Abstract
Burkholderia cenocepacia H111, which was isolated from a cystic fibrosis patient, employs a quorum-sensing (QS) system, encoded by cep, to control the expression of virulence factors as well as the formation of biofilms. The QS system is thought to ensure that pathogenic traits are expressed only when the bacterial population density is high enough to overwhelm the host before it is able to mount an efficient response. While the wild-type strain effectively kills the nematode Caenorhabditis elegans, the pathogenicity of mutants with defective quorum sensing is attenuated. To date, very little is known about the cep-regulated virulence factors required for nematode killing. Here we report the identification of a cep-regulated gene, whose predicted amino acid sequence is highly similar to the QS-regulated protein AidA of the plant pathogen Ralstonia solanacearum. By use of polyclonal antibodies directed against AidA, it is demonstrated that the protein is expressed in the late-exponential phase and accumulates during growth arrest. We show that B. cenocepacia H111 AidA is essential for slow killing of C. elegans but has little effect on fast killing, suggesting that the protein plays a role in the accumulation of the strain in the nematode gut. Thus, AidA appears to be required for establishing an infection-like process rather than acting as a toxin. Furthermore, evidence is provided that AidA is produced not only by B. cenocepacia but also by many other strains of the Burkholderia cepacia complex.
Taxonomic studies of the past few years have shown that Burkholderia cepacia-like organisms comprise a very heterogenous group of strains, collectively referred to as the Burkholderia cepacia complex (BCC). Strains of the BCC are ubiquitously distributed in nature and have been isolated from soil, water, plants, and industrial settings. Some BCC strains have enormous biotechnological potential and have been used for bioremediation of recalcitrant xenobiotics, plant growth promotion, and biocontrol purposes. At the same time, however, BCC strains have emerged as opportunistic pathogens of humans (13, 22, 39, 56). These strains can cause life-threatening lung infections in patients requiring mechanical ventilation and in individuals with chronic granulomatous disease or cystic fibrosis (CF). The clinical outcome of infection with BCC in CF patients is variable and unpredictable, ranging from asymptomatic carriage to a fulminant and fatal pneumonia, the so-called “cepacia syndrome” (29). Although all nine B. cepacia genomovars described to date have been isolated from CF patients, Burkholderia multivorans (genomovar II) and Burkholderia cenocepacia (genomovar III) are most commonly found in clinical samples (12). It is thought that the same strains that occur in the environment are also capable of infecting CF patients, and thus no judgment on the pathogenic potential of a strain can be made on the basis of its genomovar status. In contrast to Pseudomonas aeruginosa, very little is known about the pathogenic mechanisms and virulence determinants of BCC strains.
Work of the past few years has shown that B. cenocepacia utilizes an N-acyl homoserine lactone (AHL)-dependent quorum-sensing (QS) system to express certain genes in a cell density-dependent manner. This regulatory system consists of the AHL synthase CepI, which directs the synthesis of N-octanoylhomoserine lactone (C8-HSL) and, as a minor by-product, N-hexanoylhomoserine lactone (C6-HSL), and the transcriptional regulator CepR (21, 36). At low population densities, cells produce basal levels of AHLs via the activity of the CepI synthase. As the cell density increases, the diffusible AHL signal molecule accumulates in the growth medium. On reaching a critical threshold concentration, the AHL binds to the cognate LuxR-type receptor protein, which in turn leads to induction or repression of target genes. The cep system has been shown to positively regulate expression of extracellular proteases and chitinases, swarming motility, and biofilm formation (26, 36) and to repress synthesis of the siderophore ornibactin (37). Moreover, CepR tightly controls expression of cepI, creating a positive feedback loop that ensures a very rapid increase in virulence factor production once the regulatory system has been triggered. A comparison of the two-dimensional protein patterns of the B. cenocepacia strain H111 and its isogenic cepI mutant H111-I showed that 6% of the proteins detected were differentially expressed (46). Amino-terminal sequence analysis identified several proteins that were down-regulated in the cepI mutant, including a putative superoxide dismutase, a peroxidase, a FimA homologue, and a predicted ABC transporter system.
B. cenocepacia mutants with a defective cep QS system were demonstrated to be attenuated in a Caenorhabditis elegans pathogenesis model (32) as well as in a short-term mouse intranasal colonization model and a chronic agar bead infection model in rats (51). A recent study showed that the B. cepacia epidemic strain marker (BCESM), a unique 31.7-kb DNA region present in some B. cenocepacia strains that are particularly virulent, is part of a pathogenicity island (6). This island encodes an additional QS system consisting of the AHL synthase CciI and its cognate receptor protein CciR. Intriguingly, a cciI mutant was shown to be significantly less virulent in a rat model of chronic lung infection. In conclusion, these data provide convincing evidence that AHL-mediated QS plays an important role in the pathogenicity of B. cenocepacia. However, knowledge about the AHL-regulated virulence factors is still scarce. Here we report the identification of a QS-regulated gene in B. cenocepacia H111, aidA, that is required for efficient slow killing of C. elegans.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Unless otherwise stated, the strains were grown at 37°C in modified Luria-Bertani (LB) broth (5) or AB minimal medium (11) supplemented with 10 mM citrate. Solid media were routinely solidified with 1.5% agar. Growth media for examination of swarming motility contained 0.4% (wt/vol) agar (18). Antibiotics were added as required at final concentrations of 20 μg/ml for gentamicin and 10 μg/ml for chloramphenicol. Kanamycin was used at 50 μg/ml for Escherichia coli and at 100 μg/ml for B. cenocepacia. For complementation of H111-I, 200 nM C8-HSL was added. Growth of liquid cultures was monitored spectrophotometrically with an Ultrospec Plus spectrophotometer (Pharmacia, Freiburg, Germany) by measurement of optical density at 600 nm (OD600).
TABLE 1.
