Solving the complex II puzzle: SDH6 and SDH7 contribute to anchoring succinate dehydrogenase to the inner mitochondrial membrane in Arabidopsis thaliana.
Abstract
The succinate dehydrogenase complex (complex II) is a highly conserved protein complex composed of the SDH1 to SDH4 subunits in bacteria and in the mitochondria of animals and fungi. The reason for the occurrence of up to four additional subunits in complex II of plants, termed SDH5 to SDH8, so far is a mystery. Here, we present a biochemical approach to investigate the internal subunit arrangement of Arabidopsis (Arabidopsis thaliana) complex II. Using low-concentration detergent treatments, the holo complex is dissected into subcomplexes that are analyzed by a three-dimensional gel electrophoresis system. Protein identifications by mass spectrometry revealed that the largest subcomplex (IIa) represents the succinate dehydrogenase domain composed of SDH1 and SDH2. Another subcomplex (IIb) is composed of the SDH3, SDH4, SDH6, and SDH7 subunits. All four proteins include transmembrane helices and together form the membrane anchor of complex II. Sequence analysis revealed that SDH3 and SDH4 lack helices conserved in other organisms. Using homology modeling and phylogenetic analyses, we present evidence that SDH6 and SDH7 substitute missing sequence stretches of SDH3 and SDH4 in plants. Together with SDH5, which is liberated upon dissection of complex II into subcomplexes, SDH6 and SDH7 also add some hydrophilic mass to plant complex II, which possibly inserts further functions into this smallest protein complex of the oxidative phosphorylation system (which is not so small in plants).
Succinate dehydrogenase (EC 1.3.5.1) is of central importance for energy metabolism in bacteria and mitochondria of eukaryotic cells. In mitochondria, it represents the complex II of the oxidative phosphorylation (OXPHOS) system (Hatefi, 1985) and is located in the inner mitochondrial membrane. Complex II participates in two major mitochondrial processes: the tricarboxylic acid cycle as well as the mitochondrial electron transfer chain (mETC). In the tricarboxylic acid cycle, it catalyzes the conversion of succinate into fumarate. Electrons originating from this reaction are inserted into the mETC and used for the reduction of ubiquinone to ubiquinol. In contrast to other protein complexes of the OXPHOS system, complex II does not contribute to the proton gradient across the inner mitochondrial membrane.
The overall structure of the succinate dehydrogenase complex is remarkably conserved in bacteria, animals, and fungi, as revealed by biochemical investigations and x-ray crystallography (Yankovskaya et al., 2003; Oyedotun and Lemire, 2004; Sun et al., 2005; Huang et al., 2006; Iverson, 2013). It is about 120 kD in size and composed of four subunits designated SDH1, SDH2, SDH3, and SDH4 (also named SDHA to SDHD in some bacteria). SDH1 is the largest subunit and includes the succinate-binding site. Electrons from succinate are accepted by a covalently bound FAD group. SDH2 carries three iron-sulfur clusters that mediate electron transfer from SDH1 to the membrane domain of complex II. Together with SDH1, it constitutes the succinate dehydrogenase domain of complex II. Both SDH1 and SDH2 are hydrophilic and protrude into the mitochondrial matrix or into the cytoplasm of the bacterial cell. The membrane attachment of SDH1 and SDH2 is mediated by two hydrophobic transmembrane proteins termed SDH3 and SDH4. Both proteins include three parallel membrane-spanning helices that together constitute a six-helix bundle. The SDH3 and SDH4 proteins form pockets for the binding of ubiquinone and a heme group. Furthermore, lipids are bound to SDH3 and SDH4 at defined positions (Iverson, 2013).
It was quite a surprise when complex II from flowering plants was found to include four additional subunits termed SDH5, SDH6, SDH7, and SDH8 (Eubel et al., 2003; Millar et al., 2004; Huang et al., 2010; Huang and Millar, 2013; Senkler et al., 2017). (Note: The designations SDH5, SDH6, SDH7, and SDH8 were introduced for the four extra subunits present in complex II of flowering plants [Eubel et al., 2003; Millar et al., 2004]. At a later stage, the same designations also were introduced for complex II assembly factors in fungi and mammals [Hao et al., 2009, and other publications, reviewed in van Vranken et al., 2015]. The two sets of SDH5 to SDH8 proteins are not related.) The extra subunits have been characterized in Arabidopsis (Arabidopsis thaliana), bean (Phaseolus vulgaris), potato (Solanum tuberosum), and rice (Oryza sativa). Except for SDH8, genes encoding these subunits are present in the genomes of all flowering plants. SDH5 is a hydrophilic subunit, while the SDH6, SDH7, and SDH8 proteins are more hydrophobic. Sequence analyses so far have not revealed any ideas on the functions of the additional SDH proteins. Possibly, they insert an extra function into complex II in flowering plants (Millar et al., 2004). Extra functional modules of OXPHOS complexes also have been described for the complex I of plants and protozoans, which includes a carbonic anhydrase domain absent in bacteria, fungi, and animals (Sunderhaus et al., 2006; Gawryluk and Gray 2010). Furthermore, the mitochondrial processing peptidase is inserted into complex III of the mETC in plants (Braun et al., 1992). In the algal genus Polytomella, large additional subunits of mitochondrial ATP synthase form the scaffold for a stable dimerization of the enzyme complex (Villavicencio-Queijeiro et al., 2009).
The SDH subunits of complex II are encoded by nuclear genes in most flowering plants. In Arabidopsis, some SDH proteins occur in isoforms (Figueroa et al., 2001, 2002; Klodmann et al., 2011; Welchen et al., 2011). Two isoforms are present for SDH1, SDH3, and SDH7, and three isoforms are present for SDH2. In contrast, SDH4, SDH6, and SDH8 are encoded by single-copy genes. Overall, the SDH subunits are encoded by 13 genes in Arabidopsis.
The physiological role of complex II in plants so far is only partly understood. Knock out of genes encoding SDH1 or SDH2 caused a failure of gametophyte development (León et al., 2007; Huang and Millar, 2013). Down-regulation of SDH genes led to increased succinate levels as well as altered concentrations of organic acids and amino acids (Araújo et al., 2011; Fuentes et al., 2011). It was shown recently that complex II is a site of reactive oxygen species production in plants (Gleason et al., 2011; Jardim-Messeder et al., 2015). Optimal function of SDH1 is required for salicylic acid-mediated defense gene expression in Arabidopsis (Gleason et al., 2011).
Biochemically, complex II from flowering plants only has been characterized by analyses of mitochondrial fractions using two-dimensional (2D) blue native (BN) SDS-PAGE (Eubel et al., 2003; Millar et al., 2004; Huang et al., 2010; Klodmann et al., 2011). Solubilization of mitochondrial membranes with Triton X-100 or dodecylmaltoside (DDM) causes complete dissociation of Arabidopsis complex II (Eubel et al., 2003). This is in contrast to the situation in fungi and mammals, which include DDM-stable forms of complex II (Schägger and von Jagow, 1991; Schägger and Pfeiffer, 2000). Only membrane solubilization by digitonin is compatible with complex II stabilization in plants. In fact, complex II from plants is exceptionally unstable and has, to our knowledge, never been isolated successfully for any plant species.
We report here a novel gel-based approach to elucidate the topological arrangement of the eight-subunit complex II of plants. Complex II from Arabidopsis is carefully destabilized by low-concentration detergent treatment. Subsequently, dissection products are analyzed by a multidimensional gel electrophoresis system and subunits of complex II subcomplexes are identified by mass spectrometry. We show that the plant-specific SDH6 and SDH7 proteins are tightly bound to SDH3 and SDH4 and contribute to the membrane domain of mitochondrial complex II from Arabidopsis. Structural modeling suggests that SDH3 and SDH4 proteins are truncated in flowering plants. We present evidence that SDH6 and SDH7 substitute membrane-spanning helices absent in the SDH3 and SDH4 proteins in plants. Phylogenetic analyses reveal the acquisition of the additional SDH subunits during the evolution of the green lineage.
RESULTS
Complex II of Arabidopsis Can Be Dissected into Two Subcomplexes Termed IIa and IIb
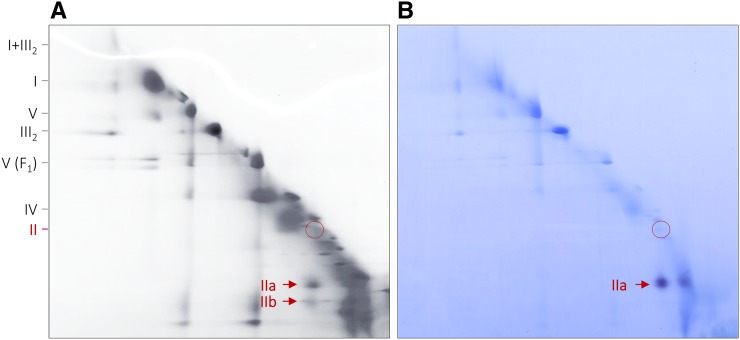
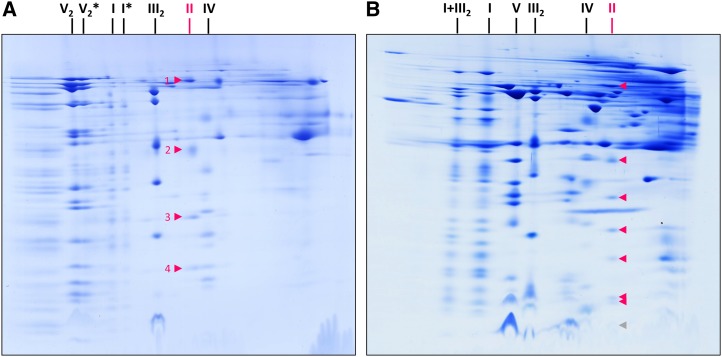
2D blue native/blue native (BN/BN) PAGE allows analyzing the subcomplex structure of protein complexes and protein supercomplexes (Schägger and Pfeiffer, 2000; Sunderhaus et al., 2007). To investigate membrane-bound complexes and supercomplexes, membrane solubilization is carried out using a very mild detergent (e.g. digitonin). First-dimension BN PAGE is carried out in the presence of the same detergent. Second-dimension BN PAGE is carried out in the presence of a slightly harsher detergent (e.g. DDM). On the resulting 2D gel, protein complexes likewise stable in the presence of both detergents end up on a diagonal line. In contrast, protein complexes destabilized in the presence of the second detergent are dissected into subcomplexes that migrate below the diagonal line. This experimental system was employed to investigate Arabidopsis complex II. While complex II of Arabidopsis is known to be stable in the presence of digitonin, it is dissected into two subcomplexes in the presence of DDM (Fig. 1A). Indeed, the complex II holo complex is hardly detectable on the resulting 2D BN/BN gel. The two subcomplexes migrate in close proximity on the 2D gel.
