Reciprocal swap experiments suggest that the class specificity of CESA proteins extends throughout the protein and the degree of specificity differs greatly between different classes of CESA.
Abstract
The cellulose synthase complex (CSC) exhibits a 6-fold symmetry and is known as a “rosette.” Each CSC is believed to contain between 18 and 24 CESA proteins that each synthesize an individual glucan chain. These chains form the microfibrils that confer the remarkable structural properties of cellulose. At least three different classes of CESA proteins are essential to form the CSC. However, while organization of the CSC determines microfibril structure, how individual CESA proteins are organized within the CSC remains unclear. Parts of the plant CESA proteins map sufficiently well onto the bacterial CESA (BcsA) structure, indicating that they are likely to share a common catalytic mechanism. However, plant CESA proteins are much larger than the bacterial BcsA protein, prompting the suggestion that these plant-specific regions are important for interactions between CESA proteins and for conferring CESA class specificity. In this study, we have undertaken a comprehensive analysis of well-defined regions of secondary cell wall CESA proteins, with the aim of defining what distinguishes different CESA proteins and hence what determines the specificity of each CESA class. Our results demonstrate that CESA class specificity extends throughout the protein and not just in the highly variable regions. Furthermore, we find that different CESA isoforms vary greatly in their levels of site specificity and this is likely to be determined by the constraints imposed by their position within the CSC rather than their primary structure.
Cellulose plays a central role in determining the mechanical properties of plant cell walls. It is important for both wall strength and rigidity and consequently plays an essential role in many aspects of plant growth (Delmer, 1999; Somerville, 2006). Cellulose is synthesized at the plasma membrane by a large protein complex, known as the cellulose synthase complex (CSC), which moves through the plane of the plasma membrane, simultaneously extruding many individual β-1,4-Glc chains. These chains are hydrogen bonded together to form the cellulose microfibril (Nishiyama, 2009). It is this microfibril that imparts the remarkable structural properties of cellulose that have led to it being so widely used by the plants. More recently, there has been increasing focus on using cellulose as an environmentally friendly and renewable source of biomass feedstock for biofuels (Carroll and Somerville, 2009) and other chemicals, which has stimulated interest in understanding how cellulose is synthesized.
Several studies have visualized the CSC using electron microscopy of freeze fractured samples. The complex exhibits a 6-fold symmetry and is known as a “rosette” (Mueller and Brown, 1980; Haigler and Brown, 1986; Kimura et al., 1999). Despite the fact that cellulose synthase activity may be assayed in crude extracts (Lai-Kee-Him et al., 2002), purification of an active CSC remains elusive. However, the crystal structure of the bacterial cellulose synthase, bacterial CESA (BcsA)/BcsB, together with the nascent glucan chain has recently been solved, providing enormous insights into the mechanism catalyzing the synthesis of individual glucan chains (Morgan et al., 2013). Parts of the plant CESA proteins map sufficiently well onto the bacterial structure, indicating that they are likely to share a common catalytic mechanism (Vergara and Carpita, 2001; Sethaphong et al., 2013; Olek et al., 2014). However, many important questions regarding CSC organization and composition remain unanswered. In particular, the crucial question of how individual catalytic subunits are organized within the CSC and the nature of the relationship between the CSC and the structure of the microfibril that it produces remains unresolved (Guerriero et al., 2010; Kumar and Turner, 2015a).
The Arabidopsis (Arabidopsis thaliana) genome contains 10 isoforms of the CesA proteins (Richmond, 2000) that may be classified based upon where they function. Genetic and biochemical studies have shown that both primary cell wall and secondary cell wall (SCW) CSCs contain at least three different isoforms of the CESA proteins. The SCW CSC contains AtCESA4, 7, and 8 (also known as IRX5, 3, and 1, respectively; Taylor et al., 2003), while the primary cell wall CSC is comprised of AtCESA1, 3, and 6 (Persson et al., 2007). Further experiments have established that AtCESA6 is partially redundant with AtCESA2, 5, and 9 (Desprez et al., 2007; Persson et al., 2007). The function of AtCESA10 remains unclear. Based on their conserved sequence, it is widely assumed that all CESAs catalyze cellulose synthesis, and three different CesA isoforms are required for the assembly of a fully functional CSC (Gardiner et al., 2003; Taylor et al., 2003). Several hypothetical models for organization of the CSC have been proposed by taking into account biochemical evidence supporting three different CESAs to be present in a 1:1:1 stoichiometry (Gonneau et al., 2014; Hill et al., 2014), the 6-fold symmetry of the rosette complex, the number of glucan chains in the cellulose microfibrils being estimated to be 18 to 24 (Kennedy et al., 2007; Fernandes et al., 2011; Newman et al., 2013; Thomas et al., 2013), the size of the complex based on freeze fracture and electron microscopy, and the assumption that all CESA subunits are catalytically active most of the time. The simplest model has each lobe of the CSC containing three different CESAs (Newman et al., 2013), but there is little direct experimental evidence to favor any one model. Recent biochemical and structural analysis using small angle x-ray scattering (SAXS) of the central catalytic domain suggests that the proteins exist in vitro as a dimer, with each capable of binding UDP-Glc to create two cellulose chains (Olek et al., 2014). Although other biochemical data support dimerization of the CESA proteins (Atanassov et al., 2009a; Olek et al., 2014), it is hard to reconcile CESA dimerization with a CSC making 18 or even 24 glucan chains unless not all chains are active.
When the first plant CESA proteins were identified in cotton (Gossypium hirsutum), it became apparent that while they shared homology with the bacterial BcsA, plant CESA proteins were much larger (Pear et al., 1996). Of particular interest are two plant specific regions that fall within the central catalytic domain. The hypervariable region, also known as the class-specific region (CSR), varies among paralogous CESAs but not among orthologous CESAs (Vergara and Carpita, 2001). The plant-conserved region is conserved among all higher plant CESAs (Vergara and Carpita, 2001). Neither of these regions map onto the crystal structure of the BcsA/B (Sethaphong et al., 2013; Olek et al., 2014). One suggestion is that these regions form loops that could be required for interactions between different CESA proteins (Sethaphong et al., 2013). More recently, the application of SAXS has suggested a role for the CSR in homodimerization (Olek et al., 2014), while an alternative study suggests that the catalytic domain of AtCESA1 forms a trimer (Vandavasi et al., 2015). None of these studies, however, provides information on what determines CESA class specificity.
While it is well documented that at least three different CESA proteins are required for cellulose synthesis and for correct assembly and localization of the CSC (Taylor et al., 2000; Gardiner et al., 2003; Persson et al., 2007), experimental evidence to elucidate what actually determines CESA class specificity remains limited. In an earlier study, Wang et al. (2006) used a swap between two halves of CESA1 and CESA3 to elucidate whether specificity was determined by either the N-terminal regions up to the first transmembrane regions or the C-terminal (CT) regions constituting the rest of the proteins. They concluded that partial complementation of the cesa1rsw1 mutant required the C-terminal catalytic region of CESA1. A similar result was obtained with cesa3rsw5, suggesting that CESA specificity lay in the large catalytic portion of the protein and the short CT region.
More recently, a comprehensive bioinformatic analysis of 82 CESA proteins from 11 different plants species looking at sequence conservation and species diversity made a number of clear predictions (Carroll and Specht, 2011). In particular, their data suggested that the extent of class specificity varied among different CESA proteins. For example, the study predicted that among secondary cell CesA proteins CESA7 exhibited very high class specificity in the N-terminal region, while CESA4 did not (Carroll and Specht, 2011).
In this study, we have undertaken a comprehensive analysis of a series of well-defined regions within the CESA proteins, with the aim of experimentally verifying what determines SCW CESA class specificity. We performed a comprehensive series of reciprocal swaps between regions of AtCESA4, 7, and 8 that constitute the CSC in SCWs. The data obtained have given us important insights into what determines the specificity of classes of CESA proteins that have important implications for how these proteins function within the CSC.
