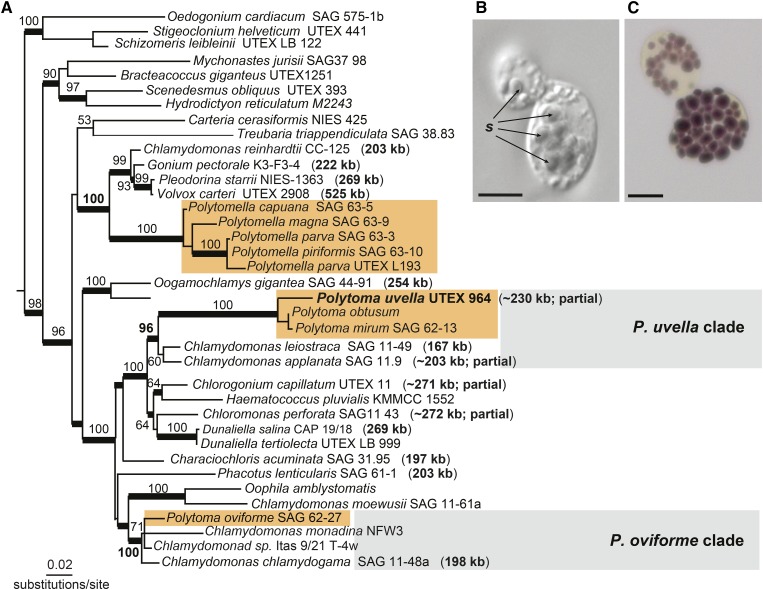
The plastid genome of the nonphotosynthetic green alga Polytoma uvella has a reduced coding capacity but is bloated with repeats. This is in striking contrast with its colorless relative, Polytomella, that lost its plastid genome.
Abstract
The loss of photosynthesis is frequently associated with parasitic or pathogenic lifestyles, but it also can occur in free-living, plastid-bearing lineages. A common consequence of becoming nonphotosynthetic is the reduction in size and gene content of the plastid genome. In exceptional circumstances, it can even result in the complete loss of the plastid DNA (ptDNA) and its associated gene expression system, as reported recently in several lineages, including the nonphotosynthetic green algal genus Polytomella. Closely related to Polytomella is the polyphyletic genus Polytoma, the members of which lost photosynthesis independently of Polytomella. Species from both genera are free-living organisms that contain nonphotosynthetic plastids, but unlike Polytomella, Polytoma members have retained a genome in their colorless plastid. Here, we present the plastid genome of Polytoma uvella: to our knowledge, the first report of ptDNA from a nonphotosynthetic chlamydomonadalean alga. The P. uvella ptDNA contains 25 protein-coding genes, most of which are related to gene expression and none are connected to photosynthesis. However, despite its reduced coding capacity, the P. uvella ptDNA is inflated with short repeats and is tens of kilobases larger than the ptDNAs of its closest known photosynthetic relatives, Chlamydomonas leiostraca and Chlamydomonas applanata. In fact, at approximately 230 kb, the ptDNA of P. uvella represents the largest plastid genome currently reported from a nonphotosynthetic alga or plant. Overall, the P. uvella and Polytomella plastid genomes reveal two very different evolutionary paths following the loss of photosynthesis: expansion and complete deletion, respectively. We hypothesize that recombination-based DNA-repair mechanisms are at least partially responsible for the different evolutionary outcomes observed in such closely related nonphotosynthetic algae.
The transition from a photosynthetic to a nonphotosynthetic lifestyle has occurred many times and in disparate lineages throughout eukaryotic evolution (Figueroa-Martinez et al., 2015). With few exceptions (Gornik et al., 2015), nonphotosynthetic algae and plants have a plastid and an associated genome, which is typically small, compact, and has reduced coding capacity (Table I). For example, the nonphotosynthetic parasitic green alga Helicosporidium sp. has one of the smallest and most compact plastid DNAs (ptDNAs) observed from a protist: 37.4 kb, approximately 95% coding, and no genes related to photosynthesis (de Koning and Keeling, 2006). Even more reduced ptDNA architectures have been found in the colorless, plastid-bearing apicomplexans, such as Plasmodium falciparum (34.2 kb; Wilson et al., 1996) and Eimeria tenella (34.7 kb; Cai et al., 2003), causative agents of malaria and coccidiosis, respectively. The most extreme example of plastid genomic reduction is the complete loss of ptDNA, as has been observed in certain apicomplexan parasites (Janouškovec et al., 2015), the parasitic land plant Rafflesia lagascae (Molina et al., 2014), and the colorless genus Polytomella (Smith and Lee, 2014), a close relative of the model green alga Chlamydomonas reinhardtii (family Chlamydomonadaceae, class Chlorophycea, Chlorophyta; Smith and Lee, 2014).
Table I. Plastid genome features of diverse nonphotosynthetic algae.
| Species | GenBank Accession No. | Size | Protein-Coding Genes | Coding DNAa | AT-Coding Regions | AT-Noncoding Regions | Average Intergenic Distanceb |
|---|---|---|---|---|---|---|---|
| kb | % | % | % | bp | |||
| Polytoma uvella | KX828177.1 (this work) | ∼230 | 25 | 26.8 | 64.4 | 80.8 | 2,967 |
| Prototheca wickerhamiic | KJ001761.1 | 55.6 | 40 | 83.17 | 66 | 82.7 | 134.6 |
| Helicosporidium sp.c | NC_008100.1 | 37.4 | 26 | 95.2 | 72.4 | 86.7 | 34.2 |
| Euglena longa | NC_002652.1 | 73.3 | 46 | 84.5 | 76.1 | 85.4 | 136.45 |
| Choreocolax polysiphoniaec | NC_026522.1 | 90.2 | 71 | 70.5 | 75.7 | 88.8 | 272 |
| Cryptomonas paramecium | NC_013703.1 | 77.7 | 82 | 87.7 | 60.7 | 70 | 84.3 |
| Plasmodium falciparumc | LN999985.1 | 34.2 | 30 | 95.8 | 85.4 | 94 | 21.6 |
| Eimeria tenellac | NC_004823.1 | 34.7 | 28 | 94.5 | 78.6 | 93.3 | 29.9 |
The percentage of coding DNA includes intronic regions, tRNA, and rRNA genes.
tRNA- and rRNA-coding regions were considered as genes in our calculations of intergenic distances.
Parasitic or pathogenic species.
