The Arabidopsis MYB107 transcription factor regulates the expression of suberin biosynthetic genes in developing seeds, thereby controlling the synthesis and assembly of seed coat suberin.
Abstract
Suberin, a lipophilic polymer deposited in the outer integument of the Arabidopsis (Arabidopsis thaliana) seed coat, represents an essential sealing component controlling water and solute movement and protecting seed from pathogenic infection. Although many genes responsible for suberin synthesis are identified, the regulatory components controlling its biosynthesis have not been definitively determined. Here, we show that the Arabidopsis MYB107 transcription factor acts as a positive regulator controlling suberin biosynthetic gene expression in the seed coat. MYB107 coexpresses with suberin biosynthetic genes in a temporal manner during seed development. Disrupting MYB107 particularly suppresses the expression of genes involved in suberin but not cutin biosynthesis, lowers seed coat suberin accumulation, alters suberin lamellar structure, and consequently renders higher seed coat permeability and susceptibility to abiotic stresses. Furthermore, MYB107 directly binds to the promoters of suberin biosynthetic genes, verifying its primary role in regulating their expression. Identifying MYB107 as a positive regulator for seed coat suberin synthesis offers a basis for discovering the potential transcriptional network behind one of the most abundant lipid-based polymers in nature.
The seed coat is an outer covering of the seed that protects the embryo and other seed components against diverse environmental agents, including biotic and abiotic stresses (Mohamed-Yasseen et al., 1994). The mature seed coat of Arabidopsis (Arabidopsis thaliana) is composed mostly of five cell layers of maternal origin. During development, its outer integument (the outer two cell layers) undergoes extensive secondary thickening and becomes a sclerotic layer (Beeckman et al., 2000; Moïse et al., 2005), where suberin, a lipophilic polymer, is deposited and functions as the primary sealing component, contributing to the seed coat’s impermeability to water and nutrients (Molina et al., 2006, 2008). The defect of suberin biosynthesis in some mutant lines, such as glycerol-3-phosphate acyltransferase5-1 (gpat5-1), renders highly permeable seed coats, which is correlated with decreased dormancy and capacity of the seeds to germinate after storage (Beisson et al., 2007).
Suberin and cutin are two types of apoplastic polymers responsible for the sealing property of plant tissues. Chemically, both polymers are glycerol-based, aliphatic polyesters impregnated with waxes (Franke et al., 2005; Molina et al., 2006). Suberin differs from cutin mainly as it consists of more abundant aromatic components (Bernards and Razem, 2001) and the longer chain-length fatty acyl constituents (Moire et al., 1999; Graça et al., 2002; Li-Beisson et al., 2013). So far, several sets of analogous enzymes and/or proteins involved in suberin and cutin biosynthesis have been characterized, including the β-ketoacyl-CoA synthase (KCS) responsible for fatty acid elongation (Franke et al., 2009; Lee et al., 2009), the cytochrome P450 CYP77A, CYP86A, and CYP86B family members involved in fatty acid oxygenation (Wellesen et al., 2001; Xiao et al., 2004; Rupasinghe et al., 2007; Höfer et al., 2008; Compagnon et al., 2009; Li-Beisson et al., 2009), the long-chain acyl-CoA synthases for fatty acid activation (Schnurr et al., 2004; Bessire et al., 2007; Lü et al., 2009), the fatty acyl-CoA reductases (FARs) for fatty acid reduction (Domergue et al., 2010), and the enzymes responsible for subsequent acyltransfer to glycerol-3-phosphate (i.e. the GPATs; Beisson et al., 2007; Li et al., 2007; Li-Beisson et al., 2009). In addition, a few close homologs of BAHD acyltransferases, namely HYDROXYCINNAMOYL-COENZYME A:ω-HYDROXYACID O-HYDROXYCINNAMOYL TRANSFERASE/ALIPHATIC SUBERIN FERULOYL TRANSFERASE (HHT/ASFT; Gou et al., 2009; Molina et al., 2009), DEFECTIVE IN CUTIN FERULATE (DCF; Rautengarten et al., 2012), and FATTY ALCOHOL:CAFFEOYL-COENZYME A CAFFEOYL TRANSFERASE (FACT; Kosma et al., 2012), were characterized for incorporating aromatics into suberin, cutin, and/or some of suberin-associated waxes. Specifically, HHT/ASFT acts as the key enzyme transferring ferulate to aliphatic compositions of suberin, yielding suberin aromatic esters (Gou et al., 2009; Molina et al., 2009). Disrupting suberin biosynthetic genes, as in the mutants gpat5-1, far1far4far5, or reduced levels of wall-bound phenolics1-1 (rwp1-1)/asft, enhances seed coat permeability (Beisson et al., 2007; Gou et al., 2009; Molina et al., 2009; Vishwanath et al., 2013), supporting the primary barrier function of suberin in the seed coat.
Despite the core reactions in suberin and cutin biosynthesis being comparable, the deposition of two polyesters and the expression of their corresponding biosynthetic genes are highly tissue/organ specific. In Arabidopsis, cutin, as a major structural component of cuticles, is presented on the outermost epidermal cell walls of virtually every aerial organ, whereas suberin is found primarily in the outer integument layer of the seed coat and in the peridermal cells of mature root and the endodermal cells of young root (Pollard et al., 2008; Nawrath et al., 2013). Consistently, nearly the entire set of suberin biosynthetic genes identified in Arabidopsis display preferential expression in the seed and/or root (Pollard et al., 2008; Schreiber, 2010; Beisson et al., 2012; Andersen et al., 2015), whereas the genes required for cuticle formation typically are expressed in the epidermis of stem, leaf, and/or flower (Wellesen et al., 2001; Li et al., 2007; Li-Beisson et al., 2009). This strict tissue-specific distribution suggests the existence of a sophisticated regulation mechanism at the transcription level that governs the polyester’s synthesis and deposition processes. Several transcription factors (TFs) were implicated in the formation of cuticle (wax and cutin) in Arabidopsis, including members of the ethylene-response factor family, WAX INDUCER1/SHINE1 (WIN1/SHN1), SHN2, and SHN3 (Aharoni et al., 2004; Broun, 2004; Broun et al., 2004; Kannangara et al., 2007; Shi et al., 2011), and the MYB TFs MYB106 and MYB16 (Aharoni et al., 2004; Broun, 2004; Cominelli et al., 2008; Yeats and Rose, 2013). However, the regulatory mechanism and the related TFs specifically involved in the tissue/organ-specific biosynthesis of suberin have not been definitively determined. In this study, we identified a MYB TF, MYB107, that functions as a positive regulator specifically controlling suberin gene expression in developing seeds of Arabidopsis and, consequently, affecting seed coat permeability.
RESULTS
Altered Seed Coat Permeability of the myb107 Mutant
Previous studies from our and other groups (Gou et al., 2009; Rautengarten et al., 2012) and in silico gene expression data (Supplemental Fig. S1) revealed that Arabidopsis HHT and DCF, the two phylogenetically related BAHD genes responsible for suberin and cutin aromatic synthesis, displayed distinct tissue-specific expression patterns. HHT is expressed predominantly in developing seeds and roots, whereas DCF is expressed mainly in epidermis of aerial tissues. In attempting to identify the regulatory components governing their tissue-specific expression, we used HHT/AFST (At5g41040) and DCF (At3g48720) genes as the baits to search for their coexpression partners. Among a set of recognized genes (Supplemental Table S1), we found that HHT exhibited high coexpression with a few annotated TF genes, including one encoding a MYB domain-containing protein, MYB107 (Dubos et al., 2010). Intriguingly, in silico gene expression data also indicate that MYB107 expresses primarily in developing seeds (Supplemental Fig. S2) and coexpresses with most of the suberin biosynthetic genes (Supplemental Table S2), which indicates its involvement in suberin synthesis.
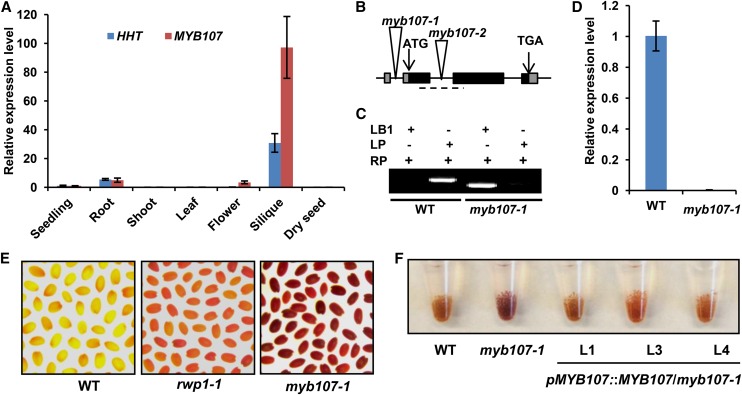
Quantitative real-time (qRT)-PCR further revealed that the transcripts of both MYB107 and HHT were strikingly high in siliques but nearly absent in shoots and leaves, confirming their tissue-specific expression patterns and their coexpression (Fig. 1A). To assess MYB107 functions, two homozygous T-DNA insertion mutants were isolated: SAIL_242_B04, with T-DNA inserted in the 5′ untranslated region (designated myb107-1); and SALK_203615, with T-DNA in the first intron of the coding region (designated myb107-2; Fig. 1B). Those insertions led to substantial reduction of MYB107 transcripts in both mutant lines (Fig. 1C; Supplemental Fig. S3A). Under normal growth conditions, the mutant (represented by myb107-1) was morphologically identical to the Columbia-0 (Col-0) wild type throughout the entire developmental process (Supplemental Fig. S4, A and B); neither its fertility nor seed setting was affected, except that its mature seeds were slightly darker than those of the wild type (Supplemental Fig. S4C), a feature reminiscent of the suberin biosynthetic mutant gpat5-1 (Beisson et al., 2007). Interestingly, compared with the wild type and the HHT mutant rwp1-1 (Gou et al., 2009), the seed coat permeability of both myb107-1 and myb107-2 exhibited substantial increase; much stronger red coloration was developed after their mature seeds were stained with tetrazolium salt, a cation dye that normally cannot penetrate the Arabidopsis seed coat (Fig. 1E; Supplemental Fig. S3B). Because myb107-1 and myb107-2 show similar phenotypes, only myb107-1 was further characterized for the following studies (for simplicity, myb107-1 is referred to as myb107 hereafter).
