Abstract
Cationic antimicrobial peptides constitute part of the innate immune system and provide an essential role in the defense against infection. At present there is a paucity of information regarding the antimicrobial profile of the chicken (Gallus gallus). Using in silico studies, an expressed sequence tag (EST) clone was identified which encodes a novel cationic antimicrobial peptide, chicken liver-expressed antimicrobial peptide 2 (cLEAP-2). The predicted amino acid sequence composed a prepropeptide, and the active peptide contained four conserved cysteine amino acids. The gene was localized to chromosome 13, and analysis of the genome revealed three exons separated by two introns. The cLEAP-2 gene was expressed in a number of chicken epithelial tissues including the small intestine, liver, lung, and kidney. Northern analysis identified liver-specific cLEAP-2 splice variants, suggesting some degree of tissue-specific regulation. To investigate whether cLEAP-2 expression was constitutive or induced in response to microbial infection, 4-day-old birds were orally infected with Salmonella. Analyses of cLEAP-2 expression by semiquantitative reverse transcription-PCR indicated that cLEAP-2 mRNA was upregulated significantly in the small intestinal tissues and the liver, indicative of direct and systemic responses. The antimicrobial activity of cLEAP-2 against Salmonella was analyzed in vitro with a time-kill assay and recombinant cLEAP-2. Interestingly Salmonella enterica serovar Typhimurium SL1344 showed increased susceptibility to the active cationic peptide (amino acids 37 to 76) compared to S. enterica serovar Typhimurium C5 and Salmonella enteritidis. Taken together, these data suggest that cationic cLEAP-2 is part of the innate host defense mechanisms of the chicken.
Endogenous cationic peptides play an important role in innate immunity, and to date, hundreds of natural peptides have been isolated from vertebrate, invertebrate, and plant species. Unlike many natural antimicrobials synthesized via specialized metabolic pathways, these peptides, ranging in size from 1.5 to 10 kDa, are expressed from individual genes and produced by proteolytic processing from precursor molecules. Their primary role is defense against microbial attack, and the cationic peptides are capable of killing gram- and gram-positive bacteria, fungi, eukaryotic parasites, and enveloped viruses (27).
The amino acid sequences of known antimicrobials are diverse. However, the peptides have been divided into groups broadly based on their known or predicted three-dimensional structures, with the most important difference being the presence or absence of cysteines (13). The first group comprises molecules with a linear, α-helical peptide structure without cysteines and includes numerous frog peptides, including the magainins and bombinins. Peptides with a β-sheet structure stabilized by two or more disulfide bonds constitute another group and include the α- and β-defensins. A further group includes peptides with a predominance of one or more amino acids, whereas molecules derived from larger peptides comprise another class and include lactoferricin, a digestion product of lactoferrin. Recently, a cationic peptide, liver-expressed antimicrobial peptide 2 (LEAP-2), which appears unique regarding its primary structure and disulfide motif, has been identified (14, 16).
Underlying all classes of antimicrobials is the ability of each peptide to adopt a shape in which clusters of hydrophobic and cationic amino acids are spatially organized in particular sections of the molecule. This structural conformation is key to their mechanism of action. In general, the peptides function by binding to the negatively charged surface components of bacterial membranes via electrostatic charge interactions, integrating into the membrane and causing it to depolarize and collapse. However, some peptides are multifunctional. For example, PR-39, a proline-arginine-rich peptide found in the pig intestine, stops bacterial synthesis and can induce wound healing (4, 6). Human β-defensins have also been shown to have a chemoattractant activity for immature dendritic cells and memory T cells, suggesting that these peptides also function as an important link between the innate and adaptive systems (24, 25).
Antimicrobials are generally expressed at sites exposed to multiple microbes including organs such as the gastrointestinal tract (GI) and skin. The importance of these peptides in protecting GI surfaces against pathogens is shown by matrilysin-deficient mice. These mice lack the proteolytic enzymes necessary for processing cryptidins, the α-defensins of murine GI cells, and consequently lack functional cryptidins. This results in their increased susceptibility to infection by orally administered Salmonella (22). Interestingly, mice engineered to synthesize human defensin 5, in addition to their natural complement of gut antimicrobials, show increased resistance to orally administered Salmonella enterica serovar Typhimurium. This raises the possibility that the natural array of GI antimicrobial peptides synthesized in each species may play a key role in determining the pathogenicity of a particular microbe in that species (19).
