containing protein |
| VIT_10s0003g00260.1 | DNAJ heat shock family protein |
| VIT_08s0007g00130.1 | Heat shock 70-kD protein4 |
| VIT_12s0035g01910.1 | 22-kD heat shock protein (AtHsp22.0) |
| VIT_13s0019g03000.1 | 18.1-kD class I heat shock protein (18.1-kD heat shock protein; AtHsp18.1) |
| VIT_13s0019g03090.1 | 17.6-kD class I heat shock protein3 (17.6-kD heat shock protein3; AtHsp17.6C) |
| VIT_13s0019g02740.1 | 18.1-kD class I heat shock protein (18.1-kD heat shock protein; AtHsp18.1) |
| VIT_13s0019g02780.1 | 18.1-kD class I heat shock protein (18.1-kD heat shock protein; AtHsp18.1) |
| VIT_13s0019g02840.1 | 17.4-kD class I heat shock protein (17.4-kD heat shock protein1; AtHsp17.4A) |
| VIT_13s0019g03170.1 | 18.1-kD class I heat shock protein (18.1-kD heat shock protein; AtHsp18.1) |
| VIT_16s0022g00510.1 | 23.6-kD heat shock protein, mitochondrial (AtHsp23.6) |
| VIT_16s0050g01150.1 | Heat shock protein90-1 (AtHSP90.1, AtHsp90-1; heat shock protein81-1, Hsp81-1; heat shock protein83) |
| VIT_17s0000g07190.1 | Chaperone protein ClpB1 (ATP-dependent Clp protease ATP-binding subunit ClpB homolog 1)(Heat shock protein101) |
| VIT_18s0041g00280.1 | T-complex protein11 |
| VIT_18s0089g01270 | 22-kD heat shock protein (AtHsp22.0) |
| VIT_19s0090g00420.1 | Translation initiation factor SUI1 family protein |
| VIT_19s0015g00130.1 | Ser acetyltransferase1, chloroplastic (AtSAT-1; EC 2.3.1.30; AtSERAT2;1; SAT-p) |
| VIT_19s0085g01050.1 | 17.6-kD class I heat shock protein3 (17.6-kD heat shock protein3; AtHsp17.6C) |
| VIT_02s0154g00480.1 | 23.6-kD heat shock protein, mitochondrial (AtHsp23.6) |
| VIT_04s0008g01490.1 | 17.6-kD class II heat shock protein (17.6-kD heat shock protein; AtHsp17.6) |
| VIT_04s0008g01500.1 | 17.6-kD class II heat shock protein (17.6-kD heat shock protein; AtHsp17.6) |
| VIT_04s0008g01520.1 | 17.6-kD class II heat shock protein (17.6-kD heat shock protein; AtHsp17.6) |
| VIT_04s0008g01530.1 | 17.6-kD class II heat shock protein (17.6-kD heat shock protein; AtHsp17.6) |
| VIT_04s0008g01590.1 | 17.6-kD class II heat shock protein (17.6-kD heat shock protein; AtHsp17.6) |
| VIT_04s0008g01510.1 | 17.6-kD class II heat shock protein (17.6-kD heat shock protein; AtHsp17.6) |
| VIT_06s0004g04470.1 | Heat shock 70-kD protein1 |
| VIT_06s0004g06010.1 | Ser/Arg-rich splicing factor SR45a |
| VIT_07s0005g01080.1 | GATA transcription factor |
| VIT_08s0058g00210.1 | 17.6-kD class I heat shock protein3 (17.6-kD heat shock protein3; AtHsp17.6C) |
| VIT_08s0007g00130.1 | Heat shock 70-kD protein4 |
| VIT_08s0007g04000.1 | Response to oxidative stress |
| VIT_09s0002g04490 | Pyridoxal 5′-phosphate synthase-like subunit PDX1.2 (AtPDX1.2, AtPDX1;3) |
| VIT_09s0018g00310 | ABC transporter |
| VIT_00s0494g00030.1 | Gal-binding protein |
| VIT_00s1490g00010.1 | 5′-Adenylylsulfate reductase1, chloroplastic |
| VIT_00s0181g00080.1 | F-box protein |
| VIT_16s0098g01060.1 | 25.3-kD heat shock protein, chloroplastic (AtHsp25.3) |
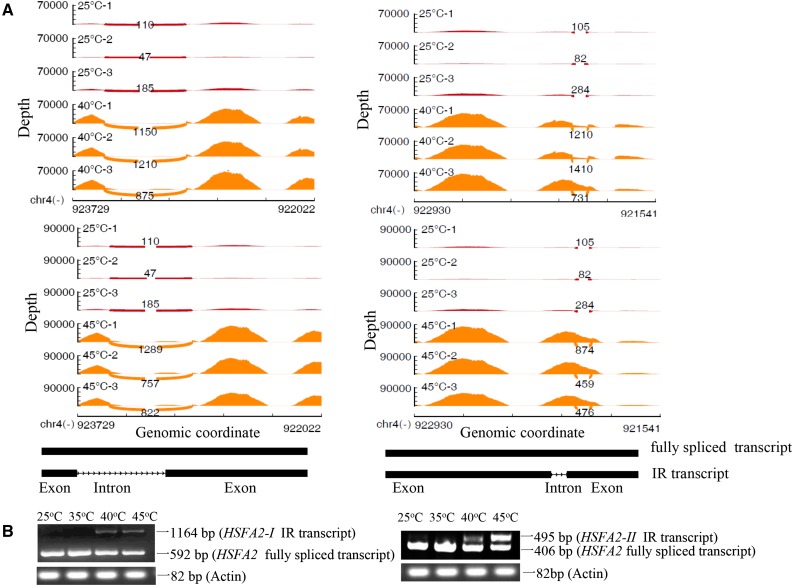
From the above data, one of the important changes of the transcriptome under different temperatures was AS, in which IR events occurred most. We integrated splicing transcriptome and protein expression data and found 14 co-up-regulated proteins and IR events under different temperatures (Fig. 8). Co-up-regulated proteins and IR events between 40°C and 45°C included HSFA2, HSP90.1, HSP25.3, clpB1 (HSP101), and HSP70.4 (Table II). For 40°C and 45°C, HSFA2 produced two transcripts with IR events that are shown in Figure 9. The model graphs of the IR events of these four HSP genes are shown in Supplemental Figures S9 to S12. These results also are validated by qRT-PCR in Supplemental Figure S13. However, there were no co-up-regulated proteins and IR events between 35°C and 45°C or between 35°C and 40°C.
Table II. Co-up-regulated IR events and proteins with differential expression in grape leaves under 40°C and 45°C compared with 25°C.
| Gene Identifier | Description |
|---|---|
| VIT_16s0050g01150.1 | Heat shock protein90.1 |
| VIT_16s0098g01060.1 | 25.3-kD heat shock protein, chloroplastic (AtHsp25.3) |
| VIT_17s0000g07190.1 | Chaperone protein ClpB1 (HSP101) |
| VIT_04s0008g01110.1 | Heat stress transcription factor A-2 (AtHsfA2, AtHsf-04) |
| VIT_08s0007g00130.1 | Heat shock 70-kD protein4 |
Figure 9.