In addition to Polytomella, the Chlamydomonadaceae includes a number of other nonphotosynthetic species in the genus Polytoma (Fig. 1A; Nedelcu, 2001; Nakada et al., 2008; Figueroa-Martinez et al., 2015). Despite being classified in the same family with the polyphyletic genus Chlamydomonas (Pröschold et al., 2001; Nakada et al., 2008) and having very similar names and free-living heterotrophic lifestyles, it is well documented that Polytomella and Polytoma lost photosynthesis independently of one another, and unlike the former, members of the latter genus are known to encode at least some plastid genes (Nedelcu, 2001; Figueroa-Martinez et al., 2015). Furthermore, phylogenetic analyses using either nuclear 18S rRNA (Pröschold et al., 2001; Nakada et al., 2008; Figueroa-Martinez et al., 2015) or plastid 16S rRNA (Nedelcu, 2001; Vernon et al., 2001; Figueroa-Martinez et al., 2015) and elongation factor Tu (tufA; Vernon et al., 2001) sequences have consistently resolved two distinct Polytoma groups: the Polytoma uvella clade and the Polytoma oviforme clade. Clearly, the two Polytoma lineages have evolved from different photosynthetic chlamydomonadalean lineages (Nedelcu, 2001; Figueroa-Martinez et al., 2015).
Figure 1.
A, Maximum likelihood tree estimated from a concatenated data set of 18S rRNA (nuclear) and 16S rRNA (plastid) sequences. Numbers near branches represent RAxML (GTRGAMMA substitution model) bootstrap values greater than 50%. Thick branches are supported by posterior probabilities of 0.95 or greater. Branch lengths are proportional to the number of substitutions per site. The sizes of the plastid genomes (including partial sequences) are indicated in parentheses. Colored rectangular boxes highlight three independent nonphotosynthetic lineages. The absence of plastid 16S rRNA sequences of the five Polytomella spp. was treated as missing data during the phylogenetic estimation. B, Light microscopy image of P. uvella cells, with starch grains (s) appearing as opaque solid bodies inside the plastids. C, P. uvella cells treated with Lugol’s reagent (aqueous solution of elemental iodine and potassium iodide used for starch detection). The arrangement of the starch grains stained with iodine (dark-purple bodies) maps to the cup-like shape of the single plastid. The presence of starch granules in plastids of different Polytoma spp. has been reported in previous studies (Lang, 1963; Siu et al., 1976; Gaffal and Schneider, 1980). Bars = 5 μm.
Consistent with previous studies, our maximum likelihood analysis of concatenated nuclear 18S rRNA and plastid 16S rRNA sequences (Fig. 1A) recovered P. uvella (strain UTEX 964), the focus of this study, within a clade containing the photosynthetic taxa Chlamydomonas leiostraca and Chlamydomonas applanata (96% bootstrap support), whereas P. oviforme branched with Chlamydomonas chlamydogama and Chlamydomonas monadina NFW3 (100% bootstrap support). Also, the different Polytomella spp. (no data for the plastid 16S rRNA set) were in the same clade with C. reinhardtii and Volvox carteri (100% bootstrap support) separated from the two Polytoma branches. A phylogenetic analysis of a broader taxon sampling of chlamaydomonadalean 18S rRNA sequences recovered a consistent tree topology (Supplemental Fig. S2; Figueroa-Martinez et al., 2015). Thus, diverse phylogenetic evidence strongly suggests that photosynthesis was lost at least three independent times in the Chlamydomonadaceae: twice within the Polytoma genus and once in Polytomella (Nedelcu, 2001; Nakada et al., 2008; Figueroa-Martinez et al., 2015).
Given the relatively close phylogenetic affiliation to C. reinhardtii and their independent divergence from photosynthetic lineages, Polytoma and Polytomella represent an outstanding duo for studying independent losses of photosynthesis (Fig. 1A; Nakada et al., 2008; Figueroa-Martinez et al., 2015). Moreover, both Polytoma and Polytomella are free living, making them distinct from other well-studied groups of colorless algae, most of which lost photosynthesis as a consequence of parasitic, pathogenic, or symbiotic lifestyles, such as the trebouxiophycean green algae Helicosporidium sp. and Prototheca wickerhamii (Nadakavukaren and McCracken, 1977; Tartar et al., 2002) and the red alga Choreocolax polysiphoniae (Salomaki et al., 2015).
Although several Polytoma spp., including P. uvella and P. oviforme, are known to possess ptDNA, the size and content of their plastid genomes have not yet been explored. One might expect Polytoma spp. to have highly reduced plastid genomes, similar to those of Helicosporidium sp. and P. wickerhamii, or genomes that are on route to being entirely lost, following in the footsteps of Polytomella. However, here we show that the opposite is true. The ptDNA of P. uvella is not only larger than those of its photosynthetic counterparts in C. leiostraca and C. applanata, it is the largest ptDNA observed from a colorless, plastid-bearing eukaryote. This finding raises questions about how and why the plastid genomes of two closely related lineages, Polytoma and Polytomella, could have followed such divergent evolutionary paths after losing photosynthesis: genome expansion versus complete deletion.
RESULTS AND DISCUSSION
Unprecedented Plastid Genome Inflation in a Nonphotosynthetic Alga
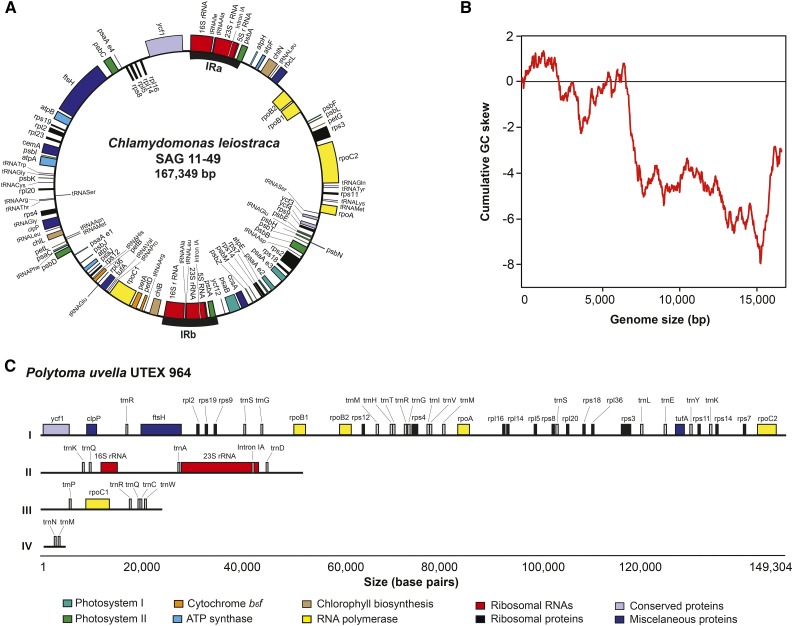
To better understand the evolutionary loss of photosynthesis in free-living algae, we sequenced the entire C. leiostraca plastid genome and large amounts of the P. uvella ptDNA. The C. leiostraca ptDNA (167 kb; Fig. 2A) was easily assembled from paired-end whole-genome shotgun Illumina data (Table II) and is smaller than the partial ptDNA sequence of its close relative C. applanata (203 kb; Fig. 1A; Lemieux et al., 2015). The ptDNAs of these two photosynthetic species have almost identical gene contents and organizations, and their overall architectures are similar to those reported from other photosynthetic chlamydomonadalean algae (Supplemental Fig. S1; Supplemental Table S1). A GC skew analysis indicates that the C. leiostraca ptDNA replicates bidirectionally (i.e. theta replication; Fig. 2B).
