An Arabidopsis MAPK kinase kinase (MAPKKK), AIK1, modulates ABA signaling by sequentially relaying and amplifying intracellular signals through the AIK1-MKK5-MPK6 cascade.
Abstract
The mitogen-activated protein kinase (MAPK) cascade is an evolutionarily conserved signal transduction module involved in transducing extracellular signals to the nucleus for appropriate cellular adjustment. This cascade essentially consists of three components: a MAPK kinase kinase (MAPKKK), a MAPK kinase, and a MAPK, connected to each other by the event of phosphorylation. Here, we report the characterization of a MAPKKK, ABA-INSENSITIVE PROTEIN KINASE1 (AIK1), which regulates abscisic acid (ABA) responses in Arabidopsis (Arabidopsis thaliana). T-DNA insertion mutants of AIK1 showed insensitivity to ABA in terms of both root growth and stomatal response. AIK1 functions in ABA responses via regulation of root cell division and elongation, as well as stomatal responses. The activity of AIK1 is induced by ABA in Arabidopsis and tobacco (Nicotiana benthamiana), and the Arabidopsis protein phosphatase type 2C, ABI1, a negative regulator of ABA signaling, restricts AIK1 activity by dephosphorylation. Bimolecular fluorescence complementation analysis showed that MPK3, MPK6, and AIK1 interact with MKK5. The single mutant seedlings of mpk6 and mkk5 have similar phenotypes to aik1, but mkk4 does not. AIK1 was localized in the cytoplasm and shown to activate MKK5 by protein phosphorylation, which was an ABA-activated process. Constitutively active MKK5 in aik1 mutant seedlings complements the ABA-insensitive root growth phenotype of aik1. The activity of MPK6 was increased by ABA in wild-type seedlings, but its activation by ABA was impaired in aik1 and aik1 mkk5 mutants. These findings clearly suggest that the AIK1-MKK5-MPK6 cascade functions in the ABA regulation of primary root growth and stomatal response.
The phytohormone abscisic acid (ABA) is involved in a plant’s response to environmental stresses and plays crucial roles in regulating stomatal movement, seed germination, vegetative growth, and development (Zhu, 2002; Cutler et al., 2010; Kim et al., 2010; Melcher et al., 2010; Duan et al., 2013). Previous studies have revealed the core ABA signaling components that enable plants to cope with decreased water availability. It has been determined that ABA is sensed by ABA receptors such as PYR/PYL/RCARs (pyrabactin resistance/pyrabactin resistance-like/regulatory component of ABA receptors; Ma et al., 2009; Park et al., 2009). The ABA-binding proteins PYR/PYL/RCARs interact with PP2Cs (type 2C protein phosphatases, such as ABI1 and ABI2). As a downstream regulatory event in ABA signaling, the PP2Cs function as negative regulators of the ABA pathways, whereas SnRK2s (SNF1-related protein kinases) act as positive regulators of ABA signaling (Ma et al., 2009; Park et al., 2009). ABA binding to PYR/PYL/RCARs inhibits the activity of PP2Cs, and this inactivation of PP2Cs leads to the accumulation of active SnRK2s (Umezawa et al., 2009; Vlad et al., 2009). SnRK2s activate the expression of downstream ABA-responsive genes via the regulation of ABA-responsive element-binding factors (AREB or ABF; e.g. AREB1/ABF2, AREB2/ABF4, and ABF3; Choi et al., 2000; Uno et al., 2000; Fujii et al., 2007; Umezawa et al., 2009; Vlad et al., 2009).
Increasing evidence suggests that protein kinases play an important role in ABA signaling. The Arabidopsis (Arabidopsis thaliana) CDK (cyclin-dependent kinase) inhibitor KRP1 (KIP-related protein 1) is highly induced by ABA, leading to the inhibition of cell division (Wang et al., 1998; Schnittger et al., 2003). ABA inhibits cell elongation in the elongated zone of the roots, and the disruption of PERK4 (Pro-rich extensin-like receptor kinase 4) was shown to decrease this inhibition by regulating the ABA-induced activation of Ca2+ channels (Bai et al., 2009). Ca2+-dependent protein kinases (CDPK/CPK; e.g. AtCPK3, AtCPK4, AtCPK6, AtCPK10-13, AtCPK21, AtCPK23, etc.; Mori et al., 2006; Yu et al., 2007; Zhu et al., 2007; Negi et al., 2008; Zou et al., 2010; Geiger et al., 2011; Brandt et al., 2012, 2015; Lynch et al., 2012; Boudsocq and Sheen, 2013; Demir et al., 2013; Ronzier et al., 2014) function as central regulators of Ca2+-mediated ABA and stress responses. For example, AtCPK3 and AtCPK6 were shown to phosphorylate the major guard cell anion channel SLAC1 (slow anion channel-associated 1; Negi et al., 2008; Brandt et al., 2012), whereas AtCPK4 and AtCPK11 both phosphorylate two basic Leu zipper-type transcription factors, ABF1 and ABF4 (Zhu et al., 2007; Lynch et al., 2012). The snrk2.2 snrk2.3 double mutant showed strong ABA-insensitive phenotypes in seed germination and root growth inhibition and a greatly reduced level of a 42-kD kinase activity capable of phosphorylating peptides from ABF transcription factors (Fujii et al., 2007). The areb1 areb2 abf3 triple mutant displays reduced drought tolerance and water content, the triple mutant is more resistant to ABA than other single and double mutants with respect to primary root growth, and the stress-responsive gene expression is remarkably impaired in the triple mutant (Yoshida et al., 2010). The areb1 areb2 abf3 abf1 quadruple mutant shows increased sensitivity to drought and reduced sensitivity to ABA in primary root growth compared with the areb1 areb2 abf3 triple mutant, and the SnRK2-mediated gene (such as RD29B and MYB41) expression is more impaired in the areb1 areb2 abf3 abf1 quadruple mutant than in the areb1 areb2 abf3 triple mutant (Yoshida et al., 2015).
The mitogen-activated protein kinase (MAPK) cascade is an evolutionarily conserved signal transduction module involved in transducing extracellular signals to the nucleus for appropriate cellular adjustment. A MAPK cascade is minimally composed of MAP kinase kinase kinases (MAPKKKs/MEKKs/MKKKs), MAP kinase kinases (MAPKKs/MEKs/MKKs), and MAP kinases (MAPKs/MPKs), which sequentially phosphorylate the corresponding downstream substrates (Tena et al., 2001; Ichimura et al., 2002; Mishra et al., 2006; Rodriguez et al., 2010; Xu and Zhang, 2015). Sequence and functional analyses of the Arabidopsis genome have revealed that there are 20 MAPKs, 10 MAPKKs, and 80 MAPKKKs, with a similar repertoire of genes observed in other plant genomes (Ichimura et al., 2002; Hamel et al., 2006; Colcombet and Hirt, 2008). In plants, MAPK cascades have been shown to be involved in signaling pathways and activated by abiotic stresses such as cold, salt, touch, wounding, heat, UV, osmotic shock, and heavy metals (Tena et al., 2001; Zhang et al., 2006; Colcombet and Hirt, 2008; Liu, 2012). Recent studies have suggested that MAPK cascades are involved in several ABA responses, including antioxidant defenses, guard cell signaling, seed germination, and plant growth (Liu and Zhang, 2004; Xing et al., 2008; Jammes et al., 2009; Zong et al., 2009; Wang et al., 2010; Zhang et al., 2010; Huang et al., 2014; Danquah et al., 2015; Lee et al., 2015; Matsuoka et al., 2015). AtMKK1 and AtMPK6 were reported to link each other and mediate ABA-induced H2O2 scavenger CAT1 (catalase 1) gene expression and H2O2 production (Xing et al., 2008). Two other MAPK members, AtMPK9 and AtMPK12, were shown to regulate guard cell signaling in response to ABA (Jammes et al., 2009), and they function upstream of S-type anion channel activation and downstream of reactive oxygen species production, cytosolic alkalization, and [Ca2+]cyt elevation in guard cell methyl jasmonate signaling (Khokon et al., 2015). A phosphoproteomic approach revealed that SnRK2 promotes ABA-induced activation of the AtMPK1 and AtMPK2 (Umezawa et al., 2013). Even though it has been recently reported that MAPKKK17/18-MKK3-MPK1/2/7/14 is an ABA-activated MAPK cascade (Danquah et al., 2015) and that MAPKKK18 controls the plant growth by adjusting the timing of senescence via its protein kinase activity in ABA-dependent matters (Matsuoka et al., 2015), the role of MAPKKKs in regulating ABA responses, such as stomatal behavior and root growth inhibition, remains largely unknown.
In this study, we found that the Arabidopsis ABA-INSENSITIVE PROTEIN KINASE1 (AIK1) gene AT3G50310, encoding MAPKKK20, plays an important role in regulating cell division and cell elongation in the primary root meristematic and elongation areas and that AIK1 is a positive regulator of ABA-induced stomatal closure. Genetic and biochemical analyses indicate that the intracellular signals are relayed and amplified through sequential phosphorylations of the AIK1-MKK5-MPK6 cascade in response to ABA, regulating the root architecture and stomatal responses.
RESULTS
AIK1 Encodes MKKK20 and Its Mutants Are Insensitive to ABA in Terms of Root Growth
To identify new modulators of ABA signaling, some ABA-responsive genes were selected from microarray data by using the AtGenExpress Visualization Tool (http://www.weigelworld.org/resources; Kilian et al., 2007), and 50 protein kinase genes that were significantly induced (more than 5-fold) by ABA were randomly selected from the ABA-responsive genes (Bai et al., 2014). T-DNA insertion mutants of these kinase genes were obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003) for genetic analysis in a root-bending assay under the application of ABA (Murphy and Taiz, 1995; Wu et al., 1996). One of the T-DNA insertion mutants, designated aik1-1, was chosen for detailed characterization because of its insensitivity to ABA in terms of the root growth of seedlings, while ABA greatly inhibited root growth in wild-type seedlings (Supplemental Fig. S1, A and B).
AIK1 encodes a member of the MAPKKK family (AT3G50310, MKKK20). N-terminal GFP-fused AIK1 protein was shown to be localized in the cytoplasm of the protoplasts (Supplemental Figure S2A). We used the β-glucuronidase (GUS) reporter gene to determine the expression patterns of AIK1 in detail. An AIK1 promoter-GUS fusion construct was transformed into wild-type Arabidopsis (Columbia). Histochemical staining revealed that GUS was expressed more or less in every plant part, including leaf, stem, root, flower, and silique (Supplemental Fig. S2B). This result is consistent with real-time quantitative reverse transcriptase PCR (qRT-PCR) detection results with the AIK1 expression level being highest in the roots (Supplemental Fig. S2C). AIK1 is expressed in the root meristematic and elongation zones. Indeed, the transcript of AIK1 was found to be induced by ABA (Supplemental Fig. S2D). These results indicate that AIK1 may participate in regulating ABA response in root development.
Apart from aik1-1 (Salk_049616), an independent T-DNA insertion line (Salk_124398, named aik1-2) was also obtained (Supplemental Fig. S3A). aik1-1 and aik1-2 were inserted upstream of the ATG initiation codon by T-DNA into the 340- and 235-bp locus, respectively (Supplemental Fig. S3B). RT-PCR analysis showed that AIK1 was knocked down in the homozygous aik1 lines (Supplemental Fig. S3, C and D), and the ABA-induced expression level of AIK1 in the wild-type plants was severely disrupted in the aik1 mutants (Supplemental Fig. S3E).
