WSL4 physically interacts with CER2-like protein, catalyzing the elongation of very-long-chain fatty acids longer than 24 carbons, and is involved in the synthesis of cuticular wax in rice leaf.
Abstract
Cuticular waxes are complex mixtures of very-long-chain fatty acids (VLCFAs) and their derivatives, forming a natural barrier on aerial surfaces of terrestrial plants against biotic and abiotic stresses. In VLCFA biosynthesis, β-ketoacyl-CoA synthase (KCS) is the key enzyme, catalyzing the first reaction in fatty acid elongation and determining substrate specificity. We isolated a rice (Oryza sativa) wax crystal-sparse leaf 4 (WSL4) gene using a map-based cloning strategy. WSL4 is predicted to encode a KCS, a homolog of Arabidopsis (Arabidopsis thaliana) CER6. Complementation of the mutant wsl4-1 with WSL4 genomic DNA rescued the cuticular wax-deficient phenotype, confirming the function of WSL4. The load of wax components longer than 30 carbons (C30) and C28 were reduced markedly in wsl4-1 and wsl4-2 mutants, respectively. Overexpression of WSL4 increased the cuticular wax load in rice leaves. We further isolated a cofactor of WSL4, OsCER2, a homolog of Arabidopsis CER2, by coimmunoprecipitation and confirmed their physical interaction by split-ubiquitin yeast two-hybrid experiments. Expression of WSL4 alone in elo3 yeast cells resulted in increased C24 but did not produce VLCFAs of greater length, whereas expressing OsCER2 alone showed no effect. Coexpression of WSL4 and OsCER2 in elo3 yeast cells yielded fatty acids up to C30. OsCER2 with a mutated HxxxD motif (H172E, D176A, and D176H) interrupted its interaction with WSL4 and failed to elongate VLCFAs past C24 when expressed with WSL4 in elo3 yeast cells. These results demonstrated that WSL4 was involved in VLCFA elongation beyond C22 and that elongation beyond C24 required the participation of OsCER2.
The aerial organ surface of terrestrial plants is covered by a hydrophobic cuticle layer preventing nonstomatal water loss (Reicosky and Hanover, 1978), lessening UV irradiation damage (Barnes et al., 1996), and acting as a barrier against bacterial and fungal pathogen invasion (Jenks et al., 1994; Eigenbrode and Espelie, 1995). The cuticle layer contains two major components, cutin polymer matrix and cuticular wax (intracuticular and epicuticular wax). Cutin is a cross-linked polymer, consisting primarily of hydroxyl and hydroxyl-epoxy fatty acid polyesters. Cuticular waxes are complex organic mixtures of very-long-chain fatty acids (VLCFAs), predominantly of chain lengths of 26 to 34 carbons, and their derivatives, including aldehydes, alcohols, alkanes, ketones, and wax esters.
Wax biosynthesis begins with de novo-synthesized C16 and C18 fatty acids within the leucoplasts. C16 and C18 fatty acids are then elongated to VLCFAs by the fatty acid elongase (FAE) complex, consisting of β-ketoacyl-CoA synthase (KCS), β-ketoacyl-CoA reductase, β-hydroxy acyl-CoA dehydratase, and enoyl-CoA reductase, on the endoplasmic reticulum (ER). VLCFA elongation involves a four-step reaction cycle: First, the condensation of C16 and C18 acyl-CoA with malonyl-CoA is catalyzed by KCS, yielding β-ketoacyl-CoA; second, the reduction of β-ketoacyl-CoA is catalyzed by β-ketoacyl-CoA reductase; third, the resulting β-hydroxy acyl-CoA is dehydrated by β-hydroxy acyl-CoA dehydratase; and fourth, the enoyl acyl-CoA is reduced by enoyl-CoA reductase. Each cycle results in an acyl-CoA with a two-carbon extension (Kunst and Samuels, 2009). The generated VLCFA-CoAs are then thiolysed to yield free fatty acids or are used further in an acyl reduction (alcohol-forming) pathway, yielding primary alcohols, or a decarbonylation (alkane-forming) pathway, yielding aldehydes and alkanes (Kunst and Samuels, 2003; Samuels et al., 2008).
During the fatty acid elongation cycle, condensation of VLCFAs, catalyzed by KCS, is the first committed step in each elongation process. The Arabidopsis genome contains at least 21 putative KCS genes, and many of them have been characterized (Joubès et al., 2008). Although some Arabidopsis KCSs are functionally redundant, each KCS possesses a unique substrate specificity for acyl-CoAs of different carbon chain lengths, according to their roles in the biosynthesis of lipids (Samuels et al., 2008; Chen et al., 2011; Haslam and Kunst, 2013). FAE1/KCS18 is expressed exclusively in seeds, catalyzing the biosynthesis of C20 and C22 VLCFAs for storage lipids (James et al., 1995). KCS2/DAISY and KCS20 are functionally redundant in the elongation of VLCFAs up to C22, which are required for cuticular wax and root suberin biosynthesis (Lee et al., 2009; Franke et al., 2009). KCS5/CER60 and KCS6/CER6/CUT1 are involved in the elongation of fatty acyl-CoAs longer than C24 VLCFAs for production of cuticular waxes in epidermis and pollen coat lipids (Millar et al., 1999; Fiebig et al., 2000; Hooker et al., 2002). Moreover, the morphological structure and stress responses in plants are affected by KCSs such as KCS10/FDH (Yephremov et al., 1999; Pruitt et al., 2000; Joubès et al., 2008) and KCS13/HIC (Gray et al., 2000).
As recently reported regarding the elongation of VLCFAs, KCS is not the only determining factor; cofactors are also involved in the process. CER2-LIKE proteins, acting as cofactors with KCS6/CER6 and KCS5/CER60, are involved in the elongation of C28 fatty acids, with CER2 and CER2-LIKE2 contributing to VLCFA elongation to C30 and CER2-LIKE1/CER26 facilitating the elongation of VLCFAs up to C34 (Haslam et al., 2012, 2015; Pascal et al., 2013). However, the molecular mechanism underlying the CER2-LIKE modification in chain lengths remains largely unknown.
Like eceriferum mutants having glossy stems in Arabidopsis (Arabidopsis thaliana; McNevin et al., 1993), mutations affecting wax biosynthesis in rice can lead to a wax crystal-sparse leaf (WSL) phenotype, with more hydrophilic leaves. Thus, such mutants are ideal for investigating the cuticular wax biosynthetic mechanism. WSL1 was predicted to encode a KCS catalyzing the biosynthesis of C20 to C24 VLCFAs. The wsl1 mutant showed substantially reduced total cuticular wax load on the leaf blades and C20-C24 VLCFA precursors (Yu et al., 2008). Rice (Oryza sativa) cuticular wax contains predominantly C28-C32 VLCFAs and their derivatives; thus, many KCS genes are presumed to be associated with wax biosynthesis in rice. Thirty-four putative KCS genes have been annotated in the rice genome (http://rice.plantbiology.msu.edu/); however, only OsWSL1 (Yu et al., 2008) has been characterized.
Here, we identified two wsl mutants and report the isolation and functional characterization of WSL4 in rice wax biosynthesis. WSL4 encodes a KCS that is largely similar to CER6 and HvKCS6 (Millar et al., 1999; Weidenbach et al., 2014). The wsl4-1 and wsl4-2 mutants showed marked reductions in contents of wax components longer than C28 and C26, respectively, whereas overexpression of WSL4 increased the quantity of C30-C34 fatty acids in rice leaves. We also isolated a cofactor of WSL4/OsKCS6, OsCER2 (a homolog of Arabidopsis CER2), and confirmed their direct interaction in VLCFA elongation.