Characterization of two HSFA2 transcripts with differential IR events in grape leaves under 40°C and 45°C compared with 25°C. A, Sashimi plots (stand alone) for alternatively spliced exon and flanking exons in samples. Per-base expression is plotted on the y axis of the Sashimi plot, with genomic coordinates on the x axis. Arcs represent splice junctions connecting exons and display the number of reads split across the junction (junction depth). Each treatment has three replicates. 25°C-1, -2, and -3 represent three replicates of the control 25°C treatment for 2 h. 40°C-1, -2, and -3 represent three replicates of the 40°C treatment for 2 h. 45°C-1, -2, and -3 represent three replicates of the 45°C treatment for 2 h. mRNA isoforms quantified are shown at bottom (exons in black and introns as lines with arrowheads). B, Semiquantitative RT-PCR analysis of the expression of HSFA2 IR splice variants in grape leaves under 25°C, 35°C, 40°C, and 45°C for 2 h. The forward and reverse primers were designed from the upstream and downstream exons of the retention intron based on each IR event, respectively.
DISCUSSION
Photosynthesis may be inhibited in grapevines under high temperature. The O-J-I-P test is a powerful tool for evaluating photosynthetic activity in grape leaves (Xu et al., 2014). We investigated the chlorophyll a fluorescence parameter Fv/Fm in grape leaves under different high temperatures (35°C, 40°C, and 45°C) versus a control temperature (25°C). The results indicated that 40°C and 45°C treatments inhibited leaf photosynthesis but 35°C had very little influence (Fig. 1). Compared with the control at 25°C, 35°C treatment resulted in about 300 genes changing significantly, but 40°C and 45°C led to significant changes of about 4,000 genes (Fig. 2). These results also indicated that 35°C had almost no influence on grape leaves; however, 40°C and 45°C treatments influenced or damaged grape leaves. Integrated analyses at the transcriptomic and proteomic levels were performed to understand the underlying mechanisms of cellular responses, and significant insights into gene functions were revealed.
HSPs Are Involved in Heat Tolerance in Grape Leaves through Up-Regulating Transcriptional and Translational Levels
Heat stress often causes protein denaturation in plants. Therefore, maintaining proteins in their functional conformations is particularly important for plant survival under heat stress (Lee and Vierling, 2000). High temperature can trigger some mechanisms of defense such as the production of HSPs, which are classified into five major families: HSP100 (Clp), HSP90, HSP70 (DnaK), HSP60 (chaperonin and GroEL), and small HSPs (sHSPs). Protein folding is mainly mediated by HSP90s, HSP70s, and HSP60s, whereas sHSPs are involved in protection against protein aggregation and HSP100s in the resolubilization of protein aggregates (Kotak et al., 2007). Changes in gene expression at the transcriptional and/or translational level are thought to be a fundamental mechanism in plant response to environmental stresses. While previous studies focused mostly on HSP change at the transcriptional level under heat stress, very little evidence was reported at both transcriptional and translational levels. In this study, we first found that 11 HSPs were co-up-regulated at the transcriptional and translational levels in grape leaves under only 40°C and 45°C high temperatures compared with the control (Fig. 8; Table I). They mainly included sHSPs (HSP17.4, HSP17.6, HSP17.6C, HSP18.1, HSP22, HSP23.6, HSP40, and HSP25.3), HSP70 (HSP70-1 and HSP70-4), HSP90-1, and HSP101. These results further corroborated previous findings showing that HSPs were not induced by a temperature such as 35°C (Vierling, 1991; Tanaka et al., 2000).
The sHSPs cannot refold nonnative proteins, but they can bind to partially folded or denatured substrate proteins, preventing irreversible unfolding or incorrect protein aggregation. In rice seedlings, the expression of HSP17.4 was up-regulated during the seedling and anthesis stages in response to heat stress (Chen et al., 2014). In Arabidopsis, up-regulated levels of HSP17.6 were detected under heat stress (Sagor et al., 2013). Györgyey et al. (1991) isolated cDNA clones (MsHSP18-1) from alfalfa (Medicago sativa) and found that MsHSP18-1 transcription was induced by elevated temperature. The mRNA levels of HSP22 cDNAs increased after hot water treatment in grapefruit (Citrus × paradisi) peel tissue (Rozenzvieg et al., 2004). Three-week-old Arabidopsis plants had increased expression of AtHSP23.6 following exposure to heat stress (Rhoads et al., 2005). Liu et al. (2012) found the transcriptional levels of HSP17.6, HSP22, and HSP40 to be increased significantly in grape leaves under a high temperature (45°C). This study identified seven co-up-regulated sHSPs at transcriptional and translational levels in grape leaves under 40°C and 45°C. They were HSP17.4, HSP17.6, HSP17.6C, HSP18.1, HSP22, HSP23.6, HSP25.3, and HSP40 and may play important roles in the heat tolerance of grape.
In almost all organisms, HSP70 functions as a chaperone for newly synthesized proteins to prevent their accumulation as aggregates, and so they fold in a proper way during the transfer to their final location. A study on Arabidopsis indicated the necessity of HSP70 in the stroma of the chloroplast for the differentiation of germinating seeds and their heat tolerance (Su and Li, 2008). There have been some studies on HSP70 at the transcriptional and translational levels. Experiments using the mRNA transcription inhibitor (actidione D) or protein synthesis inhibitor (cycloheximide) demonstrated that the accumulation of HSP70 by heat acclimation was controlled at both transcriptional and translational levels. The class HSP90 shares roles with other classes. As a molecular chaperone, HSP90 can bind HSP70 in many chaperone complexes and has an important role in signaling protein function and trafficking (Pratt and Toft, 2003). In Arabidopsis, there were some indications that cytoplasmic HSP90 inhibited HSFs in the absence of heat stress, but under heat stress this role was suspended temporarily, so that HSFs were active (Yamada et al., 2007). Under normal conditions, HSF1 activity was suppressed by interaction with HSP70 and HSP90 (HSP70/90; Anckar and Sistonen, 2011). Heat stress treatment caused the dissociation of HSP70/90 from HSFs because denatured proteins competitively interacted with the HSPs. One unique function of HSP100 was the reactivation of aggregated proteins (Parsell and Lindquist, 1993) by resolubilization of nonfunctional protein aggregates, and it also helped to degrade irreversibly damaged polypeptides (Bösl et al., 2006). HSP100 activation required the simultaneous binding of multiple HSP70 partners, restricting high HSP100 activity to the surface of protein aggregates and ensuring HSP100 substrate specificity. Heat shock induced an increase in HSP70, HSP90, and HSP101 protein expression in creeping bentgrass (Agrostis stolonifera; Wang et al., 2014). In our study, HSP70.1, HSP70.4, HSP90.1 and HSP101 were co-up-regulated at transcriptional and translational levels under high temperatures, indicating that they can form HSP70/90 and HSP100/70 complexes. Therefore, these HSPs likely played crucial roles in protecting grape leaves from heat stress. Apparently, separate transcriptomic or proteomic analysis was not sufficient to understand the underlying genetic and molecular mechanisms.
