In-depth investigation for CCRs from Sorghum bicolor provides information about the critical residuesfor substrate preference among CCRs.
Abstract
Cinnamoyl-coenzyme A reductase (CCR) catalyzes the reduction of hydroxycinnamoyl-coenzyme A (CoA) esters using NADPH to produce hydroxycinnamyl aldehyde precursors in lignin synthesis. The catalytic mechanism and substrate specificity of cinnamoyl-CoA reductases from sorghum (Sorghum bicolor), a strategic plant for bioenergy production, were deduced from crystal structures, site-directed mutagenesis, and kinetic and thermodynamic analyses. Although SbCCR1 displayed higher affinity for caffeoyl-CoA or p-coumaroyl-CoA than for feruloyl-CoA, the enzyme showed significantly higher activity for the latter substrate. Through molecular docking and comparisons between the crystal structures of the Vitis vinifera dihydroflavonol reductase and SbCCR1, residues threonine-154 and tyrosine-310 were pinpointed as being involved in binding CoA-conjugated phenylpropanoids. Threonine-154 of SbCCR1 and other CCRs likely confers strong substrate specificity for feruloyl-CoA over other cinnamoyl-CoA thioesters, and the T154Y mutation in SbCCR1 led to broader substrate specificity and faster turnover. Through data mining using our structural and biochemical information, four additional putative CCR genes were discovered from sorghum genomic data. One of these, SbCCR2, displayed greater activity toward p-coumaroyl-CoA than did SbCCR1, which could imply a role in the synthesis of defense-related lignin. Taken together, these findings provide knowledge about critical residues and substrate preference among CCRs and provide, to our knowledge, the first three-dimensional structure information for a CCR from a monocot species.
Cinnamoyl-CoA reductase (CCR; EC 1.2.1.44) catalyzes the first committed reaction toward the biosynthesis of monolignols in plants. It hydrogenates hydroxycinnamoyl-CoA thioesters, producing the corresponding hydroxycinnamaldehydes and CoASH in an NADPH-dependent reaction. CCR activity was first reported by Wengenmayer et al. (1976) in soybean (Glycine max) suspension cultures; the cloning of a CCR cDNA, from Eucalyptus gunnii, was first reported by Lacombe et al. (1997). The position of CCR in the phenylpropanoid pathway gives it a pivotal role in directing metabolic flux toward flavonoids and stilbenes or monolignols, hydroxycinnamic acids, and, depending on the species, hydroxycinnamoyl esters. The most common monolignols are p-coumaryl, coniferyl, and sinapyl alcohols. They are directed to the secondary cell walls, where they undergo oxidative coupling to form lignin polymers. The structures in lignin derived from the aforementioned monolignols are referred to as p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) residues (Ralph et al., 2004). Lignin fills the voids left between the complex network formed by cellulose microfibrils and hemicellulosic polysaccharides in secondary cell walls. The addition of lignin provides hydrophobicity to xylem cells and improves the mechanical strength of cells, providing structural support. Lignin also can be formed as a mechanical barrier in response to fungal infections or attacks from insects (Vanholme et al., 2010).
The cloning of E. gunnii CCR enabled the cloning of several of its orthologs. This resulted in the identification of two CCR genes in maize (Zea mays; Pichon et al., 1998) and Arabidopsis (Arabidopsis thaliana; Lauvergeat et al., 2001). Expression studies revealed that, in both species, CCR1 is involved in lignification of stem tissues, whereas CCR2 is involved in lignification in response to attack by pathogens. In barrel clover (Medicago truncatula), however, the different CCR paralogs appear to have distinct substrate specificity, with CCR1 displaying a preference for feruloyl-CoA and CCR2 for p-coumaroyl-CoA and caffeoyl-CoA (Zhou et al., 2010). The role of CCR in lignification has been investigated with the use of mutants and transgenics. The vascular tissue of transgenic tobacco (Nicotiana tabacum), in which CCR was down-regulated, had thinner walls, brown vascular tissue, reduced lignin concentration, an increase in the S-G ratio, and displayed reduced growth and tissue abnormalities (Piquemal et al., 1998). The secondary cell walls in these plants contained tyramine ferulate, which was hypothesized to be a sink for the accumulated feruloyl-CoA esters (Ralph et al., 1998). The Arabidopsis irregular xylem4 mutant, identified based on its collapsed xylem phenotype, was shown to have a defective CCR1 gene (Jones et al., 2001). This phenotype could be reproduced by transgenic down-regulation of CCR1 (Goujon et al., 2003). Excess feruloyl-CoA resulting from reduced CCR1 expression was converted to feruloyl malate (Derikvand et al., 2008). Two independent Arabidopsis ccr1 mutants contained less lignin than the wild-type control. The lignin of these mutants had a higher proportion of H residues and a lower proportion of S residues (Van Acker et al., 2013). In contrast, in maize, analysis of the transposon insertion mutant Zmccr1−, which had 31% residual CCR1 expression, showed slightly reduced lignin concentration in stem tissue, a slight increase in the S-G ratio, and a reduction in H residues. In addition, the organization of the sclerenchyma fibers was affected. The expression of several cell wall-related genes was decreased in this mutant, while several genes involved in flavonoid biosynthesis showed higher expression levels (Tamasloukht et al., 2011). Despite species-specific differences, taken together, these data show a pivotal role of CCR in phenylpropanoid metabolism and in ensuring the integrity of secondary cell walls.
While the role of lignin is important for the survival of a plant, lignin also reduces the efficiency of the industrial use of plant biomass. The presence of lignin reduces the digestibility of forage crops (Jung and Buxton, 1994; Fontaine et al., 2003) and impedes the enzymatic saccharification of biomass to fermentable sugars that can be fermented to renewable fuels and chemicals (Chen and Dixon, 2007; Van Acker et al., 2013). As a consequence, a thermochemical pretreatment is necessary to reduce the recalcitrance of the plant cell wall to cellulolytic enzymes (Hu and Ragauskas, 2012; Leu et al., 2013). The phenolic compounds derived from lignin during this process also are harmful to the microbes used during the fermentation (Ximenes et al., 2011).
The C4 grass sorghum (Sorghum bicolor) is receiving considerable attention as a lignocellulosic feedstock for the production of renewable fuels and chemicals, in part because of its ability to grow under harsh conditions, which include low-fertility soils, high temperatures, and extended periods of drought (Farré and Faci, 2006; Wang et al., 2014). Manipulation of lignin concentration and lignin subunit composition in sorghum through the incorporation of certain brown midrib (bmr) mutations had been shown to result in improved rumen digestibility (Porter et al., 1978) and greater efficiency of the enzymatic saccharification of sorghum biomass (Saballos et al., 2008; Dien et al., 2009) Given the important role of lignin for the survival of the plant, there is an inherent risk of reduced yields or lower biomass quality associated with the reduction in lignin concentration and/or change in lignin composition. These effects can be balanced with the use of specific mutant alleles, as has been shown for the Bmr12 gene, which encodes caffeic acid O-methyltransferase (Bout and Vermerris, 2003; Sattler et al., 2012). In this instance, the bmr12-34 allele reduced lignin concentration to a level that was intermediate between wild-type sorghum and the bmr12-ref allele, which is a null allele, while still offering the same benefit of enhanced efficiency of enzymatic saccharification (Sattler et al., 2012).
Manipulation of the monolignol biosynthetic genes, such as cinnamyl alcohol dehydrogenase (Bmr6; Saballos et al., 2009; Sattler et al., 2009) and 4-coumarate-CoA ligase (Bmr2; Saballos et al., 2012), also improved biomass conversion in sorghum (Saballos et al., 2008; Scully et al., 2016). In order to maximize the toolkit for the manipulation of cell wall composition and the redirection of metabolic flux to secondary metabolites with health-promoting properties, it is important to have a detailed understanding of substrate specificity and catalytic mechanisms of the enzymes involved in the biosynthesis of monolignols. We have recently reported detailed structural and catalytic analyses of hydroxycinnamoyltransferase (Walker et al., 2013), caffeic O-methyltransferase (Green et al., 2014), and cinnamoyl-CoA O-methyltransferase (Walker et al., 2016). Although previous studies on CCR have provided significant insight on the structure-function and kinetics of this enzyme (Pan et al., 2014), the structural basis for substrate selection and preference of CCR remains largely unknown. In this study, we provide detailed biochemical and structural findings that support our proposal for differentiating substrate selectivity among CCRs. Knowledge from this study is expected to facilitate the engineering of sorghum as an improved livestock forage or bioenergy feedstock.