RESULTS
Identification of Wax Crystal-Sparse Leaf Mutants
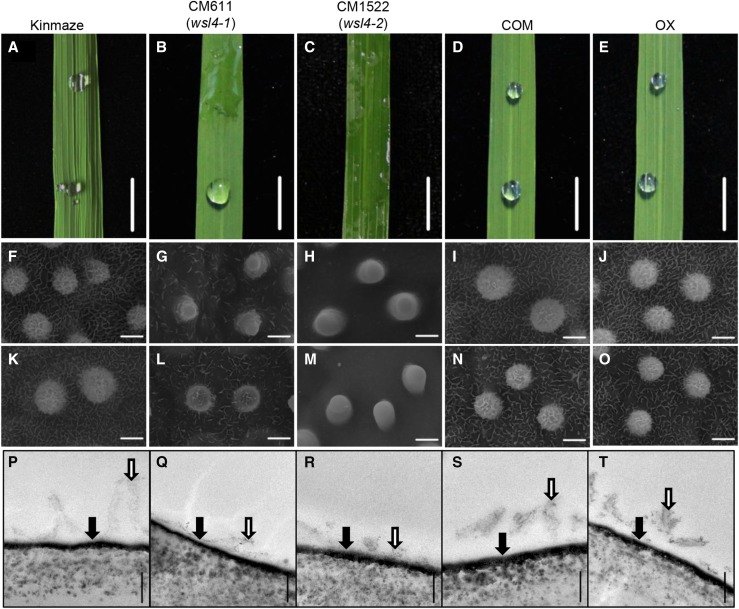
Two mutant lines (CM611 and CM1522) were searched in Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/). The mutants were derived from fertilized egg-cell treatment with 1-methyl-1-nitrosourea of rice (O. sativa cv Kinmaze). Unlike the beads of water that formed on the leaf surface of the wild type, water molecules spread out on the leaves of CM611 and CM1522, showing a WSL phenotype (Fig. 1). Notably, leaves of CM611 displayed a sectored-wetting appearance, whereas leaves of CM1522 were fully wet (Fig. 1). The phenotypes of CM611 and CM1522 were similar to those of wsl mutants with altered leaf wax coatings (Yu et al., 2008; Mao et al., 2012; Gan et al., 2016). Scanning electron microscopy revealed that the platelet-like wax crystals were distributed compactly on the surface of wild-type leaves, whereas only a few wax crystals were observed on the surface of CM611 and CM1522 leaves (Fig. 1). Ultrastructural analysis of the leaf cuticle by transmission electron microcopy showed that the cuticle in the wild-type leaf appeared to be divided into an inner opaque layer (cuticular layer) and an outer translucent layer (cuticular wax) with short clubs (Fig. 1). The short clubs reflected the crystalloid platelets, arranged vertically on the surface of the cuticle membrane visualized by scanning electron microscopy (Fig. 1). In CM611 and CM1522, the thickness of the opaque layer did not differ from that of Kinmaze, but the transparent layer appeared smoother, with fewer short clubs (Fig. 1). These results suggest that the two mutants lacked particular components of the epicuticular wax on the leaves.
Figure 1.
Characterization of the wax-crystal sparse leaf mutants. A to E, Water retention behavior on leaves of rice. Cuticular wax crystals formed on adaxial surface (F–J) and abaxial surface (K–O) of the leaf blade by scanning electron microscopy and transmission electron microscopy (P–T). COM, Complementation line, OX, overexpression line. Bars = 1 cm (A–E), 2 μm (F–O), and 200 nm (P–T). The cuticular layer is indicated by the solid arrow; the cuticular wax is indicated by the open arrow.
Analysis of Cuticular Wax Composition in wsl Mutants
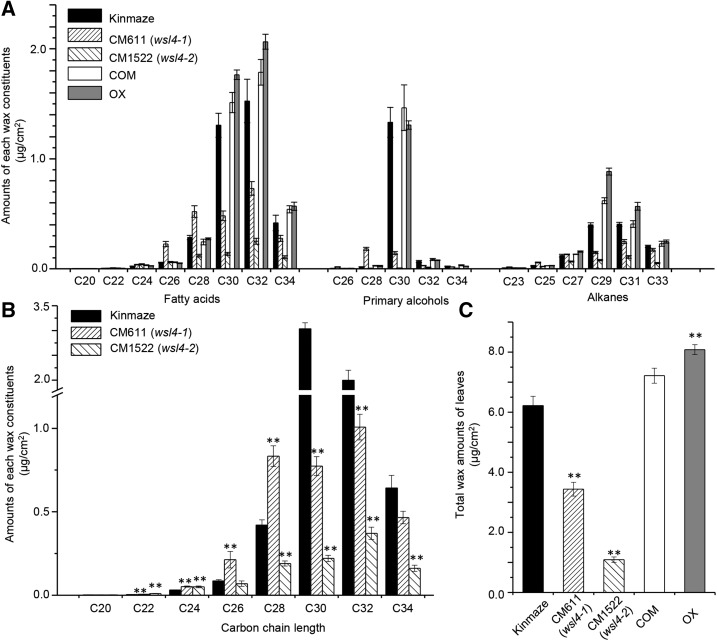
Depletion of wax crystals in the leaves of CM611 and CM1522 suggested that the content and constitution of the cuticular wax might have been changed. Wax was extracted from leaves of CM611, CM1522, and Kinmaze and subjected to gas chromatography-mass spectrometry (GC-MS). Compared with Kinmaze, CM611 and CM1522 leaf blades showed total wax loads that were reduced by 44.9 and 82.5%, respectively (Table I; Figure 2C). In CM611, the contents of VLCFAs, primary alcohols, and alkanes were reduced, by 37.0, 73.1, and 33.4%, respectively (Table I). Further analysis revealed markedly reduced levels of C30 and C32 VLCFAs, C30 and C32 primary alcohols, and C29 and C31 alkanes, with increased levels of C22-C28 VLCFAs, C26 and C28 primary alcohols, and C23 and C25 alkanes (Fig. 2A). The contents of the most abundant components (C30 and C32 monomers) were decreased significantly, whereas those of components shorter than C30 (C22-C28) were increased in CM611 (Fig. 2B), indicating that CM611 was interrupted in VLCFA carbon-chain elongation from C28 to C30 during epicuticular wax synthesis.
Table I. Cuticular wax composition and loads in rice leaf blades.
Data are mean ± se from five biological replicates. COM, Complementation line; OX, WSL4 overexpression line.
| Fatty Acids | Alcohols | Alkane | Total Wax | |
|---|---|---|---|---|
|
μg/cm2 |
μg/cm2 |
μg/cm2 |
μg/cm2 |
|
| Kinmaze | 3.617 ± 0.392 | 1.440 ± 0.151 | 1.160 ± 0.055 | 6.219 ± 0.311 |
| CM611 (wsl4-1) | 2.279 ± 0.212 | 0.387 ± 0.015 | 0.772 ± 0.037 | 3.430 ± 0.228 |
| CM1522 (wsl4-2) | 0.725 ± 0.075 | 0.031 ± 0.006 | 0.336 ± 0.030 | 1.090 ± 0.088 |
| COM | 4.184 ± 0.219 | 1.611 ± 0.223 | 1.419 ± 0.074 | 7.214 ± 0.251 |
| OX | 4.754 ± 0.125 | 1.436 ± 0.038 | 1.892 ± 0.073 | 8.082 ± 0.164 |
Figure 2.