AS Acts as an Important Posttranscriptional Regulation in Grape Leaf Response to High Temperature, Especially for Protein Folding and Proteolysis
Plants tightly respond to temperature changes by reorganizing their gene transcription and protein translation and acquire a defense capability. While different levels of gene expression are known to be responsive to heat stress, gene regulation at the posttranscriptional level is less understood. Recently, increasing evidence has shown that AS is a critical posttranscriptional event and plays an important role in plant stress responses (Staiger and Brown, 2013). Recent studies using massively parallel RNA sequencing revealed that a large percentage of genes in Arabidopsis undergo AS, which potentially could significantly increase the plasticity of the transcriptome and proteome diversity (Mastrangelo et al., 2012). AS is a process in which two or more different transcripts are generated from the same pre-mRNA molecule using different splicing sites. Although AS of some stress-responsive genes has been reported, large-scale or genome-wide studies of AS dynamics under heat stress conditions have not been conducted. In this study, high temperature caused AS changes of about 70% of the genes in grape leaves, indicating that AS may be an important style of posttranscriptional regulation in grape leaf response to high temperature. These marked AS changes under high temperature could provide molecular plasticity for the plants to adapt to stress conditions. However, whether AS can be differentially regulated remains unclear. In this study, according to the criterion (FDR ≤ 0.01) for differential splicing events, compared with 25°C, pre-mRNA splicing responded rapidly in grape leaves at 35°C, and 206 differential splicing events occurred. When temperature changed to 40°C, differential splicing events reached 1,075. Under 45°C, total differential splicing events were 748 (Fig. 3; Supplemental Table S2). These results indicated that pre-mRNA splicing in grape cells responded drastically when temperature reached 40°C. However, increasing temperature did not further raise the occurrence of differential splicing events, which may be associated with more serious damage at 45°C than 40°C to grape leaves. This result also implies that it may be relatively difficult for grape leaves to modulate gene function under serious stress at 45°C. It is possible that 45°C may cause the decrease of enhancer activity or the increase of repressor activity for AS, allowing splicing regulation of specific gene transcripts to become less active. Our RNA sequencing data showed that MXE events occurred only for a few, while ES, IR, A5SS, and A3SS events are found universally with transcripts under ambient temperature (25°C) in grape leaf cells. When grape leaves were at different high temperatures, the AS events were differentially regulated, with IR events the most abundant followed by ES events (Fig. 3; Supplemental Table S2). Therefore, IR and ES events are the most important posttranscriptional regulation at the AS level during the response of grape cells to heat stress. Moreover, with the increasing temperature, the occurrence of up-regulated IR events increased (Fig. 4).
The processing of AS depends on a large ribonucleoprotein complex, the spliceosome, which consisted of a core of five small nuclear ribonucleic proteins, called U1, U2, U4, U5, and U6. U1 and U2 recognize specific sequences in the intronic region. Conformational changes of the spliceosome and sequential phosphodiester transfer reactions remove the introns and mediate the joining of exons (Capovilla et al., 2015). The choice of splice sites is directed to a large extent by RNA-binding proteins, predominantly Ser/Arg (SR) proteins and heterogenous nuclear ribonucleoproteins, which promote and block binding of the spliceosome, respectively. SR proteins recognize splicing enhancers and recruit U2 auxiliary factor and U1 (Kornblihtt et al., 2013). High temperature promotes the expression of active isoforms of the splicing factor SR30 (Filichkin et al., 2010) but reduces the active isoform of SR34 (Lazar and Goodman, 2000). Palusa et al. (2007) reported that additional transcripts appeared or the expression level decreased in SR30 and SR1/SR34 in heat-treated Arabidopsis seedlings. Ous study showed that protein levels of SR45, SR30, SR34, and U1A in grape leaves rose gradually as ambient temperature increased (Fig. 7). This result may explain why more AS events occurred under high temperatures in grape leaves. The AS pattern of several SR proteins has been shown to have obviously changed under various abiotic stress conditions, including temperature stress, high salinity, and high-light irradiation (Isshiki et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Duque, 2011). However, in our study, SR genes themselves did not occur significantly in AS events. A few studies also showed that overexpression of certain splicing factors could increase plant tolerance to salt and other stresses (Chang et al., 2014). We predict that regulating the expression of some SR genes or other splicing factors may increase plant tolerance to high temperature by enhancing the correct splicing of heat tolerance genes.
If the increase in AS is merely a nonspecific consequence of stress damage, a random distribution would be expected among genes. However, our data, along with previous reports, demonstrated that the genes associated with stress responses tended to be alternatively spliced under stress conditions (Figs. 4 and 5). It is known that some abiotic stresses can activate the expression of a large number of plant stress-responsive genes that are not expressed or are expressed at lower levels under normal nonstressful conditions (Xiong et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). With the simultaneous production of a large amount of these stress-inducible pre-mRNAs, cells would need to immediately recruit a significant amount of splicing and other factors for their cotranscriptional or posttranscriptional processing. In our study, high temperature resulted in many HSPs occurring as AS of the IR type: for example, CPHSC70-2, HSC70-7, HSP90.1, HSP101, HOT1, HSP21, CPN20, and HSC70-5 (Fig. 4; Supplemental Data Set S1). These results also demonstrated that IR could be a key AS that results in the up-regulation of HSPs at the transcriptional level.
To date, there have only been a few reports about AS of HSPs in plants. Lund et al. (2001) observed that forms of HSP22 resulted from alternative intron splicing in maize (Zea mays). Zhang et al. (2011) reported that 13 HSP genes had one transcript each, two genes had two transcripts each, and two had three transcripts each in bovine. Heat stress-AS occurs in transcripts for specific functions. In this study, heat stress-induced IR events are mainly for protein folding (HSPs) and heat stress-induced ES events are mainly for protein degradation (peptidases; Figs. 4 and 5). These results suggested that the occurrence of differential AS events under high temperature was not a random process. Rather, it was heavily influenced by heat stress. Our study showed that HSP90.1, HSP25.3, HSP101, and HSP70.4 transcription in grape leaves under both 40°C and 45°C generated splicing variants of IR (Supplemental Figs. S9–S12). Moreover, there were co-up-regulated IR events and proteins with differential expression for the four genes under 40°C and 45°C (Table II). However, it is possible that these HSP IR transcripts were degraded posttranscriptionally. Kalyna et al. (2012) suggested that AS frequently generates nonsense mRNA with premature termination codons (PTCs), which are rapidly degraded by the cellular nonsense-mediated mRNA decay machinery in many cases. This study also showed that transcripts containing PTCs are often readily detectable and can contribute significantly to steady-state transcript levels of genes. This type of AS is often referred to as unproductive AS for adaptation to developmental demands and/or plant responses to environmental stresses (Reddy et al., 2013). Some PTC+ mRNAs escape nonsense-mediated mRNA decay and produce truncated proteins that may be missing key functional domains (Kalyna et al., 2012). In this study, we detected IR transcripts of these HSPs by RT-PCR (Supplemental Fig. S13), and transcript expression levels of these HSPs increased (Table II). In addition, we found that intron sequences of these IR transcripts of HSPs included termination codons (Supplemental Data Set S5). Therefore, it is impossible for IR transcripts of these HSPs to translate into HSPs, but it is possible that they encoded some truncated proteins that function in the response of plants to the environment (Liu et al., 2013). In our proteomics data presented here, peptides corresponding to intron sequences of IR transcripts were not detected. Therefore, we cannot confirm that these IR transcripts were translated into truncated proteins. These results indicated that IR events of these HSPs should contribute to the increase of HSP transcription. However, the translation of these HSPs was conserved and the increase of HSP proteins under 40°C and 45°C should result from fully spliced transcripts.