RESULTS
Global Structure
Recombinant SbCCR1 (Sobic.007G141200) in complex with NADP+ was crystallized in the P32 space group, and its three-dimensional structure was determined at 2.9 Å resolution (Table I). The asymmetric unit of the crystal lattice was composed of two monomers arranged in a noncrystallographic, 2-fold manner with limited intermolecular interaction. Calculations through PISA (Krissinel and Henrick, 2007), which evaluates interactions between neighboring monomers in crystal lattices for the purpose of predicting biologically relevant oligomeric states, indicated that SbCCR1 exists as a monomer in solution (solvation free energy gain = 0.8 kcal mol−1 and interface complexation significance score = 0.0).
Table I. X-ray diffraction data and refinement statistics for SbCCR1 (PDB identifier 5TQM).
r.m.s.d., Root-mean-square deviation.
| Parameter | Recombinant SbCCR1 in Complex with NADP+ |
|---|---|
| Space group | P32 |
| Cell dimensions | |
| a, b, c (Å) | 72.194, 72.194, 123.033 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50.0–2.70 (2.434–2.35) |
| Rsym | 0.155 |
| I/σI | 24.2 (1.51) |
| Completeness (%) | 98.66 (0.98) |
| Redundancy | 5.2 |
| Refinement | |
| Resolution | 41.018–2.9 |
| Unique reflections | 15,697 (1,423) |
| Rwork/Rfree | 0.2280/0.2733 |
| No. of atoms | |
| Macromolecules | 9,796 |
| Ion | 0 |
| Ligand | 210 |
| Water molecules | 0 |
| B factors | |
| Protein | 78.91 |
| Ligand ion | 86.73 |
| Water | 0.00 |
| r.m.s.d. | |
| Bond length (Å) | 0.003 |
| Bond angle (°) | 0.708 |
| Ramachandran angles | |
| Favored | 94.75 |
| Outliers | 0.48 |
| Clash score | 2.25 |
The overall structure of SbCCR1 was composed of two domains: an N-terminal domain harboring a typical Rossmann fold and a C-terminal domain of mixed α/β-fold, which was similar to the structures of CCRs from the dicotyledonous plants Petunia hybrida and M. truncatula (Pan et al., 2014). The two domains were located on two distinct lobes that sandwiched the adjacent substrate and NADP(H)-binding pockets (Fig. 1).
Figure 1.
Ribbon diagram of the global structure of SbCCR1 in complex with NADP+ and manually docked feruloyl-CoA. The N and C termini of the enzyme are marked N and C, respectively. The dashed curves at both chain termini stand in place of residues that could not be modeled in the experimental electron density map due to a disorder. NADP+, which is depicted with tan carbons in a stick model, was positioned according to its location in the experimental complex. Manually docked feruloyl-CoA is depicted with gray carbons in a stick model as well. During a reaction, the thioester bond of the substrate is positioned just above the nicotinamide ring of NADPH to promote hydride transfer. In this model, both lobes of the enzyme contribute residues to binding CoA, which may be accommodated by a conformational shift upon binding of the substrate. Molecular graphics images were produced using the UCSF Chimera package.
In order to identify closely related structural homologs in the Protein Data Bank (PDB), a Dali search (Holm and Sander, 1993) was performed using the atomic coordinates of SbCCR1. The highest match was with a CCR from P. hybrida (PDB identifier 4R1S), with a Z score of 48.19, followed by a CCR2 from M. truncatula (PDB identifier 4R1U) and a cinnamyl alcohol dehydrogenase2 (PDB identifier 4QTZ) from M. truncatula, having respective Z scores of 43.4 and 41.1. A dihydroflavonol reductase from Vitis vinifera (PDB identifier 2C29) also was highly similar, with a Z score of 39.7, which is further addressed in “Discussion.” The next most closely related structures are methylglyoxal/isovaleral reductase Gre2 from Saccharomyces cerevisiae (PDB identifier 4PVC), a vestitone reductase from Medicago sativa, and an aldehyde reductase2 from the yeast Sporobolomyces salmonicolor (PDB identifier 1Y1P), all of which had appreciably lower Z scores of 35.2, 34.5, and 32.5, respectively.
A BLAST search (Altschul et al., 1997) to identify proteins with similar amino acid sequences in the PDB revealed that CCR from P. hybrida (PDB identifier 4R1S) showed the highest identity (75%) to SbCCR1, followed by CCR2 from M. truncatula (PDB identifier 4R1U; 71%), cinnamyl alcohol dehydrogenase2 from M. truncatula (PDB identifier 4QTZ; 49%), and dihydroflavonol reductase from V. vinifera (PDB identifier 2C29; 41%). According to the BLAST output, the other enzymes identified with the Dali search showed no significant sequence similarity.
NADPH-Binding Pocket
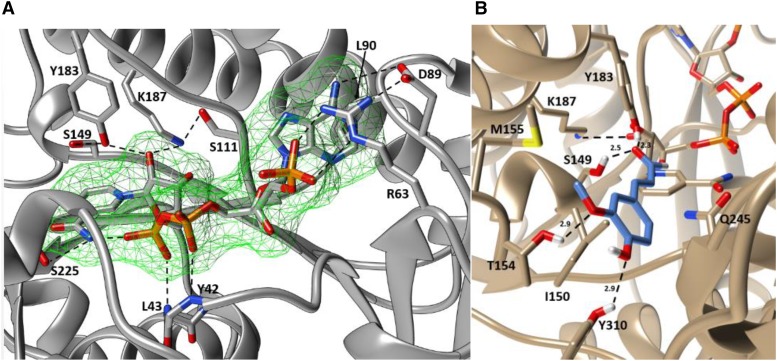
From the early stages of structural refinement, the Fo-Fc map of the SbCCR1/NADP+ complex revealed clear electron density for one NADP+ molecule buried in a deep pocket on the N-terminal domain. This binding region was composed predominantly of a six-stranded, parallel β-sheet (in the order of β3-β2-β1-β4-β5-β6; Fig. 2A), and its location opposes the putative hydroxycinnamoyl-CoA-binding pocket (Fig. 1). The diphosphate group of the bound NADP+ molecule was within hydrogen-bonding distance from the backbone secondary amines of both Tyr-42 and Ile-43. In addition, the positive charge on NADP+ appeared to be compensated through the macroelectric dipole moment of the adjacent α-helix (α1), which spans between Tyr-42 and Gly-55, through N-capping (Pereira et al., 2001). The adenine ring of NADP+ primarily interacted with Arg-63 of β1 and Asp-89 of β2, while the adenosine ribose was bound solely through a salt bridge between its 2′-phosphoryl group and the side chain of Arg-63. In short, the guanidinium group of Arg-63 contributed to adenine binding through a cation-π interaction with the six-membered heterocyclic ring (Fig. 1), and the carboxyl group of Asp-89 was hydrogen bonded to an N6 hydrogen of the ring. The side chain of Leu-90 also appeared to engage in hydrophobic interaction with the entire ring.
Figure 2.
A, The observed NADP+ in the binding pocket of SbCCR1. NADP+ and all interacting residues are represented as stick models. The backbone of SbCCR1 is represented as a ribbon diagram, and dashed lines represent hydrogen bonds or ionic interactions. All residues that contribute to NADP+ binding are labeled according to their single-letter abbreviations and numbered according to sequence positions. The catalytic triad, composed of Ser-149, Tyr-183, and Lys-187, is in close proximity to the nicotinamide ring and serves to promote hydride transfer to hydroxycinnamoyl-CoA substrates. B, Coniferaldehyde docked into the putative phenylpropanoid-binding region of SbCCR1. The backbone of SbCCR1 is represented by a ribbon diagram, with protruding side chains that contribute to coniferaldehyde binding modeled as sticks. Coniferaldehyde, which is the product of the reaction with the preferred substrate feruloyl-CoA, is shown in gray. Kinetics experiments with T154A and Y310F mutants revealed that these two residues are critical for binding the phenylpropanoid portion of feruloyl-CoA. Molecular graphics images were produced using the UCSF Chimera package.
The nicotinamide ring of NADP+ was oriented roughly in parallel to a short stretch of protein backbone, which included Pro-210 through Val-213, presumably providing stability through hydrophobic interactions (Fig. 1). In addition, the nicotinamide carboxamide was in close proximity to the backbone amine of Val-213 and the most proximal internal phosphoryl group of the ligand, which suggests that these interactions also contribute to its binding.