Bacterial strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Description or sequence (5′→3′) | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| MT102 | araD139 (ara-leu)7697 Δlac thi hsdR | Laboratory collection |
| HB101 | recA thi pro leu HsdR−M+ Smr | 31 |
| JM105 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/thi rpsL (Strr) endA sbcB15 sbcC hsdR4 (rK− mK−) Δ(lac-proAB) | 59 |
| P. putida F117 | AHL-negative derivative of P. putida IsoF; PpuI− | 52 |
| B. cenocepaciaa | ||
| H111 | CF isolate from a patient at the Medizinische Hochschule Hannover (1993), genomovar III | 21, 47 |
| H111-I | Kmr; cepI::Km mutant of H111 | 26 |
| H111-R | Kmr; cepR::Km mutant of H111 | 26 |
| H111-A | Kmr; aidA::Km mutant of H111 | This study |
| Plasmids | ||
| pAS-C8 | pBBR1MCS-5 carrying PcepI-gfp(ASV)-Plac-cepR; Gmr, fluorescent AHL sensor plasmid | 52 |
| pBAH27 | pBBR1MCS-5 containing the cepR gene of B. cenocepacia H111 | 26 |
| pBAH44 | pBBR1MCS ligated to an 840-bp fragment containing aidA | This study |
| pBAH52 | pEX19 ligated to a 2.3-kb fragment containing aidA and its flanking regions | This study |
| pBAH54 | Gmr Kmr; pEX19Gm derivative for inactivation of aidA | This study |
| pBAH62 | pBBR1MCS ligated to a 2.3-kb fragment containing aidA and aidA′ | This study |
| pBBR1MCS | Cmr; broad-host-range cloning vector | 34 |
| pBBR1MCS-5 | Gmr; broad-host-range cloning vector | 33 |
| pEX19Gm | Gmr; oriT+sacB+; gene replacement vector | 25 |
| pJBA89 | Apr; pUC18 Not-luxR-Pluxl-RBSII-gfp(ASV)-T0-T1 | 4 |
| pMH94 | Apr Kmr; delivery vector for mini-Tn5 TelrkilA telAB-PA1/04/03-gfp mut3-T0-T1 | M. Hentzer and M. R. Parsek, unpublished |
| pRK600 | Cmr; ColE1oriV; RK-2Mob+ RK2-Tra+; helper plasmid in triparental matings | 17 |
| PCR primersb | ||
| aidA-koIII-v | GGAATTCACGGCAAGGTGATCGAACTG | |
| aidA-koIV-r | ACGCATGCCGGTCGAGATCCAGTATTAC | |
| Km-v-Sac | GGAGCTCACCAGATGCGCGATTTCACC | |
| Km-r-Bam | GGGATCCAAGCCGCCGTCCCGTCAAG | |
| aidA2+P-V | CCCTCGAGTGCTGCACCGGCTCTGGAC | |
| aidA2-R | CCAAGCTTCGTGCCGCGATCAGTTATGG |
For further B. cepacia complex strains, see Table 2.
Restriction sites introduced by the primers are underlined.
Conjugative plasmid transfer.
Plasmids were delivered to B. cenocepacia by triparental mating as described previously (17). Briefly, donor and recipient strains as well as the helper strain E. coli HB101(pRK600) were grown at 37°C overnight in 5 ml of LB medium supplied with the appropriate antibiotics. Following subculturing to an OD600 of 0.9, the cells from 2 ml of culture were harvested, washed, and resuspended in 200 μl of LB medium. Donor and helper cells (100 μl each) were mixed and incubated for 30 min at room temperature. Then 200 μl of the recipient cells was added, and the mixture was spot-inoculated onto the surfaces of prewarmed LB agar plates. After overnight incubation at 37°C, the cells were scraped off and resuspended in 1 ml of 0.9% NaCl. Serial dilutions were plated on PIA medium (Becton Dickinson Biosciences, Sparks, Md.) containing antibiotics for counterselection of donor, helper, and untransformed recipient cells.
DNA manipulations.
Cloning, restriction enzyme analysis, and transformation of E. coli were performed essentially as described previously (50). PCR was performed using TaKaRa rTaq DNA polymerase (TaKaRa Shuzo, Shiga, Japan) or Herculase (Stratagene, La Jolla, Calif.). Plasmid DNA was isolated with the QIAprep Spin Miniprep kit, and chromosomal DNA from B. cenocepacia was purified with the DNeasy tissue kit. DNA fragments were purified from agarose gels by using the QIAquick gel extraction kit (all kits are from QIAGEN, Hilden, Germany).
Construction of a B. cenocepacia H111 aidA mutant.
A defined aidA mutant was constructed by the gene replacement method described by Hoang et al. (25). To this end, a 2.3-kb fragment of H111 chromosomal DNA containing aidA and its flanking region was PCR amplified by using primers aidA-koIII-v and aidA-koIV-r (Table 1). The PCR product was digested with EcoRI and SphI and ligated to the gene replacement vector pEX19Gm (25), giving rise to plasmid pBAH52. Thereafter, a 345-bp fragment was excised from the aidA gene by digestion of pBAH52 with SacI and BamHI. The npt gene from transposon Tn903 (43), which confers resistance to kanamycin, was PCR amplified as a 1.4-kb fragment by using primers Km-v-Sac and Km-r-Bam. The amplicon was digested with BamHI and SacI and ligated into pBAH52 cut with the same enzymes. The final construct, which was designated pBAH54, was transferred to B. cenocepacia H111, and integrants were selected on PIA medium containing kanamycin. To screen for gene replacement mutants, Kmr clones were tested for gentamicin sensitivity, because the gentamicin resistance gene is lost in the case of a double-crossover event. One mutant, which was designated B. cenocepacia H111-A, was chosen, and the correct genetic structure of the strain was confirmed by Southern blot analysis.
Complementation of H111-A.
For complementation with aidA, a DNA segment containing the aidA gene together with its putative promoter region was PCR amplified by using primers aidA2+P-V and aidA2-R (Table 1). Following digestion with HindIII and XhoI, the PCR fragment was inserted into the broad-host-range vector pBBR1MCS cut with the same enzymes. The resulting plasmid was designated pBAH44 (aidA+). For complementation with both aidA and aidA′, a 2.3-kb DNA fragment was PCR amplified by using the primer pair consisting of aidA-koIII-v and aidA-koIV-r. The amplicon was digested with EcoRI and SphI and ligated into the broad-host-range vector pBBR1MCS cut with the same enzymes, yielding plasmid pBAH62 (aidA+ aidA′+).
Protein analysis.
Protein concentrations were determined according to the method of Bradford (8) by using the Coomassie Plus protein assay reagent (Pierce, Bonn, Germany) according to the manufacturer's instructions. The protein concentration was calculated by using bovine serum albumin as a standard.
