Vernalization-induced VIN3 enhances the PRC2 activity through its histone binding motif, PHD-finger domain, to mediate proper vernalization response in Arabidopsis.
Abstract
Vernalization is a response to winter cold to initiate flowering in spring. VERNALIZATION INSENSITIVE3 (VIN3) is induced by winter cold and is essential to vernalization response in Arabidopsis (Arabidopsis thaliana). VIN3 encodes a PHD-finger domain that binds to modified histones in vitro. An alteration in the binding specificity of the PHD-finger domain of VIN3 results in a hypervernalization response. The hypervernalization response is achieved by increased enrichments of VIN3 and trimethylation of Histone H3 Lys 27 at the FLC locus without invoking the increased enrichment of Polycomb Repressive Complex 2. Our result shows that the binding specificity of the PHD-finger domain of VIN3 plays a role in mediating a proper vernalization response in Arabidopsis.
Some plants need an exposure to winter cold to be competent to initiate floral transition in the following spring. This phenomenon is known as vernalization (Chouard, 1960; Bernier et al., 1981). Vernalization results in the repression of a floral repressor, FLOWERING LOCUS C (FLC), in Arabidopsis (Arabidopsis thaliana), and the stable repression of FLC permits the activation of downstream floral activators to initiate flowering (Lee et al., 2000; Helliwell et al., 2006; Searle et al., 2006; Michaels and Amasino, 1999, 2001). The transcriptional level of FLC is tightly controlled by a number of chromatin-modifying complexes and therefore correlates with various histone modifications (Amasino and Michaels, 2010; Bratzel and Turck, 2015; Shafiq et al., 2014). Stable changes in histone modification signatures at FLC chromatin provide the molecular explanation how the stable nature of transcriptional repression of FLC is achieved by vernalization.
Before vernalization, FLC chromatin is enriched with active histone marks, including trimethylation of Histone H3 Lys 4 (H3K4me3) and trimethylation of Histone H3 Lys 36 (H3K36me3; Sung et al., 2006; Greb et al., 2007; Yang et al., 2014). These active histone marks are generally correlated with active transcription by RNA Polymerase II (Pol II) (Choi et al., 2011; Barski et al., 2007) and mediated by activating chromatin-modifying complexes, including the EFS-H3K4/H3K36 methyltransferase (Zhao et al., 2005; Kim et al., 2005; Ko et al., 2010). Upon vernalization, these active histone marks at FLC chromatin decrease, and the level of decrease correlates with the level of transcriptional repression of FLC. On the contrary, the enrichment of repressive histone marks, such as dimethylation of Histone H3 Lys 9 (H3K9me2) and trimethylation of Histone H3 Lys 27 (H3K27me3), increases at FLC chromatin by vernalization (Kim and Sung, 2013; Sung and Amasino, 2004; Bastow et al., 2004; Yang et al., 2014). Other FLC-clade members also undergo similar changes in their chromatin signatures by vernalization, contributing to the fine-tuning of vernalization response in Arabidopsis (Sheldon et al., 2009; Kim and Sung, 2013).
A distinct characteristic of vernalization is that vernalization is a delayed response to temperature changes. Changes in histone modifications at FLC chromatin correlate with the level of FLC repression and occur over an extended period of cold exposure, usually several weeks (Kim and Sung, 2013). The requirement for an extended period of cold exposure renders plants able to respond only to winter cold, but not to temperature fluctuations.
VERNALIZATION INSENSITIVE3 (VIN3) is a protein only produced during winter cold. The induction kinetics of VIN3 by cold correlate with the degree of the vernalization response (Sung and Amasino, 2004). Little change of repressive histone marks occur at FLC chromatin in vin3 mutants during exposure to cold (Kim and Sung, 2013; Sung and Amasino, 2004). VIN3 forms a complex with Polycomb Repressive Complex 2 (PRC2), which catalyzes trimethylation of H3 Lys-27, a repressive histone mark, and triggers the stable repression of FLC (De Lucia et al., 2008; Wood et al., 2006). VIN3 is required for the PRC2-mediated H3K27 methylation at FLC and FLC clade (Kim and Sung, 2013; Sung and Amasino, 2004; De Lucia et al., 2008). VIN3 is a PHD-finger domain protein that preferentially recognizes H3K9me2, a repressive histone mark (Kim and Sung, 2013). VIN3 also physically interacts with its related protein, VIN3-LIKE1 (VIL1)/VERNALIZATION5 (VRN5; Sung et al., 2006; Greb et al., 2007). VIL1 also biochemically copurifies with PRC2 core components and contributes to the vernalization-mediated FLC repression (De Lucia et al., 2008). Although VIN3 plays an essential role in repression of FLC during vernalization, in part through the recognition of a repressive histone mark at FLC chromatin, how VIN3 coordinates the proper epigenetic switch at FLC in response to long-term cold is not well understood.
In this report, we found that an altered binding preference of the PHD-finger domain of VIN3 results in hypervernalization responses through the enhanced accumulation of H3K27me3 at FLC chromatin. Our results suggest that the histone binding property of VIN3 provides another layer of regulatory modes to ensure proper vernalization response in Arabidopsis.
