Constitutive expression of SAUR genes increases cell wall extensibility and promotes auxin-independent elongation growth in Arabidopsis and tomato.
Abstract
The plant hormone indole-3-acetic acid (IAA or auxin) mediates the elongation growth of shoot tissues by promoting cell expansion. According to the acid growth theory proposed in the 1970s, auxin activates plasma membrane H+-ATPases (PM H+-ATPases) to facilitate cell expansion by both loosening the cell wall through acidification and promoting solute uptake. Mechanistically, however, this process is poorly understood. Recent findings in Arabidopsis (Arabidopsis thaliana) have demonstrated that auxin-induced SMALL AUXIN UP RNA (SAUR) genes promote elongation growth and play a key role in PM H+-ATPase activation by inhibiting PP2C.D family protein phosphatases. Here, we extend these findings by demonstrating that SAUR proteins also inhibit tomato PP2C.D family phosphatases and that AtSAUR19 overexpression in tomato (Solanum lycopersicum) confers the same suite of phenotypes as previously reported for Arabidopsis. Furthermore, we employ a custom image-based method for measuring hypocotyl segment elongation with high resolution and a method for measuring cell wall mechanical properties, to add mechanistic details to the emerging description of auxin-mediated cell expansion. We find that constitutive expression of GFP-AtSAUR19 bypasses the normal requirement of auxin for elongation growth by increasing the mechanical extensibility of excised hypocotyl segments. In contrast, hypocotyl segments overexpressing a PP2C.D phosphatase are specifically impaired in auxin-mediated elongation. The time courses of auxin-induced SAUR expression and auxin-dependent elongation growth were closely correlated. These findings indicate that induction of SAUR expression is sufficient to elicit auxin-mediated expansion growth by activating PM H+-ATPases to facilitate apoplast acidification and mechanical wall loosening.
The plant hormone indole-3-acetic acid (IAA or auxin) regulates numerous aspects of plant growth and development (Enders and Strader, 2015). Of particular note, auxin promotes the cell expansion underlying shoot growth. Indeed, the growth promoting properties of auxin provided the basis for its discovery (Went and Thimann, 1937). Despite detailed elucidation of the auxin signaling pathway leading to changes in gene expression (Chapman and Estelle, 2009), the mechanisms by which auxin promotes cell expansion are still uncertain.
The acid growth theory is a long-standing hypothesis for auxin-mediated cell expansion (Rayle and Cleland, 1970, 1980, 1992; Hager, 2003). Physiological studies correlating auxin-induced elongation growth with apoplastic acidification led to the hypothesis that auxin activates plasma membrane H+-ATPases (PM H+-ATPases). Reduction in apoplastic pH activates expansins and perhaps other cell wall modification enzymes to make cells amenable to expansion growth (McQueen-Mason et al., 1992; Cosgrove, 2016). Furthermore, PM H+-ATPase activation also results in plasma membrane hyperpolarization, thus promoting solute and water uptake to provide the intracellular turgor increase that drives cell expansion. However, this model lacked both strong genetic support and an understanding of the molecular components linking auxin to PM H+-ATPase activation.
Crucial insight into this process was provided when Takahashi et al. (2012) demonstrated that auxin promotes phosphorylation of the penultimate Thr residue within the autoinhibitory domain of Arabidopsis (Arabidopsis thaliana) PM H+-ATPases. Phosphorylation of this residue (corresponding to Thr-947 of ARABIDOPSIS H+-ATPASE2 [AHA2]) coincides with 14-3-3 protein binding and activation of the H+ pump (Fuglsang et al., 1999; Kinoshita and Shimazaki, 1999; Jelich-Ottmann et al., 2001). Furthermore, the kinetics of auxin-induced Thr-947 phosphorylation correlated closely with the elongation kinetics of auxin-treated hypocotyl segments, strongly suggesting that auxin promotes cell expansion by posttranslationally activating PM H+-ATPases.
Recent findings suggest that SMALL AUXIN UP-RNA (SAUR) genes function as key signaling pathway outputs that promote auxin-mediated elongation growth via control of H+ pumps. SAURs comprise the largest family of auxin-induced genes, with 79 members in Arabidopsis and comparable numbers in other species (Ren and Gray, 2015). The expression of many, but not all SAUR genes, is rapidly induced by auxin. In general, the auxin-induced SAURs tend to be most highly expressed in shoots, whereas many nonresponsive and auxin-repressed family members are preferentially expressed in roots (Paponov et al., 2008). Additionally, the expression of many auxin-induced SAUR genes correlates with active cell expansion (McClure and Guilfoyle, 1987; Franklin et al., 2011; Chae et al., 2012; Spartz et al., 2012; Stamm and Kumar, 2013; Sun et al., 2016).
SAURs encode small (typically 10–20 kD) proteins containing a highly conserved SAUR domain of ∼60 amino acids. These proteins are unique to plants and contain no obvious motifs suggestive of a biochemical function. Several SAUR proteins are highly unstable and appear to be subject to ubiquitin-mediated proteolysis (Knauss et al., 2003; Chae et al., 2012; Spartz et al., 2012; Li et al., 2015). However, the addition of an epitope or GFP tag can dramatically stabilize SAUR proteins. Expression of these stabilized SAUR fusion proteins in Arabidopsis confers several phenotypes indicative of increased cell expansion. For example, plants expressing AtSAUR63-GFP exhibit increased hypocotyl, petal, and stamen length (Chae et al., 2012), while overexpression of GFP-AtSAUR19 confers increased hypocotyl and leaf size, altered tropic response, and defects in apical hook maintenance (Spartz et al., 2012; Vi et al., 2013). Furthermore, plants overexpressing any of several additional SAURs, including AtSAUR14, 36, 41, 50, 76, 77, and 78, have also been reported to display a long hypocotyl phenotype (Kong et al., 2013; Stamm and Kumar, 2013; Li et al., 2015; Sun et al., 2016).
We have recently demonstrated that GFP-AtSAUR19 Arabidopsis seedlings exhibit increased PM H+-ATPase activity (Spartz et al., 2014). Consistent with this observation, GFP-AtSAUR19 seedlings exhibit striking phenotypic similarity to OPEN STOMATA2 (ost2) mutants (Merlot et al., 2007), which carry a dominant, gain-of-function mutation in AHA1 that confers constitutive PM H+-ATPase activity. In addition to the aforementioned cell expansion phenotypes, both GFP-AtSAUR19 transgenics and ost2-2 mutants exhibit reduced apoplastic pH, constitutive defense gene expression, and hypersensitivity to drought and toxic cations, such as Li+, all of which can be linked to elevated PM H+-ATPase activity (Merlot et al., 2007; Haruta et al., 2010; Haruta and Sussman, 2012; Spartz et al., 2014). Biochemically, the elevated PM H+-ATPase activity of GFP-AtSAUR19 plants appears to be the result of increased phosphorylation of the critical regulatory phosphosite, Thr-947 within the autoinhibitory domain. AtSAUR19, as well as several additional SAUR proteins, physically interact with D-clade type 2C protein phosphatases (PP2C.D) and inhibit PP2C.D activity both in vivo and in vitro (Spartz et al., 2014). Our prior work has demonstrated that PP2C.D proteins negatively regulate both the activity and Thr-947 phosphorylation status of PM H+-ATPases. Consistent with these findings, Arabidopsis plants overexpressing PP2C.D1 exhibit opposite phenotypes compared to ost2-2 and GFP-AtSAUR19 seedlings, including reductions in cell elongation, PM H+-ATPase activity, Thr-947 phosphorylation, and Li+ sensitivity (Spartz et al., 2014; Haruta et al., 2015). Together, these data led us to propose a model whereby AtSAUR19 and other auxin-induced SAURs are key signaling outputs that inhibit PP2C.D phosphatase activity, thereby activating PM H+-ATPases to promote auxin-mediated cell expansion.