Figure 1.
Characterization of the myb107 mutant. A, Relative expression levels of MYB107 and HHT in different tissues. The expression levels of both genes in seedlings were set as 1. B, Diagram of the T-DNA insertion mutant of MYB107. The triangles indicate the sites of T-DNA insertion. The dashed line marks the region for qRT-PCR analysis of MYB107 gene expression in D. C, Genotyping of the T-DNA insertion mutant by PCR using T-DNA left-border primer (LB1) and two pairs of MYB107-specific primers (LP and RP). D, Relative expression levels of MYB107 in siliques of the wild type (WT) and myb107-1 mutant revealed by qRT-PCR. E, myb107-1, rwp1-1, and wild-type seeds stained with tetrazolium salt for 12 h. F, Tetrazolium-stained seeds of the wild type, myb107-1, and three individual transgenic lines of myb107-1 harboring the pMYB107::MYB107 construct. The data in A and D represent means ± sd from three experimental repeats.
To determine whether the increased permeability of myb107 is attributable to the compromised MYB107 gene expression, we first examined the seed coat permeability of a population of the segregated progeny of a myb107 heterozygous mutant line (myb107/MYB107) using Tetrazolium Red. Out of 22 progeny, only seeds from six homozygous plants exhibited enhanced red staining, whereas heterozygous and non-T-DNA insertion seeds did not (Supplemental Fig. S5), demonstrating a strict cosegregation of the T-DNA insertion with the enhanced seed coat permeability. Next, myb107 was complemented with a genomic DNA fragment spanning the MYB107 native promoter and its full-length gene. A higher abundance of MYB107 transcripts was detected in the siliques of the complementation lines compared with myb107 (Supplemental Fig. S6A); correspondingly, the mature seeds of the complementation lines showed abrogated Tetrazolium Red staining similar to the wild type (Fig. 1F). These results indicate that MYB107 is essential for maintaining seed coat impermeability.
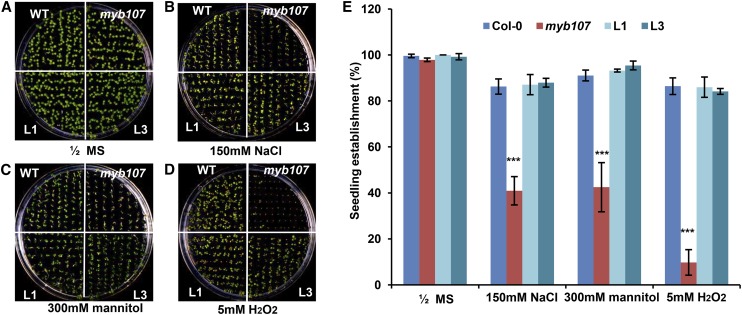
We then examined whether the altered seed coat permeability of myb107 leads to alteration of its seed physiological property under salinity, osmotic, and oxidative conditions that mimic environmental stresses. While no significant germination differences were observed for the seeds of myb107, the wild type, and the complementation lines placed on plain one-half-strength Murashige and Skoog (MS) medium (Fig. 2, A and E), the percentage of seedling establishment of myb107 was notably lower than that of the wild type in all treatments (Fig. 2, B–E). Particularly, when myb107 seeds were grown on the medium supplemented with 5 mm H2O2, the seedling establishment was reduced almost 80% compared with that of the wild type (Fig. 2, D and E). Conversely, the complementation lines exhibited a rescued seedling establishment similar to the wild type in all treatments (Fig. 2, B–E). These data indicate that down-regulation of MYB107 raises the susceptibility of seeds to abiotic stresses.
Figure 2.
Characterization of the myb107 mutant under abiotic stresses. A to D, Seven-day-old seedlings of the wild type (WT), myb107, and two complementation lines of myb107 harboring pMYB107::MYB107 (L1 and L3) germinated on a plain one-half-strength MS plate (A) and plates containing 150 mm NaCl (B), 300 mm mannitol (C), and 5 mm H2O2 (D). E, Percentage of the established seedlings in A to D. The data represent means ± sd of three replicates with approximately 80 seeds for each repeat. Asterisks indicate statistical differences compared with the wild type (Student’s t test, ***P < 0.001).
Decrease of Suberin Monomers in the myb107 Seed Coat
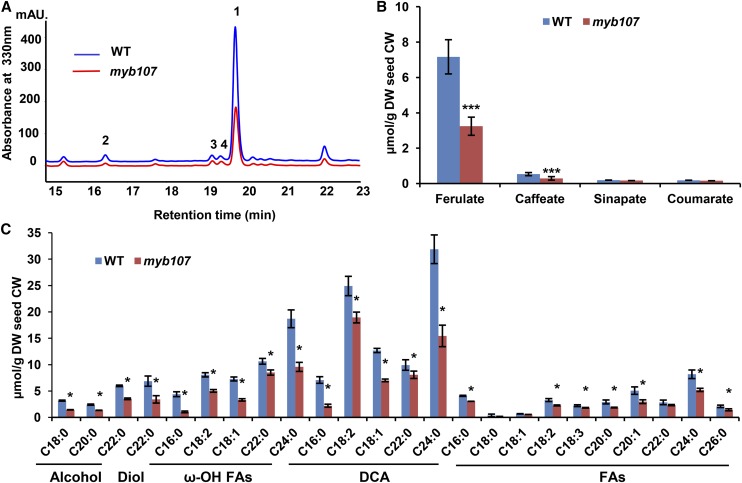
We then tested whether the enhanced permeability of the seed coat of myb107 was due to the defect in suberin synthesis. HPLC analysis of the depolymerized seed coat cell wall revealed that the content of ferulate and caffeate esters, the primary phenolics in seed coat suberin (Gou et al., 2009; Molina et al., 2009; Kosma et al., 2012), was reduced up to 50% to 60% in myb107 seeds compared with those of the wild type (Fig. 3, A and B). Conversely, the accumulation levels of both ferulate and caffeate were restored close to those of the wild type in the complementation lines (Supplemental Fig. S6B).
Figure 3.
Aromatic and aliphatic monomers of seed coat polyesters by HPLC and gas chromatography-mass spectrometry (GC-MS). A, Portion of UV light-HPLC profiles of the phenolics from seed coat suberin of the wild type (WT) and myb107. Numbers represent the identified phenolics: 1, ferulate; 2, caffeate; 3, sinapate; 4, coumarate. B, Contents of aromatic compounds in seed coat suberin quantified by HPLC. C, Contents of aliphatic monomers, including fatty alcohols, diols, ω-hydroxy fatty acids (ω-OH FAs), dicarboxylic acids (DCA), and fatty acids (FAs), in cell wall residues of seeds. The data represent means ± sd from three biological replicates. Asterisks indicate statistical differences compared with the wild type (Student’s t test, ***P < 0.001 [B] and *P < 0.05 [C]). CW, Cell wall; DW, dry weight.
GC-MS resolved a set of aliphatic monomers from seed polyester methanolysates. In general, nearly all the detected aliphatics in the seed polyesters of myb107 showed substantial reduction compared with the wild type; in particular, the dominant compositions of C24:0 dicarboxylic acid and C24:0 ω-hydroxy acid displayed up to a 50% decrease (Fig. 3C; Supplemental Fig. S7). These data suggest that MYB107 affects the synthesis of both suberin aromatics and aliphatics in the seed coat.
However, when suberin methanolysates from roots were examined, there was no difference in the content of resolved aromatics between myb107 and wild-type plants (Supplemental Fig. S8A). Similarly, the abundance of all the detected aliphatic monomers remained unchanged in myb107 root polyesters compared with the wild type (Supplemental Fig. S8B). Consistently, no difference was observed when wild-type and myb107 roots were stained with Sudan Black B (Supplemental Fig. S8C), a lipophilic dye used for the histochemical detection of suberin and associated waxes (Robb et al., 1991). These data suggest that disrupting MYB107 does not affect root suberin synthesis.