Protein samples were separated on a sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) gel (35), and bands were visualized by Coomassie blue staining (Serva Blue G; Serva, Heidelberg, Germany). Antibodies were handled according to standard procedures (24). For Western blotting, protein fractions or whole-cell proteins were separated on an SDS-15% PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore, Eschborn, Germany). The membrane was probed with the anti-AidA antibodies, and detection reactions were performed with alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Sigma, Deisenhofen, Germany) according to the recommendations of the manufacturer (Roche, Penzberg, Germany).
Fractionation of cell proteins.
A 200-μl volume of an overnight culture was harvested by centrifugation (5,000 × g, 5 min). The supernatant was retained, and the cells were washed twice with 500 μl of phosphate-buffered saline (PBS) and finally resuspended in 200 μl of PBS. Cell lysis was performed by sonication (twice, for 1 min each time) and checked microscopically. Cell debris was removed by centrifugation (10,000 × g, 10 min), and the supernatant containing the intracellular proteins was retained. Surface proteins were isolated by using the protocol of Kawai et al. (30) with minor modifications. Cells from 500 μl of a culture grown to an OD600 of 3 in LB medium were harvested, washed in PBS, and resuspended in 100 μl of 0.25% SDS. Following incubation at room temperature for 3 min and centrifugation at 6,000 × g for 5 min, the supernatants containing the cell surface proteins were collected.
Purification of AidA and generation of anti-AidA antibodies.
For purification of AidA, the B. cenocepacia wild-type strain H111 was grown on agar plates. After overnight incubation, cell material was scraped off the plates and surface proteins were extracted as described above. The samples were separated by preparative SDS-15% PAGE on a prep cell (model 491; Bio-Rad, Munich, Germany), and the fractions obtained were analyzed by SDS-PAGE as described above. Those fractions containing pure AidA were pooled, concentrated by ammonium sulfate precipitation, and resuspended in PBS (24). The sample was then applied to a PD10 column (Amersham Biosciences, Freiburg, Germany) to remove residual SDS. Purified protein (1 mg) was used to raise rabbit polyclonal antibodies (pab productions, Hebertshausen, Germany).
Protein identification and database searches.
For identification of proteins by N-terminal sequencing, protein samples were separated by SDS-PAGE, followed by semidry electroblotting to a PVDF membrane and staining with Coomassie blue. The excised piece of the PVDF membrane was subjected to automated Edman degradation on a gas phase sequencer (model 477A; Applied Biosystems, Darmstadt, Germany) according to the manufacturer's instructions. The amino-terminal sequence obtained was used for a TBLASTN search at the Sanger Centre, Cambridge, United Kingdom (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/b_cenocepacia) to identify the corresponding open reading frame (ORF) in the B. cenocepacia J2315 genome. The deduced protein sequence of this ORF was then compared to other sequences in GenBank by use of the online BLAST search engine at the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Nematode killing assays.
Nematode killing assays were performed essentially as described by Köthe et al. (32). Briefly, overnight cultures of the B. cenocepacia test strains were adjusted to a density of about 1.3 × 104 to 1.5 × 104 CFU/ml, and 100 μl of the suspensions was plated on 6.0-cm-diameter nematode growth medium (NGM) agar plates for slow-killing assays or on peptone-glucose-sorbitol (PGS) agar plates for fast-killing assays. After 24 h of incubation, approximately 25 hypochlorite-synchronized L4 larvae were used to inoculate the plates. The actual number of worms was determined by using a Stemi SV6 microscope (Zeiss, Oberkochen, Germany) at a magnification of ×50. Plates were then incubated at 23°C and scored for live worms at various time points. Nematodes were considered dead when they failed to respond to touch. All experiments were carried out five to eight times, and E. coli OP50 was used as a control in the assays.
Detection and quantification of AHLs.
Production of AHLs was investigated by cross-streaking the strains against E. coli JM105 harboring the fluorescent sensor plasmid pJBA89 (4). After overnight incubation at 30°C, fluorescence was detected by illumination with blue light by using an HQ 480/40 filter (AHF-Analysentechnik, Tübingen, Germany) in combination with a halogen lamp (Volpi, Schlieren, Switzerland) as a light source in a dark box equipped with a light-sensitive camera (Hamamatsu Photonics, Herrsching, Germany) with a Pentax CCTV camera lens and an HQ 535/20 filter (AHF-Analysentechnik). For quantifying AHL production, we used Pseudomonas putida F117 harboring the sensor plasmid pAS-C8 (52). Ten microliters of filter-sterilized culture supernatants was added to 100 μl of an exponential culture of the sensor strain in the wells of a FluoroNunc Polysorp microtiter dish (Nunc, Wiesbaden, Germany). Following incubation at 30°C for 6 h, the expression of the gfp reporter gene was measured with a Lambda Fluoro 320 Plus reader (Bio-Tek Instruments, Winooski, Vt.).
Phenotypical characterization of the aidA mutant.
Biofilm formation in microtiter plates was tested as described previously (45) with a few modifications (27). For comparing biofilm structures, bacteria were grown at 30°C in the wells of black 96-well optical microtiter plates (Nunc) filled with 100 μl of ABC minimal medium. After 1, 2, and 3 days, respectively, the medium with the free-swimming cells was removed, and the cells adhering to the walls of the wells were stained with the live/dead BacLight Bacterial Viability kit (Molecular Probes, Leiden, Netherlands). Biofilm structures were then inspected with a confocal laser scanning microscope (100× lens objective; Zeiss). Swarming motility was observed as described by Eberl et al. (18) on ABC agar plates supplemented with 0.1% Casamino Acids and solidified with 0.4% agar.