Compositional analysis of cuticular waxes in rice leaves. A, Content of each wax component in rice leaves. B, Amount of all components with equal carbon chain lengths. C, Total amount of wax in leaf blades. Data are means ± se of five biological replicates. **P < 0.01. COM and OX, as in Figure 1.
Compared with Kinmaze, CM1522 leaves showed reduced contents of VLCFAs, primary alcohols, and alkanes, by 80.0, 97.8, and 71.0%, respectively (Table I; Figure 2C). The contents of C28-C34 VLCFAs, C28-C34 primary alcohols, and C27-C33 alkanes were reduced greatly, whereas those of C22 and C24 VLCFAs increased greatly in CM1522 leaf blades (Fig. 2A). The contents of VLCFA derivatives longer than C26 (C28-C34) were decreased, by ∼55, 92, 81, and 76%, respectively. In contrast, levels of C22 and C24 VLCFA derivatives (C22 and C24) were 30 and 55% higher, respectively, in CM1522 than in Kinmaze leaf blades (Fig. 2B), suggesting that carbon-chain elongation beyond C26 was blocked in CM1522.
CM611 and CM1522 Are Allelic Mutants
For genetic analysis, CM611 was crossed with Kinmaze. All F1 progeny showed the wild-type phenotype, whereas in an F2 population of 98 plants, 22 exhibited the WSL phenotype, satisfying a 3:1 ratio (P > 0.05). Thus, the WSL phenotype in CM611 was controlled by a single recessive gene.
To determine the genetic relationship between CM611 and CM1522, we crossed the lines reciprocally. All reciprocal F1 individuals displayed a sectored-wetting phenotype, identified with CM611 (Supplemental Fig. S1). Sectored-wetting phenotype (like CM611) and fully wetting phenotype (like CM1522) segregation was observed in the F2 generation, but no wild-type phenotype was observed. Thus, CM611 and CM1522 were allelic mutants. Given that CM611 and CM1522 showed a wax crystal-sparse phenotype, like the wsl1, wsl2, and wsl3 mutants characterized previously (Yu et al., 2008; Mao et al., 2012; Gan et al., 2016), the candidate gene was designated WSL4, with CM611 and CM1522 renamed as wsl4-1 and wsl4-2, respectively.
Map-Based Cloning of WSL4
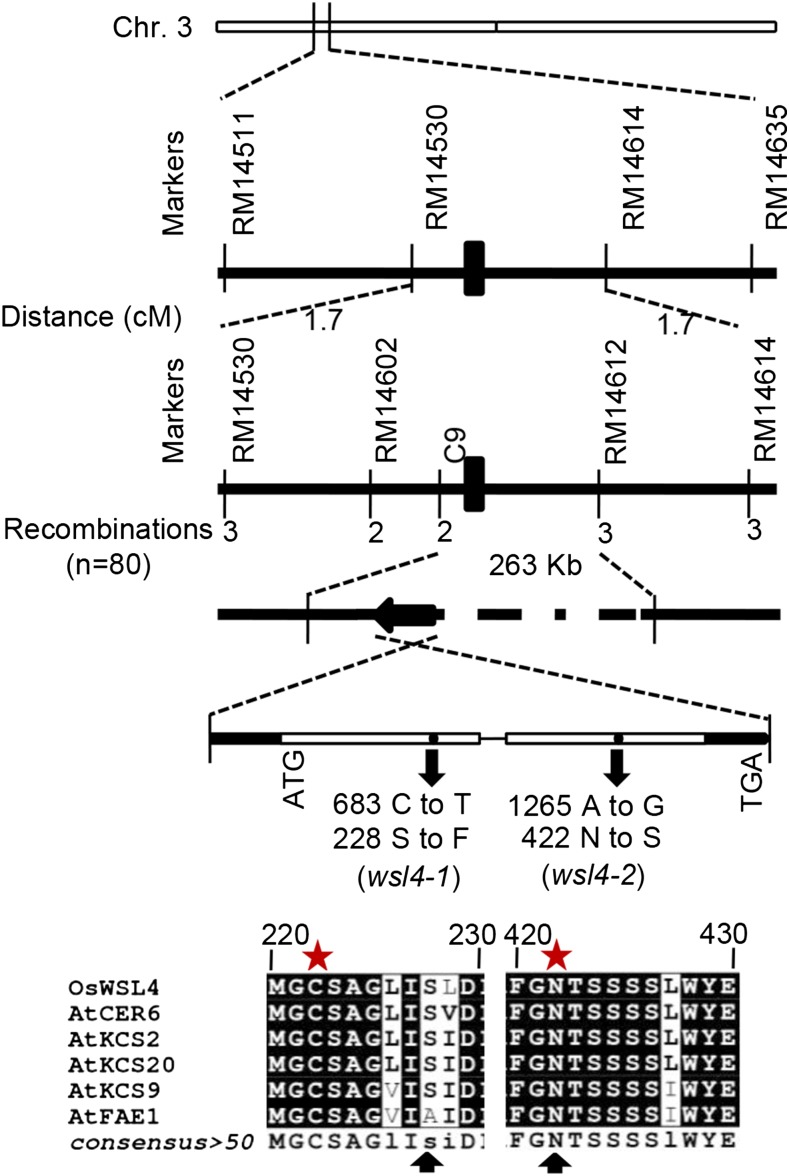
To clone WSL4, we generated a mapping population by crossing wsl4-1 with 9311 (indica). WSL4 was mapped initially to chromosome 3, between simple sequence repeat markers RM14511 and RM14635. With 80 homozygous F3 recessive individuals, the WSL4 gene was further fine-mapped between markers RM14530 and RM14614 with a genetic distance of 1.7 centimorgans. Between the two markers, we developed an additional InDel marker (C9) and mapped the gene between C9 and RM14612 (Fig. 3; Supplemental Table S1). According to the Rice Genome Annotation Project Database (http://rapdb.dna.affrc.go.jp/), in total, 40 open reading frames were predicted within the 263-kb candidate chromosomal region (Fig. 3). Two candidate genes that might be involved in wax biosynthesis were amplified and sequenced. After sequencing, a single nucleotide mutation, from C to T at position 683 downstream of the translation start site, was found in the open reading frame of Os03g0220100 in wsl4-1. This mutation (683 C to T) resulted in an amino acid substitution (Ser228Phe). The sequencing of the Os03g0220100 cDNA in wsl4-2 showed a single-base mutation (1265 A to G), resulting in a single amino acid substitution (Asn422Ser; Fig. 3).
Figure 3.
Mapping-based cloning and structure of WSL4. The schematic diagram depicts the exon (empty box), intron (black line), and untranslated region (solid black box) of WSL4. Amino acid sequence depicting the changes due to point mutations in the two mutants (wsl4-1 and wsl4-2; black arrows). Asterisks indicate the conserved active-site residues in Arabidopsis KCSs.
To confirm that the wax crystal-sparse leaf phenotype of wsl4-1 was caused by the single-nucleotide mutation of WSL4, we next performed a complementation test. A plasmid of a 5,219-bp WSL4 genomic DNA fragment containing the native promoter and terminator was introduced into wsl4-1. We obtained more than 40 independent transgenic plants. All plants showed the wild-type phenotype, with the cuticular wax crystals on leaf blades resembling those of the wild type (Fig. 1). The significant reductions in C30, C32, and C34 waxes in wsl4-1 leaves were rescued and elevated C22, C24, C26, and C28 wax monomer levels recovered, to wild-type levels, in the complementation line (Fig. 2A). These results confirmed that the single-nucleotide mutation in WSL4 was responsible for the wax crystal-sparse phenotype.