The Ultrastructures of Seed Coat Suberin and Surface Cuticles
Examining the epidermal surface of different organs of the wild type and myb107 by scanning electron microscopy (SEM), we found that both wild-type and myb107 seeds displayed the same preserved surface structure with the thick cell walls of columella (Supplemental Fig. S9, A and B). Similarly, there were no notable morphological changes on the leaf surface, trichomes, the nanoridges of petals, and the cuticular wax crystals distributed on stem surfaces (Supplemental Fig. S9, C–E). Furthermore, transmission electron microscopy (TEM) of the leaf epidermal cells of the wild type and myb107 revealed similar electron-dense cuticular layers on the outer surfaces of the cells and the identical cuticular ledges (projections) surrounding the stomatal pores (Supplemental Fig. S9, F and G). Consistently, when myb107 and wild-type plants were stained with Toluidine Blue, an aqueous dye monitoring plants with defective surface cuticles (Tanaka et al., 2004), no obvious difference was observed (Supplemental Fig. S10). These data suggest that cuticular layers of the aerial epidermis were not defective in myb107.
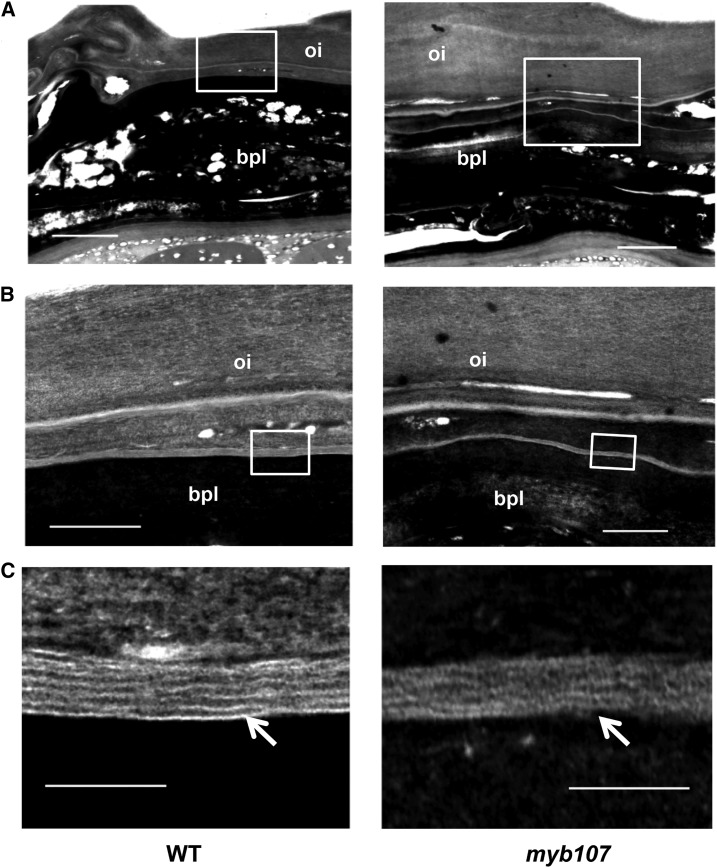
However, when the ultrastructures of the seed coats of myb107 and the wild type were examined by TEM, the finely arranged and largely continuous lamellar structures of suberin were found near the brown pigment layer of the wild-type seed coat, as reported previously (Yadav et al., 2014), where they showed regular alternating light- and dark-staining bands with the light bands spaced around 7 to 10 nm apart (Fig. 4), which were predicted to represent the aromatic and aliphatic domains of suberin, respectively (Bernards, 2002; Graça and Santos, 2007). In contrast, the lamellation of the suberin layer was largely disordered, with seemingly shorter, thicker, but discontinuous light-staining strands in the myb107 mutant seed coat (Fig. 4C). This observation suggests that knocking down MYB107 impairs the seed coat suberin ultrastructural arrangement.
Figure 4.
TEM observation of suberin ultrastructures in seed coats of the wild type (WT) and myb107. Mature seeds were ultrathin sectioned, and the sections (90–100 nm) were treated with 10% H2O2 and then stained with 1% aqueous uranyl acetate and Sato lead. Enlarged images from the squared areas in A are shown in B, and enlarged images from the squared areas in B are shown in C. bpl, Brown pigment layer; oi, outer integument. Arrows point to suberin lamellae in C. Bars = 2 µm (A), 500 nm (B), and 100 nm (C).
Alteration of Suberin Biosynthetic Gene Expression
The observed drastic effects of MYB107 on seed coat suberin synthesis and assembly prompted us to assess the potential function of MYB107 as a putative TF in regulating suberin biosynthetic gene expression. First, we explored the genes misregulated in myb107 by RNA sequencing (RNA-seq) analysis. Since MYB107 is expressed dominantly in siliques (Fig. 1A), RNAs from mixed siliques of different developmental stages were extracted and used in the study. Analyzing RNA-seq data sets revealed that most genes known to be involved in the biosynthesis of suberin and its associated waxes were down-regulated in the myb107 mutant to near or above 2-fold; they include not only HHT but also the genes FACT, CYP86A1, CYP86B1, FAR1, FAR4, and FAR5 (Table I; Supplemental Data Set S1). In contrast, the transcript abundance of cutin biosynthetic genes, such as CYP86A2, CYP86A8, CYP77A6, DCF, GPAT6, and GPAT8, showed no significant changes (Table I; Supplemental Data Set S1).
Table I. Comparisons of the expression levels of suberin and cutin biosynthetic and deposition-related genes in the wild type (Col-0) and myb107, showing the fold change of gene expression in myb107 versus the wild type quantified by RNA-seq and qRT-PCR.
For repeat 1 and repeat 2 of the RNA-seq and qRT-PCR experiments, RNAs from mixed siliques of different growth stages were used. For repeat 3 of the qRT-PCR experiment, RNAs from stage III developing seeds were used. Fold change values less than 0.5 are highlighted in boldface. ND, Not determined.
| Gene Name | Arabidopsis Genome Initiative No. | Pathways |
RNA-Seq (myb107/Wild Type) |
qRT-PCR (myb107/Wild Type) |
|||
|---|---|---|---|---|---|---|---|
| Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | Repeat 3 | |||
| BAHD acyltransferase | |||||||
| HHT/ASFT | AT5G41040 | Suberin | 0.59 | 0.48 | 0.48 | 0.63 | 0.15 |
| FACT | AT5G63560 | Suberin | 0.10 | 0.11 | 0.39 | 0.39 | 0.06 |
| DCF | AT3G48720 | Cutin | 0.95 | 0.59 | 1.18 | 1.74 | 0.73 |
| DCR | AT5G23940 | Cutin | 1.15 | 1.17 | 0.99 | 0.80 | 1.02 |
| Cytochrome P450 monooxygenase | |||||||
| CYP86A1 | AT5G58860 | Suberin | 0.49 | 0.17 | 0.49 | 0.57 | 0.59 |
| CYP86B1 | AT5G23190 | Suberin | 0.38 | 0.46 | 0.39 | 0.32 | 0.25 |
| CYP86A2 | AT4G00360 | Cutin | 0.89 | 0.87 | 0.76 | 0.76 | 0.63 |
| CYP86A8 | AT2G45970 | Cutin | 0.79 | 0.71 | 0.85 | 0.60 | 0.65 |
| CYP77A6 | AT3G10570 | Cutin | 1.16 | 0.92 | 1.22 | 0.60 | 1.23 |
| Long-chain acyl-CoA synthetase | |||||||
| LACS1 | AT2G47240 | Cutin | 1.00 | 0.92 | ND | ND | ND |
| LACS2 | AT1G49430 | Cutin/suberin | 0.57 | 0.62 | 0.40 | 0.45 | 0.49 |
| Glycerol-3-phosphate acyltransferase | |||||||
| GPAT5 | AT3G11430 | Suberin | 0.84 | 0.60 | 0.68 | 1.12 | 0.36 |
| GPAT4 | AT1G01610 | Cutin/suberin | 0.95 | 0.49 | 0.71 | 0.43 | 0.45 |
| GPAT6 | AT2G38110 | Cutin | 0.84 | 1.02 | 0.83 | 0.63 | 1.02 |
| GPAT8 | AT4G00400 | Cutin | 0.91 | 1.64 | 0.98 | 0.83 | ND |
| Alcohol-forming fatty acyl-CoA reductase | |||||||
| FAR1 | AT5G22500 | Suberin | 0.33 | 0.53 | 0.20 | 0.37 | 0.08 |
| FAR4 | AT3G44540 | Suberin | 0.24 | 0.23 | 0.28 | 0.36 | 0.15 |
| FAR5 | AT3G44550 | Suberin | 0.57 | 0.59 | 0.36 | 0.67 | 0.28 |
| 3-Ketoacyl-CoA synthase | |||||||
| KCS2 | AT1G04220 | Wax/suberin | 1.00 | 0.85 | 0.78 | 0.73 | 1.06 |
| KCS20 | AT5G43760 | Wax/suberin | 0.82 | 0.79 | 0.81 | 0.70 | 0.58 |
| ABC transporter | |||||||
| ABCG2 | AT2G37360 | Suberin | 0.53 | 0.72 | ND | ND | ND |
| ABCG6 | AT5G13580 | Suberin | 1.55 | 1.21 | 0.93 | 0.65 | 0.86 |
| ABCG20 | AT3G53510 | Suberin | 0.86 | 0.80 | 0.56 | 0.58 | 0.84 |
| ABCG11 | AT1G17840 | Cutin | 0.92 | 1.02 | 1.43 | 0.68 | 0.64 |
In general, qRT-PCR analyses of two independent biological repeats using RNAs extracted from the mixed siliques (repeats 1 and 2 in Table I) validated the transcriptional changes of those suberin biosynthetic genes found in the RNA-seq data sets (Table I). LACS2 and GPAT4 that encode enzymes known for cutin synthesis also appeared to be down-regulated in myb107 (Table I), which agrees with their proposed additional roles in suberin biosynthesis (Li-Beisson et al., 2013). The expression of KCS2 and KCS20, however, was not affected in myb107. KCS2 and KCS20 are the fatty acid elongases functionally redundant in the two-carbon elongation to very-long-chain fatty acids that is required for the biosynthesis of cuticular waxes and root suberin (Franke et al., 2009; Lee et al., 2009). The recently identified suberin ATP-binding cassette transporter genes ABCG2, ABCG6, and ABCG20 (Yadav et al., 2014) did not experience significant down-regulation in myb107 (Table I), suggesting that MYB107 does not affect the expression of genes involved in the process of polyester monomer transport/deposition. In addition, genes involved in seed coat pigmentation, namely the TRANSPARENT TESTA (TT) genes (Debeaujon et al., 2000; Appelhagen et al., 2014), also did not show significant changes (Supplemental Table S3; Supplemental Data Set S1).