RESULTS
Altered Binding Preference of VIN3 PHD-Finger Domain in Vitro
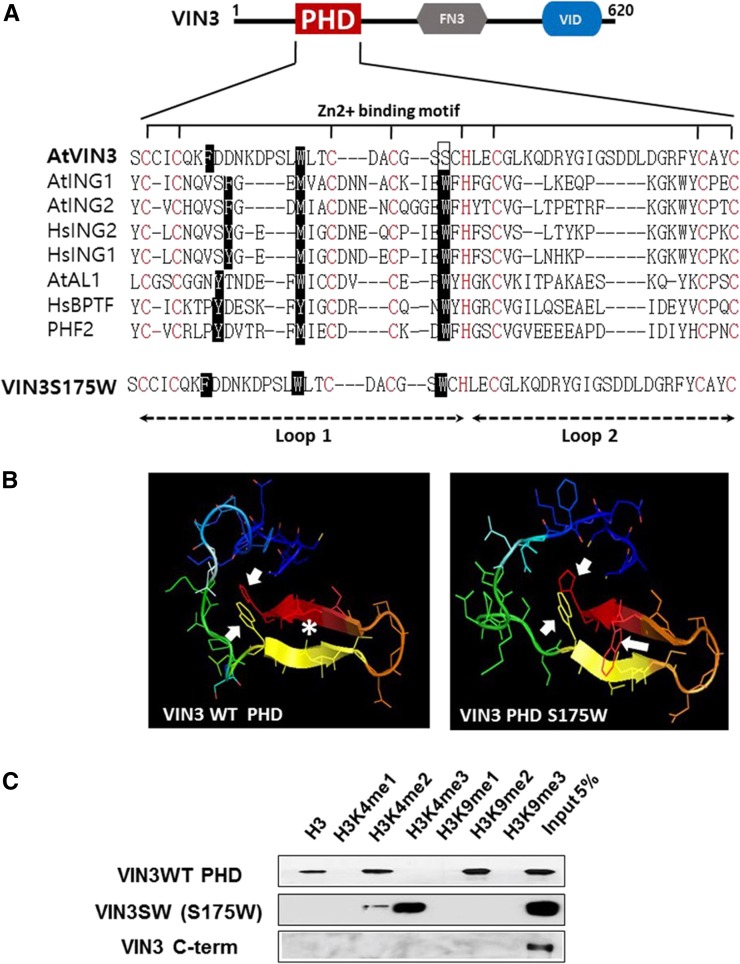
The PHD-finger domain of VIN3 contains two hydrophobic aromatic ring residues that form a putative histone-binding pocket (Figs. 1A; Supplemental Fig. S1). Most PHD-finger domains that have binding specificity for trimethylated histone tails contain at least three hydrophobic residues within the Loop 1 (Fig. 1A). Computer modeling shows that VIN3-PHD finger domain forms a histone binding cage with only two hydrophobic aromatic residues that is capable of binding up to two methylated Lys residues (Fig. 1B). Consistent with the model, the VIN3-PHD finger domain preferentially binds to dimethylated H3K9 (a repressive histone mark), although it also exhibits binding to unmodified histone H3 and dimethylated H3K4 in vitro (Fig. 1C; Kim and Sung, 2013). The majority of PHD finger proteins recognize an active histone mark, H3K4me3, via a binding pocket with three hydrophobic residues within the PHD-finger domain (Peña et al., 2006; Shi et al., 2006; Taverna et al., 2006; Matthews et al., 2007; Musselman and Kutateladze, 2011). However, the VIN3-PHD finger domain lacks the third hydrophobic residue, Trp (W). Instead, the VIN3-PHD finger domain contains a Ser (Ser-175) residue at the position of Trp (Fig. 1A). This difference (W→S) is found in all VIN3 family of proteins in Arabidopsis and in other plant species (Supplemental Fig. S1). Because all known H3K4me3 binding PHD-finger domains contain Trp at the position of VIN3 Ser-175, we substituted VIN3 Ser-175 to Trp (hereafter referred to VIN3SW; Fig. 1A). A predicted structure of VIN3SW-PHD finger domain indicates that the histone binding pocket is capable of recognizing three methylated Lys residues. (Fig. 1B). Indeed, in vitro histone peptide binding assays showed that the binding preference of VIN3SW shifted toward H3K4me3 (an active histone mark; Fig. 1C). A similar alteration in the binding preference is also observed in VIL1/VRN5-PHD finger domain (Supplemental Fig. S1B), indicating that the Ser residue is important for specifying the binding preference of VIN3 family of proteins. This result shows that a single amino acid substitution results in a major change of the binding preference of VIN3-PHD finger domain.
Figure 1.
An alteration in the PHD-finger domain of VIN3 results in the change in the histone binding specificity. A, Multiple alignments of PHD-finger domains between known H3K4me3-binding PHD-finger domains and VIN3 (AtVIN3). A modified VIN3, in which Ser at 175 residue is substituted to Trp residue, is shown at the bottom. B, Predicted three-dimensional structures of PHD-finger domain of VIN3WT (wild type) and VIN3SW (modified). Arrows indicate aromatic ring residues forming histone binding pockets, and the asterisk indicates a polar charged Ser-175 residue of VIN3. C, In vitro histone peptides binding assay using His-tagged recombinant VIN3WT and VIN3SW proteins. The full list of synthesized histone peptides used in this study is described in Supplemental Table S2.
Accelerated Vernalization Response by Altered Histone Binding Preference of VIN3
Among the components of PRC2 that trigger vernalization-mediated H3K27 methylation at FLC, VIN3 is the only component that is induced by cold temperature (De Lucia et al., 2008; Kim and Sung, 2013). Therefore, it is reasonable to speculate that ectopic expression of VIN3 would enable a vernalization response without cold, including the enrichment of H3K27 methylation at FLC by PRC2. However, the ectopic expression of VIN3 alone does not result in any alteration in vernalization response or FLC expression as determined by flowering time, although it is capable of complementing vin3 mutants (Fig. 2; Lee et al., 2015).