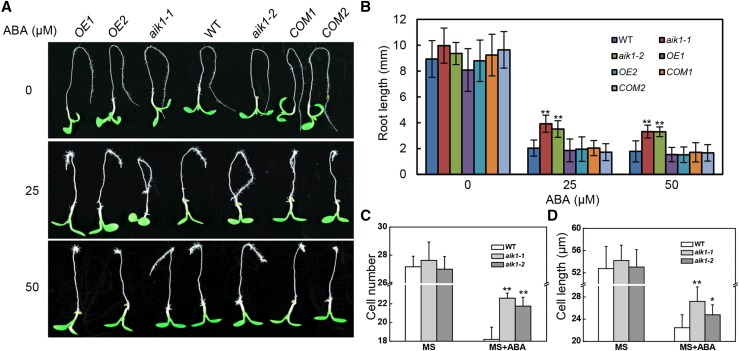
Both aik1-1 and aik1-2 were insensitive to ABA in terms of the root growth with a dosage-dependent manner (Fig. 1, A and B; Supplemental Fig. S4, A and B). This insensitivity of aik1 to ABA was quantified by measuring the root elongation of seedlings by transferring them to Murashige and Skoog (MS) medium agar plates with or without ABA. After treatment for 3 d with different concentrations of ABA, the elongated root length was measured. At 50 μm ABA, elongated aik1-1 (3.52 ± 0.66 mm) roots and elongated aik1-2 (3.27 ± 0.48 mm) roots were longer than elongated wild-type roots (1.54 ± 0.50 mm; Fig. 1B; Student’s t test, P < 0.01). At 25 μm ABA, the aik1 roots were also longer than the wild-type roots (Fig. 1B). These results suggest that AIK1 functions in primary root growth.
Figure 1.
aik1 mutants show decreased sensitivity to ABA with respect to root tip growth. A, Decreased sensitivity of root growth to ABA. Five-day-old seedlings were transferred to MS medium supplemented with 25 μm ABA or 50 μm ABA. Pictures were taken 3 d later. OE1 and OE2 are AIK1-overexpressing transgenic lines. COM1 and COM2 are aik1 complementary transgenic plants. B, Effect of ABA on root growth. Root length was measured 3 d after the seedlings had been transferred to medium containing ABA. Values are the means of ∼100 seedlings (±sd) from three independent experiments. Asterisks indicate significant differences from the wild type at **P < 0.01 by Student’s t test. C, Average numbers of cells in an array along the root axis from the tip to the elongation zone of plants treated with 25 μm ABA. Values are means ± sd from groups of five seedlings. Asterisks indicate significant differences from the wild type at **P < 0.01 by Student’s t test. D, Average cell length of the elongation zone. Five-day-old seedlings were transferred to MS medium supplemented with 25 μm ABA for 3d. Data are shown as means ± sd (n = 15 plants). Asterisks indicate significant differences from the wild type at *P < 0.05 and **P < 0.01 by Student’s t test.
To confirm this conclusion, transgenic seedlings of overexpression AIK1 in wild-type and complementation AIK1 driven by native AIK1 promoter in aik1 mutants were also tested in root-bending assay under ABA treatment. T3 progeny of hygromycin-resistant transformants were examined for AIK1 expression level (Supplemental Fig. S5A) and subjected to ABA treatment; overexpression of AIK1 lines and the complementation lines showed a similar phenotype to the wild type in terms of root growth (Fig. 1, A and B). However, when 5-d-old seedlings were transferred to MS medium containing ABA and incubated for a longer time, seedlings overexpressing AIK1 gradually exhibited yellow and eventually bleached leaves (∼25 d) compared to the wild-type seedlings (Supplemental Fig. S5B). This is consistent with the fact that deficiency in AIK1 resulted in a decreased sensitivity to ABA during Arabidopsis root growth.
To determine whether the longer root elongation morphology of aik1 under ABA treatment could be the result of a disruption in cell division, cell elongation, or both, we measured the number and size of root tip cells in wild-type and aik1-1 seedlings loaded with the FM4-64 probe after ABA treatment under a laser scanning confocal microscope (LSCM). The wild-type plants showed a shorter distance from the root tips to the maturation zone than the aik1 mutants (Supplemental Fig. S6, A–C). The data indicated that ABA inhibited the growth of the elongation zone and decreased the total number of root cells from the tip to the elongation zone in both wild-type and aik1 seedlings (Fig. 1, C and D), but aik1 roots had much longer and more pronounced elongation zones than the wild type. The wild-type plants showed shorter elongation zones (Supplemental Fig. S6C; Student’s t test, P < 0.05) and crowded root hairs at the primary root tips (Supplemental Fig. S6A, green arrowheads). In addition, the total number of cells from root tip to elongation zone was greater in aik1 than in wild-type seedlings (Fig. 1C). The epidermal cells in the elongation zone of mutant roots were significantly longer than those in the wild-type roots; cell length reached an average of 27.20 ± 2.62 μm for aik1-1 and 24.78 ± 1.80 μm for aik1-2 plants compared with 22.46 ± 2.32 μm for wild-type seedlings (Fig. 1D; Student’s t test, P < 0.05). These results suggest that the decreased sensitivity to ABA during root tip growth in the absence of AIK1 manifests as increased cell length in the elongation area and an increased number of cells from the root tip to the elongation zone.
Deficiency of AIK1 Impairs Stomatal Responses under Dehydration and Application of ABA
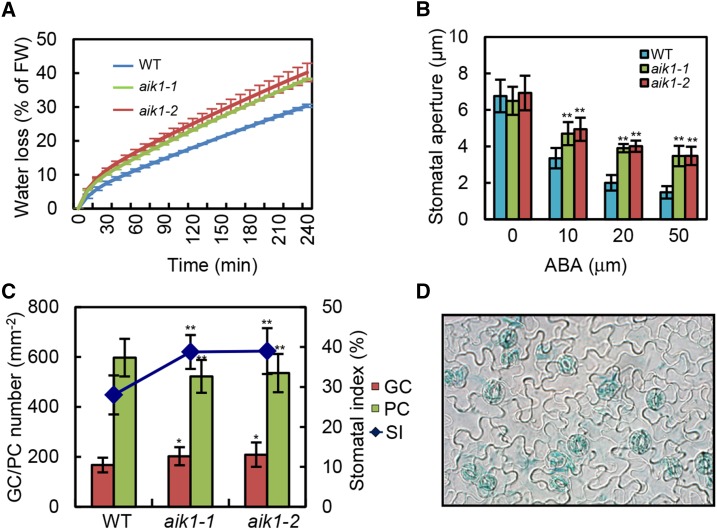
To test whether mutation of AIK1 disturbs stomatal behavior, we also compared the rates of water loss in aik1 mutants and wild-type plants under dehydrated conditions. The data indicate that the rate of water loss in aik1 mutants was about 10% greater than that in wild-type plants (Fig. 2A). When both wild-type and aik1-1 mutants alternated under light and dark conditions, significant changes were observed in the stomatal conductance (gs). Specifically, when transitioning from dark to light, after just 30 min of light exposure, both plants showed increased gs, with aik1 mutants showing a much steeper increase (222.33 ± 17.75 mmol m−2 s−1) compared to the wild type (188.36 ± 13.37 mmol m−2 s−1; Supplemental Fig. S7; Student’s t test, P < 0.05).
Figure 2.
AIK1 mutations impair ABA-induced stomatal closure. A, Analysis of transpirational water loss. Four-week-old rosette leaves were detached and allowed to dry at room temperature for the indicated periods. Water loss is presented as a percentage of initial fresh weight (FW; n = 5 per sample), and three independent experiments were repeated. B, Effects of ABA on stomatal closure in the wild type and aik1 mutants. Stomatal apertures were measured on epidermal peels of the wild type, aik1-1, and aik1-2. Values are means ± sd of 50 measurements from three independent experiments. Asterisks indicate significant differences from the wild type at **P < 0.01 by Student’s t test. C, Stomatal index (SI) of the wild type and aik1. Representative curve shows the epidermis of the abaxial surface of rosette leaves from the wild type, aik1-1, and aik1-2. Change in stomatal index and the number of guard cells (GCs) and pavement cells (PCs) per mm2 of the leaves, ±sd determined from leaves of at least 15 individual wild-type and mutant plants. Five independent counts were performed on each leaf. Asterisks indicate significant differences from the wild type at *P < 0.05 and **P < 0.01 by Student’s t test. D, Expression of AIK1 promoter-GUS in guard cells. GUS activity in guard cells of wild-type plants expressing the GUS reporter gene under the control of the AIK1 promoter is shown.
Measurements of the stomatal aperture showed that the degree of opening of the aik1 mutants’ stomata without ABA treatment was consistent with that seen in the wild-type plants (Fig. 2B). However, the stomatal closing in aik1 plants was less sensitive to ABA than that in wild-type plants at 10, 20, and 50 μm ABA (Fig. 2B; Student’s t test, P < 0.01). The average sizes of the stomatal aperture of the wild type were 3.35 ± 0.60 μm, 2.00 ± 0.42 μm, and 1.48 ± 0.34 μm, or 49.5, 29.6, and 21.9% of the control value, at 2 h after treatment with 10, 20, and 50 μm ABA, respectively; those of aik1-1 were 4.70 ± 0.63 μm, 3.90 ± 0.23 μm, and 3.47 ± 0.56 μm, or 72.3, 60.0, and 53.4% of the control value, at the same time point.
In addition, the aik1 mutants exhibited slightly greater stomatal density (Fig. 2C; Student’s t test, P < 0.05) and significantly reduced pavement cell density (Fig. 2C; Student’s t test, P < 0.01) compared to wild-type plants. Therefore, the stomatal index was significantly increased, by 10.8 and 11.0%, in aik1-1 and aik1-2 (Student’s t test, P < 0.01), respectively. The total epidermis cell numbers (including guard cells and pavement cells) are the same between wild-type and aik1 mutants (Supplemental Fig. S8). Consistent with a role in stomatal signal transduction, high levels of GUS expression were detected in the guard cells of AIK1 promoter-GUS transgenic plants (Fig. 2D). Together, these data suggest that the AIK1 protein is involved in the modulation of ABA-mediated stomatal behavior, the level of leaf stomatal density, and the size of stomatal aperture and, as a consequence, the extent of transpirational water loss.