RESULTS
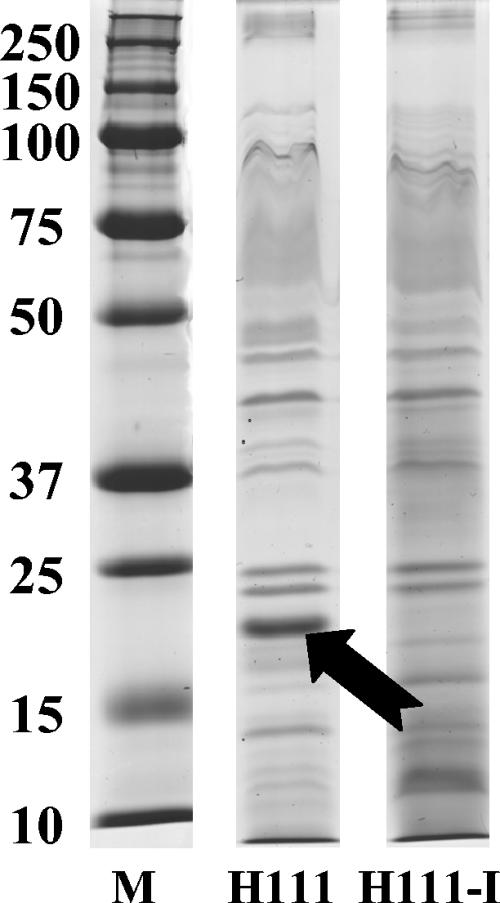
Expression of AidA is regulated by the cep QS system. In a recent study, Huber et al. analyzed the surface proteins of B. cenocepacia H111 and various transposon insertion mutants with defective biofilm formation (27). We noticed that a band of approximately 19 kDa, which is abundant in the wild type, is missing in the cepR mutant m64 and is greatly reduced in mutants MA18 and MA24, which bear the transposon in suhB and yciL, respectively. Given that all three mutants show defective production of C8-HSL (27), we hypothesized that expression of this protein may be regulated by the cep QS system. To test this idea, we analyzed the surface proteins of the cepI mutant H111-I. The 19-kDa band was absent in the mutant, and this defect could be fully restored by addition of 200 nM C8-HSL to the culture medium (Fig. 1; also data not shown), indicating that expression of this protein is QS regulated. To ascertain the identity of this band, the protein was purified and the N-terminal amino acid sequence was determined: SRVTDVLVSFDTETILKKYPNP. Searching the B. cenocepacia J2315 genome database (http://www.sanger.ac.uk/Projects/B_cenopacia/) allowed the identification of an ORF with an identical N-terminal sequence. This ORF is predicted to encode an 18.1-kDa protein that has 53% identity (67% similarity) with AidA of the plant pathogen Ralstonia solanacearum AW1. aidA (autoinducer dependent) is located upstream of the sol QS locus of the organism and has been shown to be regulated by the LuxR homologue SolR (20). However, no function has been assigned to this gene so far. These results are in full agreement with those of a recent proteomics study, which identified AidA among those proteins that were missing in the cepI mutant H111-I compared to the wild-type strain H111 (46).
FIG. 1.
Surface proteins of B. cenocepacia H111 and the cepI mutant H111-I. Surface proteins were extracted by treatment of cells with 0.25% SDS, and samples were separated on an SDS-15% PAGE gel. The band missing in H111-I is marked with an arrow. Lane M, protein markers (in kilodaltons).
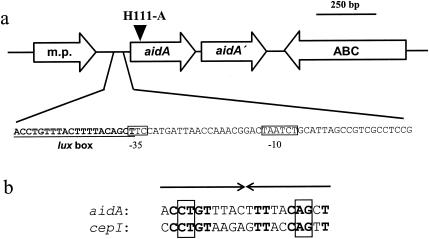
On the basis of the B. cenocepacia J2315 genome sequence, we PCR amplified the DNA regions flanking aidA. Sequence analysis of the aidA downstream region revealed an ORF, tentatively designated aidA′, that is predicted to encode a second AidA homologue that has 31% identity (51% similarity) with AidA of B. cenocepacia H111 and 27% identity (46% similarity) with AidA of R. solanacearum AW1 (Fig. 2). It is tempting to speculate that this second copy of aidA may have arisen from a gene duplication event. Downstream of aidA′ and oriented in the opposite direction is an ORF that is highly homologous to periplasmic binding proteins of various ABC transporters. The aidA promoter region contains putative −10 and −35 E. coli consensus sequences (Fig. 3). Most importantly, the −35 position is adjacent to a 20-bp imperfect palindrome with a high similarity to lux box-like elements, in particular with the operator sequence present upstream of cepI (36).
FIG. 2.
Comparison of the AidA homologues of B. cenocepacia H111 (AidA and AidA′) and R. solanacearum AW1 (AidA-R). Shown on a black background are identical amino acids; shown on a grey background are similar amino acids.
FIG. 3.
(a) Genetic organization of the aidA locus of B. cenocepacia. Genes are represented by open arrows pointing in the direction of transcription. Shown below is the nucleotide sequence of the aidA upstream region of B. cenocepacia H111. Putative −35 and −10 promoter sequences are boxed, and an inverted repeat that exhibits similarity to lux box sequences is underlined. The solid arrowhead above the diagram marks the location of the npt cassette in the aidA mutant H111-A. m.p., predicted membrane protein; ABC, putative ABC transporter component, substrate binding protein. (b) Nucleotide sequence comparison of the lux box-like sequence upstream of aidA and the operator region of cepI (36). Boldfaced nucleotides, identical bases. Boxed sequences represent the minimal consensus defined for operators of QS-regulated genes in P. aeruginosa (58).
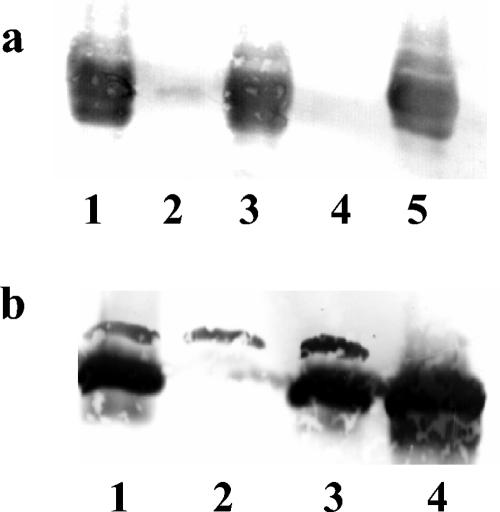
A rabbit antiserum directed against purified AidA was prepared as described in Materials and Methods. The results of Western blotting on samples from cultures of the wild-type strain H111, the cepR mutant H111-R, the complemented cepR mutant H111-R(pBAH27), and the cepI mutant H111-I in the presence or absence of 200 nM C8-HSL are shown in Fig. 4a. Expression of AidA was virtually abolished in both QS mutants but could be completely restored when the mutant H111-I was grown in the presence of 200 nM C8-HSL or when the mutant H111-R was complemented with the wild-type cepR allele on a plasmid. In conclusion, our data provide clear evidence that expression of AidA in B. cenocepacia H111 is regulated by the cep QS system.