Figure 2.
A and C, Maps of the plastid genomes of C. leiostraca (A) and P. uvella (C). The C. leiostraca plastid genome (ptDNA) is depicted as a circular mapping molecule. In the case of the P. uvella ptDNA, the four individual scaffolds assembled (4.2, 23.9, 52.5, and 149.5 kb) are indicated (I–IV, respectively). The color code indicates the functional classification of the genes contained in both genomic sequences. B, The cumulative GC skew analysis of the C. leiostraca ptDNA reveals a bimodal profile typical of bidirectional theta replication.
Table II. Basic statistics of the Illumina assemblies from P. uvella and C. leiostraca.
Numbers in parentheses indicate the number of genes/sequences considered for each estimation.
| Parameter | P. uvella | C. leiostraca |
|---|---|---|
| No. of Illumina paired-end reads (100 bp) | 3.4 × 107 × 2 | 4.0 × 107 × 2 |
| No. of contigs of 500 nucleotides or greatera | 15,240 | 10,222 |
| Average contig sizea | 4,531 nucleotides | 9,122 nucleotides |
| Largest contiga | 102,330 nucleotides | 167,968 nucleotides |
| Plastid genome cumulative size | ∼230 kb | 167.4 kb |
| Illumina average coverage (reads per nucleotide ± sd) | ||
| Plastid protein-coding genes | 5,776 ± 601 (25) | 6,239 ± 620 (65)b |
| Plastid rRNA genes | 5,917 ± 716 (2)c | 11,851 ± 1,311 (9)d |
| Plastid tRNA genese | 6,274 ± 1,230 (27) | 6,282 ± 828 (23) |
| Plastid scaffolds/complete genome | 6,662 ± 441 (4)f | 6,753 ± 766 |
| Mitochondrial protein-coding genes | 5,813 ± 970 (7) | 8,326 ± 228 (7) |
| Mitochondrial rRNA genes | 6,997 ± 911 (2)g | 8,320 ± 617 (2) |
| Mitochondrial complete genome | 6,047 ± 1,268 | 8,163 ± 955 |
| Nuclear sequences | 38 ± 8 (20) | 67 ± 23 (10) |
Using Ray assembler with a k-mer size of 31. bCoverage calculated in protein-coding genes present in the long and short single-copy regions. cOnly considering plastid rRNA-coding regions (see Supplemental Fig. S3). dGenes localized in the C. leiostraca ptDNA inverted-repeat regions: three rRNAs, two tRNAs, and four open reading frames (ORFs). etRNA genes located in the long and short single-copy regions. fThe average coverage estimations by read mapping in the P. uvella ptDNA intragenic regions are ambiguous given the presence of numerous short repeats. gOnly considering mitochondrial rRNA-coding regions.
Unlike that of C. leiostraca, the P. uvella ptDNA assembly was fraught with challenges because the genome is dense with short repeats. The assembly of paired-end Illumina data from P. uvella resulted in 25 contigs containing typical plastid-localized genes, but most of these contigs were short (average size, approximately 4 kb) and contained a single or a partial gene sequence, bordered by direct or inverted AT-rich repeats. Numerous attempts to bridge the contigs by PCR using diverse primer sets based on conserved P. uvella ptDNA-coding regions were unsuccessful. However, real-time sequencing (PacBio) of P. uvella DNA ultimately allowed us to bridge the 25 Illumina contigs into four scaffolds of 4.2 , 23.9, 52.5, and 149.3 kb (Fig. 2C), giving a cumulative length of 229.9 kb.
Illumina read coverage of coding regions was similar among the four scaffolds (approximately 5,700 reads per nucleotide; Table II; Supplemental Table S3) and was larger than that of known nucleus-located genes and large nuclear contigs (approximately 38 reads per nucleotide; Table II; Supplemental Table S3). Similar read coverage values were estimated when considering the complete sequences of the Polytoma ptDNA scaffolds (approximately 5,600 reads per nucleotide; Table II). No nuclear or mitochondrial sequences were found adjacent to ptDNA genes on the PacBio reads. Additionally, the AT content of the four scaffolds containing plastid genes (77%) is different from that of nuclear (42%) and mitochondrial (45%) sequences. Together, these data suggest that the putative plastid contigs represent bona fide ptDNA and components of a single, intact plastid genome rather than nuclear or mitochondrial ptDNA-like sequences, which are known to be very rare, if not absent, in green algae (Smith, 2011; Smith et al., 2011). Nevertheless, the highly repetitive intergenic ptDNA flanking these scaffolds prevented us from accurately linking them together, which is a recurring theme in chlamydomonadalean plastid genomics (Del Vasto et al., 2015).
Remarkably, the ptDNA of P. uvella is larger than those of C. leiostraca and C. applanata. To the best of our knowledge, this is the first example of a nonphotosynthetic plant or alga having a bigger plastid genome than its closest known photosynthetic relative(s). If anything, previous work has proven that the forfeiting of photosynthesis almost always results in a reduction of plastid genome size (Yan et al., 2015; Naumann et al., 2016). To put this in perspective, the P. uvella ptDNA is at least 6 times larger than that of Helicosporidium sp. (de Koning and Keeling, 2006), despite both algae having near identical gene contents (Table III), and 12 times bigger than that of the nonphotosynthetic orchid Epipogium roseum (Schelkunov et al., 2015).
Table III. Gene content in plastid genomes of nonphotosynthetic algae.
Pu, P. uvella; He, Helicosporidium sp.; Pw, P. wickerhamii; El, E. longa; Cp, C. paramecium; Pf, P. falciparum; Co, C. polysiphoniae.