Conversely, when transcripts of suberin biosynthetic genes, represented by FAR1 and FACT, were examined in complementation lines of myb107, we found that their defective expression in siliques could be restored to levels even slightly higher than those of the wild type (Supplemental Fig. S11A), confirming that MYB107 is sufficient for activating seed suberin biosynthetic gene expression.
Coexpression of MYB107 with Seed Suberin Biosynthetic Genes in a Temporal Manner
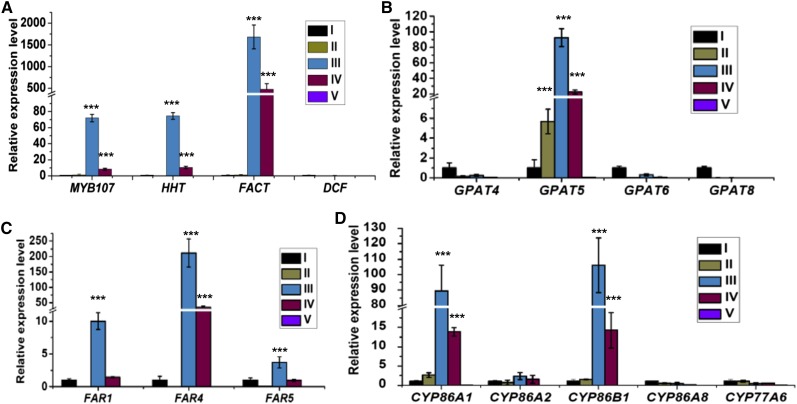
In the above RNA-seq and qRT-PCR experiments using RNAs from mixed siliques, the detected changes in expression levels of suberin biosynthetic genes in myb107 were somehow less substantial. We suspected that MY107 may temporally regulate suberin biosynthetic gene expression at a specific seed developmental stage and that the real differences might have been diluted when RNAs of the mixed siliques were used. Therefore, we dissected MYB107 expression patterns at different seed developmental stages and examined its potential correlation with other suberin biosynthetic genes. Total RNAs were extracted from seeds at five defined developmental stages (Supplemental Fig. S12): stages I (1–5 d after flowering [DAF]), II (6–10 DAF), III (11–15 DAF), IV (16–20 DAF), and V (mature seeds). qRT-PCR analyses revealed that the relative expression level of MYB107 was extremely low at stages I and II, surged to its maximum at stage III, declined at stage IV, and was nearly absent at stage V (Fig. 5A). Coincidently, nearly all the known suberin biosynthetic genes, including HHT, FACT, GPAT5, FAR1, FAR4, FAR5, CYP86A1, and CYP86B1, showed an essentially identical expression pattern to that of MYB107 (Fig. 5). In contrast, although cutin-like polyesters were proposed to occur in the seed coat of Arabidopsis (Molina et al., 2008), the expression of cutin biosynthetic genes, including DCF, GPAT4, GPAT6, GPAT8, CYP86A2, CYP86A8, and CYP77A6, exhibited no obvious expression pattern like that of MYB107 (Fig. 5). These data indicate that MYB107 coexpresses temporally with suberin biosynthetic genes but not cutin biosynthetic genes during seed development and that stage III (11–15 DAF) of developing seeds is a crucial period for the onset of suberin biosynthetic gene expression.
Figure 5.
Expression patterns of MYB107 and suberin/cutin biosynthetic genes during seed development. Developing siliques and mature seeds were grouped into five different developmental stages. The relative expression levels of MYB107 and three BAHD family genes (A), four GPAT genes (B), three FAR genes (C), and five CYP genes (D) at each developmental stage (I–V) are shown. UBIQUITIN10 (UBQ10) was used as the reference gene for the normalization. The expression level of each gene in stage I of developing seeds was set as 1. All data represent means ± sd of three experimental repeats. Asterisks indicate statistical differences of each stage compared with that at stage I (Student’s t test, ***P < 0.001).
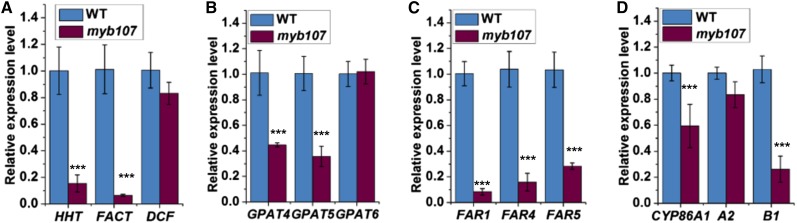
Subsequently, we undertook a third repeat of the qRT-PCR analysis using RNAs from the seeds at stage III of both the wild type and myb107. As expected, most of the known suberin biosynthetic genes were found more significantly down-regulated in myb107 (Fig. 6; repeat 3 in Table I) compared with the data from two previous repeats. Among them, the reductions of FARs, FACT, and HHT transcripts were most profound, showing more than 80% decline (Fig. 6; Table I). In contrast, expression of the genes involved in cutin biosynthesis, such as DCF, CYP86A2, and GPAT6, was not changed substantially (Fig. 6; Table I). Taken together, these data offer evidence that MYB107 regulates suberin biosynthetic gene expression in Arabidopsis developing seeds in a temporal manner.
Figure 6.
Expression of suberin/cutin biosynthetic genes in seed developmental stage III of the wild type (WT) and myb107. The relative expression levels of three BAHD family genes (A), three GPAT genes (B), three FAR genes (C), and three CYP genes (D) at seed developmental stage III are shown. UBQ10 was used as the reference gene for the normalization. The expression level of each gene in the wild type was set as 1. All data represent means ± sd of three experimental repeats. Asterisks indicate statistical differences of each stage compared with that at stage I (Student’s t test, ***P < 0.001).