FIG. 4.
Expression of AidA is controlled by the cep QS system. The B. cenocepacia wild-type strain H111 and various isogenic mutants were grown in LB medium overnight. Total-cell proteins were separated by SDS-PAGE, and AidA was visualized by immunoblotting with antisera directed against the protein. (a) Lanes: 1, H111; 2, H111-I (cepI); 3, H111-I (cepI) plus 200 nM C8-HSL; 4, H111-R (cepR); 5, H111-R(pBAH27). (b) Lanes: 1, H111; 2, H111-A (aidA); 3, H111-A(pBAH44); 4, H111-A(pBAH62).
Cellular localization of AidA.
A recent proteome analysis of the QS regulon of B. cenocepacia H111 identified AidA as an AHL-regulated protein in the intracellular, extracellular, and surface-bound protein fractions (46). To determine the cellular localization of AidA, we performed Western blotting on samples that were fractionated as described in Materials and Methods. Quantification of the amounts of AidA in the different cell fractions revealed that the largest amount of AidA (approximately 85%) was detected in the intracellular fraction and only minor amounts were observed in the surface fraction (approximately 5%) and the culture supernatant (approximately 10%). We also performed immunofluorescence microscopy on B. cenocepacia H111 cells but were unable to detect AidA on the bacterial cell surface (data not shown). These results, together with the fact that AidA does not possess an identifiable signal sequence, strongly suggest that AidA is in fact an intracellular protein. Hence, the minor amounts of the protein present in the extracellular and surface-bound cell fractions are likely due to cell lysis and/or contamination during the separation of cellular compartments.
AidA is expressed in the late-exponential phase and accumulates during growth arrest.
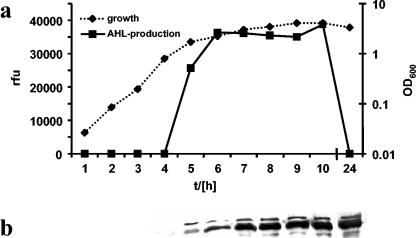
To investigate expression of AidA in more detail, we determined the amounts of the protein in cells along the growth curve. In parallel, we measured the concentrations of AHLs in the culture supernatant by use of the green fluorescent protein (GFP)-based AHL sensor F117(pAS-C8).
When cells were grown in LB medium, expression of AidA was very low during early-exponential growth but was greatly stimulated in the transition to stationary phase (Fig. 5). Importantly, at this time point, production of AHLs was also boosted, indicating that expression of AidA is tightly regulated by the cep QS system. Similar results were obtained when the experiment was performed in ABC minimal medium, except that induction of both AHL production and AidA synthesis occurred at a lower optical density (data not shown). Addition of 200 nM C8-HSL to the medium affected neither the time point of AidA induction nor the eventual concentration of the protein in cells (data not shown). In this context it is also worth noting that AidA appears to be a remarkably stable protein; the greatest amounts were observed in stationary-phase cells (see Fig. 5, 24-h time point), when AHLs were no longer detectable in the culture supernatant.
FIG. 5.
Expression of AidA is growth phase dependent. The B. cenocepacia wild-type strain H111 was grown in LB medium, and samples were taken along the growth curve. AHL concentrations were quantified by mixing 10 μl of cell-free culture supernatant with 100 μl of an exponentially growing culture of the GFP-based AHL biosensor P. putida(pAS-C8). Following incubation for 4 h, relative fluorescence units (rfu) were measured with a fluorimeter (a). Expression of AidA was assessed by normalizing samples to an OD600 of 1 and using 10 μl for Western blotting with anti-AidA antibodies (b).
Construction and characterization of a defined B. cenocepacia H111 aidA mutant.
In order to investigate the role of AidA, we constructed a site-directed insertion mutation in the aidA gene by using a gene replacement method (for details, see Materials and Methods) (Fig. 3). The genetic structure of the mutant, which was designated H111-A, was confirmed by Southern blot analysis (data not shown). B. cenocepacia H111-A no longer produced AidA, unless plasmid pBAH44 (aidA+) or pBAH62 (aidA+ aidA′+) was introduced into the strain (Fig. 4b). The growth rate of the aidA mutant H111-A was indistinguishable from that of the wild type in LB medium as well as in ABC minimal medium. We also performed a competition assay, in which the wild-type strain H111 and the isogenic aidA mutant H111-A were grown together as a mixed culture in rich LB medium as well as in ABC minimal medium. Both strains grew equally well in the two media, and the viable counts of the strains remained stable during extended periods of starvation (up to 6 weeks in ABC medium). These results show that the mutant has no competitive disadvantage compared to the wild type.
It has been shown previously that the formation of a mature biofilm is controlled by the cep QS system in B. cenocepacia H111 (26). We therefore investigated whether AidA plays a role in biofilm formation. However, the biofilms formed by the wild-type strain H111 and the aidA mutant H111-A in the wells of microtiter dishes (see Materials and Methods for details) were virtually indistinguishable in terms of biofilm thickness and structure and exhibited similar biofilm formation kinetics (data not shown), indicating that AidA is not required for biofilm formation. Another QS-regulated phenotype in B. cenocepacia H111 is swarming motility (26). However, the aidA mutant colonized the surfaces of appropriate swarming plates as efficiently as the wild type. Likewise, no differences in the production of extracellular proteases or siderophores, both known QS-controlled functions in B. cenocepacia (26, 36), could be observed between H111 and H111-A (data not shown).
AidA of B. cenocepacia H111 is involved in killing of C. elegans.
It has been shown previously that the cep QS system of B. cenocepacia H111 is required for efficient killing of the nematode C. elegans (32). However, the cep-regulated virulence factors that are responsible for nematode pathogenicity are unknown. Depending on the culture conditions, B. cenocepacia H111 kills the nematode by one of two distinct mechanisms. On a high-osmolarity medium (PGS), the nematodes are killed within 24 h by the action of a yet unidentified extracellular toxin (fast killing). On NGM, killing occurs over the course of 2 days and involves the accumulation of bacteria in the intestinal lumen of C. elegans (slow killing).