Figure 1.
Detection of complex II in mitochondrial fractions from Arabidopsis by BN/BN PAGE and complex II activity staining. BN/BN PAGE was carried out as outlined in “Materials and Methods.” A, Coomassie Blue-stained 2D gel. B, Parallel 2D gel used for complex II activity staining. The identities of the separated protein complexes and supercomplexes are given to the left: I, complex I; II, complex II; III2, dimeric complex III; IV, complex IV; V, complex V; V(F1), F1 part of complex V; I+III2, supercomplex formed by complexes I and III2. The position of intact complex II is given by the red circles, and the positions of two complex II subcomplexes (IIa and IIb) are indicated by red arrows.
Subcomplex IIa Exhibits in Gel Succinate Dehydrogenase Activity
The larger subcomplex, which was termed subcomplex IIa, exhibits succinate dehydrogenase activity upon analysis by an in gel activity assay (Fig. 1B). In contrast, the smaller subcomplex, termed subcomplex IIb, does not exhibit succinate dehydrogenase activity. A faint activity signal also is visible on the diagonal line, indicating that a small proportion of the holo complex is already dissected into the two subcomplexes upon digitonin solubilization. A similar observation has been described previously by in gel complex II activity staining using one-dimensional (1D) BN PAGE (Huang et al., 2010; Sunderhaus et al., 2010).
The gel spots of subcomplexes IIa and IIb were analyzed by mass spectrometry to investigate their subunit compositions (Supplemental Fig. S1). As expected from the in gel activity assay, SDH1 and SDH2 had highest intensities in the gel spot of subcomplex IIa. In contrast, subunits SDH3, SDH4, SDH6, and SDH7 had highest intensities in the gel spot of subcomplex IIb (subunits SDH5 and SDH8 were not identified in either of the two subcomplexes). However, SDH1 and SDH2 also were detected in subcomplex IIb, and SDH3, SDH6, and SDH7 were detected in subcomplex IIa, probably caused by overlap of the two complex II subcomplex spots on the 2D gel. Therefore, we decided to optimize our gel electrophoresis strategy for further analysis of Arabidopsis complex II.
Complex II Subcomplexes Can Be Separated by Three-Dimensional BN/BN SDS-PAGE
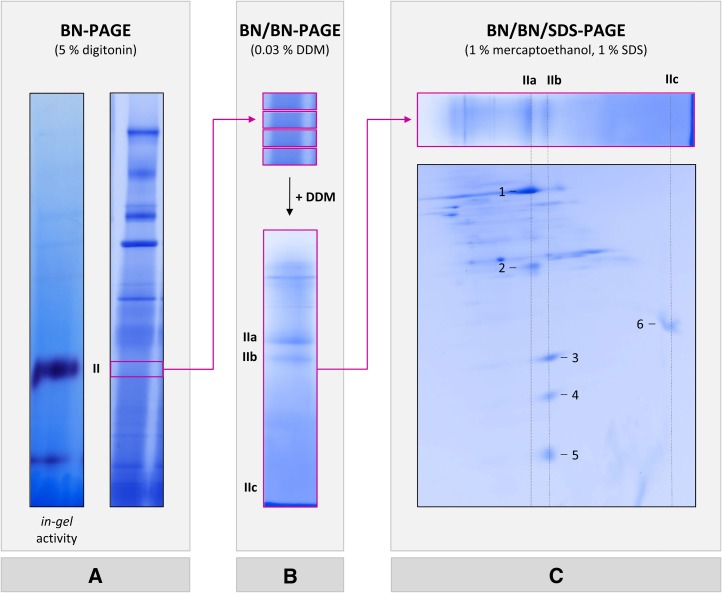
Complex II was further analyzed by combining three gel dimensions (Fig. 2). First, digitonin-solubilized mitochondrial fractions were separated by 1D BN PAGE using a 4.5% to 16% (v/v) polyacrylamide gradient gel. Since the complex II band is not visible upon Coomassie Blue staining, its position in the BN gel stripe was identified by the succinate dehydrogenase in gel activity assay (Fig. 2A). Next, gel pieces including complex II, as identified by activity staining, were cut out on parallel BN gel stripes, which were neither fixed nor stained. Gel pieces were incubated with 0.03% (w/v) DDM to dissect complex II into subcomplexes. Four gel pieces including dissected complex II were subsequently stacked and transferred onto a second-dimension BN gel to increase the protein amount. Subcomplex separation took place on a 5% to 20% (v/v) polyacrylamide gradient gel. Using this second-dimension BN PAGE nicely allows separating subcomplexes IIa and IIb (Fig. 2B). Finally, subunits of complex II subcomplexes were separated by a third gel dimension, which was carried out in the presence of SDS (Fig. 2C).
Figure 2.
Experimental strategy for analyzing complex II subcomplexes from Arabidopsis. A, Protein complexes of isolated mitochondria were separated by 1D BN PAGE and either complex II activity stained (left) or Coomassie Blue stained (right). The region of complex II as identified by activity staining (boxed in magenta) was cut out on parallel BN gel stripes that were neither fixed nor stained. B, Four BN gel pieces including native complex II were incubated with 0.03% (w/v) DDM and subsequently stacked on top of a second BN gel. This second BN PAGE dimension was used to separate complex II subcomplexes. C, The resulting gel stripe (boxed in magenta) was incubated with 1% (w/v) SDS and 1% (v/v) 2-mercaptoethanol, and subunits of complex II subcomplexes were subsequently separated in the orthogonal direction on a third gel dimension, which was carried out in the presence of SDS. Magenta arrows indicate the work flow. The position of native complex II and the positions of subcomplexes IIa, IIb, and IIc are indicated to the left or above the gel stripes. Proteins analyzed by mass spectrometry are numbered (for results, see Table I).
Subunits SDH3, SDH4, SDH6, and SDH7 Constitute the Membrane Domain
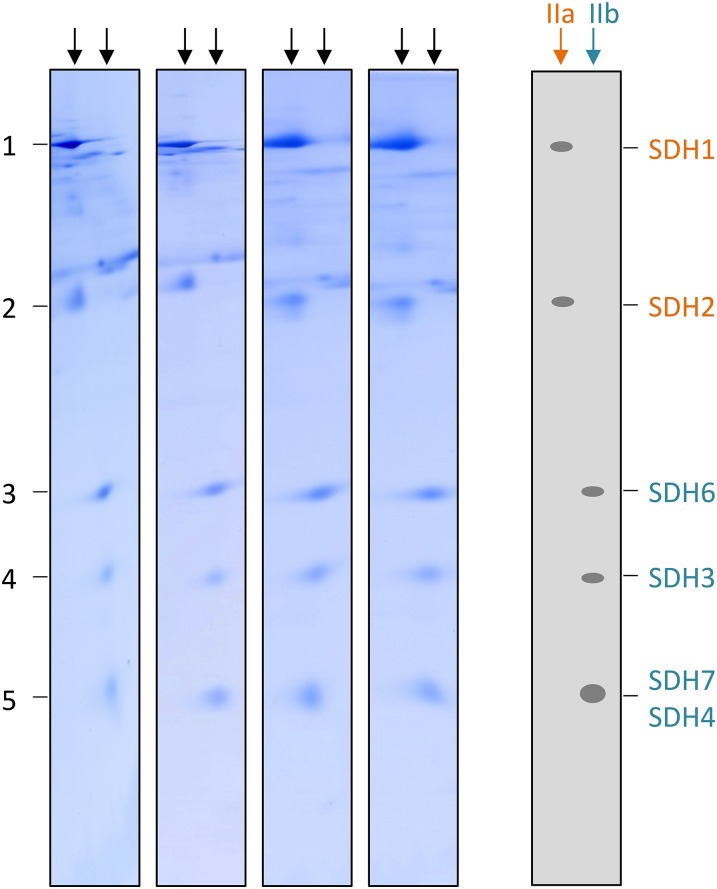
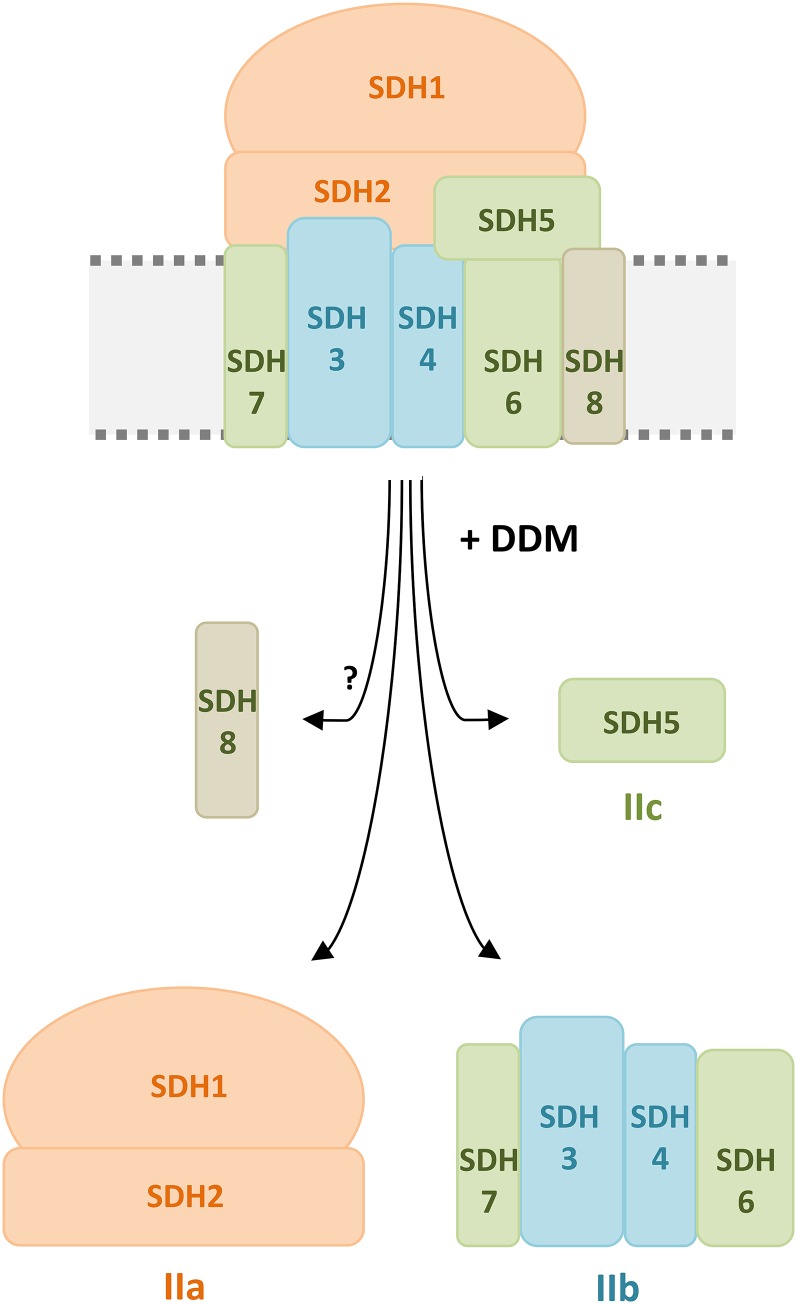
On the resulting SDS gel, subcomplex IIa reproducibly is separated into two and subcomplex IIb into three protein spots (Figs. 2C and 3). Another protein spot present in the low-molecular-mass region of the second-dimension BN gel is detectable on the SDS gel and termed subcomplex IIc (Fig. 2C). Mass spectrometry was finally employed for protein identification (Table I; Supplemental Table S2). The results revealed that subcomplex IIa represents the succinate dehydrogenase domain of complex II, which consists of SDH1 and SDH2. Subcomplex IIb includes the membrane anchor subunits SDH3 and SDH4. Interestingly, SDH6 and SDH7 form part of this domain. Finally, subcomplex IIc includes the plant-specific subunit SDH5. We conclude that SDH5 probably is located at the interface of the succinate dehydrogenase and membrane domains and that it is liberated from complex II by destabilization using DDM. SDH8 was not detected by our three-dimensional (3D) PAGE approach.