RESULTS
Knockout Mutants in CESA4, CESA7, and CESA8 All Exhibit Similar Phenotypes
We used cesa4irx5-4, cesa7irx3-7, and cesa8irx1-7 for all transformations. These mutants harbor T-DNA insertions within exons but not close to the beginning or end of the gene (Supplemental Fig. S1). Consequently, these mutants are unlikely to produce any functional CESA protein, thereby avoiding compounding effects caused by residual proteins. These mutants all exhibit at least 4 clear phenotypes, including: dark green leaves and inflorescence stems; reduced plant height, lower cellulose content (Supplemental Fig. S2) and collapsed xylem. In this study, we focused on cellulose content as this represents the most quantitative and direct method of assessing the severity of the mutant phenotype. However, we also measured plant height, which appears to be a good indicator of cellulose content. There is a clear correlation between cellulose content and plant height, but complementation of the plant height defect generally tended to be greater than the complementation of the cellulose content. Therefore, in some cases the increases in plant height were statistically significant even though the differences in cellulose content were not.
This study involved an analysis of 116 genotypes, making it impractical to grow all the lines at the same time. Instead, data were collated from four separately grown batches of plants. To facilitate comparisons between batches, a common set of control genotypes, composed of Col-0 wild type and the three irx knockout mutants, was always included. Plant height was recorded for all experiments when the plants were between 7 and 8 weeks old, after which samples were collected for cellulose analysis. Data for the Col-0 wild type and the three mutants across the four experiments are shown in Supplemental Figure S2. Across experiments, Col-0 wild type measured 38 to 43 cm in plant height and 31% to 37% cellulose, while the mutants varied between 10 and 16 cm for plant height and between 8% and 12% for cellulose content (Supplemental Fig. S2). Available evidence suggests that null cesa7 mutations contain very little, if any, cellulose in the secondary cell wall (Ha et al., 2002). A comparison of the cellulose content and overall growth suggests that cesa4irx5-4, cesa7irx3-7, and cesa8irx1-7 have equally severe phenotypes, and consequently all three are likely to possess little, if any, cellulose in the SCW (Supplemental Fig. S2).
To normalize the data across experiments, for every experiment we first subtracted the cellulose content (or plant height) of the mutant from both the Col-0 wild type and all swap lines to obtain a corrected value that represented the increase in cellulose content (or plant height) compared to the mutant baseline. We then expressed the cellulose content for each line as a percentage of corrected Col-0 wild type. This made Col-0 wild type as 100% for all experiments while the mutants were all 0%.
CESA4, CESA7, and CESA8 Knockout Mutants Can Only Be Complemented by Respective Wild-Type Genes
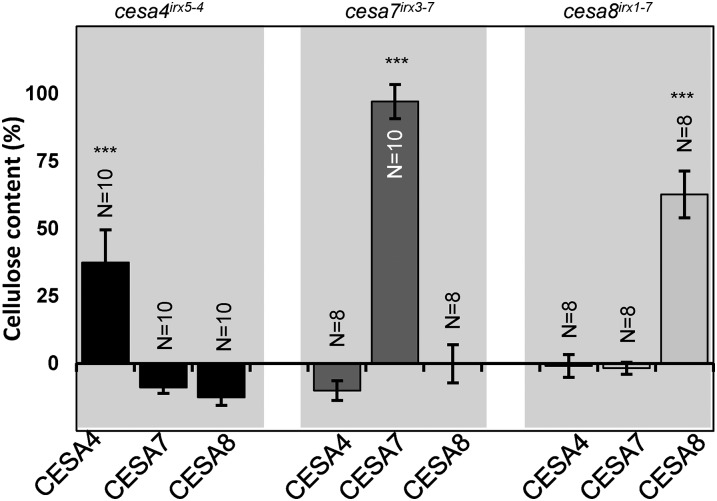
To assess the redundancy among SCW CESA proteins, we first transformed the cesa4irx5-4, cesa7irx3-7, and cesa8irx1-7 mutants with the wild-type AtCESA4, 7, and 8 genes. Only the corresponding wild-type genes were able to rescue the mutant phenotypes (Fig. 1; Supplemental Fig. S3). While CESA7 showed close to 100% complementation, CESA4 and CESA8 showed reduced levels of complementation. For both of these genes, we were able to identify some lines that exhibited full complementation (Supplemental Fig. S4), and the lower mean cellulose content was the result of greater variation between lines (Supplemental Fig. S4). The basis of this variability was unclear, but it was considered in the interpretation of subsequent experiments.
Figure 1.
Cellulose content of the cesa mutants transformed with wild-type (WT) CESA genes. Error bars are SEM. Significance levels from univariate ANOVA between the genotype and the mutant background are shown: *** Significant at 0.001, ** significant at 0.01, and * significant at 0.05.
To determine whether the smaller proportion of fully complemented lines we obtained with CESA4 and CESA8 were a result of the CESA7 promoter, we repeated the experiment using CESA4 and CESA8 under their own promoters. CESA4 and 8 also exhibited variable complementation with their native promoters (Supplemental Fig. S4). The CESA7 promoter gave comparable levels of complementation for CESA4 and CESA8 to that seen with their native promoters and was subsequently used for all further constructs.
Using Bioinformatics Analysis to Identify Sequences Likely to Be Involved in Class Specificity
Having established that CESA proteins are nonredundant, we wanted to investigate whether the specificity of SCW CESA proteins lay in any particular region of the protein. To identify likely regions that may determine class specificity, we first identified the CESA proteins from 43 plant species from Phytozome v11 (Goodstein et al., 2012). A total of 449 curated CESA sequences were identified, as described in “Materials and Methods.” These sequences were then placed into one of six classes (CESA1, 3, 6, and CESA4, 7, and 8; Supplemental Fig. S5). In this analysis, the lower plant CESAs formed a class of their own.
Having categorized the CESA sequences into classes, we repeated the class specificity analysis of Carroll and Specht (2011), who used the BLOSUM62 substitution matrix to determine the conservation and class specificity at each position in the CESA alignment (Supplemental Fig. S6). This analysis was in broad agreement with the previous analysis of Carroll and Specht (2011), suggesting that the previously described variable regions (VR1 and VR2) exhibit high levels of class specificity. Furthermore, the short C-terminal tail of CESA4 and CESA7 showed high class specificity, while only CESA7 appeared to exhibit high levels of class specificity over most of the much larger N-terminal region up to the first transmembrane domain (Supplemental Fig. S6). Not surprisingly, the two regions that are highly conserved in all CESA proteins (CR1 and CR2) were predicted to have little or no class specificity.
Using the bioinformatics analysis (Supplemental Fig. S6) and previously described structural features of CESA proteins (Pear et al., 1996; Delmer, 1999; Kumar and Turner, 2015a), we divided each CESA protein into nine distinct regions (Supplemental Figs S7 and S8). From the N to C terminus, these regions are as follows: a short N terminus (NT) prior to the zinc finger, the Cys-rich zinc RING finger domain (ZN), VR1, transmembrane helices 1-2 (TM1), conserved region 1 (CR1), VR2, conserved region 2 (CR2), transmembrane helices 3-8 (TM2), and the CT (Supplemental Figs. S7 and S8). These regions correspond closely to those originally described for cotton GhCESA1 (Pear et al., 1996; Delmer, 1999), and the boundaries largely define regions that are predicted to have either high or low class specificity (Supplemental Fig. S6). The region that has been described as the plant-conserved region corresponds to the middle of our CR1 region, while the CSR corresponds to the majority of VR2, plus a few amino acids at the beginning of CR2 (Vergara and Carpita, 2001). An alignment of CESA proteins from different species suggests that the CSR is composed of a variable region that gives it specificity and a more conserved region. We considered it was more appropriate to include the latter as part of the conserved region, because despite not being present in the bacterial sequence, it is well conserved among all plant CESAs (Kumar and Turner, 2015a; Supplemental Fig. S8). This boundary also fits with predictions based upon bioinformatics analysis (Supplemental Fig. S6).
