Abstract
A variety of organisms have evolved mechanisms to detect and respond to light, in which the response is mediated by protein structural changes following photon absorption. The initial step is often the photo-isomerization of a conjugated chromophore. Isomerization occurs on ultrafast timescales, and is substantially influenced by the chromophore environment. Here we identify structural changes associated with the earliest steps in the trans to cis isomerization of the chromophore in photoactive yellow protein. Femtosecond, hard X-ray pulses emitted by the Linac Coherent Light Source were used to conduct time-resolved serial femtosecond crystallography on PYP microcrystals over the time range from 100 femtoseconds to 3 picoseconds to determine the structural dynamics of the photoisomerization reaction.
Trans-cis isomerization constitutes a major class of chemical reactions of critical importance to biology, where for example light-dependent isomerization of a retinal chromophore underlies vision (1). Since isomerization occurs on the femtosecond (fs) to picosecond (ps) time scale, ultrafast time-resolved methods are necessary to follow the reaction in real time. The spectral response after photon absorption reveals the dynamics of the molecules involved (2-5) but does not directly observe the associated structural changes, which have to be inferred by computational approaches (6). Until recently it has been impossible to directly determine the structure of molecules on ultrafast time scales. With the recent availability of hard X-ray pulses on the fs time scale emitted by free electron laser (FEL) sources such as the Linac Coherent Light Source (LCLS), the ultrafast fs to ps time scale has become experimentally accessible (7-11). Photochemical reactions (12) are initiated by photon absorption, which promotes electrons into the excited state. Thereafter, the nuclei experience - and the structure evolves on - the excited state potential energy surface (PES) (13, 14). The shape of the surface controls the subsequent nuclear dynamics. After returning to the ground state PES, the reaction continues and is driven thermally. Although structures of longer-lived excited state intermediates have been characterized with ~100 ps time resolution at synchrotrons (15-19), the fs structural dynamics of ultrafast photochemical reactions can only be investigated at an X-ray FEL (11). The photoactive yellow protein (PYP) is an ideal macromolecular system with which to investigate ultrafast trans to cis isomerization. Its chromophore, p-coumaric acid (pCA), can be photoexcited by absorbing a photon in the blue region of the spectrum. Upon photon absorption PYP enters a reversible photocycle involving numerous intermediates (Fig. 1A). The primary photochemical event that controls entry into the photocycle is isomerization of pCA about its C2=C3 double bond (see Fig. 1B for the pCA geometry). The pCA chromophore remains electronically excited for a few hundred fs (3, 5, 20). Excited state dynamics is thought to drive the configurational change from trans to cis (3, 21). The chromophore pocket within the PYP protein is sufficiently flexible to allow certain relatively large atomic displacements, but also imposes structural constraints that may affect the pathway and dynamics of isomerization (22, 23). In particular, the pCA chromophore is constrained by a covalent bond to the Cys69 side chain of PYP (Fig. 1B), by unusually short hydrogen bonds between its phenolate oxygen and nearby glutamate and tyrosine side chains (24), and by a hydrogen bond between the carbonyl oxygen of its tail and the main chain amide of Cys69.
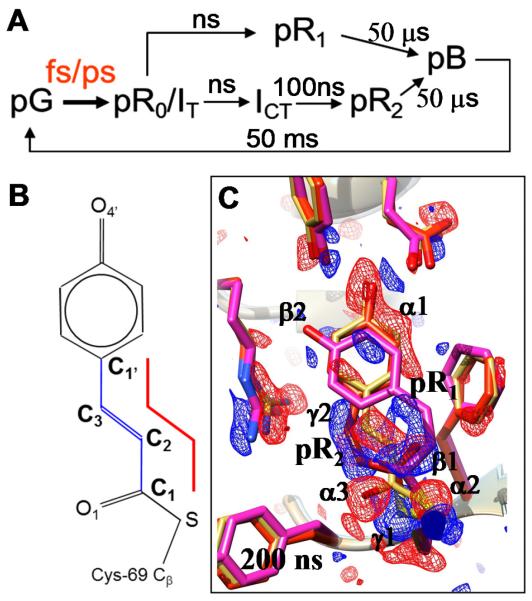
Figure 1.
A. The PYP photocycle from the perspective of a time-resolved crystallographer. Approximate time scales are given. The fs/ps time scale (in red) is structurally charted in this paper. B. The chemical structure of the pCA chromophore. The red line marks the four atoms that define the torsional angle ϕtail about the C2=C3 bond. C. Results of the positive control experiment at a 200 ns time delay. Reaction initiated by fs laser pulses. Negative (red) and positive (blue) DED features on the −3σ/3σ level. A mixture of the pR1 (magenta) and pR2 (red) structures is present. Main signature of pR1: features β1 and β2. Main signature for pR2: features γ1 and γ2. Structure of PYPref (dark) in
Previously, we showed that time-resolved pump-probe serial femtosecond crystallography (TR-SFX) could be successfully carried out on PYP on the ns to microsecond (μs) time scales. Difference electron density (DED) maps of very high quality, which compare the structures before (dark) and after (light) absorption of a photon (25), were obtained to near-atomic (1.6 Å) resolution. These experiments used a nanosecond (ns) laser pulse to initiate isomerization and subsequent structural changes. An overall reaction yield as high as 40% (25) could be reached. However, achieving fs time resolution requires that a fs pump laser pulse be used, which restricts the reaction yield to the much lower value of the primary quantum yield (around 10%) and correspondingly reduces the structural signal. The energy of fs pulses i.e. the number of photons per pulse must also be limited to avoid damaging effects from their significantly higher peak power. Here, we present results of TR-SFX experiments covering the time range from 100 fs to 3 ps. We directly follow the trans-cis isomerization of the pCA chromophore and the concomitant structural changes in its protein environment in real time. Full details of the experiment and data analysis are provided in the Supplementary Materials (SM). Light-initiated structural changes in PYP were investigated at the Coherent X-ray Imaging (CXI) instrument of the LCLS (26). Electronic excitation was initiated in microcrystals of PYP by fs pump laser pulses (λ=450 nm). Permanent bleaching of the chromophore was avoided by limiting the laser pulse energy to 0.8 mJ/mm2 (5.7 GW/mm2). Laser pulse duration, spectral distribution and phase were characterized by ‘Second Harmonic Generation Frequency Resolved Optical Gating’ (SHG-FROG) (27). The pulse duration was 140±5 fs and had both positive group delay dispersion and third order dispersion to maximize the conversion to the excited state (28). Offline spectroscopic experiments on thin crushed crystals of PYP had established that photoexcitation with fs laser pulses under comparable conditions could be as high as 10% without inducing damage (SM). The structural changes induced by the laser pulse were probed with 40 fs X-ray FEL pulses at 9 keV (1.36 Å). Both the pump-probe and the reference X-ray diffraction data were collected at the full 120 Hz pulse repetition rate of the LCLS to a resolution of 1.6 Å and 1.5 Å, respectively. To address concerns that the detector response might be influenced by the stray light of the intense fs laser pulse, the reference data were collected as a negative time delay, where the fs laser pulse arrived 1 ps after the X-ray pulse.