In this study, we extend our previous findings by taking advantage of a tomato (Solanum lycopersicum) seedling system. We find that constitutive GFP-AtSAUR19 expression in tomato confers phenotypes similar to those previously described in Arabidopsis (Spartz et al., 2012, 2014) and demonstrate that constitutive GFP-AtSAUR19 expression bypasses the requirement for auxin to promote elongation of excised hypocotyl segments. Additionally, we provide evidence suggesting that like auxin treatment, GFP-AtSAUR19 expression alters biomechanical properties of tomato hypocotyls that may play a role in facilitating elongation growth.
RESULTS AND DISCUSSION
GFP-AtSAUR19 Functions in Tomato
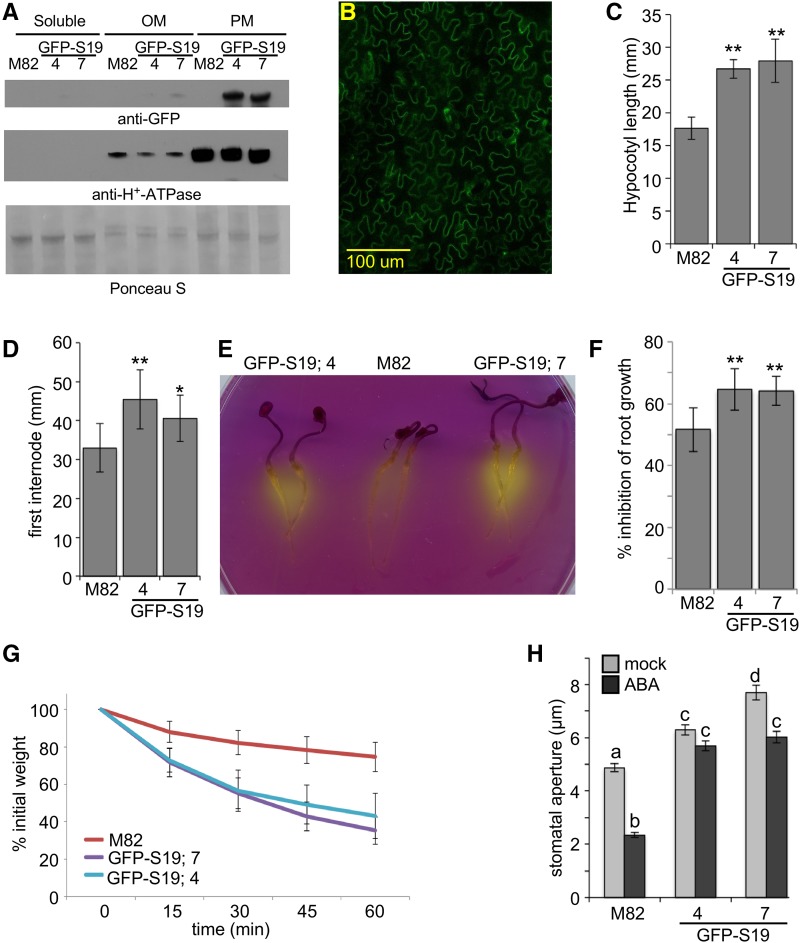
Two representative transgenic tomato lines expressing a GFP-tagged Arabidopsis SAUR19 transgene under the control of the CaMV 35S promoter (35S:GFP-AtSAUR19) were characterized. GFP-AtSAUR19 expression was confirmed by both immunoblot and confocal analyses (Fig. 1, A and B). Per our findings in Arabidopsis (Spartz et al., 2012), the GFP-AtSAUR19 protein from tomato was highly enriched in plasma membrane fractions and localized primarily to the cell periphery consistent with a plasma membrane localization. Light-grown seedlings of both transgenic lines displayed increases in hypocotyl and internode lengths compared to the wild-type M82 parental line (Fig. 1, C and D; Supplemental Fig. S1). GFP-AtSAUR19 expression in Arabidopsis confers several phenotypes indicative of elevated PM H+-ATPase activity, including increased medium acidification, hypersensitivity to Li+ and other toxic cations, and drought hypersensitivity. Likewise, GFP-AtSAUR19 expression in tomato conferred increased medium acidification in bromocresol purple assays, hypersensitivity to Li+ in root growth inhibition assays, and phenotypes indicative of drought hypersensitivity, including accelerated water loss of excised leaves and pronounced wilting upon exposure to reduced humidity (Fig. 1, E–G; Supplemental Fig. 2). To examine the basis of the drought hypersensitivity of GFP-AtSAUR19 tomatoes, we measured guard cell apertures of leaf epidermal peels subjected to mock and 10 μm abscisic acid (ABA) treatments. Like ost2-2 mutants of Arabidopsis, GFP-AtSAUR19 expression conferred an increase in stomatal aperture and a reduction in ABA-mediated pore closure (Fig. 1H). Thus, our results indicate that like Arabidopsis, GFP-AtSAUR19 overexpression confers increased PM H+-ATPase activity in tomato.
Figure 1.
35S:GFP-AtSAUR19 expression in tomato. A, Five micrograms of soluble, lower phase-enriched other membrane (OM) or upper phase-enriched plasma membrane (PM) fractions prepared from 8-d-old M82 or 35S:GFP-AtSAUR19 (GFP-S19) seedlings were immunoblotted with α-GFP and α-AHA antibodies. A section of the Ponceau S-stained blot is shown as a loading control. GFP-S19.4 and GFP-S19.7 are two independent transgenic lines expressing the 35S:GFP-AtSAUR19 transgene. B, Confocal image of GFP-AtSAUR19 tomato leaf epidermal cells. C and D, Mean (±sd) hypocotyl lengths of 8-d-old seedlings (n ≥ 15; C) and first internode lengths of 27-d-old plants (n ≥ 12; D). Asterisks indicate significant difference from the wild type: **P ≤ 0.01 and *P ≤ 0.05. E, Three-day-old seedlings were transferred to plates containing the pH indicator dye, bromocresol purple, and incubated an additional 24 h. F, Percentage of root growth inhibition ± sd by 12.5 mm LiCl. **P ≤ 0.01. G, Kinetics of water loss in leaf detachment assays. Data depict relative mean weights of ≥24 leaves/genotype ± sd. Both GFP-AtSAUR19 transgenic lines exhibit significant differences from M82 at all time points after t = 0 (P ≤ 0.05). H, Mean stomatal apertures (±se) of epidermal peels obtained from 27-d-old tomato leaves incubated in opening buffer ± 10 μm ABA for 2 h. Different letters above bars indicate significant (P ≤ 0.01) difference when analyzed by one-way ANOVA with Tukey’s HSD test.