Multiple CESA Protein Regions Confer Class Specificity
To investigate if single regions of CESA7 can be substituted by those of CESA4 and 8, we made a comprehensive series of swap constructs. All constructs are named according to the following convention CESAXREGION_CESAX, where CESAX denotes the CESA protein contributing to the majority of the protein (recipient CESA) and REGION_CESAX denotes the region name and the CESA contributing the region being swapped (donor CESA). A graphical representation of every construct is presented in Supplemental Figure S9.
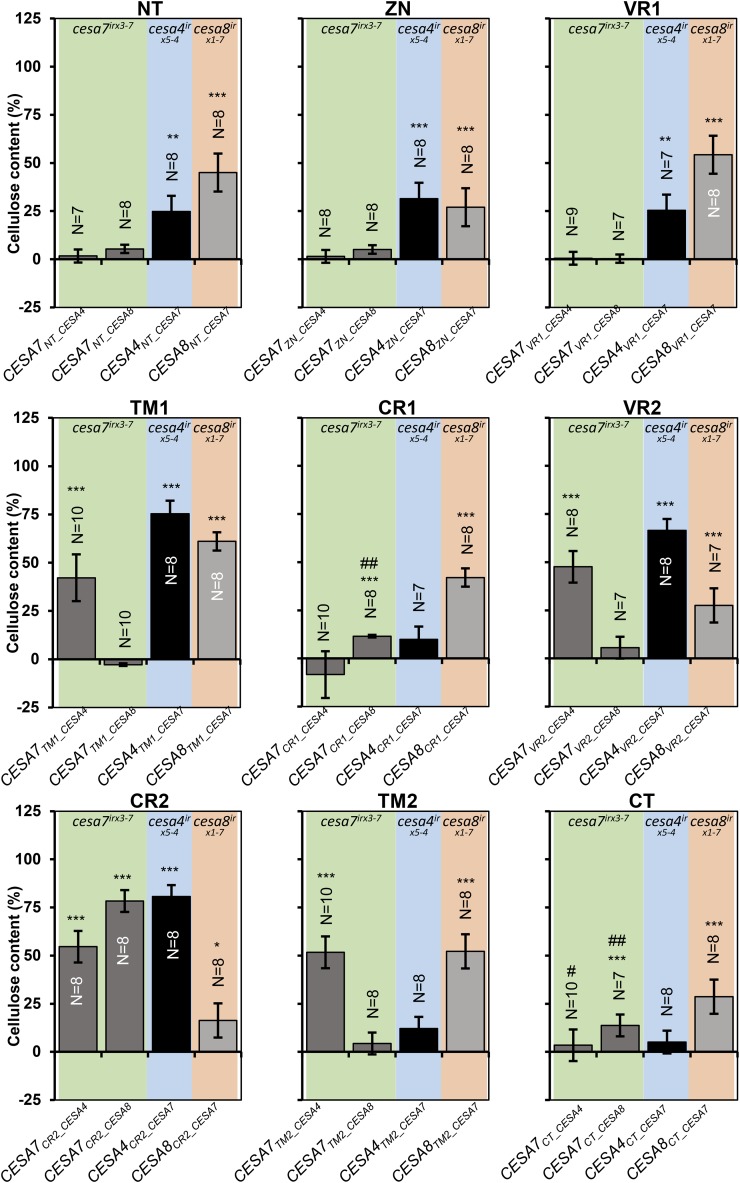
We introduced individual regions from CESA4 or CESA8 into the corresponding position in CESA7 (Supplemental Fig. S9) and transformed the constructs into cesa7irx3-7. In addition, we carried out the reciprocal experiment and swapped each region in CESA7 with the corresponding region from both CESA4 and CESA8 (Supplemental Fig. S9). These constructs were then transformed into cesa4irx5-4 and cesa8irx1-7 mutants, respectively, and the plants were analyzed for cellulose content and plant height (Figs. 2 and 3). CR2 is the most highly conserved region, with only six amino acid differences between CESA7 and CESA4 and only eight differences between CESA7 and CESA8. It is not surprising that this region exhibited the highest degree of functional equivalence among the three proteins. CR2 from both CESA4 and CESA8 exhibited good complementation when swapped into CESA7, as did the converse experiment when CR2 from CESA7 was swapped into CESA4. When CR2 from CESA7 was swapped into CESA8, the level of complementation was much less, but both cellulose content and plant height were still significantly higher than in the mutant (Figs. 2 and 3). The TM1 region also exhibited good complementation in three of the four swaps. All other swaps gave varying levels of complementation among the four different swaps. In contrast to CR2, complementation with the CR1 swaps was relatively poor, with only CESA8CR1_CESA7 giving 40% complementation of the cellulose defect, while the other 3 swaps gave little or no increase in cellulose content or plant height (Figs. 2 and 3). Similarly, the short CT region composed of only 19 to 21 amino acids exhibited a high degree of class specificity, exhibiting only low, if any, levels of complementation when swapped between CESAs (Figs. 2 and 3). We also found that some VR2 swaps gave reasonable complementation even though by definition there is little sequence conservation in this region (Supplemental Figs. S6 and S8). To some degree, VR2 was interchangeable between CESA7 and CESA4 even though CESA4 contains a distinctive Lys-rich 19-amino acid insertion relative to CESA7 and CESA8. These results demonstrate that making predictions about domain function based upon sequence conservation is not always reliable.
Figure 2.
Cellulose content of single swap constructs transformed into the recipient cesa mutant backgrounds. Error bars are SEM. Significance levels from univariate ANOVA between the genotype and the mutant background are shown *** Significant at 0.001, ** significant at 0.01, * significant at 0.05, # significant differences in plant height in Figure 3, ## plant height differences not significant in Figure 3.
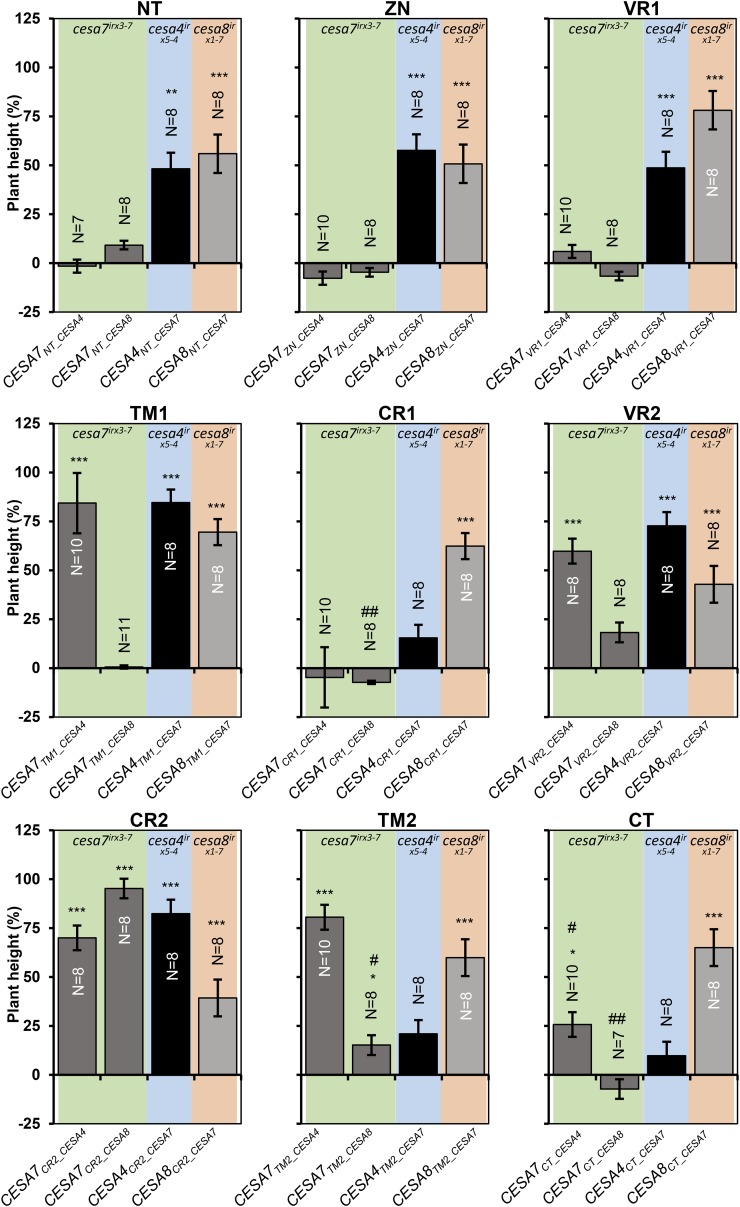
Figure 3.