Some Important Transcript Factors Could Be Involved in Heat Tolerance in Grape Leaves through Up-Regulation at Transcriptional and Translational Levels
In this study, using iTRAQ analysis, protein changes across four different temperatures were studied to elucidate a response network specific to heat stress in grape leaves (Fig. 6; Supplemental Figs. S4–S7). The AS and the proteomic data were integratively analyzed. The results revealed that co-up-regulated genes at IR splicing and protein translation under different high temperatures included HSFA2 (Fig. 9; Table II). A study in Arabidopsis showed that HSFA2 plays an important role among the 21 HSFs in both basal and acquired thermotolerance (von Koskull-Döring et al., 2007). Recently, it was reported that high temperature induces HSFA2 to generate a splice variant in Arabidopsis (Sugio et al., 2009), suggesting that high temperature also regulates HSFA2 expression at the posttranscriptional level in plants. Liu et al. (2013) suggested a novel mechanism underlying the self-regulation of HSFA2 expression through a small truncated HSFA2 isoform encoded by this new splice variant. Cheng et al. (2015) found that OsHSFA2d RNA could be alternatively spliced within the conserved introns of the DNA-binding domain coding region and that these splicing events could introduce one or two small exons with premature stop codons, resulting in three AS products, OsHSFA2dI, OsHSFA2dII, and OsHSFA2dIII. A similar phenomenon also was observed for HSF1 in alfalfa (He et al., 2007), HSFA1d and HSFA2 in Arabidopsis (Sugio et al., 2009), and HSFA2a2 in the genus Potamogeton (Amano et al., 2012). These findings indicate that the AS of the HSF intron might serve as a conserved mechanism at the posttranscriptional level for HSF expression regulation in plants. Although Potenza et al. (2015) and Vitulo et al. (2014) reported the AS occurring in grape, IR events of HSFA2 expression in grape were not reported. Our study found that HSFA2 expression in grape leaves under 40°C and 45°C generated two IR splicing variants (Fig. 9), which could introduce two new truncated proteins. ES and A3SS events of HSFA2 also were detected; however, they exhibited no significant differences among different high temperatures (35°C, 40°C, and 45°C) compared with the control temperature (25°C). Therefore, IR should be an important posttranscriptional regulation for HSFA2, which make HSFA2 play a key function in the grape response to high temperature.
In our study, in addition to HSPs, co-up-regulated genes and proteins between 40°C and 45°C also included the GATA transcription factor (Table I). GATA can specifically bind 5′-GATA-3′ or 5′-GAT-3′ motifs within gene promoters and may be involved in the regulation of some light-responsive genes. Only one co-up-regulated gene and protein (MBF1c) was found under 35°C, 40°C, and 45°C treatments (Fig. 8). MBF1 is very interesting, as it is a highly conserved protein that functions as a non-DNA-binding transcriptional coactivator involved in different developmental and metabolic pathways in different organisms, ranging from yeast to humans (Takemaru et al., 1998; Kabe et al., 1999; Liu et al., 2003). At present, there are only a few studies of MBF1. In the model plant Arabidopsis, MBF1 was encoded by three different genes, MBF1a, MBF1b, and MBF1c (Tsuda et al., 2004). Suzuki et al. (2008) reported that MBF1c protein accumulated rapidly and was localized to nuclei during heat stress in Arabidopsis. It was required for thermotolerance and functions upstream of salicylic acid (SA), trehalose, ethylene, and PATHOGENESIS-RELATED PROTEIN1. In contrast, MBF1c was not required for the expression of transcripts encoding HSFA2 and various HSPs. However, Suzuki et al. (2011) suggested that MBF1c in Arabidopsis functions as a transcriptional regulator that binds DNA and controls the expression of 36 different transcripts during heat stress, including the important transcriptional regulator DRE-BINDING PROTEIN2A, two HSFs (HSFB2B and HSFB2A), and several zinc finger proteins. Moreover, the DNA-binding domain of MBF1c had a dominant-negative effect on heat tolerance when constitutively expressed in plants. Yan et al. (2014) isolated a full-length MBF1 cDNA sequence (VvMBF1) from V. vinifera × Vitis amurensis that was up-regulated in the leaves of grape plants following both drought and abscisic acid treatments. Carbonell-Bejerano et al. (2013) reported that berry MBF1a and MBF1c were up-regulated under high temperature when grape ‘Muscat Hamburg’ plants growing from cuttings were exposed to two different environmental regimes, 20°C/15°C (day/night) and 30°C/25°C (day/night), at the onset of ripening (veraison). Wang and Li (2006a) reported that exogenous SA pretreatment decreased thiobarbituric acid-reactive substances content and relative electrolyte leakage in grape leaves under heat stress, indicating that SA can induce intrinsic heat tolerance in grape. In addition, heat acclimation (38°C) induced an increase of free SA concentration in grape leaves (Wang and Li, 2006b). In this study, high temperature under 40°C and 45°C increased the expression of SALICYLIC ACID-BINDING PROTEIN2 by 860- and 3,100-fold, respectively, compared with the control (data not shown). These results indicated that it was possible that MBF1c modulates heat tolerance through controlling the SA concentration in grape. In addition, SA was recently proposed to play an important role in thermotolerance in other plant species (Larkindale and Knight, 2002; Martel and Qaderi, 2016). Previous studies have shown that HSFs play an important role in thermotolerance in plants and other organisms, regulating HSPs as well as different acclimation and detoxification proteins (von Koskull-Döring et al., 2007). Therefore, MBF1c should be a key regulator that controls the HSF-HSP pathway and the SA pathway in the thermotolerance of grape. Among different temperature treatments, there is no occurrence of differential AS in the MBF1c gene. Therefore, MBF1c could be involved in heat tolerance in grape leaves through up-regulating transcriptional and translational levels.