To assess whether fs laser pulses excited a sufficiently large number of molecules under these experimental conditions, we first performed a positive control experiment with a 200 ns pump-probe time delay, where large structural differences between the light and dark states have been well characterized (25, 29). From the pump-probe TR-SFX data and the reference data, DED maps were calculated (SM). Fig. 1C shows that the fs laser pulses are able to initiate sufficient entry into the photocycle to produce strong, chemically meaningful features. The 200 ns DED map is essentially identical to maps determined earlier at both the LCLS (25) and at BioCARS (29) at a time delay of 1 μs, and can be interpreted with the same mixture of intermediates, pR1 and pR2. The extent of reaction initiation is 12.6 % as determined by fitting a calculated ‘pR1&pR2 minus pG’ difference map to the 200 ns DED map, a value which agrees with the maximum extent of excitation determined spectroscopically (7 – 10%). The fs time scale was explored by using nominal settings for the time delay of 300 fs and 600 fs. The timing jitter between the 140 fs laser pump and 40 fs X-ray probe pulses is ~280 fs (8). The jitter was measured for every X-ray pulse by a timing tool (30, 31), which was combined with adjustments that take longer-term experimental drift into account (see SM). Thus, each individual diffraction pattern was associated with a definite “time stamp”. However, due to the drift, the time stamps were non-uniformly distributed in time (Fig. S1). Since the quality of structure amplitudes and of the DED maps derived from them depends on the number of diffraction patterns, indexed, time-stamped diffraction patterns were binned into 8 different pump-probe delays with about the same number of patterns (40,000) in each bin , spanning the time range from 100 to 1000 fs (Tab. S1B). A set of diffraction patterns at a time delay of 3 ps was also collected. Since the jitter and drift are much smaller than the delay, time stamping was not necessary for the 3 ps or the 200 ns delays. The values of R-split for all datasets is 7.5 – 9.9% which indicates the high quality of the diffraction data, and results in DED maps of comparable, good quality for all delays. Maps at 7 time delays are shown in Fig. 2. Visual inspection of these maps reveals an important qualitative result. The features in all maps at delays less than 500 fs are similar (compare Fig. 2, A-C); and features in all maps at delays greater than 700 fs are also similar (compare Fig. 2, D-G), but differ from those in the first set. Consequently, there must be a structural transition between the 455 fs and 799 fs time delays that gives rise to the two distinct sets of features. To identify with more precision the time delay at which this transition occurs, the time-stamped diffraction patterns were re-binned into 16 narrower time bins with about 20,000 patterns in each bin (Tab. S1A). The resultant time series of 16 DED maps in the fs time range (together with the map for the 3 ps time delay) were subjected to singular value decomposition (SVD; Fig. S2B) (32). The volume occupied by the pCA chromophore, by the Cys69 sulfur and the Glu46 carboxyl was included in the analysis. When a time series exhibits a change, a corresponding change should be even more readily recognizable in the right singular vectors (rSVs). This change is evident in the magnitude of both the first and second rSVs around 550 fs (red arrow in Fig. S2B). The substantial increase in the magnitude of the first right singular vector after 155 fs (Fig. S2B) shows the earliest (fastest) evolution of the structure after excitation. We tentatively associate the structural transition at around 550 fs, qualitatively evident by inspection of the DED maps and more quantitatively in their SVD analysis, with the trans to cis isomerization of the pCA chromophore. The transition occurs within ~180 fs (Fig. S2B), but its exact duration needs to be further established. Rate kinetics would require that after a ~500 fs dwell time the transition time would be stretched beyond the bandwidth limited rate. Yet the observed transition time matches the experimental bandwidth of 3.15 THz. Therefore the ensemble phase relation imparted by the optical pulse appears to be maintained for the duration of the dwell time, which may be supported by coherent motion. Although no oscillatory motion was detected in the TR-SFX data (they may be masked by the non-uniform data sampling), the time delay is however within the vibrational dephasing time of the PYP S1 state (3) and ground state modes in proteins (33). We further propose that at ~550 fs the system lies at or very close to a conical intersection (20) (Fig. S8), a branch point from which molecules either continue towards the cis configuration and enter the photocycle, or revert to the trans configuration and return to the resting (dark) state.
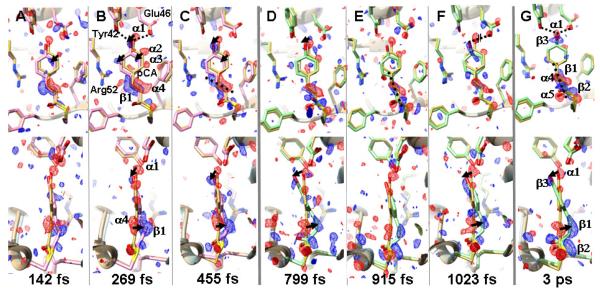
Figure 2.
Trans to Cis isomerization in PYP. Weighted DED maps in red (−3 σ) and blue (3σ); front (upper) and side view (lower). Each map is prepared from about the same number of diffraction patterns, except the 3 ps map (see Tab. S1 B-C). The reference, dark structure is shown in yellow throughout; structures before the transition and still on the electronic excited state PES are shown in pink; structures after the transition and on the electronic ground state PES are shown in light green. Important negative difference density features are denoted α, positive features as β in panels B and G. Pronounced structural changes are marked by arrows. A-C: time-delays before the transition. A. Twisted trans at 142 fs, ϕtail 154°. B. Twisted trans at 269 fs, ϕtail 140° some important residues are marked; dotted lines in B: hydrogen bond of the ring hydroxyl to Glu46 and Tyr42. C. Twisted trans at 455 fs, ϕtail 144°; dotted line in C: direction of C2=C3 double bond. D-G: time delays and chromophore configuration after the transition. D. Early cis at 799 fs, ϕtail 50°. E. Early cis at 915 fs; dotted line in E: direction of C2=C3 double bond. F. Early cis at 1023 fs; for E and F ϕtail ~65°. G. 3 ps delay; dashed line: direction of C2=C3 double bond, feature β1; ϕtail is 35°.
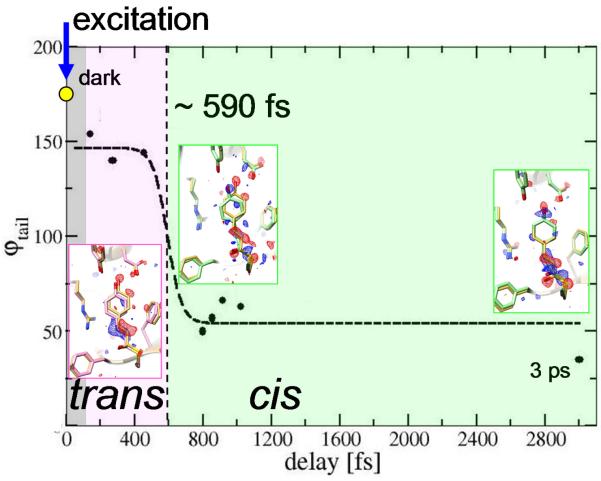
To identify the isomerization, refined structures before and after the transition are required. Initially, date in bins with 40,000 indexed diffraction patterns each were used, and preliminary PYP structures refined against these data. Refinement details are in the SM. The 3 bins with the shortest delays can be interpreted with chromophores in a twisted trans configuration (Fig. 2 A-C). After 700 fs the configuration is near cis (Fig. 2 D-E). The time-course of the refined ϕtail torsional angles can be fit with a transition time identical to that observed in the second rSV (Fig. 3). We took advantage of the similarity of the DED maps for extended time ranges before and after the transition to further increase the accuracy of the refined structures. We combined the diffraction patterns into two bins: the fast time scale (100-400 fs with 81,237 patterns) and a slower time scale (800-1200 fs with 157,082 patterns) (Tab. S1C). We refined the structure denoted PYPfast against the 100-400 fs data, and that denoted PYPslow against the 800-1200 fs data. The refinement statistics are presented in Tab. S2. The DED maps are shown in Fig. 3, inserts (see also Fig. S9B,D), with the corresponding, refined structures of PYPfast and PYPslow in pink and light green, respectively. The 3ps DED map and the refined PYP3ps structure are shown in Fig. 2G. We used as many diffraction patterns as possible to refine PYPslow (Fig. S12 B,D) and PYP3ps because at the transition, roughly 30% of the excited molecules return directly to the dark state, no longer contribute to the DED maps and reduce the signal. We emphasize that refinement of transient structures populated on an ultrafast timescale is challenging, since these structures are very far from equilibrium and likely to be highly strained. Restraints in standard libraries are derived from structures at equilibrium and are therefore not applicable. In order to provide restraints more appropriate for this refinement, we employed excited state quantum mechanics/molecular mechanics (QM/MM) calculations on PYP (20, 34) (SM). In addition, we employed an iterative procedure, in which improved difference phases ϕΔF,calc were obtained and used with observed difference structure factor amplitudes during refinement (SM). The structural results of the refinement are summarized in Tab. 1. For the shortest time delays (up to about 450 fs), the PYP chromophore tail adopts a highly strained, twisted trans configuration, in which the C1-C2=C3-C1’ torsional angle ϕtail (shown by the red line spanning these four atoms in Fig. 1B) is ~140°. The position of the C2=C3 double bond in PYPfast is displaced by ~1Å behind the chromophore plane (loosely defined by the Cys69 sulfur, the tail carbonyl oxygen and the atoms of the phenyl ring; Fig. 2A-C). Hydrogen bonds to Glu46 and Tyr42, which are unusually short in the reference (dark) structure (24), are substantially elongated from 2.5 Å to 3.4 Å (Tab. 1). This structure is primed for the transition to cis. During the structural transition, substantial rotation about the double bond takes place. The head of the chromophore pivots about tail atom C2 and thereby aligns the C2=C3 bond along the tail axis. Simultaneously, the head rotates about the C3-C1’ single bond. (The complex motions can be effectively illustrated by using an educator’s stick model set, see Fig. S3). The phenolate oxygen (Fig. 1B, O4’) moves even further away (3.6 Å, Tab. 1) from Glu46 (Fig. 2D-F and Fig. S9C-D), thereby breaking the hydrogen bond. At time delays longer than about 700 fs, ϕtail has decreased to ~50° (PYPslow, Fig. 3), which is characteristic of a cis configuration. PYPslow relaxes further towards the 3 ps structure (PYP3ps), in which the hydroxyl oxygen of the head re-establishes its hydrogen bond with Glu46 (Fig. 2G). ϕtail changes slightly to ~35°. The PYP3ps structure is already very similar to the early structures derived with 100 ps time resolution by independent, synchrotron-based approaches (Tab. 1; PDB entries 4I38 and 4B90) (22, 23), and has evolved only slightly from PYPslow by establishing shorter hydrogen bonds to Tyr42 and Glu46.
Figure 3.
Trans to cis isomerization in PYP. Pink: twisted trans on excited state PES; light green: cis on ground state PES. Torsional angle ϕtail (solid spheres) from structural refinement at various delays (see also Tab. S3). Gray region: not time-resolved. Dashed line: fit with eqn. S2, with a transition time of about 590 fs (see also Fig. S2). Inserts: structures of PYPfast (pink), PYPslow and PYP3ps (light green), and dark state structure PYPref in yellow. Difference electron density in red (−3σ) and blue (3σ).