Arabidopsis SAUR Proteins Inhibit Tomato PP2C.D Phosphatases
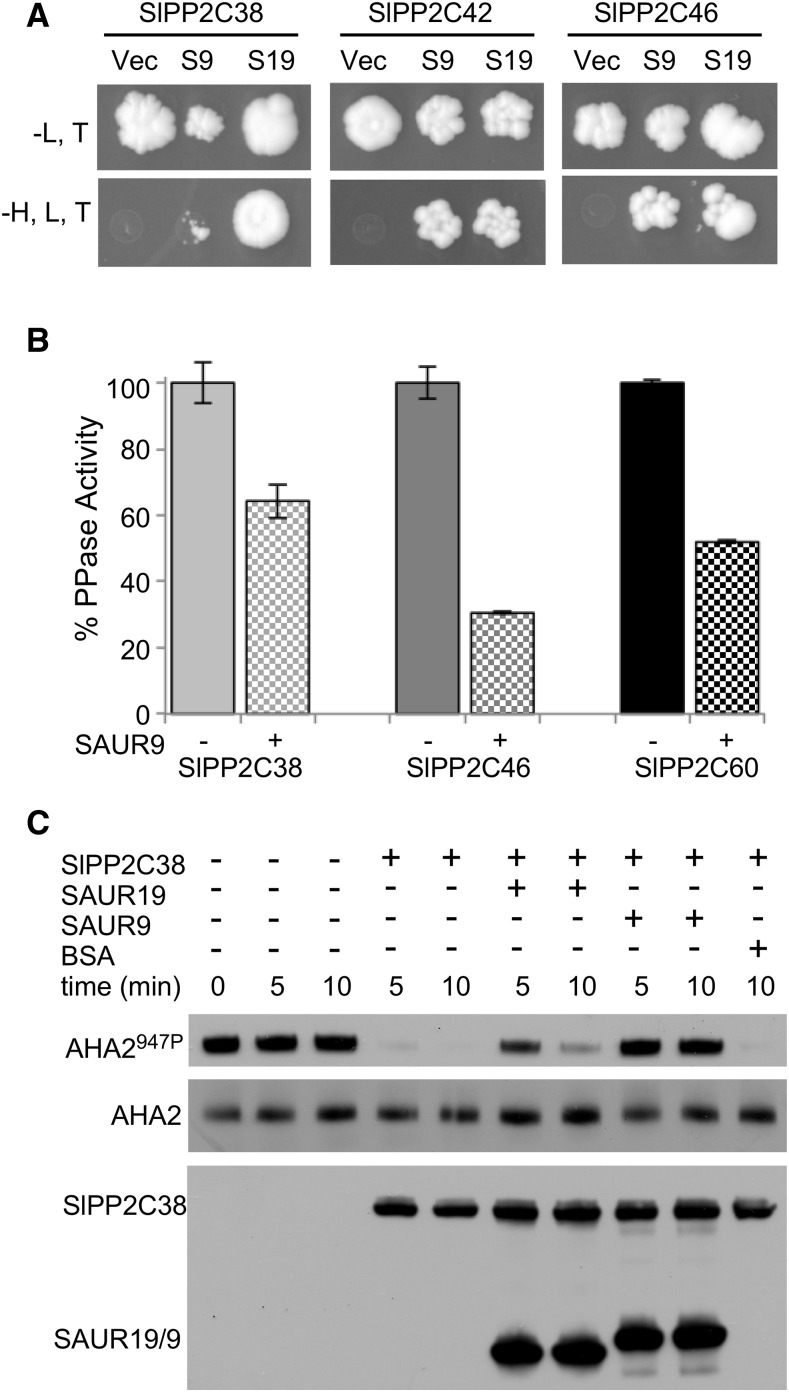
Our prior work in Arabidopsis demonstrated that AtSAUR19, as well as several additional SAURs, specifically interact with D-clade PP2C protein phosphatases and negatively regulate their enzymatic activity (Spartz et al., 2014). A search of the NCBI protein database identified several tomato PP2C proteins bearing high similarity to the Arabidopsis PP2C.D clade. We isolated the full-length cDNAs for tomato SlPP2C38 (XP_004239681.1), SlPP2C42 (XP_004241785.1), SlPP2C46 (XP_004231056.1), and SlPP2C60 (XP_004248644.1) and generated constructs to test for possible interactions with Arabidopsis SAUR proteins. SlPP2C38, 42, and 46 all interacted with AtSAUR19 and AtSAUR9 in yeast two-hybrid assays (Fig. 2A). We were unable to obtain yeast transformants expressing SlPP2C60, preventing us from testing this tomato PP2C.D family member.
Figure 2.
Arabidopsis SAUR proteins interact with tomato PP2C.D phosphatases. A, Yeast cells harboring LexA DNA binding domain-AtSAUR9 or AtSAUR19 and GAL4 activation domain-SlPP2C fusion constructs were spotted onto –leu, trp and –leu, trp, his plates. Vector control (Vec) is the empty LexA DNA binding domain vector. B, 6xHis-SlPP2C.D proteins were purified from Escherichia coli and tested in phosphatase assays employing pNPP. Values indicate the mean relative activities ± sd of three assays. C, In vitro phosphatase assay examining SlPP2C38-mediated dephosphorylation of yeast-expressed AHA2. Where indicated, recombinant AtSAUR19, AtSAUR9, or commercial BSA was added to a 4-fold molar excess relative to SlPP2C38. AHA2 Thr-947 phosphorylation status was assessed by GST-14-3-3 far-western blotting. SlPP2C38 and AtSAUR9/19 abundance was determined by detection of the S-tag.