In summary, we have provided data to suggest that grape leaves respond rapidly to temperature greater than 35°C simultaneously at transcriptional, posttranscriptional, and translational levels. AS was an important posttranscriptional regulation, and IR events were the most abundant up- and down-regulated AS events followed by ES. Proteomic analyses showed that protein levels of RNA-binding proteins SR45, SR30, and SR34 and nuclear ribonucleic protein U1A in grape leaves gradually rose as ambient temperatures increased. These results also explained why AS events occurred more under high temperature in grape leaves. HSPs were involved in heat tolerance in grape leaves through up-regulating mRNA levels, IR event occurrence, and translational levels. The important transcript factor MBF1c was involved in heat tolerance in grape through up-regulating transcriptional and translational levels, but HSFA2 was involved in heat tolerance by modulating AS. Further investigations are needed to determine how temperature signaling regulates splicing activity and how the splicing machinery distinguishes transcript species.
MATERIALS AND METHODS
Plant Materials and Heat Treatments
One-year-old ‘Jingxiangyu’ grapevines (Vitis vinifera) were planted in pots, then grown in a greenhouse at 70% to 80% relative humidity at 18°C to 25°C, with the maximum photosynthetically active radiation at approximately 1,000 μmol photons m−2 s−1. When the sixth leaves (from base to apex) of grapevines became mature, all grapevines were divided into four groups and acclimated for 2 d in a controlled-environment room (70% average relative humidity, 25°C/18°C [12-h/12-h] day/night cycle, and photosynthetically active radiation at 800 μmol m−2 s−1). On day 3, the grapevines were subjected to the following treatments: (1) the plants of the control group were maintained at the optimal day/night temperature (25°C/18°C) in the above growth room; (2) the plants of the treatment groups were exposed to 35°C, 40°C, or 45°C from 11:30 am to 1:30 pm (the conditions were the same as the control, except for temperature). The fourth to sixth leaves (from base to apex) of each plant were detached at 1:30 pm (the end of the heat stress treatment). Each biological replicate included three plants, and three replicates were used for both the three treatments and the control. Leaves were frozen in liquid nitrogen immediately and stored at −80°C for further analysis.
Analysis of Chlorophyll Fluorescence Parameters
The O-J-I-P test was measured by a Handy Plant Efficiency Analyzer (Hansatech Instruments) after the leaves (using the fifth leaf from the base) were adapted for 15 min in the dark when heat treatments ended. Five independent replicates were used in three treatments and controls, and each replicate consisted of a plant. The chlorophyll a fluorescence transient was induced by a saturating photon flux density at 3,000 μmol photons m−2 s−1, provided by an array of six light-emitting diodes (peak of 650 nm). The fluorescence signals were recorded within a time span from 10 μs to 1 s, with a data acquisition rate of 10 μs for the first 2 ms and every 1 ms thereafter. The parameters Fv and Fm were measured, and Fv/Fm, which represents the intrinsic efficiency of PSII photochemistry (the ratio of variable and maximum fluorescence of PSII in the dark-adapted state), was calculated.
RNA Isolation and mRNA Sequencing
Total RNA was isolated from grape leaves by use of the RNAprep Pure Plant Kit (polysaccharides and polyphenolics rich; Tiangen) and quantified with the Nanodrop system (Thermo Fisher Scientific). DNase digestion was performed during total RNA isolation to remove genomic DNA. A total of 3 μg of RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly(T) oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction buffer (5×). First-strand cDNA was synthesized using random hexamer primer and M-MuLV reverse transcriptase (RNaseH−). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, deoxyribonucleotide triphosphates with dTTP were replaced by dUTP. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments with the right length, the library fragments were purified with the AMPure XP system (Beckman Coulter). Then, 3 µL of USER Enzyme (New England Biolabs) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, products were purified (AMPure XP system), and library quality was assessed on an Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit version 3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on the Illumina HiSeq platform, and paired-end reads were generated.
Sequencing Data Processing and Differential Expression Analysis
Adaptor sequences and low-quality sequences were first removed, and then clean reads were mapped to the grape reference genome (IGGP_12x) using TopHat (version 2.1.0) with Ensembl gene annotation as transcript index and fr-firststrand given to –library-type (Kim et al., 2013). New transcripts were assembled using Cufflinks (version 2.2.1; Trapnell et al., 2013). Raw read counts for each gene were calculated using HTSeq (version 0.6.0; Anders et al., 2015). Gene expression normalization among samples was performed using DESeq2. Then, differentially expressed genes were screened with fold change ≥ 2, FDR ≤ 0.01, and P < 0.05 (Love et al., 2014).
AS Analysis
AS events including ES, IR, MXE, A5SS, and A3SS were detected and quantified using rMATS with self-assembled transcript annotation (version 3.0.9; Shen et al., 2014). In brief, AS analysis was performed as shown in Supplemental Figure S14. IR is used as an example to illustrate the method of AS events. First, the exon inclusion level was represented by the count of uniquely mapped reads to the IR isoform or the ES isoform. The count of reads mapped to the IR isoform was represented by I. The count of reads mapped to the ES isoform was represented by S. Then, total count reads mapped to the IR isoform or ES isoform would be I + S, denoted as n. Second, as both isoforms had different lengths, isoform-specific read counts were normalized before calculating the intron inclusion levels by the lengths of isoform-specific effective segments. For a segment with length l and reads with length r, the effective length equaled the number of unique reads with intervals of l – r + 1. For IR, the effective length of the IR isoform was set as ll and that of the ES isoform as ls. After normalizing by effective length, the intron inclusion level could be denoted as α = (I/ll)/(I/ll + S/ls). Then, the proportion of reads from the IR isoform would be P = llα/[llα + ls(1 − α)]. Third, the comparative model to filter out differential IR events was constructed using rMATS. Other types of AS events also can be modeled by this framework, with details illustrated in Supplemental Figure S15 from Shen et al. (2014). The inclusion level of each candidate splicing event was calculated using reads mapped to splicing junctions. Differential splicing events under 35°C, 40°C, and 45°C compared with 25°C were selected with FDR ≤ 0.01. Sashimi plots used to show splicing events were generated by MISO (version 0.5.3; Katz et al., 2010).
Protein Sample Preparation
Grape samples (approximately 2 g each) were ground in liquid nitrogen, and the resulting powder was precipitated with 10% (w/v) TCA in cold acetone containing 0.07% (v/v) β-mercaptoethanol at −20°C overnight. Protein mixtures were denatured using 6 m guanidine-HCl in 50 mm Tris-HCl, pH 8, and then in-solution digested. Disulfide bonds were reduced by the addition of 20 mm DTT and incubation at 60°C for 30 min. After cooling to room temperature, 40 mm iodoacetamide was added, and the samples were kept in the dark for 1 h to block free Cys. The samples were dialyzed against 2 m urea and 50 mm NH4HCO3 for 2 h twice. Trypsin was added to the samples at 1:50 (w/w, trypsin:sample) and incubated at 37°C overnight. The digested peptides were further filtered with Amicon ultra centrifugal filters (10-kD MWKO; Merck Millipore) and were dried under vacuum extensively to remove residual primary amine from reagents. Samples were labeled with iTRAQ 4plex reagent (ABSciex) according to manufacturer’s instructions and combined after labeling and dried.