Plant height of single swap constructs transformed into the recipient cesa mutant backgrounds. Error bars are SEM. Significance levels from univariate ANOVA between the genotype and the mutant background are shown: *** Significant at 0.001, ** significant at 0.01, and * significant at 0.05. #, Cellulose content differences not significant in Figure 2; ##, significant differences in cellulose content in Figure 2.
One of the most important overall trends that emerged from this analysis was the difference between complementation when CESA7 was the recipient and when CESA7 was the donor for the swaps. This is particularly true at the N-terminal region of the protein where the NT, ZN, and VR1 regions from CESA7 all gave good complementation when either CESA4 or CESA8 was the recipient, but the reciprocal swaps in which the same regions from both CESA4 and CESA8 were swapped into CESA7 gave no complementation. With the exception of CR2, this pattern seemed to be a general trend along the protein.
No Single CESA Region Is Sufficient to Alter Class Specificity
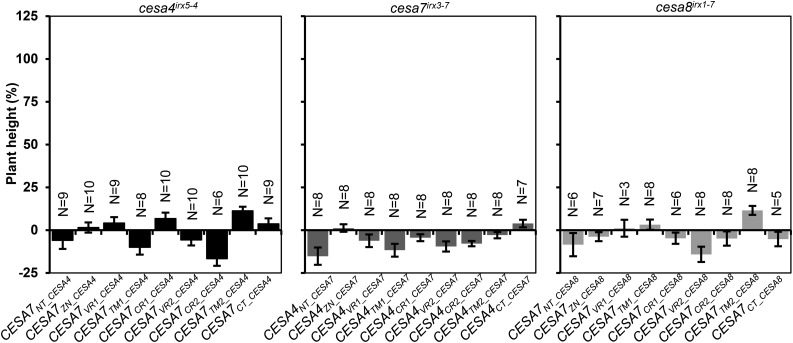
To determine whether any of the regions studied would be sufficient to alter the class specificity of another CESA, we also transformed the single region constructs into the plants lacking the donor CESA. For instance, the CESA7REGION_CESA4 constructs, where CESA7 was used as the recipient and CESA4 as the donor, were transformed into cesa4irx5-4. We measured plant height as our most sensitive measure of complementation. None of the plants exhibited any significant complementation (Fig. 4), indicating that no single CESA region tested here was able to alter the class specificity of any of the three SCW CESA proteins.
Figure 4.
Plant height of single swap constructs transformed into the donor cesa mutant backgrounds. Error bars are SEM.
Swapping Multiple Regions May Increase the Extent to Which CESA May Be Interchanged
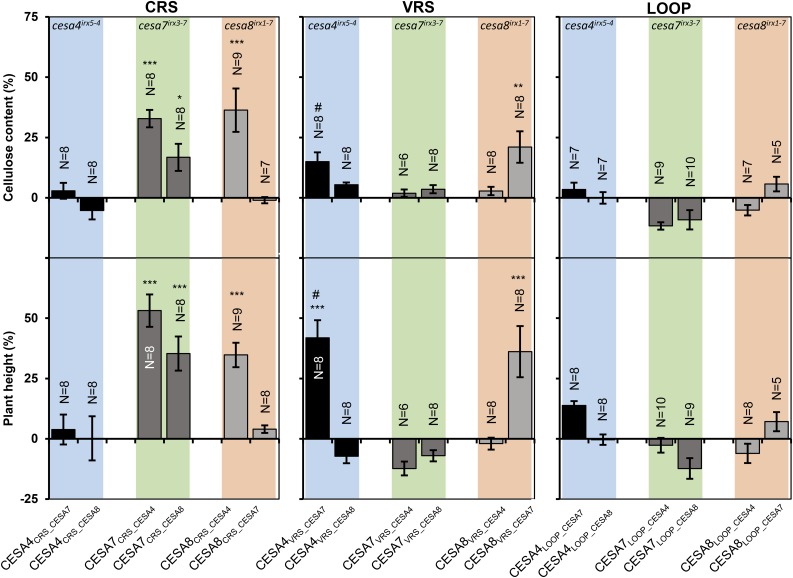
Next we investigated if some of the regions acted as a unit within the CESA protein. Functional interactions between regions could mean that swapping only a single region might be detrimental to their function. We generated three reciprocal sets of region swap constructs by swapping either both conserved regions (CR1 and CR2, termed CRS), both VR1 and VR2 (termed VRS), or the central catalytic loop between the transmembrane domains, (CR1, VR2, and CR2, termed LOOP). These swaps were generated in all possible combinations between CESA4, CESA7, and CESA8 (Supplemental Fig. S9) and transformed into backgrounds lacking the recipient CESA. For CRS swaps, we observed weak complementation when CESA7 was the recipient of both CESA4 and CESA8 donors (CESA7CRS_CESA4, CESA7CRS_CESA8; Fig. 5), whereas the single region swaps in which only CR1 from CESA4 or CESA8 was used as the donor gave no complementation (Figs. 2 and 3), suggesting that CR2 may help CR1 to function as a donor. For the VRS swaps, there was significant complementation for CESA4CRS_CESA7 and CESA8CRS_CESA7 (Fig. 5). However, this level of complementation was lower than that achieved by either of the single VR1 or VR2 swaps from the corresponding proteins (Fig. 3). None of the LOOP swaps gave any significant complementation (Fig. 5).
Figure 5.
Multiregion swap constructs transformed into recipient cesa mutant background. Error bars are SEM. Significance levels from univariate ANOVA between the genotype and the mutant background are shown: *** Significant at 0.001, ** significant at 0.01, and * significant at 0.05. # Complementation of plant height significant, but complementation of cellulose content not significant.
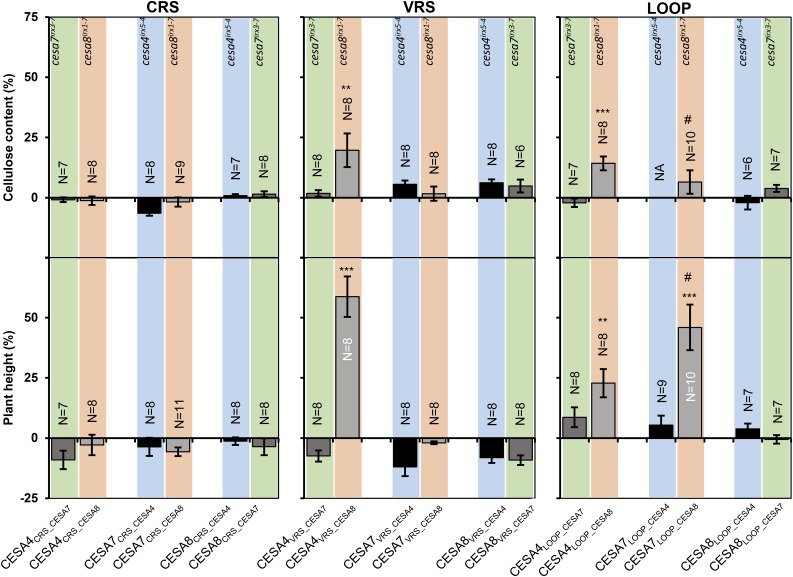
Can Multiregion Swaps Alter CESA Class Specificity?