Hydroxycinnamoyl-CoA-Binding Pocket
The substrate-binding domain of the SbCCR1 was surrounded by two groups of α-helices, and the floor of the substrate-binding pocket was largely composed of β-strands. The first helix group included α5, α6, and α7, and the second helix group was composed of α8, α10, and α12 (Fig. 1). The two groups were observed to be on the opposing lobes, surrounding a group of β-strands (β8, β10, β11, and β13) that provided structural integrity for the putative phenylpropanoid-binding region. β8 ran perpendicular to β10, β11, and β13 and was sandwiched between the triad of strands and the nicotinamide ring of NADP+. β9 and β12 were in an antiparallel arrangement that formed a solvent-exposed wall distal to the nicotinamide ring.
To investigate specific interactions between SbCCR1 and the phenylpropanoid constituents of hydroxycinnamoyl-CoA substrates, numerous unsuccessful soaking and cocrystallization attempts were made using either feruloyl-CoA or coniferaldehyde. Consequently, molecular docking approaches were employed using these same compounds. The docking calculations positioned the aldehydic group of coniferaldehyde over the A-face of the nicotinamide ring of NADP+ in the trans-configuration (Fig. 2B), which allows pro-R hydride transfer from the C4 atom of NADPH to the reactive carbonyl of the substrate(s). The distance between the aldehydic C9 atom of coniferaldehyde and the C4 atom of the nicotinamide ring was 2.3 Å. The lipophilic body of the ligand sat atop the side chains of Ile-150 and Val-211. The 3-methoxy and para-hydroxyl groups of the phenylpropanoid ring extended to 2.9 Å of the Thr-154 and Tyr-310 hydroxyl groups, respectively. Met-155, which is completely conserved among known CCRs, appeared to be in a suitable position to provide hydrophobic interaction with the 3-methoxy group on the ring of the ligand.
Isothermal Titration Calorimetry for Wild-Type and Mutant Forms of SbCCR1
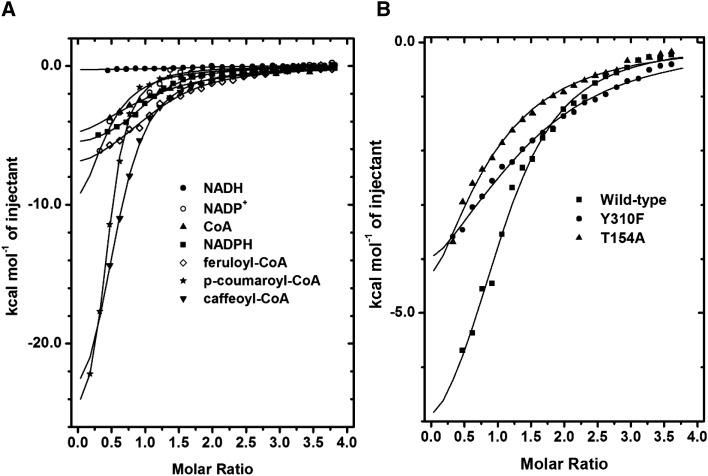
Isothermal titration calorimetry (ITC) was used to determine the thermodynamic parameters for the association of potential substrates, products, and substrate/product analogs. For these experiments, ligand solutions were injected into a calorimetric cell containing protein solution, and consequent changes in thermodynamic states of the cell solution were recorded. A large amount of heat was released (ΔH = −6.1 kcal mol−1) when NADPH was used as the titrant. This enthalpic change was accompanied by a modest entropic contribution to binding (ΔS = 3.2 cal mol−1 K−1), resulting in a Kd of 6.5 μm (Fig. 3A; Table II). No significant thermodynamic changes were observed when NADH was used as the titrant, which indicated that the ligand did not bind to SbCCR1. CoA bound to SbCCR1 with a Kd value of 25 μm, but SbCCR1 did not show significant affinity for ferulic acid (Fig. 3A; Table II). Feruloyl-CoA bound to SbCCR1 with a Kd of 13 μm in the absence of NADP+, with ΔH and ΔS of −8.6 kcal mol−1 and −6.4 cal mol−1 K−1, respectively. However, feruloyl-CoA bound only weakly or not at all in the presence of NADP+. Caffeoyl-CoA displayed slightly higher affinity (Kd = 7.2 μm) but similarly had little affinity for the enzyme-NADP+ complex.
Figure 3.
A, ITC curves for wild-type SbCCR1 upon titration with various ligands. As expected, significant enthalpic events were observed when the enzyme was titrated with NADP+, NADPH, CoA, and feruloyl-CoA. Surprisingly, reasonable affinity was observed for p-coumaroyl-CoA and caffeoyl-CoA as well. B, ITC comparison between wild-type SbCCR1 and mutants T154A and Y310F upon titration with feruloyl-CoA. Significant heat release was observed upon titration of the wild-type enzyme (squares) compared with titrations of T154A (triangles) or Y310F (circles).
Table II. Thermodynamic properties of interaction between SbCCR1 and various ligands.
ND, Not detected.
| Ligand | Kd | ΔH | ΔS |
|---|---|---|---|
| μm | kcal mol−1 | cal mol−1 K−1 | |
| Feruloyl-CoA (apo form) | 13 ± 1.7 | −8.6 ± 0.6 | −6.4 |
| Caffeoyl-CoA (apo form) | 7.2 ± 0.5 | −29 ± 1.9 | −72.8 |
| p-Coumaroyl-CoA | 2.1 ± 0.2 | −28 ± 0.9 | 23.5 |
| NADPH | 6.5 ± 0.6 | −6.1 ± 0.2 | 3.16 |
| NADP+ | 14 ± 4.7 | −3.1 ± 1.0 | 2.01 |
| CoA | 25 ± 4.0 | −7.2 ± 1.0 | −3.14 |
| NADH | ND | ND | ND |
| Ferulic acid | ND | ND | ND |
| Feruloyl-CoA (NADP+ bound) | ND | ND | ND |
| Caffeoyl-CoA (NADP+ bound) | ND | ND | ND |
To confirm the involvement in binding of Thr-154 and Tyr-310, which were postulated from the molecular docking results to interact with functional groups on the phenolic ring of feruloyl-CoA through hydrogen bonding, site-directed mutagenesis was employed to generate SbCCR1 mutants T154A and Y310F. The mutant proteins were expressed at similar levels to wild-type SbCCR1, and both were stable. Based on ITC data analysis, purified T154A and Y310F proteins displayed significantly lower affinity for feruloyl-CoA compared with the wild type (Fig. 3B; Table III), which supports that both residues contribute to binding feruloyl-CoA.
Table III. Thermodynamic properties of interaction between wild-type SbCCR1 or mutants T154A and Y310F and feruloyl-CoA.
| Plant | Kd | ΔH | ΔS |
|---|---|---|---|
| μm | kcal mol−1 | cal mol−1 K−1 | |
| Wild type | 13 ± 2 | −8.6 ± 0.6 | −6.4 |
| T154A | 37 ± 4 | −8.4 ± 1.6 | −8.1 |
| Y310F | 46 ± 6 | −6.8 ± 0.6 | −3.1 |
Identification of and Fold-Recognition Modeling for CCR Candidates in Sorghum
The amino acid sequence of SbCCR1 was used as a BLAST (Altschul et al., 1997) query to search the sorghum proteome (Phytozome database; https://phytozome.jgi.doe.gov/) for putative CCRs or CCR-like enzymes. The refined search yielded four additional candidates, Sobic.004G065600, Sobic.002G146000, Sobic.010G066000, and Sobic.003g116800, all of which contained the currently accepted CCR signature sequence NWYCY. To begin investigating whether these enzymes were true CCRs, the amino acid sequences were first aligned using Clustal Omega and then truncated to the N and C termini of SbCCR1 observed in the crystal structure (Fig. 4). The truncated sequences were submitted to the fold-recognition server Phyre2 using the SbCCR1 sequence as a control, and the resulting three-dimensional structures of these four candidates were superimposed. Extensive alignment was observed between each candidate and SbCCR1, with r.m.s.d. values ranging from 0.25 to 0.31 Å. The putative catalytic residues, which included Ser-149, Tyr-183, and Lys-187 of SbCCR1, were entirely conserved (Fig. 4) and in the proper positions for catalysis in all four CCR candidates.
Figure 4.