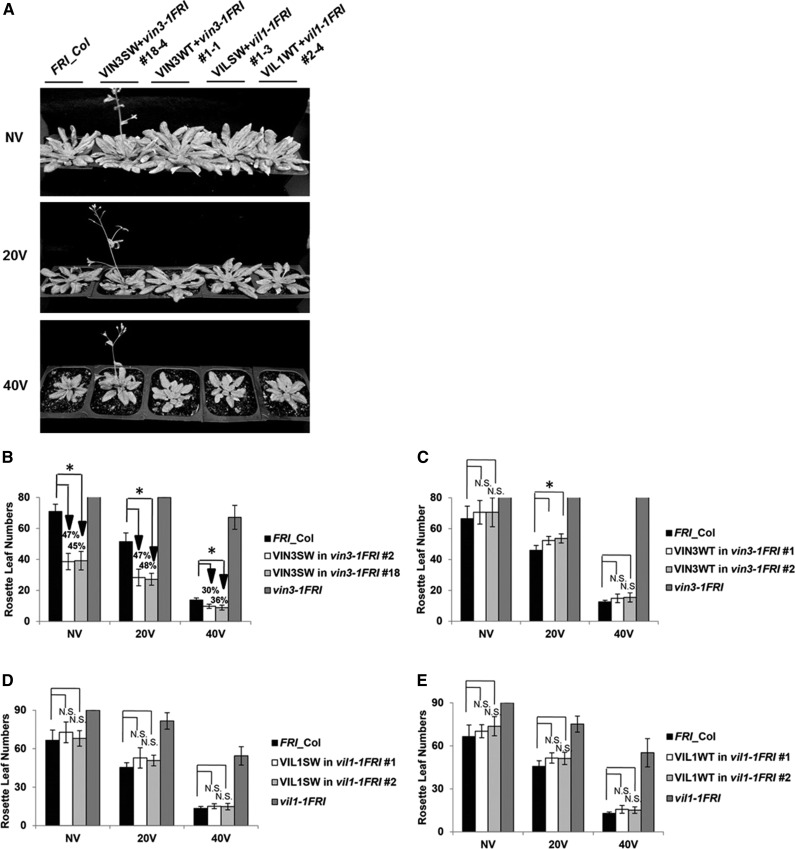
Figure 2.
The hypervernalization response in VIN3SW transgenic lines. A, Accelerated flowering is observed in VIN3SW transgenic plants in vin3-1FRI mutant background in all vernalization time points. NV, Nonvernalized; 20V, 20 d vernalized; 40V, 40 d vernalized. Plants were grown under long-day conditions (16 h light and 8 h dark) after vernalization treatment. B, Flowering times of two representative homozygous transgenic plants of VIN3SW in vin3-1FRI after various durations of vernalization treatment. Earlier flowering are indicated by downward arrows and changes in the percentage compared to the wild-type plants (FRI_Col). C, Flowering times of two representative transgenic plants of VIN3WT in vin3-1FRI after various durations of vernalization treatment. B and C, FRI_Col (wild type) and vin3-1FRI mutants are used as controls for vernalization response. D, Flowering times of two representative homozygous transgenic plants of VIL1SW in vil1-1FRI after various durations of vernalization treatment. E, Flowering times of two representative homozygous transgenic plants of VIL1WT in vil1-1FRI after various durations of vernalization treatment. D and E, FRI_Col (wild type) and vil1-1FRI mutants are used as controls for vernalization response. B to D, Asterisks indicate statistically significant differences in flowering times compared to the wild-type (FRI_Col) plants. N.S., Not significant (Student’s t test, P < 0.01; n = 12).
H3K4me3 is generally associated with the active transcription and this modification is enriched at FLC chromatin prior to vernalization (Pien et al., 2008; Tamada et al., 2009; Yu and Michaels, 2010; Yang et al., 2014). The wild-type VIN3 protein does not bind to H3K4me3 in vitro, and a predicted model indicates that the wild-type VIN3-PHD finger lacks an essential hydrophobic residue to recognize H3K4me3 (Fig. 1, B and C). Thus, we reasoned that H3K4me3-enriched FLC chromatin might interfere with the function of VIN3. To address the effect of the histone binding preference of VIN3, we introduced the wild-type VIN3 (hereafter referred to VIN3WT) and the VIN3SW under the control of the constitutive 35S promoter into vin3 mutant background (Fig. 2; Supplemental Fig. S2). We evaluated the flowering behavior of transgenic lines and nontransgenic wild-type plants with different periods of vernalization treatment (Fig. 2). Interestingly, we observed accelerated flowering in VIN3SW transgenic lines in all tested periods of vernalization treatment compared to nontransgenic plants (Fig. 2, A and B; Supplemental Fig. S2C). Furthermore, this accelerated flowering is observed only in VIN3SW transgenic lines, but not in VIN3WT transgenic lines (Fig. 2, A and B; Supplemental Fig. S2C). This is not due to the difference in the level of expression of transgenes because both transgenic lines constitutively express VIN3 at a level up to 100-fold compared to the maximum level of expression of VIN3 in nontransgenic wild type (Supplemental Fig. S2, A and B). Similar accelerated flowering was also observed when the VIN3SW transgene was introduced into wild-type plants, further confirming the biological effect of the amino acid change (Supplemental Fig. S3).
We also introduced the wild-type VIL1 (VIL1WT) and VIL1SW (in which a conserved Ser-102 residue is substituted with Trp in its PHD-finger domain) transgenes driven by the constitutive 35S promoter into vil1 mutants (Fig. 2, A, D, and E; Supplemental Fig. S2). Although both VIL1WT and VIL1SW can rescue vil1 mutants, none of them shows a significant difference in flowering time under different periods of vernalization treatment compared to the wild type (Fig. 2, A, D, and E). Therefore, the binding preference of the VIL1 PHD finger does not affect vernalization response. On the other hand, accelerated flowering observed in VIN3SW suggests that the binding preference of VIN3 plays roles in the repression of FLC and proper vernalization response.