Nuclear Localization and Interaction of MYB107 with Suberin Biosynthetic Gene Promoters
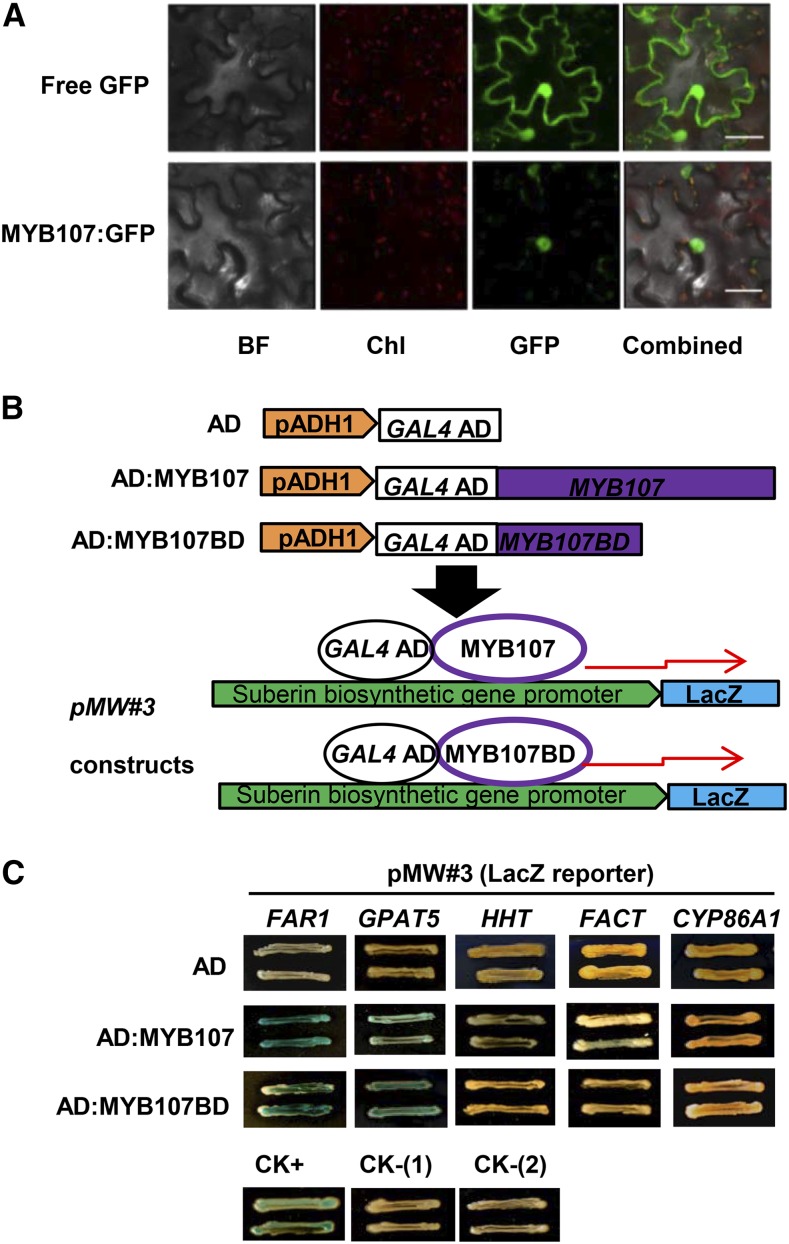
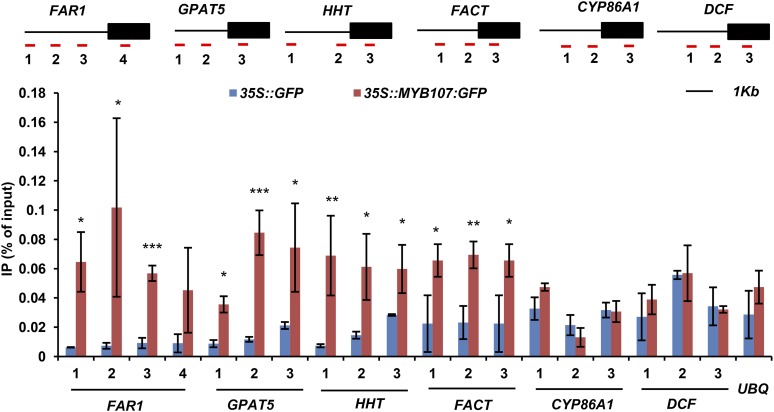
In agreement with its proposed function as a TF, we found that MYB107 localized in the nucleus when the p35S::MYB107:GFP construct was transiently expressed in the leaf cells of Nicotiana tabacum (Fig. 7A). To detect whether MYB107 indeed activates suberin biosynthetic gene expression, we examined the interaction of MYB107 and suberin biosynthetic gene promoters via yeast one-hybrid (Y1H) assay and chromatin immunoprecipitation (ChIP) coupled with quantitative PCR (qPCR) analysis. For the Y1H analysis, the full-length promoters of the selected representative suberin biosynthetic genes FAR1, GPAT5, CYP86A1, HHT, and FACT were placed upstream of the LacZ reporter gene to create the promoter::reporter constructs. Both the full-length MYB107 and its DNA-binding domain (MYB107BD) were hybridized with the yeast GAL4 activation domain (AD) and then transferred into the yeast cells harboring the promoter constructs (Fig. 7B). Yeast harboring an N-terminal fragment of the mutator-like transposase FHY3 (FHY3N) and its targeting promoter of FHL (pFHL; Lin et al., 2007) was used as the positive control. We found that yeast harboring AD:MYB107 (or AD:MYB107BD) and pFAR1::LacZ or pGPAT5::LacZ yielded obvious positive color reaction in β-Gal assays; the yeast cells containing AD:MYB107 (or AD:MYB107BD) and the promoters of HHT and FACT produced a relatively weak coloration; while yeast containing AD:MYB107 (or AD:MYB107BD) and pCYP86A1::LacZ or pFHL::LacZ constructs, or harboring the AD alone with any of promoter::reporter constructs, did not yield any coloration (Fig. 7C). These data suggest the potential interactions of MYB107 with the promoters of FAR1, GPAT5, HHT, and FACT. To validate the in vitro Y1H results, we performed ChIP coupled with qPCR using Arabidopsis seedlings transiently expressing MYB107:GFP and free GFP (as the control) against anti-GFP antibody. The resulting DNAs were amplified against the primers complementary to the sequences of promoters and/or coding regions of FAR1, GPAT5, HHT, FACT, CYP86A1, and DCF genes (Fig. 8). The qPCR data indicated that the promoter regions of FAR1, GPAT5, HHT, and FACT were significantly enriched in the MYB107:GFP-mediated immunoprecipitates relative to the immunoprecipitates of the free GFP control (Fig. 8). No significant enrichment was detected for the promoter regions of CYP86A1 and DCF (Fig. 8). These results are consistent with Y1H assays and indicate that MYB107 does bind specifically to the promoters of particular suberin biosynthetic genes in vivo.
Figure 7.
Subcellular localization of MYB107 and its physical interactions with the suberin biosynthetic gene promoters. A, Subcellular localization of the MYB107:GFP fusion protein. Confocal images of free GFP (top row) and MYB107:GFP (bottom row) in tobacco leaf cells are displayed. BF, Bright field; Chl, chlorophyll autofluorescence; GFP, GFP fluorescence; Combined, the merged images. Bars = 25 µm. B, Diagram of the expression constructs of MYB107 and the promoters of suberin biosynthetic genes used in the Y1H assay. The full-length MYB107 and MYB107 DNA-binding domains (MYB107BD) were fused with the yeast GAL4 activation domain (AD). The suberin biosynthetic gene promoters were placed to drive a LacZ reporter gene. C, Color reaction of yeast cells harboring AD:MYB107 or AD:MYB107BD with the promoter::LacZ construct. The images were captured after growing the yeast for 8 d on an SC(−Trp,−Ura) plate containing 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal). CK+, AD:FHY3N+pFHL::LacZ; CK-(1), AD:MYB107+pFHL::LacZ; CK-(2), AD:MYB107BD+pFHL::LacZ.
Figure 8.
ChIP-qPCR assays of MYB107-DNA complexes. The scheme of the primer design for each gene is shown at top, in which black lines represent the promoter region and black boxes represent the gene-coding region. Red lines beneath the gene structures with numbers mark the locations of amplicons amplified by ChIP-qPCR. UBQ10 (UBQ) was used as a negative control. Two biological replicates were performed; the data presented represent means ± sd of three technical repeats from one representative biological replicate. Asterisks indicate statistical differences compared with the free GFP control (Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001). IP, Immunoprecipitate.
DISCUSSION
Suberin is an essential physical barrier contributing to seed coat impermeability (Pollard et al., 2008; Nawrath et al., 2013). A defect of suberin synthesis changes the seed coat sealing property and lowers seed viability and tolerance to biotic and abiotic stresses (Beisson et al., 2007; Gou et al., 2009; Molina et al., 2009; Vishwanath et al., 2013). While the knowledge on suberin synthesis and deposition is relatively advanced, the regulatory mechanisms underlying seed coat suberin formation remain elusive. In this study, we identified and defined a TF, MYB107, a predicated R2R3-MYB family member of subgroup 10 (Dubos et al., 2010), as a positive regulator controlling suberin synthesis in the Arabidopsis seed coat. MYB107 coexpresses with a set of suberin biosynthetic genes in a temporal manner during seed development (Fig. 5). Down-regulation of MYB107 suppresses the expression of most known suberin biosynthetic genes (Fig. 6; Table I). Consequently, the suberization of the myb107 seed coat is impaired, which is demonstrated by the drastic reduction of suberin aliphatic and aromatic monomers and the altered lamellation of the suberin layer in outer integument of the seed coat (Figs. 3 and 4). These chemical and structural changes ultimately result in the enhanced seed coat permeability and increased abiotic stress susceptibility of the myb107 seed (Figs. 1 and 2; Supplemental Fig. S3). Conversely, complementation of MYB107 expression in myb107 restores suberin biosynthetic gene expression, defective suberin synthesis, seed coat impermeability, and abiotic stress tolerance (Figs. 1 and 2; Supplemental Fig. S6). Moreover, both in vitro and in vivo studies show that MYB107 interacts directly with the promoters of at least four suberin biosynthetic genes, GPAT5, FAR1, HHT, and FACT, although the promoters of the latter two genes exhibited relatively weak interactions with MYB107 in the Y1H assay (Fig. 7). GPAT5 is critical for the proper synthesis of suberin and associated waxes via the formation of sn-2 monoacylglycerol intermediates (Beisson et al., 2007); FAR1 is the fatty acyl-CoA reductase responsible for producing 22:0 primary fatty alcohol in suberized tissues; while both HHT and FACT are aromatic acyltransferases responsible for the formation of feruloyl and caffeoyl esters, respectively, in suberin or some of the suberin-associated waxes (Gou et al., 2009; Molina et al., 2009; Domergue et al., 2010; Kosma et al., 2012; Vishwanath et al., 2013). All these data suggest that MYB107 functions as a positive regulator dominating the expression of suberin biosynthetic genes and, thus, the synthesis and assembly of both suberin aliphatics and aromatics in the Arabidopsis seed coat (Supplemental Fig. S13).