AIK1 Possesses Protein Kinase Activity Regulated by Protein Phosphatase 2C
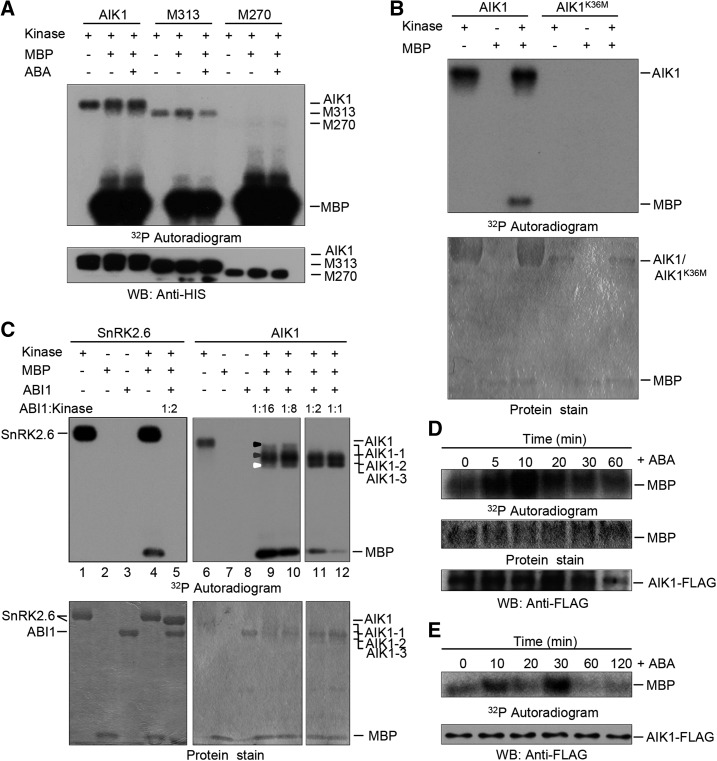
The AIK1 protein sequence was analyzed using a protein sequence analysis and classification tool (InterPro, http://www.ebi.ac.uk/interpro/). The N terminus of AIK1 was shown to be a kinase domain, including an ATP binding site and an active site, and the C terminus of AIK1 may be a regulatory domain (Supplemental Fig. S9). Therefore, HIS-tagged AIK1 and two truncated segments of AIK1, M313 (1–313 amino acids) and M270 (1–270 amino acids), were prepared and verified using anti-HIS antibodies (Supplemental Fig. S9; Fig. 3A). We analyzed the kinase activity of AIK1 with myelin-basic protein (MBP) as a substrate for the assay. As shown in Figure 3A, AIK1 kinase activity was at very high levels in terms of autophosphorylation and MBP phosphorylation. The fragments of AIK1, M313 and M270, also showed similar patterns of phosphorylation but low levels of autophosphorylation, which indicate that the C-terminal region of AIK1 is associated with autophosphorylation activity. To further test AIK1 kinase activity, an inactive form of AIK1 (AIK1K36M) was generated by site-specific mutation in a kinase-dead mutant (Supplemental Fig. S9). As shown in Figure 3B, AIK1K36M shows no protein kinase activity.
Figure 3.
AIK1 possesses protein kinase activity regulated by ABA signaling pathway. A, In vitro analysis of the kinase activity of recombinant AIK1 protein purified from E. coli. Fragments M313 and M270 of AIK1 correspond to amino acids 1 to 313 and 1 to 270, respectively. The recombinant AIK1 and the fragments were used to carry out both autophosphorylation and phosphorylation of the MBP substrate. Immunoblotting anti-HIS antibodies were performed to show the loading of HIS-tagged AIK1, M313, and M270, respectively. B, The mutation of Lys-36 (K36) of AIK1 to Met (M) resulted in the kinase activity lost. C, ABI1 inactivates AIK1 autophosphorylation and transphosphorylation in vitro, with MBP as a substrate and SnRK2.6 as a positive control. Equal amounts of recombinant AIK1 and SnRK2.6 were incubated at the indicated concentrations with MBP, γ-32P-ATP, and ABI1. For lanes 9 to 12, the ratios of AIK1:ABI1 were 16:1, 8:1, 2:1, and 1:1, respectively; for lane 5, the ratio of SnRK2.6:ABI1 was 2:1. The black, gray, and white triangles show the different phosphorylation modification of AIK1: AIK1-1, AIK1-2, and AIK1-3, respectively. D, ABA induced the activation of AIK1 in Arabidopsis. Total proteins were prepared from 2-week-old 35S:AIK1-FLAG plants treated with 100 μm ABA for the indicated time points and incubated with anti-FLAG agarose beads. Equal amounts of AIK1-FLAG were incubated in the kinase reaction mixture containing MBP as a substrate. After the phosphorylation reaction, the samples were resolved using SDS-PAGE and subjected to autoradiography. Radioactivity is shown in the top panel. Coomassie Brilliant Blue-stained MBP is shown in the middle panel. AIK1 was monitored by immunoblot analysis, using the anti-FLAG tag antibody in the lowest panel. E, ABA-induced activation of AIK1 in tobacco. FLAG-AIK1 was immunoprecipitated from the protein extracts in 100 μm ABA-treated tobacco leaves. The tobacco leaves were transiently transformed with agrobacteria carrying the 35S:AIK1-FLAG vector. Equal amounts of the immunoprecipitates were incubated in the kinase reaction mixture containing MBP as a substrate. After the phosphorylation reaction, the samples were resolved using SDS-PAGE and subjected to autoradiography. The phosphorylation of MBP is shown in the upper panel. The AIK1 was monitored by immunoblot analysis using the anti-FLAG tag antibody in the lower panel.
ABI1, a type 2C protein phosphatase, which contains a putative MAPK interaction motif, is a negative regulator of ABA signaling (Gosti et al., 1999; Schweighofer et al., 2007). To further analyze the role of AIK1 in ABA signaling, we performed an in vitro kinase assay using HIS-AIK1 and HIS-ABI1. AIK1 activity was monitored using MBP as a substrate. Remarkably, AIK1 exhibited reduced kinase activity, with the observed MBP phosphorylation ratios of ABI1/AIK1 increasing from 1:16 to 1:1 (Fig. 3C). Deactivation of AIK1 (AIK1*: AIK1-1, AIK1-2, and AIK1-3, dephosphorylated form of AIK1) by ABI1 resulted in a faster mobilization band on the Coomassie Brilliant Blue staining gel. We speculate that this variance in mobilization observed for AIK1 and AIK1* may be due to the dephosphorylation of AIK1 by ABI1. Thus, the liquid chromatography-tandem mass spectrometry (LC-MS/MS) data from tryptic digests of AIK1 and three AIK1* bands indicated that nearly all phospho-Tyr, phospho-Thr, and phospho-Ser were detected, and phosphorylated amino sites were gradually decreased with the increase of ABI level in the reaction mixture (Supplemental Table S2). An equal amount of SnRK2.6 was used as a positive control. As shown in Figure 3C (left panel), ABI1 can completely to deactivate SnRK2.6 at a ratio of 1:2. The dephosphorylated form of SnRK2.6 had a faster mobilization band in the Coomassie Brilliant Blue staining gel. These data suggest that there may be many phosphorylation sites in AIK1, and the deactivation of AIK1 by ABI1 is the downstream event of ABA signaling.
To analyze the effects of ABA treatment on AIK1 activity, we performed in vivo phosphorylation assay experiments using 35S:AIK1-FLAG transgenic Arabidopsis and tobacco (Nicotiana benthamiana) plants transiently transformed with Agrobacterium tumefaciens carrying the 35S:AIK1-FLAG vector. Two-week-old 35S:AIK1-FLAG Arabidopsis seedlings were treated with ABA, FLAG-AIK1 was purified from protein extract with anti-FLAG agarose beads, and equal amounts of AIK1-FLAG were incubated in the kinase reaction mixture containing MBP as a substrate. Phosphorylation of MBP by FLAG-AIK1 was activated at 10 min after the treatment with ABA (Fig. 3D). The same result was obtained from 6-week-old tobacco plants transiently transformed with agrobacteria carrying the 35S:AIK1-FLAG vector. Compared to FLAG-AIK1 from leaves without ABA treatment, the phosphorylation of MBP by AIK1 was significantly activated after 10 min of ABA treatment, peaked at 30 min, and then decreased (Fig. 3E). However, no obvious autophosphorylation activity of AIK1 was found in the assays, for little amount of AIK1 was used in these experiments (Fig. 3, D and E). The inconsistent time course for ABA-activated AIK1 in Arabidopsis and tobacco suggests different ABA-regulated patterns in these two species of plants. These results clearly indicate that the kinase activity of AIK1 is triggered by ABA treatment in planta.
AIK1 Interacts with MKK5 and Is Required for MKK5 Activation in ABA Signaling in Terms of Root Growth
On the basis of sequence homology of the kinase domains to mammalian MAPKKs, there are at least 10 putative Arabidopsis MAPKKs. To identify the possible downstream targets that can be activated by and interact with AIK1 in the MAPK cascade in ABA-regulated root growth response, several biochemical and genetic screen approaches were employed.
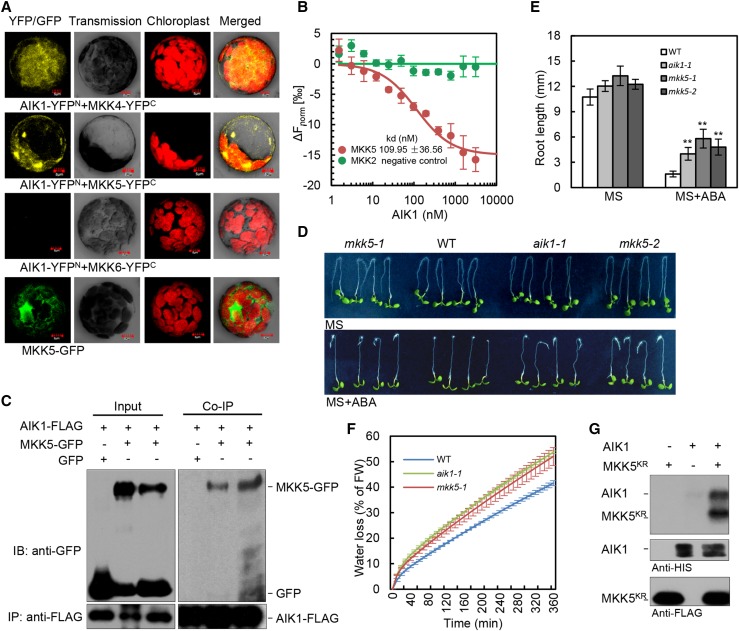
In the first set of experiments, we searched for an AIK1 interaction partner from among 10 MKKs using a bimolecular fluorescence complementation approach (Walter et al., 2004). AIK1 was fused to the N-terminal fragment of YFP (AIK1-YFPN), while MKK1 to MKK10 were fused to the C-terminal fragment of YFP (MKK1-YFPC to MKK10-YFPC). As Figure 4A shows, YFP fluorescence was only observed in the cytoplasm of Arabidopsis mesophyll protoplasts cotransformed with AIK1-YFPN and MKK4-YFPC or AIK1-YFPN and MKK5-YFPC. No interaction was found between AIK1 and other members of MKKs (e.g. MKK6-YFPC, which was used as a negative control; Fig. 4A). Protein interaction between AIK1 and MKK5 was confirmed using microscale thermophoresis (MST; Wienken et al., 2010). The data indicate that AIK1 bound to MKK5 with a dissociation constant (Kd) of 109.95 ± 36.56 nm (Fig. 4B), which demonstrated a high affinity of AIK1 for MKK5. In contrast, MKK2 as a negative control did not show similar interaction with AIK1. We further examined the possible interaction in vivo by using coimmunoprecipitation assay in tobacco leaves expressing MKK5-GFP and AIK1-FLAG. The data show that MKK5 was pulled down by AIK1 (Fig. 4C). Together, these results indicate that MKK5 of the MKK family might be the substrate of AIK1.
Figure 4.