Hyper-Repression of FLC in VIN3SW Transgenic Lines
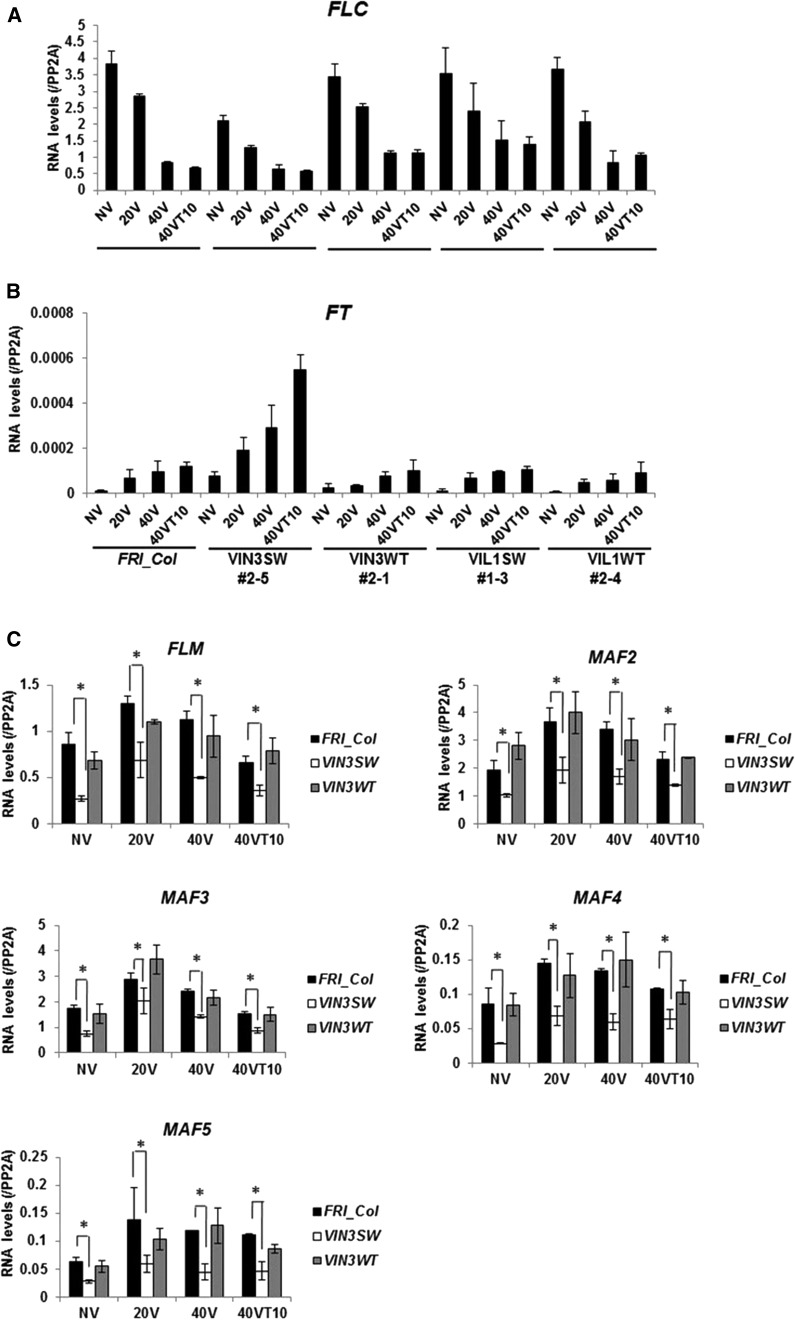
To determine the molecular basis of accelerated flowering observed in VIN3SW plants, we evaluated mRNA expression of FLC during the course of vernalization. The different degrees of more rapid flowering time over different periods of vernalization treatment are correlated with the levels of FLC expression in all plants (Fig. 3). Particularly, FLC mRNA transcripts in VIN3SW showed significant decreases in all periods of vernalization treatment relative to the wild type, even in the absence of vernalization (Fig. 3A). This result shows that an altered histone binding preference of VIN3-PHD finger domain is capable of repressing FLC, regardless of vernalization treatment. However, it should also be noted that the degree of FLC repression in VIN3SW without vernalization treatment is not at the same level as in fully vernalized wild-type plants, suggesting that other vernalization-specific events must occur to achieve the full vernalization-mediated FLC repression.
Figure 3.
A, The levels of FLC mRNA in FRI_Col (WT) and transgenic plants during vernalization. B, The levels of FT mRNA in FRI_Col (WT) and transgenic plants during vernalization. C, The levels of FLC clade genes, FLM and MAF2 to MAF5, in FRI_Col (WT) and transgenic plants during vernalization. Statistically significant changes are indicated by asterisks (Student’s t test, P < 0.01; n = 3). N.S., Not significant. A to C, PP2A (At1g13320) is used as a control for qRT-PCR analyses. Data (relative levels; mean ± sd of quantitative RT-PCR; n = 3). NV, Nonvernalized; 20V, 20 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature.
FLC is a MADS-box transcription factor that directly represses expressions of floral activators, including FLOWERING LOCUS T (FT; Amasino and Michaels, 2010; Michaels, 2009; Michaels and Amasino, 1999; Searle et al., 2006). Therefore, we examined the level of FT over different periods of vernalization treatment (Fig. 3B). Overall, the levels of FT in all plants show negative correlations with FLC expression as expected. Interestingly, FT is hyperexpressed in VIN3SW lines in all periods of vernalization treatment (Fig. 3B). This is consistent with observed earlier flowering of VIN3SW in all periods of vernalization treatment compared to the wild-type and VIN3WT transgenic plants (Fig. 2B). VIN3 is also required to repress other FLC clade (FLM and MAF2 to MAF5) by vernalization through the PRC2-mediated H3K27 methylation, similar to the FLC repression by vernalization (Kim and Sung, 2013), and other FLC clade members also act as floral repressor through the repression of floral activators (Gu et al., 2013). Consistent with the role of VIN3 in the repression of the FLC clade, the mRNA levels of all FLC clade members also decreased in VIN3SW over the course of vernalization treatment (Fig. 3C), contributing to the higher level of FT in VIN3SW.