The wsl4-1 and wsl4-2 Mutations Occurred in a Highly Conserved Region among KCSs
The coding sequence of WSL4 contains 1,482 nucleotides, encoding a polypeptide of 494 amino acids. BLAST analysis revealed that WSL4 shared 79 and 90% amino acid sequence identities with CER6/CUT1/KCS6 and HvKCS6, respectively (Fiebig et al., 2000; Weidenbach et al., 2014). The amino acid sequence of WSL4 was compared with that of reported KCSs in Arabidopsis. The Asn422Ser replacement in WSL4 in the wsl4-2 is found in a region highly conserved among VLCFA-condensing enzymes, whereas the Ser228Phe altered in WSL4 in the wsl4-1 was moderately conserved (Fig. 3). The conserved Cys-222, His-389, and Asn-422 residues have been reported to be putative catalytic residues in FAE1/KCS18; substitution of these residues reduced the activity of the enzyme greatly (Ghanevati and Jaworski, 2002). The Ser228Phe mutation was close to Cys-222 residue, whereas the Asn422Ser mutation was exactly the Asn-422 residue of WSL4 in wsl4-1 and wsl4-2, respectively (Fig. 3; Supplemental Fig. S2). Thus, the single-nucleotide polymorphisms of wsl4-1 and wsl4-2 might suppress the activity of the condensing enzyme to different levels and then generate divergence in the WSL phenotypes in rice leaves.
Spatial Expression and Subcellular Localization of WSL4
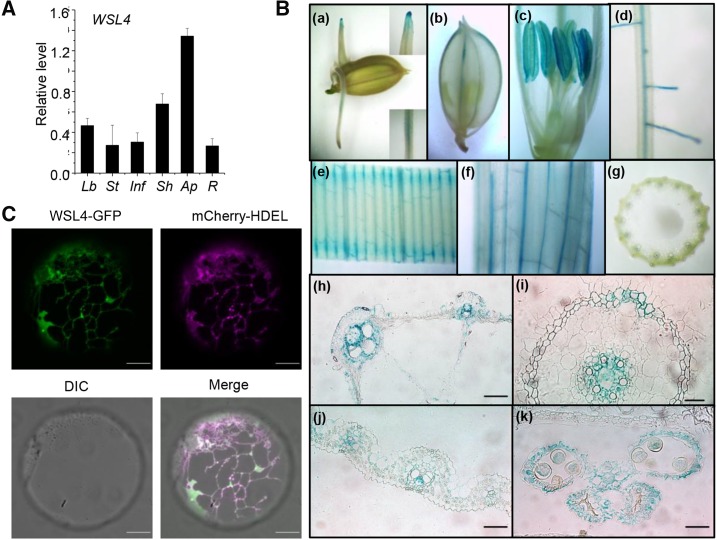
We investigated the expression patterns of WSL4 by RT-PCR and quantitative PCR (qPCR) analysis. The results showed that the WSL4 transcript was detected in all tissues, including root, leaf blade, inflorescence, stem, and sheath, with a high level in the aerial part of the seedling (Fig. 4A; Supplemental Fig. S3). To investigate the tissue expression pattern of WSL4, we generated transgenic rice lines expressing the β-glucuronidase (GUS) reporter gene under the control of the WSL4 5′ promoter region. GUS expression was detected in root, leaf blade, sheath, stem, inflorescence, glumes, lemma, and anther, but not in stigma papillae, consistent with the RT-PCR and qPCR analyses (Fig. 4A; Supplemental Fig. S3). Cross sections of anthers revealed that GUS was expressed in pollen and anther epidermal cells, but not in tapetum cells. Strong GUS signals were detected in cortex cells, the vascular cylinder of the root, and in the vascular bundles of leaves, sheaths, and stems (Fig. 4B). The expression pattern of WSL4 suggested that WSL4 was involved in reproductive and vegetative organ development processes as well as epicuticular wax formation in rice.
Figure 4.
Expression profiles of WSL4. A, qPCR analysis of WSL4 mRNA expression. Lb, Leaf blade; St, stem; Inf, inflorescence; Sh, sheath; Ap, aerial part of 2-week-old seedling; R, root from 2-week-old seedling. Data are means ± se of three biological replicates. B, Spatial expression pattern of the WSL4 gene in transgenic rice plants harboring the WSL4 promoter fused to the GUS gene. a, Germinating seed; b, spikelet; c, anther; d, root; e, young leaf; f, leaf sheath; g, cross section of stem; h, cross section of leaf sheath; i, cross section of anther; j, cross section of young leaf; and k, cross section of root. Bars = 100 μm (h–k). C, Subcellular localization of WSL4. The green and magenta signals obtained with confocal microscopy indicated fusion proteins WSL4-GFP and mCherry-HDEL (ER marker protein), respectively. The overlap of GFP and mCherry fluorescent signals is indicated in merged images. Bars = 10 μm.
To identify the subcellular localization of WSL4, we fused GFP to the C terminus of WSL4 and expressed the construct transiently in rice protoplasts. Coexpression of WSL4 with the ER marker mCherry-HDEL showed that the green fluorescence of WSL4-GFP merged well with the magenta fluorescence of mCherry-HDEL, indicating that WSL4 was located in the ER (Fig. 4C).
WSL4 Overexpression Increased C30-C34 VLCFAs
To further analyze the function of WSL4, we introduced the gene into Kinmaze under the control of the maize ubiquitin-1 promoter (Supplemental Fig. S4). The hydrophobic leaf phenotype and wax crystal formed on transgenic leaves were similar to those of the wild-type control and the WSL4 rescued lines (Fig. 1). However, the wax load in the transgenic leaf was 30.3% higher than in the wild type (Fig. 2C; Table I). The contents of C30-C34 VLCFAs, C32 primary alcohols, and C27-C33 alkanes were increased in transgenic lines (Fig. 2A). The profile of wax components in the WSL4 overexpression line was comparable with that of the WSL4 rescued line. Thus, an elevated expression level of WSL4 enhanced the synthesis of C30 fatty acids, which were subsequently elongated to longer fatty acids, leading to enhanced conversion of fatty acids to their derivatives.
Detection and Isolation of Cofactors Interacting with WSL4 in Rice
The wax compositions of the wsl4-1 and wsl4-2 mutants suggested that WSL4 could act as a condensing enzyme in the elongation of VLCFAs longer than C26, even to C30. To assess whether WSL4 was sufficient for C26 elongation to C30, we expressed WSL4 in yeast. GC-MS analysis revealed only trace amounts of C28 fatty acids and no C30 fatty acids. The composition of fatty acids in WSL4-transformed yeast cells was equivalent to that with the empty vector (Supplemental Fig. S5). The trace amount of C28 fatty acids reflected the background in the wild-type yeast strain (Oh et al., 1997). Meanwhile, expressing WSL4 alone in elo3 yeast cells, a mutant with a defect in the synthesis of VLCFAs beyond C24, resulted in elevated accumulation of C24 fatty acids, but no longer fatty acid was detected (Supplemental Fig. S5). Thus, WSL4 alone may have no biochemical function in elongation of VLCFAs beyond C24 in vivo, suggesting that other partner proteins may be required for WSL4 activity.