AIK1 interacts with and phosphorylates MKK5 and participates in the process involved in the ABA regulation of root growth and stomatal closure. A, AIK1 interactions with MKK4 and MKK5. Epifluorescence, transmission, chloroplast autofluorescence, and merged images of Arabidopsis mesophyll protoplasts cotransfected with constructs expressing different fusion proteins. Vectors expressing AIK1-YFPN and MKK6-YFPC were used as negative controls. B, Dissociation curve for the interaction of AIK1 with MKK5. Quantification of binding affinity between AIK1 and MKK5 was obtained by MST analysis. MKK2 was used as a negative control. Error bars represent ±sd of three independent measurements. C, Interaction of AIK1 and MKK5 proteins in vivo. The total protein extracts from tobacco leaves cotransfected with 35S:AIK1-FLAG/35S:GFP or 35S:AIK1-FLAG/35S:MKK5-GFP were immunoprecipitated with anti-FLAG Sepharose beads. The proteins from crude lysate (left, input) and immunoprecipitated proteins (right) were detected with anti-FLAG and anti-GFP antibodies. D and E, Effects of ABA on root growth of the wild type, aik1, and mkk5 mutants. Five-day-old seedlings were transferred to MS medium supplemented with or without 50 μm ABA. Root length was measured 3 d after the seedlings had been transferred to ABA-containing medium. Values are the means of ∼40 seedlings (±sd) from three independent experiments. Asterisks indicate significant differences from control at **P < 0.01 by Student’s t test. F, Analysis of transpirational water loss. Approximately 4-week-old rosette leaves were detached and allowed to dry at room temperature for the indicated periods. Water loss is presented as a percentage of initial fresh weight (n = 5 per sample), and three independent experiments were repeated. G, AIK1 can phosphorylate MKK5KR in vitro. AIK1 and MKK5KR were detected by immunoblotting using HIS and FLAG antibodies.
To explore which MKK is involved in response to ABA in terms of root growth, we performed quantitative ABA response screens of MKK loss-of-function mutants collected from various T-DNA insertion resources (Alonso et al., 2003; Supplemental Fig. S10A). The T-DNA insertion sites are shown in Supplemental Figure S10B; both mkk4 and mkk5 were gene down-regulated mutants (Supplemental Fig. S10, C and D). The wild-type and various mutant plants were grown under the same conditions for 5 d and then transferred to MS medium containing ABA for 3 d. Similar to aik1 mutants, the mkk5 mutant plants showed ABA-insensitive phenotypes in terms of primary root growth (Fig. 4, D and E). By contrast, mkk4 mutant seedlings showed an ABA-sensitive phenotype in terms of primary root growth, similar to the wild type (Supplemental Fig. S10, E and F). In addition, the rate of water loss of mkk5 mutants is higher than that of the wild type (Fig. 4F), and the expression level of MKK5 was induced by ABA in both the wild type and aik1 (Supplemental Fig. S10 G). These results were consistent with aik1 mutants, suggesting that MKK5 and AIK1 might have similar functions in ABA responses.
The direct physical interaction between AIK1 and MKK5 and the genetic evidence on AIK1 and MKK5 prompted us to test whether MKK5 is a substrate for AIK1. To test whether AIK1 can phosphorylate MKK5, an inactive form of FLAG-MKK5 was generated by site-specific mutation, replacing the conserved Lys residue in the kinase ATP-binding loop with an Arg: MKK5KR (K99R; Ren et al., 2002). HIS-tagged AIK1 and FLAG -tagged MKK5KR were expressed in Escherichia coli BL21 (DE3) cells, and the purified proteins were verified using anti-HIS and anti-FLAG antibodies. In vitro kinase assays were subsequently performed using the AIK1 and MKK5KR proteins. As Figure 4G shows, MKK5KR was phosphorylated by AIK1, suggesting that MKK5 is indeed a substrate of AIK1. These findings suggest that AIK1 is the protein kinase responsible for the phosphorylation of MKK5 in regulating ABA responses.
Functional Complementation of the ABA-Insensitive Root Growth in aik1 by Constitutively Active MKK5 in aik1 Mutant Plants
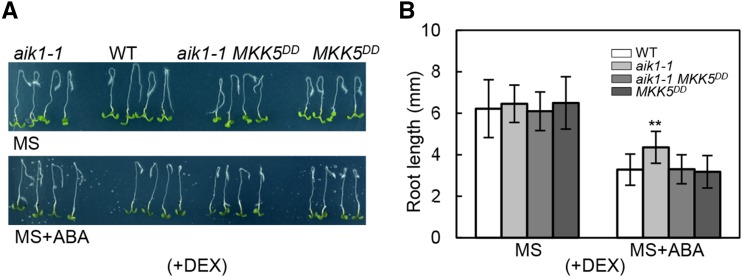
In Arabidopsis, MKK5DD is the DEX (dexamethasone)-induced constitutively active mutant (Ren et al., 2002). It has been also suggested that MKK5DD transgenic lines may induce gene silencing (Liu and Zhang, 2004). In fact, the expression level of MKK5 showed decreased expression in the MKK5DD (without DEX) seedlings compared with the wild-type plants, which is similar to a knockdown allele of MKK5. However, no difference was found in the MKK9 expression level between the MKK5DD and wild-type seedlings (Supplemental Fig. S11, A and B). Thus, MKK5DD lines showed insensitivity of primary root growth up to 50 μm of ABA in a root bending assay on MS medium without DEX (Supplemental Fig. S11, C and D). Upon ABA treatment, the elongated length of MKK5DD roots was nearly 3 times greater than that of wild-type seedlings (Supplemental Fig. S11D); the distance from the root tip to the elongation zone of MKK5DD was longer than in wild-type seedlings after exposure to ABA (Supplemental Fig. S11, E–G), and the difference of the root length between MKK5DD and the wild type was mainly because of the different elongation zone lengths (Supplemental Fig. S11G). We also compared the rates of water loss in MKK5DD with wild-type plants under dehydrated conditions; this rate in MKK5DD was greater than that in wild-type plants, similar to the findings in aik1 and mkk5 mutants (Figs. 2A and 4F; Supplemental Fig.S11H). These results indicate that the reduced expression of MKK5 in MKK5DD induced the ABA-insensitive root growth and a higher water transpiration rate.
Initially, constitutively active MKK5 in MKK5DD by exposure to 0.2 μm DEX for 2 d showed an ABA-inhibited root growth phenotype, similar to that in the wild type (Supplemental Fig. S11, I and J). However, when the plants grew on DEX-containing MS medium for 12 d, the MKK5DD plants were smaller than the wild-type seedlings. The long-lasting activation of MKK5 even led to yellowish leaves of the seedling and finally to the death of plants when they grew on the ABA- and DEX-containing MS medium (Supplemental Fig. S11K), which was similar to the case for seedlings overexpressing AIK1 (Supplemental Fig. S5B).
To further analyze whether MKK5 is a downstream component of AIK1 in ABA signaling, the single aik1-1 mutant was crossed with MKK5DD, and PCR screening and hygromycin-resistant or anti-FLAG western blotting (Ren et al., 2002) were performed to obtain an aik1-1 MKK5DD homozygous mutant plants, in which MKK5 could be constitutively activated by DEX. DEX-treated aik1-1 MKK5DD seedlings showed high level of expression of the active MKK5DD (Supplemental Fig. S11L). We then analyzed the ABA responses in aik1-1, MKK5DD, and aik1-1 MKK5DD after the constitutive activation of MKK5 in MKK5DD and aik1-1 MKK5DD plants. Compared with the wild-type plants, aik1-1 showed ABA-insensitive primary root growth. By contrast, MKK5DD and aik1-1 MKK5DD showed ABA-inhibited root morphology, similar to that in the wild type (Fig. 5, A and B), which indicates that the constitutively active MKK5 in aik1-1 rescued the ABA-insensitive root growth phenotype. This genetic evidence strongly suggests that the significant function of MKK5 is downstream of AIK1 in ABA-regulated root growth in Arabidopsis.
Figure 5.
Constitutively active MKK5 in aik1 complements the ABA-insensitive root growth of aik1. A and B, Five-day-old seedlings of the wild type, aik1-1, aik1-1 MKK5DD, and MKK5DD were transferred to 0.2 μm DEX-containing MS medium for 2 d to activate MKK5 in aik1-1 MKK5DD and MKK5DD seedlings and then the plants (with DEX, +DEX) were transferred to 0.02 μm DEX-containing MS medium supplemented with 50 μm ABA. Pictures were taken and the elongated root length was determined 3 d later. Values are the means of ∼40 seedlings. Asterisks indicate significant differences from the wild type at **P < 0.01 by Student’s t test.
AIK1-MKK5 Functions Upstream of MPK6
On the basis of several in vitro and in vivo studies, the downstream targets of MKK5 are believed to be MPK3 and MPK6 (Wang et al., 2007, 2010; Liu et al., 2008) and is involved in regulating stomatal development and patterning, ethylene signaling during the cell death process, and nitric oxide production and signal transduction in response to hydrogen peroxide during Arabidopsis root development. However, whether MPK6 and MPK3 are downstream of AIK1-MKK5 in the response to ABA was unclear. We analyzed their interactions using a bimolecular fluorescence complementation approach. MKK5 was fused to the C-terminal fragment of YFP (MKK5-YFPC), whereas MPK3 and MPK6 were fused to the N-terminal fragment of YFP (MPK3-YFPN and MPK6-YFPN). YFP fluorescence was observed in the cytoplasm of Arabidopsis mesophyll protoplasts cotransformed with MKK5-YFPC and MPK3-YFPN or MKK5-YFPC and MPK6-YFPN. MKK5-YFPC and YFPN did not show any fluorescence signal as negative controls (Supplemental Fig. S12A). This result suggests that MKK5 associates with MPK3 and MPK6 in vivo. The expression level of MPK3 and MPK6 is also induced by ABA in wild-type and aik1-1 seedlings (Supplemental Fig. S12, B and C), similar to the expression pattern of MKK5 (Supplemental Fig. S10G).
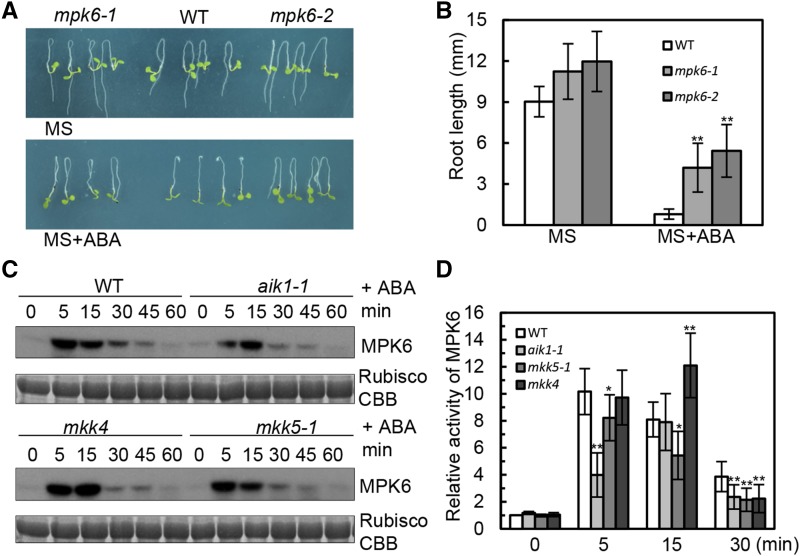
To test whether MPK3 and MPK6 have functions similar to those of AIK1 and MKK5 in ABA-regulated root growth and development, T-DNA insertion knockout mutants of MPK3 and MPK6 were used for genetic phenotype analysis. The 5-d-old wild-type and various mutant seedlings were transferred to MS medium containing ABA for 3 d. Both mpk3 and mpk6 were insensitive to ABA in terms of primary root growth, in that the elongated root length was clearly greater than in the wild type in the ABA-containing medium (Fig. 6, A and B; Supplemental Fig. S12, D and E). aik1 was used as a control. Under ABA treatment, the relative root length of mpk6 mutants was shown to be significantly insensitive to ABA compared with the wild type (Supplemental Fig. S12F).