Since altering a single CESA region is not sufficient to alter CESA class specificity, we tested whether a combination of multiple CESA regions might be sufficient. Consequently, we transformed all three sets of multiregion constructs into the donor CESA mutant background (Fig. 6). No combination involving the CRS exhibited any significant complementation. For the VRS swaps, CESA4VRS_CESA8 gave good complementation of plant height and significant complementation of the cellulose content when transformed into cesa8irx1-7. No other combination showed significant complementation. For the LOOP constructs, both CESA7LOOP_CESA8 and CESA4LOOP_CESA8 gave significant complementation of the plant height defect of cesa8irx1-7; however, the reciprocal constructs using the LOOP region from CESA4 or CESA7 with CESA8 as the recipient were not able to complement cesa4irx5-4 or cesa7irx3-7, respectively. Indeed, no other combination showed any signs of complementation (Fig. 6).
Figure 6.
Multiregion swap constructs transformed into donor cesa mutant background. Error bars are SEM. Significance levels from univariate ANOVA between the genotype and the mutant background are shown: *** Significant at 0.001, ** significant at 0.01, and * significant at 0.05. # Complementation of plant height significant, but complementation of cellulose content not significant.
Analysis of Protein Expression in Swap Lines
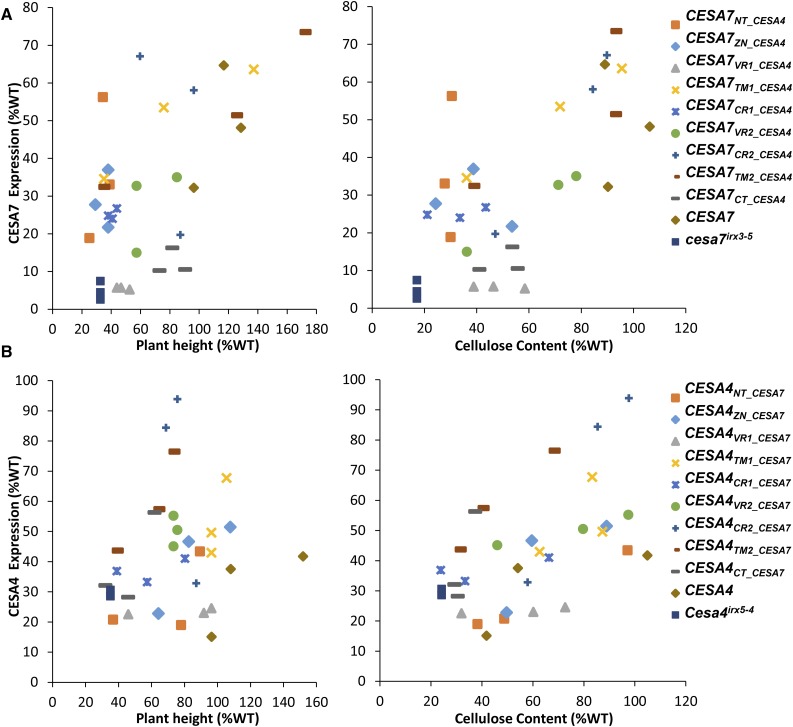
To test if the chimeric proteins were being expressed in the region swap lines, we performed a quantitative protein expression analysis in two full series of swaps, analyzing CESA7 expression in CESA7REGION_CESA4 lines and CESA4 expression in CESA4REGION_CESA7 lines (Fig. 7). Quantitative western-blot analysis was performed using LI-COR IR dye labeled secondary antibodies. To control the loading variation between various samples, blots were simultaneously probed with a normalization antibody that is raised against the chaperonin HSP73 (Supplemental Fig. S10). Expression levels for the majority of swap constructs varied among the independent lines. Also, while none of the transgenic lines reached the level of expression in Col-0 wild type, almost all had expression levels above the background mutants (Fig. 7). Furthermore, for the majority of swap constructs, there was no correlation between the expression levels and the cellulose content. For example, CESA7NT_CESA4 exhibited around 30% cellulose content, similar to that of the mutant, while protein levels varied from 20% to 60% of wild-type levels. Two lines containing the wild-type CESA7 showed almost complete complementation of the irx phenotype even though there was a 2-fold difference in their expression.
Figure 7.
Analysis of protein expression in the swap constructs. Level of expression was measured for CESA7 (A) or CESA4 (B) using quantitative western-blot analysis. All extracts were also simultaneously probed with anti-HSP73 antibody to normalize for loading variations. HSP73 normalized signals were then expressed as percentage of Col-0 wild type (WT) and plotted against plant height or cellulose content. Western-blot images used in calculating the normalized protein expression are shown in Supplemental Figure S10. CESA7 and CESA4 antibodies used in these experiments were raised against the VR1 domain, which would mean that any expression in CESA7VR1_CESA4 and CESA4VR1_CESA7 is the background level of expression.
DISCUSSION
The irx mutants display large decreases in cellulose content along with other phenotypes, including decreased plant height and reduced growth. We transformed the various swap constructs in these mutant backgrounds and analyzed their complementation of the plant height and cellulose content phenotype. This is a large-scale study involving a large number of genotypes. To overcome the practical difficulties with such a large experiment, we have made certain logical assumptions. All analyses were done at the T1 stage, and we measured multiple independent transformants. This can lead to variation in the level of complementation between independent transformants, which could be because of a number of factors, including variable expression levels. However, by looking for statistically significant levels of complementation based upon eight independent lines, we are able to determine whether any complementation observed is significant.
The swap constructs are designed solely to assay whether particular regions are interchangeable among CESAs, and the conclusions are drawn from results where complementation is observed. Where a construct does not complement, we are unable to say where the problem occurs, as the chimeric proteins may prevent complementation by interfering with protein stability, protein folding, or interactions between CESA proteins and the activity of CSC. Recent data from SAXS analysis does suggest that various domains of the CESA are able to fold independently (Olek et al., 2014; Vandavasi et al., 2015). Furthermore, in some cases, such as with VR2 swaps, there are a very large number of amino acid changes, yet they are still able to complement the mutant phenotype, suggesting they are able to fold normally despite differences in their primary structure.