Table 1.
Geometry of PYP structures. The PYPfast structure was refined using a data bin spanning 100-400 fs with 81327 snapshots, and the PYPslow structure from a bin spanning 800 – 1200 fs with 157082 snapshots (Tab. S1b). Structures of IT, pR0 and pB1 from Protein Data Bank, code listed in brackets (22, 23, 47). Uncertainties of the torsional angles can be estimated to be +/− 20° by displacing the 4 atoms that define the angle with the coordinate error (0.2 Å).
| PYPref (dark) |
PYPfast | PYPslow | PYP3ps | PYP200ns (fs-laser) pR1/pR2 |
IT (4I38) |
pR0 (4B90) |
pB1 (1TS0) |
|
|---|---|---|---|---|---|---|---|---|
| Time Delay | 0 | 100- 400 fs |
800 - 1200 fs |
3 ps | 200 ns | 100 ps |
100 ps | ms |
| Torsional Angles [°] | ||||||||
|
C1-C2=C3-C1’
(ϕtail) |
172 | 136 | 53 | 35 | 3/−8 | 90 | 33 | −27 |
| O1-C1-C2=C3 | −15 | −21 | 28 | 30 | 12/−6 | 11 | 29 | −10 |
| CB-S-C1-C2 | −185 | −171 | −164 | −137 | 163/−165 | −136 | −123 | 180 |
| Hydrogen bonds [Å] | ||||||||
|
pCA-O4’ -
Glu46-Oε |
2.50 | 3.40 | 3.60 | 2.94 | 4.97/2.88 | 2.73 | 2.73 | 8.03 |
|
pCA-O4’ -
Tyr42-Oη |
2.54 | 2.92 | 2.63 | 2.88 | 2.97/2.66 | 2.57 | 2.59 | 5.19 |
|
pCA-O1 –
Cys69-N |
2.77 | 3.11 | 2.50 | 3.12 | 3.37/4.29 | 3.04 | 3.05 | 2.88 |
| others | ||||||||
| <pCA>a [Å] | 0 | 0.66 | 0.78 | 0.60 | 1.55/0.81 | 0.67 | 0.68 | 2.39 |
| <global>b [Å] | 0 | 0.20 | 0.19 | 0.24 | 0.13 | 0.13 | 0.19 | 0.17 |
|
Radius of
gyration c [Å] |
13.32 | 13.33 | 13.30 | 13.34 | 13.29 | - nd - | - nd - | - nd- |
| Volume [Å3] | 17831 | 17856 | 17833 | 17838 | 17672 | 17830 | 17683 | 17807 |
| ΔV to dark [Å3] | 0 | 25 | 2 | 7 | −159 | −1 | −148 | −24 |
|
Photoactivation
Yield [%]d |
- na - | 15.2 | 9.6 | 10.1 | 12.5c | (5%)e | (10%)e | (10%)e |
Mean displacement of equivalent chromophore atoms relative to dark (SM).
Mean displacement of equivalent cα atoms relative to dark (SM).
See SM for the calculation.
Determined by by fitting calculated DED maps to the experimental DED maps in the chromophore region.
Estimate
The structures derived from the refinements confirm that the transition at around 550 fs is indeed associated with a trans to cis isomerization. Theoretical considerations (20) (Fig. S8) suggest that during isomerization the PYP chromophore relaxes through a conical intersection between the electronically excited state PES and the ground state PES. Accordingly, structures between 100 and 400 fs can be identified as electronically excited, whereas the structures at time delays > 700 fs can be identified with the electronic ground state. In both the excited and ground states, structural changes i.e. translation of atoms may also have occurred. Our experiments identify the ultrafast dynamics of both the excited state structures and the ground state structures (Figs. 2-3). Since we restricted our pump laser pulses to moderate power, we avoid damaging nonlinear effects (e.g. two photon absorption) and most excited molecules populate the excited state surface S1 (5). Part of the stored energy is used to rapidly displace the chromophore by about 0.7 Å within the crowded molecular environment in the interior of PYP (Fig. 2A, Tab. 1). If this initial displacement is complete after 250fs the chromophore must have experienced an acceleration of ~2×1015 m/s2 and attains a final velocity of 500 m/s (SM). Fig. 1B shows that 9 carbon atoms, two oxygens and 7 hydrogen atoms (molecular mass = 147 g/mol) are displaced. During the first few hundred fs the force on the chromophore is ~500 pN which is enormous compared to forces in single molecules at thermal equilibrium which are usually only a few pN (35). The origin of the force is due to the change of the potential energy surface when the chromophore is excited to the electronic excited state which affects the intra and intermolecular interactions of the chromophore as also inferred from ultrafast Raman spectroscopy(3). The energy required to displace the chromophore is ~0.2 eV which is ~10% of the blue photon energy (2.76 eV) that starts the reaction. It appears that by rapidly evolving down the excited state PES, part of the photon energy is initially converted into kinetic energy which is then released by collision of the chromophore atoms with the surrounding protein atoms comprising the chromophore pocket. The excited chromophore loses 0.12 eV energy by intramolecular vibrational energy redistribution on the sub-100 fs time scale (39) which can be roughly estimated from the Stokes shift by comparing absorption and fluorescence spectra(3). Accordingly,~85% of the photon energy remains stored as strain and electronic excitation in the chromophore before isomerization occurs. On passing through the conical intersection (20), the molecules either revert towards the initial dark state (30% of the excited molecules, Tab. 1, see also Tab. S3) or continue relaxing towards the cis isomer (70%), gradually releasing the excess energy as heat. Because the chromophore pocket tightly restricts the chromophore head displacements, further structural changes must be volume-conserving i.e. they minimize the volume swept out by the atoms as they move. Accordingly, the chromophore performs the complex motions described above (Fig. S3). Although the energy stored in the chromophore is sufficient to break the hydrogen bonds (~0.1 eV), the spatial constraints imposed by the chromophore pocket direct the reformation of the hydrogen bonding network at longer time delays (Tab. 1). This is a ‘macromolecular cage effect’ reminiscent of the ‘solvent cage effect’ in liquid chemical dynamics (36). The ‘macromolecular cage’ in PYP, however, is soft enough to allow certain specific, relatively large (up to 1.3 Å, Tab. 1) structural changes. This contrasts with crystals of small molecules, where the stronger crystal lattice constraints usually do not allow such large displacements. Hence, biological macromolecular crystallography aimed at elucidating biological function may also provide insight into the reaction mechanisms of small molecules.
To assess global conformational changes of PYP on the fs time scale, we calculated the radius of gyration Rg from each refined structure (SM). Rg fluctuates by only 0.2% in all structures from 200 fs to 200 ns (Tab. 1). An increase of Rg by up to 1 Å determined by others using X-ray scattering in solution upon photo-dissociation of CO from CO-myoglobin in solution (9) is not observed in our PYP crystals. Concomitant, systematic large volume changes are also not apparent in PYP crystals over the first 3 ps that our data span. Our data show no evidence for a protein quake (9, 10, 37), characterized by an ultrafast and large change in Rg that occurs significantly before a large volume change. The reason for this is unclear and will require further experiments.
Ultrafast fluorescence and transient absorption spectroscopy of PYP has shown that excited state decay is multi-phasic (3, 5, 38). The fast (sub-ps) time constants are significantly more productive in creating the cis-like photoproduct than the slow (ps) time constants; the long-lived excited state population primarily decays back to the ground state (5, 39). With excitation at 450 nm, at least 50% of the total isomerization yield is generated with a dominant ~600 fs time constant (5), which agrees with our observation of a transition at ~550 fs. It should be noted that a ‘ground state intermediate’ with a 3-6 ps life time has been proposed by ultrafast spectroscopy (39). However under the conditions employed here, the peak concentration of this intermediate is expected to be small (5). In contrast to spectroscopic techniques that reported vibrational coherence with 50 cm−1 and 150 cm−1 frequency (3, 40), we could not unambiguously detect oscillations in our data (see above). Intense femtosecond optical pumping of PYP crystals generates both excited state and ground state vibrational coherences within the 3.15 THz experimental bandwidth(41). It will be an important goal of future experiments to structurally characterize these coherences using fs TR-SFX. Nevertheless, our data show that before 400 fs there are large distortions corresponding to a Franck-Condon (FC) excited state (42). The nuclear dynamics of the FC excited state at 100-200 fs agrees with the conclusions from ultra-fast spectroscopy (3, 42-45) that also suggest a distortion of the C2=C3 double bond on similar timescales, as in the PYPfast structure. The isomerization at 550 fs through the conical intersection between the excited state and ground state PES is in reasonable agreement with the timescales for isomerization reported by others (3, 5, 42, 46). After passing through the conical intersection, the chromophore is cis-like and still highly strained. The transiently-broken hydrogen bond is reestablished quickly as the structure relaxes, exemplified by the PYP3ps structure (Fig. 3). Further relaxation on the ground state PES completes the initial phase of the isomerization.