| Parameter | Gene | Pu | He | Pw | Ela | Cpb | Pfb | Co |
|---|---|---|---|---|---|---|---|---|
| Ribosomal large subunit | rpl1 | x | x | x | ||||
| rpl2 | x | x | x | x | x | x | ||
| rpl3 | x | x | ||||||
| rpl4 | x | x | x | |||||
| rpl5 | x | x | x | x | x | x | ||
| rpl6 | x | x | x | |||||
| rpl11 | x | x | ||||||
| rpl12 | x | x | x | x | x | |||
| rpl13 | x | x | ||||||
| rpl14 | x | x | x | x | x | x | x | |
| rpl16 | x | x | x | x | x | x | x | |
| rpl18 | x | x | ||||||
| rpl19 | x | x | x | |||||
| rpl20 | x | x | x | x | x | x | ||
| rpl21 | x | x | ||||||
| rpl22 | x | x | ||||||
| rpl23 | x | x | x | x | x | |||
| rpl24 | x | |||||||
| rpl27 | x | x | ||||||
| rpl29 | x | x | ||||||
| rpl31 | x | x | ||||||
| rpl32 | x | x | x | |||||
| rpl33 | x | |||||||
| rpl34 | x | |||||||
| rpl35 | x | |||||||
| rpl36 | x | x | x | x | x | x | x | |
| Ribosomal small subunit | rps2 | x | x | x | x | x | ||
| rps3 | x | x | x | x | x | x | x | |
| rps4 | x | x | x | x | x | x | x | |
| rps5 | x | x | x | |||||
| rps6 | x | |||||||
| rps7 | x | x | x | x | x | x | x | |
| rps8 | x | x | x | x | x | x | x | |
| rps9 | x | x | x | x | x | |||
| rps10 | x | x | ||||||
| rps11 | x | x | x | x | x | x | x | |
| rps12 | x | x | x | x | x | x | x | |
| rps13 | x | x | ||||||
| rps14 | x | x | x | x | x | x | ||
| rps16 | x | x | ||||||
| rps17 | x | x | ||||||
| rps18 | x | x | x | |||||
| rps19 | x | x | x | x | x | x | x | |
| rps20 | x | x | ||||||
| Transcription/translation | rpoA | x | x | x | x | x | ||
| rpoB1 | x | x | x | x | x | x | x | |
| rpoB2 | x | |||||||
| rpoC1 | x | x | x | x | x | x | x | |
| rpoC2 | x | x | x | x | x | x | x | |
| infA | x | |||||||
| infB | x | |||||||
| infC | x | |||||||
| tsf | x | x | ||||||
| tufA | x | x | x | x | x | x | x | |
| ATP synthase | atpA | x | x | |||||
| atpB | x | x | ||||||
| atpD | x | |||||||
| atpE | x | x | ||||||
| atpF | x | |||||||
| atpG | x | |||||||
| atpH | x | x | ||||||
| atpI | x | x | ||||||
| Other proteins | accA | x | ||||||
| accB | x | |||||||
| accD | x | x | ||||||
| acpP | x | x | ||||||
| cbbX | x | |||||||
| cemA | x | |||||||
| chlI | x | |||||||
| clpC | x | x | ||||||
| clpP | x | x | x | |||||
| cysT | x | x | ||||||
| dnaB | x | |||||||
| dnaK | x | x | ||||||
| fabH | x | |||||||
| ftsH | x | x | x | x | ||||
| groEL | x | |||||||
| hisS | x | |||||||
| ilvB | x | x | ||||||
| ilvH | x | x | ||||||
| minD | x | |||||||
| odpA | x | |||||||
| odpB | x | |||||||
| petF | x | x | ||||||
| psbA | x | |||||||
| rbcL | x | x | ||||||
| rbcS | x | |||||||
| rne | x | |||||||
| roaA | x | |||||||
| secA | x | x | ||||||
| secY | x | x | ||||||
| sufB | x | |||||||
| sufC | x | |||||||
| tatC | x | |||||||
| tilS | x | x | ||||||
| trpA | x | |||||||
| trpG | x | |||||||
| ycf1 | x | x | x | |||||
| ycf13 | x | |||||||
| ycf16 | x | |||||||
| ycf19 | x | |||||||
| ycf20 | x | |||||||
| ycf24 | x | x | ||||||
| ycf29 | x | |||||||
| ycf67 | x | |||||||
| Ribosomal RNAs | rrnF | x | x | x | x | x | ||
| rrnL | x | x | x | x | x | x | x | |
| rrnS | x | x | x | x | x | x | x | |
| Transfer RNAs | 27 | 25 | 27 | 28 | 29 | 34 | 24 |
Secondary plastid of green algal origin. bSecondary plastid of red algal origin.
We did not identify a quadripartite structure in the P. uvella plastid genome. Large inverted repeats are found in the ptDNAs from close photosynthetic relatives of P. uvella but are absent in some nonphotosynthetic algae, including Helicosporidium sp. and P. wickerhamii (de Koning and Keeling, 2006; Yan et al., 2015). Read coverage of the plastid rRNA-coding regions from P. uvella (approximately 5,900 reads per nucleotide; for details, see Table II) was similar to that of ptDNA protein- and tRNA-coding regions (approximately 5,700 and 6,300 reads per nucleotide, respectively), suggesting that the quadripartite structure might have been lost. In contrast to P. uvella, the read coverage of the C. leiostraca plastid rRNA-coding regions (approximately 11,800 reads per nucleotide) almost doubles the values estimated for plastid protein-coding regions (approximately 6,200 reads per nucleotide; Table II), which is consistent with the quadripartite structure of the C. leiostraca ptDNA (Fig. 2A). However, in P. uvella, we were not able to bridge the contig containing the rRNA-coding regions to any other contigs, leaving open the possibility that the genome does indeed have a quadripartite structure, which, if proven true, would make it much larger than our current assessment.
Long AT-Rich Repeats Permeate throughout the P. uvella Plastid Genome
The large size of the P. uvella ptDNA is a reflection of expanded intergenic regions, which on average are 2.9 kb and in some cases exceed 10 kb, making them at least 29 times larger than those from other nonphotosynthetic algae (Table I). Comprehensive analyses of the AT-rich intergenic regions of the P. uvella ptDNA, including six-frame translations and subsequent BLAST searches (see “Materials and Methods”), did not reveal obvious pseudogenes. The vast majority of the few BLASTP and PSI-BLAST hits were very short, fragmented, and gapped matches to hypothetical proteins but not to products of typical green algal plastid genes.
The P. uvella ptDNA has an overall noncoding content of 73%, which is on par with other bloated plastid genomes, including those of the green algae V. carteri (Smith and Lee, 2010) and Floydiella terrestris (Brouard et al., 2010), two of the largest ptDNAs on record, but is in stark contrast to the compact ptDNAs typical of most colorless algae (Table I). At approximately 5% noncoding, the plastid genomes of Helicosporidium sp. and the apicomplexan parasites P. falciparum and E. tenella (Wilson et al., 1996; Cai et al., 2003) are paragons of compactness, as are those of P. wickerhamii (approximately 17% noncoding; Yan et al., 2015), the euglenophyte Euglena longa (approximately 16% noncoding; Gockel and Hachtel, 2000), and the cryptophyte Cryptomonas paramecium (approximately 12% noncoding; Donaher et al., 2009).
The global AT content of the P. uvella ptDNA is approximately 77%, reaching 81% in the intergenic regions. The high AT content and large size of the intergenic regions may explain our initial failed attempts to bridge the plastid contigs using a standard PCR protocol rather than one designed specifically for AT-rich DNA (Su et al., 1996). In addition to being bigger and more AT rich, the P. uvella plastid genome is also more repeat rich than its photosynthetic peers. Whole-genome dot-plot analyses (Fig. 3) showed that the P. uvella ptDNA has more repetitive DNA than those of C. leiostraca and C. applanata and even Dunaliella salina, which is renowned for being dense with repeats (Smith et al., 2010).
Figure 3.
Sequence self-similarity plots of plastid genomes of C. leiostraca (complete sequence; A), C. applanata (partial sequence; B), P. uvella (one scaffold of 149.5 kb; C), and D. salina (complete sequence; D). Individual dot plots were calculated for each genomic sequence using the dottup application (EMBOSS suite) defining a word size of 15 nucleotides.