In vitro phosphatase assays employing recombinant SlPP2C38, 46, and 60 and p-nitrophenylphosphate (pNPP) as a substrate revealed that all three PP2C.D proteins exhibited phosphatase activity. Since our previous work demonstrated that AtSAUR9 strongly inhibited several Arabidopsis PP2C.D enzymes in this assay (Spartz et al., 2014), we used AtSAUR9 in phosphatase inhibition assays with the three tomato PP2C.D proteins. AtSAUR9 significantly inhibited all three phosphatases (Fig. 2B), demonstrating that per our findings in Arabidopsis, SAUR proteins can inhibit the enzymatic activity of tomato PP2C.D phosphatases. In pNPP assays employing AtSAUR19, however, we failed to detect significant inhibition of the three tested SlPP2C.D family members (Supplemental Fig. S3). This may suggest that AtSAUR19 cannot inhibit SlPP2C38, 46, or 60 but does not preclude the possibility that other SlPP2C.D family members may be inhibited, similar to our prior findings in Arabidopsis where AtSAUR19 inhibited AtPP2C.D1, but not D2, D4, or D5 (Spartz et al., 2014). Alternatively, assays employing a chemical substrate may not accurately reflect biological activities on protein substrates. To investigate this possibility, we employed a previously described (Spartz et al., 2014) in vitro phosphatase assay that uses yeast-expressed AHA2 that is phosphorylated on Thr-947. In this assay, SlPP2C38 efficiently catalyzed dephosphorylation of AHA2 (Fig. 2C). Both AtSAUR19 and AtSAUR9 inhibited SlPP2C38-mediated dephosphorylation, whereas an equivalent amount of BSA control protein did not. This finding clearly demonstrates that AtSAUR19 can inhibit a tomato PP2C.D family member. As our genetic studies in Arabidopsis demonstrated that SAURs and PP2C.D proteins act antagonistically to regulate PM H+-ATPase activity and plant growth (Spartz et al., 2014), it seems likely that the phenotypes conferred by GFP-AtSAUR19 overexpression in tomato are also the result of reduced PP2C.D phosphatase activity.
Constitutive GFP-AtSAUR19 Expression Bypasses the Requirement for Auxin in Elongation Growth
Generations of plant physiologists have employed excised coleoptile/hypocotyl elongation assays to investigate auxin-mediated growth. Briefly, coleoptile, hypocotyl, or stem segments from the growing region are decapitated and incubated briefly in hormone-free media to deplete endogenous auxin (Takahashi et al., 2012). Segments are then transferred to media containing auxin, and elongation growth is measured over time. This simple assay and variations thereof provided much of the foundational data leading to the acid growth theory. More recent efforts employing this assay coupled with molecular analyses of Arabidopsis hypocotyls have demonstrated that auxin promotes phosphorylation of the penultimate Thr residue of PM H+-ATPases with kinetics similar to those observed for elongation growth (Takahashi et al., 2012). Given that SAUR gene expression is strongly auxin-inducible and our previous findings that GFP-AtSAUR19 expression leads to increases in elongation and PM H+-ATPase phosphorylation and activity, we investigated the possibility that SAUR gene expression is sufficient to mediate this classic growth response. High-resolution quantification of elongation growth of Arabidopsis and tomato hypocotyl segments was conducted using a computer-controlled bank of CCD cameras to collect images of hypocotyl segments on the surface of agar plates, and a custom image analysis software tool was developed to automate quantification of elongation growth.
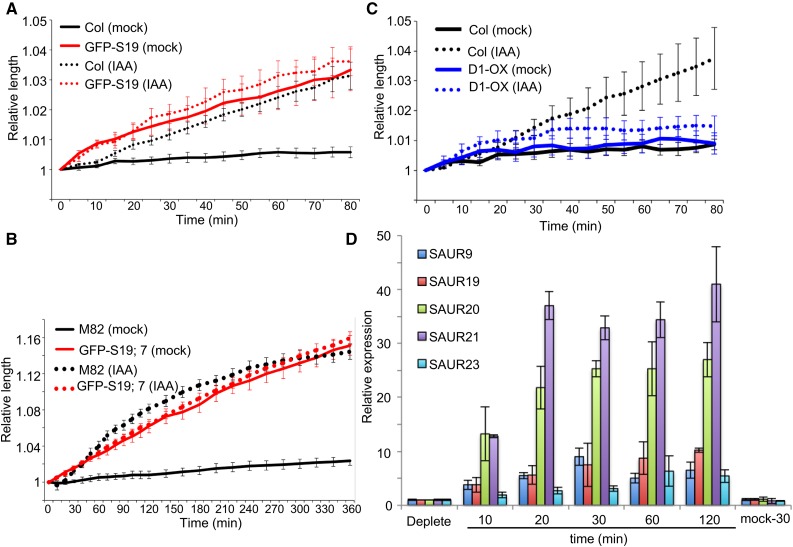
As reported previously (Takahashi et al., 2012), auxin promoted the elongation of auxin-depleted hypocotyl segments following a lag time of ∼10 min in Arabidopsis (Fig. 3A) and 20 min in tomato (Fig. 3B). GFP-AtSAUR19 hypocotyl segments exhibited comparable elongation to wild-type controls on media containing IAA. However, a lag time preceding elongation was not apparent. Furthermore, while the wild-type hypocotyls displayed minimal elongation on hormone-free media, GFP-AtSAUR19 hypocotyls elongated to the same extent as observed in the presence of IAA (Fig. 3, A and B). These findings suggest that constitutive GFP-AtSAUR19 expression bypasses the requirement for auxin in elongation growth. In contrast to the constitutive elongation response conferred by GFP-AtSAUR19 expression, hypocotyls obtained from Arabidopsis seedlings overexpressing AtPP2C.D1 (D1-OX) exhibited minimal elongation on both control and auxin supplemented media (Fig. 3C). However, D1-OX hypocotyl elongation in response to the fungal toxin fusicoccin was comparable to the wild type, indicating that AtPP2C.D1 overexpression does not preclude expansion growth per se, but rather prevents auxin-dependent growth (Supplemental Fig. S4). These findings are consistent with our prior work demonstrating that AtSAUR19 and AtPP2C.D1 act antagonistically to regulate elongation growth and that AtPP2C.D1 overexpression confers a reduction in PM H+-ATPase activity (Spartz et al., 2014).
Figure 3.
Auxin-independent elongation of GFP-AtSAUR19 hypocotyl segments. A, Elongation of 5-mm auxin-depleted Arabidopsis hypocotyl segments on medium ±10 μm IAA. B, Elongation of 8-mm auxin-depleted tomato hypocotyl segments on medium ±10 μm IAA. C, Elongation of auxin-depleted Arabidopsis hypocotyl segments on medium ±10 μm IAA. For Arabidopsis assays, hypocotyl segments were prepared from 3- to 4-d-old etiolated Col and GFP-AtSAUR19 seedlings. For the D1-OX line, however, seedlings were grown for 11 d in order to obtain hypocotyl segments that were long enough to conduct the assay. Tomato hypocotyl segments were excised from 6-d-old etiolated seedlings. All data points represent the mean relative segment length ± se from a minimum of five hypocotyls/genotype/treatment. All assays were repeated at least three times. D, qRT-PCR analysis of auxin-depleted Arabidopsis hypocotyl segments treated with 10 μm IAA. Values indicate the mean relative expression (±sd) from three biological replicates.