Figure 6.
AIK1 regulates ABA-inhibited root growth by activating MPK6. A and B, Five-day-old seedlings were transferred to MS medium supplemented with 50 μm ABA. Pictures were taken 3 d later. Values are the means of ∼40 seedlings. Asterisks indicate significant differences from the wild type at **P < 0.01 by Student’s t test. C, In-gel kinase assays of wild-type, aik1, mkk4, and mkk5 plants under 100 μm ABA treatment. Equal amounts of the samples were resolved on SDS-PAGE and stained with Coomassie Brilliant Blue (CBB). The large subunit of Rubisco is shown as the loading control for equal protein amounts in the wild-type and mutant plants. D, MPK6 is activated by ABA. Relative MPK6 activities in the wild type, aik1-1, mkk5, and mkk4; the seedlings were exposed to 100 μm ABA. The activity of MPK6 in the wild type (0 min) was used as a control and was set as 1. Biological replications (n = 3) show the reduced activity of MPK6 in aik1-1 and mkk5 at 5 and 15 min. Error bars indicate ±sd for the different mutants. Asterisks indicate significant differences from the wild type (0 min) at *P < 0.05 and **P < 0.01 by Student’s t test.
To test whether AIK1-MKK5 is also required for the activation of MPK6 and MPK3 in response to ABA, we used an in-gel kinase assay with MBP as the embedded kinase substrate to measure the activity of these two kinases in the wild-type and mutant plants. As shown in Figure 6C, the application of 100 μm ABA resulted in the strongest activation of MPK6 in wild-type seedlings, but no clear MPK3 activity was observed. The NaCl-treated mpk3 and mpk6 seedlings were used as controls to confirm the position of the MPK6 in the 32P autoradiogram film (Supplemental Fig. S12G). Compared with wild-type plants, the activity of MPK6 had no significant difference in the mkk4 single mutant at different time points, but was clearly reduced in aik1 and mkk5 mutants when the seedlings were exposed to ABA for 5 and 30 min (Fig. 6D). The transient activation of MPK6 as shown in Figure 6D may be due to that MAPK signaling strength and/or duration could be tightly controlled. Consistent with the wild type, the primary root growth of mkk4 mutant seedlings was inhibited by ABA; however, the root length of aik1 and mkk5 plants was insensitive to ABA compared to the wild type (Figs. 1A and 4D; Supplemental Fig. S10E), suggesting that MKK5 and AIK1 are required for the ABA activation of MPK6, but MKK4 is not. These findings indicate that the AIK1-MKK5-MPK6 cascade is activated by ABA and regulates primary root growth and development.
AIK1 Genetically and Enzymatically Interacts with MKK5 and MPK6 in the ABA Regulation of Root Growth
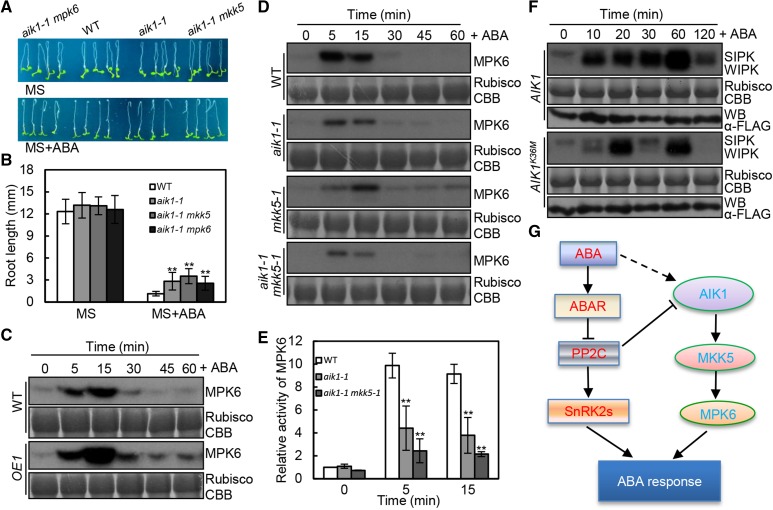
Since MKK5-MPK6 was found to be molecularly connected to AIK1 in terms of the ABA response, we wanted to determine the physiological significance of such an interaction. We generated two sets of double mutants, namely, aik1 mpk6 and aik1 mkk5, through genetic crosses and examined the morphology of the mutant seedlings in response to ABA. After transfer to ABA-containing medium for 3 d, the root growth of aik1 mkk5 was more insensitive to ABA than that of the aik1 single mutant. Although aik1 mpk6 mutant plants displayed similar root length to the aik1 single mutant, the seedling growth of aik1 mpk6 mutants exhibit a more severe insensitivity to ABA compared to the single mutants (Supplemental Fig. S13, A and B). Thus, these data suggest that MPK6 functions downstream of AIK1 and MKK5 in the regulation of ABA-mediated root growth (Fig. 7, A and B; Supplemental Fig. S13).
Figure 7.
Model of the AIK1-MKK5-MPK6 cascade involved in the ABA regulation of root growth and stomatal responses. A and B, Root morphology of wild-type, aik1-1, aik1-1 mpk6, and aik1-1 mkk5 plants. Five-day-old seedlings were transferred to MS medium supplemented with 50 μm ABA. Pictures were taken 3 d later. Asterisks indicate significant differences from the wild type at **P < 0.01 by Student’s t test. C, In-gel kinase assays of wild-type and AIK1 overexpressing plants (OE1) treated with 100 μm ABA. Equal amounts of the samples were resolved on SDS-PAGE and stained with Coomassie Brilliant Blue. The large subunit of Rubisco is shown as the loading control for equal protein amounts in the wild-type and transgenic plants. D, In-gel kinase assays of wild-type, aik1-1, mkk5, and aik1-1 mkk5 plants treated with 100 μm ABA. E, MPK6 is activated by ABA. Relative MPK6 activities in the wild type, aik1-1, and aik1-1 mkk5-1; the seedlings were exposed to 100 μm ABA. The activity of MPK6 in the wild type (0 min) was used as a control, which is set as 1. Biological replications (n = 3) show the reduced activity of MPK6 in aik1-1 and aik1-1 mkk5-1 at 0, 5, and 15 min. Error bars indicate sd for the different mutants. Asterisks indicate significant differences from the wild type at *P < 0.05 and **P < 0.01 by Student’s t test. F, Tobacco leaves were transiently transformed with agrobacteria carrying the 35S:AIK1-FLAG vector or the 35S:AIK1K36M-FLAG vector control. The activation of endogenous MAPKs was determined by an in-gel kinase assay. Equal amounts of the samples were resolved on SDS-PAGE and stained with Coomassie Brilliant Blue (CBB). The large subunit of Rubisco is shown as the loading control for equal protein amounts in transgenic plants. Immunoblotting of anti-FLAG antibodies was performed to show the equal amounts of FLAG-AIK1 and FLAG-AIK1K36M in samples. G, Working model of the crosstalk between AIK1-MKK5-MPK6 cascade and ABA signal transduction in regulating ABA responses.
To further determine whether AIK1 and MKK5 were involved in the activation of MPK6 by ABA, endogenous kinase activity of MPK6 in the wild type, AIK1 overexpression transgenic seedlings, and aik1 knockdown mutants was determined after ABA treatment using in-gel kinase assay. Before ABA treatment, the MPK6 activity was hardly detected both in wild-type and AIK1 overexpression (OE1) seedlings (Fig. 7C). In response to ABA, a transient (for 5 and 15 min) increase in MPK6 activity was observed in the wild-type and AIK1-OE1 seedlings. More importantly, compared with the wild type, the AIK1 overexpression lines exhibit a more significant increase in MPK6 activity (Fig. 7C). By contrast, in aik1, mkk5, and aik1 mkk5 mutants, there are more significant decreases in MPK6 activity compared with that in the wild type upon exposure to ABA (Fig. 7, D and E). Since the activation of MPK6 in aik1 and aik1 mkk5 was still observed, there might be AIK1 knockdown effects and/or MAPKKK redundant function involved in the response to ABA.
Meanwhile, under ABA treatment, the AIK1-dependent MPK activity was also tested in tobacco plants because salicylic acid-induced protein kinase and wound-induced protein kinase are the orthologs of AtMPK6 and AtMPK3, respectively. As shown in Figure 7F, overexpressing AIK1 in tobacco increased the activity of salicylic acid-induced protein kinase and wound-induced protein kinase under ABA treatment. However, the inactive form of AIK1 (K36M), used as a control, shows a very weak activation. Taking all our findings together, we present a model describing the ABA-activated MAPK cascade and how it regulates root growth and stomatal responses (Fig. 7G).
DISCUSSION
In this study, we revealed that AIK1 plays an important role in the primary root growth and guard cell signaling in response to ABA. We have shown that the mutation of AIK1 leads to insensitivity to ABA in terms of guard cell signaling, primary root growth, and development in Arabidopsis. Further experiments have indicated that, upon exposure to ABA, the stomatal closing in aik1 plants was less sensitive to ABA than that in wild-type plants, the cell numbers from root tip to the elongation zone were greater in aik1 than in wild-type seedlings, and the cell length in the elongation zone of aik1 was longer than in wild-type seedlings. We have also shown that the intracellular signals were relayed and amplified through sequential phosphorylations of the AIK1 (MKKK20)-MKK5-MPK6 cascade, in response to ABA for regulating primary root growth (Fig. 7G).
AIK1 (MKKK20) Is an Important Modulator of ABA Signaling
It has been documented that several components of distinct MAPK signaling cascades involved in transcriptional regulation, protein accumulation and stability, as well as kinase activity are induced by ABA, indicating that MAPK pathways play an important role in ABA signaling in Arabidopsis (Xing et al., 2008; Jammes et al., 2009, 2011; Salam et al., 2013; Danquah et al., 2014, 2015; Han et al., 2014; López-Bucio et al., 2014; Lee et al., 2015; Matsuoka et al., 2015). However, there are few reports concerning the involvement of MAPKKK in the modulation of ABA signaling. Here, we reported that the transcriptional level of AIK1 was up-regulated by ABA, and down-regulation of AIK1 in aik1 mutants resulted in primary root growth that was insensitive to ABA (Fig. 1A). AIK1 mutations have been shown to cause ABA-insensitive stomatal closing and rapid water loss resulting from transpiration (Fig. 2, A and B). This finding was supported by the data showing that AIK1 expression occurred preferentially in guard cells (Fig. 2D). More importantly, the phosphorylation activity of AIK1 is activated by ABA in planta (Fig. 3, D and E). We conclude that AIK1 mediates a primary ABA-dependent response in Arabidopsis.
In the absence of an activating signal, the constituents of the AIK1 cascade return to an inactive dephosphorylated state, suggesting a role for phosphatases in the regulation of the MAPK pathway. Supporting this conclusion is the observation that PP2C ABI1 inactivates AIK1 kinase activity for MBP, at least in vitro (Fig. 3C). Similarly, ABI1 interacts with MAPKKK18 and inhibits its activity of by promoting proteasomal degradation of MAPKKK18 (Mitula et al., 2015). These observations raise the possibility that the broad substrate specificity of PP2C (ABI1, ABI2, or HAB1) for MAPKKKs contributes to the generalized maintenance of MAPKKKs in an inactive state when lacking activation signal (e.g. ABA).