Multiple sequence alignment of the amino acid sequences for SbCCR1 and several related enzymes. Included in the alignment are a CCR1 from P. hybrida, CCR2s from M. truncatula and sorghum, a dihydroflavonol reductase from V. vinifera, and five CCRs or CCR-like enzymes from sorghum. Mining of sorghum proteomic data using the sequence of SbCCR1 as a query revealed four additional CCR candidates. Among the four additional candidates, high CCR activity could only be confirmed for SbCCR2. Residues that are completely conserved among all enzymes in the alignment are marked with asterisks. Residues highlighted in yellow are in β-strands, while those in red are in α-helices. Residues involved in binding of substrates or cosubstrates are in boldface and underlined. The CCR signature sequence NWYCY is outlined in a box. The N and C termini of CCR candidates from sorghum were truncated to closely match the experimentally observed termini of SbCCR1. The colon (:) and the period (.) indicate conservation of strongly similar properties and conservation of weakly similar properties, respectively.
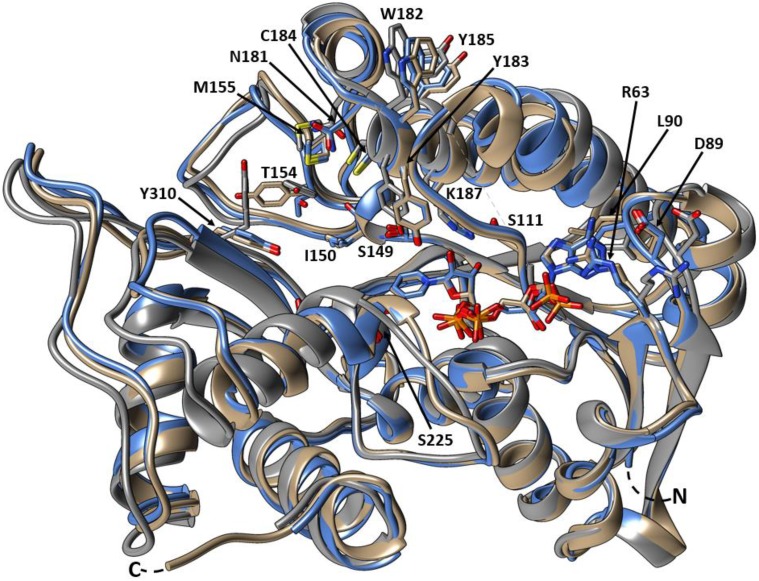
Structural Comparisons between SbCCR1, PhCCR1, and MtCCR2
To structurally compare SbCCR1, PhCCR1 (PDB identifier 4R1S), and MtCCR2 (PDB identifier 4R1U), the atomic coordinates of those three crystal structures were submitted for tertiary structure alignment using the software program COOT. The superimposed structures depicted conservation of the residues involved in phenylpropanoid and NADP(H) binding as well as conservation of the Ser-Tyr-Lys catalytic triad (Figs. 4 and 5). The only significant difference is that Thr-154 of SbCCR1 is replaced with Tyr in PhCCR1. As expected due to isoform difference and the absence of NADP(H) in the MtCCR2 structure, the alignment between SbCCR1 and MtCCR2 yielded a higher r.m.s.d. value (1.31 Å) than the alignment between SbCCR1 and PhCCR1 (0.68 Å).
Figure 5.
Superimposed tertiary structures of SbCCR1 and other CCRs. Depicted are SbCCR1 (blue), PhCCR1 (tan), and MtCCR2 (gray). Catalytic and phenylpropanoid-binding residues, as well as NADP+ from the SbCCR1 and PhCCR1 models, are represented as sticks. Conserved residues are represented according to residue type and sequence position in SbCCR1. Molecular graphics images were produced using the UCSF Chimera package.
Enzyme Kinetics
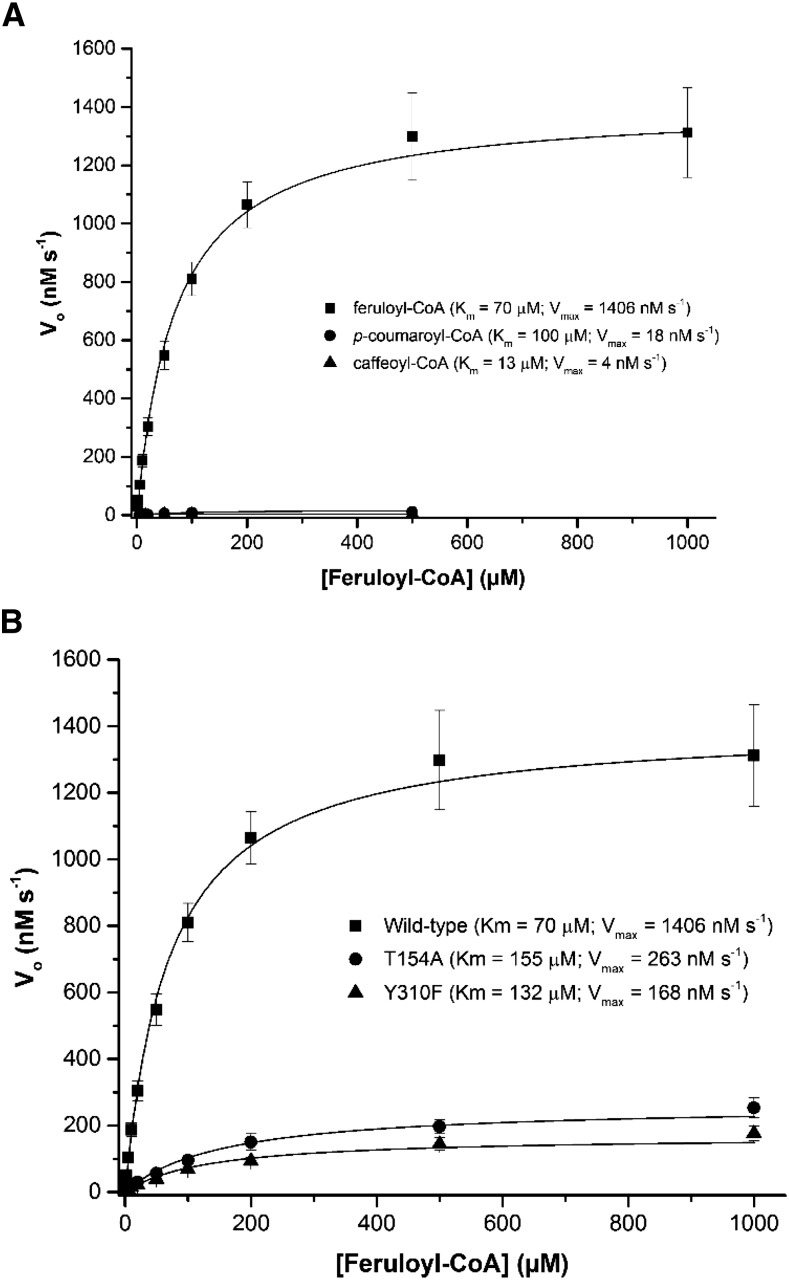
Enzyme kinetic assays with the wild-type SbCCR1 were carried out in the presence of various hydroxycinnamoyl-CoAs, including feruloyl-CoA, caffeoyl-CoA, and p-coumaroyl-CoA. While low activity was observed with caffeoyl-CoA and p-coumaroyl-CoA, much higher activity was observed for feruloyl-CoA (Fig. 6A). In addition, catalytic activities of three of its mutant forms, T154A, T154Y, and Y310F, were assessed with feruloyl-CoA or p-coumaroyl-CoA (only T154A and T154Y). Both T154A and Y310F mutants exhibited Vmax values for feruloyl-CoA that were approximately 5-fold lower than that of the wild-type enzyme as well as comparatively reduced catalytic efficiencies and higher Km values (Fig. 6B). However, the T154Y mutant exhibited 4.9- and 144-fold increases in catalytic efficiency for feruloyl-CoA and p-coumaroyl-CoA, respectively, over those of wild-type SbCCR1 (Table IV).
Figure 6.
A, Michaelis-Menten curves for wild-type SbCCR1 in the presence of three hydroxycinnamoyl-CoA substrates. All curves were constructed using initial rate measurements. For each reaction, the concentration of NADPH was held constant at 1 mm and the concentrations of hydroxycinnamoyl-CoA substrates were varied. SbCCR1 is highly active in the presence of feruloyl-CoA but minimally active with caffeoyl- or p-coumaroyl-CoA. B, Michaelis-Menten curves for wild-type SbCCR1 and two mutant forms, T154A and Y310F, in the presence of feruloyl-CoA. The two mutants were generated and tested for activity. Significant loss of activity was observed with either mutant. Given the locations of the two residues within the substrate-binding pocket, these data indicate that both residues are involved in binding feruloyl-CoA. The data were processed using Origin Pro 2015.