Figure 3.
Subunit composition of complex II subcomplexes IIa and IIb in Arabidopsis. Subcomplexes of complex II were analyzed by BN/BN SDS-PAGE in four replicates. Subunits of subcomplexes IIa (orange) and IIb (turquoise) were identified by mass spectrometry (Table I). Identities of the complex II subunits are given to the right.
Table I. Proteins identified by mass spectrometry.
Further details on the measurements are presented in Supplemental Table S2.
| Spota | Accession No.b | Name | Massc | Scored | Peptidese | Coveragef |
|---|---|---|---|---|---|---|
| kD | % | |||||
| 1 | At5g66760 | SDH1-1 | 69.6 | 1,988 | 44 | 57.1 |
| 2 | At3g27380 | SDH2-1 | 31.2 | 1,194 | 26 | 64.5 |
| 2 | At5g40650 | SDH2-2 | 31.1 | 1,093 | 24 | 62.9 |
| 3 | At1g08480 | SDH6 | 15.8 | 356 | 12 | 51.4 |
| 4 | At4g32210 | SDH3-2 | 23.4 | 132 | 3 | 19.7 |
| 5 | At3g47833 | SDH7-1 | 10.3 | 162 | 3 | 14.0 |
| 5 | At5g62575 | SDH7-2 | 10.9 | 146 | 3 | 18.2 |
| 5 | At2g46505 | SDH4 | 16.8 | 91 | 4 | 23.2 |
| 6 | At1g47420 | SDH5 | 28.1 | 730 | 10 | 48.0 |
Spot number as indicated in Figures 2 and 3 and Supplemental Table S2. bAccession numbers according to TAIR (https://www.arabidopsis.org/). cCalculated molecular mass of the precursor protein. dMASCOT reliability score. eNumber of unique peptides. fCoverage of the protein sequence by peptides.
The SDH3 and SDH4 Subunits of Arabidopsis Lack Membrane-Spanning Helices Essential for the Anchoring of SDH1 and SDH2
SDH6, SDH7, and also SDH8 (which was not identified in the course of our investigation) are predicted to include membrane-spanning helices, which supports their localization within the membrane domain of complex II (Supplemental Fig. S2). In contrast, SDH5 is a soluble protein. Since the subunit composition of the membrane domain of complex II is highly conserved in all bacteria and mitochondria from animals and fungi analyzed so far (it only consists of SDH3 and SDH4), we wondered why this domain includes at least two further subunits in flowering plants. SDH3 and SDH4 are known to be evolutionarily related and belong to a small protein family (sequence identity between SDH3 and SDH4 is 24% in Escherichia coli, 14% in chicken [Gallus gallus], and 15% in yeast; see Supplemental Fig. S3). Both proteins are very hydrophobic and have molecular masses of about 12 to 15 kD. Analysis of complex II structure in E. coli, pig (Sus scrofa), and chicken using x-ray crystallography revealed that both proteins have a highly conserved structure that consists of three membrane-spanning helices each (Yankovskaya et al., 2003; Sun et al., 2005; Huang et al., 2006). The resulting six-helix bundle forms defined pockets for the binding of heme and ubiquinone. At the same time, the six-helix bundle forms the interface for the binding of the succinate dehydrogenase domain constituted by SDH1 and SDH2.
We used the SDH3, SDH4, SDH5, SDH6, SDH7, and SDH8 sequences of Arabidopsis for structure prediction analyses. All six proteins are encoded by the nuclear genome and posttranslationally transported into the mitochondria. The N termini of the mature proteins have been determined previously (the N termini of SDH3, SDH5, SDH7, and SDH8 were identified by Edman degradation [Millar et al., 2004], and the N termini of SDH1, SDH2, SDH4, and SDH6 can be deduced from the sequences of semitryptic peptides identified by shotgun proteome projects [Baerenfaller et al., 2008; Huang et al., 2009]; for details, see Supplemental Fig. S4). The calculated molecular masses of mature SDH1 to SDH8 proteins from Arabidopsis are summarized in Table II. The calculated molecular masses of the mature SDH subunits nicely correspond to their apparent masses on SDS gels as determined previously (Millar et al., 2004). Furthermore, the sum of the molecular masses of the eight subunits corresponds exactly to the mass of the holo complex II (160 kD) as determined previously (Klodmann et al., 2011).
Table II. Calculated and native masses of complex II subunits and subcomplexes in Arabidopsis.
n.i., Not identified.
| Subunit | Accession No.a | Calculated Mass Precursorb | Calculated Mass Matureb,c | Apparent Massd | Holo Complex II | Subcomplex IIa | Subcomplex IIb | Subcomplex IIc |
|---|---|---|---|---|---|---|---|---|
| kD | kD | kD | ||||||
| SDH1 | At5g66760 | 69.656 | 66.043 | 65 | + | + | ||
| At2g18450 | 69.363 | 65.745 | ||||||
| SDH2 | At3g27380 | 31.171 | 28.253 | 29 | + | + | ||
| At5g40650 | 31.141 | 28.143 | ||||||
| At5g65165 | 34.983 | n.i. | ||||||
| SDH3 | At5g09600 | 23.454 | 12.172 | 12 | + | + | ||
| At4g32210 | 23.454 | 12.172 | ||||||
| SDH4 | At2g46505 | 16.842 | 9.181 | 6 | + | + | ||
| SDH5 | At1g47420 | 28.106 | 18.417 | 18 | + | + | ||
| SDH6 | At1g08480 | 15.813 | 15.682 | 15 | + | + | ||
| SDH7 | At3g47833 | 10.298 | 7.095 | 7 | + | + | ||
| At5g62575 | 10.860 | 7.485 | ||||||
| SDH8 | At2g46390 | 4.884 | 4.884 | 5 | + | n.i. | ||
| Σ calculated mass (kD)e | 161.7 | 161.7 | 94.3 | 44.1 + SDH8f | 18.4 | |||
| Σ apparent mass (kD)e | 157 | 157 | 94 | 40 + SDH8f | 18 |
Accession numbers according to TAIR (https://www.arabidopsis.org/). Note that two isoforms are present for SDH1, SDH3, and SDH7 and three isoforms are present for SDH2. bMolecular masses were calculated using the ProtParam tool (http://web.expasy.org/protparam/). cFor definition of the mature sequences, see Supplemental Figure S4. dApparent molecular masses as determined by Millar et al. (2004). eMolecular masses for the holo complex and subcomplexes IIa, IIb, and IIc were determined considering their subunit compositions. Subunit masses were summed using the first isoform given in this table. fNote that inclusion of SDH8 in subcomplex IIb is not clear.