CESA7 Exhibits the Greatest Class Specificity
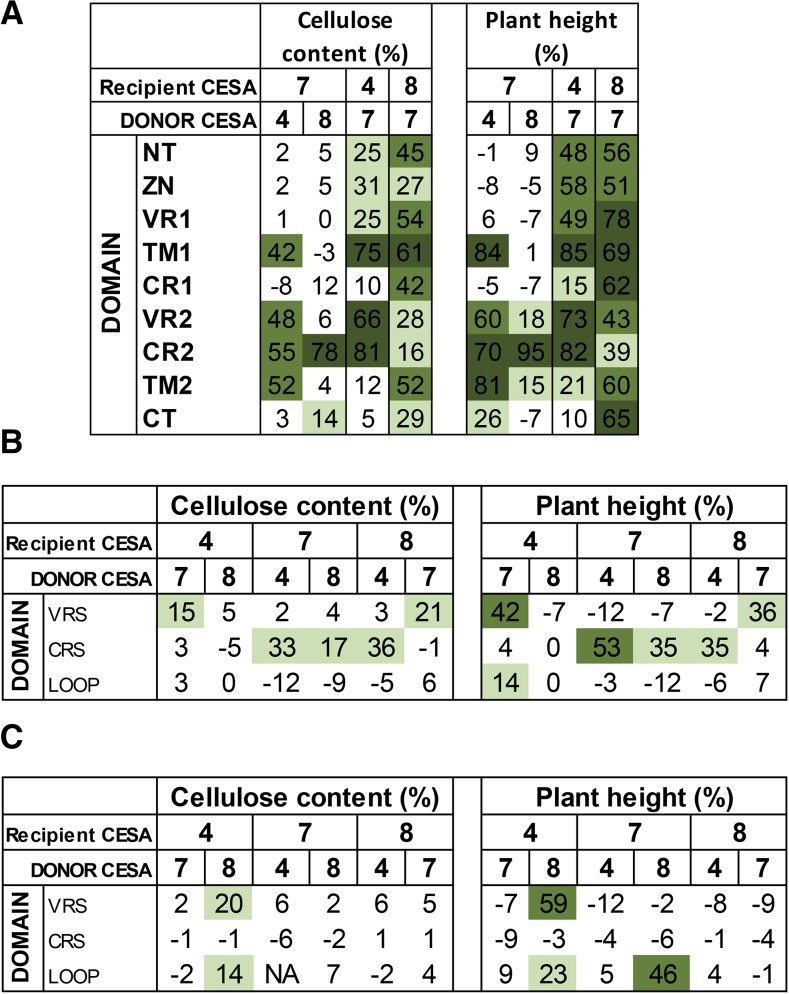
To facilitate easier understanding of the results of this study, the results shown in Figures 2, 3, 5, and 6 have been summarized diagrammatically in Figure 8. One of the most obvious observations is that CESA7 has the highest degree of class specificity among the SCW CESA proteins. In particular, CESA7 exhibits particularly high specificity at its N terminus. None of the three N-terminal domains up to the first transmembrane domain (NT, ZN, and VR1) of CESA7 can be substituted with the homologous regions from CESA4 or CESA8. In contrast, when CESA7 is used as the donor for these three regions, significant complementation is seen using both CESA4 and CESA8 as recipients. This is consistent with the bioinformatics analysis (Supplemental Fig. S6; Carroll and Specht, 2011) that predicts high class specificity at the N terminus for CESA7, but not CESA4 or 8. These results are perhaps most striking for the short NT region. CESA7 has the longest NT and is the only SCW CESA that remains functional when YFP is fused to its N terminus (Atanassov et al., 2009a). So while this longer NT may help accommodate the fluorescent proteins, the sequence itself is essential for the proper functioning of CESA7. In contrast, CESA4 and CESA8 have shorter NT regions (8 and 22 amino acids, respectively) that will not accommodate a fluorescent tag but can be functionally substituted by the longer (36-amino acid) NT region from CESA7. The results from swapping the CT region are also in good agreement with the bioinformatics analysis, which suggests CESA8 has the lowest specificity in this region (Fig. 8A; Supplemental Fig. S6). Reciprocal CT region swaps of CESA4 and CESA7 show little complementation, while CESA8CT_CESA7 complements both the cellulose content and plant height of cesa8irx1-7 mutants.
Figure 8.
Cellulose content and plant height data for all sets of swaps performed in this study. A, Single swap constructs in recipient CESA mutant background (also see Figs. 2 and 3). B, Multiregion swap constructs in recipient CESA mutant background (also see Fig. 5). C, Multiregion swap constructs in donor CESA mutant background (also see Fig. 6). Color coding is as follows: dark green, >60%; green, 40% to 60%; light green, 12.5% to 40%; white, <12.5%.
Not surprisingly, CR1 is predicted to have low class specificity, while VR2 is predicted to have the highest level of class specificity (Supplemental Fig. S6). In contrast, our data suggest that the CR1 region of CESA4 or CESA8 cannot complement this region in CESA7 (CESA7CR1_CESA4 and CESA7CR1_CESA8), despite the obvious high degree of sequence homology in this region (Supplemental Fig. S8), indicating there is a degree of class specificity in this region. In contrast, while the VR2 region is predicted to have the highest level of class specificity, CESA7 VR2 swaps from both CESA4 and CESA8 show significant complementation. These results are the opposite of those predicted by the bioinformatics analysis. However, it is consistent with a recent analysis of the CSR, a region that contains VR2, which demonstrated that the CSR was interchangeable between CESA1 and CESA3 (Sethaphong et al., 2016). Furthermore, a group of four cysteines in a six-amino acid stretch of this region of CESA7 can be substituted with a 12-amino acid sequence containing six cysteines from CESA4 (Kumar et al., 2016). Therefore, although the sequence analysis suggests this region forms part of the so called class-specific region (Vergara and Carpita, 2001), the functional analysis suggests this region does little to determine class specificity within the secondary wall CESAs. This region could, however, provide specificity between the primary wall and secondary wall CESAs.
Overall, there is a clear general trend: with the possible exception of the CT, all other regions function as well or better when CESA7 is the donor and CESA4 and CESA8 the recipients than in the reverse swaps when CESA7 is the recipient (Fig. 8A). All these data are consistent with CESA7 exhibiting a high degree of class specificity that extends throughout most, if not all, of the protein.
CESA8 Exhibits the Lowest Class Specificity
Using donor regions from CESA7, significant increases in cellulose content were observed in all constructs when CESA8 was used as the recipient. This is in stark contrast to the reciprocal swaps described above in which CESA7 was the recipient. This conclusion is supported by the multiregion swaps (Fig. 8C and discussed below) in which CESA8 is the only protein for which it is possible to alter class specificity. A previous study has shown that cesa8irx1-7 mutants can be partially complemented with CESA1 (Carroll et al., 2012), a CESA essential for primary cell wall biosynthesis, even though there is no obvious relationship between CESA1 and CESA8 in terms of their primary amino acid sequences. We consider this result to support our findings, as it demonstrates that the low site specificity of CESA8 allows another CESA to function in its place. Only CESA1 and CESA3 were tested by Carroll et al. (2012), but we predict that other CESAs may also function to complement cesa8 mutants.
As the recipient, CESA4 behaved in an intermediate manner. When CESA4 was used as the recipient, there was no significant increase in cellulose content when CR1, TM2, and CT were swapped for the corresponding CESA7 regions; however, all other regions exhibited some degree of complementation (Fig. 8A).
Functionally, CESA7 Appears More Similar to CESA4 Than to CESA8
In our experiments, regions of CESA7 have been swapped with those from CESA4 and CESA8. Of the nine regions tested, it was only for CR2 where both CESA4 and CESA8 were able to function. This is consistent with the very high sequence conservation in this area in which there are only six amino acids different between CESA7 and CESA4 and eight amino acids different between CESA7 and CESA8. There were no regions in which CESA8 was functional as the donor while CESA4 was not. In contrast, there were three regions, TM1, TM2, and VR2, that were functional in CESA7 with CESA4 as donor, but not with CESA8 as donor (Fig. 8A). These data suggest that in these regions, CESA4 is functionally more closely related to CESA7.
Changing More Than One Region May Improve Their Ability to Function in Another CESA
As sections of both CR1 and CR2 map onto the structure of BcsA/B and both regions are closely associated with one another (Morgan et al., 2013; Sethaphong et al., 2013), it could be possible that changing both CR1 and CR2 would improve their ability to function as donor regions. When the CRS swaps were examined, some complementation was observed using CESA4 or CESA8 as the donor and CESA7 as the recipient; however, the level of complementation was less than that observed using CR2 alone. The fact that CRS swaps exhibit some degree of complementation when the CR1 swap does not may suggest some interaction between these regions. This is consistent with structural analysis of bacterial CESA, which demonstrates that the four structurally conserved catalytic motifs (Saxena et al., 1995; Delmer, 1999; Carpita, 2011) localize to the active site (Morgan et al., 2013). Since two of these motifs are found in CR1 and the other two are found in CR2, these regions must interact. While partial complementation was observed for some of the combinations involving VRS swaps, the levels were lower than in either VR1 or VR2 swaps. None of the LOOP swaps showed any complementation. These data suggest that while the CRS may act as functional unit, VRS and LOOP do not.