Table IV. Kinetic values for wild-type SbCCR1 in the presence of three hydroxycinnamoyl-CoA substrates.
| Substrate | Kcat | Km | Kcat/Km |
|---|---|---|---|
| s−1 | μm | μm−1 s−1 | |
| Feruloyl-CoA | 3.96 | 70 | 0.0566 |
| p-Coumaroyl-CoA | 0.05 | 100 | 0.0005 |
| Caffeoyl-CoA | 0.01 | 14 | 0.0008 |
Among the four additional sorghum genes with similarity to SbCCR1 identified through our BLAST search, Sobic.004G065600, Sobic.002G146000, and Sobic.010G066000 were isolated and could be expressed at levels sufficient for enzyme kinetic assays. As shown in Figure 7 and Table V, Sobic.004G065600 displayed a substantial increase in activity over SbCCR1 with p-coumaroyl-CoA, but its activity in the presence of feruloyl-CoA was relatively diminished. Because of its demonstrated CCR activity, we named this protein SbCCR2. The enzymes encoded by Sobic.002G146000 and Sobic.010G066000 were far less active overall than SbCCR1 or SbCCR2 under the assay conditions used and, thus, will be referred to in this report using only their gene identifiers until there is more clarity about their biological roles.
Figure 7.
A, Reaction velocities in the presence of feruloyl-CoA or p-coumaroyl-CoA for several forms of CCR. Sobic.002G146000 and Sobic.010G066000 are represented using the last five numbers of their gene identifiers. B, Thr-154 and Tyr-310 in SbCCR1 are shown binding functional groups of coniferaldehyde. C, Fold-recognition model of the active site of Sobic.004G065600 (SbCCR3). The bar graph was generated using Excel 2013 (Microsoft). Molecular graphics images were produced using the UCSF Chimera package.
Table V. Kinetic values for wild-type and mutant forms of SbCCR1, SbCCR2, Sobic.002G146000, and Sobic.010G066000 in the presence of feruloyl-CoA or p-coumaroyl-CoA.
ND, Not determined.
| Plant | Kcat (s−1) |
Km (μm) |
Kcat/Km (μm−1 s−1) |
|||
|---|---|---|---|---|---|---|
| Feruloyl-CoA | p-Coumaroyl-CoA | Feruloyl-CoA | p-Coumaroyl-CoA | Feruloyl-CoA | p-Coumaroyl-CoA | |
| Wild type | 3.96 | 0.05 | 70.2 | 100 | 0.0566 | 0.0005 |
| T154A | 0.74 | 0.02 | 155 | 70 | 0.0048 | 0.0003 |
| Y310F | 0.47 | ND | 132 | ND | 0.0036 | ND |
| T154Y | 17.24 | 8.42 | 63 | 113 | 0.2736 | 0.0745 |
| SbCCR2 | 3.24 | 0.50 | 247 | 14 | 0.0131 | 0.0358 |
| Sobic.002G146000 | 0.05 | 0.09 | 7.0 | 52 | 0.0072 | 0.0017 |
| Sobic.010G066000 | 0.04 | 0.00 | 8.0 | 5.0 | 0.0049 | 0.0000 |
DISCUSSION
Active Site, Kinetics of Enzyme Reaction, and Catalytic Mechanism
Our kinetic data indicate that SbCCR1 strongly prefers feruloyl-CoA as a substrate over the two other tested hydroxycinnamoyl-CoA thioester compounds (Figs. 6A and 7). Based on this substrate preference, the substrate-binding pocket was reexamined in order to pinpoint specific binding interactions between SbCCR1 and the phenylpropanoid moiety of feruloyl-CoA.
Although it was not the highest matches in our Dali and BLAST searches, dihydroflavonol reductase (DFR) from V. vinifera, which catalyzes the NADPH-dependent reduction of dihydroquercetin, is the only one that offers the ternary complex structure with its substrate (Trabelsi et al., 2008). Given that this dihydroflavonol and coniferaldehyde share critical aspects of chemical architecture, and that SbCCR1 contains functional analogs of DFR substrate-binding residues, we deemed the DFR an appropriate model for comparisons. The structural data for this reductase (PDB identifier 2C29) in complex with its substrate indicated that the enzyme uses the polar side chains of two residues (Asn-133 and Asn-227) for binding two hydroxyl groups located at the 3 and 4 positions on the aromatic ring of the substrate (Trabelsi et al., 2008). The backbone α-carbons of SbCCR1 and the dihydroflavonol reductase superimposed well, yielding an r.m.s.d. value of 1.38 Å. Comparisons of the overlapped structures showed that the side chains of Thr-154 and Tyr-310 of SbCCR1 were at nearly equivalent positions in space to the side chains of Asn-133 and Asn-227 in the dihydroflavonol reductase, which suggest that Thr-154 and Tyr-310 could be their respective functional analogs. Our molecular docking results for coniferyl aldehyde and kinetic profiling of the T154A, T154Y, and Y310F mutants indicated substantial involvement of these two residues in binding feruloyl-CoA. Overall, these structural and kinetic data, coupled with knowledge of the substrate-binding mode of a close structural homolog, strongly suggest that Thr-154 and Tyr-310 in SbCCR1 are directly involved in the binding of feruloyl-CoA.
In the proposed binding mechanism, the hydroxyl proton of Thr-154 interacted with the 3-methoxy oxygen of the ligand while the para-hydroxyl group established a hydrogen bond with the hydroxyl side chain of Tyr-310, as shown in the docked complex (Fig. 3C). This mechanism is consistent with the previous finding that substrate analogs bearing a 4-methoxy substitution resulted in comparatively less inhibition than endogenous substrate (Baltas et al., 2005). Given the close proximity of Gln-245 to the bound feruloyl-CoA, its side chain also could interact with the para-hydroxyl group of the substrate, which is held in the required conformer through a hydrogen bond with the hydroxyl group of Tyr-310. A Q245L mutant was generated to test whether this residue is essential for enzyme activity, but the mutant protein was not expressed at a level sufficient for protein purification. Nevertheless, kinetic profiling of a CCR1 with an amino acid substitution at Gln-245 will be needed to further increase the understanding of binding interactions between CCR1 and hydroxycinnamoyl-CoA substrates in lieu of a complex crystal structure.
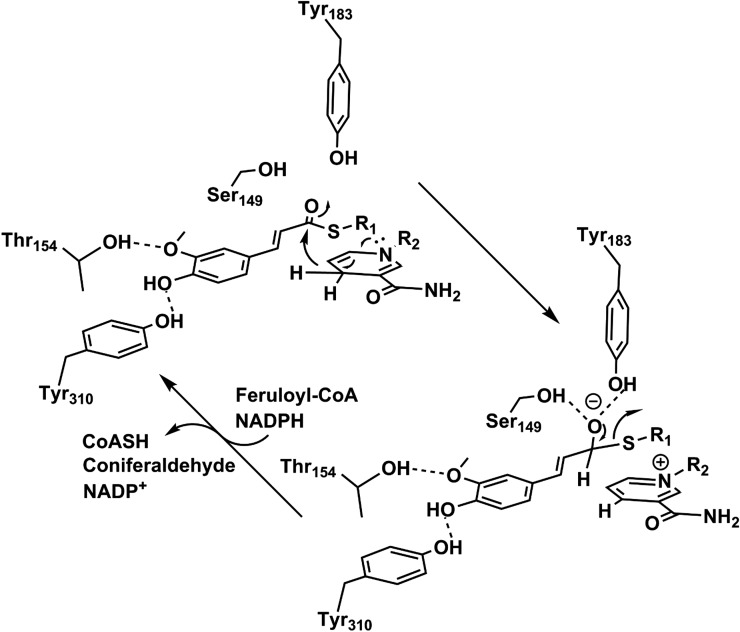
New information from this study significantly augments knowledge of the reaction mechanism for CCRs and related reductases. NADPH and feruloyl-CoA associate with their binding pockets, which are located nearest the N- and C-terminal domains, respectively. Tyr-310 binds the para-hydroxyl of feruloyl-CoA, while Thr-154 binds the 3-methoxy group of this molecule (Fig. 8). Considering the apparent overlap of the substrates while they are both bound, they are predicted to follow an ordered, sequential mechanism, where NADPH binds first and is followed by the hydroxycinnamoyl-CoA substrate. In the first catalytic step, pro-R hydride transfer occurs from the C4 atom of NADPH to the reactive thioester carbonyl. The resulting oxyanion is temporarily stabilized by the oxyanion hole established from the side chain hydroxyl groups of Ser-149 and Tyr-183. Collapse of the tetrahedral intermediate is then followed by C-S bond cleavage and protonation of the CoA thiolate. As indicated by our ITC data, in the presence of NADP+, there is very low affinity for the CoA ester compounds, which precludes the formation of a nonproductive complex.