Analysis of High-pH Reverse-Phase HPLC and Nano-LC-MS/MS
iTRAQ-labeled total lysate (400 µg) was solubilized in 200 µL of buffer A (20 mm ammonium formate, pH 10) and separated on an Xbridge column (Waters; C18; 3.5 µm, 2.1 × 150 mm) using a linear gradient of 1% B min−1, then from 2% to 45% B (20 mm ammonium formate in 90% acetonitrile, pH 10) at a flow rate of 200 µL min−1 using the same Gilson system as described above. Fractions were collected at 1-min intervals and dried under vacuum.
Nano-LC-MS/MS was performed using a Dionex rapid-separation liquid chromatography system interfaced with a QExactive HF (Thermo Fisher Scientific). Samples were loaded onto a self-packed 100-µm × 2-cm trap packed with Magic C18AQ, 5 µm, 200 A (Michrom Bioresources), and washed with buffer A (0.2% formic acid) for 5 min at a flow rate of 10 µL min−1. The trap was brought in line with the homemade analytical column (Magic C18AQ; 3 µm, 200 A, 75 µm × 50 cm), and peptides were fractionated at 300 nL min−1 with a multistep gradient (4% to 15% buffer B [0.16% formic acid and 80% acetonitrile] during 20 min, then 15%–25% B during 40 min, followed by 25%–50% B for 30 min). Mass spectrometry data were acquired using a data-dependent acquisition procedure with a cyclic series of a full scan acquired with a resolution of 120,000 followed by tandem mass spectrometry (MS/MS) scans (30% collision energy in the HCD cell) with resolution of 30,000 of the 20 most intense ions with dynamic exclusion duration of 30 s.
Protein Data Analysis
iTRAQ quantification was performed with MaxQuant software version 1.5.3.8 (Cox and Mann, 2008; Cox et al., 2011) using a grape database (UniProt UP000009183; 29,907 protein sequences). MS/MS searches for the proteome data sets were performed with the following parameters: oxidation of Met and iTRAQ 4plex label on the N terminus of peptides as variable modifications; iTRAQ 4plex label on Lys as fixed modification; and carbamidomethylation as fixed modification. Trypsin/P was selected as the digestion enzyme, and a maximum peptide charge was set to six and two missed cleavages per peptide. Values of ±7 and 20 ppm were used as tolerances for precursor and product ions, respectively. Peptide and protein FDRs were set to 1%. Only proteins with at least two peptides were considered as reliably identified. Labeled protein quantification was switched on, and unique and razor peptides were considered for quantification with a minimum ratio count of 2. MS/MS identifications were transferred by b and y ions. The quantification was based on the extracted reporter intensity. The ratio between samples was calculated using reporter ion intensity and normalized to the median ratio of all identified spectra that fit certain criteria: peptide belongs to the grape database, and peptide FDR ≤ 0.01. Pairwise median ratios (35°C/25°C, 40°C/25°C, and 45°C/25°C) of individual proteins were calculated by the Doby package under the R environment using all spectra belonging to a protein that fit the criteria above and the sum of reporter ion intensity of both channels greater than 10,000. After the R analysis, proteins were further filtered by the number of unique peptides (≥1) and MS/MS counts (≥2) matched to each protein.
GO Analysis
GO analyses were performed using in-house scripts. The significance of enrichment of differentially expressed genes or differentially spliced genes in each GO term were determined at a threshold of adjusted P ≤ 0.05 using Fisher’s exact test together with the Benjamini-Hochberg FDR procedure. Visualizations of enriched GO terms were generated using R and Excel.
Quantitative Reverse Transcription-PCR Analysis
cDNA (20 μL volume) syntheses were performed using 800 ng of total RNA, an oligo(dT), and the HiScript Q RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme). qRT-PCR analysis involved the use of the AceQ qPCR SYBR Green Master Mix (without ROX; Vazyme). Primers designed by DNAMAN 8.0 (Adobe) are listed in Supplemental Table S3. qRT-PCR was performed in triplicate. VvACT2 (EC969944) was used as an internal control for normalization.
RT-PCR Analysis of IR Splicing
The RT-PCR splicing analysis of selected genes (HSP90-1, HSP25.3, HSP101, HSP70-4, and HSFA2) was performed in a total volume of 20 μL of cDNA. The first-strand cDNA was used for RT-PCR assays. The cycling conditions were 98°C for 3 min, followed by 35 cycles of a 10-s denaturation at 98°C, a 5-s annealing at 58°C to 61°C, and a 90-s extension at 72°C, with a final 2-min extension at 72°C. The PCR amplicons were analyzed using 2% agarose electrophoresis. The amount of first-strand cDNA used for the RT-PCR was determined by the expression of VvACTIN7. The forward and reverse primers were designed from the upstream and downstream exons of the introns based on each IR event, respectively. These primers are listed in Supplemental Table S4. The resulting RT-PCR products were cloned into pMD18T vector (Takara) and sequenced. All PCRs were performed using Ex-Taq polymerase (Takara).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under GEO accession number GSE89113.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Cluster analysis for all transcripts in grape leaves under four different temperatures (25°C, 35°C, 40°C, and 45°C).
Supplemental Figure S2. Summary number of differentially expressed transcripts in grape leaves under 35°C, 40°C, and 45°C compared with 25°C at a fold change greater than 2 and FDR less than 0.05.
Supplemental Figure S3. Flow chart of RNA sequencing and quantitative proteome for grape leaves.
Supplemental Figure S4. Venn diagram of nonredundant UniProt-annotated proteins identified with at least two unique peptides (protein and peptide FDR < 0.01) in grape leaves.
Supplemental Figure S5. Up- and down-regulated biological pathways of differentially expressed proteins in grape leaves under 35°C compared with 25°C.
Supplemental Figure S6. Up- and down-regulated biological pathways of differentially expressed proteins in grape leaves under 40°C compared with 25°C.
Supplemental Figure S7. Up- and down-regulated biological pathways of differentially expressed proteins in grape leaves under 45°C compared to 25°C.
Supplemental Figure S8. Linear correlation analysis (r2 = 0.79) between qRT-PCR and RNA-sequencing results for 35 genes in grape leaves under four temperatures (25°C, 35°C, 40°C, and 45°C).
Supplemental Figure S9. Sashimi plots of HSP90.1 transcripts with an IR event in grape leaves under 40°C and 45°C compared with 25°C.
Supplemental Figure S10. Sashimi plots of HSP25.3 transcripts with an IR event in grape leaves under 40°C and 45°C compared with 25°C.
Supplemental Figure S11. Sashimi plots of HSP101 transcripts with an IR event in grape leaves under 40°C and 45°C compared with 25°C.
Supplemental Figure S12. Sashimi plots of HSP70.4 transcripts with an IR event in grape leaves under 40°C and 45°C compared with 25°C.
Supplemental Figure S13. RT-PCR analysis of IR splice variants of HSP90.1, HSP25.3, HSP101, and HSP70.4 in grape leaves under 25°C, 35°C, 40°C, and 45°C for 2 h.