Upon treatment with ABA, the cell number from the root tip to the elongation zone was shown to be greater in aik1 than in wild-type seedlings (Fig. 1C), and the cell length in the elongation zone of aik1 was longer than that in wild-type seedlings (Fig. 1D). It has been reported that an Arabidopsis receptor kinase, PERK4, mediates cell elongation not cell division in ABA-inhibited root growth (Bai et al., 2009). It has also been shown that one of the CDK inhibitors, ICK1/KRP1, functions to reduce cell division rate through decrease of endoreduplication (Wang et al., 2000; Schnittger et al., 2003). Therefore, it is important to analyze the relationship between AIK1 and PERK4 or KRP1, which will further reveal regulatory mechanisms controlling AIK1 activity in response to ABA signal. This may improve understanding of the roles of the MAPK cascade in ABA-modulated root growth.
Consistent with increased cell number in roots, the number of stomata in aik1 was also increased compared with that in the wild type (Fig. 2C). Loss-of-function mutants of YODA (YDA), a MAPKKK, have clustered stomata, whereas gain-of-function mutation of YDA results in no stomatal differentiation (Bergmann et al., 2004). Both gain- and loss-of-function data also show the involvement of YDA in the regulation of localized cell proliferation and inflorescence architecture, which further shapes the morphology of plant organs (Meng et al., 2012). Therefore, AIK1 is another MAPKKK involved in the regulation of plant development. Alternatively, there may be some overlap of developmental regulation between AIK1 and YDA in stomatal development. However, this overlapping function in ABA responses needs to be further tested.
MKK5-MPK6 Are the Downstream Targets of AIK1 in ABA Signaling
An analysis of protein interactions showed that only MKK4 and MKK5 could interact with AIK1 in vivo (Fig. 4, A and C). In addition, AIK1 can phosphorylate MKK5 in vitro (Fig. 4G). AIK1 and MKK5 were shown to be localized in the cytoplasm (Fig. 4A; Supplemental Fig. S2A), and their expression levels were up-regulated by ABA (Supplemental Figs. S2D and S10G), which indicated that MKK5 may be the substrate of AIK1 in response to ABA. The aik1 and the mkk5 T-DNA insertion mutants exhibited similar morphology in terms of their stomatal response and root growth (Figs. 1A, 2A, 4D, and 4F). Constitutively active MKK5 in the aik1 mutant plants complemented the ABA-insensitive primary root growth phenotype in aik1 (Fig. 5, A and B). MKK4 and MKK5 share high sequence similarity, with 78% identity at the protein sequence level. MKK4 and MKK5 together with MPK3 and MPK6 were also previously shown to be involved downstream of several MAPKKKs and to function in plant responses to environmental stimuli (Tena et al., 2001; Zhang and Klessig, 2001; Nakagami et al., 2005; Pedley and Martin, 2005). MKK4 and MKK5 are functionally redundant in stress-responsive signaling pathways; they are regulators of stomatal development and patterning (Yang et al., 2001; Ren et al., 2002; Wang et al., 2007). However, it is likely that MKK4 and MKK5 have different functions in regulating primary root growth and development; upon the disruption of these genes, only mkk5 exhibited a similar phenotype to aik1 (Supplemental Fig. S10E).
The observation that ABA-induced increase in MPK6 kinase activity was clearly decreased in aik1 and mkk5 mutants and increased in AIK1 overexpression lines suggests that AIK1 and MKK5 are required for ABA-triggered activation of MPK6 (Figs. 6, C and D, and 7, D and E). The activity of MPK6 was increased in the wild type and mkk4 upon treatment with ABA, but the activity of MPK6 was not consistent with those in aik1 and mkk5 when treated with ABA (Fig. 6, C and D); this result indicates that MKK4 may not be involved in the ABA activation of MPK6 and AIK1-dependent MAPK cascade pathway. Interestingly, the expression levels of MKK5 and MPK6 were up-regulated in wild-type seedlings when exposed to ABA, and the disruption of AIK1 increased their transcriptional levels (Supplemental Figs. S10G and S12C); this indicates that AIK1 may affect MKK5 and MPK6 activities at the transcriptional level or by posttranscriptional protein regulation. The similar morphologies of the aik1, mkk5, and mpk6 mutants and the phosphorylation analysis suggest that the AIK1-MKK5-MPK6 cascade pathway regulates ABA-responsive primary root growth and development.
The direct activation of components of MAPK cascade by ABA has been identified in several stress responses (Xing et al., 2008; Jammes et al., 2009). Recently, a complete ABA-activated MAPK cascade of MAPK has been established, in which MAPKKK17/18-MKK3-MPK1/2/7/14 module might contribute to a robust signal to modulate ABA-dependent responses under stress conditions (Danquah et al., 2015). Our module of AIK1-MKK5-MPK6 may present another modulation mechanism in regarding root growth governed by ABA signaling. Interestingly, AIK1 overexpression seedlings gradually exhibited yellow and eventually bleached leaves (∼25 d) compared to the wild-type seedlings (Supplemental Fig. S5B), and ABA-induced leaf senescence was accelerated in 35S:MAPKKK18 (Matsuoka et al., 2015). AIK1 has redundant functions with MAPKKK18 in ABA-regulated senescence. However, they apparently do not have completely overlapping functions in the ABA-triggered MAPK cascade because AIK1 does not interact with MKK3 (Fig. 4A), and MAPKKK18 also did not interact with MKK5 (Matsuoka et al., 2015). The diversity of MAPK pathway components might help plant to adapt to different environmental conditions.
One question that arises in this context is why MPK6 activity was only slightly decreased in aik1 and mkk5 mutants upon exposure to ABA. The main reason for this may be the multifunctional nature of MPK6, which plays essential roles in plant stress responses, growth, and development (Nakagami et al., 2005; Xing et al., 2008, 2009; Schikora et al., 2011; Dutilleul et al., 2012; Xu et al., 2015; Xu and Zhang, 2015). Actually, MPK6 is a common target among several MAPK cascades (Wang et al., 2007; Meng et al., 2012; Smékalová et al., 2014). Another possibility is the mutants of AIK1 and MKK5 used for phenotype analysis are the knockdown mutants (the aik1 knockout mutant probably is lethal). Therefore, there may have been small amounts of AIK1 and MKK5 functioning in ABA-responsive root growth regulation in these mutants. In addition, MAPK signaling in plants involves the redundancy of signaling components, antagonism among distinct pathways, and both positive and negative regulatory mechanisms, so there may be other components participating in ABA regulation of plant growth, development, and environmental responses. For example, earlier reports demonstrated that MPK9 and MPK12 redundantly function as positive regulators downstream of reactive oxygen species production and [Ca2+]cyt oscillation in the ABA-induced stomatal closure (Jammes et al., 2009; Salam et al., 2013). Although AIK1 and MKK5 could modulate both root growth and stomatal response, MPK6 just has a significant effect on root growth. It would be possible that there may be other downstream MAPKs (e.g. MPK9 or 12) of AIK1-MKK5 to regulate ABA signaling in guard cells. Therefore, evidence for direct interaction of MPK9/12 with MKK5, which acts as a downstream component of AIK1-MKK5 to regulate ABA-induced stomatal closure, is crucial and of great interest.
Moreover, considering the hypersensitivity of ABA or the tolerance of ABA to the loss of function of one or several ABA signaling transduction constituents, it will be interesting to develop conditional rescue systems and partial loss-of-function materials using overexpression of dominant-negative and RNA interference lines containing aik1 and other components of MAPK cascade (e.g. MKK5 and/or MPK6). The combination of this genetic analysis with a biochemical analysis of the protein interaction network of MAPKKK and MKK should further clarify whether they indeed represent a novel ABA signaling cascade and shed much needed light on the currently poorly understood plant MAPK composition in ABA signal transduction.
MATERIALS AND METHODS
Plant Materials, Mutant Screens, and Characterization and Generation of Double Mutants
Seedlings were grown vertically on 0.5× MS media (Murashige and Skoog, 1962) supplemented with 0.6% (w/v) agarose for 10 d in controlled environmental conditions of 21°C and a 16-h-light/8-h-dark photoperiod. The illumination intensity was 150 μmol m−2 s−1 (Holman et al., 2010). Plants were transferred to soil for 12 to 15 d for other experiments in controlled environmental conditions of 21°C and a 12-h-light/12-h-dark photoperiod, with the illumination intensity is 150 μmol m−2 s−1.
The T-DNA insertion lines for AIK1 used in this study were aik1-1 (Salk_049616) and aik1-2 (Salk_124398). The T-DNA insertion lines for MKKs used in this study were mkk4 (Salk_058307), mkk5-1 (Salk_047797), and mkk5-2 (Salk_050700). The T-DNA insertion lines for MPK3 and MPK6 were mpk3 (Salk_100651; Xu et al., 2008), mpk6-3 (Salk_127507), and mpk6-2 (Salk_073907; Wang et al., 2010). The double mutant of AIK1 and MKK5 used in this study was aik1-1 mkk5-1, and the double mutant of AIK1 and MPK6 used in this study was aik1-1 mpk6-3. The primers to check the T-DNA insertions were designed by the SIGnAL iSect tool (http://singal.salk.edu/tdnaprimers.2.html), and PCR was performed using genomic DNA from seedlings (Supplemental Table S1). To generate double mutants, the alleles aik1-1 and aik1-2 were both crossed with mkk5-1, creating aik1-1 mkk5-1 and aik1-2 mkk5-1. Because similar results were obtained with these double mutants, we present data only from the analysis of aik1-1 mkk5-1.
To generate the overexpression and complementation transgenic lines, the coding sequence of wild-type AIK1 was placed under the control of the constitutive 35S promoter of cauliflower mosaic virus and introduced into Arabidopsis (Arabidopsis thaliana; Columbia) for overexpression, and the coding sequence of AIK1 under the control of the native AIK1 promoter (1,507 bp, upstream of the initiation codon ATG of AIK1) was introduced into aik1 mutant plants for complementation lines by Agrobacterium tumefaciens-mediated transformation. T3 progeny of hygromycin-resistant transformants were used for experiments.
For gene expression analysis, RNA was extracted from 2-week-old seedlings grown on MS plates, and RNA was purified by RNA pure kit from Tiangen. Reverse transcription was performed using the M-MLV RTase cDNA synthesis kit from Promega. Real-time PCR was performed on the cDNA reverse transcribed from three independent RNA samples using Takara SYBR Premix Ex Taq II RR820A. The cDNA was diluted 10-fold and then used as a template for qRT-PCR amplification. qRT-PCR was performed with the Applied Biosystems 7500 Real Time PCR system using SYBR Green to monitor double-stranded DNA products. ACTIN2 was used as an internal control. All experiments on gene expression analysis were repeated at least three times. Primer sequences for the PCR are presented in Supplemental Table S1.
Measurement of Root Tip and Cell Lengths
For cell membrane dye analysis, 4- to 5-d-old seedlings were grown on MS plates with 1.2% (w/v) agarose and transferred to ABA-containing 1.2% MS plates for 3 to 4 d and to 1.2% MS plates without ABA as a control. The seedlings were stained with FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide] (Invitrogen F34653) according to procedures previously described (Levesque et al., 2006; Brunoud et al., 2012).