Supplementary Material
Figure S1. Snapshot density (diffraction patterns per 50 fs interval). The influence of a substantially varying number of patterns per bin on data quality is avoided when bins with approximately equal number of patterns per bin are used (e.g. red squares illustrate use of 16 bins; see Tab. S1. Unavoidably, these 16 bins are of non-uniform width in time).
Figure S2. A. Torsional angle ϕtail from refinement at various time delays (from bins containing 40 k diffraction patterns, see also Tab. S2B and S5). Dashed line: fit by eqn. S2 with a 590 fs transition. B. Right singular vectors (rSV) from the singular value decomposition of the 17 difference electron density maps on the fs (with 20 k diffraction patterns in each bin) to 3 ps time scale. Black dots: rSV1; black line: fit with exponential (eqn. S1) with characteristic time τ = 155 fs; blue and red dashed lines: fit of two separate parts of the rSV1 with exponentials. Red arrow: transition. Red squares: rSV2, dashed black line: fit by an exponential function (eqn. S2) with a 560 fs transition. Circle: time point near the assumed conical intersection. Green triangles: rSV3. The values of rSV4 and rSV5 are not significant, and displayed by thin dashed dotted and dashed double dotted lines. Blue bell-shaped curve: duration of the transition, from 470 to 650 fs. Inset: magnitude of the singular values.
Figure S3. Schematic view of the trans to cis transition at 550 fs. The rotations can be easily understood using an educator’s stick model set for chemical structures. A. PYPfast structure. To produce PYPfast from the reference, PYPref, grab the C2=C3 double bond with the left hand and the C1-S bond with the right hand. Rotate the double bond clockwise by about 40°. The C1-C2 single bond points now backwards as observed, the configuration is still trans. Compared to the dark state, the hydrogen bonds to Glu46 and Tyr42 are elongated. To further the isomerization (and reach B), continue rotating the double bond clockwise by an additional 90° by keeping the atom C2 location fixed. The entire head-structure pivots about (the fixed) atom C2. Since the head cannot swing out, concerted anti-clockwise rotations about the C1-C2 and the C3-C1’ single bonds are necessary to minimize the volume swept out by the atoms that move during isomerization; see arrows in A. B. PYPslow structure (800-1200 fs), cis. The hydrogen bond to Glu46 is broken (head displacement exaggerated for clarity).
Figure S4. The solvent accessible surface (blue) of PYP3ps.
Figure S5. Entry into the photocycle in PYP crystals as evident by optical absorption changes initiated by 900 fs (A-B), 300 fs, and 100fs (C-D) laser pulses. Pump-probe delay 180μs for A-B, 1 ms for C-D. A. PYP absorption spectra of the dark state, following a sequence of 10 pump-probe cycles with 900 fs pulses at increasing laser pulse energy densities (shown in the legend). Irreversible bleaching (damage), as evident by the reduced OD of the dark state, occurs at 2.2mJ/mm2. B. Light-dark difference absorption spectra at various laser pulse energy densities. Spectra are recorded by accumulating 10 pump-probe cycles. Extent of (reversible) bleach at 450 nm is roughly proportional to number of molecules in photocycle. At 1.2 mJ/mm2 about 5% of molecules can be pumped into the photocycle. C. Permanent single-shot photo-bleaching (damage) measured after 100 fs (red squares) and 300 fs (blue diamonds) laser pulses of various energies (spot size 80 μm). D. Extent of reaction initiation with 100 fs and 300 fs laser pulses measured as the pB concentration at 1ms as the reversible ΔA450nm/A450nm. 100 fs laser pulses with 2 μJ (energy density: 400 μJ/mm2) result in up to 6% reaction initiation with small permanent damage.
Figure S6. A. A diagram of the SHG-FROG used to characterize the 450nm femtosecond pulses incident on PYP crystals. The incoming pulse (black) is split into two replicas that travel down individual paths (shown in blue and red for clarity) until they are overlapped in a space and time in the 20 μm thin BBO crystal. The resulting FROG signal (pink) is measured using the UV spectrometer. B. The experimentally measured FROG traces for the 140 fs stretched pulse. C. Reconstructed FROG traces. The “G error” for the fitting was <1%. The high order structure seen at the top of the reconstruction that is not seen in the experimental data is the result of fluctuations in the shot-to-shot pulse shape. This is commonly seen in multi-shot FROG measurements and does not greatly impact the recovered pulse length or phase. D-E. FROG reconstruction of the temporal and the spectral intensity (red) of the reconstructed pulse incident on the PYP microcrystals with the retrieved phase indicated (blue). High order time-structure is a reconstruction artifact due to beam intensity instability (85, 86). The pulses were found to be 140±5 fs long with a GDD of ≈1000fs2 and TOD ≈+1300fs3.
Figure S7. A. PYP configurations along the QM/MM trajectory. The atoms included in the QM region are shown in ball-and-stick representation and modeled at the CASSCF(12,11)/cc-pVDZ level of ab initio theory. The rest of the system, consisting of the protein, water, Cl− and Na+ counter ions (most of which is not shown here) are modeled with the Amber03 force field. B. Snapshots from a QM/MM trajectory after photo-excitation at t = 0 fs (Tab. S4, trajectory a). C. Torsional angles as a function of time in this trajectory. Left yellow shaded region, dynamics on the excited state potential energy surface. Vertical dashed line (surface hop): seam that separates excited state structural dynamics (left) from ground state dynamics (right). D. Color code for the torsional angles of the pCA chromophore in C.
Figure S8. Transition through conical intersection as conceived from theoretical consideration (lower scheme) and viewed from the perspective of the TR-SFX experiment (structures above).
Figure S9. Trans to Cis isomerization in PYP. Direct comparison of difference maps and structures of individual time-points determined from 40,000 diffraction patterns with those determined from averaging diffraction pattern in larger time-bins (compare Tab. S1 B-C). The 269 fs, 799 fs and 3 ps delays (panels A,C and E) delays are also shown in Fig. 2 in the main text. Weighted DED maps in red (−3 σ) and blue (3σ); front (upper) and side view (lower). The reference, dark structure is shown in yellow throughout; structures before the transition and on the electronic excited state PES are shown in pink; structures after the transition and on the electronic ground state PES are shown in green. Important negative difference density features are denoted α, positive features as β. Pronounced structural changes are marked by arrows. A. Representative time-delay (269 fs) before the transition; some important residues are marked. Dotted lines: hydrogen bonds of the ring hydroxyl to Glu46 and Tyr42. B. Chromophore configuration (PYPfast) by averaging 100 fs to 400 fs pump-probe delays. C. Representative time delay (799 fs) after the transition. D. Structure PYPslow on longer fs times obtained by averaging the 800 fs to 1200 fs pump-probe delays. E. 3 ps chromophore configuration. Dashed lines in A, D and E: direction of feature β1.
Figure S10. Difference-difference maps (residual maps): Calculated ∣Fcalc(t)∣-∣Fcalc(dark)∣ difference maps were subtracted from the observed ∣Fobs(t)∣-∣Fobs(dark)∣ difference maps. Pink: PYPfast; green: PYPslow and PYP3ps; yellow: PYPref. Residual features (blue: positive, red negative) on the +/−3 σ level. A and B. Fast and slow fs time scale, residual maps using the correct, refined models that correspond to the time-scales, twisted-trans for A, and near-cis for B. C and D. Residual maps using interchanged, deliberately incorrect models, near-cis for C and twisted-trans for D. The chromophore tail is much cleaner in A and B, but strong residual features denoted α and β in C and D indicate mismatch. Note: remaining features near the sulfur emerge from its strong electron density; its position is difficult to model exactly. E. Data binned from 400-800 fs across the transition. Feature β shows some mixing-in of the PYPfast structure in this time range. F. residual map at 3 ps time delay.
Figure S11. PYPref (yellow), PYPfs,fast (pink) and the theoretical PYP model from QM/MM simulations (orange) overlaid on the 269 fs difference map (red/blue −3σ/4σ). A. front view, B. side view. Nearby amino acid residues are marked in A. Arrow in B: similar chromophore tail displacements for the observed and theoretical PYP structures.
Figure S12. Full view of the DED maps (contour: red/green −3σ/3σ) at various time-ranges. The maps are overlaid on PYPref (yellow) and the refined structures. Contrary to all other figures that depict PYP structures, the view is from the opposite (back) of the chromophore, hence the Arg52 is on the right. This orientation allows for a detailed view of the positive feature β1 which indicates the configuration of the chromophore and the strong feature on the Cys69 sulfur (β2) that is also associated with the cis configuration. A. On the fast time scale, feature β1 is strongly bent, β2 is absent. The DED map is interpreted with a trans configuration (structure in pink). B. The slow time range, feature β1 is aligned along the tail axis, which indicates a cis configuration (structure in green). C. At the 3 ps delay feature β1 is similar to B (structure also in green). D. Consistency check by randomly selecting ~80,000 snapshots from the 157,000 snapshots in the bin. DED map with features β1 and β2 and structure (green) essentially the same as in B.