Swapping Multiple Regions Allows Limited Alteration in Class Specificity
No single region used in this study is sufficient to change class specificity (Fig. 4). However, by swapping more than one region, it is possible to alter class specificity, albeit to a very limited extent (Figs. 6 and 8C). One model of CESA structure places the CSR and plant-conserved regions in the central domain (Supplemental Fig. S7) forming structures that loop away from the catalytic site, suggesting that they may have a role in the interactions between different CESAs (Sethaphong et al., 2013). Both of these regions are contained within the LOOP region. When CESA8 was used as a LOOP donor and CESA4 the recipient (CESA4LOOP_CESA8), limited complementation of cesa8irx1-7 was obtained in terms of both cellulose content and plant height. When CESA7 was used as the recipient (CESA7LOOP_CESA8), significant complementation of cesa8irx1-7 was seen with plant height, but the small increases in cellulose content were not significant. No complementation was observed with the other four combinations of the LOOP swaps (Fig. 8C). Even greater complementation of cesa8irx1-7 was observed with the VRS swap, when CESA8 acted as the donor and CESA4 the recipient (CESA4VRS_CESA8). For remaining VRS constructs and all CRS constructs, no evidence for alteration in class specificity was observed (Fig. 8C).
The results from these multiregion swap experiments reinforce the evidence that different CESAs have differing degrees of constraints because of their position within the CSC. The single region swaps involving CESA8 as the recipient gave the highest levels of complementation. For the multiregion swaps, partial complementation of cesa8irx1-7 is possible with CESA4 containing only the two variable regions or only the LOOP of CESA8 (Fig. 8C). All these data are consistent with CESA8 having the lowest level of class specificity that allows the protein to accommodate regions from other CESAs.
CONCLUSION
Based on our analyses, we conclude that individual regions do not confer class specificity and that class specificity can extend throughout the CESA proteins. We found very large differences in the degree to which different CESAs exhibit class specificity. The functional analyses presented here are in broad agreement with predictions made based on bioinformatics analyses of amino acid conservation and divergence of CESA from 43 higher plant species (Supplemental Fig. S6; Carroll and Specht, 2011). As predicted, the functional analysis presented here suggests that CESA7 exhibits the highest degree of class specificity. The large difference in class specificity between CESA7 and CESA4 exists for nearly all of the regions we have studied that span the entire length of the protein and makes it unlikely that this difference is solely due to chance and is therefore the result of some kind of selective pressure. This most likely suggests that CESA7 holds a highly constrained position within the CSC. In contrast, CESA8 appears to exhibit very low class specificity, because every region from CESA7 will function in CESA8 to some extent. A variety of cartoon models has been generated to suggest how three different CESAs may be accommodated within a CSC that exhibits six distinct lobes (Doblin et al., 2002; Fernandes et al., 2011). Our data suggest that CESA7 is more highly constrained. These constraints could result from CESA7 occupying a position closer to the center of the CSC where it is closely surrounded by other CESAs. Alternatively, CESA7 could occupy a more peripheral position where its structure is constrained by its interactions with a variety of other proteins. The converse would apply to CESA8, where a lack of intimate association with other CESAs or other proteins allows greater promiscuity in its structure. Our study provides a framework that can be used for future experiments to test these ideas.
MATERIALS AND METHODS
Plant Material
SCW CESA T-DNA mutants cesa4irx5–4 (SALK_084627), cesa7irx3–7 (GABI_819B03), and cesa8irx1–7 (GABI_339E12) were obtained from NASC and homozygous plants were identified. These mutants have been previously described in Kumar and Turner (2015b).
DNA Cloning
cDNA fragments, containing the coding sequence and 3′UTRs of AtCESA4, 7, and 8, were cloned into a gateway entry vector pDONOR/pZeo by a BP clonase reaction to produce entry clones for CESA4, 7, and 8 (Atanassov et al., 2009a). These entry clones were used as plant-conserved region templates to amplify swap fragments, and the coordinates for these regions for CESA4, 7, and 8 are presented in Supplemental Table S1. For each region swap, three PCR fragments were amplified. These fragments were named A, B, and C. The primers used for these three amplifications were MF+BR, BF+CR, and CF+MR, respectively. A full list of all plant-conserved region primer sequences and templates used is presented in Supplemental Table S2. Fragments were gel extracted with the Qiagen gel extraction kit and then combined in an overlap extension reaction (Atanassov et al., 2009b), which was then purified. The overlap extension product was then used in an LR reaction with a Gateway destination vector, p3HC (pIRX3::GW, hygromycin plant selection, based on pCB1300 backbone). The positive selection of gateway technology meant that only the clones containing all fragments in the correct order would survive (Atanassov et al., 2009b). For each swap construct, plasmids were extracted with the Qiagen plasmid miniprep kit and sequenced fully to verify that no mutations had occurred. Sequenced plasmids were transformed into the Agrobacterium strain, pGV3101, which were then transformed into Arabidopsis (Arabidopsis thaliana) plants using the floral dip method (Clough and Bent, 1998).
Bioinformatic Analysis of CESA Sequences
A total of 66 GT2 (glycosyl transferase family 2) family protein sequences was used as a query to search the Phytozome database (Goodstein et al., 2012; 42 GT2 family proteins from Arabidopsis, BcsA sequences from 15 different bacterial species, and 9 chitin synthase sequences from 9 different fungal species). We also extracted all protein sequences from Phytozome that contained the PFAM domains PF00535 (GT 2) or PF03552 (cellulose synthase). Combining the results from these searches, a total of 2,385 unique loci were identified. One representative protein sequence per locus was included in the analysis. These protein sequences were aligned with either ClustalX2 (Larkin et al., 2007) or Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Phylogenetic trees were constructed with either ClustalX2 or FastTree (Price et al., 2010). The overall GT2 tree contained 2,385 protein sequences, which were then divided into various clades: CESAs and various CSL clades based on known members of the clades. For the purposes of this study, the 576 sequences belonging to the CESA subtree were extracted and realigned. Since a number of genes are still poorly annotated, we manually curated the alignment and all sequences that introduced large insertions into the global alignment were excluded from the analysis. This resulted in a final set of 449 CESA proteins from 43 different plant species. A new phylogenetic tree was produced from this curated set (Supplemental Fig. S5) that was used for classifying CESA sequences into six main classes (CESA1, 3, 6, and CESA4, 7, and 8), while the CESA sequences from Physcomitrella and Selaginella formed a clade of their own. The tree in Supplemental Fig. S5 was then rooted to this clade.
Once the CESA sequences were placed in one of the six classes, the global alignment was used for assigning a class specificity and conservation score at each position. The algorithm used for calculating the specificity score was the same as described in Carroll and Specht (2011). Class specificity scores for CESA4, 7, and 8 classes were plotted to produce Supplemental Figure S6.
Plant Growth and Analysis
T1 seeds were harvested from dipped Arabidopsis plants and selected on half-strength MS plates containing 35 µg/mL hygromycin. After growing for 7 d on plates in an incubator, 8 to 10 independent lines for each construct were transplanted into a 1:1:5 mixture of perlite, vermiculite, and compost. Plants were grown for a further 6 weeks on soil under long-day conditions (16 h day/8 h night, 22°C/18°C temperature, and 80% humidity). Col-0 wild type and the cesa mutants were grown on plates without any selection before being transplanted. A vector-only control for Col-0 wild type was included in one of the four experiments, and no differences were found in the growth patterns or cellulose content as compared to Col-0 wild-type grown on nonselection plates. Plant height measurements were taken when plants were 7 weeks old, after which 50-mm pieces from the primary inflorescence stem starting at 5 mm above the base were harvested and stored in 70% ethanol for analysis of cellulose content as described (Kumar and Turner, 2015b). T2 seeds were collected from the secondary inflorescences that were left intact.