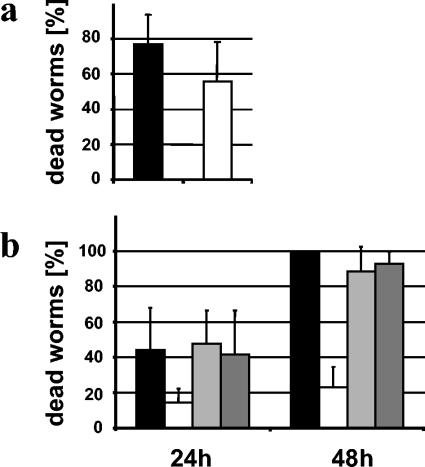
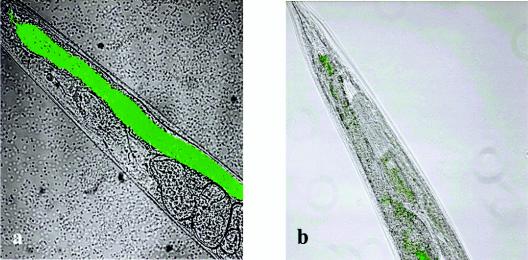
We tested the aidA mutant H111-A in the two nematode-killing models. While no significant difference between H111-A and the wild-type strain H111 was observed in the fast killing of C. elegans, the virulence of the mutant was attenuated in the slow-killing model (Fig. 6). When grown on wild-type cells, all nematodes were killed within 48 h, compared to 24% when they were grown on H111-A lawns. Upon prolonged incubation, the nematodes resumed reproduction when the aidA mutant was used as a food source. Importantly, when H111-A was complemented with plasmid pBAH44 (aidA+) or pBAH62 (aidA+ aidA′+), killing rates were rescued to wild-type levels (Fig. 6). These results show that AidA is not required for toxin production but may be important for the persistence of B. cenocepacia H111 in the nematode gut, as slow killing resembles an infection-like process during which the bacteria accumulate in the lumen of the intestine. To further address this issue, we constructed an H111-A strain expressing Aequoria victoria GFP. Microscopic inspection of nematodes after 24 h of feeding on tagged cells revealed that, in contrast to the wild type, the aidA mutant H111-A did not colonize the worm gut (Fig. 7). Because the grinder in the pharynx of the nematode is a major barrier to the entry of bacteria into the intestine, we investigated whether the attenuated pathogenicity of H111-A is uniquely a problem of penetration. To this end, we performed slow-killing experiments with the C. elegans phm-2 mutant, which has a nonfunctional grinder. As with the wild-type nematode, we were unable to detect GFP-expressing H111-A in the intestinal lumen of the phm-2 mutant (data not shown), indicating that AidA may not be essential for passing the grinder but rather is required for persisting in the nematode gut.
FIG. 6.
Killing of nematodes by B. cenocepacia H111 (solid bars), the aidA mutant H111-A (open bars), and the complemented aidA mutants H111-A(pBAH44) (aidA+) (light shaded bars) and H111-A(pBAH62) (aidA+ aidA′+) (dark shaded bars) under conditions of fast killing (a) and slow killing (b). Each data point represents the mean and standard deviation from eight (fast killing) or five (slow killing) independent trials.
FIG. 7.
The ability of B. cenocepacia H111 to accumulate in the C. elegans intestine is dependent on AidA. Nematodes were fed on GFP-tagged derivatives of the B. cenocepacia wild-type strain H111 (a) or the aidA mutant H111-A (b). Worms were inspected with a confocal laser scanning microscope, and transmission images were merged with epifluorescence micrographs showing bacterial GFP fluorescence. The entire lumen of a nematode fed on GFP-tagged wild-type H111 was filled with green fluorescent cells (a). In contrast, only a few GFP-tagged cells were observed in the nematode gut when the aidA mutant H111-A (b) was used as a food source.
Production of AidA is widespread among members of the BCC.
Because in B. cenocepacia strain H111 AidA is an important virulence factor for nematode killing, we were interested to know if other strains of the BCC produce this protein. To this end, representatives of all nine currently described genomovars plus a strain belonging to the proposed genomovar X (12) were tested for AidA production by Western blotting (Table 2). Given that expression of AidA is stringently controlled by QS in B. cenocepacia H111, we also tested the strains for AHL production in cross-streaking experiments using the GFP-based AHL biosensor E. coli JM105(pJBA89). Positive signals with anti-AidA antibodies were obtained for many strains of the BCC; only for two genomovars, namely, Burkholderia anthina (genomovar VIII; two strains) and Burkholderia ubonensis (genomovar X; one strain), could no AidA-positive strain be identified (Table 2). In the case of B. multivorans, five out of seven strains tested were AidA negative. Likewise, two of the three Burkholderia ambifaria (genomovar VII) strains did not produce AidA. In the remaining genomovars, the great majority of strains tested were AidA positive, although with some strains only weak signals were obtained. With strains from B. cenocepacia, Burkholderia vietnamiensis (genomovar V), Burkholderia dolosa (genomovar VI), and Burkholderia pyrrocinia (genomovar IX), synthesis of AidA corresponded well with the production of AHL signal molecules. Within the other genomovars, this correlation was less clear. We also tested six randomly chosen strains of other Burkholderia species with varying phylogenetic distances from the BCC strains (12), including Burkholderia tuberum, Burkholderia phymatum, Burkholderia gladioli, Burkholderia phenazinium, Burkholderia fungorum, and Burkholderia terricola, for production of AidA. None of the strains investigated tested positive for AidA (data not shown).
TABLE 2.