Table S1. TR-SFX data statistics. Resolution 1.6 Å; data in the highest resolution bin (1.6 - 1.67 Å) in brackets. A. Statistics of the data sorted into time bins with 32,690 snapshots with Bragg reflections (hits)/bin (approximately 20,000 indexed snapshots/bin). Times shown are the average of the time stamps in each bin and their standard deviation. B. Data statistics of bins with 65,380 snapshots/bin (approximately 40,000 indexed snapshots/bin). C. Statistics of the individual −1ps, 3 ps and 200 ns time points and when snapshots with time-stamps 100 – 400 fs and 800 – 1200 fs are combined.
Table S2. Difference maps and refinement for selected structures.
Table S3. Torsional angles and distances from structural refinement at various, individual time delays.
Table S4. Global structural changes [Å] as measured by averaging the distances between cα atoms of equivalent residues.
Table S5. Excited state lifetimes (2rd column), and products (3th column) in the excited state QM/MM trajectories.
Acknowledgements
This work is supported by NSF-STC “BioXFEL” (NSF-1231306), by NIH grants R01GM095583 (PF), R01EY024363 (KM) and R24GM111072 (VS, RH and KM), Helmholtz Association “Virtual Institute Dynamic Pathways” (HC), and NSF-0952643 (MS). KP is partly supported by NSF-1158138 (to Dilano Saldin and MS) and BMBF 05K14CHA (to HC). JvT acknowledges support from the Engineering and Physical Sciences Research Council via grant agreement EP/M000192/1. GG and DM are supported by the Academy of Finland, DO by BMBF project 05K13GUK, and MM by the European Union through grant FP7-PEOPLE-2011-ITN NanoMem. Use of the Linac Coherent Light Source (LCLS), SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract DE-AC02-76SF00515. Part of this work was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory (LLNL) under contract DE-AC52-07NA27344, and M. F. was supported by LLNL Lab-Directed Research and Development Project 012-ERD-031. C.G. thanks the PIER Helmholtz Graduate School for financial support. Parts of the sample injector used at LCLS for this research were funded by the NIH grant P41GM103393, formerly P41RR001209. We thank the Moscow State University supercomputing center and the CSC–IT Center for Science, Finland, for computing resources. We thank Dr. Mark Hunter for valuable discussions, Dr. Timo Graen for help with the computer simulations and Chufeng Li for assistance with injectors. The PYPref, PYPfast, PYPslow, PYP3ps and PYP200ns structures are deposited in the protein data bank together with their respective weighted difference structure factor amplitudes under accession codes 5HD3, 5HDC, 5HDD, 5HDS and 5HD5, respectively.
Footnotes
Author Contributions.
M.S. prepared the proposal with input from J.J.vT., K.M., V.S., J.C.H.S., H.N.C., A.O. and P.F.; A.A., S.B., M.L., J.S.R. and J.E.K. operated the CXI instrument including the time-tool and the fs-laser; K.P., A.B., J.T., S.B., T.A.W., N.Z., O.Y. and T.D.G. analyzed the SFX data. C.D.M.H and J.J.vT. set up the FROG at the CXI instrument; G.G. and D.M. performed QM/MM calculations; J.T., J.B., D.O., P.L.X., C.G., C.K. and M.S. prepared protein and grew nano- and microcrystals; D.DeP., C.K., C.C., S.R-C., J.D.C., M.M., G. K., and U.W. provided and operated the injector system; M.F., R.F., M.S., J.T., P.F., D.O. and C.G. wrote the electronic log; M.F., M.S, J.T., J.S.R., J.J.vT. and K.M. discussed fs laser excitation; J.T., M.S, V.S, R.H, C.D.M.H. and J.J.vT. performed preliminary ultra-fast experiments on crystals; M.S. calculated and analyzed the difference maps; M.S., K.P., K.M., G.G., P.F. and J.J.vT. wrote the manuscript with improvements from all authors.
References
- 1.Wald G. Molecular basis of visual excitation. Science. 1968;162:230–239. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 2.Mizutani Y, Kitagawa T. Direct observation of cooling of heme upon photodissociation of carbonmonoxy myoglobin. Science. 1997;278:443–446. doi: 10.1126/science.278.5337.443. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura R, Hamada N, Ichida H, Tokunaga F, Kanematsu Y. Coherent oscillations in ultrafast fluorescence of photoactive yellow protein. The Journal of chemical physics. 2007;127:215102. doi: 10.1063/1.2802297. published online EpubDec 7. [DOI] [PubMed] [Google Scholar]
- 4.Champion PM. Chemistry. Following the flow of energy in biomolecules. Science. 2005;310:980–982. doi: 10.1126/science.1120280. published online EpubNov 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lincoln CN, Fitzpatrick AE, van Thor JJ. Photoisomerisation quantum yield and non-linear cross-sections with femtosecond excitation of the photoactive yellow protein. Physical chemistry chemical physics : PCCP. 2012;14:15752–15764. doi: 10.1039/c2cp41718a. published online EpubDec 5. [DOI] [PubMed] [Google Scholar]
- 6.Warshel A. Bicycle-pedal model for the first step in the vision process. Nature. 1976;260:679–683. doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- 7.Minitti MP, Budarz JM, Kirrander A, Robinson JS, Ratner D, T.J L, Zhu D, Glownia J, Kozina M, Lemke H, Sikorski M, Feng Y, Nelson S, Saita K, Stankus B, Northey T, Hastings JB, Weber PM. Imaging Molecular Motion: Femtosecond X-ray Scattering of an Electrocyclic Chemical Reaction. Physical review letters. 2015;114:1–5. doi: 10.1103/PhysRevLett.114.255501. [DOI] [PubMed] [Google Scholar]
- 8.Glownia JM, Cryan J, Andreasson J, Belkacem A, Berrah N, Blaga CI, Bostedt C, Bozek J, DiMauro LF, Fang L, Frisch J, Gessner O, Guhr M, Hajdu J, Hertlein MP, Hoener M, Huang G, Kornilov O, Marangos JP, March AM, McFarland BK, Merdji H, Petrovic VS, Raman C, Ray D, Reis DA, Trigo M, White JL, White W, Wilcox R, Young L, Coffee RN, Bucksbaum PH. Time-resolved pump-probe experiments at the LCLS. Optics express. 2010;18:17620–17630. doi: 10.1364/OE.18.017620. published online EpubAug 16. [DOI] [PubMed] [Google Scholar]
- 9.Levantino M, Schiro G, Lemke HT, Cottone G, Glownia JM, Zhu D, Chollet M, Ihee H, Cupane A, Cammarata M. Ultrafast myoglobin structural dynamics observed with an X-ray free-electron laser. Nat Commun. 2015;6:6772. doi: 10.1038/ncomms7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnlund D, Johansson LC, Wickstrand C, Barty A, Williams GJ, Malmerberg E, Davidsson J, Milathianaki D, DePonte DP, Shoeman RL, Wang DJ, James D, Katona G, Westenhoff S, White TA, Aquila A, Bari S, Berntsen P, Bogan M, van Driel TB, Doak RB, Kjaer KS, Frank M, Fromme R, Grotjohann I, Henning R, Hunter MS, Kirian RA, Kosheleva I, Kupitz C, Liang MN, Martin AV, Nielsen MM, Messerschmidt M, Seibert MM, Sjohamn J, Stellato F, Weierstall U, Zatsepin NA, Spence JCH, Fromme P, Schlichting I, Boutet S, Groenhof G, Chapman HN, Neutze R. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nature methods. 2014;11:923–926. doi: 10.1038/Nmeth.3067. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barends TRM, Foucar L, Ardevol A, Nass K, Aquila A, Botha S, Doak B, Falahati K, Hartmann E, Hilpert M, Heinz M, Hoffmann MC, Koefinger J, Koglin JE, Kovacsova G, Liang M, Milathianaki D, Lemke H, Reinstein J, Roome CM, Shoeman RL, Williams GJ, Burghardt I, Hummer G, Boutet S, Schlichting I. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015 doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- 12.Coppens P. Perspective: On the relevance of slower-than-femtosecond time scales in chemical structural-dynamics studies. Structural Dynamics. 2015;2 doi: 10.1063/1.4913362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zewail AH. Femtochemistry: Atomic-Scale Dynamics of the Chemical Bond Using Ultrafast Lasers (Nobel Lecture) Copyright((c)) The Nobel Foundation 2000. We thank the Nobel Foundation, Stockholm, for permission to print this lecture. Angew Chem Int Ed Engl. 2000;39:2586–2631. doi: 10.1002/1521-3773(20000804)39:15<2586::aid-anie2586>3.0.co;2-o. published online EpubAug 4. [DOI] [PubMed] [Google Scholar]
- 14.Gao M, Lu C, Jean-Ruel H, Liu LC, Marx A, Onda K, Koshihara SY, Nakano Y, Shao X, Hiramatsu T, Saito G, Yamochi H, Cooney RR, Moriena G, Sciaini G, Miller RJ. Mapping molecular motions leading to charge delocalization with ultrabright electrons. Nature. 2013;496:343–346. doi: 10.1038/nature12044. published online EpubApr 18. [DOI] [PubMed] [Google Scholar]
- 15.Techert S, Schotte F, Wulff M. Picosecond X-ray diffraction probed transient structural changes in organic solids. Physical review letters. 2001;86:2030–2033. doi: 10.1103/PhysRevLett.86.2030. [DOI] [PubMed] [Google Scholar]
- 16.Kim CD, Pillet S, Wu G, Fullagar WK, Coppens P. Excited-state structure by time-resolved X-ray diffraction. Acta crystallographica. Section A, Foundations of crystallography. 2002;58:133–137. doi: 10.1107/s0108767301017986. [DOI] [PubMed] [Google Scholar]