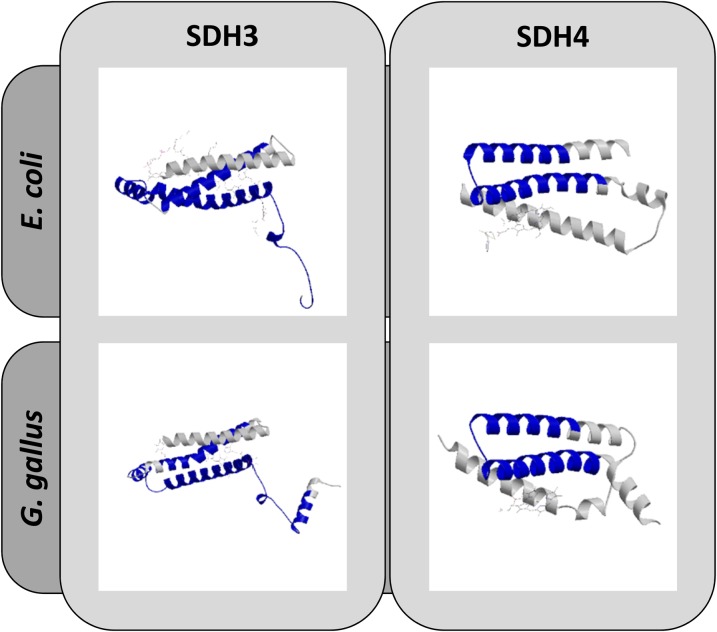
Arabidopsis SDH3 (12.2 kD) and SDH4 (9.2 kD) are smaller than their counterparts in bacteria, fungi, and animals. Furthermore, in comparison with SDH3 and SDH4 from other organisms, they have reduced numbers of transmembrane helices (Supplemental Fig. S5). Arabidopsis SDH3 only has two instead of three membrane-spanning helices, which was reported previously by Adams et al. (2001). Similarly, SDH4 only includes one sequence stretch having a clear potential to form a transmembrane helix. Alignments of SDH3 and SDH4 sequences from Arabidopsis with corresponding sequences from E. coli, yeast, and chicken revealed the absence of the C-terminal helix in Arabidopsis SDH3 and the absence of the N-terminal one- to two-transmembrane helices in Arabidopsis SDH4 (Supplemental Fig. S6). This result was confirmed by comparative structure prediction analyses using the SWISS-MODEL software package (Fig. 4). Interestingly, absent helices in SDH3 and SDH4 contribute to SDH3/SDH4 interaction with the SDH1 subunits in E. coli, yeast, and chicken (Supplemental Figs. S7 and S8). This may cause the decreased stability of complex II from flowering plants if compared with complex II from fungi and mammals. In flowering plants, DDM solubilization of mitochondrial membranes causes dissociation of SDH3/SDH4 from SDH1/SDH2. In contrast, DDM does not affect SDH1/SDH2/SDH3/SDH4 interaction in fungi and mammals (Schägger and von Jagow, 1991).
Figure 4.
Structural modeling of SDH3 and SDH4 from Arabidopsis. 3D structure models were deduced from protein homology modeling using reference structures of SDH3 and SDH4 from E. coli (RCSB 2ACZ) and chicken (RCSB 2H88). The predicted Arabidopsis structures (given in blue) and the reference structures (given in gray) are superimposed. The analysis was carried out with SWISS-MODEL.
SDH6 and SDH7 Substitute Membrane-Spanning Helices Lacking in SDH3 and SDH4
Since the six-helix bundle is a highly conserved structural motif in the membrane domain of complex II in bacteria, fungi, and animals essential for ubiquinone and heme binding as well as for the anchoring of SDH1 and SDH2 to the bacterial or inner mitochondrial membrane, we had a closer look at structural motifs within the SDH6, SDH7, and SDH8 subunits from flowering plants that could replace the missing helices in SDH3 and SDH4 (the Arabidopsis SDH5 sequence was not included in this investigation because it is very hydrophilic and lacks membrane-spanning helices; see Supplemental Fig. S2). Comparative structural modeling of the SDH6, SDH7, und SDH8 subunits using SWISS-MODEL did not give reliable results. Therefore, we used the C-terminal sequences of SDH3 from E. coli and chicken that are missing in SDH3 of Arabidopsis as well as the N-terminal sequences of SDH4 in E. coli and chicken missing in Arabidopsis SDH4 for pairwise sequence alignments with the SDH6, SDH7, and SDH8 proteins. Indeed, the transmembrane regions of SDH6 and SDH7 exhibit some sequence identity to the SDH3 and SDH4 regions missing in Arabidopsis (Supplemental Fig. S9). Sequence identity is low but in the same range as reported previously for the SDH3 and SDH4 proteins from different organisms (rather the structural motifs are highly conserved in SDH3 or SDH4 proteins). Based on the alignments (Supplemental Fig. S9), it cannot be determined at present whether SDH6 corresponds to the C terminus of E. coli SDH3 or the N terminus of E. coli SDH4. Considering the level of truncation with respect to the SDH3 and SDH4 subunits from other organisms, we postulate that Arabidopsis SDH7 corresponds to the C terminus of SDH3 from E. coli and that Arabidopsis SDH6 corresponds to the N terminus of E. coli SDH4.
SDH5, SDH6, and SDH7 Are Stoichiometric Subunits of Plant Complex II
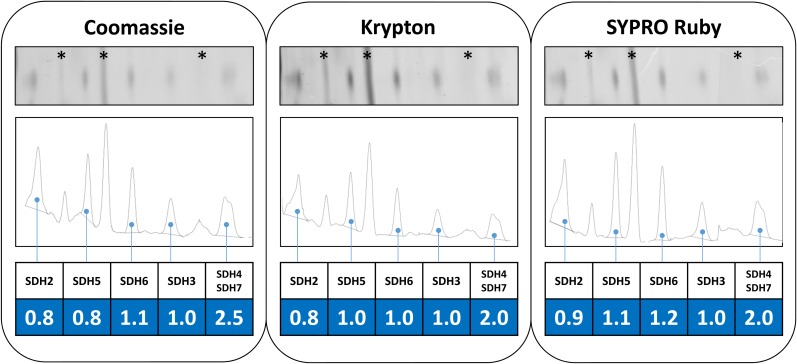
If SDH6 and SDH7 contribute membrane-spanning helices missing in SDH3 and SDH4, they must represent stoichiometric subunits of plant complex II. This is supported by experimental findings. The sum of the calculated molecular masses of the eight subunits of Arabidopsis is 161.7 kD (Table II), which perfectly matches the mass of the holo complex (Klodmann et al., 2011). This, and the very similar intensities of the corresponding spots upon Coomassie Blue staining (Figs. 2 and 3), strongly suggest that complex II particles of Arabidopsis consist of one copy of each of the eight subunits. To further investigate the stoichiometry of the SDH subunits in Arabidopsis, spot intensities on 2D BN SDS gels were evaluated by fluorescent staining. A gel stripe including complex II subunits was incubated with Coomassie Blue, Krypton, or SYPRO Ruby. Gels were scanned using a laser scanner, and protein spots representing complex II subunits were quantified densitometrically. Finally, the ratio of peak area to molecular mass was calculated for all SDH subunits using their mature molecular masses. The results were normalized with respect to SDH3 because it represents a core subunit present once per monomeric complex II particle (Yankovskaya et al., 2003; Sun et al., 2005). SDH1 could not be quantified due to high background on the 2D BN SDS gels in the region of this protein (data not shown). Results indicate that the SDH2, SDH5, SDH6, SDH3, and SDH4/SDH7 proteins (the SDH4 and SDH7 proteins overlap on the 2D BN SDS gel stripes, forming only one spot) occur in a 1:1:1:1:2 stoichiometry (Fig. 5). Considering that the latter spot includes two proteins (SDH4 and SDH7) and that SDH3 and SDH4 have a 1:1 stoichiometry in all organisms investigated so far, our results indicate a 1:1:1:1:1:1 stoichiometry for SDH2, SDH5, SDH6, SDH3, SDH4, and SDH7. Note that the results differ slightly between the three fluorophores because their binding depends to a certain degree on the chemical properties of proteins. This especially has been reported for Coomassie Blue. In contrast, Krypton efficiently binds to basic as well as hydrophobic amino acids and has been found to nicely allow the estimation of absolute protein quantities. Indeed, the occurrence of one copy of each of the SDH subunits per complex II particle is especially clear and convincing using this fluorescent dye. We conclude that, based on stoichiometric considerations, SDH6 and SDH7 can substitute missing transmembrane helices of SDH3 and SDH4.
Figure 5.
Stoichiometry of complex II subunits in Arabidopsis. Isolated mitochondria from Arabidopsis were solubilized by 5% digitonin, and mitochondrial proteins were separated by 2D BN SDS-PAGE. Stripes of the 2D gels including complex II (left, large subunits; right, small subunits) were stained using colloidal Coomassie Blue (left), Krypton fluorescent protein stain (middle), or SYPRO Ruby protein stain (right). Gel stripes were scanned using a Typhoon laser scanner, and proteins were quantified by densitometric measurements using the ImageJ software tool. The peak area-to-molecular mass ratio was calculated for SDH2, SDH5, SDH6, and SDH3 as well as for the protein spot including SDH4 and SDH7 (these two subunits overlap on the gel stripes). Results were normalized with respect to SDH3. Resulting stoichiometries are given at the bottom (highlighted in blue). Subunits not belonging to complex II are marked by asterisks.
The Number of Transmembrane Helices of SDH3 and SDH4 Decreased While the Number of Complex II Subunits Increased during Plantae Evolution
SDH5, SDH6, and SDH7 also have been biochemically characterized in rice (Huang et al., 2010; Huang and Millar, 2013). Furthermore, the rice genome encodes two highly similar homologs of SDH8 (Q0DF13 and Q5VS45). In accordance with the findings in Arabidopsis, rice SDH6 and SDH7 are predicted to include membrane-spanning helices, whereas rice SDH3 and SDH4 have reduced numbers of transmembrane regions (Supplemental Fig. S10). Meanwhile, complete genome sequences for numerous other plants have been determined and can be used for phylogenetic analyses (Michael and Jackson, 2013). We used the plant genomics resource Phytozome (https://phytozome.jgi.doe.gov/; Goodstein et al., 2012), which currently includes 58 sequenced and annotated green plant genomes (database release version 11, May 2016), the genome sequences of the gymnosperm genera Picea and Pinus (http://congenie.org; Sundell et al., 2015), the algae database TaxoBLAST (https://giavap-genomes.ibpc.fr/cgi-bin/AlgoBLAST/algoBlast.php), and the Joint Genome Institute algae data set (http://genome.jgi.doe.gov/pages/blast-query.jsf?db=Algae; Grigoriev et al., 2012; Nordberg et al., 2014) to systematically analyze the occurrence of SDH subunits in the green lineage.
Genes encoding SDH5, SDH6, and SDH7 are present in all sequenced angiosperms and gymnosperms (Table III; Supplemental Table S1). In contrast, genes encoding SDH8 have been found only in the angiosperm clades Brassicaceae and in monocotyledonous plants. SDH5, SDH6, and SDH7 also are present in Amborella trichopoda, a species placed at or near the base of the flowering plant lineage (Soltis and Soltis, 2013), the moss Physcomitrella patens, and the liverwort Marchantia polymorpha. We conclude that the occurrence of at least three of the extra SDH subunits is conserved in Embryophyta. In contrast, genes encoding SDH5, SDH6, and SDH7 could not be identified in the genomes of Chlorophyta (e.g. Chlamydomonas reinhardtii; Supplemental Table S1). To exclude that corresponding genes were overlooked due to a low degree of sequence conservation, mitochondria were isolated from Polytomella spp., a close relative of C. reinhardtii. Analysis of corresponding protein fractions by 2D BN SDS-PAGE and mass spectrometry revealed the presence of a classical four-subunit complex II including SDH1, SDH2, SDH3, and probably SDH4 (Fig. 6; Table IV).