A ruler was imaged alongside the samples and used to calibrate measurements. The root tip length for wild-type and aik1 seedlings, with or without ABA treatment, was measured under a confocal microscope (Zeiss 710). The root tip refers to the portion of the root from the root apex to the beginning of the primary root tip fibril. Root cells were stained with 1 μm FM4-64 for 1 to 3 min, and then the lengths of epidermal cells at the root tip were measured as described earlier (Lally et al., 2001; Bai et al., 2009) using LSCM. Epidermal cells in an array from the tip to the elongation region were counted.
Bioassays of Water Loss
For water loss measurement, rosette leaves of wild-type and mutant plants were detached, placed in weighing dishes, and incubated on the laboratory bench. The data were collected by water loss measurement system software (Sartorius SQP). Losses in fresh weight were monitored at the indicated times. Water loss is expressed as the percentage of initial fresh weight.
Estimation of Stomatal Conductance, Stomatal Aperture, and Density
Gas exchange measurement was performed on the aerial tissues of about 25-d-old seedlings using a portable gas exchange fluorescence system (GFS-3000; Heinz Walz) equipped with a 3010-A Arabidopsis chamber (Hashimoto-Sugimoto et al., 2016). The GFS-3000 system was connected to a computer with data acquisition software (GFS-Win V3.51). The cuvette for Arabidopsis conditions was set at a light intensity of 200 μmol m−2 s−1, provided by a special artificial light (LED array/PAM fluorometer 3055-FL, optional; Heinz Walz), with relative humidity and air temperature set at 50% and 22°C, respectively. The leaf area was estimated by cellSens Dimension software (Olympus). Measurements were made every 60 s. Details of replication are shown in the figure legend.
Epidermal strips were peeled from Arabidopsis rosette leaves of 4-week-old wild-type and mutant plants, and the chlorophyll was brushed away. These epidermal strips were then floated in the stomatal-opening solution, containing 10 mm KCl, 50 μm CaCl2, and 10 mm MES (pH 6.15), under strong light for about 90 μmol m−2 s−1 for 2 h (Miao et al., 2006). The strips were then incubated in 10, 20, and 50 μm ABA, and the stomatal apertures were measured after 2 h. Controls (absolute ethyl alcohol instead of ABA added) were performed in parallel. Pictures were taken by a LSCM FV1000 (Olympus). Three independent experiments were performed, with stomata in 50 pictures measured each time.
A. tumefaciens-mediated Transformation
A. tumefaciens-mediated transient transformation experiments in tobacco (Nicotiana benthamiana) were performed as previously described (Yang et al., 2001). Six-week-old tobacco plants grown at 25°C in a growth room programmed for a 14-h-light/10-h-dark cycle were used for experiments. The tobacco leaves were infiltrated with agrobacteria cells that carry p1307-AIK1-cFLAG and p1307-AIK1K36M-cFLAG constructs, and the transgenic tobacco leaves were sprayed with 100 μm ABA 4 d later.
Coimmunoprecipitation Assay
The total proteins were extracted from tobacco leaves expressing 35S:AIK1-FLAG/35S:GFP or 35S:AIK1-FLAG/35S:MKK5-GFP constructs with A. tumefaciens-mediated transient transformation experiments in tobacco plants. The total proteins were incubated with anti-FLAG agarose beads, and proteins bound to the beads were detected with anti-GFP antibody (Sigma-Aldrich).
Histochemical Detection of GUS Activity
For the GUS assays, excised tissues from two independent transgenic lines that contained the AIK1 promoter-GUS construct were tested according to previously described (Song et al., 2005).
Bimolecular Fluorescence Complementation Assay
To measure in vivo interactions, the coding regions of AIK1, MKK1-MKK10, MPK3, and MPK6 were amplified by PCR (Fast Pfu DNA polymerase; Novoprotein) with primers that contained appropriate restriction sites, and the amplified fragments were inserted into the plasmids pSPYNE and pSPYCE, which contain DNA encoding the N-terminal and C-terminal regions of YFP, respectively (Walter et al., 2004), to form pSPYNE-AIK1, pSPYCE-MKK1-10, pSPYNE-MPK3, and pSPYNE-MPK6, respectively. According to the previous protocols (Walter et al., 2004), protoplasts isolated from Arabidopsis leaves were transformed with the following combinations of plasmids: pSPYNE-AIK1 and pSPYCE-MKK1-10, and pSPYCE-MKK5 and pSPYCE-MPK3/6. For the GFP constructs, the coding regions of AIK1 and MKK5 were amplified by PCR and inserted into the modified plasmid pHBT-GFP-NOS (Sheen, 2001) to form pAIK1-GFP and pMKK5-GFP, respectively. The protoplast transient expression assay was performed as described (Sheen, 2001). After incubation for 16 to 20 h, the fluorescence of the protoplasts was measured with a FV1000 LSCM (Olympus). All figures show representative images from three independent experiments. Primer sequences for the PCR are presented in Supplemental Table S1.
Fluorescence Microscopy
The fluorescence was observed with laser scanning confocal microscope (LSCM FV1000 [Olympus] and LSCM 710 [Zeiss]). The fluorescence were excited at 488 nm and detected at 500 to 560 nm for GFP or excited at 514 nm and detected at 530 to 570 nm for YFP; autofluorescence of chloroplasts was detected at 650 to 750 nm. FM4-64 was excited at 514 nm and detected at 600 to 700 nm. Transmitted field was collected simultaneously for merging images.
Recombinant Protein Expression in Escherichia coli
For in vitro protein expression, the DNA regions containing the coding region of AIK1 and MKK5 were amplified and inserted in frame into the plasmid pET-28a (Novagen). Mutations of MKK5 were introduced by the Fast Mutagenesis System (TransGen Biotech). pET-AIK1, pET-M313 (1–313 amino acids), pET-M270 (1–270 amino acids), and a construct that expressed FLAG-MKK5KR (K99R; Ren et al., 2002) were introduced into E. coli BL21 (DE3) cells. The recombinant HIS-tagged proteins were purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s protocol. Primer sequences for the PCR are presented in Supplemental Table S1.
Phosphorylation Assay
The in vitro kinase assay was performed as described previously (Liu and Zhang, 2004). Recombinant HIS-tagged AIK1, M313, and M270 (10 μg) were used to phosphorylate MBP or recombinant protein FLAG-MKK5KR purified from E. coli (1:10 = enzyme:substrate ratio) with 50 μm ATP and γ-32P-ATP (0.1 μCi per reaction) in 50 mL of reaction buffer (20 mm HEPES, pH 7.5, and 10 mm MgCl2). The reactions were stopped by the addition of SDS loading buffer after incubation at 30°C for 30 min. The phosphorylated MKK5KR was visualized by autoradiography after separation on a 10% SDS polyacrylamide gel. After electrophoresis, the gel was exposed to a Kodak X-Omat film for 10 h. For enzymes used in the immunoblot assay, phosphorylation was performed without the addition of γ-32P-ATP. The phosphorylation of the substrates was visualized and quantified with a Bioimaging Analyzer Tool (ImageJ, http://rsbweb.nih.gov/ij/). The data were analyzed according to the tutorial in ImageJ: Using ImageJ to Quantify Gel Images.
For dephosphorylation assay of AIK1 and SnRK2.6 by ABI1, HIS-AIK1 (78.7 nm) and HIS-SnRK2.6 (78.7 nm) were preincubated with different ratios of HIS-ABI1 protein (119–434 amino acids) in 25 mm Tris-Cl, pH 7.4, 12 mm MgCl2, and 0.1 mm EGTA for 30 min at room temperature. This was followed by 30 min incubation at 30°C with MBP (2 μg), 50 μm ATP, and γ-32P-ATP (0.1 μCi per reaction) in 50 mL of reaction buffer. The reactions were stopped by the addition of SDS loading buffer.
LC-MS/MS Analysis
Phosphorylation reaction mixtures were separated by SDS-PAGE (10% gel); bands corresponding to AIK1, AIK1-1, AIK1-2, and AIK1-3 proteins were excised from the Coomassie Brilliant Blue-stained gel and subjected to in-gel digestion using trypsin (Promega) to specially digest the carboxyl end of the peptide bond in Arg and Lys overnight. The reaction was quenched by the addition of 0.1% formic acid (DIMA Technology) to consume the remaining trypsin. Ten microliters of the resulting peptide mixtures was subjected directly to LC-MS/MS analyses. LC-MS/MS analysis was performed using a hybrid mass spectrometer Q-Exactive (Thermo Scientific) and an ultimate 3000-UHPLC system (Dionex) The generated peptides were applied to a reverse phase trap column (Symmetry C18; 5 μm, 750 μm × 2 cm; Thermo Scientific) connected to an analytical column (Venusi I × BPC, C18, 5 μm, 150A, 750 μm × 10 cm; Agela Technologies) in vented configuration using nano-T coupling union. For phosphopeptide identifications, the same parameters were applied to phosphorylation of Ser/Thr/Tyr residues. Acceptance parameters were as follows: score, 22 and 15; E-value, 0.01 and 0.05 for protein and peptide identifications, respectively.
Protein Extraction and Immunoblotting Analysis
Total protein was extracted from Arabidopsis seedlings and tobacco leaves by grinding with plastic pestles in extraction buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerophosphate, 10 mm DTT, 1 mm PMSF, 5 μg/mL leupeptin, 5 μg/mL aprotinin, and 5% glycerol). After centrifugation at 4°C, 10,000g for 30 min, supernatants were transferred into clean tubes. The protein concentration was determined using the protein assay kit (Bio-Rad) using bovine serum albumin as the standard. For western-blot analysis, 7.5 μg of total protein per lane was separated on a 10% SDS-PAGE gel. After electrophoresis, the proteins were electrotransferred onto nitrocellulose membranes. After blocking for 2 h in TBST buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Tween 20) with 5% fat-free dried milk at room temperature, the membranes were incubated with anti-FLAG or anti-HIS antibody in mouse as first antibody (Sigma-Aldrich; 1:10,000 dilution) overnight at 4°C. Following three washes with TBST buffer, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG as secondary antibody (Sigma-Aldrich; 1:10,000 dilution). After three washes with TBST buffer, the membranes were then visualized using an enhanced Lumi-Light Western Blotting substrate kit (Roche) following the manufacturer’s instructions.
In-Gel Kinase Activity Assay
The in-gel kinase activity assay was performed using a previously described method (Wang et al., 2010; Ren et al., 2002). In brief, an aliquot containing 7.5 μg of protein extract was electrophoresed on 10% SDS-polyacrylamide gels embedded with 0.1 mg/mL MBP in separating gel as a substrate for kinase. After electrophoresis, the SDS was removed from the gel by washing with washing buffer (25 mm Tris-HCl, pH 7.5, 0.5 mm DTT, 0.1 mm Na3VO4, 5 mm NaF, 0.5 mg/mL bovine serum albumin, and 0.1% Triton X-100) three times for 30 min each at room temperature. The proteins were then renatured in 25 mm Tris-HCl, pH 7.5, 1 mm DTT, 0.1 mm Na3VO4, and 5 mm NaF at 4°C overnight with three changes of the buffer. The gel was incubated at room temperature in 100 mL of reaction buffer (25 mm Tris-HCl, pH 7.5, 2 mm EGTA, 12 mm MgCl2, 1 mm DTT, and 0.1 mm Na3VO4) for 30 min. Phosphorylation was performed for 1.5 h at room temperature in 30 mL reaction buffer that contained 200 nm ATP and 50 μCi of γ-32P-ATP (3 000 μCi/mm). The reaction was stopped by transferring the gel into a solution of 5% trichloroacetic acid (w/v) and 1% sodium pyrophosphate (w/v). The unincorporated radioactivity was subsequently removed by washing the gel for 5 h at room temperature with five changes. The gel was dried on Whatman 3MM paper and subjected to autoradiography. Prestained size markers (NEB) were used to calculate the sizes of the kinases.