The determined regulatory role of MYB107 in the seed coat appears specific for suberin biosynthetic genes. MYB107 coexpresses dominantly with genes involved in suberin biosynthesis but not those for cutin synthesis or for the formation of seed coat pigmentation (Supplemental Table S2). The down-regulation of MYB107 essentially did not affect the expression of specific cutin biosynthetic genes or TT genes (Table I; Fig. 6; Supplemental Table S3). Furthermore, the myb107 mutant showed no obvious morphological and ultrastructural alterations associated with the cuticles (Supplemental Figs. S9 and S10). Its seeds also have no tt phenotype (Supplemental Fig. S4C), as seen in other tt mutants (Debeaujon et al., 2000; Appelhagen et al., 2014). These data suggest that MYB107 controls seed coat permeability by specifically regulating suberin biosynthesis but not other elements contributing to the seed-sealing property. They also are consistent with the previous discovery that the synthesis of suberin and cutin, the two analogous polyesters derived from fatty acids, is under distinct transcriptional controls (Molina et al., 2008). Such regulatory distinction might be the key determinant in establishing the unique tissue- and/or organ-specific distribution of the two chemically analogous lipid-based polymers.
Arabidopsis seed coat and root peridermal or endodermal cells are the major sites of suberization. While several lines of evidence suggest that MYB107 is a regulator that controls the onset of programmed expression of suberin biosynthetic genes during seed development, knocking down MYB107 has little effect on suberin gene expression in roots (Supplemental Fig. S11B). The synthesis of suberin aliphatics and aromatics in the roots of myb107 was not impaired (Supplemental Fig. S8). These data indicate that MYB107 does not dominate suberin synthesis in roots. The exact reasons for MYB107 exhibiting no function on root suberin synthesis remain unclear. This may indicate the existence of additional redundant TF(s) in roots.
The down-regulation of MYB107 affects the expression of a set of suberin biosynthetic genes; moreover, MYB107 directly targets the promoters of a few suberin biosynthetic genes (Figs. 7 and 8). This result implies that MYB107 regulates suberin biosynthetic genes both directly and indirectly. The mode of MYB107 action is reminiscent of the TFs involved in cutin synthesis (Yeats and Rose, 2013) and in secondary cell wall formation (Zhao and Dixon, 2011). The TFs regulating cuticle formation appear to form a regulatory cascade. WIN1/SHN1 regulates the expression of a set of cutin and/or wax biosynthetic genes accompanied by the effect of wax load and cutin synthesis (Broun et al., 2004; Kannangara et al., 2007); meanwhile, WIN1/SHN1 and its homologous members can directly activate promoters of several cutin biosynthetic genes (Kannangara et al., 2007; Shi et al., 2011). In addition, two MYB TFs, MYB106 and MYB16, that were implicated in cuticle development were demonstrated to act upstream of WIN1/SHN1 (Oshima et al., 2013). Similarly, in a hierarchical regulatory network for secondary cell wall formation, the NAC-type TFs VND6/7 and NST1-3 act as the top master switches controlling the bottom transcriptional activators or repressors and, meanwhile, directly targeting some of the specific lignin or cellulose biosynthetic genes (Zhao and Dixon, 2011). The temporal expression pattern of MYB107 in Arabidopsis developing seeds indicates that MYB107 may position in an unidentified regulatory cascade or network and itself is developmentally controlled by upstream regulators; meanwhile, it also may control and/or coordinate the actions of additional TFs to regulate suberin biosynthetic gene expression (Supplemental Fig. S13). Indeed, a few other TF genes, such as the WRKY, bZIP, and MYB family members, were found to be either coexpressed with MYB107 or cosuppressed in myb107 (Supplemental Table S2; Supplemental Data Set S1). These include MYB41, a TF that was defined recently as the suberin regulator likely controlling the stress-induced suberin synthesis in roots (Kosma et al., 2014).
CONCLUSION
Several lines of evidence from bioinformatics, molecular genetics, biochemistry, microscopic, and physiological studies suggest that Arabidopsis MYB107 positively regulates the expression of a set of suberin (but not cutin) biosynthetic genes in developing seeds, by which it controls the synthesis and assembly of seed coat suberin, thus affecting the sealing property of the seed coat. MYB107 exerts its regulatory role directly and indirectly on suberin biosynthetic genes, suggesting that it positions in an undefined regulatory network or cascade. Identifying MYB107 as a positive regulator for suberin synthesis in seeds offers a basis for further discovering the potential transcriptional network behind one of the most abundant lipid-based polymers in nature and for further elucidating its interplay and coordination with other transcriptional regulators in polyester synthesis, deposition, and assembly.
MATERIALS AND METHODS
Arabidopsis Plant Growth, Mutant Genotyping, and Staining
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized with 20% (v/v) bleach for 5 min. Seeds were stratified at 4°C for 2 to 3 d and germinated on a one-half-strength MS medium plate (Murashige and Skoog, 1962) containing 0.7% agar and 1% Suc. Seven-day-old seedlings were transferred to soil (Metro-Mix 200; SunGro) or to a hydroponic device (Tocquin et al., 2003) and grown in growth chambers under photoperiodic cycles of 16 h of light and 8 h of dark at 22°C. The T-DNA insertion mutant lines myb107-1 (SAIL_242_B04) and myb107-2 (SALK_203615) were obtained from the Arabidopsis Biological Resource Center and genotyped with the primers listed (Supplemental Table S4). The Tetrazolium Red staining of seeds and the Sudan Black B staining of root lipid polyesters were conducted for 12 and 1 h, respectively, following described methods (Beisson et al., 2007). To characterize leaf cuticles, 10-d-old seedlings or 35-d-old plants were stained with 0.05% Toluidine Blue solution for 15 min or 2 h, respectively, as described (Li et al., 2007).
RNA Extraction and qRT-PCR Analysis
Plant tissues, including 10-d-old plate-grown seedlings and 8-week-old hydroponic-grown roots, stems, leaves, flowers, and siliques, were collected for RNA extraction. Siliques at different growth stages of the soil-grown plants also were collected and dissected to obtain young seeds for RNA extraction, except that the siliques in stage I (instead of young seeds) were used directly for RNA extraction (Supplemental Fig. S12).
Total RNAs were extracted from collected plant materials using the Trizol reagent (Invitrogen) as instructed by the manufacturer, except that a Trizol-based two-step method was specifically used for extracting RNAs from the siliques and seeds (Meng and Feldman, 2010). M-MuLV reverse transcriptase (New England Biolabs) was used to synthesize cDNA from the mRNAs. qRT-PCR was performed by following the defined standards and guidelines (Udvardi et al., 2008) and using the primers listed (Supplemental Table S4). SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used for the reaction. The cycle threshold value was calculated by the CFXManager Software version 3.0 (Bio-Rad). Relative gene expression levels were calculated by following the documented standard comparative cycle threshold method (i.e. 2−ΔΔCt method; Livak and Schmittgen, 2001). Briefly, either the housekeeping gene UBQ10 or TUBULIN2 was used as the reference gene. The expression level for a given gene was first normalized to that of either UBQ10 or TUBULIN2 (only for Fig. 1D) and then was normalized to the expression level of the same gene in the defined control sample. The data were presented as the fold change.
Complementation of the myb107 Mutant
A 5,705-bp genomic fragment of the MYB107 gene, including the 3,839-bp native promoter sequence upstream of the start codon ATG and the 176-bp 3′ untranslated region sequence downstream of the stop codon TAG, was amplified using the primers PCR8LB-PMYB107-1-F and PCR8RB-MYB107-ORF-3UTR-R (Supplemental Table S4) and was cloned into the pCR8/GW/TOPO entry vector by the ligation-independent cloning method (Jeong et al., 2012). The genomic piece was then subcloned into the binary pGWB501 (Nakagawa et al., 2007) by LR reaction (Invitrogen) to generate the native promoter-driven MYB107 expression construct. Full-length cDNA of MYB107 (with or without the stop codon) was amplified with the primers ATTB1-MYB107-F and ATTB1-MYB107-R2 or ATTB1-MYB107-R1 (Supplemental Table S2) and cloned into the Gateway entry vector pDONR207 by the BP reaction (Invitrogen). They were then subcloned into pMDC32 and pGWB405 (Nakagawa et al., 2007) vectors, respectively, to generate the 35S promoter-driven MYB107 or MYB107:GFP fusion construct. The gene expression constructs were transformed into the myb107 mutant or the wild type by the floral dip method. T2 mature seeds of individual transgenic lines were stained with Tetrazolium Red as described above to check the seed coat permeability. Two-week-old seedlings, 3-week-old roots, and mixed siliques of different developmental stages of the individual T2 transgenic lines were used for RNA extraction and qRT-PCR analysis of MYB107 expression levels. Mature seeds of two individual lines were used to measure the suberin monomers.
Analyses of Suberin Monomers
For compositional determination and measurement of suberin, 500 mg of mature (dry) seeds was used to prepare cell wall residues following the described protocol (Li-Beisson et al., 2013). For depolymerization, we followed the previously described method of Franke et al. (2005) with some modifications. Briefly, 10 mg of cell wall residues was mixed with 2 mL of boron trichloride/methanol (12%, w/w; Sigma) in a screw-cap Teflon-sealed tube and incubated at 80°C for 2 h. At this step, 100 nmol of chrysin was added as the internal standard for HPLC analysis, and 5 µg heptadecanoic acid (C17) and 10 µg of 1-tricosanol (C23) were added as the internal standards for GC-MS. Two milliliters of water was added to terminate the reaction, and 3 mL of chloroform was used to extract the hydrolysates twice. The extracts were pooled and divided into two portions and then dried under a stream of N2 gas. One portion of residue was dissolved in 250 µL of 80% methanol, and 30 µL was injected for HPLC analysis. Another portion was dissolved in 50 µL of pyridine, further derivatized with 50 µL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma) at 70°C for 1 h, and then dried under a stream of N2 gas. The dried residues were dissolved in 100 µL of heptane:toluene (1:1) for GC-MS analysis.