At present, there is a paucity of information regarding the avian antimicrobial profile and the potential role these peptides play in microbial pathogenicity and colonization of the avian GI tract. This is especially important, as birds are universally recognized as a major reservoir of human enteropathogens but are themselves often asymptomatic. To date, four antimicrobial peptides, termed gallinacin 1 and 2 and turkey heterophil peptides 1 and 2, have been characterized from avian heterophils (5, 8). All are homologous to β-defensins and show activity against Salmonella enteritidis and Campylobacter jejuni. A third chicken antimicrobial, gallinacin 3, encoding a defensin-like antimicrobial peptide, has been reported in chicken epithelial tissues, including the colon and kidney, although its antimicrobial profile is not known (28).
We report here on the expression and antimicrobial activity of the novel avian antimicrobial peptide, chicken LEAP-2 (cLEAP-2). We show cLEAP-2 gene expression to be induced in avian tissues, including the small intestine, in response to Salmonella infection and provide in vitro evidence for selective cLEAP-2 antimicrobial activity against Salmonella strains.
MATERIALS AND METHODS
Livestock.
The poultry (Goldline) used in the experiments were supplied by the Comparative Biology Centre, Newcastle University. All birds were sacrificed by cervical dislocation. The in vivo bacterial challenge experiment was carried out under United Kingdom Animal Project license no. PPL60/2702.
Molecular analyses.
The expressed sequence tag (EST) clone chEST497m18, encoding cLEAP-2, was obtained from MRC Geneservice United Kingdom. Forward and reverse primers were designed to amplify coding regions of the cLEAP-2 cDNA. Primers 1F (5′-CTCATACTGTGAAGATGCAC-3′) and 3R (5′-CCGTTCTAAGGAAGCAGC-3′) were designed to correspond to the exon 1 and 3 cLEAP-2 sequences, respectively, and amplified nucleotides 65 to 298 of the EST 497m18 cDNA, which encoded the putative cLEAP-2 prepropeptide.
Tissue expression. (i) RT-PCR.
Total RNA was extracted from chick tissues snap-frozen in liquid nitrogen. Initially, the tissues were ground to a fine powder, and the RNA was extracted by using RNAwiz (Ambion). To reduce degradation, RNase inhibitor (Invitrogen) was added to each sample (1 U/μg of RNA) before storage at −80°C. All samples were pretreated, before reverse transcription (RT), with DNase (Promega) at a concentration of 1 U/μg of RNA. RT was performed at 42°C for 1 h, followed by heat inactivation for 5 min at 95°C. The cDNAs were amplified in sequential cycles. The annealing conditions consisted of an initial 1-min denaturation step at 95°C; 35 cycles of 1 min at 95°C, 1 min at 55°C, 1 min of 72°C; and a final extension step of 72°C for 12 min. The RT products were resolved by electrophoresis with either 1 or 1.5% Tris-borate-EDTA agarose gels with added ethidium bromide and photographed under UV illumination. In control samples, reverse transcriptase was omitted to demonstrate that PCR amplification was not due to contamination with genomic DNA. Each RT product was verified by DNA sequencing.
(ii) Semiquantitative RT-PCR.
Coamplification of cLEAP-2 and 18S rRNA was carried out by using the GeneAmp RNA PCR core kit (Applied Biosystems). The optimum conditions were established according to the manufacturer's instructions by using the cLEAP-2-specific primers and 18S rRNA-specific primers (Ambion). The PCR cycle number was limited to within an experimentally determined linear range of amplification (21). Products were separated by using 1% Tris-borate-EDTA agarose gels with added ethidium bromide. Densitometry was performed by using a GDS 5000 gel documentation system (UV Products; Cambridge, United Kingdom).
(iii) Northern blot analysis.
Poly(A+) RNA was extracted from avian tissues by using the NucleoTrap kit (Clontech). Two micrograms from each tissue was electrophoresed on a formaldehyde gel, blotted onto nylon, and fixed by UV cross-linking. The hybridization probes were a 150-bp EcoRI cDNA fragment encoding the cLEAP-2 propeptide and a 1,150-bp DNA clone (EST 116g5 purchased from MRC Geneservice United Kingdom) encoding chicken glyceraldehyde 3-phosphate dehydrogenase (cGAPDH). The cDNAs were labeled with [α-32P]CTP by using the Megaprime DNA labeling system (Amersham). For each probe, the membrane was prehybridized at 68°C in Quickhyb buffer (Stratagene), hybridized for 1 h with the probe, and washed at high stringency. The membrane was analyzed by using a Packard Instant Imager.
Peptide expression.