Distribution of AidA among B. cepacia complex strainsa
| Speciesb | Strainc | Origind | AHL productione | AidA signale |
|---|---|---|---|---|
| B. cepacia (genomovar I) | LMG 1222T | Allium cepa | + | + |
| LMG 6963 | Soil | + | − | |
| LMG 18821 | CF patient | + | − | |
| B. multivorans (genomovar II) | H59 | CF patient | − | − |
| H107 | CF patient | (+) | + | |
| H115 | CF patient | − | − | |
| H158 | CF patient | − | − | |
| LMG 13010 | CF patient | − | − | |
| LMG 16660 | CF patient | − | + | |
| LMG 17588 | Soil | − | − | |
| LMG 18822 | CF patient | (+) | + | |
| LMG 18945 | CF patient | − | − | |
| B. cenocepacia (genomovar III) | H111 | CF patient | + | + |
| H147 | CF patient | + | + | |
| LMG 6981 | Bronchial washings | + | + | |
| LMG 6988 | Leg wound | + | + | |
| LMG 6993 | Soil | + | + | |
| LMG 14271 | CF patient | + | + | |
| LMG 16656 | CF patient | + | + | |
| LMG 16659 | CF patient | + | + | |
| LMG 18826 | CF patient | − | − | |
| LMG 18863 | CF patient | + | + | |
| B. stabilis (genomovar IV) | H118 | CF patient | + | + |
| H134 | CF patient | + | − | |
| H145 | CF patient | + | − | |
| H162 | CF patient | + | + | |
| H173 | CF patient | − | − | |
| H175 | CF patient | + | + | |
| H177 | CF patient | + | + | |
| H177E | Water outlet | + | + | |
| H193 | CF patient | + | + | |
| LMG 6997 | Ear | + | + | |
| LMG 14291 | CF patient | + | + | |
| B. vietnamiensis (genomovar V) | LMG 6998 | Blood | + | + |
| LMG 6999 | Neck abscess | + | + | |
| LMG 10929T | Rice rhizosphere | + | + | |
| LMG 18836 | CGD patient | + | + | |
| B. dolosa (genomovar VI) | LMG 18941 | CF patient | + | + |
| LMG 18942 | CF patient | + | + | |
| LMG 18943 | CF patient | + | + | |
| LMG 21443 | Alysicarpus glumaceus, root nodule | + | + | |
| LMG 21819 | CF patient | + | + | |
| LMG 21820 | CF patient | + | + | |
| B. ambifaria (genomovar VII) | LMG 17828 | Corn roots | + | + |
| LMG 19182T | Pea rhizosphere | + | − | |
| LMG 19467 | CF patient | + | − | |
| B. anthina (genomovar VIII) | LMG 20983 | CF patient | + | − |
| LMG 21821 | CF patient | + | − | |
| B. pyrrocinia (genomovar IX) | LMG 14191T | Soil | − | − |
| LMG 21822 | Cornfield soil | + | + | |
| LMG 21823 | Water | + | + | |
| B. ubonensis (genomovar X)* | LMG 20358T | Surface soil | − | − |
Strains were tested for AidA by Western blotting of cell proteins obtained from 5 μl of an overnight culture and separated on an SDS-15% polyacrylamide gel. AHL production was tested by cross-streaking strains against the monitor strain E. coli JM105(pJBA89).
*, the taxonomic position of B. ubonensis within the B. cepacia complex has not been fully established (12).
Strains were obtained from the Laboratorium voor Microbiologie Gent Culture Collection, Universiteit Gent, Gent, Belgium (LMG), or the Medizinische Hochschule Hannover, Hanover, Germany (H).
CGD, chronic granulomatous disease.
+, clear signal; (+), weak signal; −, no signal.
DISCUSSION
A large number of factors appear to contribute to the pathogenicity of BCC strains. Previous work has identified potential virulence factors, including a type III secretion system (44, 54), cable pili (49, 53), flagella (55), an extracellular polysaccharide (10), production of a melanin pigment (60), at least four iron-chelating siderophores (16), and extracellular hydrolytic enzymes (14, 28, 42, 57). Furthermore, some strains have the ability to survive within respiratory epithelial cells (9, 41), murine macrophages (48), and amoebae (40).
Recent work has established that the cep QS system contributes to the virulence of B. cenocepacia in a wide range of hosts, including the plant alfalfa (7), the nematode C. elegans (32), and murine species (51). The importance of AHL-mediated cell-to-cell signaling for the virulence of B. cenocepacia was further corroborated by the study of Baldwin et al. (6), who showed that the BCESM is a genomic island that contains an additional QS locus, cci, that is involved in the regulation of pathogenic traits. However, at present very little is known about the QS-regulated virulence factors that are responsible for the pathogenicity of the organism in the various models. Recent work has demonstrated that expression of a zinc metalloprotease, ZmpA, is regulated by CepIR (51). By using an agar bead infection model in rats, it was shown that this extracellular protease contributes to the virulence of some but not all B. cenocepacia strains, indicating that in addition to ZmpA, other, yet unknown virulence factors are involved in the pathogenesis of this bacterium (14).
The nematode C. elegans has emerged as a highly valuable model for the study of bacterial pathogenicity (for reviews see references 1 and 19). It has been shown previously that B. cenocepacia H111, which was isolated from a CF patient, effectively kills C. elegans (32). Depending on the medium used for growth of B. cenocepacia H111, two different modes of killing were observed. On a high-osmolarity medium (PGS), the worms were killed within 24 h due to the action of a diffusible extracellular toxin (fast killing). When worms were grown on NGM, the killing occurred over the course of 1 to 3 days (slow killing) and involved the accumulation of bacteria in the intestinal lumen of the worm, resembling an infection-like process. While the cep QS system of B. cenocepacia H111 was found to be essential for the killing of C. elegans under both slow- and fast-killing conditions, the type II secretion machinery was not required for nematode killing. Hence, neither extracellular protease nor lipase, both of which are secreted via the type II pathway, contributes to the virulence of B. cenocepacia H111 in the C. elegans model.
Our study identified a novel virulence factor, AidA, which plays an important role in the slow killing of C. elegans. Given that the aidA mutant H111-A was only slightly attenuated under fast-killing conditions, it is unlikely that this gene encodes a toxic factor; rather, aidA is probably required for survival and/or proliferation in the nematode intestine. Two additional lines of evidence support this hypothesis. First, microscopic investigations showed that, in contrast to the wild type, H111-A was unable to colonize the intestinal lumen of C. elegans, even when a nematode mutant with a defective grinder was used as the host. Second, when a recombinant E. coli strain expressing AidA was used as a food source for C. elegans, the life span of the nematode was the same as that of nematodes fed the background E. coli strain (data not shown). The aidA gene was originally identified in R. solanacearum AW1 and was shown to be regulated by the sol QS system, to which the gene is also physically linked (20). Because AHLs were not required for the production of virulence factors in culture or for virulence in planta, it was concluded that the sol QS system (and consequently AidA) is not essential for plant pathogenicity. To our knowledge there are no reports that R. solanacearum is pathogenic for hosts other than plants. However, in a recent study, Couillaut and Ewbank (15) showed that the well-known plant pathogens Agrobacterium tumefaciens and Erwinia carotovora subsp. carotovora kill C. elegans. Given that R. solanacearum is well adapted to survive in soil in the absence of host plants (23), it is tempting to speculate that AidA enables the bacterium to survive ingestion by predatory nematodes. It is also noteworthy that in the case of B. cepacia ATCC 25416 (genomovar I), the cep QS system has been demonstrated to be involved in the pathogenicity of the bacterium toward onions (2). While our data show that AidA is an essential factor in slow killing of C. elegans, additional work will be required to determine if this protein also contributes to the virulence of B. cenocepacia in other hosts.