Table III. Interdependence of the occurrence of SDH5 to SDH8 and the number of membrane-spanning helices in SDH3 and SDH4.
Searches for homologs were performed by BLASTp using the amino acid sequences of mature SDH3 to SDH8 from Arabidopsis (Supplemental Fig. S4). Databases used were the Charophyta protein database (https://giavap-genomes.ibpc.fr/cgi-bin/AlgoBLAST/algoBlast.php) and the plant genome database (https://phytozome.jgi.doe.gov/pz/portal.html). Searches for homologs in M. viride and N. hyalina were carried out by tBLASTn using EST databases at the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Homologs of SDH8 in monocots were identified using the sequence of SDH8 from rice (UniProt identifier Q0DF13). + indicates the occurrence of homologs (for details, see Supplemental Table S1; Supplemental Fig. S11), and − indicates the absence of homologs. Accession numbers of homologs are given in Supplemental Figure S11.
| Organism | Clade | SDH5a | SDH6a | SDH7a | SDH8a | SDH3 (No. of Transmembrane Helices)b | SDH4 (No. of Transmembrane Helices)b |
|---|---|---|---|---|---|---|---|
| Escherichia coli | Prokaryota | − | − | − | − | P69054 (3) | P0AC44 (3) |
| Mesostigma viride | Charophyta | + | + | − | − | G8DKD7 (3) | G8DKD0 (2) |
| Klebsormidium flaccidum | Charophyta | + | + | − | − | AJF36733.1 (3) | AJF36745.1 (2) |
| Chlorokybus atmophyticus | Charophyta | + | + | − | − | YP_001315099.1 (3) | YP_001315098.1(2) |
| Nitella hyalina | Charophyta | + | + | + | − | YP_006073043.1 (3) | YP_006073042.1 (2) |
| Physcomitrella patens | Bryophyta | + | + | + | − | Q1XGB9 (3) | Q1XGC0 (2) |
| Marchantia polymorpha | Marchantiophyta | + | + | + | − | P35721 (3) | NP_054431.1 (2) |
| Amborella trichopoda | Basal angiosperm | + | + | + | − | V9VIA6 (2) | V9VF77 (2) |
| Oryza sativa | Monocot | + | + | + | + | Q6ZH92 (1) | Q942X4 (2) |
| Zea mays | Monocot | + | + | + | + | A0A096QEW9 (1) | C4J8S0 (2) |
| Medicago truncatula | Dicot | + | + | + | − | G7JDI5 (2) | G7J5Z9 (2) |
| Arabidopsis thaliana | Dicot | + | + | + | + | A8MSF5 (2) | Q941A0 (1) |
Occurrence of SDH5 to SDH8 subunits in Streptophyta (land plants and Charophyta) and in E. coli. bUniProt identifiers (if UniProt identifiers are not available, alternative accessions are given, such as GenBank identifier [K. flaccidum] and NCBI reference sequence [C. atmophyticus, N. hyalina, and M. polymorpha]). The number of transmembrane helices in SDH3 and SDH4 was determined using the TMHMM server version 2.0 (Supplemental Figs. S5 and S10). For details, see Supplemental Figure S12. The threshold for helix prediction was set to 0.3.
Figure 6.
Comparative analysis of OXPHOS complexes from Polytomella spp. and Arabidopsis. Isolated mitochondria from Polytomella spp. (A) and Arabidopsis suspension cell culture (B) were solubilized by 5% digitonin, and resulting protein fractions were separated by 2D BN SDS-PAGE. Gels were stained using colloidal Coomassie Blue. The identity of OXPHOS complexes is given above the gels. SDH subunits are marked by arrowheads. Complex II subunits from Polytomella spp. were identified by mass spectrometry (Table IV). Complex II subunits from Arabidopsis were identified previously (Klodmann et al., 2011); protein spots represent (from top to bottom) SDH1, SDH2, SDH5, SDH6, SDH3, and SDH7/SDH4 (SDH8 is not visible but most likely is present in the dye front at the position indicated by the gray arrowhead in B). Identities of OXPHOS complexes are given above the 2D gels: I, complex I; I*, subcomplex of complex I; II, complex II; III2, dimeric complex III; IV, complex IV; V, monomeric complex V; V2, dimeric complex V; V2*, subcomplex of dimeric complex V; I+III2, supercomplex composed of complex I and dimeric complex III.
Table IV. Polytomella spp. proteins identified by mass spectrometry.
Further details on the measurements are presented in Supplemental Table S2.
| Spota | Accession Nob | Name | Massc | Scored | Peptidese | Coveragef |
|---|---|---|---|---|---|---|
| kD | % | |||||
| 1 | EDP09580 | SDH1 | 68.8 | 376 | 11 | 12.6 |
| 2 | EDP08267 | SDH2 | 32.3 | 295 | 10 | 19.6 |
| 3 | EDP09244 | SDH3 | 19.5 | 84 | 2 | 8.2 |
| 4 | EDP09690g | SDH4 | 9.3 | – | – | – |
Spot number as indicated in Figure 6. bAccession numbers according to ChlamyRein version 3.1 (http://genome.jgi.doe.gov/chlamy/chlamy.info.html). cCalculated molecular mass of the precursor protein. dMASCOT reliability score. eNumber of unique peptides. fCoverage of the protein sequence by peptides. gPeptides of Polytomella spp. SDH4 could not be identified based on the SDH4 sequence from C. reinhardtii (EDP09690).
To obtain further insights into complex II evolution, additional algae genomes were analyzed with respect to genes encoding SDH5, SDH6, and SDH7 (Supplemental Table S1). Genomes of Charophyta encode SDH5 and SDH6 but lack genes encoding SDH7 (Table III; Supplemental Table S1; Supplemental Fig. S13). Only the Charophyta species Nitella hyalina, which is especially closely related to Embryophyta (Adl et al., 2012; Leliaert et al., 2012), includes a gene coding for an SDH7 homolog. We conclude that SDH7 probably evolved later than SDH5 and SDH6. Sequences resembling Arabidopsis SDH6 were found in Cryptophyta, Glaucophyta, and Rhodophyta but are absent in Chlorophyta and the clades of the SAR supergroup (which includes, besides others, multicellular brown algae). Consequently, SDH6 probably was present in the last common ancestor of land plants, Cryptophyta, Glaucophyta, and Rhodophyta and subsequently got lost in Chlorophyta and algae of the SAR supergroup (or it evolved several times). We conclude that the occurrence of SDH6 preceded the occurrence of SDH7. At the same time, truncation of SDH4 preceded the truncation of SDH3 during Streptophyta (Embryophyta and Charophyta) evolution (Table III). We speculate that the emergence of SDH6 and SDH7 genes was caused by duplications of the genes encoding SDH3 and SDH4 and subsequent truncations. At a later stage in evolution, the presence of SDH6 and SDH7 in plants was the prerequisite for SDH3 and SDH4 truncation. However, a reliable model for complex II evolution in plants will have to await extended genome sequence information on the various clades of Plantae.
DISCUSSION
The subunit composition of plant complex II so far is mysterious. In contrast to the well-conserved succinate dehydrogenase complexes present in bacteria, animals, and fungi, which are composed of the SDH1, SDH2, SDH3, and SDH4 subunits, the complex of plants includes up to four additional proteins termed SDH5, SDH6, SDH7, and SDH8 (Eubel et al., 2003; Millar et al., 2004). Besides plants, only the complex II of trypanosomes has been reported to contain additional subunits, including a split version of SDH2 (Morales et al., 2009; Harada et al., 2013). Mitochondrial processes are known to be highly derived in trypanosomes due to far-ranging adaptive processes that take place upon living in host species. In analogy, far-ranging molecular changes were necessary for the adaptation of aquatic photosynthetic organism to terrestrial life. We applied a biochemical approach to understand the subunit arrangement within plant complex II. Treatment of digitonin-solubilized Arabidopsis complex II by low concentrations of DDM allowed its dissection into three subcomplexes. A 3D electrophoresis system was used to unravel the subunit composition of these subcomplexes (Fig. 7). Subcomplex IIa includes SDH1 and SDH2 and represents the succinate dehydrogenase domain. Subcomplex IIb consists of SDH3, SDH4, SDH6, and SDH7. All four subunits include transmembrane helices and together form the membrane domain of complex II, which is involved in electron transfer onto ubiquinone and which attaches the succinate dehydrogenase domain to the inner mitochondrial membrane. Structural modeling reveals that Arabidopsis SDH3 and SDH4 are truncated and lack sequence regions including one and two of their transmembrane helices, respectively, which are highly conserved in other groups of organisms. Evidence is presented that the SDH6 and SDH7 subunits of plant complex II represent the missing segments of SDH3 and SDH4.
Figure 7.
Controlled disassembly of complex II from Arabidopsis by detergent treatment. The scheme shows complex II disassembly induced by incubation with 0.03% (w/v) DDM. Upon detergent treatment, the complex is split into three subcomplexes termed IIa, IIb, and IIc. The largest subcomplex (IIa; orange) consists of the matrix-exposed subunits SDH1 and SDH2. A smaller subcomplex (IIb; turquoise) represents the membrane anchor of complex II. It consists of SDH3 and SDH4 as well as the plant-specific subunits SDH6 and SDH7 (green). The smallest subcomplex (IIc; green) consists of the plant-specific and hydrophilic subunit SDH5, which forms neither part of subcomplex IIa nor IIb. The localization of the plant-specific subunit SDH8 (beige) is not known. Note that not all subunit interactions shown in the scheme have been experimentally proven. SDH7 is suggested to interact with SDH3, and SDH6 is suggested to interact with SDH4, based on evolutionary considerations (Table III). Furthermore, SDH5 occurred during evolution in parallel with SDH6 and possibly is involved in its integration into complex II. However, SDH5 also could be positioned at another location.