Figure 8.
Proposed catalytic reaction mechanism of SbCCR1. Using the reduction potential of NADPH, SbCCR1 executes the hydrogenation of hydroxycinnamoyl-CoA substrates to form hydroxycinnamyl aldehyde and CoASH. Thr-154 and Tyr-310, which are involved in substrate binding, are shown forming hydrogen bonds to functional groups around the aromatic ring.
CCR Signature Residues and Residue Criteria for Substrate Specificity
To date, all confirmed CCRs share the 181NWYCY185 sequence motif. Although the central Tyr of this motif (Tyr-183 in SbCCR1) is known to be involved in enzyme catalysis (Jörnvall et al., 1995), the roles for three of the remaining residues appear to be structural in nature by positioning the phenolic side chain of Tyr-183. In SbCCR1, the side chain of Trp-182 is engaged in hydrophobic interactions with nearby Pro-117 and Tyr-185. In addition, the Trp-182 side chain was hydrogen bonded to the side chain of Lys-177. Because the side chains of Tyr-185 and Pro-117 both interact favorably with the Trp ring through face-to-face or face-to-edge interactions (Samanta et al., 2000), it is likely that substitution of Trp-182 with another residue will destabilize the α-helix (α7) that contains the conserved NWYCY motif, which, in turn, could directly affect both the catalysis and binding of the CoA group. The electron density map of SbCCR1 indeed reveals a face-to-face interaction between the pyrrole portion of the indole ring of Trp-182 and the Pro-117 side chain, while the orientation of the Tyr-185 side chain facilitates a face-to-edge interaction with the indole side chain of Trp-182. In addition, Cys-184 is in close proximity to Cys-176, and it has been shown that the two residues contribute to cystine formation under oxidizing conditions (Pan et al., 2014). Given that a 40% loss of activity was observed under these conditions, it seems that reduction of CCR in this region affords flexibility that allows for tighter hydroxycinnamoyl-CoA binding. The role of Asn-181 is less apparent, but it may be involved in the binding of hydroxycinnamoyl-CoA substrates.
The two CCRs that have been identified in several species, CCR1 and CCR2, are currently distinguished by expression differences (Pichon et al., 1998; Lauvergeat et al., 2001) or substrate specificity (Zhou et al., 2010). However, the lack of knowledge about the signature sequence for differentiating the two isoforms based on substrate preference, which is not strictly consistent with their genetic identities, underscores the need for criteria to recognize a functional dichotomy at the protein level (Escamilla-Treviño et al., 2010; Zhou et al., 2010). Convoluting matters is that the fundamental basis for substrate specificity has remained unknown so far. As shown in Figures 6A and 7, our data demonstrate that SbCCR1 strongly prefers feruloyl-CoA over p-coumaroyl- or caffeoyl-CoA. Through analysis of activity data from the mutant enzyme (Fig. 6B) and ITC results (Fig. 3), Thr-154 in SbCCR1 is clearly involved in directing affinity for feruloyl-CoA. A consistent feature of CCRs possessing Thr at this position is that they display a much higher preference for feruloyl-CoA than for other CCRs. Although PhCCR1, MtCCR1, MtCCR2, Sobic.002G146000, and SbCCR2 have a Tyr at this position and display higher activity for feruloyl-CoA, the disparities between their catalytic rates and efficiencies with feruloyl-CoA and p-coumaroyl-CoA or caffeoyl-CoA are far less than in in vitro observations for PvCCR1a (Escamilla-Trevino et al., 2010) and SbCCR1. The latter two enzymes have Thr at the equivalent position. Consistent with this observation, the T154Y SbCCR1 mutant exhibited substantially broader substrate specificity and a higher turnover rate. The fact that SbCCR1 is far more active with feruloyl-CoA implies that the other two hydroxycinnamoyl-CoA substrates bind to SbCCR1 in such a way that their reactive thioester bonds are not in position for efficient hydride transfer from NADPH. If this is true, then the physical premise for substrate specificity may not be due only to differences in phenylpropanoid-binding residues but also in the proximity of the conserved Tyr (Tyr-310 of SbCCR1) to the nicotinamide ring.
Our structurally guided search for other CCRs among sorghum genomic data yielded four more candidates each containing the NWYCY sequence, which were expected to display CCR activity. Sobic.002G146000 is an ortholog of ZmCCR2 (Pichon et al., 1998) and, according to the MOROHOSHI transcriptome database (Makita et al., 2015), was expressed robustly in sorghum roots, albeit at lower levels than SbCCR1. Sobic.003g116800 also was expressed in roots, but only under nitrogen stress. The remaining two genes, SbCCR2 and Sobic.010G066000, displayed low expression in all conditions and tissues examined and are not likely to play a significant role in the lignification of the secondary cell wall. By employing the same method as for SbCCR1, we were able to purify recombinant SbCCR2, Sobic.002G146000, and Sobic.010G066000. Although Sobic.002G146000 and Sobic.010G066000 displayed low activity for both feruloyl-CoA and p-coumaroyl-CoA, SbCCR2 showed substantially higher activity with p-coumaroyl-CoA relative to SbCCR1. SbCCR2 contains Tyr at the position of Thr-154 in SbCCR1; thus, their activities are coincidentally consistent with those from other CCRs that possess Tyr at the equivalent position to Thr-154. Given these consistencies, it is possible that having Thr at this position drives a strong preference for feruloyl-CoA, whereas the presence of Tyr (or potentially residues other than Thr) affords CCRs broader reductive capacity. p-Coumaroyl-CoA is a precursor to a wide range of phenylpropanoid compounds that have been implicated in plant defense. Therefore, the observed substrate promiscuity of SbCCR2 may be relevant to plant defenses against pathogens, which include the production of defense lignin at the site of attack. Broadening substrate specificity to include significant activity toward p-coumaroyl-CoA, which as a precursor also for flavonoids and stilbenes is quite abundant, will enable the more rapid generation of monolignols. Furthermore, lignin containing p-coumaryl alcohol is more heavily cross-linked, because both C3 and C5 of the phenolic ring can participate in radical coupling reactions with the growing lignin polymer. Consistent with this hypothesis, defense lignin formed in response to pathogen attack has been shown to have a higher proportion of H units in a number of different species (Lange et al., 1995; Pomar et al., 2004; Zhang et al., 2007). The activities of Sobic.002G146000 and Sobic.010G066000 were very low for both feruloyl-CoA and p-coumaroyl-CoA, leaving open the possibility that these two putative enzymes have different substrates in vivo.
CONCLUSION
Reducing lignin content can lower the barrier to the efficient production of renewable fuels and chemicals from lignocellulosic biomass. In view of this goal, CCR activity is necessary for the synthesis of all main monolignols, which makes CCR an attractive target for mutational and transgenic approaches that can modify this step of the pathway. Through molecular docking, mutagenesis, and kinetic analyses, we have identified a unique residue, Thr-154, that is critical to the association of the phenylpropanoid portion of feruloyl-CoA, the preferred substrate of SbCCR1. Based on structural and sequence similarities to CCR1, we also have identified four additional CCRs or CCR-like enzymes in sorghum and, following kinetic analyses of the CCRs encoded by the two most highly expressed SbCCR genes, identified a signature residue for substrate specificity that can be exploited in genome-editing approaches. The detailed structural and other mechanistic knowledge forms the basis for engineering monolignol biosynthetic enzymes that display altered substrate specificity, have reduced substrate or product inhibition, or have greater velocities, so that metabolic flux can be modulated. The potential of this approach is illustrated by the single amino acid substitution T154Y in SbCCR1, leading to faster turnover and broader substrate specificity. Taken together, these findings provide structural information, experimental evidence for binding residue involvement, and information about previously unidentified CCR genes in sorghum.
MATERIALS AND METHODS
Chemicals and Enzymes
Chemicals were obtained from Sigma-Aldrich or Fisher Scientific. Crystallization screens were obtained from Hampton Research and Qiagen.
Cloning and Enzyme Purification
cDNAs encoding SbCCR1 (Sobic.007G141200), SbCCR2 (Sobic.004G065600), Sobic.002G146000, and Sobic.010G066000 from sorghum (Sorghum bicolor) were ligated downstream of the sequence encoding the 6× His tag of pET30a (EMD Millipore) for SbCCR1 or pET45a for Sobic.002G146000 and SbCCR2 and introduced in Escherichia coli Rosetta (DE3) cells (EMD Millipore) via heat shock transformation.