To compare the kinetics of the induction of SAUR gene expression with elongation, we conducted qRT-PCR analysis examining the expression of AtSAUR19 and several related SAURs in wild-type Arabidopsis hypocotyl segments prepared exactly as we used for the elongation assays. Expression of AtSAUR19 and AtSAUR9 was ∼4-fold higher following a 10-min auxin treatment, while AtSAUR20 and AtSAUR21 were up-regulated greater than 10-fold. The expression of all four SAUR genes continued to increase at the 20-min time point and remained elevated throughout the 2-h time course (Fig. 3D). Thus, the auxin induction of SAUR expression correlates well with the onset of auxin-mediated elongation growth. Likewise, auxin-induced expression of SAUR gene expression in tomato has also been found to occur with very rapid kinetics (Wu et al., 2012), including in assays employing auxin-depleted hypocotyl segments (Zurek et al., 1994).
The above findings are consistent with the hypothesis that SAURs are the primary auxin effectors that drive elongation growth. A recent study investigating hypocotyl elongation during shade avoidance supports this notion (Procko et al., 2016). This study found that auxin signaling in the hypocotyl epidermis plays a particularly important role in hypocotyl elongation. Remarkably, 23 of the 29 auxin-related genes that were induced by shade and enriched in the epidermal layer were SAUR genes. This includes AtSAUR19, which was specifically expressed in the epidermis. Second, in an independent study employing our 35S:GFP-AtSAUR19 Arabidopsis line, Fendrych et al. (2016) recently reported auxin-independent apoplastic acidification and hypocotyl elongation. This study further demonstrated that activation of PM H+-ATPases is sufficient to trigger elongation growth, providing strong support for our model that auxin-induced SAUR expression results in PM H+-ATPase activation to promote cell expansion.
That said, our findings contradict some prior studies. First, in elongation assays with excised soybean (Glycine max) hypocotyls, auxin was reported to elicit a biphasic growth response, with the early elongation response suggested to occur independent of changes in gene expression (Vanderhoef et al., 1976a, 1976b). As the auxin-mediated elongation response in segment assays is more robust in tomato than Arabidopsis, part of our rationale for generating GFP-AtSAUR19 tomato plants was to explicitly test this hypothesis and elucidate the role of AtSAUR19 in the two stages of auxin-induced elongation. However, we failed to observe biphasic growth in response to auxin. Rather, with wild-type tomato hypocotyls, we observed a 20-min lag phase followed by ∼60 min of maximal elongation and then a slow decline (Fig. 3B). We also note that other studies have failed to obtain evidence of a biphasic elongation response to auxin with Arabidopsis hypocotyl segments (Takahashi et al., 2012; Fendrych et al., 2016). Whether this discrepancy is due to differences in species or assay conditions in unclear. Second, hypocotyl segments prepared from tir1 afb1 afb2 afb3 or tir1 afb2 afb3 seedlings were reported to elongate in response to auxin, albeit with delayed kinetics (Schenck et al., 2010; Fendrych et al., 2016). These seedlings lack three to four members of the six-member family of nuclear TIR1/AFB auxin receptors and exhibit a strong reduction in auxin-inducible gene expression (Dharmasiri et al., 2005), therefore suggesting that auxin-mediated SAUR expression may not be required for hypocotyl elongation. However, recent work suggests that TIR1/AFB receptor activity is indeed required for expansion growth. For example, elongation is clearly impaired in hypocotyl segments prepared from seedlings expressing a dominant axr3-1 mutant allele of the IAA17 auxin coreceptor (Fendrych et al., 2016). Second, by taking advantage of the preferential affinity of the AFB5 receptor for the synthetic auxin, picloram (Walsh et al., 2006; Prigge et al., 2016), Fendrych et al. (2016) demonstrated that afb5 hypocotyls exhibited diminished elongation in response to picloram. Furthermore, cycloheximide treatment of wild-type hypocotyl segments completely abolished auxin-dependent elongation, demonstrating that changes in gene expression are clearly needed for this growth response. Together, these findings suggest that TIR1/AFB-mediated changes in gene expression are required for expansion growth but that redundancy within the TIR1/AFB receptor family prevents the aforementioned triple and quadruple mutants from exhibiting a dramatic elongation defect in response to IAA. The fact that we observe constitutive, auxin-independent elongation in 35S:GFP-AtSAUR19 hypocotyls demonstrates that elevated SAUR expression is sufficient to promote hypocotyl elongation. It therefore seems highly plausible that in wild-type plants, auxin signaling through the TIR1/AFB receptor system results in elevated SAUR expression, leading to PM H+-ATPase activation and expansion growth via an acid growth mechanism.
GFP-AtSAUR19 Overexpression Alters the Mechanical Properties of Tomato Hypocotyls
A fundamental tenant of the acid growth theory is that apoplastic acidification leads to cell wall loosening (Rayle and Cleland, 1992). Auxin treatment has been shown to increase the extensibility of cell walls (Heyn, 1931; Tagawa and Bonner, 1957; Cleland, 1967; Daniel et al., 1989; Nakahori et al., 1991). The underlying basis of this effect is unclear. Consistent with the acid growth hypothesis, apoplastic acidification following PM H+-ATPase activation confers an increase in α-expansin activity (Cosgrove, 2016). However, additional mechanisms have also been proposed, including auxin-induced expression of expansins and other genes encoding wall modification enzymes (Catalá et al., 1997, 2000), increased xyloglucan metabolism (Labavitch and Ray, 1974; Talbott and Ray, 1992), and auxin-mediated cytoskeletal reorganization (Sassi and Traas, 2015).
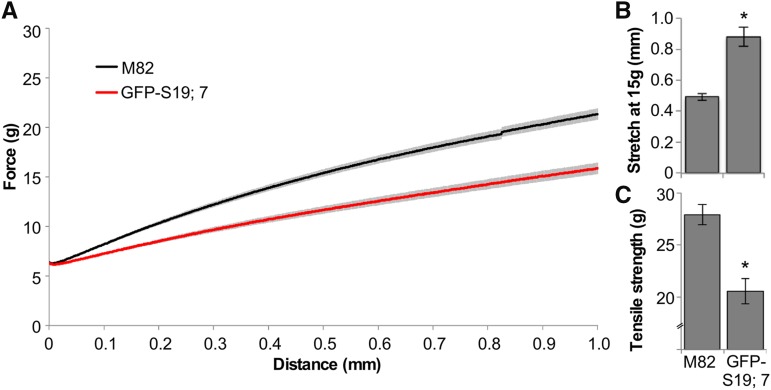
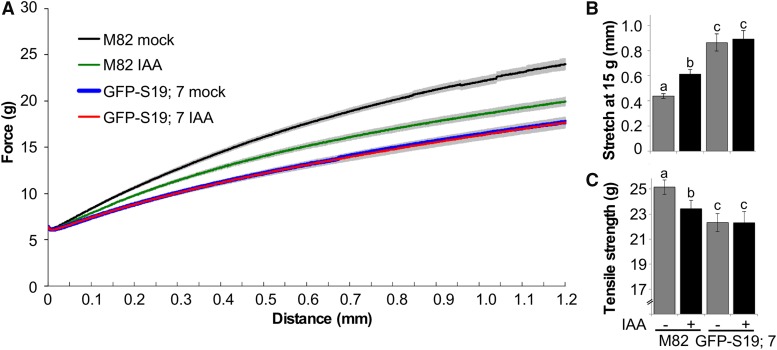
The larger size of tomato hypocotyls compared to Arabidopsis enabled us to examine the possibility that GFP-AtSAUR19 affected the mechanical properties of hypocotyls in a manner that might facilitate elongation growth. Hypocotyls isolated from etiolated wild-type and GFP-AtSAUR19 seedlings were clamped and mechanically extended at a rate of 1 mm min−1 using a TA.XTplus texture analyzer. Striking differences in the extension curves were apparent between genotypes, with GFP-AtSAUR19 hypocotyls displaying an ∼2-fold increase in extensibility as well as a reduction in tensile strength (Fig. 4; Supplemental Fig. S4).