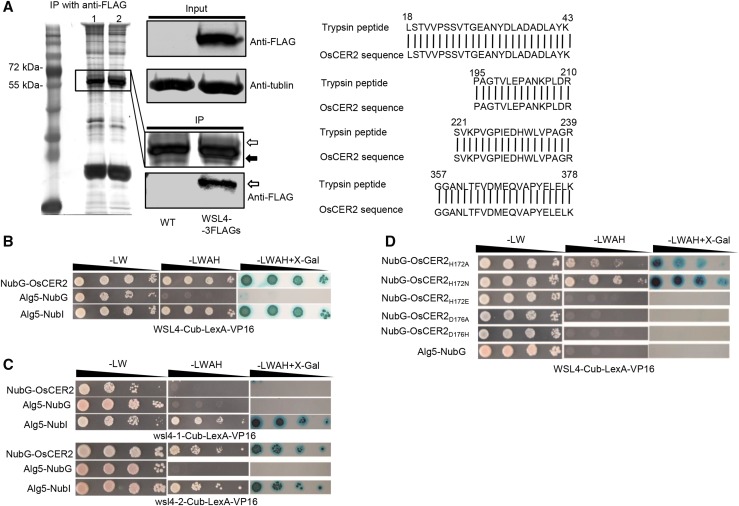
To search for cofactors interacting with WSL4 in rice, we performed coimmunoprecipitation (co-IP) experiments using WSL4-3FLAG-overexpressing rice calli. The callus cell extract was incubated with anti-FLAG antibody linked beads, and the wild-type extract was used as a control. After SDS-PAGE and staining, a unique band of ∼55 kD was detected, with no band observed at the equivalent position in the control lane. Liquid chromatography-tandem mass spectrometric analysis of the band showed that the tryptic peptides aligned well with the primary sequence deduced from Os04g0611200 (Fig. 5A). The amino acid sequence of Os04g0611200 shared 31.77% identity with that of CER2 (Supplemental Fig. S6). Thus, the gene was named OsCER2.
Figure 5.
Interaction assays between WSL4 and OsCER2. A, Isolation of OsCER2 from rice calli. Co-IP performed in transgenic rice calli expressing WSL4-3FLAGs. Plant proteins were separated by SDS-PAGE; the unique band (solid arrow) was excised and analyzed by LC-MS. WSL4-3FLAGs was detected by immunoblotting using an anti-FLAG primary antibody (open arrow). Antitubulin was used as a loading control. Lane 1, wild type; lane 2, expressed WSL4-3FLAGs. B to D, SUY2H assay of WSL4 and OsCER2 with WSL4-, wsl4-1-, and wsl4-2-Cub-LexA-VP16 as bait, and NubG-OsCER2 and NubG-mutated-OsCER2 as prey. Alg5-NubI and Alg5-NubG expression plasmids were used as positive and negative controls, respectively. Yeast transformants were spotted on control medium (–LW) and selective medium (–AHLW; OD yeast cells: 100, 10−1, 10−2, 10−3).
OsCER2 was expressed in leaves, sheaths, stems, inflorescences, and seedlings (shoots and roots; Supplemental Fig. S3). Coexpression of OsCER2-GFP with mCherry-HDEL showed that the green fluorescence of OsCER2-GFP almost completely merged with the red fluorescence of mCherry-HDEL (Supplemental Fig. S7), indicating that the OsCER2 protein was distributed on the ER. The ER localization of WSL4 and OsCER2 suggested that they might act synergistically for carbon-chain elongation in fatty acid synthesis.
To confirm the interaction between WSL4 and OsCER2, we performed a split-ubiquitin yeast two-hybrid (SUY2H) assay with WSL4-Cub-LexA-VP16 as the bait and NubG-OsCER2 as the prey, using WSL4-Cub-LexA-VP16 coexpressed with Alg5-NubI and Alg5-NubG as positive and negative controls, respectively. The yeast coexpressing WSL4-Cub and NubG-OsCER2 was able to grow on the selective medium (-Ade/-His/-Leu/-Trp) and showed β-galactosidase activity (Fig. 5B). The yeast coexpressing wsl4-2-Cub with NubG-OsCER2 showed similar results with that from yeast coexpressing WSL4 and OsCER2; however, the yeast coexpressing wsl4-1-Cub with NubG-OsCER2 did not grow under selection (Fig. 5C). These data further confirmed that WSL4 interacted directly with OsCER2 and that the Ser-228 residue in WSL4 was important for the interaction.
WSL4 Interacts with OsCER2, Catalyzing Elongation of C24 to C30 Fatty Acids in Yeast Cells
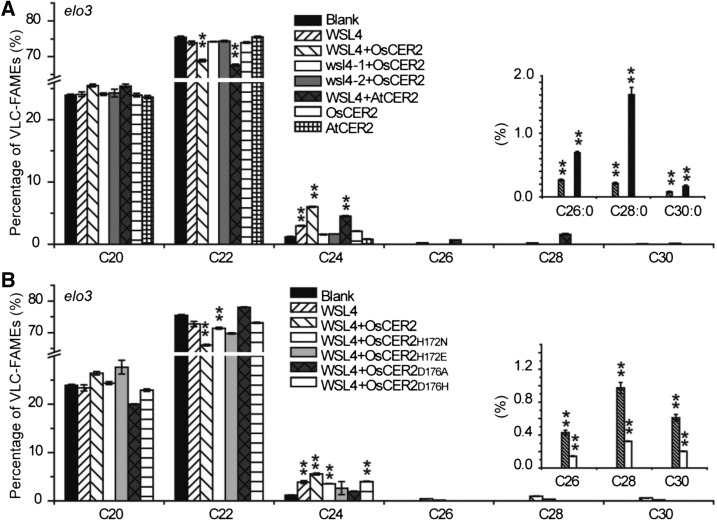
To investigate whether OsCER2 had VLCFA elongation activity, we expressed OsCER2 in yeast cells and analyzed the fatty acid composition by GC-MS. The fatty acid profile of yeast cells transformed with OsCER2 was equivalent to that of cells transformed with the empty vector (Supplemental Fig. S5). Thus, OsCER2 was not sufficient for VLCFA elongation past C26, like the CER2-LIKEs reported in Arabidopsis (Haslam et al., 2015). However, in the strain expressing both WSL4 and OsCER2, we detected a 5-fold increase in the yield of C28 fatty acids versus the strain expressing WSL4 relative to the vector control and a large amount of C30 fatty acids (Supplemental Fig. S5). We further coexpressed WSL4 and OsCER2 in elo3 yeast cells. Coexpression of WSL4 and OsCER2 in elo3 yeast cells yielded C30 VLCFAs (Fig. 6A). Coexpression of WSL4 with AtCER2 in elo3 yeast cells also produced notable amounts of C28 and C30 fatty acids. It is notable that AtCER2 was more active than OsCER2 with WSL4 (Fig. 6A). In contrast, there was no change in the fatty acid profiles of yeast cells (wild type or elo3) coexpressing wsl4-1 plus OsCER2, nor wsl4-2 plus OsCER2 (Fig. 6A; Supplemental Fig. S5). These results indicated that the functional combination of WSL4 and OsCER2 was necessary and sufficient for the elongation of C24 fatty acids to C30.
Figure 6.
GC-MS profiles of fatty acid methyl esters from elo3 knockout mutant yeast cells. Gas chromatographs of the saturated fatty acid methyl esters from transformed yeast cells expressing the empty vector, WSL4, OsCER2, AtCER2, WSL4 and OsCER2, wsl4-1 and OsCER2, wsl4-2 and OsCER2, and WSL4 and AtCER2 are displayed in A. WSL4, OsCER2, and mutated OsCER2s are displayed in B. Data are means ± se of three biological replicates. **P < 0.01. n-Nonadecanoic acid was used as an internal standard.