The SDH3 and SDH4 Subunits
The SDH3 and SDH4 proteins constitute a small protein family (Supplemental Fig. S3). The identity of their amino acid sequences is low. However, their 3D structures are well conserved, as revealed by x-ray crystallography of complex II from E. coli and animal mitochondria. Both proteins include three parallel transmembrane helices that form a six-helix bundle important for ubiquinone and heme binding and for attaching the succinate dehydrogenase domain to the bacterial or the inner mitochondrial membrane. Some additional members of this protein family form part of the bacterial quinol:fumarate reductases that are structurally and functionally related to complex II (Hägerhäll and Hederstedt, 1996; Lemos et al., 2002; Iverson, 2013). Interestingly, the SDH3 protein additionally was reported to constitute a subunit of the preprotein translocase of the inner mitochondrial membrane (the TIM complex) in yeast (Gebert et al., 2011). Taken together, the members of the SDH3/SDH4 protein family represent important building blocks for anchoring more hydrophilic proteins to the bacterial and the inner mitochondrial membranes. The structural prerequisite is a highly conserved six-helix bundle structure. This structure is truncated in flowering plants and some trypanosomes.
The SDH6 and SDH7 Subunits
The SDH6 and SDH7 subunits were discovered in Arabidopsis and also have been biochemically characterized for bean, potato, and rice (Eubel et al., 2003; Millar et al., 2004; Huang et al., 2010). Homologous proteins are encoded by the genomes of all Embryophyta (Table III; Supplemental Table S1; Supplemental Fig. S11). In contrast, genomes of Chlorophyta lack genes coding for SDH6 and SDH7, and the genomes of Charophyta encode SDH6 (and additionally SDH5) but not SDH7. The function of SDH6 and SDH7 so far was unclear. Here, we present evidence that both proteins substitute sequence regions absent in plant SDH3 and SDH4 and participate in forming the six-helix bundle conserved in the membrane domains of complex II in all organisms investigated. However, the size of mature SDH6 and SDH7 proteins (15.7 and 7.1 kD) much extends the regions to be substituted in SDH3 and SDH4. In fact, both proteins also comprise hydrophilic sequence regions that might be involved in extra functions of complex II in plants.
The SDH5 Subunit
The SDH5 subunit is liberated during the dissection of plant complex II into the succinate dehydrogenase and membrane domains and most likely is located at the interface of these domains. It clearly is hydrophilic, and its sequence does not resemble any other known protein sequence. SDH5 is present in all Streptophyta (Charophyta and Embryophyta; Table III; Supplemental Table S1; Supplemental Fig. S11). Together with the hydrophilic regions of SDH6 and SDH7, it might constitute an extra domain of plant complex II involved in a so far unknown additional function. Indeed, the overall size of the complex II monomer, which is in the range of 160 kD in Arabidopsis (Table II), clearly is larger than that of complex II in bacteria and in the mitochondria of animals and fungi.
The SDH8 Subunit
Being only 4.9 kD in size, the SDH8 subunit is the smallest known component of any of the five OXPHOS complexes in plants. Biochemically, it has been described only in Arabidopsis (Millar et al., 2004), but homologs also are encoded by the genomes of other Brassicaceae and monocotyledoneous plants (Supplemental Table S1). SDH8 migrates close to the dye front during electrophoresis and hardly can be stained by Coomassie Blue. Its N-terminal sequence has been determined by Edman degradation. SDH8 so far has not been identified by mass spectrometry. It is predicted to contain one membrane-spanning helix. Upon trypsination, it is cleaved in three very short peptides and one hydrophobic peptide of 2,978 D (Supplemental Fig. S14), which possibly is difficult to detect by mass spectrometry. In the course of our investigation, SDH8 was not found. It currently cannot be excluded that SDH8 does not represent a true subunit of complex II. However, it is equally likely that SDH8 was hidden in the dye front of our 3D BN/BN SDS-PAGE system and, therefore, not visible. It possibly contributes to the membrane anchor domain of complex II. Further insights will require extended biochemical analyses.
Evolution of Mitochondrial Complex II
Mitochondria descended from endosymbiotic bacteria that were related to α-proteobacteria. It can be assumed that mitochondria originally included a four-subunit complex II like present-day proteobacteria. During establishment of the endosymbiosis, most bacterial genes migrated into the nucleus (but a few were retained in the mitochondrial genomes present in all eukaryotic cells). Interestingly, some mitochondrial genomes include genes encoding subunits of complex II, such as the mitochondrial genomes of the red alga Porphyra purpureacodes and the zooflagellate Reclinomonas americana, which comprise genes for SDH2, SDH3, and SDH4 (Burger et al., 1996). The mitochondrial genome of the liverwort M. polymorpha encodes SDH3 and SDH4 (Oda et al., 1992). Some mitochondrial genomes of higher plants include pseudogenes encoding complex II subunits. For instance, the mitochondrial genomes of Arabidopsis and some other plants include a nonfunctional SDH4 gene (Giegé et al., 1998). A systematic analysis of mitochondrial genomes from angiosperms revealed that several plant species even include active sdh3 and sdh4 genes in their mitochondrial genomes (Adams et al., 2001; Adams and Palmer, 2003). In evolutionary terms, this indicates that the transfer of complex II genes to the nucleus is an ongoing process (Adams et al., 2001; Knoop, 2012).
Gene transfer from mitochondria to the nucleus requires the acquisition of extra sequences that encode information for transporting the corresponding proteins back into the mitochondria. This process is based on cleavable or noncleavable mitochondrial presequences. Of the eight complex II subunits from Arabidopsis, the SDH1, SDH2, and SDH4 subunits have cleavable presequences of typical length and amino acid composition (Supplemental Fig. S4). The SDH6 and SDH8 subunits lack cleavable presequences and, therefore, must include some internal targeting information (however, in the case of SDH6, the N-terminal Met is removed). The SDH3 and SDH5 subunits comprise presequences of extraordinary length (105 amino acids in SDH3 and 89 amino acids in SDH5). Most interestingly, the presequence of SDH5 is very similar to the N-terminal sequence of the carbonic anhydrase subunit 2 of mitochondrial complex I of Arabidopsis (Supplemental Fig. S15). Furthermore, the long N-terminal extension of SDH3 very much resembles the amino acid sequence of mitochondrial HSP70 (At5g09590), which has been reported previously (Adams et al., 2001; Liu et al., 2009; Supplemental Fig. S16). These findings reflect that presequence acquisitions for the SDH proteins represent comparatively recent events.
Origin of the Plant-Specific SDH Subunits
Complex II of flowering plants includes truncated versions of SDH3 and SDH4 and at the same time comprises SDH6 and SDH7 subunits, which resemble the missing parts of plant SDH3 and SDH4. The origin of SDH6 and SDH7 could be explained by duplications of the genes encoding SDH3 and SDH4 and subsequent truncations. In general, mitochondrial OXPHOS complexes acquired several new subunits at an early stage of the evolution of the eukaryotic cell, which possibly was the basis for the remodeling of the original subunits (the original subunits became slightly more hydrophilic; this possibly facilitated their import into mitochondria after their genes had been transferred to the nucleus; van der Sluis et al., 2015). It will require more extensive genome analyses to better understand complex II evolution in plants. Furthermore, it is unclear at present why new SDH subunits only were acquired in the mitochondria of plants and of trypanosomes. Gene knockout experiments might give further insights into the role of the extra complex II subunits in some groups of organisms. However, deletion of these genes most likely will cause the absence of complex II, which is not compatible with plant life (León et al., 2007). It is clear that the assembly process of complex II in plants should follow unique routes, since it requires the integration of twice as many subunits as in other groups of organisms.
CONCLUSION
Using controlled destabilization of Arabidopsis complex II and subunit analysis of the generated dissection products (Supplemental Fig. S17), we present insights into the subunit arrangement within this eight-subunit complex. We suggest a structural model as given in Figure 8. However, besides substituting transmembrane helices missing in plant SDH3 and SDH4, the four supernumerary proteins present in plant complex II contribute some additional hydrophilic mass. As a consequence, complex II from plants, which is comparatively large, might include extra functions. This should be addressed by future investigations.
Figure 8.
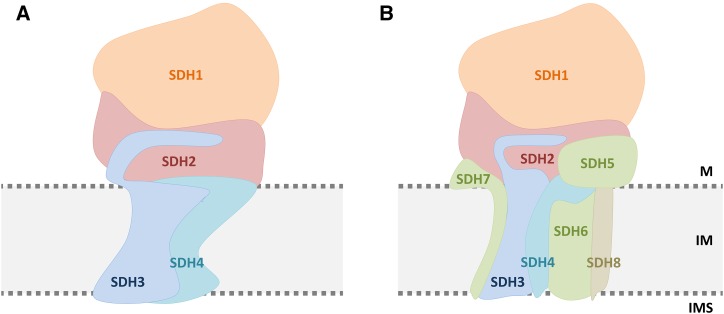
Subunit arrangement within mitochondrial complex II of chicken (A) and Arabidopsis (B). The subunit model for chicken is taken from the crystal structure (Huang et al., 2006); the model for Arabidopsis is based on the biochemical results presented here (Figs. 2 and 3) as well as on amino acid sequence analyses (Fig. 4; Supplemental Figs. S2 and S5). The top part of the complex, which protrudes into the mitochondrial matrix (M), includes SDH1 (orange) and SDH2 (red). This domain contains the succinate-binding site and includes covalently bound FAD in SDH1 as well as three iron-sulfur clusters in SDH2. The bottom part of the complex anchors SDH1 and SDH2 to the inner mitochondrial membrane (IM). Besides SDH3 and SDH4 (blue), it includes the plant-specific subunits SDH6 and SDH7 (green). SDH7 is suggested to interact with SDH3, and SDH6 is suggested to interact with SDH4, based on evolutionary considerations (Table III). The localization of SDH5 (green), which also is a plant-specific component, is probably at the interface between the succinate dehydrogenase and the membrane domain but also could be at another position. The localization of SDH8 (beige), which is only 4.9 kD in size and includes one membrane-spanning helix, is not known. IMS, Mitochondrial intermembrane space.