Microscale Thermoporesis Analysis of AIK1 and MKK5
Specific interaction between AIK1 and MKK5 was measured by the MST method (Duhr and Braun, 2006; Seidel et al., 2013; Ribeiro et al., 2014). Unlabeled AIK1 was titrated into a fixed concentration of fluorescent bodipy MKK5 (nm; Monolith NT Protein Labeling Kit RED-NHS), and the mixture of labeled and unlabeled molecules was centrifuged at 10,000g for 5 min. The labeled MKK2 was used as a negative control. Binding reactions were carried out in buffer containing 25 mm HEPES, pH 7.5, and 150 mm NaCl. Samples were loaded in NT.115 standard treated capillaries (Nanotemper Technologies) immediately after blending well to avoid unspecific adsorption. Before MST measurement, the reaction was incubated at room temperature for 10 min mounted in the monolith NT.115 apparatus (Nanotemper Technologies). The data for MST analysis were recorded at 25°C using the red LED at 60% (green filter; excitation 515–525 nm and emission 560–585 nm) and infrared laser power at 50%. Data analyses were performed with NT Analysis software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AIK1/MKKK20, AT3G50310; MKK2, AT4G29810; MKK4, AT1G51660; MKK5, AT3G21220; MKK6, AT5G56580; MKK9, AT1G73500; MPK6, AT2G43790; MPK3, AT3G45640; ABI1, AT4G26080; and ACTIN2, AT3G18780.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Screening for the aik1-1 mutant.
Supplemental Figure S2. The AIK1 protein localization in cells and gene expression in different tissues.
Supplemental Figure S3. Screening and identification of the AIK1 mutants.
Supplemental Figure S4. aik1 plants were insensitive to ABA in root growth with a dose-dependent manner.
Supplemental Figure S5. Overexpression of AIK1 showed sensitive to ABA in the inhibition of plant growth and development.
Supplemental Figure S6. AIK1 negatively regulates root cell division and cell elongation.
Supplemental Figure S7. Stomatal conductance in response to light and dark in wild-type and aik1 mutant plants.
Supplemental Figure S8. Total cell numbers of the leaf epidermis in wild-type and aik1 mutants.
Supplemental Figure S9. The sequence analysis of AIK1 protein by InterPro (http://www.ebi.ac.uk/interpro/) and sketch map of the expressed proteins of AIK1.
Supplemental Figure S10. Decreased sensitivity of root growth to ABA in aik1 and mkk5 mutants, but not mkk4.
Supplemental Figure S11. Decreased sensitivity to ABA in root growth and water loss in MKK5DD plants.
Supplemental Figure S12. Decreased sensitivity of root growth to ABA in mpk3 and mpk6 mutants.
Supplemental Figure S13. Comparative analysis of aik1-1, mkk5, and mpk6 single and double mutants in response to ABA in terms of seedling growth.
Supplemental Table S1. List of the primers used in all experiments.
Supplemental Table S2. LC-MS/MS assay of the phosphorylation sites of AIK1.
Glossary
- ABA
abscisic acid
- MAPK
mitogen-activated protein kinase
- MAPKKK
MAP kinase kinase kinase
- MAPKK
MAP kinase kinase
- MS
Murashige and Skoog
- LSCM
laser scanning confocal microscope
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MST
microscale thermophoresis
- DEX
dexamethasone
Footnotes
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bai L, Ma X, Zhang G, Song S, Zhou Y, Gao L, Miao Y, Song CP (2014) A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. Plant Cell 26: 1497–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP (2009) Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J 60: 314–327 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Munemasa S, Wang C, Nguyen D, Yong T, Yang PG, Poretsky E, Belknap TF, et al. (2015) Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4: e03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Frei Dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, et al. (2015) Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82: 232–244 [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32: 40–52 [DOI] [PubMed] [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D, Kreuzer I (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110: 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhr S, Braun D (2006) Why molecules move along a temperature gradient. Proc Natl Acad Sci USA 103: 19678–19682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Benhassaine-Kesri G, Demandre C, Rézé N, Launay A, Pelletier S, Renou JP, Zachowski A, Baudouin E, Guillas I (2012) Phytosphingosine-phosphate is a signal for AtMPK6 activation and Arabidopsis response to chilling. New Phytol 194: 181–191 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Maierhofer T, Al-Rasheid KA, Scherzer S, Mumm P, Liese A, Ache P, Wellmann C, Marten I, Grill E, Romeis T, Hedrich R (2011) Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci Signal 4: ra32. [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Han S, Wang CW, Wang WL, Jiang J (2014) Mitogen-activated protein kinase 6 controls root growth in Arabidopsis by modulating Ca2+ -based Na+ flux in root cell under salt stress. J Plant Physiol 171: 26–34 [DOI] [PubMed] [Google Scholar]
- Hashimoto-Sugimoto M, Negi J, Monda K, Higaki T, Isogai Y, Nakano T, Hasezawa S, Iba K (2016) Dominant and recessive mutations in the Raf-like kinase HT1 gene completely disrupt stomatal responses to CO2 in Arabidopsis. J Exp Bot 67: 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman TJ, Wilson MH, Kenobi K, Dryden IL, Hodgman TC, Wood AT, Holdsworth MJ (2010) Statistical evaluation of transcriptomic data generated using the Affymetrix one-cycle, two-cycle and IVT-Express RNA labelling protocols with the Arabidopsis ATH1 microarray. Plant Methods 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li CY, Qi Y, Park S, Gibson SI (2014) SIS8, a putative mitogen-activated protein kinase kinase kinase, regulates sugar-resistant seedling development in Arabidopsis. Plant J 77: 577–588 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, et al. ; MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F, Yang X, Xiao S, Kwak JM (2011) Two Arabidopsis guard cell-preferential MAPK genes, MPK9 and MPK12, function in biotic stress response. Plant Signal Behav 6: 1875–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokon MA, Salam MA, Jammes F, Ye W, Hossain MA, Uraji M, Nakamura Y, Mori IC, Kwak JM, Murata Y (2015) Two guard cell mitogen-activated protein kinases, MPK9 and MPK12, function in methyl jasmonate-induced stomatal closure in Arabidopsis thaliana. Plant Biol (Stuttg) 17: 946–952 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH (2001) Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13: 1317–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lee MH, Kim JI, Kim SY (2015) Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol 56: 84–97 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Y, Xu J, Su T, Liu G, Ren D (2008) Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res 18: 422–432 [DOI] [PubMed] [Google Scholar]
- Liu Y. (2012) Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio JS, Dubrovsky JG, Raya-González J, Ugartechea-Chirino Y, López-Bucio J, de Luna-Valdez LA, Ramos-Vega M, León P, Guevara-García AA (2014) Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J Exp Bot 65: 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T, Erickson BJ, Finkelstein RR (2012) Direct interactions of ABA-insensitive(ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol Biol 80: 647–658 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Matsuoka D, Yasufuku T, Furuya T, Nanmori T (2015) An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol Biol 87: 565–575 [DOI] [PubMed] [Google Scholar]
- Melcher K, Zhou XE, Xu HE (2010) Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr Opin Struct Biol 20: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NS, Tuteja R, Tuteja N (2006) Signaling through MAP kinase networks in plants. Arch Biochem Biophys 452: 55–68 [DOI] [PubMed] [Google Scholar]
- Mitula F, Tajdel M, Cieśla A, Kasprowicz-Maluśki A, Kulik A, Babula-Skowrońska D, Michalak M, Dobrowolska G, Sadowski J, Ludwików A (2015) Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol 56: 2351–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, Kwak JM, Schroeder JI (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murphy A, Taiz L (1995) A new vertical mesh transfer technique for metal-tolerance studies in Arabidopsis (ecotypic variation and copper-sensitive mutants). Plant Physiol 108: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Ribeiro EdeA Jr., Pinotsis N, Ghisleni A, Salmazo A, Konarev PV, Kostan J, Sjöblom B, Schreiner C, Polyansky AA, Gkougkoulia EA, et al. (2014) The structure and regulation of human muscle α-actinin. Cell 159: 1447–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Ronzier E, Corratgé-Faillie C, Sanchez F, Prado K, Brière C, Leonhardt N, Thibaud JB, Xiong TC (2014) CPK13, a noncanonical Ca2+-dependent protein kinase, specifically inhibits KAT2 and KAT1 shaker K+ channels and reduces stomatal opening. Plant Physiol 166: 314–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam MA, Jammes F, Hossain MA, Ye W, Nakamura Y, Mori IC, Kwak JM, Murata Y (2013) Two guard cell-preferential MAPKs, MPK9 and MPK12, regulate YEL signalling in Arabidopsis guard cells. Plant Biol (Stuttg) 15: 436–442 [DOI] [PubMed] [Google Scholar]
- Schikora A, Schenk ST, Stein E, Molitor A, Zuccaro A, Kogel KH (2011) N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol 157: 1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schöbinger U, Hülskamp M (2003) Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, et al. (2013) Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59: 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Smékalová V, Luptovčiak I, Komis G, Šamajová O, Ovečka M, Doskočilová A, Takáč T, Vadovič P, Novák O, Pechan T, et al. (2014) Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol 203: 1175–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP (2010) Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1: 100. [DOI] [PubMed] [Google Scholar]
- Wu ZY, Chen J, Zhu MJ (1996) Identification of the photoaffinity-labeled abscisic acid binding proteins from maize root microsome. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 28: 694–696 [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J (2008) AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J 54: 440–451 [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J (2009) AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol Biol 70: 725–736 [DOI] [PubMed] [Google Scholar]
- Xu DB, Chen M, Ma YN, Xu ZS, Li LC, Chen YF, Ma YZ (2015) A G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PLoS One 10: e0116385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20: 56–64 [DOI] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38: 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yu XC, Zhu SY, Gao GF, Wang XJ, Zhao R, Zou KQ, Wang XF, Zhang XY, Wu FQ, Peng CC, Zhang DP (2007) Expression of a grape calcium-dependent protein kinase ACPK1 in Arabidopsis thaliana promotes plant growth and confers abscisic acid-hypersensitivity in germination, postgermination growth, and stomatal movement. Plant Mol Biol 64: 531–538 [DOI] [PubMed] [Google Scholar]
- Zhang A, Zhang J, Ye N, Cao J, Tan M, Zhang J, Jiang M (2010) ZmMPK5 is required for the NADPH oxidase-mediated self-propagation of apoplastic H2O2 in brassinosteroid-induced antioxidant defence in leaves of maize. J Exp Bot 61: 4399–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhang T, Liu Y, Yang T, Zhang L, Xu S, Xue L, An L (2006) Diverse signals converge at MAPK cascades in plant. Plant Physiol Biochem 44: 274–283 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong XJ, Li DP, Gu LK, Li DQ, Liu LX, Hu XL (2009) Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 229: 485–495 [DOI] [PubMed] [Google Scholar]
- Zou JJ, Wei FJ, Wang C, Wu JJ, Ratnasekera D, Liu WX, Wu WH (2010) Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol 154: 1232–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]