The HxxxD Motif in OsCER2 Is Essential for Its Interaction with WSL4
According to the characterization of AtCER2-LIKEs, the 172HxxxD176 catalytic motif is not required for CER2 function (Haslam et al., 2012). We used site-directed mutagenesis to replace the catalytic His-172 in OsCER2 with Ala, Asn, and Glu, and to replace Asp-176 with Ala and His. SUY2H assays with WSL4 as the bait and mutated OsCER2 proteins as prey showed that the replacement of H172A and H172N did not prevent the interaction between WSL4 and OsCER2, whereas OsCER2s with H172E, D176A, and D176H mutations failed to interact with WSL4 (Fig. 5D).
The site-mutated OsCER2s were coexpressed with WSL4 in elo3 cells. Fatty acid analyses showed that coexpressing WSL4 and OsCER2H172N yielded C30 VLCFAs. However, the fatty acid profiles of the yeast cells coexpressing mutated OsCER2s (H172E, D176A, and D176H, alleles with disrupted protein-protein interactions) with WSL4 were equivalent to that expressing WSL4 alone, with only increased C24 fatty acids, but no VLCFAs beyond C24 (Fig. 6B). These results demonstrated that the HxxxD motif in OsCER2 was required for its interaction with WSL4 in VLCFA elongation.
DISCUSSION
The synthesis of VLCFAs relies on the fatty acid elongase system, in which KCSs catalyze the first reaction in fatty acid elongation and determine the chain length of substrates and products. Because of the large number of annotated KCS genes in plants (21 in Arabidopsis and 34 in rice), determining the metabolic function of a specific KCS is complex. Although the functions of several Arabidopsis KCSs have been identified, we cannot deduce the functions of rice KCSs by homology analysis alone. In this study, we isolated the rice WSL4 gene, which encodes a KCS, via a map-based cloning strategy. Genetic complementation confirmed that a single-base mutation in WSL4 was responsible for the observed wax-sparse leaf phenotype in wsl4-1 and wsl4-2 mutants.
An important characteristic of KCSs is their substrate specificity. The amino acid sequence of WSL4 shares high identity with that of CER6/KCS6; however, their substrates are not identical. CER6 is responsible for VLCFA elongation beyond C24 in Arabidopsis (Millar et al., 1999). The wsl4-1 and wsl4-2 mutants showed decreased contents of VLCFAs longer than C28 and C26, respectively, and their derivatives (Fig. 2B), indicating that WSL4 acted as the major condensing enzyme for VLCFA elongation past C26. The involvement of WSL4 in the synthesis of wax compounds with more than 26 carbons was reconfirmed by the restoration of wax biosynthesis in plants expressing WSL4 in a wsl4-1 mutant background (Fig. 2). Expression of WSL4 alone in elo3 yeast cells resulted in increased C24 fatty acids and decreased C22 fatty acids, but did not produce fatty acids beyond C24 (Fig. 6). Coexpression of WSL4 with OsCER2 or AtCER2 in elo3 yeast cells produced VLCFAs up to C30 (Fig. 6). These results indicated that WSL4 was involved in VLCFA elongation beyond C22, to C30. This was supported by the observation that the fatty acid profile of WSL4-overexpressing rice calli showed increased C24-C30 fatty acids (Supplemental Fig. S8). Amounts of C24 fatty acids were not reduced in wsl4-1 and wsl4-2 mutants (Fig. 2). This may be explained by the functional redundancy of KCSs, such as WSL1, which has been predicted to encode a KCS catalyzing the elongation of VLCFAs from C20 to C24 (Yu et al., 2008). The functional redundancy of KCSs in the elongation of VLCFAs might also be responsible for the observation that wax components longer than C26 were not abolished in wsl4-2 (Fig. 2). Similar phenomena were reported in Arabidopsis, where CER6 and CER60 are involved in the elongation of fatty acyl-CoAs longer than C24 VLCFAs for cuticular waxes in epidermis and pollen coat lipids, with CER6/KCS6 as the major condensing enzyme, while CER60 made a smaller contribution to the synthesis of stem and pollen surface lipids due to its very low expression level and/or differing tissue specificity (Millar et al., 1999; Fiebig et al., 2000; Hooker et al., 2002).
Although wsl4-1 and wsl4-2 are allelic mutants, their wax compositions were quite different, reflecting the distinct roles of the mutated residues. Ghanevati and Jaworski (2002) identified Cys-223, His-391, and Asn-424 as key active site residues in KCS18/FAE1. KCS18/FAE1 with the Asn424His substitution lost its enzyme activity for condensation, whereas that with the Asn424Asp substitution retained 20% activity (Ghanevati and Jaworski, 2002). The corresponding Cys, His, and Asn in WSL4 are Cys-222, His-389, and Asn-422, respectively. Ser228Phe in WSL4 in wsl4-1 was not in the highly conserved domain; the mutation was in close proximity to the active site Cys-222 (Supplemental Fig. S2). Although the replacement of Ser228Phe in WSL4 in wsl4-1 was predicted to be in the interior of the protein based on the structures of homologous KCSs, a Kyte-Doolittle hydrophobicity analysis showed that the substitution of hydrophilic Ser for hydrophobic Phe (Ser228Phe) enhanced the hydrophobicity of the region covering the Cys-222 residue (Supplemental Fig. S9), which might affect its binding to acyl-CoA substrates (Trenkamp et al., 2004) or reduce WSL4 activity. Furthermore, homology model structure analysis of WSL4 showed that the Ser-228 residue is close to the surface residues of Ile-227, Asp-230, and Leu-231 of the protein (Supplemental Fig. S10). The replacement of Ser228Phe might change the interaction domain of WSL4 affecting its interaction with OsCER2 or the structure stability of WSL4. The SUY2H assay confirmed that the Ser228Phe mutagenesis in WSL4 in wsl4-1 interrupted the association with OsCER2 (Fig. 5). These results indicated that the Ser-228 residue may play an important role in the interaction between WSL4 and OsCER2, affecting the elongation of VLCFAs beyond C24. The wsl4-2 mutagenesis site was exactly the Asn-422 residue (Supplemental Fig. S2). However, the substitution of Asn for Ser (Asn422Ser) in WSL4 in the wsl4-2 did not affect its interaction with OsCER2 (Fig. 5C). These observations suggested that the substitution of Asn422Ser generated an inactive WSL4. Like that in Arabidopsis, rice may contain redundant CER2-LIKE proteins. It is possible that wsl4-1 with partial function might interact with other CER2-LIKE proteins producing VLCFAs longer than C28, which might be responsible for the fact that wax load and wax crystal in wsl4-1 were more than that in wsl4-2, and wsl4-1 showed a sectored-wetting leaf phenotype, whereas leaves of wsl4-2 were fully wet (Fig. 1; Table I). The relationships between amino acid residues and domains of KCSs and their substrate specificity remain to be clarified.
Plant KCS enzymes expressed in yeast cells can work with other subunits of the yeast FAE complex to produce specific acyl-CoAs (Millar and Kunst, 1997). When expressed alone in wild-type or elo3 yeast cells, WSL4 was not able catalyze the synthesis of VLCFAs longer than C24 (Fig. 6), indicating that WSL4 itself was not sufficient for VLCFA elongation beyond C24. We obtained a CER2-like protein (OsCER2) through co-IP with WSL4 and confirmed their physical interaction in SUY2H assays (Fig. 5). Both WSL4 and OsCER2 were localized to the ER membrane (Fig. 4D; Supplemental Fig. S7), consistent with CER6/KCS6 and CER2 (Joubès et al., 2008; Haslam et al., 2012). The physical interaction of WSL4 and OsCER2 might provide an explanation for OsCER2 being located on the ER, although CER2-LIKE proteins do not have predicted transmembrane domains or an ER targeting signal (Haslam et al., 2012).