MATERIALS AND METHODS
Cultivation of an Arabidopsis Cell Suspension Culture
An Arabidopsis (Arabidopsis thaliana) Columbia-0 callus culture was established according to May and Leaver (1993). Callus was used to establish a cell suspension culture as described previously (Sunderhaus et al., 2006).
Isolation of Mitochondria
Preparation of mitochondria from the cell suspension culture was carried out by differential centrifugation and Percoll density gradient centrifugation as outlined by Werhahn et al. (2001). Isolated organelles were stored at −80°C.
2D BN/BN PAGE
2D BN/BN PAGE was performed as described by Sunderhaus et al. (2007). Mitochondrial membranes were solubilized by 10 g of digitonin per 1 g of mitochondrial protein. First-dimension BN PAGE was carried out in the presence of digitonin, and second-dimension BN PAGE was carried out in the presence of DDM.
Characterization of Complex II by 3D BN/BN SDS-PAGE
To characterize mitochondrial complex II from Arabidopsis, a 3D BN/BN SDS-PAGE system was established that combines BN/BN PAGE as described by Sunderhaus et al. (2007) and BN SDS-PAGE according to Wittig et al. (2006). All gel runs were carried out in the Protean II xi electrophoresis unit (Bio-Rad). Mitochondrial membranes were solubilized by digitonin at a concentration of 10 g of detergent per 1 g of protein (Eubel et al., 2003). First-dimension BN PAGE (4.5%–16% [v/v] acrylamide) was performed using 100 µg of mitochondrial protein (for complex II in gel activity staining or Coomassie Blue staining) or 350 µg of mitochondrial protein (for protein analysis by further gel dimensions). The gel run was performed in two steps: (1) 100 V (maximum 15 mA) for 45 min and (2) 10 mA (maximum 500 V) for 9 h. Lanes of the 1D BN gel were used for complex II activity staining (see below), Coomassie Blue staining (see below), or further electrophoretic gel dimensions for complex II analysis. For the latter approach, gel slices including complex II (as identified by parallel complex II in gel activity staining) were cut out from BN lanes that were neither fixed nor stained. The gel slices were incubated in BN cathode buffer supplemented with 0.03% (w/v) DDM for 30 min to dissect complex II into subcomplexes. Afterwards, four gel slices were stacked and transferred on top of a second-dimension BN PAGE gel (5%–20% [v/v] acrylamide). The gel run was performed in two steps: (1) 100 V (maximum 15 mA) for 1.5 h and (2) 15 mA (maximum 500 V) for 11 h. Finally, a gel lane including separated complex II subcomplexes was cut out and incubated in a solution containing 1% (v/v) 2-mercaptoethanol as well as 1% (w/v) SDS. The gel lane was transferred horizontally on top of a third gel dimension to separate subcomplex subunits using SDS-PAGE (16% [v/v] acrylamide).
Complex II in Gel Activity Staining
In gel activity staining of succinate dehydrogenase activity (complex II) was performed according to Jung et al. (2000), excluding EDTA and cyanide as suggested by Wittig et al. (2007). During the assay, the gel increases slightly in size. To allow precise matching with untreated BN lanes, the activity-stained gel was incubated in 50% (v/v) ethanol for several minutes, which causes shrinking of the gel. The reaction was stopped when the gels to be compared had equal dimensions.
Colloidal Coomassie Blue Staining
Gels were transferred into fixing solution (40% [v/v] ethanol and 10% [v/v] acetic acid) after gel runs and incubated for 3 h. Staining was done using the colloidal Coomassie Brilliant Blue G-250 protocol as described by Neuhoff et al. (1988, 1990).
Quantification of Proteins on 2D BN SDS Gels
For stoichiometry analysis of complex II subunits, isolated mitochondria from Arabidopsis were solubilized by 5% digitonin, and the resulting protein fractions were separated by 2D BN SDS-PAGE (Wittig et al., 2006). Gels were stained using colloidal Coomassie Blue (see above), Krypton fluorescent protein stain, or SYPRO Ruby protein gel stain (Thermo Fisher Scientific) according to the manufacturer’s instructions. Gels were scanned by the Typhoon laser scanner FLA7000 (GE Healthcare). Gel-scanning conditions were as follows: (1) Coomassie Blue, 633-nm laser/no filter; (2) Krypton, 532-nm laser/emission filter 580BP30; and (3) SYPRO Ruby, 532-nm laser/emission filter 610BP30. Proteins were quantified by densitometric analysis of protein spots using the ImageJ software tool (Schneider et al., 2012). Finally, the peak area-to-molecular mass ratio was calculated for complex II subunits. Values were normalized with respect to SDH3.
Analysis of Complex II from Polytomella spp.
Polytomella spp. were obtained from the Experimental Phycology and Culture Collection of Algae (SAG [http://www.uni-goettingen.de/en/45175.html]). Cultivation of Polytomella spp. was carried out as described by Brumme et al. (1998), and isolation of mitochondria from Polytomella spp. was carried out according to Dudkina et al. (2005). Mitochondrial proteins were separated by 2D BN SDS-PAGE (Wittig et al., 2006).
Protein Identification by Mass Spectrometry
For protein identification, spots were cut out from Coomassie Blue-stained gels. Tryptic in gel digestion, peptide extraction, as well as mass spectrometry were performed according to Klodmann et al. (2010). Peptides were separated using the EASY-nLC system (Proxeon; Thermo Scientific), and coupled mass spectrometry analyses were performed by using a micrOTOF-Q II mass spectrometer (Bruker). Evaluation of primary mass spectrometry data was done employing the ProteinScape software package (version 2.; Bruker), the Mascot Search Engine (Matrix Science), and the Arabidopsis protein database (www.arabidopsis.org; release TAIR 10).
Sequence Evaluation
Sequence analyses were performed for complex II subunits SDH1 to SDH8 of Arabidopsis (Supplemental Fig. S4), SDH3 (SDHC) and SDH4 (SDHD) of Escherichia coli, SDH3 and SDH4 of Saccharomyces cerevisiae, and SDH3 (SDHC) and SDH4 (SDHD) of chicken (Gallus gallus; Supplemental Fig. S18).
Presequence Determination
Information on presequences was taken from the literature. For SDH subunits of Arabidopsis, data were taken from Baerenfaller et al. (2008), Huang et al. (2009), and Millar et al. (2004) (see Supplemental Fig. S4). Presequences of S. cerevisiae SDH subunits were defined in accordance with Daignan-Fornier et al. (1994) and Bullis and Lemire (1994) (see Supplemental Fig. S18). The mature sequence of chicken SDH4 was taken from the x-ray structure (Huang et al., 2006).
Prediction of Transmembrane Helices
Transmembrane helices were predicted using TMHMM server version 2.0 at www.cbs.dtu.dk/services/TMHMM (Krogh et al., 2001). Data for the probabilities of transmembrane helices were exported into Excel (Microsoft). Transmembrane helices also were calculated with other programs (at http://topcons.cbr.su.se/; Tsirigos et al., 2015). Results very much resembled the outcome of the TMHMM predictions.
Molecular Mass Calculation
Calculation of molecular masses (and other parameters) was done using the ExPASy ProtParam tool (http://web.expasy.org/protparam/; Wilkins et al., 1999; Gasteiger et al., 2005).
Homology Modeling
Homology modeling of 3D structures was performed by the SWISS-MODEL workspace (http://swissmodel.expasy.org/; Biasini et al., 2014). Templates of interest (2ACZ for E. coli and 2H88 for chicken) were chosen for sequence similarity comparisons.
Sequence Alignments
Alignments of the amino acid sequences of mature complex II subunits were performed with the TCOFFEE tool for transmembrane proteins (PSI/TM-Coffee at http://tcoffee.crg.cat/; Di Tommaso et al., 2011). Alignment files were processed with the GeneDoc software package (Nicholas et al., 1997).
Homology Searches
Arabidopsis protein sequences homologous to SDH presequences were searched using BLASTp 2.2.8 at www.arabidopsis.org (TAIR 10 protein database). Mature amino acid sequences of Arabidopsis SDH5, SDH6, SDH7, and SDH8 were used to search homologs in other organisms. BLASTp searches (against nonredundant protein sequences) and tBLASTn searches (against ESTs) were carried out at www.ncbi.nlm.nih.gov. Homology searches for Embryophyta and Chlorophyta were performed by additional BLASTp analyses at https://phytozome.jgi.doe.gov/pz/portal.html, searches for Gymnospermae were performed at http://congenie.org, searches for Chlorophyta and Streptophyta were performed at https://giavap-genomes.ibpc.fr/cgi-bin/AlgoBLAST/algoBlast_protein.php, and searches for additional green algae were performed at http://genome.jgi.doe.gov/pages/blast-query.jsf?db=Algae. The threshold for E values was set to less than e−10.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Proteins identified in the two complex II subcomplexes separated by 2D BN/BN PAGE (Fig. 1).
Supplemental Figure S2. Prediction of transmembrane helices in the plant-specific subunits of complex II.
Supplemental Figure S3. The SDH3 and SDH4 proteins constitute a protein family.
Supplemental Figure S4. Amino acid sequences of subunits SDH1 to SDH8 in Arabidopsis.
Supplemental Figure S5. Prediction of transmembrane helices in SDH3 and SDH4 of E. coli and Arabidopsis.
Supplemental Figure S6. Sequence alignment of the membrane subunits of complex II from Arabidopsis and E. coli/chicken.
Supplemental Figure S7. Amino acid residues of functional importance in SDH3.
Supplemental Figure S8. Amino acid residues of functional importance in SDH4.
Supplemental Figure S9. Alignments of the C-terminal region of SDH3 and the N-terminal region of SDH4 from E. coli/chicken with the sequences of Arabidopsis SDH6 and SDH7.
Supplemental Figure S10. Prediction of transmembrane helices in the plant-specific subunits of complex II from rice.
Supplemental Figure S11. Occurrence of SDH5 to SDH8 in Streptophyta (land plants and Charophyta).
Supplemental Figure S12. Prediction of transmembrane helices of complex II subunits in Streptophyta.
Supplemental Figure S13. Homologs of complex II subunits in the phylogenetic tree of the green lineage.
Supplemental Figure S14. Theoretical tryptic peptides of SDH8 from Arabidopsis.
Supplemental Figure S15. The presequence of Arabidopsis SDH5 resembles the N-terminal amino acid sequence of the carbonic anhydrase subunit 2 (At1g47260) of the mitochondrial complex I.