HPLC analysis was performed with a gradient of solvent B (0.2% acetic acid in acetonitrile) in solvent A (0.2% acetic acid in water) as follows: 2 min, 15%; 35 min, 100%; 38 min, 100%; and 40 min, 15%, at a linear flow rate of 0.8 mL min−1 in a reverse-phase C18 column [Luna C18 (2), 5 μm; Phenomenex]. To quantify the content of suberin phenolics, the UV light-absorptive area of a particular peak from each sample was first normalized to that of the internal standard, then calibrated with the standard curve of ferulate, caffeate, p-coumarate, or sinapate established in the same HPLC, running using a series of concentrations of authentic chemicals after the same boron trichloride/methanol treatment and extraction.
For GC-MS, we followed the reported protocol (Franke et al., 2005). Briefly, 4-µL samples were injected via splitless mode and resolved on a DB-5MS capillary column (30 m × 0.32 mm, 0.1 mm; J&W) on an Agilent 7890A gas chromatograph equipped with an Agilent 5975C mass detector. The temperature settings were as follows: inlet, 300°C; the oven temperature program was set to 50°C for 2 min and then increased to 150°C at a rate of 10°C min−1; after maintaining it for 1 min, the temperature was increased to 310°C at the rate of 3°C min−1 and maintained for 10 min. The helium flow rate was 1.5 mL min−1. The contents of fatty acids and dicarboxylic acids were quantified using heptadecanoic acid as the standard, while ω-hydroxy fatty acids, alcohols, and diols were quantified using 1-tricosanol as the standard.
SEM and TEM Analyses
For SEM, plant tissues, including leaf, stem, flower, and silique from 5-week-old plants as well as fully matured seeds, were chemically fixed in 2.5% glutaraldehyde and 2% paraformaldehyde diluted with 0.1 m cacodylate buffer and then processed for SEM analysis. The samples were washed with the same buffer, dehydrated in a graded ethanol series, and dried with a Tousimis critical-point drying apparatus (Tousimis Research). All dried samples were mounted onto aluminum stubs, sputter coated with gold, and imaged with a Quanta 200 environmental scanning electron microscope (FEI).
For TEM, mature and dried seeds were imbibed in distilled water for 24 h and then carefully pricked with a microneedle. Plant tissues, including the pricked seeds and leaves from 5-week-old plants, were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde diluted with 0.1 m cacodylate buffer and postfixed in 1% osmium tetroxide diluted with the same buffer. Samples then were dehydrated through a graded acetone series and embedded in Spurr’s epoxy resin (Electron Microscopy Science). All the samples were sectioned with an Ultracut E microtome (Reichert-Jung, Cambridge Instruments). Thin sections (90–100 nm) were mounted onto 200-mesh copper grids, treated with 10% H2O2 for 10 min, and then stained with 1% aqueous uranyl acetate and Sato lead. Three to five sections for each sample were examined and imaged at 120 kV with a JEM-1400 transmission electron microscope (JEOL). Representative images are presented.
Abiotic Stress Treatment
Surface-sterilized wild-type (Col-0) and myb107 seeds were stratified and germinated on a one-half-strength MS plate supplemented with 150 mm NaCl, 300 mm mannitol, and 5 mm H2O2. After 7 d of growth under light, the numbers of seedlings with two fully established cotyledons were counted, and the establishment rates were calculated. Three plates were set for each treatment (as three biological replicates), and approximately 80 seeds were plated in each repeat.
RNA-Seq
The mixed siliques of 8-week-old myb107 and wild-type (Col-0) plants were collected. RNAs were extracted using the method described above. The assessment of purity and concentration of each RNA sample, and the strand-specific RNA-seq library preparation, were conducted using the Polar Genomics service following a protocol described by Zhong et al. (2011). Sequencing was done using the Illumina HiSeq2500 platform via 100-bp single-end reads of multiplexed RNA samples. RNA-seq reads first were aligned to the rRNA database (Quast et al., 2013) using Bowtie (Langmead et al., 2009), and those mapped were excluded for downstream analysis. The resulting cleaned reads were aligned to the Arabidopsis genome sequence (TAIR10) using TopHat (Trapnell et al., 2009) allowing one mismatch. Following these alignments, raw counts for each Arabidopsis gene were derived and normalized to reads per kilobase of exon model per million mapped reads. The differentially expressed genes were identified with the integrated Cuffdiff program (Langmead et al., 2009; Trapnell et al., 2010) based on a false discovery rate-adjusted P value (i.e. q value) at the cutoff of 0.1 and fold change greater than or less than 2.
Confocal Microscopy of MYB107-GFP
Both pGWB405:MYB107 (MYB107:GFP) and the empty vector pCAMBIA1302 (free GFP) were transformed into Agrobacterium tumefaciens strain GV3101. Six-week-old Nicotiana tabacum plants were infiltrated with an A. tumefaciens strain harboring each of the constructs. Fluorescence images were captured via a Leica TCS SP5 Laser Scan Confocal Microscope with excitation at 488 nm; an emission signal between 493 and 560 nm was collected for GFP fluorescence, and excitation at 496 nm and an emission signal of 630 to 720 nm were collected for chlorophyll autofluorescence.
Y1H Assay
Full-length cDNA of MYB107 (with the stop codon) was amplified using primers ATTB1-MYB107-F and ATTB1-MYB107-R2 (Supplemental Table S2). A cDNA fragment of MYB107 encoding the DNA-binding domain (MYB107BD) was amplified using primers ATTB1-MYB107-BD-F and ATTB2-MYB107-BD-R (Supplemental Table S2). Both cDNAs first were cloned into pDONR207 by the BP reaction (Invitrogen) and then subcloned into the pDESTAD-2µ vector by the LR reaction (Invitrogen) to generate the Y1H-TF prey constructs (Reece-Hoyes et al., 2011). The promoters of suberin biosynthetic genes were amplified using the primers listed (Supplemental Table S4), first cloned into pDONR P4P1R (Invitrogen) by the BP reaction, and then subcloned into the pMW#3 vector to generate the pMW#3:DNA bait constructs. The Y1H experiment was performed following the methods described (Deplancke et al., 2006; Reece-Hoyes et al., 2011) with slight modifications. Briefly, pMW#3:DNA bait constructs first were transformed and integrated into the genome of the yeast strain Y1H-aS2. The positive colonies selected from SC(−Ura) medium were used for the self-activation test by streaking them onto the SC(−Ura) plate spread with 100 µL of 30 mg mL−1 X-Gal (Sigma). The colonies showing no blue coloration after streaking for 8 d were picked for culturing and retransformed with either the TF prey constructs or pDESTAD-2µ empty vector (as the control). The resulting positive colonies selected from the SC(−Trp,−Ura) medium were restreaked onto the SC(−Trp,−Ura) plate spread with 100 µL of 30 mg mL−1 X-Gal and incubated for 8 d before then capturing the color changes. Yeast harboring a well-characterized N-terminal fragment of mutator-like transposase FHY3 (FHY3N) and its targeting promoter of FHL (pFHL; Lin et al., 2007) was used as the positive control. Meanwhile, yeast harboring MYB107 or MYB107BD with the pFHL DNA construct was used as the negative controls.
ChIP-qPCR Analysis
The pCAMBIA1302 (p35S::GFP) and pGWB405:MYB107 (p35S::MYB107:GFP) constructs were transiently transformed into 4-d-old Arabidopsis wild-type (Col-0) seedlings by A. tumefaciens-mediated transformation using vacuum infiltration (Marion et al., 2008). One gram of seedlings after transformation for 3 d was collected and cross-linked using 40 mL of cross-linking buffer (50 mm KH2PO4/K2HPO4, pH 5.8, and 1% formaldehyde) for 15 min under vacuum. The cross-linking was then quenched using quenching buffer (50 KH2PO4/K2HPO4, pH 5.8, and 0.3 m Gly) for 5 min by vacuum. The materials were ground into fine power. The chromatin isolation and nuclei lysis were performed as described previously (He et al., 2013). Chromatin was then sonicated on ice eight times for 10 s each (30-s interval) with power setting at 2 on a Fisher Scientific 550 sonic dismembrator to shear DNA into 500- to 1,500-bp fragments (He et al., 2013). After dilution to 1.5 mL using ChIP dilution buffer, 3% (45 µL) of the sheared sample was used as the input fraction. The rest was precleared with 20 µL of coated Dynabeads Protein G beads (1003D; Thermo Fisher Scientific) for 3 h, and 10 µg of Living Colors monoclonal GFP antibody (JL-8; Clontech) was added and incubated overnight. The protein-DNA complexes were captured by incubating with 30 µL of newly added coated Dynabeads Protein G beads for 2 h at 4°C. The beads were washed with low- and high-salt buffer and LiCl wash buffer and eluted with 100 µL of elution buffer as described (He et al., 2013). The elution was then incubated overnight at 65°C to reverse cross-linking. Four microliters of RNase (10 mg mL−1) was added and incubated for 1 h at 37°C, and 4 µL of proteinase K (20 mg mL−1) was added and incubated for another 2 h at 45°C. The DNA was purified with the PureLink PCR Purification Kit (K310001; Thermo Fisher Scientific). Eluted solutions were used for qPCR. UBQ10 was used as a negative control. Three or four pairs of primers were synthesized for each tested gene with matching to their promoter and/or coding regions (Fig. 8). The primer sequences are listed in Supplemental Table S4. Three biological experiments were performed. Data are normalized by dividing qPCR signal from the ChIP samples with qPCR signal from the input samples (i.e. percentage of input) following the manufacturer’s instructions (https://www.thermofisher.com/us/en/home/life-science/epigenetics-noncoding-rna-research/chromatin-remodeling/chromatin-immunoprecipitation-chip/chip-analysis.html).