Plant height (cm) and cellulose content (% cell wall) were converted into plant height (%) and cellulose content (%) to assess the level of complementation. For statistical analysis, the data were imported into IBM SPSS statistics program, and a univariate ANOVA with a lsd posthoc test was used to calculate the significance levels for the differences in the means.
Analysis of Protein Expression
Protein expression analysis was performed for two full series of single swap constructs. Three independent lines were tested for each of the swaps. A total of 16 plants for each line were grown until 5 weeks old when stems were harvested and stripped of their leaves and flowers. Stems were ground in liquid nitrogen into a fine powder. Then 50 mg of powder was homogenized in 250 µL of 1× loading buffer (Bio-Rad Laboratories) + 50 mm DTT. The mixture was heated to 95°C for 10 min and centrifuged at maximum speed for 5 min, and then the supernatant was transferred to a new tube to produce the crude extract. Then 25 µL of this crude protein extract was separated on a 7.5% SDS-PAGE gel and transferred to PVDF membranes. Membranes were blocked with LI-COR blocking buffer for 1 h, incubated with anti-CESA7 (1:2,000 dilution) or CESA4 (1:100 dilution) primary antibodies for 3 h, washed three times with 1× TBS + 0.2% Tween 20, incubated with IR Dye conjugated secondary antibodies, and washed three times with 1× TBS + 0.2% Tween 20. After the final wash with 1× TBS, membranes were air dried for 1 h. CESA4/7 antibodies were raised in sheep as described previously (Taylor et al., 2000, 2003). A loading control antibody raised in mouse against HSP73 (StressGen, 1:20,000 dilution) was multiplexed with the CESA antibodies during primary antibody incubations. Multiplexed secondary antibodies included donkey anti-goat 800CW and donkey anti-mouse 680RD (LI-COR, 1:20,000) dilution. Primary antibody incubations included 0.2% Tween 20, while the secondary antibody incubations included 0.2% Tween 20 and 0.01% SDS. Probed and dried membranes were scanned with a LI-COR Odyssey scanner using the auto settings in two channels, 800 nm for CESA antibodies (green bands in Supplemental Fig. S10) and 700 nm for HSP73 (red bands in Supplemental Fig. S10). Quantifications were performed with the ImageStudio v5.2 using the manual mode according to the manufacturer’s instructions. The background subtracted and area normalized band intensities were collected for both channels. The area around each band was used for background subtraction. Band intensities for CESA proteins were normalized with the HSP73 band intensities in respective lanes. Loading control normalized band intensities were transformed into percentage of Col-0 wild type, which was run on each blot. Final protein expression values were expressed as percent wild type.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Exon/intron structure of Arabidopsis SCW CESA genes.
Supplemental Figure S2. Phenotypes of the cesa mutants and wild-type control.
Supplemental Figure S3. Height of different cesa mutant plants transformed with wild-type CESA genes.
Supplemental Figure S4. Complementation of SCW cesa mutants using different promoters.
Supplemental Figure S5. Classification of CESA proteins into six classes.
Supplemental Figure S6. Analysis of CESA protein class specificity.
Supplemental Figure S7. Schematic representation of CESA proteins showing the location of the regions used in this study.
Supplemental Figure S8. Amino acid positions of the different CESA protein regions used in this study.
Supplemental Figure S9. Schematic representation of the swap constructs used in this study.
Supplemental Figure S10. Analysis of protein expression in the swap constructs used in this study.
Supplemental Table S1. Protein coordinates for regions of AtCESA4, 7, and 8 used in the swap constructs.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Thomas Nuhse, John Pittman, Liam Campbell, and Holly Allen for a critical reading of the manuscript. We are grateful to Andy Carroll for his help with calculating the class specificity scores.
Glossary
- CSC
cellulose synthase complex
- CSR
class-specific region
- CT
C-terminal
- SAXS
small angle x-ray scattering
- SCW
secondary cell wall
Footnotes
This work was supported by Biotechnology and Biological Sciences Research Council grant no. BB/H012923/1.
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Atanassov II, Pittman JK, Turner SR (2009a) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284: 3833–3841 [DOI] [PubMed] [Google Scholar]
- Atanassov II, Atanassov II, Etchells JP, Turner SR (2009b) A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. (2011) Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1->4)-β-D-glycans. Plant Physiol 155: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Mansoori N, Li S, Lei L, Vernhettes S, Visser RG, Somerville C, Gu Y, Trindade LM (2012) Complexes with mixed primary and secondary cellulose synthases are functional in Arabidopsis plants. Plant Physiol 160: 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60: 165–182 [DOI] [PubMed] [Google Scholar]
- Carroll A, Specht CD (2011) Understanding plant cellulose synthases through a comprehensive investigation of the cellulose synthase family sequences. Front Plant Sci 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108: E1195–E1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner JC, Taylor NG, Turner SR (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Höfte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero G, Fugelstad J, Bulone V (2010) What do we really know about cellulose biosynthesis in higher plants? J Integr Plant Biol 52: 161–175 [DOI] [PubMed] [Google Scholar]
- Ha MA, MacKinnon IM, Sturcová A, Apperley DC, McCann MC, Turner SR, Jarvis MC (2002) Structure of cellulose-deficient secondary cell walls from the irx3 mutant of Arabidopsis thaliana. Phytochemistry 61: 7–14 [DOI] [PubMed] [Google Scholar]
- Haigler CH, Brown RM (1986) Transport of rosettes from the golgi apparatus to the plasma membrane in isolated mesophyll-cells of Zinnia elegans during differentiation to tracheary elements in suspension culture. Protoplasma 134: 111–120 [Google Scholar]
- Hill JL, Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy CJ, Cameron GJ, Sturcova A, Apperley DC, Altaner C, Wess TJ, Jarvis MC (2007) Microfibril diameter in celery collenchyma cellulose: X-ray scattering and NMR evidence. Cellulose 14: 235–246 [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Turner S (2015a) Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112: 91–99 [DOI] [PubMed] [Google Scholar]
- Kumar M, Turner S (2015b) Protocol: a medium-throughput method for determination of cellulose content from single stem pieces of Arabidopsis thaliana. Plant Methods 11: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Wightman R, Atanassov I, Gupta A, Hurst CH, Hemsley PA, Turner S (2016) S-Acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353: 166–169 [DOI] [PubMed] [Google Scholar]
- Lai-Kee-Him J, Chanzy H, Müller M, Putaux JL, Imai T, Bulone V (2002) In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J Biol Chem 277: 36931–36939 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Morgan JLW, Strumillo J, Zimmer J (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SC, Brown RM Jr (1980) Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol 84: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ (2013) Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol 163: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y. (2009) Structure and properties of the cellulose microfibril. J Wood Sci 55: 241–249 [Google Scholar]
- Olek AT, Rayon C, Makowski L, Kim HR, Ciesielski P, Badger J, Paul LN, Ghosh S, Kihara D, Crowley M, et al. (2014) The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell 26: 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2010) FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. (2000) Higher plant cellulose synthases. Genome Biol 1: S3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM Jr, Fevre M, Geremia RA, Henrissat B (1995) Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J Bacteriol 177: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethaphong L, Davis JK, Slabaugh E, Singh A, Haigler CH, Yingling YG (2016) Prediction of the structures of the plant-specific regions of vascular plant cellulose synthases and correlated functional analysis. Cellulose 23: 145–161 [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P (2016) A structural study of CESA1 catalytic domain of Arabidopsis thaliana cellulose synthesis complex: evidence for CESA trimers. Plant Physiol 170: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara CE, Carpita NC (2001) Beta-D-glycan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1-->3),(1-->4)beta-D-glucan synthase. Plant Mol Biol 47: 145–160 [PubMed] [Google Scholar]
- Wang J, Howles PA, Cork AH, Birch RJ, Williamson RE (2006) Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol 142: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.