It has been reported that CER2-LIKEs do not have condensing enzyme activity, but act as cofactors of KCS in VLCFA elongation (Haslam et al., 2012, 2015; Pascal et al., 2013). However, the mechanism by which CER2-LIKEs participate in the process remains unclear. Based on sequence homology, CER2-LIKEs are predicted to be members of the BAHD acyltransferase family, which has a wide range of substrates, including malonyl-CoA (Bontpart et al., 2015). The highly conserved HxxxD motif in BAHD family members has been predicted to be a substrate-binding and catalytic site (Ma et al., 2005; Unno et al., 2007). Although CER2-LIKEs do not have BAHD acyltransferase activity (Haslam et al., 2012), they may be able to bind malonyl-CoAs. In this study, we found that the HxxxD motif in OsCER2 was essential for the interaction between OsCER2 and WSL4. Substituting the His residue with Ala or Asn did not affect the interaction between OsCER2 and WSL4 (Fig. 5). However, mutated OsCER2s with H172E, D176A, and D176H were not able to interact with WSL4 (Fig. 5D) and failed to produce VLCFAs beyond C24 when coexpressed with WSL4 in elo3 cells (Fig. 6B). It is noteworthy that coexpression of WSL4 with OsCER2 in elo3 yeast cells yield more C24 fatty acids than that expression of WSL4 alone (Fig. 6), which indicated that OsCER2 might also be involved in the elongation of C22 fatty acids to C24. Nevertheless, expression of WSL4 alone in elo3 yeast cells also increased the level of C24 fatty acids (Fig. 6), which suggested that OsCER2 is not essential for the elongation of C22 fatty acids to C24. Taken together, our findings indicate that the functional combination of WSL4 and OsCER2 was necessary and sufficient for the elongation of C24 fatty acids to C30. Based on these results, together with the fact that the BAHD family may take malonyl-CoAs as substrates (Bontpart et al., 2015), we suggest that OsCER2 may be involved in VLCFA elongation through binding and transferring the substrate of malonyl-CoA to WSL4 for condensation with VLCF-CoA. The precise function of OsCER2 in fatty acid elongation remains to be determined.
MATERIALS AND METHODS
Plant Materials
The rice (Oryza sativa) mutant lines CM611 and CM1522 were obtained from Kyushu University. Rice seeds were germinated in plastic pots. Then, 4-week-old seedlings were raised in the field to maturity.
Scanning and Transmission Electron Microscopy
Leaf blades were excised from 6-week-old plants and dried as described previously (Mao et al., 2012). The dried samples were mounted on stubs, coated with platinum, and examined by scanning electron microscopy (S-4800; Hitachi) at an accelerating voltage of 10 kV and a working distance of 30 mm. Mature expanded leaves were simultaneously cut into 5 × 2-mm samples. The samples were processed as described previously (Sturaro et al., 2005). Ultrathin sections (80 nm) were cut using an Ultracut E ultramicrotome (Leica) and mounted on copper grids. The sections were stained with uranyl acetate and lead citrate solution and observed by transmission electron microscopy (JEM-1230; Hitachi).
Cuticular Wax Analysis
The cuticular waxes of mature expanded blade leaves from tilling-stage plants were derivatized as described previously (Mao et al., 2012). The composition was analyzed using GC-TOF/MS (Leco Pegasus IV).
Fine Mapping and Isolation of WSL4
Genomic DNA samples of 80 individuals with the wax crystal-sparse leaf phenotype isolated from the F3 generation were subjected to fine mapping of WSL4 by microsatellite analysis (McCouch et al., 2002). Additional indel markers were developed based on sequence comparisons between the genomic sequences in the primary mapped regions of Kinmaze and 9311.
Gene Expression Analyses
Total RNA was extracted from leaves, leaf sheaths, stems, inflorescences of booting-stage plants, aerial parts, and roots of 2-week-old seedlings using the TRIzol RNA reagent (Invitrogen). Each RNA sample was reversed-transcribed to cDNA after DNase I treatment, according to the manufacturer’s protocol (Reverse Transcription System; Promega). The ACTIN1 gene was used as a control. PCR amplification was performed with the gene-specific primers listed in Supplemental Table S3.
For the promoter GUS assay, the 2,561-bp promoter region upstream of WSL4 was amplified from Kinmaze genomic DNA using primers (Supplemental Table S3), and the PCR product was digested with SalI/SmaI. The resulting fragment was inserted into the pGPTV-GUS vector upstream of the GUS reporter gene. The genetic transformation and histochemical analysis were performed as described previously (Tian et al., 2013).
Subcellular Localization
To assess the subcellular localization of WSL4 and OsCER2, both coding sequences were subcloned into an XbaI- and SmaI-digested pBI221 vector to produce a C-terminal GFP fusion driven by a constitutive 35S promoter. The constructs were cotransformed with an ER marker fusion (mCherry-HDEL) into rice protoplasts as described previously (Tian et al., 2013). The transformed cells were detected by confocal microscopy (Olympus FV1000 MPE) and images were captured at 488 nm for GFP excitation and 534 nm for RFP excitation.
Plastid Construction and Genetic Transformation
The WSL4 complementation vector was constructed by introducing a 5,219-bp genomic DNA fragment, including the 2,478-bp promoter region upstream of the ATG start codon, the 1,578-bp WSL4 coding region containing a 93-bp intron, and the 1,163-bp untranslated region downstream of the stop codon, into the binary vector pCAMBIA 1301. The construct was introduced into CM611 via Agrobacterium tumefaciens-mediated transformation (Tian et al., 2013).
The 1,485-bp coding sequence of WSL4 was inserted into the binary vector pCAMBIA1300 downstream of the ubiquitin promoter. The construct was transformed into wild-type rice as described above.
Interaction Analyses
For the co-IP experiment, the construct pCAMBIA 1300-Pubi-WSL4-3FLAGs was introduced into rice calli. Total protein was extracted from the transformed calli using lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100). Co-IP was performed according to the manufacturer’s protocol (FLAG immunoprecipitation kit; Sigma-Aldrich). The resin was subjected to 10% SDS-PAGE. The different bands were recovered, digested with trypsin, and subjected to MS analyses with the Triple TOF 5600 system (AB SCIEX) as described previously (Qian et al., 2015).
For yeast two-hybrid assays, WSL4 (optimized for expression in Saccharomyces cerevisiae) and OsCER2 cDNA were amplified by PCR using SfiI site-containing primers (Supplemental Table S3) with subcloning of WSL4 into the pCCW-SUC bait vector and OsCER2 into the pDSL-Nx prey vector. NMY32 cells were cotransformed with pCCW-SUC-WSL4 and pDSL-Nx-OsCER2, and pAI-Alg5-NubI (positive control) or pDL2-Alg5-NubG (negative control). Yeast transformants were selected on selective medium SD–Leu/–Trp (–LW). The interaction was assayed on SD/–Ade/–His/–Leu/–Trp (–AHLW) medium. Then, β-galactosidase activity was measured on SD–AHLW covered with X-Gal-agarose buffer (0.5% agarose, 0.5 m phosphate buffer, pH 7.0, and 0.002% X-Gal) and incubated at 37°C for 20 min.