Figure 4.
Altered mechanical properties of GFP-AtSAUR19 tomato hypocotyls. A, Mean (±se) extension curves of M82 and GFP-AtSAUR19 tomato hypocotyl segments. Six-day-old etiolated seedlings were collected and frozen at −80°C. After thawing in ice water, cotyledons and roots were removed and the apical 12 mm of the hypocotyl mechanically extended on a TA.XTplus texture analyzer as described in “Materials and Methods.” B and C, Mean extension at a force of 15g (B) and mean tensile strength (C). Error bars depict se. Asterisks indicate significant (P < 0.01) difference from M82 wild-type hypocotyls as determined using the Wilcoxon rank-sum test.
We next asked whether auxin treatment elicited similar effects on these hypocotyl physical properties. Wild-type hypocotyl segments subjected to a 45-min auxin treatment displayed a significant increase in extensibility as well as a slight reduction in tensile strength (Fig. 5). In contrast, GFP-AtSAUR19 hypocotyl segments were completely unaffected by auxin treatment. Together, these findings suggest that auxin-induced SAUR expression alters the cell wall or possibly other mechanical parameters to facilitate elongation growth. When GFP-AtSAUR19 is constitutively expressed, auxin is not required to promote either extensibility (Fig. 5) or the resulting expansion growth (Fig. 3, A and B).
Figure 5.
Auxin increases extensibility of wild-type but not GFP-AtSAUR19 hypocotyls. A, Mean (±se) extension curves of auxin- or mock-treated tomato hypocotyls. Auxin-depleted hypocotyls were treated with 10 μm IAA or solvent control for 45 min and frozen. Hypocotyl segments were thawed in ice water and extensibility assessed on a TA.XTplus texture analyzer as described in “Materials and Methods.” B and C, Mean extension at a force of 15g (B) and mean tensile strength (C). Error bars depict se. Different letters above bars indicate significant (P < 0.05) differences between samples as determined by one-way ANOVA assessed by Dunnett’s test.
CONCLUSION
Auxin regulates the expression of hundreds of genes, including many SAUR genes. However, it has long been hypothesized that at least part of auxin’s ability to promote cell expansion leading to the elongation growth of shoot organs may occur independent of auxin-mediated changes in gene expression. Consistent with this possibility, changes in ion fluxes across the plasma membrane occur within seconds of auxin application and antibodies against AUXIN BINDING PROTEIN1 (ABP1) abolish these currents (Ruck et al., 1993). However, the recent finding that Arabidopsis abp1 mutants exhibit no apparent defects in auxin signaling or elongation growth (Gao et al., 2015; Fendrych et al., 2016) raises the important question as to whether these rapid ion fluxes are in fact related to auxin-mediated cell expansion. We find that constitutive expression of AtSAUR19 bypasses the normal requirement for auxin in promoting elongation of hypocotyl segments. While we cannot exclude the possibility that very rapid ion flux contributes to auxin-mediated cell expansion, our findings demonstrate that constitutive expression of a normally auxin-induced SAUR gene is sufficient to confer organ elongation. Furthermore, in wild-type hypocotyls, the kinetics of auxin-induced SAUR expression closely parallels the kinetics of both PM H+-ATPase activation (Takahashi et al., 2012) and elongation. Together, these findings suggest that increased SAUR activity may be the primary mechanism underlying auxin-mediated elongation growth. Lastly, we find that GFP-AtSAUR19 overexpression in tomato confers a nearly identical suite of phenotypes as previously reported for Arabidopsis (Spartz et al., 2012, 2014) and that SAURs inhibit both Arabidopsis and tomato PP2C.D family phosphatases. Together with the fact that SAUR and PP2C.D genes are found throughout the plant kingdom, this suggests that the SAUR-PP2C.D regulatory module is a conserved mechanism plants employ to regulate growth and may be an attractive target for genetic modification to enhance crop production.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study were in the Col-0 ecotype and have been previously described (Franklin et al., 2011; Spartz et al., 2012, 2014). Seedlings were grown on ATS media (Lincoln et al., 1990) at 22°C. Stable transgenic tomato (cv M82) plants expressing the 35S:GFP-AtSAUR19 transgene were generated as previously described (Lor et al., 2014). Tomato seedlings were grown on 1× Murashige and Skoog (MS) medium on either agar plates or saturated paper towels under long-day (16:8 h) fluorescent lighting (80 μE m−2 s−1). For LiCl root growth inhibition assays, seedlings were germinated on MS-saturated paper towels until radical emergence and then transferred to fresh towels saturated in MS ±12.5 mm LiCl and grown an additional 4 d before measuring root lengths. Medium acidification experiments with bromocresol purple were conducted as previously described (Spartz et al., 2014). Briefly, 3- to 4-d-old light-grown tomato seedlings were transferred to bromocresol purple media and incubated 12 to 24 h prior to photographing.
Water Loss Assays and Stomatal Aperture Measurement
Leaves were excised from approximately 25-d-old tomato (Solanum lycopersicum) plants and weighed at 15-min intervals. A minimum of 24 leaves/genotype were measured. For aperture measurements, three leaves/genotype from the second leaflet were incubated under light in opening buffer (Azoulay-Shemer et al., 2015) for 2 h. Each leaf was then split down the midvein and the half leaves were transferred to fresh opening buffer containing 10 μm ABA or solvent control and incubated an additional 2 h. Stomatal apertures were examined using the medical adhesive method for intact epidermis imaging on a Leica DM5000B microscope. Thirty to forty stomatal apertures/half leaf/treatment were measured.
Yeast Two-Hybrid Assays
Tomato PP2C38, 42, 46, and 60 donor plasmids were constructed by PCR (Supplemental Table S1) using pDONOR207 (SlPP2C60) or pENTR/D-TOPO (SlPP2C38, SlPP2C42, and SlPP2C46). The lexA-based yeast two-hybrid reporter system using Gateway-ized plasmids, pBTM116 and pACT-GW, has been described previously (Weber et al., 2005). Arabidopsis AtSAUR9 and AtSAUR19 donor plasmids were described previously (Spartz et al., 2014). pBTM116-AtSAUR and pACT-SlPP2C.D plasmids were cotransformed into yeast strain L40ccU3 (Weber et al., 2005), plated onto appropriate selection media, and grown at room temperature for 3 to 6 d.