Vernalization-Mediated Enrichment of VIN3 at FLC Chromatin
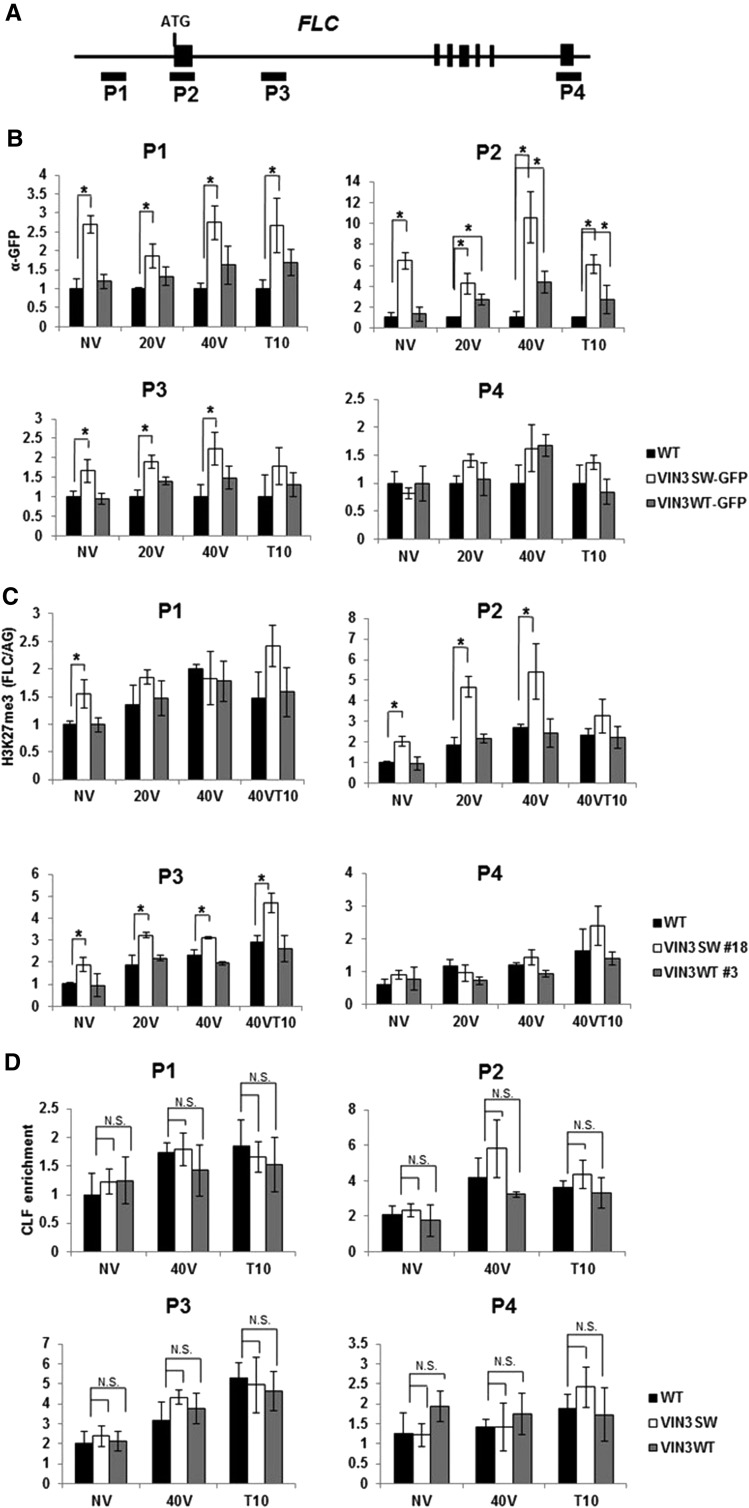
To address whether the binding preference of VIN3 plays a role in the enrichment of VIN3 at FLC chromatin, we measured the levels of VIN3 enrichment using chromatin immunoprecipitation (ChIP) assay (Fig. 4, A and B). Although both VIN3WT and VIN3SW are ectopically overexpressed (Supplemental Fig. S2), significant enrichment of VIN3 at FLC chromatin is observed only in VIN3SW transgenic lines without vernalization. The enrichment of VIN3WT is eventually observed when plants are vernalized (Fig. 4, A and B). This result indicates that the native VIN3 is not capable of enriching at FLC chromatin unless vernalized.
Figure 4.
Changes at FLC chromatin during vernalization. A, A schematic representation of genomic structure of FLC and relative positions of the primers used in ChIP assays are shown (P1 to P4). B, The levels of enrichment of VIN3 at FLC chromatin in FRI_Col (WT), VIN3WT, and VIN3SW transgenic lines during vernalization. C, The levels of enrichment of H3K27me3 in FRI_Col (WT), VIN3WT, and VIN3SW transgenic lines during vernalization. D, The levels of enrichment of CLF in FRI_Col (WT), VIN3WT, and VIN3SW transgenic lines during vernalization. B to D, Mean ± sd of quantitative PCR; n = 3. Statistically significant changes are indicated by asterisks (Student’s t test, P < 0.01). N.S., Not significant; NV, nonvernalized; 20V, 20 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. WT, FRI_Col; VIN3SW, VIN3SW in vin3-1FRI plants; VIN3WT, VIN3WT in vin3-1FRI.
On the other hand, VIN3SW accumulates at FLC chromatin even in the absence of vernalization treatment. Prior to vernalization, FLC chromatin is enriched with H3K4me3 (Supplemental Fig. S4; Yu and Michaels, 2010; Adrian et al., 2009; Kim and Sung, 2013; Ding et al., 2013; Li et al., 2016). This suggests that the H3K4me3 binding of VIN3SW allows the protein to accumulate at H3K4me3-enriched FLC chromatin and to modulate FLC repression. In addition, the lower level of H3K4me3 enrichment is observed in VIN3SW lines before vernalization, indicating that the association of VIN3 protein subsequently reduced the level of H3K4me3 at FLC chromatin. The wild-type VIN3 eventually accumulates at FLC when plants are vernalized (Supplemental Fig. S4; when the level of H3K4me3 at FLC chromatin decreases). Therefore, the wild-type VIN3 may function to monitor the status of chromatin modifications at FLC and also may contribute to the demethylation of H3K4me3 at FLC chromatin.
Vernalization-Mediated Enrichment of H3K27me3 and PRC2 at FLC Chromatin
VIN3 is required for the PRC2-mediated H3K27me3 enrichment at FLC by vernalization and forms a complex with PRC2 (Wood et al., 2006; De Lucia et al., 2008; Kim and Sung, 2013). First, we tested whether the increased enrichment of VIN3SW can trigger increased H3K27me3 enrichment at FLC chromatin. The level of H3K27me3 at FLC chromatin in VIN3SW transgenic plants is substantially higher than those in wild-type and VIN3WT plants under all conditions, including without vernalization (NV; Fig. 4C), suggesting that the increased enrichment of VIN3 can trigger hyper-H3K27me3 enrichment at FLC chromatin without vernalization. One simple explanation would be that the enrichment of VIN3SW triggers the increased enrichment of PRC2, which mediates trimethylation of H3K27 at FLC chromatin.