The accumulation of introns also can result in genome size increase. However, the survey of the P. uvella ptDNA scaffolds with RNAweasel (Lang et al., 2007) predicted only a single group IA intron (200 nucleotides) located in the putative 23S rRNA (rrnL) gene. The predicted intron is not in the same position as in the C. leiostraca or C. applanata rrnL genes (Supplemental Fig. S3A). Nevertheless, the P. uvella ptDNA region encoding the 23S rRNA is unusually long (15.8 kb; approximately 4 times larger than rrnL genes of C. leiostraca and C. applanata) because of 11 AT-rich regions that account for 81% (12.8 kb) of the rrnL gene expansion. Two AT-rich insertions are present as well in the 16S rRNA (rrnS) gene (Supplemental Fig. S3C; Nedelcu, 2001). The insertions in the P. uvella rRNA genes have no evident similarity to other chlamydomonadalean ptDNAs, and the high AT content (approximately 80%) of these insertions matches the AT levels of the ptDNA bona fide intergenic regions (Fig. 3B; Supplemental Fig. S3A). Further experimental studies are required to clarify if these insertions are part of the mature rRNAs or are just unconventional intronic sequences.
ptDNA-Coding Capacity Is Highly Conserved among Colorless Algae
Of the 69 unique protein-coding genes identified in the C. leiostraca and C. applanata plastid genomes, only 25 were found in the P. uvella ptDNA, implying that dozens of protein-coding genes were lost from this genome following the transition to an obligate heterotrophic lifestyle (Supplemental Table S1). Indeed, like the ptDNAs from other nonphotosynthetic algae, the P. uvella plastid genome does not encode any proteins involved in photosynthesis (Supplemental Table S1). In addition, at least one of the missing proteins is involved in plastid gene expression (rps2) and was potentially transferred to the nucleus or not captured in our assembly (TBLASTN searches with the C. leiostraca and C. applanata plastid Rps2 amino acid sequences against the P. uvella contigs gave no hits). Our P. uvella ptDNA assembly also contains two rRNAs (rrnL and rrnS) and 27 tRNAs able to decode 19 essential amino acids (Table III; Supplemental Table S1). No rrnF (5S rRNA) and trnF genes were detected in our four scaffolds.
The 25 proteins encoded in the P. uvella plastid genome are nearly all related to gene expression and include subunits of RNA polymerase (rpoA, rpoB2, rpoB1, rpoC1, and rpoC2), 16 ribosomal proteins, the elongation factor Tu 1 (tufA), a ClpP Ser-type peptidase (clpP; Adam et al., 2006; Andersson et al., 2009; Derrien et al., 2009), a conserved RF1 protein (ycf1), and a putative zinc metalloprotease (ftsH; Yu et al., 2004; Table III). All of these genes have been found in the plastid genomes of other colorless algae (Table III). Plastid gene content surveys of seven diverse colorless species, including P. uvella, revealed a common set of 14 genes involved in expression of the plastid genes, encoding 10 ribosomal proteins, three subunits of RNA polymerase (rpoB1, rpoC1, and rpoC2), and the translation elongation factor Tu 1.
The ptDNA of P. uvella shares very little gene synteny with either C. leiostraca or C. applanata ptDNAs (Supplemental Fig. S1), suggesting that there have been rampant plastid gene rearrangements in P. uvella after the loss of photosynthesis. This is in clear contrast to the case of the nonphotosynthetic trebouxiophyte P. wickerhamii ptDNA, which, when ignoring genes for photosynthesis, mirrors the arrangement of coding regions in the plastid genome of its close photosynthetic relative Auxenochlorella protothecoides (Yan et al., 2015).
When looking at the conserved plastid protein-coding repertoire among green algae, in addition to coding sequences involved in plastid gene expression, three genes stand out as being particularly important for nonphotosynthetic species: clpP, ftsH, and ycf1. All three genes are found in the P. uvella and P. wickerhamii ptDNAs, and the latter two also are encoded in the plastid genome of Helicosporidium sp. (Table III). The fact that P. uvella and the two trebouxiophytes, P. wickerhamii and Helicosporidium sp., lost photosynthesis independently of one another (Lewis and McCourt, 2004; Leliaert et al., 2012; Mancera et al., 2012; Figueroa-Martinez et al., 2015) suggests that these genes have been retained independently in three distinct lineages. This observation further supports the notion that clpP, ftsH, and ycf1 carry out crucial functions apart from photosynthesis. The clpP gene encodes a subunit of a ClpP peptidase, which is thought to be involved in protein homeostasis (Ramundo et al., 2014). The ftsH gene encodes a putative protease conserved in ptDNAs of diverse chlorophycean algae (Maul et al., 2002). The precise function of ycf1 is unknown (de Vries et al., 2015; Nakai, 2015), but it is believed to encode a putative membrane-anchorage and/or nucleic acid-binding protein essential for cell viability (Boudreau et al., 1997; Drescher et al., 2000; Ozawa et al., 2009). The protein YCF1 of Arabidopsis (Arabidopsis thaliana) is a putative element of the plastid TIC complex, one of the central components of the TOC/TIC system that imports proteins into the organelle from the cytosol (Kikuchi et al., 2013). However, there is low sequence identity between chlorophycean and angiosperm YCF1 proteins, and the precise function of the chlorophycean YCF1 has yet to be investigated. Interestingly, Polytomella spp., which lack a plastid genome, do not appear to encode clpP, ftsH, or ycf1 in their nuclear genomes, implying that their colorless plastids can be maintained in the absence of these genes and their protein products (Smith and Lee, 2014; Figueroa-Martinez et al., 2015).
ptDNA in Colorless Chlamydomonadalean Algae: Go Big or Go Home
A recurring theme from the study of plastid genomes in nonphotosynthetic taxa is downsizing: the pruning and purging of genes, introns, and intergenic regions and the general loss and outsourcing of functions. In some respects, the P. uvella ptDNA fits this theme: it has lost all genes related to photosynthesis and encodes a bare minimum of tRNAs and ribosomal proteins. In other respects, the P. uvella plastid genome is the antithesis of downsized, boasting more than 165 kb of noncoding DNA and countless repeats. However, the large size of this genome should come as no major surprise. If comparative genomics has taught us anything over the past four decades, it is that, given the right circumstances, virtually any type of chromosome can undergo expansion, so why not the genomes of colorless plastids? Perhaps our view of nonphotosynthetic plastid-bearing species is skewed toward parasites, which are known to have a penchant for genomic reduction (Wilson et al., 1996; de Koning and Keeling, 2006; Salomaki et al., 2015; Yan et al., 2015), although there are also many nonparasitic colorless plants and algae that have highly reduced ptDNAs (Gockel and Hachtel, 2000; Donaher et al., 2009; Schelkunov et al., 2015).