Yeast Fatty Acid Profiling
The protein coding sequence of WSL4 was codon-optimized and synthesized with 5′- and 3′-restriction sites (EcoRI and NotI, respectively). The synthesized WSL4 was subcloned into pYES2 (Invitrogen), downstream of the GAL1 promoter. The OsCER2 cDNA sequence was subcloned into EcoRI- and SpeI-digested pESC-His (Agilent), downstream of the GAL10 promoter. S. cerevisiae wild-type INVSc1 cells and BY4741 elo3 mutant cells were transformed with the different combinations and grown on synthetic complete selection medium (SC) lacking the appropriate amino acids indicated in Supplemental Table S4. The rice proteins were induced in the SC medium with 2% Gal as the only carbon source for 48 h at 30°C. Fatty acid methyl esters were prepared as described previously (Tresch et al., 2012). Samples were analyzed by GC-QqQ-MS/MS (Agilent).
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project Database or GenBank/EMBL databases under the following accession numbers: WSL4, Os03g0220100; OsCER2, Os04g0611200; and AtCER2, At4g24510.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Water retention of leaf blades in wsl4-1, wsl4-2, and their F1 generations.
Supplemental Figure S2. Amino acid sequence of WSL4.
Supplemental Figure S3. Analysis of the expression pattern of WLS4 and OsCER2.
Supplemental Figure S4. qPCR analysis of WSL4 expression in the overexpression line.
Supplemental Figure S5. Profiles of VLCFAs in yeast cells expressing WSL4 and OsCER2.
Supplemental Figure S6. Alignment of amino acid sequences of OsCER2 with AtCER2s.
Supplemental Figure S7. Subcellular location of OsCER2.
Supplemental Figure S8. Profiles of fatty acids distribution in rice organs and tissues.
Supplemental Figure S9. Hydrophilicity plot (Kyte-Doolittle) for WSL4 and wsl4-1 peptides.
Supplemental Figure S10. Three-dimensional homology modeling of WSL4.
Supplemental Table S1. Genotype for 14 markers from chromosome 3 across eight recombinants of F3 offspring originating from a wsl4-1 (japonica) × 9311 (indica) cross.
Supplemental Table S2. Mass spectrometric identification of OsCER2 peptides.
Supplemental Table S3. Primers.
Supplemental Table S4. Transgenic yeast cell lines.
Supplementary Material
Acknowledgments
We thank Dr. Toshihiro Kumamaru (Faculty of Agriculture, Kyushu University, Japan) for providing CM611 and CM1522 mutants. We also thank Dr. Jianmin Wan (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the elo3 yeast strain and Dr. Lianwei Peng (Institute of Botany, Chinese Academy of Sciences) for providing the pCCW-SUC and pDSL-Nx vectors and NMY32 yeast strain.
Glossary
- VLCFA
very-long-chain fatty acid
- KCS
β-ketoacyl-CoA synthase
- ER
endoplasmic reticulum
- GC-MS
gas chromatography-mass spectrometry
- co-IP
coimmunoprecipitation
- SUY2H
split-ubiquitin yeast two-hybrid
Footnotes
This work was supported by the Natural Science Foundation of China (no. 31570245 to L.Q.Q.).
References
- Barnes JD, Percy KE, Paul ND, Jones P, McLaughlin CK, Mullineaux PM, Creissen G, Wellburn AR (1996) The influence of UV-B radiation on the physicochemical nature of tobacco (Nicotiana tabacum L.) leaf surfaces. J Exp Bot 47: 99–109 [Google Scholar]
- Bontpart T, Cheynier V, Ageorges A, Terrier N (2015) BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol 208: 695–707 [DOI] [PubMed] [Google Scholar]
- Chen Y, Kelly EE, Masluk RP, Nelson CL, Cantu DC, Reilly PJ (2011) Structural classification and properties of ketoacyl synthases. Protein Sci 20: 1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrode SD, Espelie KE (1995) Effects of plant epicuticular lipids on insect herbivores. Annu Rev Entomol 40: 171–194 [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Höfer R, Briesen I, Emsermann M, Efremova N, Yephremov A, Schreiber L (2009) The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J 57: 80–95 [DOI] [PubMed] [Google Scholar]
- Gan L, Wang X, Cheng Z, Liu L, Wang J, Zhang Z, Ren Y, Lei C, Zhao Z, Zhu S, et al. (2016) Wax crystal-sparse leaf 3 encoding a β-ketoacyl-CoA reductase is involved in cuticular wax biosynthesis in rice. Plant Cell Rep 35: 1687–1698 [DOI] [PubMed] [Google Scholar]
- Ghanevati M, Jaworski JG (2002) Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. Eur J Biochem 269: 3531–3539 [DOI] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, van der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM (2000) The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Haslam TM, Mañas-Fernández A, Zhao L, Kunst L (2012) Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol 160: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam TM, Kunst L (2013) Extending the story of very-long-chain fatty acid elongation. Plant Sci 210: 93–107 [DOI] [PubMed] [Google Scholar]
- Haslam TM, Haslam R, Thoraval D, Pascal S, Delude C, Domergue F, Fernández AM, Beaudoin F, Napier JA, Kunst L, Joubès J (2015) ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol 167: 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker TS, Millar AA, Kunst L (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DW Jr., Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EN (1994) Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol 105: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels L (2009) Plant cuticles shine: advances in wax biosynthesis and export. Curr Opin Plant Biol 12: 721–727 [DOI] [PubMed] [Google Scholar]
- Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC (2009) Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J 60: 462–475 [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J (2005) Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem 280: 13576–13583 [DOI] [PubMed] [Google Scholar]
- Mao B, Cheng Z, Lei C, Xu F, Gao S, Ren Y, Wang J, Zhang X, Wang J, Wu F, et al. (2012) Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235: 39–52 [DOI] [PubMed] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9: 199–207 [DOI] [PubMed] [Google Scholar]
- McNevin JP, Woodward W, Hannoufa A, Feldmann KA, Lemieux B (1993) Isolation and characterization of eceriferum (cer) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome 36: 610–618 [DOI] [PubMed] [Google Scholar]
- Millar AA, Kunst L (1997) Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J 12: 121–131 [DOI] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C, Toke DA, Mandala S, Martin CE (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 272: 17376–17384 [DOI] [PubMed] [Google Scholar]
- Pascal S, Bernard A, Sorel M, Pervent M, Vile D, Haslam RP, Napier JA, Lessire R, Domergue F, Joubès J (2013) The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J 73: 733–746 [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Tian L, Qu LQ (2015) Proteomic analysis of endoplasmic reticulum stress responses in rice seeds. Sci Rep 5: 14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reicosky DA, Hanover JW (1978) Physiological effects of surface waxes I. Light reflectance for glaucous and nonglaucous Picea pungens. Plant Physiol 62: 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Sturaro M, Hartings H, Schmelzer E, Velasco R, Salamini F, Motto M (2005) Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol 138: 478–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Dai LL, Yin ZJ, Fukuda M, Kumamaru T, Dong XB, Xu XP, Qu LQ (2013) Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J Exp Bot 64: 2831–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenkamp S, Martin W, Tietjen K (2004) Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci USA 101: 11903–11908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresch S, Heilmann M, Christiansen N, Looser R, Grossmann K (2012) Inhibition of saturated very-long-chain fatty acid biosynthesis by mefluidide and perfluidone, selective inhibitors of 3-ketoacyl-CoA synthases. Phytochemistry 76: 162–171 [DOI] [PubMed] [Google Scholar]
- Unno H, Ichimaida F, Suzuki H, Takahashi S, Tanaka Y, Saito A, Nishino T, Kusunoki M, Nakayama T (2007) Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J Biol Chem 282: 15812–15822 [DOI] [PubMed] [Google Scholar]
- Weidenbach D, Jansen M, Franke RB, Hensel G, Weissgerber W, Ulferts S, Jansen I, Schreiber L, Korzun V, Pontzen R, et al. (2014) Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol 166: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, He C (2008) Wax Crystal-Sparse Leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228: 675–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.