Cultures (1.5 L) of E. coli Rosetta (DE3) strains expressing SbCCR1 or 3-L cultures of E. coli Rosetta (DE3) strains expressing SbCCR2, Sobic.002G146000, or Sobic.010G066000 were grown at 37°C in Luria-Bertani medium prior to expression of the recombinant enzymes. Strain selectivity was provided by supplementing the medium with 50 µg mL−1 kanamycin and 30 µg mL−1 chloramphenicol (SbCCR1) or 100 µg mL−1 ampicillin and 30 µg mL−1 chloramphenicol for all other CCRs. For expression and purification of His-tagged CCRs, all cultures were allowed to reach OD600 = 0.6, brought to their respective induction temperatures (20°C for SbCCR1 or 22°C for all other enzymes), and induced to express by bringing the medium to 0.5 mm IPTG and allowing it to shake for 20 h. Cells were harvested by centrifugation at 5,000g, frozen, and then suspended in 35 mL of 50 mm Tris-HCl supplemented with 300 mm NaCl and 20 mm imidazole at pH 8. Cells were lysed by sonication, and lysates were cleared by centrifugation at 15,000g. The cleared lysates were stirred into 10 mL of Ni-NTA agarose resin (Qiagen), the column was washed with 2 volumes of the lysis buffer, and recombinant enzymes were eluted with buffer containing 50 mm Tris base, pH 8, 300 mm NaCl, and 250 mm imidazole. The eluted sample was concentrated and exchanged into 20 mm Tris-HCl, pH 7.5, then applied to a 10-mL MonoQ column (GE Healthcare), and the enzyme was eluted with a 50 mm step-wise NaCl gradient in the same buffer. All CCRs eluted in approximately 150 mm NaCl, then they were buffer exchanged and concentrated into the appropriate buffer for crystallization or biochemical experiments. Purity was monitored for all protein preparations by SDS-PAGE, and protein concentrations were determined with the method of Bradford using BSA as a standard.
Protein Crystallization and Structure Determination
Crystals of SbCCR1 were grown using the hanging-drop, vapor-diffusion method. For crystallization, purified protein concentrated to 20 mg mL−1 in 20 mm Tris base, pH 7.5, 2 mm DTT, and 1 mm NADP+ was mixed with an equal volume of reservoir solution and equilibrated against the same solution at 4°C. The reservoir solution contained 100 mm Bis-Tris, pH 6.5, and 25% (w/v) PEG 3350. Crystals of the enzyme typically appeared within 5 d. Adequate cryoprotection was achieved by passing crystals through a small drop of storage buffer/mother liquor mixture (50% of each component by volume) that was brought to a final concentration of 19% (v/v) glycerol.
SbCCR1 in complex with NADP+ crystallized in the P32 space group with the asymmetric unit containing two monomers. Diffraction data were collected up to 2.9 Å resolution at the Berkeley Advanced Light Source (beam line 8.2.1). The data were processed with the HKL2000 package (Otwinowski and Minor, 1997). The statistics for the diffraction data are listed in Table I. Initial phasing of the diffraction data was conducted by molecular replacement with the PDB coordinates of CCR from Petunia hybrida (PDB identifier 4R1S) using PHENIX Phaser (Adams et al., 2002). Model building and iterative refinements were conducted using the programs COOT (Emsley et al., 2010) and PHENIX. The coordinates and diffraction data have been deposited in the PDB with identifier 5TQM.
Substrate Synthesis and Purification
Feruloyl-CoA, caffeoyl-CoA, and p-coumaroyl-CoA were synthesized according to a method described previously (Walker et al., 2013) but with slight modifications. Briefly, the substrates were prepared enzymatically by the addition of recombinant 4-coumarate ligase from sorghum to a solution containing 50 mm sodium phosphate buffered to pH 7, 800 μm CoA, 2.5 mm ATP, 2 mm ferulic, p-coumaric, or caffeic acid, and 5 mm magnesium chloride. The reactions, ranging in volume from 20 to 50 mL, were gently rocked for no less than 16 h prior to termination. Termination was achieved by thermal denaturation of the enzyme at 80°C for 10 min followed by centrifugation at 15,000g for 30 min. Solutes in the supernatant were pelleted by centrifugation under vacuum. Substrates were purified using ethanol to crystallize the remaining reaction components. The remaining fluid containing pure hydroxycinnamoyl-CoA esters was centrifuged under vacuum to pellet the products, which were stored at −20°C for later use.
Enzyme Kinetic Assays
Wild-type SbCCR1, its mutants, and all other enzymes were purified and used for steady-state kinetic experiments in the presence of the potential substrates p-coumaroyl-, caffeoyl-, and feruloyl-CoA. For experiments using varying concentrations of thioester substrate, the 70-μL reactions contained 50 mm Bis-Tris, pH 6.5, 1 mm NADPH, 1 μg of enzyme, and 1, 2, 5, 10, 20, 50, 100, 200, 500, or 1,000 μm of the thioester substrate. All reactions proceeded at 30°C and were terminated with 30 µL of 17% (v/v) trifluoroacetic acid after various incubation times in the range of 30 s (SbCCR1 and the T154Y SbCCR1 mutant) to 20 min (Sobic.002G146000, Sobic.010G066000, and the T154A SbCCR1 mutant). The reactions were centrifuged at 21,000g for 30 min at room temperature to pellet protein aggregates. A total of 50 μL of each reaction was injected onto a Luna C18(2) 5 μm, 4.6-mm × 150-mm column (Phenomenex) using the Hitachi Organizer HPLC unit, and corresponding aldehyde products from each reaction were quantified via peak integration from absorbance profiles at 333 or 346 nm. All reactions were performed in triplicate, and data were processed using Origin version 7.1 software (OriginLab).
Coniferaldehyde Docking
Molecular docking using SbCCR1 and coniferaldehyde was performed using AutoDock Vina (Trott and Olson, 2010). The files used in the calculations were prepared using AutoDock Tools 1.5.6 (Trott and Olson, 2010). The search space (x = 14 Å, y = 8 Å, and z = 22 Å) was defined considering the probable location of the phenylpropanoid-binding domain. Coniferaldehyde was constructed using the program COOT, structurally optimized using GaussView3 (Gaussian), and validated through comparison with the crystal structure of feruloyl adenylate in complex with a 4-coumarate ligase (PDB identifier 5BSV).
Site-Directed Mutagenesis and Mutant Expression
Two residues of SbCCR1, Thr-154 and Tyr-310, were mutated to Ala (A) and Phe (F), respectively, to test their involvement in binding of feruloyl-CoA. The T154A mutant was generated using the following primers: forward, 5′-TCCATCCGCGCGGTGGCCATGGACCCCAGC-3′, and reverse, 5′-GCTGGGGTCCATGGCCACCGCGCCGATGGA-3′. The Y310F mutant was generated using the following primers: forward, 5′-CGCGGAAGCAGCCGTTCAAGTTCTCGAACCAG-3′, and reverse, 5′-CTGGTTCGAGAACTTGAACGGCTGCTTCCGCG-3′. Additionally, mutant T154Y was generated to observe any changes in substrate specificity. The primer sequences used to generate the mutant are as follows: forward, 5′-CATCGGCGCGGTGTACATGGACCCCAGC-3′, and reverse, 5′-GCTGGGGTCCATGTACACCGCGCCGATG-3′. E. coli Rosetta (DE3) cells were transformed with plasmids containing the desired missense mutations in the SbCCR1 cDNA sequence, and proteins were expressed using the procedure described above.
ITC
Isothermal calorimetry titrations were performed for wild-type or mutant SbCCR1s and several ligands in an ITC200 instrument (Malvern Instruments). The protein was prepared by extensive buffer exchange (greater than 10,000-fold) into the titration buffer, which consisted of 10 mm sodium phosphate buffered to pH 7. The concentration of protein in the calorimetric titration cell was diluted to 50 μm. All titrations were performed at 25°C with a stirring speed of 750 rpm and 27 injections (1.4 µL each). All ligands were brought to a concentration of 1 mm in the titration buffer and injected into the protein solution, and the heat of binding was recorded.
Accession Numbers
Structural data from this article can be found in the Protein Data Bank under accession number 5TQM.
Glossary
- PDB
Protein Data Bank
- ITC
isothermal titration calorimetry
- r.m.s.d.
root-mean-square deviation
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB 102114, CHE 118359, and 1231085 to C.K.), the National Institutes of Health (grant no. 1R01GM11125401 to C.K.), and the M.J. Murdock Charitable Trust (to C.K.); by the Biomass Research and Development Initiative (grant no. 2011-1006-30358 to W.V.); by the U.S. Department of Energy’s Office of Energy Efficiency and Renewable Energy, Bioenergy Technologies Office (grant no. DE-PI0000031 to W.V.); and by the U.S. Department of Agriculture (National Institute of Food and Agriculture AFRI grant no. 2011-67009-30026 to S.E.S. and CRIS project grant no. 3042-21220-032-00D).