Using anti-AidA antibodies, we showed that expression of AidA is stringently controlled by the cep QS system. In the putative promoter region of aidA, we identified a 20-bp imperfect palindrome that shares a high degree of similarity with lux box-like elements, in particular with the operator sequence upstream of cepI (36). Aguilar et al. (3), using a genetic approach, identified 28 QS-regulated promoters in B. cepacia ATCC 25416 (genomovar I). In full agreement with our study, the authors demonstrated that a CepR-regulated promoter is positioned upstream of aidA. Taken together, these results strongly suggest that expression of AidA is regulated at the transcriptional level by binding of a CepR-C8-HSL complex in the promoter region of aidA.
We showed that many strains of the BCC express AidA. While for strains of B. cenocepacia, B. vietnamiensis, B. dolosa, and B. pyrrocinia, synthesis of AidA corresponded well with the production of AHL signal molecules, this correlation was less evident for the other BCC genomovars (Table 2). Previous work has shown that synthesis of AHL signal molecules is widespread among the members of B. cepacia genomovars I to VII (21, 38). In this study we provide evidence that strains belonging to genomovars VIII (B. anthina) and IX (B. pyrrocinia) also produce AHLs. No AHL synthesis was detectable with the only B. ubonensis strain tested in this study. Notably, not all Burkholderia strains that tested positive for AidA killed C. elegans, and not all strains that were pathogenic for the nematode expressed AidA, at least under the conditions tested (data not shown). These data support the view that aidA is required for persistence in the nematode gut rather than encoding a toxin, and they indicate that additional virulence factors may contribute to the differences in C. elegans pathogenicity between the various strains. Work is currently under way to identify these nematocidal determinants in B. cenocepacia H111.
Acknowledgments
We thank C. Merkl, C. Krammer, and C. Bruns for excellent technical assistance, F. Lottspeich for N-terminal sequencing of AidA, and H. P. Schweitzer and M. E. Kovach for providing plasmids.
This work was supported by the Deutsche Forschungsgemeinschaft (EB 2051/1-3).
Editor: J. B. Bliska
REFERENCES
- 1.Aballay, A., and F. M. Ausubel. 2002. Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr. Opin. Microbiol. 5:97-101. [DOI] [PubMed] [Google Scholar]
- 2.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 71:5306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, J. W., E. Altman, T. J. Beveridge, and D. P. Speert. 2003. Colonial morphology of Burkholderia cepacia complex genomovar III: implications in exopolysaccharide production, pilus expression, and persistence in the mouse. Infect. Immun. 71:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, J. D., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 12.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 13.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbett, C. R., M. N. Burtnick, C. Kooi, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149:2263-2271. [DOI] [PubMed] [Google Scholar]
- 15.Couillault, C., and J. J. Ewbank. 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70:4705-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 18.Eberl, L., M. K. Winson, C. Sternberg, G. S. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 19.Ewbank, J. J. 2002. Tackling both sides of the host-pathogen equation with Caenorhabditis elegans. Microbes Infect. 4:247-256. [DOI] [PubMed] [Google Scholar]
- 20.Flavier, A. B., L. M. Ganova-Raeva, M. A. Schell, and T. P. Denny. 1997. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:7089-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotschlich, A., B. Huber, O. Geisenberger, A. Tögl, A. Steidle, K. Riedel, P. Hill, B. Tümmler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 22.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granada, G. A., and L. Sequeira. 1983. Survival of Ralstonia solanacearum in soil, rhizosphere and plant roots. Can. J. Microbiol. 29:433-440. [Google Scholar]
- 24.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 26.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 27.Huber, B., K. Riedel, M. Köthe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 28.Hutchison, M. L., I. R. Poxton, and J. R. Govan. 1998. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect. Immun. 66:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 30.Kawai, E., H. Akatsuka, A. Idei, T. Shibatani, and K. Omori. 1998. Serratia marcescens S-layer protein is secreted extracellularly via an ATP-binding cassette exporter, the Lip system. Mol. Microbiol. 27:941-952. [DOI] [PubMed] [Google Scholar]
- 31.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 32.Köthe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 33.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 34.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 40.Marolda, C. L., B. Hauroder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 41.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKevitt, A. I., S. Bajaksouzian, J. D. Klinger, and D. E. Woods. 1989. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect. Immun. 57:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 44.Parsons, Y. N., K. J. Glendinning, V. Thornton, B. A. Hales, C. A. Hart, and C. Winstanley. 2001. A putative type III secretion gene cluster is widely distributed in the Burkholderia cepacia complex but absent from genomovar I. FEMS Microbiol. Lett. 203:103-108. [DOI] [PubMed] [Google Scholar]
- 45.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 46.Riedel, K., C. Arévalo-Ferro, G. Reil, A. Görg, F. Lottspeich, and L. Eberl. 2003. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 24:740-750. [DOI] [PubMed] [Google Scholar]
- 47.Römling, U., B. Fiedler, J. Bosshammer, D. Grothues, J. Greipel, H. von der Hardt, and B. Tümmler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 48.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 49.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 52.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acyl homoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, L., R. Z. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 54.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 71:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomich, M., C. A. Herfst, J. W. Golden, and C. D. Mohr. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect. Immun. 70:1799-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 57.Vasil, M. L., D. P. Krieg, J. S. Kuhns, J. W. Ogle, V. D. Shortridge, R. M. Ostroff, and A. I. Vasil. 1990. Molecular analysis of hemolytic and phospholipase C activities of Pseudomonas cepacia. Infect. Immun. 58:4020-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 60.Zughaier, S. M., H. C. Ryley, and S. K. Jackson. 1999. A melanin pigment purified from an epidemic strain of Burkholderia cepacia attenuates monocyte respiratory burst activity by scavenging superoxide anion. Infect. Immun. 67:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]