Supplemental Figure S16. The presequence of Arabidopsis SDH3 resembles the N-terminal amino acid sequence of mitochondrial HSP70 (At5g09590).
Supplemental Figure S17. Experimental strategy: investigation of the internal subunit arrangement of complex II by controlled destabilization of the holo complex and analysis of the subunit compositions of the generated subcomplexes.
Supplemental Figure S18. Reference sequences used for the analysis of SDH3 and SDH4 in Arabidopsis.
Supplemental Table S1. Homologs of complex II subunits SDH5 to SDH8 in the clade Chloroplastida.
Supplemental Table S2. Peptides of complex II subunits from Arabidopsis and Polytomella spp. as identified by mass spectrometry.
Supplementary Material
Acknowledgments
We thank Dagmar Lewejohann and Marianne Langer for expert technical assistance and Steffanie Fromm for critically reading the article.
Glossary
- OXPHOS
oxidative phosphorylation
- mETC
mitochondrial electron transfer chain
- 2D
two-dimensional
- BN
blue native
- DDM
dodecylmaltoside
- BN/BN
blue native/blue native
- 1D
one-dimensional
- 3D
three-dimensional
Footnotes
Articles can be viewed without a subscription.
This work was supported by SFP (Strategisches Förderprogramm) of the Faculty of Natural Sciences of Leibniz University Hannover.
References
- Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Adams KL, Rosenblueth M, Qiu YL, Palmer JD (2001) Multiple losses and transfers to the nucleus of two mitochondrial succinate dehydrogenase genes during angiosperm evolution. Genetics 158: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. (2012) The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun HP, Emmermann M, Kruft V, Schmitz UK (1992) The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J 11: 3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme S, Kruft V, Schmitz UK, Braun HP (1998) New insights into the co-evolution of cytochrome c reductase and the mitochondrial processing peptidase. J Biol Chem 273: 13143–13149 [DOI] [PubMed] [Google Scholar]
- Bullis BL, Lemire BD (1994) Isolation and characterization of the Saccharomyces cerevisiae SDH4 gene encoding a membrane anchor subunit of succinate dehydrogenase. J Biol Chem 269: 6543–6549 [PubMed] [Google Scholar]
- Burger G, Lang BF, Reith M, Gray MW (1996) Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc Natl Acad Sci USA 93: 2328–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B, Valens M, Lemire BD, Bolotin-Fukuhara M (1994) Structure and regulation of SDH3, the yeast gene encoding the cytochrome b560 subunit of respiratory complex II. J Biol Chem 269: 15469–15472 [PubMed] [Google Scholar]
- Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39: W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP (2005) Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett 579: 5769–5772 [DOI] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria: supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50: 725–734 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Jordana X (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46: 241–250 [DOI] [PubMed] [Google Scholar]
- Fuentes D, Meneses M, Nunes-Nesi A, Araújo WL, Tapia R, Gómez I, Holuigue L, Gutiérrez RA, Fernie AR, Jordana X (2011) A deficiency in the flavoprotein of Arabidopsis mitochondrial complex II results in elevated photosynthesis and better growth in nitrogen-limiting conditions. Plant Physiol 157: 1114–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In Walker JM, ed, The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp 571–607 [Google Scholar]
- Gawryluk RMR, Gray MW (2010) Evidence for an early evolutionary emergence of gamma-type carbonic anhydrases as components of mitochondrial respiratory complex I. BMC Evol Biol 10: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Gebert M, Oeljeklaus S, von der Malsburg K, Stroud DA, Kulawiak B, Wirth C, Zahedi RP, Dolezal P, Wiese S, et al. (2011) Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell 44: 811–818 [DOI] [PubMed] [Google Scholar]
- Giegé P, Knoop V, Brennicke A (1998) Complex II subunit 4 (sdh4) homologous sequences in plant mitochondrial genomes. Curr Genet 34: 313–317 [DOI] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, et al. (2012) The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40: D26–D32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägerhäll C, Hederstedt L (1996) A structural model for the membrane-integral domain of succinate:quinone oxidoreductases. FEBS Lett 389: 25–31 [DOI] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325: 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Inaoka DK, Ohmori J, Kita K (2013) Diversity of parasite complex II. Biochim Biophys Acta 1827: 658–667 [DOI] [PubMed] [Google Scholar]
- Hatefi Y. (1985) The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 54: 1015–1069 [DOI] [PubMed] [Google Scholar]
- Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA (2006) 3-Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem 281: 5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Millar AH (2013) Succinate dehydrogenase: the complex roles of a simple enzyme. Curr Opin Plant Biol 16: 344–349 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Narsai R, Eubel H, Whelan J, Millar AH (2010) Functional and composition differences between mitochondrial complex II in Arabidopsis and rice are correlated with the complex genetic history of the enzyme. Plant Mol Biol 72: 331–342 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Whelan J, Millar AH (2009) Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol 150: 1272–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson TM. (2013) Catalytic mechanisms of complex II enzymes: a structural perspective. Biochim Biophys Acta 1827: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim-Messeder D, Caverzan A, Rauber R, de Souza Ferreira E, Margis-Pinheiro M, Galina A (2015) Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytol 208: 776–789 [DOI] [PubMed] [Google Scholar]
- Jung C, Higgins CM, Xu Z (2000) Measuring the quantity and activity of mitochondrial electron transport chain complexes in tissues of central nervous system using blue native polyacrylamide gel electrophoresis. Anal Biochem 286: 214–223 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jänsch L, Braun HP (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. (2012) Seed plant mitochondrial genomes: complexity evolving. In Bock R, Knoop V, eds, Genomics of Chloroplasts and Mitochondria. Springer, Dordrecht, The Netherlands, pp 175–200 [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, de Clerck O (2012) Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 31: 1–46 [Google Scholar]
- Lemos RS, Fernandes AS, Pereira MM, Gomes CM, Teixeira M (2002) Quinol:fumarate oxidoreductases and succinate:quinone oxidoreductases: phylogenetic relationships, metal centres and membrane attachment. Biochim Biophys Acta 1553: 158–170 [DOI] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Zhuang Y, Zhang P, Adams KL (2009) Comparative analysis of structural diversity and sequence evolution in plant mitochondrial genes transferred to the nucleus. Mol Biol Evol 26: 875–891 [DOI] [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Jackson S (2013) The first 50 plant genomes. Plant Genome 6: 1–7 [Google Scholar]
- Millar AH, Eubel H, Jänsch L, Kruft V, Heazlewood JL, Braun HP (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Morales J, Mogi T, Mineki S, Takashima E, Mineki R, Hirawake H, Sakamoto K, Omura S, Kita K (2009) Novel mitochondrial complex II isolated from Trypanosoma cruzi is composed of 12 peptides including a heterodimeric Ip subunit. J Biol Chem 284: 7255–7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V, Arold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9: 255–262 [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Stamm R, Pardowitz I, Arold N, Ehrhardt W, Taube D (1990) Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11: 101–117 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. Embnew News 4: 1 [Google Scholar]
- Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I (2014) The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42: D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, et al. (1992) Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA: a primitive form of plant mitochondrial genome. J Mol Biol 223: 1–7 [DOI] [PubMed] [Google Scholar]
- Oyedotun KS, Lemire BD (2004) The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase: homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem 279: 9424–9431 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun HP (2017) The mitochondrial complexome of Arabidopsis thaliana. Plant J (in press) 10.1111/tpj.13448 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2013) Angiosperm phylogeny: a framework for studies of genome evolution. In Leitch IJ, Greilhuber J, Dolezel J, Wendel JF, eds, Plant Genome Diversity, Vol 2. Springer, Vienna, pp 1–11 [Google Scholar]
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121: 1043–1057 [DOI] [PubMed] [Google Scholar]
- Sundell D, Mannapperuma C, Netotea S, Delhomme N, Lin YC, Sjödin A, Van de Peer Y, Jansson S, Hvidsten TR, Street NR (2015) The Plant Genome Integrative Explorer Resource: PlantGenIE.org. New Phytol 208: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Sunderhaus S, Dudkina NV, Jänsch L, Klodmann J, Heinemeyer J, Perales M, Zabaleta E, Boekema EJ, Braun HP (2006) Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J Biol Chem 281: 6482–6488 [DOI] [PubMed] [Google Scholar]
- Sunderhaus S, Eubel H, Braun HP (2007) Two-dimensional blue native/blue native polyacrylamide gel electrophoresis for the characterization of mitochondrial protein complexes and supercomplexes. In Leister D, Herrmann JM, eds, Methods in Molecular Biology. Mitochondria: Practical Protocols, Humana Press, Totowa, NJ, pp 315–324 [DOI] [PubMed] [Google Scholar]
- Sunderhaus S, Klodmann J, Lenz C, Braun HP (2010) Supramolecular structure of the OXPHOS system in highly thermogenic tissue of Arum maculatum. Plant Physiol Biochem 48: 265–272 [DOI] [PubMed] [Google Scholar]
- Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43: W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, Frauenfeld J, Berninghausen O, Neupert W, Herrmann JM, Beckmann R (2015) Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol 7: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vranken JG, Na U, Winge DR, Rutter J (2015) Protein-mediated assembly of succinate dehydrogenase and its cofactors. Crit Rev Biochem Mol Biol 50: 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villavicencio-Queijeiro A, Vazquez-Acevedo M, Cano-Estrada A, Zarco-Zavala M, Tuena de Gomez M, Mignaco JA, Freire MM, Scofano HM, Foguel D, Cardol P, et al. (2009) The fully-active and structurally-stable form of the mitochondrial ATP synthase of Polytomella sp. is dimeric. J Bioenerg Biomembr 41: 1–13 [DOI] [PubMed] [Google Scholar]
- Welchen E, Klodmann J, Braun HP (2011) Biogenesis and supramolecular organization of the oxidative phosphorylation system in plants. In Kempken F, ed, Plant Mitochondria. Springer, New York, pp 327–355 [Google Scholar]
- Werhahn W, Niemeyer A, Jänsch L, Kruft V, Schmitz UK, Braun H (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: identification of multiple forms of TOM20. Plant Physiol 125: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112: 531–552 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H (2006) Blue native PAGE. Nat Protoc 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schägger H (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299: 700–704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.