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltas M, Lapeyre C, Bedos-Belval F, Maturano M, Saint-Aguet P, Roussel L, Duran H, Grima-Pettenati J (2005) Kinetic and inhibition studies of cinnamoyl-CoA reductase 1 from Arabidopsis thaliana. Plant Physiol Biochem 43: 746–753 [DOI] [PubMed] [Google Scholar]
- Bout S, Vermerris W (2003) A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol Genet Genomics 269: 205–214 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Derikvand M, Sierra JB, Ruel K, Pollet B, Do CT, Thévenin J, Buffard D, Jouanin L, Lapierre C (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Dien B, Sarath G, Pedersen J, Sattler S, Chen H, Funnel-Harris D, Nichols N, Cotta M (2009) Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor (L.) Moench) lines with reduced lignin contents. Bioenerg Res 2: 153–164 [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Treviño LL, Shen H, Uppalapati SR, Ray T, Tang Y, Hernandez T, Yin Y, Xu Y, Dixon RA (2010) Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol 185: 143–155 [DOI] [PubMed] [Google Scholar]
- Farré I, Faci J (2006) Comparative response of maize (Zea mays L.) and sorghum (Sorghum bicolor (L.) Moench) to deficit irrigation in a Mediterranean environment. Agric Water Manage 83: 135–143 [Google Scholar]
- Fontaine AS, Bout S, Barrière Y, Vermerris W (2003) Variation in cell wall composition among forage maize (Zea mays L.) inbred lines and its impact on digestibility: analysis of neutral detergent fiber composition by pyrolysis-gas chromatography-mass spectrometry. J Agric Food Chem 51: 8080–8087 [DOI] [PubMed] [Google Scholar]
- Goujon T, Ferret V, Mila I, Pollet B, Ruel K, Burlat V, Joseleau JP, Barrière Y, Lapierre C, Jouanin L (2003) Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: effects on phenotype, lignins and cell wall degradability. Planta 217: 218–228 [DOI] [PubMed] [Google Scholar]
- Green AR, Lewis KM, Barr JT, Jones JP, Lu F, Ralph J, Vermerris W, Sattler SE, Kang C (2014) Determination of the structure and catalytic mechanism of Sorghum bicolor caffeic acid O-methyltransferase and the structural impact of three brown midrib12 mutations. Plant Physiol 165: 1440–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Hu F, Ragauskas A (2012) Pretreatment and lignocellulosic chemistry. Bioenerg Res 5: 1043–1066 [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34: 6003–6013 [DOI] [PubMed] [Google Scholar]
- Jung H, Buxton D (1994) Forage quality variation among maize inbreds: relationships of cell-wall composition and in-vitro degradability for stem internodes. J Sci Food Agric 66: 313–322 [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, Boudet AM, Grima-Pettenati J (1997) Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J 11: 429–441 [DOI] [PubMed] [Google Scholar]
- Lange BM, Lapierre C, Sandermann H Jr (1995) Elicitor-induced spruce stress lignin: structural similarity to early developmental lignins. Plant Physiol 108: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Leu SY, Zhu JY, Gleisner R, Sessions J, Marrs G (2013) Robust enzymatic saccharification of a Douglas-fir forest harvest residue by SPORL. Biomass Bioenergy 59: 393–401 [Google Scholar]
- Makita Y, Shimada S, Kawashima M, Kondou-Kuriyama T, Toyoda T, Matsui M (2015) MOROKOSHI: transcriptome database in Sorghum bicolor. Plant Cell Physiol 56: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pan H, Zhou R, Louie GV, Mühlemann JK, Bomati EK, Bowman ME, Dudareva N, Dixon RA, Noel JP, Wang X (2014) Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis. Plant Cell 26: 3709–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PJ, Macedo-Ribeiro S, Párraga A, Pérez-Luque R, Cunningham O, Darcy K, Mantle TJ, Coll M (2001) Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme. Nat Struct Biol 8: 215–220 [DOI] [PubMed] [Google Scholar]
- Pichon M, Courbou I, Beckert M, Boudet AM, Grima-Pettenati J (1998) Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes. Plant Mol Biol 38: 671–676 [DOI] [PubMed] [Google Scholar]
- Piquemal J, Lapierre C, Myton K, O’Connell A, Schuch W, Grima-Pettenati J, Boudet A (1998) Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J 13: 71–83 [Google Scholar]
- Pomar F, Novo M, Bernal MA, Merino F, Barceló A (2004) Changes in stem lignins (monomer composition and crosslinking) and peroxidase are related with the maintenance of leaf photosynthetic integrity during Verticillium wilt in Capsicum annuum. New Phytol 163: 111–123 [DOI] [PubMed] [Google Scholar]
- Porter K, Axtell J, Lechtenberg V, Colenbrander V (1978) Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci 18: 205–208 [Google Scholar]
- Ralph J, Hatfield RD, Piquemal J, Yahiaoui N, Pean M, Lapierre C, Boudet AM (1998) NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc Natl Acad Sci USA 95: 12803–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz P, Marita J, Hatfield R, Ralph S, Christensen J, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70: 818–830 [DOI] [PubMed] [Google Scholar]
- Saballos A, Vermerris W, Rivera L, Ejeta G (2008) Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenerg Res 1: 193–204 [Google Scholar]
- Samanta U, Pal D, Chakrabarti P (2000) Environment of tryptophan side chains in proteins. Proteins 38: 288–300 [PubMed] [Google Scholar]
- Sattler SE, Palmer NA, Saballos A Greene AM, Xin Z, Sarath G, Vermerris W, Pedersen JF (2012) Identification and characterization of 4 missense mutations in brown midrib 12 (Bmr12): the caffeic O-methyltransferase (COMT) of sorghum. Bioenerg Res 5: 855–865 [Google Scholar]
- Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol 150: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully ED, Gries T, Funnell-Harris DL, Xin Z, Kovacs FA, Vermerris W, Sattler SE (2016) Characterization of novel Brown midrib 6 mutations affecting lignin biosynthesis in sorghum. J Integr Plant Biol 58: 136–149 [DOI] [PubMed] [Google Scholar]
- Tamasloukht B, Wong Quai Lam MS, Martinez Y, Tozo K, Barbier O, Jourda C, Jauneau A, Borderies G, Balzergue S, Renou JP, et al. (2011) Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fibre development, and global gene expression. J Exp Bot 62: 3837–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi N, Petit P, Manigand C, Langlois d’Estaintot B, Granier T, Chaudière J, Gallois B (2008) Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols. Acta Crystallogr D Biol Crystallogr D64: 883–891 [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A, Hayes R, Youn B, Vermerris W, Sattler S, Kang C (2013) Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol 162: 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AM, Sattler SA, Regner M, Jones JP, Ralph J, Vermerris W, Sattler SE, Kang C (2016) The structure and catalytic mechanism of Sorghum bicolor caffeoyl-CoA O-methyltransferase. Plant Physiol 172: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Rezenom YH, Cho KC, Tran JL, Lee DG, Russell DH, Gill JJ, Young R, Chu KH (2014) Cultivation of lipid-producing bacteria with lignocellulosic biomass: effects of inhibitory compounds of lignocellulosic hydrolysates. Bioresour Technol 161: 162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengenmayer H, Ebel J, Grisebach H (1976) Enzymic synthesis of lignin precursors: purification and properties of a cinnamoyl-CoA:NADPH reductase from cell suspension cultures of soybean (Glycine max). Eur J Biochem 65: 529–536 [DOI] [PubMed] [Google Scholar]
- Ximenes E, Kim Y, Mosier N, Dien B, Ladisch M (2011) Deactivation of cellulases by phenols. Enzyme Microb Technol 48: 54–60 [DOI] [PubMed] [Google Scholar]
- Zhang YH, Ding SY, Mielenz JR, Cui JB, Elander RT, Laser M, Himmel ME, McMillan JR, Lynd LR (2007) Fractionating recalcitrant lignocellulose at modest reaction conditions. Biotechnol Bioeng 97: 214–223 [DOI] [PubMed] [Google Scholar]
- Zhou R, Jackson L, Shadle G, Nakashima J, Temple S, Chen F, Dixon RA (2010) Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc Natl Acad Sci USA 107: 17803–17808 [DOI] [PMC free article] [PubMed] [Google Scholar]