PRC2 preoccupies FLC chromatin prior to vernalization, and its association with FLC chromatin increases during and after vernalization, which results in the increased level of H3K27me3 (De Lucia et al., 2008; Kim and Sung, 2013). However, the increased level of H3K27me3 at FLC chromatin in VIN3SW may not be due to the increased enrichment of PRC2. There is little difference in the level of enrichment of a core component of PRC2, CLF (an H3K27 methyltransferase), in the VIN3SW lines compared to nontransgenic wild type or VIN3WT, especially without vernalization (Fig. 4D). Therefore, this result indicates that the increased enrichment of VIN3 at FLC chromatin is able to enhance the catalytic activity of PRC2, methylation of H3K27, at FLC without additional recruitment of the CLF-containing PRC2.
DISCUSSION
Vernalization is a response to the long-term cold of winter. Plants have evolved intricate systems to measure the duration of cold and to monitor other seasonal changes. The induction of VIN3 itself correlates with the duration of cold and perhaps functions to ensure that the vernalization response occurs only from long-term cold (Sung and Amasino, 2004). It should be noted that the ectopic expression of VIN3SW and its increased level of enrichment at FLC do not achieve a “fully” vernalized state without vernalization, suggesting that other cold-specific events must occur to achieve the “full” vernalization response. Natural variation studies identified multiple putative cis-elements within FLC locus that are necessary to mediate proper vernalization response (Li et al., 2014; Coustham et al., 2012). In addition, several noncoding RNAs have been reported to play roles in vernalization-mediated FLC repression (Heo and Sung, 2011; Csorba et al., 2014). Therefore, the vernalization response utilizes multiple regulatory modes to ensure that vernalization occurs in response to only prolonged cold of winter and the binding preference of VIN3-PHD finger also plays an important role in mediating proper vernalization response through monitoring the status of FLC chromatin.
Unlike VIN3, the altered histone binding preference of VIL1 (VIL1SW) does not result in difference in vernalization response. The enrichment of VIL1 at FLC chromatin by vernalization depends on the presence of VIN3 (De Lucia et al., 2008). In addition, VIN3 functions to initiate the repression of FLC by vernalization (Helliwell et al., 2011), whereas VIL1 is mainly involved in the maintenance of the repressed state of FLC (Sung et al., 2006; Greb et al., 2007). Nonetheless, VIL1SW is still able to maintain the repressed state of FLC (Figs. 2D and 3A), suggesting that the histone binding preference of VIL1 is not critical for the vernalization.
VIN3 protein cannot be enriched at FLC chromatin when FLC chromatin is enriched with H3K4me3 in the absence of vernalization treatment. Vernalization-mediated FLC repression also includes the decrease in the level of H3K4me3 at FLC chromatin (Sheldon et al., 2009; Tamada et al., 2009; Kim and Sung, 2013; Yang et al., 2014). Therefore, the level of H3K4me3 enrichment at FLC chromatin must decrease to allow VIN3-PRC2 to trigger the methylation of H3K27 upon vernalization (Fig. 5). There are two classes of putative histone demethylases known in Arabidopsis, KDM1/LSD1-type demethylases and JumonjiC-domain-containing demethylases (Liu et al., 2010). KDM1/LSD1-type demethylases appear not to function in the vernalization response (Jiang et al., 2007). There are 21 JumonjiC-domain-containing histone demethylases in Arabidopsis (Lu et al., 2008), and many JumonjiC-domain-containing histone demethylases have been reported to function in a number of developmental programs in Arabidopsis (Luo et al., 2014; Chen et al., 2011). Whether active demethylation events by JumonjiC-domain-containing histone demethylases occur at FLC chromatin by vernalization remains to be addressed.
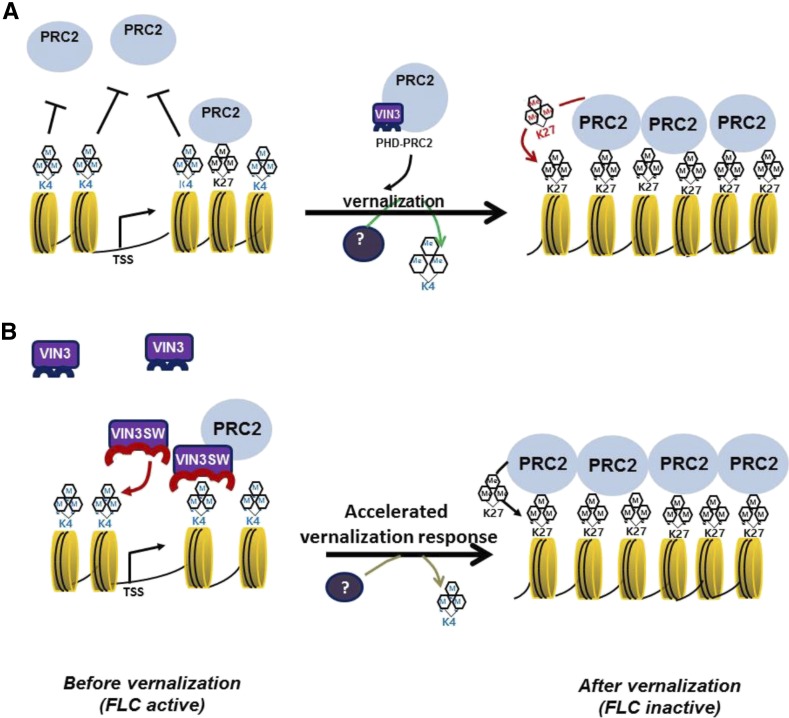
Figure 5.
Schematic models describing the role of VIN3 in vernalization response. A, Before vernalization, FLC is actively transcribed and enriched with H3K4me3 (active) mark. By vernalization, H3K4me3 is decreased, and H3K27me3 is substantially increased by VIN3-containing PRC2 complex. B, Before vernalization, VIN3SW is capable of binding to H3K4me3-enriched FLC chromatin. VIN3SW protein enhances the PRC2 activity at FLC chromatin during vernalization and results in an accelerated vernalization response.