- 17.Benedict JB, Makal A, Sokolow JD, Trzop E, Scheins S, Henning R, Graber T, Coppens P. Time-resolved Laue diffraction of excited species at atomic resolution: 100 ps single-pulse diffraction of the excited state of the organometallic complex Rh2(mu-PNP)2(PNP)2.BPh4. Chem Commun (Camb) 2011;47:1704–1706. doi: 10.1039/c0cc04997b. published online EpubFeb 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihee H, Lorenc M, Kim TK, Kong QY, Cammarata M, Lee JH, Bratos S, Wulff M. Ultrafast x-ray diffraction of transient molecular structures in solution. Science. 2005;309:1223–1227. doi: 10.1126/science.1114782. published online EpubAug 19. [DOI] [PubMed] [Google Scholar]
- 19.Neutze R, Wouts R, Techert S, Davidsson J, Kocsis M, Kirrander A, Schotte F, Wulff M. Visualizing photochemical dynamics in solution through picosecond x-ray scattering. Physical review letters. 2001;87:195508. doi: 10.1103/PhysRevLett.87.195508. [DOI] [PubMed] [Google Scholar]
- 20.Groenhof G, Bouxin-Cademartory M, Hess B, De Visser SP, Berendsen HJ, Olivucci M, Mark AE, Robb MA. Photoactivation of the photoactive yellow protein: why photon absorption triggers a trans-to-cis Isomerization of the chromophore in the protein. Journal of the American Chemical Society. 2004;126:4228–4233. doi: 10.1021/ja039557f. published online EpubApr 7. [DOI] [PubMed] [Google Scholar]
- 21.Larsen DS, Vengris M, van Stokkum IH, van der Horst MA, de Weerd FL, Hellingwerf KJ, van Grondelle R. Photoisomerization and photoionization of the photoactive yellow protein chromophore in solution. Biophysical journal. 2004;86:2538–2550. doi: 10.1016/S0006-3495(04)74309-X. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung YO, Lee JH, Kim J, Schmidt M, Moffat K, Srajer V, Ihee H. Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography. Nature chemistry. 2013;5:212–220. doi: 10.1038/nchem.1565. published online EpubMar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schotte F, Cho HS, Kaila VR, Kamikubo H, Dashdorj N, Henry ER, Graber TJ, Henning R, Wulff M, Hummer G, Kataoka M, Anfinrud PA. Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19256–19261. doi: 10.1073/pnas.1210938109. published online EpubNov 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson S, Crosson S, Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallogr D. 2004;60:1008–1016. doi: 10.1107/S090744490400616x. published online EpubJun. [DOI] [PubMed] [Google Scholar]
- 25.Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, Hunter M, Boutet S, Williams GJ, Koglin JE, Oberthuer D, Heymann M, Kupitz C, Conrad C, Coe J, Roy-Chowdhury S, Weierstall U, James D, Wang D, Grant T, Barty A, Yefanov O, Scales J, Gati C, Seuring C, Srajer V, Henning R, Schwander P, Fromme R, Ourmazd A, Moffat K, Van Thor JJ, Spence JC, Fromme P, Chapman HN, Schmidt M. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–1246. doi: 10.1126/science.1259357. published online EpubDec 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang M, Williams GJ, Messerschmidt M, Seibert MM, Montanez PA, Hayes M, Milathianaki D, Aquila A, Hunter MS, Koglin JE, Schafer DW, Guillet S, Busse A, Bergan R, Olson W, Fox K, Stewart N, Curtis R, Miahnahri AA, Boutet S. The Coherent X-ray Imaging instrument at the Linac Coherent Light Source. J Synchrotron Radiat. 2015;22:514–519. doi: 10.1107/S160057751500449X. published online EpubMay 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trebino R, DeLong KW, Fittinghoff DN, Sweetser JN, Krumbugel MA, Richman BA, Kane DJ. Measuring ultrashort laser pulses in the time-frequency domain using frequency-resolved optical gating. Review of Scientific Instruments. 1997;68:3277–3295. doi: 10.1063/1.1148286. published online EpubSep. [DOI] [Google Scholar]
- 28.Bardeen CJ, Wang Q, Shank CV. Selective Excitation of Vibrational Wave-Packet Motion Using Chirped Pulses. Physical review letters. 1995;75:3410–3413. doi: 10.1103/PhysRevLett.75.3410. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt M, Srajer V, Henning R, Ihee H, Purwar N, Tenboer J, Tripathi S. Protein energy landscapes determined by five-dimensional crystallography. Acta Crystallogr D. 2013;69:2534–2542. doi: 10.1107/S0907444913025997. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartmann N, Helml W, Galler A, Bionta MR, Grunert J, Molodtsov SL, Ferguson KR, Schorb S, Swiggers ML, Carron S, Bostedt C, Castagna JC, Bozek J, Glownia JM, Kane DJ, Fry AR, White WE, Hauri CP, Feurer T, Coffee RN. Sub-femtosecond precision measurement of relative X-ray arrival time for free-electron lasers. Nat Photonics. 2014;8:706–709. [Google Scholar]
- 31.Bionta MR, Lemke HT, Cryan JP, Glownia JM, Bostedt C, Cammarata M, Castagna JC, Ding Y, Fritz DM, Fry AR, Krzywinski J, Messerschmidt M, Schorb S, Swiggers ML, Coffee RN. Spectral encoding of x-ray/optical relative delay. Optics express. 2011;19:21855–21865. doi: 10.1364/OE.19.021855. published online EpubOct 24. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt M, Rajagopal S, Ren Z, Moffat K. Application of singular value decomposition to the analysis of time-resolved macromolecular X-ray data. Biophysical journal. 2003;84:2112–2129. doi: 10.1016/S0006-3495(03)75018-8. published online EpubMar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rector KD, Fayer MD. Myoglobin dynamics measured with vibrational echo experiments. Laser Chem. 1999;19:19–34. [Google Scholar]
- 34.Groenhof G. Introduction to QM/MM Simulations. Methods Mol Biol. 2013;924:43–66. doi: 10.1007/978-1-62703-017-5_3. [DOI] [PubMed] [Google Scholar]
- 35.Rief M, Grubmuller H. Force spectroscopy of single biomolecules. Chemphyschem. 2002;3:255–261. doi: 10.1002/1439-7641(20020315)3:3<255::AID-CPHC255>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 36.Patron F, Adelman SA. Solvent Cage Effects and Chemical-Dynamics in Liquids. Chem Phys. 1991;152:121–131. doi: 10.1016/0301-0104(91)80039-K. published online EpubApr 15. [DOI] [Google Scholar]
- 37.Genberg L, Richard L, Mclendon G, Miller RJD. Direct Observation of Global Protein Motion in Hemoglobin and Myoglobin on Picosecond Time Scales. Science. 1991;251:1051–1054. doi: 10.1126/science.1998121. [DOI] [PubMed] [Google Scholar]
- 38.Vengris M, van der Horst MA, Zgrablic G, van Stokkum IH, Haacke S, Chergui M, Hellingwerf KJ, van Grondelle R, Larsen DS. Contrasting the excited-state dynamics of the photoactive yellow protein chromophore: protein versus solvent environments. Biophysical journal. 2004;87:1848–1857. doi: 10.1529/biophysj.104.043224. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen DS, van Stokkum IH, Vengris M, van Der Horst MA, de Weerd FL, Hellingwerf KJ, van Grondelle R. Incoherent manipulation of the photoactive yellow protein photocycle with dispersed pump-dump-probe spectroscopy. Biophysical journal. 2004;87:1858–1872. doi: 10.1529/biophysj.104.043794. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mataga N, Chosrowjan H, Shibata Y, Imamoto Y, Kataoka M, Tokunaga F. Ultrafast photoinduced reaction dynamics of photoactive yellow protein (PYP): observation of coherent oscillations in the femtosecond fluorescence decay dynamics. Chem Phys Lett. 2002;352:220–225. doi: 10.1016/S0009-2614(01)01448-8. published online EpubJan 30. [DOI] [Google Scholar]
- 41.Kumar ATN, Rosca F, Widom A, Champion PM. Investigations of ultrafast nuclear response induced by resonant and nonresonant laser pulses. Journal of Chemical Physics. 2001;114:6795–6815. [Google Scholar]
- 42.Creelman M, Kumauchi M, Hoff WD, Mathies RA. Chromophore Dynamics in the PYP Photocycle from Femtosecond Stimulated Raman Spectroscopy. Journal of Physical Chemistry B. 2014;118:659–667. doi: 10.1021/jp408584v. [DOI] [PubMed] [Google Scholar]
- 43.Groot ML, van Wilderen LJ, Larsen DS, van der Horst MA, van Stokkum IH, Hellingwerf KJ, van Grondelle R. Initial steps of signal generation in photoactive yellow protein revealed with femtosecond mid-infrared spectroscopy. Biochemistry. 2003;42:10054–10059. doi: 10.1021/bi034878p. published online EpubSep 2. [DOI] [PubMed] [Google Scholar]
- 44.Heyne K, Mohammed OF, Usman A, Dreyer J, Nibbering ET, Cusanovich MA. Structural evolution of the chromophore in the primary stages of trans/cis isomerization in photoactive yellow protein. Journal of the American Chemical Society. 2005;127:18100–18106. doi: 10.1021/ja051210k. published online EpubDec 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Wilderen LJ, van der Horst MA, van Stokkum IH, Hellingwerf KJ, van Grondelle R, Groot ML. Ultrafast infrared spectroscopy reveals a key step for successful entry into the photocycle for photoactive yellow protein. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15050–15055. doi: 10.1073/pnas.0603476103. published online EpubOct 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devanathan S, Pacheco A, Ujj L, Cusanovich M, Tollin G, Lin S, Woodbury N. Femtosecond spectroscopic observations of initial intermediates in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophysical journal. 1999;77:1017–1023. doi: 10.1016/S0006-3495(99)76952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ihee H, Rajagopal S, Srajer V, Pahl R, Anderson S, Schmidt M, Schotte F, Anfinrud PA, Wulff M, Moffat K. Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. published online EpubMay 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weierstall U, Spence JC, Doak RB. Injector for scattering measurements on fully solvated biospecies. The Review of scientific instruments. 2012;83:035108. doi: 10.1063/1.3693040. published online EpubMar. [DOI] [PubMed] [Google Scholar]