Supplemental Figure S14. Flow chart of AS analysis for grape leaves under different temperatures.
Supplemental Figure S15. Schematic diagrams illustrating the read counts and effective lengths of different categories of AS events.
Supplemental Table S1. Statistics of all AS events in grape leaves under 35°C, 40°C, and 45°C compared with 25°C.
Supplemental Table S2. Statistics of all genes with significantly different AS events in grape leaves under 35°C, 40°C, and 45°C compared with 25°C.
Supplemental Table S3. qRT-PCR primers used in this study.
Supplemental Table S4. Primers of RT-PCR splicing analysis used in this study.
Supplemental Data Set S1. Enriched analysis of genes with up-regulated IR events in grape leaves under each high-temperature (35°C, 40°C, and 45°C) treatment compared with the control (25°C).
Supplemental Data Set S2. Enriched analysis of genes with down-regulated IR events in grape leaves under each high-temperature (35°C, 40°C, and 45°C) treatment compared with the control (25°C).
Supplemental Data Set S3. Enriched analysis of genes with up-regulated ES events in grape leaves under each high-temperature (35°C, 40°C, and 45°C) treatment compared with the control (25°C).
Supplemental Data Set S4. Enriched analysis of genes with down-regulated ES events in grape leaves under each high-temperature (35°C, 40°C, and 45°C) treatment compared with the control (25°C).
Supplemental Data Set S5. Sequence analysis of IR transcripts and corresponding proteins of HSP90.1, HSP25.3, HSP101, HSP70.4, and HSFA2 genes in grape leaves under 40°C and 45°C.
Supplementary Material
Acknowledgments
We thank Baicheng Wang (Institute of Botany, Chinese Academy of Sciences), Douglas D. Archbold (Department of Horticulture, University of Kentucky), Yuehua Cui (Department of Statistics and Probability, Michigan State University), and Xiaoting Qi (College of Life Science, Capital Normal University) for critical reading and valuable suggestions and Dr. Tiancong Lu (ProteinWorld Biotechnology) for good advice regarding proteomics analysis.
Glossary
- iTRAQ
isobaric tags for relative and absolute quantitation
- GO
Gene Ontology
- FDR
false discovery rate
- AS
alternative splicing
- ES
exon skipping
- IR
intron retention
- MXE
mutually exclusive exon
- A3SS
alternative 3′ splice sites
- A5SS
alternative 5′ splice sites
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- PTC
premature termination codon
- SA
salicylic acid
- MS/MS
tandem mass spectrometry
Footnotes
Articles can be viewed without a subscription.
This work was supported by the Agricultural Science and Technology Innovation Program (grant no. CAAS-ASTIP-2016-ZFRI to C.L.) and the National Natural Science Foundation of China (grant no. 31270718 to L.W.).
References
- Amano M, Iida S, Kosuge K (2012) Comparative studies of thermotolerance: different modes of heat acclimation between tolerant and intolerant aquatic plants of the genus Potamogeton. Ann Bot (Lond) 109: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L (2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80: 1089–1115 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M, Harris PJC (2013) Photosynthesis under stressful environments: an overview. Photosynthesis 51: 163–190 [Google Scholar]
- Bita CE, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokszczanin KL. (2013) Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front Plant Sci 4: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösl B, Grimminger V, Walter S (2006) The molecular chaperone Hsp104: a molecular machine for protein disaggregation. J Struct Biol 156: 139–148 [DOI] [PubMed] [Google Scholar]
- Capovilla G, Pajoro A, Immink RGH, Schmid M (2015) Role of alternative pre-mRNA splicing in temperature signaling. Curr Opin Plant Biol 27: 97–103 [DOI] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Santa María E, Torres-Pérez R, Royo C, Lijavetzky D, Bravo G, Aguirreolea J, Sánchez-Díaz M, Antolín MC, Martínez-Zapater JM (2013) Thermotolerance responses in ripening berries of Vitis vinifera L. cv Muscat Hamburg. Plant Cell Physiol 54: 1200–1216 [DOI] [PubMed] [Google Scholar]
- Chang CY, Lin WD, Tu SL (2014) Genome-wide analysis of heat-sensitive alternative splicing Physcomitrella patens. Plant Physiol 165: 826–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lin S, Liu Q, Huang J, Zhang W, Lin J, Wang Y, Ke Y, He H (2014) Expression and interaction of small heat shock proteins (sHsps) in rice in response to heat stress. Biochim Biophys Acta 1844: 818–828 [DOI] [PubMed] [Google Scholar]
- Cheng Q, Zhou Y, Liu Z, Zhang L, Song G, Guo Z, Wang W, Qu X, Zhu Y, Yang D (2015) An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol (Stuttg) 17: 419–429 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Duque P. (2011) A role for SR proteins in plant stress responses. Plant Signal Behav 6: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60: 3891–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George IS, Pascovici D, Mirzaei M, Haynes PA (2015) Quantitative proteomic analysis of Cabernet Sauvignon grape cells exposed to thermal stresses reveals alterations in sugar and phenylpropanoid metabolism. Proteomics 15: 3048–3060 [DOI] [PubMed] [Google Scholar]
- González-Schain N, Dreni L, Lawas LMF, Galbiati M, Colombo L, Heuer S, Jagadish KSV, Kater MM (2016) Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol 57: 57–68 [DOI] [PubMed] [Google Scholar]
- Greer DH, Weedon MM (2012) Modelling photosynthetic responses to temperature of grapevine (Vitis vinifera cv. Semillon) leaves on vines grown in a hot climate. Plant Cell Environ 35: 1050–1064 [DOI] [PubMed] [Google Scholar]
- Grover A, Mittal D, Negi M, Lavania D (2013) Generating high temperature tolerant transgenic plants: achievements and challenges. Plant Sci 205-206: 38–47 [DOI] [PubMed] [Google Scholar]
- Györgyey J, Gartner A, Németh K, Magyar Z, Hirt H, Heberle-Bors E, Dudits D (1991) Alfalfa heat shock genes are differentially expressed during somatic embryogenesis. Plant Mol Biol 16: 999–1007 [DOI] [PubMed] [Google Scholar]
- He ZS, Xie R, Zou HS, Wang YZ, Zhu JB, Yu GQ (2007) Structure and alternative splicing of a heat shock transcription factor gene, MsHSF1, in Medicago sativa. Biochem Biophys Res Commun 364: 1056–1061 [DOI] [PubMed] [Google Scholar]
- Isshiki M, Tsumoto A, Shimamoto K (2006) The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 18: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y, Goto M, Shima D, Imai T, Wada T, Morohashi Ki, Shirakawa M, Hirose S, Handa H (1999) The role of human MBF1 as a transcriptional coactivator. J Biol Chem 274: 34196–34202 [DOI] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40: 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165 [DOI] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, Prášil IT, Renaut J (2011) Plant proteome changes under abiotic stress: contribution of proteomics studies to understanding plant stress response. J Proteomics 74: 1301–1322 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Knight MR (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM (2000) The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol Biol 42: 571–581 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Yang KA, Hong JK, Choi JS, Yun DJ, Hong JC, Chung WS, Lee SY, Cho MJ, Lim CO (2006) Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cells. J Plant Res 119: 373–383 [DOI] [PubMed] [Google Scholar]