VIN3SW increases the enrichment of H3K27me3 without significant increases in the enrichment of CLF-containing PRC2 (Fig. 4). Similar positive effects of PHD-finger-containing proteins on the PRC2 activity have been reported in Drosophila melanogaster and human (Sarma et al., 2008; Cao et al., 2008). Alternatively, the reduced level of H3K4me3 may contribute to the more efficient deposition of H3K27me3, as we observed the reduced level of H3K4me3 at FLC chromatin in VIN3SW transgenic lines. Addressing the molecular nature of the enhancement of H3K27 methylation by VIN3 will be of importance.
Diverse histone-reader domains, including the PHD-finger domain, have been identified as components of various chromatin-modifying complexes in eukaryotes (Musselman and Kutateladze, 2011). In Arabidopsis, about 80 PHD-finger-domain-containing proteins have been identified (Lee et al., 2009). However, their roles in chromatin-modifying activities are largely not known. In particular, the VIN3 family of proteins is conserved throughout higher plants and play roles in diverse developmental programs (Zhao et al., 2010; Wang et al., 2013). Whether the histone binding preference of PHD-finger domains of the VIN3 family of proteins in other plant species functions in a similar manner remains to be addressed.
CONCLUSION
In this report, we showed that the binding preference of VIN3 protein itself provides another layer of control to ensure that the vernalization response occurs only from long-term cold (Fig. 5). The lack of enrichment of VIN3WT at FLC without vernalization indicates that other chromatin-modifying events need to happen at FLC chromatin for the VIN3 protein to trigger the repression of FLC. The VIN3SW that is capable of the binding to H3K4me3 is able to repress FLC even in the absence of vernalization treatment; thus, the removal of H3K4me3 is a likely event that needs to occur before the VIN3-containing PRC2 is able to repress FLC by depositing repressive H3K27me3 marks at FLC by vernalization. Therefore, the coordinated changes in multiple regulatory components, including chromatin modifications and the induction of histone-binding proteins and noncoding RNAs, ensure proper circumstances for the vernalization response. A flurry of histone modification changes occurs when chromatin undergoes changes in its transcriptional activity. Various histone binding proteins in chromatin-modifying complexes play diverse regulatory roles to coordinate multiple changes in chromatin modifications. Our study indicates that the binding specificity of such histone binding proteins can function to fine-tune the activity of chromatin-modifying complexes in vivo through the exclusion of chromatin substrate.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds were germinated on agar plates for 10 d in the short-day condition and then seedlings were collected or transferred to 4°C for vernalization treatment under short-day conditions. Vernalization treatments were performed as previously described (Kim et al., 2010). Flowering time was measured as a rosette leaf number at a bolting stage under the long-day condition. All plants are in the Columbia FRIsf2 background (FRI_Col) unless otherwise indicated.
Generation of Transgenic Plants
For VIN3WT-GFP and VIL1WT-GFP transgenic lines, cDNAs were cloned into the pENTR-D-TOPO cloning vector (Invitrogen). For VIN3SW-GFP and VIL1SW-GFP transgenic plants, the PCR-based site-specific mutagenesis was performed to provide point mutations within PHD-finger domain and cloned into a pENTR-d-TOPO vector. Point mutations were confirmed by sequencing. The expression cassette of each pENTR clone was transferred into the plant binary Gateway vector pB7FWG2,0 using LR reaction (Invitrogen). Each construct was transformed into vin3-1FRI, vil1-1FRI mutants and/or FRI_Col (wild type) plants. Each Construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into plants using floral dip methods (Clough and Bent, 1998). All transgenic lines were selected on selective antibiotic media and were confirmed by PCR. Flowering time was measured as a rosette leaf number at the bolting stage of T1 or T3 generation. Stable homozygous T3 transgenic plants were used for molecular experiments.
Real-Time Quantitative RT-PCR Analyses
Total RNA was isolated using Plant RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Before reverse transcription reaction, RQ1 DNase (Promega) treatment was performed for 30 min at 37°C to eliminate contaminated genomic DNA from total RNAs. First-strand cDNA synthesis was performed using 2 µg of total RNA using the M-MLV system (Promega). Real-time quantitative RT-PCR was carried out using SYBR green dye reaction mixture (Thermo Scientific) on a ViiA7 real-time PCR system (Thermo Scientific). Each sample was quantified in triplicates, and the relative transcript level of each gene was determined by normalization of the resulting expression levels compared to that of PP2A, as described (Czechowski et al., 2005). For primer sequences used in real-time RT-PCR analyses, see Supplemental Table S1.
ChIP
Whole seedlings were cross-linked in 1% formaldehyde solution and collected. ChIP assays were performed as previously described (Kim and Sung, 2010). ChIP data were quantified by qPCR, normalized to the internal reference gene (AG, At4g18960; or PP2A, At1g13320) and then the relative fold changes were calculated. Values represent the average and se of three technical replicates from two biological replicates. qPCR reaction was performed using Maxima SYBR green master mix (Thermo Scientific) in a 384-well PCR plate of ViiA 7 Real-Time PCR system (Life Technologies). Antibodies used in this study were anti-H3K27me3 (ab6002; Abcam), anti-H3K4me3 (ab1012; Abcam), anti-GFP (ab290; Abcam), and anti-CLF (Hyun et al., 2013). Primer sequences used in ChIP-qPCR are shown in Supplemental Table S1.
Western-Blot Analysis
Total protein was extracted with plant protein extraction buffer (4% SDS, 25 mm Tris-Cl, pH 8.8, 2.5% glycerol, and protease inhibitor cocktail [Roche]) from seedlings. The same amount of total proteins was loaded onto 10% SDS-PAGE gel and transferred to PVDF blotting membrane (GE Healthcare Life Science). Anti-GFP (ab290; Abcam) and anti-RPT5 (PW8375-0100; Enzo Life Sciences) were used for western-blot analyses.