Accession Numbers
Sequence data of the gene investigated in this study can be found in The Arabidopsis Information Resource under accession number At3g02940 (MYB107).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. In silico expression patterns of HHT and DCF in different tissues.
Supplemental Figure S2. In silico expression patterns of MYB107 in different tissues.
Supplemental Figure S3. Characterization of the myb107-2 mutant.
Supplemental Figure S4. Morphologies of wild-type and myb107 plants.
Supplemental Figure S5. Cosegregation of the T-DNA insertion of myb107 with the enhanced tetrazolium staining phenotype.
Supplemental Figure S6. Complementation of the myb107 mutant by pMYB107::MYB107.
Supplemental Figure S7. GC-MS profile of the aliphatic monomers from seed coat cell wall of the wild type and myb107.
Supplemental Figure S8. Aromatic and aliphatic monomers of suberin in roots.
Supplemental Figure S9. Microscopic observation of suberin ultrastructures in seed coats of the wild type and myb107.
Supplemental Figure S10. Phenotypic characterization of wild-type and myb107 plants.
Supplemental Figure S11. Expression of suberin biosynthetic genes in MYB107 complementation lines.
Supplemental Figure S12. Sample collection of Arabidopsis young seeds at the defined developmental stages.
Supplemental Figure S13. Proposed model of MYB107-mediated regulation of suberin biosynthesis.
Supplemental Table S1. Gene coexpression analysis using HHT/ASFT (At5g41040) as the bait.
Supplemental Table S2. Gene coexpression analysis using MYB107 (At3g02940) as the bait.
Supplemental Table S3. Expression of TT genes in the wild type and myb107 detected by RNA-seq.
Supplemental Table S4. PCR primer sequences used in this study.
Supplemental Data Set S1. Complete RNA-seq data set of the wild type (Col-0) and the myb107 mutant.
Supplementary Material
Acknowledgments
We thank Drs. Lifang Zhang and Doreen Ware at Cold Spring Harbor Laboratory for advice on the Y1H assay system and Jiapei Yan and Zhixue Wang at Cornell University for advice on the ChIP experiment.
Glossary
- TF
transcription factor
- qRT
quantitative real-time
- Col-0
Columbia-0
- MS
Murashige and Skoog
- GC-MS
gas chromatography-mass spectrometry
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
- RNA-seq
RNA sequencing
- DAF
days after flowering
- Y1H
yeast one-hybrid
- ChIP
chromatin immunoprecipitation
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactopyranoside
- qPCR
quantitative PCR
Footnotes
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy (grant no. DEAC0298CH10886 to C.-J.L.) and the National Science Foundation (grant no. MCB-1051675 to C.-J.L).
Articles can be viewed without a subscription.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen TG, Barberon M, Geldner N (2015) Suberization: the second life of an endodermal cell. Curr Opin Plant Biol 28: 9–15 [DOI] [PubMed] [Google Scholar]
- Appelhagen I, Thiedig K, Nordholt N, Schmidt N, Huep G, Sagasser M, Weisshaar B (2014) Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240: 955–970 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Rycke RD, Viane R, Inze D (2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113: 139–148 [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M (2012) Solving the puzzles of cutin and suberin polymer biosynthesis. Curr Opin Plant Biol 15: 329–337 [DOI] [PubMed] [Google Scholar]
- Bernards MA. (2002) Demystifying suberin. Can J Bot 80: 227–240 [Google Scholar]
- Bernards MA, Razem FA (2001) The poly(phenolic) domain of potato suberin: a non-lignin cell wall bio-polymer. Phytochemistry 57: 1115–1122 [DOI] [PubMed] [Google Scholar]
- Bessire M, Chassot C, Jacquat AC, Humphry M, Borel S, Petétot JM, Métraux JP, Nawrath C (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7: 202–209 [DOI] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53: 53–64 [DOI] [PubMed] [Google Scholar]
- Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F (2009) CYP86B1 is required for very long chain ω-hydroxyacid and α,ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol 150: 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJ (2006) Gateway-compatible yeast one-hybrid screens. Cold Spring Harb Protoc 2006: doi/10.1101/pdb.prot459 [DOI] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, et al. (2010) Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol 153: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L (2005) Apoplastic polyesters in Arabidopsis surface tissues: a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Franke R, Höfer R, Briesen I, Emsermann M, Efremova N, Yephremov A, Schreiber L (2009) The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J 57: 80–95 [DOI] [PubMed] [Google Scholar]
- Gou JY, Yu XH, Liu CJ (2009) A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc Natl Acad Sci USA 106: 18855–18860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça J, Santos S (2007) Suberin: a biopolyester of plants’ skin. Macromol Biosci 7: 128–135 [DOI] [PubMed] [Google Scholar]
- Graça J, Schreiber L, Rodrigues J, Pereira H (2002) Glycerol and glyceryl esters of ω-hydroxyacids in cutins. Phytochemistry 61: 205–215 [DOI] [PubMed] [Google Scholar]
- He Y, Sidhu G, Pawlowski WP (2013) Chromatin immunoprecipitation for studying chromosomal localization of meiotic proteins in maize. Methods Mol Biol 990: 191–201 [DOI] [PubMed] [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R (2008) The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot 59: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JY, Yim HS, Ryu JY, Lee HS, Lee JH, Seen DS, Kang SG (2012) One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol 78: 5440–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Molina I, Ohlrogge JB, Pollard M (2012) Identification of an Arabidopsis fatty alcohol:caffeoyl-coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiol 160: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Murmu J, Razeq FM, Santos P, Bourgault R, Molina I, Rowland O (2014) AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J 80: 216–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC (2009) Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J 60: 462–475 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi/10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA (2009) Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J 59: 553–564 [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD (2008) Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Meng L, Feldman L (2010) A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J 5: 183–186 [DOI] [PubMed] [Google Scholar]
- Mohamed-Yasseen Y, Barringer SA, Splittstoesser WE, Costanza S (1994) The role of seed coats in seed viability. Bot Rev 60: 426 [Google Scholar]
- Moire L, Schmutz A, Buchala A, Yan B, Stark RE, Ryser U (1999) Glycerol is a suberin monomer: new experimental evidence for an old hypothesis. Plant Physiol 119: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moïse JA, Han S, Gudynaitę-Savitch L, Johnson DA, Miki BLA (2005) Seed coats: structure, development, composition, and biotechnology. In Vitro Cell Dev Biol 41: 620–644 [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M (2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67: 2597–2610 [DOI] [PubMed] [Google Scholar]
- Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M (2009) Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol 151: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Ohlrogge JB, Pollard M (2008) Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J 53: 437–449 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway Binary Vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Schreiber L, Franke RB, Geldner N, Reina-Pinto JJ, Kunst L (2013) Apoplastic diffusion barriers in Arabidopsis. The Arabidopsis Book 11: e0167, doi/10.1199/tab.0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M (2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Ouellet M, Nafisi M, Baidoo EE, Benke P, Stranne M, Mukhopadhyay A, Keasling JD, Sakuragi Y, et al. (2012) Arabidopsis Deficient in Cutin Ferulate encodes a transferase required for feruloylation of ω-hydroxy fatty acids in cutin polyester. Plant Physiol 158: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, Walhout AJ (2011) Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat Methods 8: 1059–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb J, Lee SW, Mohan R, Kolattukudy PE (1991) Chemical characterization of stress-induced vascular coating in tomato. Plant Physiol 97: 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupasinghe SG, Duan H, Schuler MA (2007) Molecular definitions of fatty acid hydroxylases in Arabidopsis thaliana. Proteins 68: 279–293 [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L. (2010) Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci 15: 546–553 [DOI] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, Bernier G, Périlleux C (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanath SJ, Kosma DK, Pulsifer IP, Scandola S, Pascal S, Joubès J, Dittrich-Domergue F, Lessire R, Rowland O, Domergue F (2013) Suberin-associated fatty alcohols in Arabidopsis: distributions in roots and contributions to seed coat barrier properties. Plant Physiol 163: 1118–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc Natl Acad Sci USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Molina I, Ranathunge K, Castillo IQ, Rothstein SJ, Reed JW (2014) ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26: 3569–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16: 227–233 [DOI] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ (2011) High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.