Excised Hypocotyl Elongation Assays
Apical 5-mm hypocotyl segments were excised from 3- to 4-d-old etiolated Col and GFP-AtSAUR19 Arabidopsis seedlings and incubated in the dark for 30 min on KPS media containing 2.5 mm potassium phosphate (pH 6), 7.5 mm potassium chloride, 2% Suc, and 1% type E agar (Sigma-Aldrich). Due to the reduced hypocotyl elongation conferred by 35S:AtPP2C.D1 expression, seedlings were grown for 9 to 11 d in order to obtain seedlings with sufficiently long hypocotyls. Hypocotyl segments were then placed on fresh KPS plates ±10 μm IAA or 1 μm fusicoccin. Images of multiple hypocotyl segments in each frame were automatically collected at 5-min intervals over a period of 80 min using a Marlin F146B CCD camera (Allied Visions Technologies) equipped with an 18- to 108-mm zoom lens. For tomato assays, the apical 8 mm of 5-d-old etiolated hypocotyls were excised, incubated on KPS plates for 1 h, and then transferred to fresh KPS plates ±10 μm IAA. A glass slide was placed over the tomato hypocotyls to reduce their tendency to lift off of the agar surface. Digital images of the hypocotyl segments were automatically collected at 10-min intervals over a period of several hours.
Image Analysis
Confocal images of GFP-AtSAUR19 expression in tomato leaves were obtained on a Nikon C1si confocal system (Nikon USA) as previously described (Spartz et al., 2012).
Elongation of the excised Arabidopsis and tomato segments was measured from time series of digital images by custom software. For each frame in a time series, a threshold operation based on the Otsu method (Otsu, 1979) was performed to create a binary image in which the hypocotyl segments appeared as slightly curved rectangular objects with a slightly irregular contour. Measuring elongation of the hypocotyl segments required finding the midline of each segment object in each frame of the time series (Miller et al., 2007). The binary segment objects were skeletonized. Dijkstra’s algorithm (Dijkstra, 1959) was used to order the skeleton points. The midline of a rectangle determined by this method will inevitably branch at the ends. The branches were removed by a pruning step. The skeleton was connected to the two end points of the hypocotyl segment, which are defined as the two contour points having the highest curvature. The midline length at each time point was divided by the initial length to produce the results shown in Figure 3. The software package is available for download at https://bitbucket.org/uwphytomorph/phytomorph.
Protein Work
Soluble and microsomal fractions were prepared from 8-d-old tomato seedlings as previously described (Spartz et al., 2012). Microsomal fractions were subjected to two-phase partitioning (Ito and Gray, 2006), and western blots utilizing a GFP monoclonal antibody (Covance) were conducted using 5 μg of each fraction.
The 6xHIS-TRX-SlPP2C fusion proteins were made by Gateway recombination (Life Technologies) between pET32-GW (Life Technologies) and the above SlPP2C donor clones. The 6xHIS-TRX-tagged AtSAUR9 and AtSAUR19 were described previously (Spartz et al., 2014). Escherichia coli cultures expressing 6xHIS-TRX-tagged AtSAUR or 6xHIS-TRX-tagged SlPP2C proteins were lysed by a French press in lysis buffer (50 mm Tris, pH 7.6, 100 mm NaCl, 2 mm MnCl2, 10 mm MgCl2, 1 mm EDTA, and 0.25% Triton X-100) and purified over Ni-NTA agarose beads. Imidazole (200 mm) was used for elution. For phosphatase assays, SlPP2C at 0.15 μm final concentration was preincubated with 1.5 μm 6xHIS-TRX-AtSAUR protein or 6xHIS-TRX control protein for 15-min. Protein mixtures were then added to assay buffer containing 75 mm Tris pH 7.6, 10 mm MnCl2, 100 mm NaCl, 0.5 mm EDTA, and 5 mm pNPP to obtain a final volume of 100 μL. Absorbance at 405 nm was recorded every minute for 20 min on a Powerwave 340 plate reader (Biotek Instruments). Average velocities (ΔΑ402/min) were calculated within the linear range of the reaction (5–15 min) and shown as percentages of PP2C.D activity compared to control samples (6xHIS-SlPP2C + 6xHIS-TRX).
AHA2 dephosphorylation assays were conducted as described by Spartz et al. (2014) with minor modifications. Seventy-five nanograms of purified 6xHis-TRX-SlPP2C38 was preincubated for 20 min with a 4-fold molar excess of 6xHis-TRX-AtSAUR9, 6xHis-TRX-AtSAUR19, or BSA and then mixed with 5 μg of plasma membranes prepared from yeast cells expressing AHA2. Reactions were incubated at 25°C for 5 to 10 min and then stopped with the addition of SDS-PAGE sample buffer. AHA2 Thr-947 phosphorylation status was assessed by GST-14-3-3 far western blotting. SlPP2C38 and AtSAUR9/19 abundance was confirmed by probing immunoblots with an S-protein-HRP conjugate (Novagen).
qRT-PCR
Primers used for assessing gene expression are listed in Supplemental Table S1. RNA was prepared with the RNeasy Plant Mini Kit (Qiagen) from 5-d-old etiolated hypocotyl segments depleted and then treated with 10 μm auxin (or solvent control) as described above. An on-column DNase treatment was included. Two micrograms of RNA was used for cDNA synthesis with M-MLV reverse transcriptase. qRT-PCR reactions were performed on the LightCycler system (Roche Applied Sciences) using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). Expression levels were normalized to ACT7, and three biological replicates were performed.
Extension Assays
GFP-AtSAUR19 or M82 parental tomato seeds were placed on water-saturated paper towels and grown for 6 d in the dark. Seedlings with hypocotyls of 30 to 35 mm were collected and stored at −80°C. For auxin treatments, etiolated seedlings were obtained as above. Cotyledons and roots were excised, leaving 2 to 3 mm of root in order to facilitate identification of the apical and basal ends of the hypocotyl. The hypocotyls were incubated in KPS buffer in the dark for 1 h to deplete endogenous auxin and then treated with 10 μm IAA (or solvent control) for 45 min in the dark, collected, and frozen at −80°C. Hypocotyl extensibility and tensile strength were measured using a TA.XTplus texture analyzer (Stable Micro Systems) equipped with a 5-kg load cell. Frozen seedlings/hypocotyls were thawed in ice water. Rubber coated tensile grips were clamped onto each end of apical 12-mm hypocotyl segments, which were then stretched at a rate of 1 mm min−1 until breaking. Force was recorded every 5 ms once a 6 g threshold force was reached. A minimum of 15 hypocotyls/genotype/treatment were assayed and at least three biological replicates conducted.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SlPP2C38, XP_004239681; SlPP2C42, XP_004241785; SlPP2C46, XP_004231056; SlPP2C60, XM_004234474; AtSAUR9, At4g36110; and AtSAUR19, At5g18010.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. 35S:GFP-AtSAUR19 expression confers a long hypocotyl phenotype.