In Vitro Histone Peptide Pull-Down Assay
cDNAs of PHD-finger domains of VIN3 and VIL1 were amplified by RT-PCR and cloned into pENTR-D-TOPO vector and transferred into a pVP13 expression vector (Thao et al., 2004) using LR reaction (Invitrogen). Recombinant proteins of VIN3 or VIL1 with His-tag were purified using Ni-NTA column (Qiagen). Biotinylated histone peptides (Millipore; also see Supplemental Table S2) were used for the histone binding assays. After pull-down, proteins were transferred using a Mini Transblot electrophoresis transfer cell apparatus (Bio-Rad). His-tag antibody (Sc-803; Santa Cruz Biotech) was used for the western-blot analysis.
Tertiary Structure Prediction of PHD Domain
The predicted tertiary structure for PHD-finger domains was obtained using the SWISS-MODEL (http://swissmodel.expasy.org/) that simulates tertiary structure based on the reported protein structures (Arnold et al., 2006). The acquired PDB file was graphically visualized using Pymol version 1.4.0 program (www.pymol.org).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NP_200548 (Arabidopsis VIN3, AtVIN3, At5g57380), NP_189087 (Arabidopsis VIL1, AtVIL1, At3g24440), NP_974639 (Arabidopsis VIL2, AtVIL2, At4g30200), NP_179478 (Arabidopsis VIL3, AtVIL3, At2g18880), XP_015639862 (Oryza sativa VIN3, OsVIN3), XP_013629072 (Brassica oleracea VIN3, BoVIN3), AGU09528 (Arabis alpina VIN3, AaVIN3), XP_004238003 (Solanum lycopersicum VIN3, SlVIN3), XP_013727218 (Brassica napus VIN3, BnVIN3) NP_566742 (Arabidopsis ING1, AT3G24010), NP_974026 (Arabidopsis ING2, At1g54390), NP_937862 (Homo sapiens ING1 isoform A, HsING1), NP_001555 (H. sapiens ING2 isoform 1, HsING2), NP_196180 (Arabidopsis AL1, At5g05610), NP_872579 (H. sapiens BPTF, HsBPTF), and NP_005383 (H. sapiens PHF2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PHD-finger domains of VIN3 family proteins.
Supplemental Figure S2. Characterization of VIN3SW, VIN3WT, VIL1SW, and VIL1WT transgenic lines.
Supplemental Figure S3. The introduction of VIN3SW transgene into FRI_Col wild-type plants also triggers accelerated flowering.
Supplemental Figure S4. Changes in H3K4me3 at FLC chromatin during vernalization.
Supplemental Table S1. Oligonucleotide used in this study
Supplemental Table S2. Biotinylated histone peptides used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Hong Qiao and members of Sung lab for helpful discussion.
Glossary
- ChIP
chromatin immunoprecipitation
Footnotes
This work was funded by a grant from the National Institutes of Health (GM100108).
Articles can be viewed without a subscription.
References
- Adrian J, Torti S, Turck F (2009) From decision to commitment: the molecular memory of flowering. Mol Plant 2: 628–642 [DOI] [PubMed] [Google Scholar]
- Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bernier G, Kinet J-M, Sachs RM. 1981. The Physiology of Flowering. CRC Press, Boca Raton, FL [Google Scholar]
- Bratzel F, Turck F (2015) Molecular memories in the regulation of seasonal flowering: from competence to cessation. Genome Biol 16: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y (2008) Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 28: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hu Y, Zhou DX (2011) Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim Biophys Acta 1809: 421–426 [DOI] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P. (1960) Vernalization and its relations to dormancy. Annu Rev Plant Physiol 11: 191–238 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C (2012) Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337: 584–587 [DOI] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111: 16160–16165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Kim SY, Michaels SD (2013) FLOWERING LOCUS C EXPRESSOR family proteins regulate FLOWERING LOCUS C expression in both winter-annual and rapid-cycling Arabidopsis. Plant Physiol 163: 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17: 73–78 [DOI] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES (2011) Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6: e21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y (2013) The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140: 156–166 [DOI] [PubMed] [Google Scholar]
- Jiang D, Yang W, He Y, Amasino RM (2007) Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19: 2975–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S (2010) The plant homeo domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc Natl Acad Sci USA 107: 17029–17034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S (2013) Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25: 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Zografos BR, Sung S (2010) Mechanisms underlying vernalization-mediated VIN3 induction in Arabidopsis. Plant Signal Behav 5: 1457–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, Noh B, Noh YS (2010) Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J 29: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yun JY, Zhao W, Shen WH, Amasino RM (2015) A methyltransferase required for proper timing of the vernalization response in Arabidopsis. Proc Natl Acad Sci USA 112: 2269–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Lee D, Chung WI, Kwon CS (2009) Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J 58: 511–524 [DOI] [PubMed] [Google Scholar]
- Li P, Filiault D, Box MS, Kerdaffrec E, van Oosterhout C, Wilczek AM, Schmitt J, McMullan M, Bergelson J, Nordborg M, Dean C (2014) Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev 28: 1635–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang D, Fu X, Luo X, Liu R, He Y (2016) Coupling of histone methylation and RNA processing by the nuclear mRNA cap-binding complex. Nat Plants 2: 16015. [DOI] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X (2008) Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50: 886–896 [DOI] [PubMed] [Google Scholar]
- Luo M, Hung FY, Yang SG, Liu XC, Wu KQ (2014) Histone lysine demethylases and their functions in plants. Plant Mol Biol Report 32: 558–565 [Google Scholar]
- Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. (2007) RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. (2009) Flowering time regulation produces much fruit. Curr Opin Plant Biol 12: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG (2011) Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res 39: 9061–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D (2008) Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 28: 2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiq S, Berr A, Shen WH (2014) Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol 201: 312–322 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Finnegan EJ, Peacock WJ, Dennis ES (2009) Mechanisms of gene repression by vernalization in Arabidopsis. Plant J 59: 488–498 [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM (2006) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC, Amasino RM (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL (2004) Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics 5: 267–276 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu J, Qian Q, Xue HW (2013) LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol Plant 6: 514–527 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C (2014) Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24: 1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Michaels SD (2010) The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol 153: 1074–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20: 935–947 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7: 1256–1260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.