There is a strong possibility that P. uvella has a predisposition toward plastid genomic inflation, which might have existed long before its ancestor lost photosynthesis. For instance, P. uvella has many close relatives with enormous plastid genomes (Fig. 1A) abounding with repeats. Even the ptDNAs of C. leiostraca and C. applanata (P. uvella’s close relatives) are by no means small, and one of them is relatively repeat dense (Fig. 3). Thus, it is conceivable that the most recent photosynthetic ancestor of the P. uvella clade (Fig. 1A) had a large ptDNA and that the tendency toward genomic expansion merely persisted in P. uvella, despite ongoing gene deletions associated with the loss of photosynthesis. The repeat-rich intergenic ptDNA in P. uvella may have even mediated gene losses, as is known to occur in other systems (Ogihara et al., 1992; Phadnis et al., 2005).
The roots of organelle genomic expansion are complicated and multifaceted (Smith, 2016). There is increasing evidence that DNA maintenance machineries play a key role in organelle genomic architecture, including genome size (Smith and Keeling, 2015). Recent studies detected what appear to be different types of double-strand break DNA repair occurring in organelles, some of which might be lengthening noncoding regions (Christensen, 2013, 2014). For example, the expansion of Arabidopsis and D. salina organelle genomes can be partly explained by break-induced replication (Christensen, 2013; Del Vasto et al., 2015), a recombination-dependent DNA-repair mechanism that is fundamental to genomic integrity during replication but can cause genomic rearrangements and insertion/deletion mutations (Bosco and Haber, 1998; Maréchal and Brisson, 2010). Break-induced replication can be effective at using short (less than 30 nucleotides) repeats to repair double-stranded breaks, thus promoting rearrangements and insertions or deletions in repeat-rich organelle genomes (Maréchal and Brisson, 2010). The large number of short repeats in the Polytoma ptDNA (Fig. 3) and the lack of gene synteny with its close relatives (Supplemental Fig. S1) are consistent with genomic rearrangement and expansion via recombination-based and error-prone DNA-repair mechanisms.
Whatever the forces behind ptDNA inflation in P. uvella, they do not appear to act on the mitochondrial genome, which is small (17.4 kb) and relatively compact (approximately 30% noncoding; Del Vasto et al., 2015). Perhaps the efficiency of the plastid recombination-based DNA-repair mechanisms potentially contributing to genomic rearrangements and expansion took over the molecular systems that counteract repeat-mediated recombination events (e.g. RECA-like proteins, RECG helicases, MutS homologs, and DNA-binding proteins of the Whirly family; Inouye et al., 2008; Cappadocia et al., 2010; Xu et al., 2011; Odahara et al., 2015), compromising the stability of the oganelle genome. These mechanisms limiting the impact of repeat-mediated DNA repair probably became relatively inefficient in the P. uvella plastid after facing relaxed pressures following the loss of photosynthesis.
Certain P. uvella plastid-encoded proteins appear to have undergone higher rates of amino acid substitution compared with those of C. leiostraca and C. applanata. Pairwise alignments of various nonribosomal plastid proteins (Supplemental Table S2) show that the sequence identity between orthologs of C. leiostraca and C. applanata is, on average, higher than the equivalent pairwise comparison with P. uvella. In fact, the plastid-encoded proteins from both C. leiostraca and C. applanata show greater sequence identity with the distantly related C. reinhardtii than they do with the more closely related P. uvella. It is plausible that reshaped (e.g. relaxed) selective forces acting after the loss of photosynthesis underlie the apparent accelerated amino acid substitution rates in the protein-coding regions of the P. uvella plastid genome.
At first glance, P. uvella and Polytomella appear to reveal diametrically opposed paths following the loss of photosynthesis: a large ptDNA in the former and no ptDNA in the latter. However, the evolutionary processes leading to these different events are not mutually exclusive and can occur in parallel. The loss of a plastid genome centers on coding DNA and involves the deletion of genes and the outsourcing of ptDNA-dependent pathways to other genetic compartments (Barbrook et al., 2006; Smith and Lee, 2014). Conversely, the expansion of a plastid genome acts on noncoding DNA, whereby error-prone DNA maintenance processes or selfish elements, for example, result in insertions in intergenic DNA. Therefore, the increase in noncoding DNA in a plastid genome does not preclude that genome from ultimately being lost. In fact, as noted above, repeat-rich noncoding DNA may even promote gene loss. In other words, there is no reason to assume that the nonphotosynthetic ancestor of Polytomella did not have a large, repeat-rich ptDNA or that P. uvella will not eventually lose its plastid genome. What is clear is that some chlamydomonadalean algae, whether they are photosynthetic or nonphotosynthetic, have a remarkable tendency toward extremes in organelle genome size.
Although most explanations for different genomic architectures are centered around defective DNA replication and repair mechanisms or relaxed selection on specific cellular functions (resulting in an increased propensity for higher mutation and recombination rates), other factors related to less obvious or understood aspects of life history, ecology, and population size/structure should not be overlooked. Polytoma and Polytomella are both unicellular free-living nonphotosynthetic algae, but they are likely to differ in other aspects that might have influenced the evolution of their organelle genomes. The Polytomella-Polytoma comparison, particularly with respect to nucleus-encoded plastid-targeted proteins, should allow further studies aimed at a better understanding of the interaction between mechanistic, neutral, and selective forces that can shape genome evolution.
MATERIALS AND METHODS
Algal Cultures
Cultures of Chlamydomonas leiostraca (strain SAG 11-49) were maintained in standard Volvox medium (Kirk et al., 1999) under constant shaking (200 rpm). Polytoma uvella (strain UTEX 964) was grown in Polytomella medium (0.2% sodium acetate, 0.1% yeast extract, and 0.1% tryptone) with no shaking. Algal cultures were grown in illuminated growth chambers at 18°C under a 16-h-light/8-h-dark cycle.
DNA Extraction
Cells were harvested from 250-mL cultures in exponential growth phase by centrifugation at 4,500 rpm for 10 min. Pellets were washed three times with saline-EDTA (50 mm Tris-HCl, pH 8, 50 mm NaCl, and 5 mm EDTA), resuspended in the same buffer, and then incubated in the presence of proteinase K (33 μg mL−1) and 0.5% SDS for 1 h at 50°C under constant shaking. DNA was extracted by standard phenol-chloroform procedures and recovered by precipitation with 3 m NaOAc (pH 7) and 95% ethanol. Nucleic acid pellets were washed with 75% ethanol and resuspended in TE buffer (pH 7.6). Semipurified fractions were then incubated with RNase A (25 μg μL−1) for 1 h at 37°C, followed by precipitation of polysaccharides with 7 m NH4OAC. Purified DNA was then recovered by precipitation with 95% ethanol and resuspended in TE buffer (pH 7.6).