- Liu GT, Ma L, Duan W, Wang BC, Li JH, Xu HG, Yan XQ, Yan BF, Li SH, Wang LJ (2014) Differential proteomic analysis of grapevine leaves by iTRAQ reveals responses to heat stress and subsequent recovery. BMC Plant Biol 14: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GT, Wang JF, Cramer G, Dai ZW, Duan W, Xu HG, Wu BH, Fan PG, Wang LJ, Li SH (2012) Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol 12: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun N, Liu M, Liu J, Du B, Wang X, Qi X (2013) An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol 162: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QX, Jindra M, Ueda H, Hiromi Y, Hirose S (2003) Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development 130: 719–728 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL (2008) Prioritizing climate change adaptation needs for food security in 2030. Science 319: 607–610 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AA, Rhoads DM, Lund AL, Cerny RL, Elthon TE (2001) In vivo modifications of the maize mitochondrial small heat stress protein, HSP22. J Biol Chem 276: 29924–29929 [DOI] [PubMed] [Google Scholar]
- Martel AB, Qaderi MM (2016) Does salicylic acid mitigate the adverse effects of temperature and ultraviolet-B radiation on pea (Pisum sativum) plants? Environ Exp Bot 122: 39–48 [Google Scholar]
- Mastrangelo AM, Marone D, Laidò G, De Leonardis AM, De Vita P (2012) Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci 185-186: 40–49 [DOI] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy AS (2007) Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Pillet J, Egert A, Pieri P, Lecourieux F, Kappel C, Charon J, Gomès E, Keller F, Delrot S, Lecourieux D (2012) VvGOLS1 and VvHsfA2 are involved in the heat stress responses in grapevine berries. Plant Cell Physiol 53: 1776–1792 [DOI] [PubMed] [Google Scholar]
- Potenza E, Racchi ML, Sterck L, Coller E, Asquini E, Tosatto SCE, Velasco R, Van de Peer Y, Cestaro A (2015) Exploration of alternative splicing events in ten different grapevine cultivars. BMC Genomics 16: 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 228: 111–133 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, White SJ, Zhou Y, Muralidharan M, Elthon TE (2005) Altered gene expression in plants with constitutive expression of a mitochondrial small heat shock protein suggests the involvement of retrograde regulation in the heat stress response. Physiol Plant 123: 435–444 [Google Scholar]
- Rozenzvieg D, Elmaci C, Samach A, Lurie S, Porat R (2004) Isolation of four heat shock protein cDNAs from grapefruit peel tissue and characterization of their expression in response to heat and chilling temperature stresses. Physiol Plant 121: 421–428 [Google Scholar]
- Sagor GHM, Berberich T, Takahashi Y, Niitsu M, Kusano T (2013) The polyamine spermine protects Arabidopsis from heat stress-induced damage by increasing expression of heat shock-related genes. Transgenic Res 22: 595–605 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S, Jamshed M, Samuel MA (2013) Proteomics approaches advance our understanding of plant self-incompatibility response. J Proteome Res 12: 4717–4726 [DOI] [PubMed] [Google Scholar]
- Salazar-Parra C, Aguirreolea J, Sanchez-Diaz M, Irigoyen JJ, Morales F (2010) Effects of climate change scenarios on Tempranillo grapevine (Vitis vinifera L.) ripening: response to a combination of elevated CO2 and temperature, and moderate drought. Plant Soil 337: 179–191 [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA 111: E5593–E5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkel K. (2006) Thermostability of photosynthesis of Vitis aestivalis and Vitis vinifera. J Am Soc Hortic Sci 131: 476–483 [Google Scholar]
- Staiger D, Brown JWS (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Dreos R, Aparicio F, Maule AJ (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R (2008) The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283: 9269–9275 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R (2011) Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J 66: 844–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NH, Kalyna M, Marquez Y, Barta A, Brown JWS (2012) Alternative splicing in plants: coming of age. Trends Plant Sci 17: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Harashima S, Ueda H, Hirose S (1998) Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol 18: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nishiyama Y, Murata N (2000) Acclimation of the photosynthetic machinery to high temperature in Chlamydomonas reinhardtii requires synthesis de novo of proteins encoded by the nuclear and chloroplast genomes. Plant Physiol 124: 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Tsuji T, Hirose S, Yamazaki K (2004) Three Arabidopsis MBF1 homologs with distinct expression profiles play roles as transcriptional co-activators. Plant Cell Physiol 45: 225–231 [DOI] [PubMed] [Google Scholar]
- Vierling E. (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- Vitulo N, Forcato C, Carpinelli EC, Telatin A, Campagna D, D’Angelo M, Zimbello R, Corso M, Vannozzi A, Bonghi C, et al. (2014) A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol 14: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad M (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61: 199–223 [Google Scholar]
- Wang K, Zhang X, Goatley M, Ervin E (2014) Heat shock proteins in relation to heat stress tolerance of creeping bentgrass at different N levels. PLoS ONE 9: e102914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LJ, Li SH (2006a) Salicylic acid-induced heat or cold tolerance in relation to Ca2+ homeostasis and antioxidant systems in young grape plants. Plant Sci 170: 685–694 [Google Scholar]
- Wang LJ, Li SH (2006b) Thermotolerance and related antioxidative enzyme activities induced by heat acclimation or salicylic acid in grape leaves (Vitis vinifera L. cv. Jingxiu). Plant Growth Regul 48: 137–144 [Google Scholar]
- Wang XC, Li Q, Jin X, Xiao GH, Liu GJ, Liu NJ, Qin YM (2015) Quantitative proteomics and transcriptomics reveal key metabolic processes associated with cotton fiber initiation. J Proteomics 114: 16–27 [DOI] [PubMed] [Google Scholar]
- Wu Z, Yuan X, Li H, Liu F, Wang Y, Li J, Cai H, Wang Y (2015) Heat acclimation reduces postharvest loss of table grapes during cold storage: analysis of possible mechanisms involved through a proteomic approach. Postharvest Biol Technol 105: 26–33 [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu G, Liu G, Yan B, Duan W, Wang L, Li S (2014) Comparison of investigation methods of heat injury in grapevine (Vitis) and assessment to heat tolerance in different cultivars and species. BMC Plant Biol 14: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M (2007) Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem 282: 37794–37804 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yan Q, Hou H, Singer SD, Yan X, Guo R, Wang X (2014) The grape VvMBF1 gene improves drought stress tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult 118: 571–582 [Google Scholar]
- Zhang B, Peñagaricano F, Driver A, Chen H, Khatib H (2011) Differential expression of heat shock protein genes and their splice variants in bovine preimplantation embryos. J Dairy Sci 94: 4174–4182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.