- 49.Hart P, Boutet S, Gabriella C, Dubrovin M, Duda B, Fritz D, Haller G, Herbst R, Herrmann S, Kenney C, Kurita N, Lemke H, Messerschmidt M, Nordby M, Schafer D, Swift M, Weaver M, Williams G, Zhu D, Van Bakel N, Morse J. The CSPAD megapixel x-ray camera at LCLS. Proc. SPIE. 2012;8504:85040C. [Google Scholar]
- 50.Hutchison CDM, Tenboer J, Kupitz C, Moffat K, Schmidt M, van Thor JJ. Photocycle Populations with Femtosecond Excitation of Crystalline Photoactive Yellow Protein. J Chem Phys Lett. 2016 (submitted) [Google Scholar]
- 51.Barty A, Kirian RA, Maia FRNC, Hantke M, Yoon CH, White TA, Chapman H. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J Appl Crystallogr. 2014;47:1118–1131. doi: 10.1107/S1600576714007626. published online EpubJun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White TA. Post-refinement method for snapshot serial crystallography. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369:20130330. doi: 10.1098/rstb.2013.0330. published online EpubJul 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White TA, Kirian RA, Martin AV, Aquila A, Nass K, Barty A, Chapman HN. CrystFEL: a software suite for snapshot serial crystallography. J Appl Crystallogr. 2012;45:335–341. doi: 10.1107/S0021889812002312. published online EpubApr. [DOI] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/Jcc.20084. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 55.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta crystallographica. Section D, Biological crystallography. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 56.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt M. Biological and medical physics, biomedical engineering. In: Zinth W, Braun M, Gilch P, editors. Structure based enzyme kinetics by time-resolved X-ray crystallography, in: ultrashort laser pulses in medicine and biology. Springer; Berlin ; New York: 2008. ISSN 1618-7210. c2008, Germany. [Google Scholar]
- 59.Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D. 2015;70:136–153. doi: 10.1107/S1399004714021683. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta crystallographica. Section D, Biological crystallography. 1996;52:842–857. doi: 10.1107/S0907444995016477. published online EpubJul 1. [DOI] [PubMed] [Google Scholar]
- 61.Parak F, Hartmann H, Schmidt M, Corongiu G, Clementi E. The hydration shell of myoglobin. European biophysics journal : EBJ. 1992;21:313–320. doi: 10.1007/BF00188343. [DOI] [PubMed] [Google Scholar]
- 62.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. Journal of molecular biology. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 63.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–562. doi: 10.1093/nar/gkq395. published online EpubJul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. Journal of computational chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. published online EpubDec. [DOI] [PubMed] [Google Scholar]
- 65.Purwar N, Tenboer J, Tripathi S, Schmidt M. Spectroscopic studies of model photo-receptors: Validation of a nanosecond time-resolved micro-spectrophotometer design using photoactive yellow protein and alfa-phycoerythrocyanin. International Journal of Molecular Sciences. 2013;14:17. doi: 10.3390/ijms140918881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manz J, Naundorf H, Yamashita K, Zhao Y. Quantum model simulation of complete S-0 -> S-1 population transfer by means of intense laser pulses with opposite chirp. Journal of Chemical Physics. 2000;113:8969–8980. [Google Scholar]
- 67.Tannor DJ, Rice SA. Control of selectivity of chemical reaction via control of wave packet evolution. J. Chem. Phys. 1985;83:5013–5018. [Google Scholar]
- 68.Ruhman S, Kosloff R. Application of Chirped Ultrashort Pulses for Generating Large-Amplitude Ground-State Vibrational Coherence - a Computer-Simulation. J Opt Soc Am B. 1990;7:1748–1752. [Google Scholar]
- 69.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of computational chemistry. 2003;24:1999–2012. doi: 10.1002/jcc.10349. published online EpubDec. [DOI] [PubMed] [Google Scholar]
- 70.Becke AD. A New Mixing of Hartree-Fock and Local Density-Functional Theories. Journal of Chemical Physics. 1993;98:1372–1377. [Google Scholar]
- 71.Lee CT, Yang WT, Parr RG. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron-Density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 72.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges - the Resp Model. J Phys Chem-Us. 1993;97:10269–10280. [Google Scholar]
- 73.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chemical reviews. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 74.Yamaguchi S, Kamikubo H, Kurihara K, Kuroki R, Niimura N, Shimizu N, Yamazaki Y, Kataoka M. Low-barrier hydrogen bond in photoactive yellow protein. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:440–444. doi: 10.1073/pnas.0811882106. published online EpubJan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jorgensen WL, Chandrasekar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 76.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. Journal of Chemical Physics. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 77.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. Journal of computational chemistry. 1997;18:1463–1472. [Google Scholar]
- 78.Miyamoto S, Kollman PA. Settle - an Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. Journal of computational chemistry. 1992;13:952–962. [Google Scholar]
- 79.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- 80.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of chemical theory and computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 81.Roos BO, Taylor PR, Siegbahn PEM. A complete active space SCF method (CASSCF) using a density matrix formulated super-Cl approach. Chem. Phys. 1980;48:157–173. [Google Scholar]
- 82.Boggio-Pasqua M, Burmeister CF, Robb MA, Groenhof G. Photochemical reactions in biological systems: probing the effect of the environment by means of hybrid quantum chemistry/molecular mechanics simulations. Physical chemistry chemical physics : PCCP. 2012;14:7912–7928. doi: 10.1039/c2cp23628a. published online EpubJun 14. [DOI] [PubMed] [Google Scholar]
- 83.Fabiano E, Groenhof G, Thiel W. Approximate switching algorithms for trajectory surface hopping. Chem Phys. 2008;351:111–116. doi: 10.1016/j.chemphys.2008.04.003. published online EpubJul 3. [DOI] [Google Scholar]
- 84.Granovsky AA. Extended multi-configuration quasi-degenerate perturbation theory: The new approach to multi-state multi-reference perturbation theory. Journal of Chemical Physics. 2011;134 doi: 10.1063/1.3596699. published online EpubJun 7 (Artn 21411310.1063/1.3596699) [DOI] [PubMed] [Google Scholar]
- 85.Trebino R. personal communication.
- 86.Gu X, Xu L, Kimmel M, Zeek E, O'Shea P, Shreenath AP, Trebino R, Windeler RS. Frequency-resolved optical gating and single-shot spectral measurements reveal fine structure in microstructure-fiber continuum. Optics letters. 2002;27:1174–1176. doi: 10.1364/ol.27.001174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Snapshot density (diffraction patterns per 50 fs interval). The influence of a substantially varying number of patterns per bin on data quality is avoided when bins with approximately equal number of patterns per bin are used (e.g. red squares illustrate use of 16 bins; see Tab. S1. Unavoidably, these 16 bins are of non-uniform width in time).
Figure S2. A. Torsional angle ϕtail from refinement at various time delays (from bins containing 40 k diffraction patterns, see also Tab. S2B and S5). Dashed line: fit by eqn. S2 with a 590 fs transition. B. Right singular vectors (rSV) from the singular value decomposition of the 17 difference electron density maps on the fs (with 20 k diffraction patterns in each bin) to 3 ps time scale. Black dots: rSV1; black line: fit with exponential (eqn. S1) with characteristic time τ = 155 fs; blue and red dashed lines: fit of two separate parts of the rSV1 with exponentials. Red arrow: transition. Red squares: rSV2, dashed black line: fit by an exponential function (eqn. S2) with a 560 fs transition. Circle: time point near the assumed conical intersection. Green triangles: rSV3. The values of rSV4 and rSV5 are not significant, and displayed by thin dashed dotted and dashed double dotted lines. Blue bell-shaped curve: duration of the transition, from 470 to 650 fs. Inset: magnitude of the singular values.