ptDNA Sequencing, Assembly, Annotation, and Comparative Analyses
Paired-end libraries (approximately 450-bp insert length) prepared from total DNA samples were sequenced in the Roy J. Carver Center for Genomics of the University of Iowa and Genome Quebec at McGill University using Illumina technology (Hi-Seq 2500). A total of 68.6 × 106 and 78 × 106 100-bp reads were obtained from P. uvella and C. leiostraca, respectively. Illumina reads from each strain were assembled with Ray version 2.2.0 (Boisvert et al., 2010) using k-mer lengths of 21 and 31. Four PacBio SMRT cells from total Polytoma uvella DNA were produced at Genome Quebec. PacBio raw sequences were corrected and assembled using the RS HGAP Assembly.2 protocol from the SMRT Analysis Software version 2.2.0 (Pacific Biosciences). Contigs encoding typical plastid genes were initially identified with the Automatic Annotation tool of Geneious version R8 (Kearse et al., 2012) using as a reference the ptDNA gene repertoires of Chlamydomonas applanata, Chlamydomonas reinhardtii, Volvox carteri, and Dunaliella salina. After preliminary annotation, each coding region was revised using BLAST searches (TBLASTX and TBLASTN) in public repositories, followed by pairwise alignments with orthologous sequences and final manual refinements. ORFs (more than 100 encoded amino acids) were identified with the ORF-finding function of Geneious version R8 using the universal and bacterial genetic codes. To explore the presence of pseudogenes, we performed similarity searches in our local plastid genome database using as alternative queries the nucleotide sequences of the P. uvella ptDNA intergenic regions (BLASTX cutoff E value ≤ 0.1), the corresponding six-frame conceptual translations (11,198 sequences), and predicted ORFs larger than 45 bp (2,410 sequences). Conceptual intergenic regions with AT content from 60% to 70% (lower than the average AT content in the intergenic regions) were analyzed closer using the Web-based PSI-BLAST (cutoff E value of 10; threshold of 0.05). Additionally, we used the coding regions and ORFs (encoding products of more than 100 amino acids) of the C. leiostraca and C. applanata ptDNAs as queries to perform BLASTN and TBLASTN searches (cutoff E value ≤ 10−5) in the P. uvella ptDNA intergenic spaces. tRNA genes were predicted with the tRNAscan-SE Search Server (Schattner et al., 2005), and introns were identified with RNAweasel (Lang et al., 2007). Corrected PacBio reads were used as references to map ptDNA fragments generated from the Illumina data (i.e. contigs from Ray). Alignments of complete, or nearly complete, plastid genomes were prepared with MAUVE version 2.3.1 (Darling et al., 2004) implemented in Geneious using default parameters. Genomic dot plots were generated with dottup of the EMBOSS suite (Rice et al., 2000) using a tuple size (-wordsize option) of 15 nucleotides.
Multiple Sequence Alignments and Phylogenetic Analyses
Multiple alignments of 16S rRNA and 18S rRNA sequences were generated with MAFFT version 7 (Katoh and Standley, 2013). Resulting amino acid matrices were manually edited with the Geneious version 8 graphical interphase to discard ambiguous and poorly aligned regions. Maximum likelihood trees of the 16S rRNA data (1,227 nucleotides) and the concatenated 16S rRNA and 18S rRNA set (2,799 nucleotides) were estimated with RAxML version 7.2.6 (Stamatakis, 2006) considering the GTR model and GAMMA distribution. Branch support was assessed with 1,000 bootstrap replicates. Bayesian posterior probabilities were calculated with MrBayes 3.2.1 considering the GTR substitution model and running two independent Metropolis-coupled Markov Chains Monte Carlo for 2.5 million generations. Trees were sampled every 100 generations, and posterior probabilities were estimated discarding the first 5,000 sampled trees. Pairwise alignments of nonribosomal proteins were prepared as well with MAFFT version 7 considering the BLOSUM62 substitution matrix. Protein sequence identity was estimated from pairwise raw alignments. All sequence alignments are available upon request.
Accession Numbers
The ptDNA sequences of C. leiostraca and P. uvella were deposited in GenBank with the accession numbers KX828176.1 and KX828177.1, respectively
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of the plastid genomes of C. leiostraca, C. applanata, and P. uvella.
Supplemental Figure S2. Maximum likelihood tree estimated from 18S rRNA sequences of diverse chlamydomonadalean taxa.
Supplemental Figure S3. Multiple alignment of plastid 23S rRNA (rrnL) genes from diverse chlamydomonadalean algal species and GC content in the plastid genes encoding the 23S rRNA (rrnL) and 16S rRNA (rrnS) of P. uvella.
Supplemental Table S1. Gene content of plastid genomes from Polytoma uvella and diverse photosynthetic algae of the order Chlamydomonadales.
Supplemental Table S2. Pairwise sequence identity between nonribosomal plastid-encoded proteins.
Supplemental Table S3. Illumina read coverage in coding regions of plastid and nuclear contigs of P. uvella.
Supplementary Material
Glossary
- ptDNA
plastid DNA
- ORF
open reading frame
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant no. 402421–2011 to A.R.-P.), the Canada Foundation for Innovation (grant no. 28276 to A.R.-P.), the New Brunswick Innovation Foundation (grant no. RIF2012–006 to A.R.-P.), and the Postdoctoral Fellowship Program from the Consejo Nacional de Ciencia y Tecnología, Mexico (to F.F.-M.).
Articles can be viewed without a subscription.