Supplemental Figure S2. 35S:GFP-AtSAUR19 expression confers drought hypersensitivity.
Supplemental Figure S3. SlPP2C38, 46, or 60 pNPP phosphatase assays with AtSAUR19.
Supplemental Figure S4. Fusicoccin promotes elongation of 35S:AtPP2C.D1 hypocotyl segments.
Supplemental Figure S5. Raw TA.XTplus data.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Ted Labuza (University of Minnesota) for providing the TA.XTplus texture analyzer, Dr. Cindy Tong (University of Minnesota) for providing tensile grips, and the College of Biological Sciences Imaging Center for assistance with confocal microscopy. W.M.G. thanks Dr. Michael Sussman (University of Wisconsin-Madison) for hosting him during a 2015 sabbatical when this work was initiated.
Glossary
- pNPP
p-nitrophenylphosphate
- MS
Murashige and Skoog
- ABA
abscisic acid
Footnotes
Articles can be viewed without a subscription.
References
- Azoulay-Shemer T, Palomares A, Bagheri A, Israelsson-Nordstrom M, Engineer CB, Bargmann BO, Stephan AB, Schroeder JI (2015) Guard cell photosynthesis is critical for stomatal turgor production, yet does not directly mediate CO2- and ABA-induced stomatal closing. Plant J 83: 567–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá C, Rose JK, Bennett AB (1997) Auxin regulation and spatial localization of an endo-1,4-beta-D-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J 12: 417–426 [DOI] [PubMed] [Google Scholar]
- Catalá C, Rose JK, Bennett AB (2000) Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiol 122: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Cleland R. (1967) Extensibility of isolated cell walls: Measurement and changes during cell elongation. Planta 74: 197–209 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2016) Catalysts of plant cell wall loosening. F1000Res 5: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel SG, Rayle DL, Cleland RE (1989) Auxin physiology of the tomato mutant diageotropica. Plant Physiol 91: 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dijkstra EW. (1959) A note on two problems in connexion with graphs. Numer Math 1: 269–271 [Google Scholar]
- Enders TA, Strader LC (2015) Auxin activity: Past, present, and future. Am J Bot 102: 180–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J (2016) TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, Wigge PA, Gray WM (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG (1999) Binding of 14-3-3 protein to the plasma membrane H(+)-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J Biol Chem 274: 36774–36780 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA 112: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. (2003) Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res 116: 483–505 [DOI] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem 285: 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Gray WM, Sussman MR (2015) Regulation of the plasma membrane proton pump (H(+)-ATPase) by phosphorylation. Curr Opin Plant Biol 28: 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sussman MR (2012) The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol 158: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn ANJ. (1931) Der mechanismus der Zellstreckung. Recl Trav Bot Neerl 28: 113–244 [Google Scholar]
- Ito H, Gray WM (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol 142: 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelich-Ottmann C, Weiler EW, Oecking C (2001) Binding of regulatory 14-3-3 proteins to the C terminus of the plant plasma membrane H+-ATPpase involves part of its autoinhibitory region. J Biol Chem 276: 39852–39857 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki Ki (1999) Blue light activates the plasma membrane H(+)-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L (2003) The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278: 23936–23943 [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhu Y, Gao C, She W, Lin W, Chen Y, Han N, Bian H, Zhu M, Wang J (2013) Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol 54: 609–621 [DOI] [PubMed] [Google Scholar]
- Labavitch JM, Ray PM (1974) Relationship between promotion of xyloglucan metabolism and induction of elongation by indoleacetic acid. Plant Physiol 54: 499–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZG, Chen HW, Li QT, Tao JJ, Bian XH, Ma B, Zhang WK, Chen SY, Zhang JS (2015) Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci Rep 5: 12477. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lor VS, Starker CG, Voytas DF, Weiss D, Olszewski NE (2014) Targeted mutagenesis of the tomato PROCERA gene using transcription activator-like effector nucleases. Plant Physiol 166: 1288–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, Giraudat J, Leung J (2007) Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ND, Parks BM, Spalding EP (2007) Computer-vision analysis of seedling responses to light and gravity. Plant J 52: 374–381 [DOI] [PubMed] [Google Scholar]
- Nakahori K, Kate K, Okamoto H (1991) Auxin changes both the extensibility and the yield threshold of the cell wall of Vigna hypocotyls. Plant Cell Physiol 32: 121–129 [Google Scholar]
- Otsu N. (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9: 62–66 [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Greenham K, Zhang Y, Santner A, Castillejo C, Mutka AM, O’Malley RC, Ecker JR, Kunkel BN, Estelle M (2016) The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 6: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland R (1970) Enhancement of wall loosening and elongation by acid solutions. Plant Physiol 46: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE (1980) Evidence that auxin-induced growth of soybean hypocotyls involves proton excretion. Plant Physiol 66: 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE (1992) The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol 99: 1271–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck A, Palme K, Venis MA, Napier RM, Felle HH (1993) Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J 4: 41–46 [Google Scholar]
- Sassi M, Traas J (2015) When biochemistry meets mechanics: a systems view of growth control in plants. Curr Opin Plant Biol 28: 137–143 [DOI] [PubMed] [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H (2010) Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol 152: 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P, Kumar PP (2013) Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep 32: 759–769 [DOI] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa T, Bonner J (1957) Mechanical properties of the Avena coleoptile as related to auxin and to ionic interactions. Plant Physiol 32: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T (2012) Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbott LD, Ray PM (1992) Changes in molecular size of previously deposited and newly synthesized pea cell wall matrix polysaccharides: effects of auxin and turgor. Plant Physiol 98: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhoef LN, Stahl CA, Lu TY (1976a) Two elongation responses to auxin respond differently to protein synthesis inhibition. Plant Physiol 58: 402–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhoef LN, Stahl CA, Williams CA, Brinkmann KA (1976b) Additional evidence for separable responses to auxin in soybean hypocotyl. Plant Physiol 57: 817–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vi SL, Trost G, Lange P, Czesnick H, Rao N, Lieber D, Laux T, Gray WM, Manley JL, Groth D, Kappel C, Lenhard M (2013) Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc Natl Acad Sci USA 110: 13994–13999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Bernhardt A, Dieterle M, Hano P, Mutlu A, Estelle M, Genschik P, Hellmann H (2005) Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol 137: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went FW, Thimann KV (1937) Phytohormones. Macmillan, New York [Google Scholar]
- Wu J, Liu S, He Y, Guan X, Zhu X, Cheng L, Wang J, Lu G (2012) Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 509: 38–50 [DOI] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD (1994) Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol 104: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.