Figure S3. Schematic view of the trans to cis transition at 550 fs. The rotations can be easily understood using an educator’s stick model set for chemical structures. A. PYPfast structure. To produce PYPfast from the reference, PYPref, grab the C2=C3 double bond with the left hand and the C1-S bond with the right hand. Rotate the double bond clockwise by about 40°. The C1-C2 single bond points now backwards as observed, the configuration is still trans. Compared to the dark state, the hydrogen bonds to Glu46 and Tyr42 are elongated. To further the isomerization (and reach B), continue rotating the double bond clockwise by an additional 90° by keeping the atom C2 location fixed. The entire head-structure pivots about (the fixed) atom C2. Since the head cannot swing out, concerted anti-clockwise rotations about the C1-C2 and the C3-C1’ single bonds are necessary to minimize the volume swept out by the atoms that move during isomerization; see arrows in A. B. PYPslow structure (800-1200 fs), cis. The hydrogen bond to Glu46 is broken (head displacement exaggerated for clarity).
Figure S4. The solvent accessible surface (blue) of PYP3ps.
Figure S5. Entry into the photocycle in PYP crystals as evident by optical absorption changes initiated by 900 fs (A-B), 300 fs, and 100fs (C-D) laser pulses. Pump-probe delay 180μs for A-B, 1 ms for C-D. A. PYP absorption spectra of the dark state, following a sequence of 10 pump-probe cycles with 900 fs pulses at increasing laser pulse energy densities (shown in the legend). Irreversible bleaching (damage), as evident by the reduced OD of the dark state, occurs at 2.2mJ/mm2. B. Light-dark difference absorption spectra at various laser pulse energy densities. Spectra are recorded by accumulating 10 pump-probe cycles. Extent of (reversible) bleach at 450 nm is roughly proportional to number of molecules in photocycle. At 1.2 mJ/mm2 about 5% of molecules can be pumped into the photocycle. C. Permanent single-shot photo-bleaching (damage) measured after 100 fs (red squares) and 300 fs (blue diamonds) laser pulses of various energies (spot size 80 μm). D. Extent of reaction initiation with 100 fs and 300 fs laser pulses measured as the pB concentration at 1ms as the reversible ΔA450nm/A450nm. 100 fs laser pulses with 2 μJ (energy density: 400 μJ/mm2) result in up to 6% reaction initiation with small permanent damage.
Figure S6. A. A diagram of the SHG-FROG used to characterize the 450nm femtosecond pulses incident on PYP crystals. The incoming pulse (black) is split into two replicas that travel down individual paths (shown in blue and red for clarity) until they are overlapped in a space and time in the 20 μm thin BBO crystal. The resulting FROG signal (pink) is measured using the UV spectrometer. B. The experimentally measured FROG traces for the 140 fs stretched pulse. C. Reconstructed FROG traces. The “G error” for the fitting was <1%. The high order structure seen at the top of the reconstruction that is not seen in the experimental data is the result of fluctuations in the shot-to-shot pulse shape. This is commonly seen in multi-shot FROG measurements and does not greatly impact the recovered pulse length or phase. D-E. FROG reconstruction of the temporal and the spectral intensity (red) of the reconstructed pulse incident on the PYP microcrystals with the retrieved phase indicated (blue). High order time-structure is a reconstruction artifact due to beam intensity instability (85, 86). The pulses were found to be 140±5 fs long with a GDD of ≈1000fs2 and TOD ≈+1300fs3.
Figure S7. A. PYP configurations along the QM/MM trajectory. The atoms included in the QM region are shown in ball-and-stick representation and modeled at the CASSCF(12,11)/cc-pVDZ level of ab initio theory. The rest of the system, consisting of the protein, water, Cl− and Na+ counter ions (most of which is not shown here) are modeled with the Amber03 force field. B. Snapshots from a QM/MM trajectory after photo-excitation at t = 0 fs (Tab. S4, trajectory a). C. Torsional angles as a function of time in this trajectory. Left yellow shaded region, dynamics on the excited state potential energy surface. Vertical dashed line (surface hop): seam that separates excited state structural dynamics (left) from ground state dynamics (right). D. Color code for the torsional angles of the pCA chromophore in C.
Figure S8. Transition through conical intersection as conceived from theoretical consideration (lower scheme) and viewed from the perspective of the TR-SFX experiment (structures above).
Figure S9. Trans to Cis isomerization in PYP. Direct comparison of difference maps and structures of individual time-points determined from 40,000 diffraction patterns with those determined from averaging diffraction pattern in larger time-bins (compare Tab. S1 B-C). The 269 fs, 799 fs and 3 ps delays (panels A,C and E) delays are also shown in Fig. 2 in the main text. Weighted DED maps in red (−3 σ) and blue (3σ); front (upper) and side view (lower). The reference, dark structure is shown in yellow throughout; structures before the transition and on the electronic excited state PES are shown in pink; structures after the transition and on the electronic ground state PES are shown in green. Important negative difference density features are denoted α, positive features as β. Pronounced structural changes are marked by arrows. A. Representative time-delay (269 fs) before the transition; some important residues are marked. Dotted lines: hydrogen bonds of the ring hydroxyl to Glu46 and Tyr42. B. Chromophore configuration (PYPfast) by averaging 100 fs to 400 fs pump-probe delays. C. Representative time delay (799 fs) after the transition. D. Structure PYPslow on longer fs times obtained by averaging the 800 fs to 1200 fs pump-probe delays. E. 3 ps chromophore configuration. Dashed lines in A, D and E: direction of feature β1.
Figure S10. Difference-difference maps (residual maps): Calculated ∣Fcalc(t)∣-∣Fcalc(dark)∣ difference maps were subtracted from the observed ∣Fobs(t)∣-∣Fobs(dark)∣ difference maps. Pink: PYPfast; green: PYPslow and PYP3ps; yellow: PYPref. Residual features (blue: positive, red negative) on the +/−3 σ level. A and B. Fast and slow fs time scale, residual maps using the correct, refined models that correspond to the time-scales, twisted-trans for A, and near-cis for B. C and D. Residual maps using interchanged, deliberately incorrect models, near-cis for C and twisted-trans for D. The chromophore tail is much cleaner in A and B, but strong residual features denoted α and β in C and D indicate mismatch. Note: remaining features near the sulfur emerge from its strong electron density; its position is difficult to model exactly. E. Data binned from 400-800 fs across the transition. Feature β shows some mixing-in of the PYPfast structure in this time range. F. residual map at 3 ps time delay.
Figure S11. PYPref (yellow), PYPfs,fast (pink) and the theoretical PYP model from QM/MM simulations (orange) overlaid on the 269 fs difference map (red/blue −3σ/4σ). A. front view, B. side view. Nearby amino acid residues are marked in A. Arrow in B: similar chromophore tail displacements for the observed and theoretical PYP structures.
Figure S12. Full view of the DED maps (contour: red/green −3σ/3σ) at various time-ranges. The maps are overlaid on PYPref (yellow) and the refined structures. Contrary to all other figures that depict PYP structures, the view is from the opposite (back) of the chromophore, hence the Arg52 is on the right. This orientation allows for a detailed view of the positive feature β1 which indicates the configuration of the chromophore and the strong feature on the Cys69 sulfur (β2) that is also associated with the cis configuration. A. On the fast time scale, feature β1 is strongly bent, β2 is absent. The DED map is interpreted with a trans configuration (structure in pink). B. The slow time range, feature β1 is aligned along the tail axis, which indicates a cis configuration (structure in green). C. At the 3 ps delay feature β1 is similar to B (structure also in green). D. Consistency check by randomly selecting ~80,000 snapshots from the 157,000 snapshots in the bin. DED map with features β1 and β2 and structure (green) essentially the same as in B.
Table S1. TR-SFX data statistics. Resolution 1.6 Å; data in the highest resolution bin (1.6 - 1.67 Å) in brackets. A. Statistics of the data sorted into time bins with 32,690 snapshots with Bragg reflections (hits)/bin (approximately 20,000 indexed snapshots/bin). Times shown are the average of the time stamps in each bin and their standard deviation. B. Data statistics of bins with 65,380 snapshots/bin (approximately 40,000 indexed snapshots/bin). C. Statistics of the individual −1ps, 3 ps and 200 ns time points and when snapshots with time-stamps 100 – 400 fs and 800 – 1200 fs are combined.
Table S2. Difference maps and refinement for selected structures.
Table S3. Torsional angles and distances from structural refinement at various, individual time delays.
Table S4. Global structural changes [Å] as measured by averaging the distances between cα atoms of equivalent residues.
Table S5. Excited state lifetimes (2rd column), and products (3th column) in the excited state QM/MM trajectories.