References
- Adam Z, Rudella A, van Wijk KJ (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Andersson FI, Tryggvesson A, Sharon M, Diemand AV, Classen M, Best C, Schmidt R, Schelin J, Stanne TM, Bukau B, et al. (2009) Structure and function of a novel type of ATP-dependent Clp protease. J Biol Chem 284: 13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S (2006) Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci 11: 101–108 [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Haber JE (1998) Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150: 1037–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E, Turmel M, Goldschmidt-Clermont M, Rochaix JD, Sivan S, Michaels A, Leu S (1997) A large open reading frame (orf1995) in the chloroplast DNA of Chlamydomonas reinhardtii encodes an essential protein. Mol Gen Genet 253: 649–653 [DOI] [PubMed] [Google Scholar]
- Brouard JS, Otis C, Lemieux C, Turmel M (2010) The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol 2: 240–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Fuller AL, McDougald LR, Zhu G (2003) Apicoplast genome of the coccidian Eimeria tenella. Gene 321: 39–46 [DOI] [PubMed] [Google Scholar]
- Cappadocia L, Maréchal A, Parent JS, Lepage E, Sygusch J, Brisson N (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22: 1849–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC. (2013) Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol 5: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC. (2014) Genes and junk in plant mitochondria: repair mechanisms and selection. Genome Biol Evol 6: 1448–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Keeling PJ (2006) The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vasto M, Figueroa-Martinez F, Featherston J, González MA, Reyes-Prieto A, Durand PM, Smith DR (2015) Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biol Evol 7: 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B, Majeran W, Wollman FA, Vallon O (2009) Multistep processing of an insertion sequence in an essential subunit of the chloroplast ClpP complex. J Biol Chem 284: 15408–15415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Sousa FL, Bölter B, Soll J, Gould SB (2015) YCF1: a green TIC? Plant Cell 27: 1827–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaher N, Tanifuji G, Onodera NT, Malfatti SA, Chain PSG, Hara Y, Archibald JM (2009) The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: reduction, compaction, and accelerated evolutionary rate. Genome Biol Evol 1: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Figueroa-Martinez F, Nedelcu AM, Smith DR, Reyes-Prieto A (2015) When the lights go out: the evolutionary fate of free-living colorless green algae. New Phytol 206: 972–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffal KP, Schneider GJ (1980) Morphogenesis of the plastidome and the flagellar apparatus during the vegetative life cycle of the colourless phytoflagellate Polytoma papillatum. Cytobios 27: 43–61 [PubMed] [Google Scholar]
- Gockel G, Hachtel W (2000) Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist 151: 347–351 [DOI] [PubMed] [Google Scholar]
- Gornik SG, Febrimarsa, Cassin AM, MacRae JI, Ramaprasad A, Rchiad Z, McConville MJ, Bacic A, McFadden GI, Pain A, et al. (2015) Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc Natl Acad Sci USA 112: 5767–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye T, Odahara M, Fujita T, Hasebe M, Sekine Y (2008) Expression and complementation analyses of a chloroplast-localized homolog of bacterial RecA in the moss Physcomitrella patens. Biosci Biotechnol Biochem 72: 1340–1347 [DOI] [PubMed] [Google Scholar]
- Janouškovec J, Tikhonenkov DV, Burki F, Howe AT, Kolísko M, Mylnikov AP, Keeling PJ (2015) Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA 112: 10200–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kirk MM, Stark K, Miller SM, Müller W, Taillon BE, Gruber H, Schmitt R, Kirk DL (1999) regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126: 639–647 [DOI] [PubMed] [Google Scholar]
- Lang BF, Laforest MJ, Burger G (2007) Mitochondrial introns: a critical view. Trends Genet 23: 119–125 [DOI] [PubMed] [Google Scholar]
- Lang NJ. (1963) Electron-microscopic demonstration of plastids in Polytoma. J Eukaryot Microbiol 10: 333–339 [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O (2012) Phylogeny and molecular evolution of the green algae. CRC Crit Rev Plant Sci 31: 1–46 [Google Scholar]
- Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M (2015) Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol 15: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM (2004) Green algae and the origin of land plants. Am J Bot 91: 1535–1556 [DOI] [PubMed] [Google Scholar]
- Mancera N, Douma LG, James S, Liu S, Van A, Boucias DG, Tartar A (2012) Detection of Helicosporidium spp. in metagenomic DNA. J Invertebr Pathol 111: 13–19 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N (2010) Recombination and the maintenance of plant organelle genome stability. New Phytol 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14: 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA, et al. (2014) Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol Biol Evol 31: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakavukaren MJ, McCracken DA (1977) An ultrastructural survey of the genus Prototheca with special reference to plastids. Mycopathologia 61: 117–119 [DOI] [PubMed] [Google Scholar]
- Nakada T, Misawa K, Nozaki H (2008) Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol 48: 281–291 [DOI] [PubMed] [Google Scholar]
- Nakai M. (2015) The TIC complex uncovered: the alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim Biophys Acta 1847: 957–967 [DOI] [PubMed] [Google Scholar]
- Naumann J, Der JP, Wafula EK, Jones SS, Wagner ST, Honaas LA, Ralph PE, Bolin JF, Maass E, Neinhuis C, et al. (2016) Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol Evol 8: 345–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM. (2001) Complex patterns of plastid 16S rRNA gene evolution in nonphotosynthetic green algae. J Mol Evol 53: 670–679 [DOI] [PubMed] [Google Scholar]
- Odahara M, Masuda Y, Sato M, Wakazaki M, Harada C, Toyooka K, Sekine Y (2015) RECG maintains plastid and mitochondrial genome stability by suppressing extensive recombination between short dispersed repeats. PLoS Genet 11: e1005080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Terachi T, Sasakuma T (1992) Structural analysis of length mutations in a hot-spot region of wheat chloroplast DNAs. Curr Genet 22: 251–258 [DOI] [PubMed] [Google Scholar]
- Ozawa S, Nield J, Terao A, Stauber EJ, Hippler M, Koike H, Rochaix JD, Takahashi Y (2009) Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21: 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Sia RA, Sia EA (2005) Analysis of repeat-mediated deletions in the mitochondrial genome of Saccharomyces cerevisiae. Genetics 171: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Marin B, Schlösser UG, Melkonian M (2001) Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152: 265–300 [DOI] [PubMed] [Google Scholar]
- Ramundo S, Casero D, Mühlhaus T, Hemme D, Sommer F, Crèvecoeur M, Rahire M, Schroda M, Rusch J, Goodenough U, et al. (2014) Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 26: 2201–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Salomaki ED, Nickles KR, Lane CE (2015) The ghost plastid of Choreocolax polysiphoniae. J Phycol 51: 217–221 [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelkunov MI, Shtratnikova VY, Nuraliev MS, Selosse MA, Penin AA, Logacheva MD (2015) Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol 7: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu C, Swift H, Chiang K (1976) Characterization of cytoplasmic and nuclear genomes in the colorless alga Polytoma. I. Ultrastructural analysis of organelles. J Cell Biol 69: 352–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. (2011) Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol Evol 3: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. (2016) The mutational hazard hypothesis of organelle genome evolution: ten years on. Mol Ecol 25: 3769–3775 [DOI] [PubMed] [Google Scholar]
- Smith DR, Crosby K, Lee RW (2011) Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol Evol 3: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ (2015) Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA 112: 10177–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW (2010) Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol 27: 2244–2256 [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW (2014) A plastid without a genome: evidence from the nonphotosynthetic green alga Polytomella. Plant Physiol 164: 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW, Cushman JC, Magnuson JK, Tran D, Polle JEW (2010) The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol 10: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Su XZ, Wu Y, Sifri CD, Wellems TE (1996) Reduced extension temperatures required for PCR amplification of extremely A+T-rich DNA. Nucleic Acids Res 24: 1574–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartar A, Boucias DG, Adams BJ, Becnel JJ (2002) Phylogenetic analysis identifies the invertebrate pathogen Helicosporidium sp. as a green alga (Chlorophyta). Int J Syst Evol Microbiol 52: 273–279 [DOI] [PubMed] [Google Scholar]
- Vernon D, Gutell RR, Cannone JJ, Rumpf RW, Birky CW Jr (2001) Accelerated evolution of functional plastid rRNA and elongation factor genes due to reduced protein synthetic load after the loss of photosynthesis in the chlorophyte alga Polytoma. Mol Biol Evol 18: 1810–1822 [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, et al. (1996) Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol 261: 155–172 [DOI] [PubMed] [Google Scholar]
- Xu YZ, Arrieta-Montiel MP, Virdi KS, de Paula WBM, Widhalm JR, Basset GJ, Davila JI, Elthon TE, Elowsky CG, Sato SJ, et al. (2011) MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23: 3428–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wang Y, Murakami T, Shen Y, Gong J, Jiang H, Smith DR, Pombert JF, Dai J, Wu Q (2015) Auxenochlorella protothecoides and Prototheca wickerhamii plastid genome sequences give insight into the origins of non-photosynthetic algae. Sci Rep 5: 14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.