To determine the antimicrobial activities of the cLEAP-2 peptides, DNA fragments encoding the putative cLEAP-2 propeptide (amino acids 25 to 76) and mature peptide (amino acids 37 to 76) were amplified by PCR. Primers 1F′-3R′ and 2F′-3R′ amplified the DNA sequences encoding the propeptide and mature peptides, respectively, and are as follows: 1F′, 5′-CGCGGATCCCTGCACCAACCAC-3′; 2F′, 5′-CGCGGATCCATGACGCCTTTCTGG-3′; 3R′, 5′-GGGGTACCTCACTCGGAGGCCGTTCTAAG-3′. The forward primers were engineered to contain BamHI restriction sites, and the reverse primer was engineered to contain a KpnI restriction site. The restriction sites are underlined. Each fragment was cloned into the pRSETA expression vector (Invitrogen) and transformed into Escherichia coli Origami B::pLysS by selection with 50 μg of ampicillin/ml and 34 μg of chloramphenicol/ml. Freshly transformed recombinant clones were cultured at 37°C, in the appropriate antibiotics, until turbid and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Two hours after induction, the cells were harvested by centrifugation, resuspended in 20 mM Tris (pH 8) and 1 mM Tris (2-carboxyethyl) phosphine (Sigma), and lysed by sonication. The soluble and insoluble fractions were recovered, and the protein concentrations were determined and either analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 15% (wt/vol) polyacrylamide gels or used for studies of antimicrobial activity.
Western analyses.
Bacterial extracts (30 μg of protein per lane) containing either no cLEAP-2 peptide, the propeptide, or peptide were separated on an SDS-15% polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Hybond P; Amersham). The membrane was incubated overnight at 4°C in a blocking solution of phosphate-buffered saline (PBS), 0.1% (vol/vol) Tween 20, and 5% (wt/vol) milk powder containing a 1:5,000 dilution of antipolyhistidine antibody (Sigma). The membrane was washed in PBS-Tween 20 (0.1%), a chemiluminescence detection reaction was performed by using ECL Plus (Amersham), and the membrane was exposed to X-ray film according to the manufacturer's recommendations.
Antimicrobial assay.
Salmonella strains were cultured until turbid at 37°C in 20 ml of Luria broth containing 0.05 M glucose. Of each culture, 250 μl was subcultured into a fresh 20 ml of Luria broth with 0.05 M glucose and incubated for 3 h at 37°C. After the second incubation, 20 μl of each culture was diluted to 2 ml in PBS. Ten microliters of each peptide extract (20 μg/ml) or PBS was added to 90 μl of diluted culture, vortexed, and incubated for 3 h at 37°C. The suspensions were sequentially diluted to 10−4 in PBS, and each dilution was plated onto blood agar. All plates were incubated overnight at 37°C, and the colonies were counted. The number of CFU resulting from a 3-h incubation with either propeptide or mature peptide were presented relative to the control incubations with a peptide-free E. coli extract. Ten microliters of 200-mg/ml magainin (Sigma) was used as the antimicrobial peptide positive control. cLEAP-2 extracts tested for antimicrobial activity were solubilized in PBS. Assays were performed with the following strains: S. enterica serovar Typhimurium phoP (2), S. enterica serovar Typhimurium C5 (12), S. enterica serovar Typhimurium SL1344 (10), and S. enteritidis (gift of A. Khan, University of Newcastle, Newcastle, United Kingdom).
Experimental infections.
Two groups of 5-day-old female chicks were orally gavaged with 0.1 ml of inoculum containing approximately 5 × 106 organisms of either S. enteritidis or S. enterica serovar Typhimurium SL1344. Age-matched, noninfected control birds were housed under similar environmental conditions, and 0.1 ml of PBS was introduced by gavage. Chicks were sacrificed on the fourth day after infection. Tissue samples were collected, snap-frozen in liquid nitrogen, and stored at −80°C until analyzed.
Statistics.
Data are expressed as means ± standard errors. Statistical analyses were performed by one-way analysis of variance, followed by the Dunnett's multiple-comparison test for unpaired data. A difference with a P value of <0.05 was considered statistically significant.
RESULTS
Gene identification, location, and expression analyses.
BLASTp searches of the chicken EST intestinal database (3) identified two ESTs, 497m18 (633 bp) and 148o1 (616 bp), whose predicted amino acid sequence showed homology to that of the human LEAP-2 (hLEAP-2) antimicrobial peptide (Fig. 1). Using the BLAST (tblastn) nucleic acid alignment program and the DNA sequence from the chEST clone 497m18, cLEAP-2 was identified on chromosome 13 of the Gallus gallus genome. The gene comprises three exons and two introns of approximately 0.6 and 0.95 kb, respectively.
FIG. 1.
Comparison of the amino acid structures of LEAP-2 homologues from the chicken (Fig. 3), mouse, and human. The putative propeptide region is underlined, the predicted endoprotease cleavage site is shown in italics, and the mature peptide is shown below, after the hyphen. The pattern of cysteine amino acids is conserved between all three homologues.
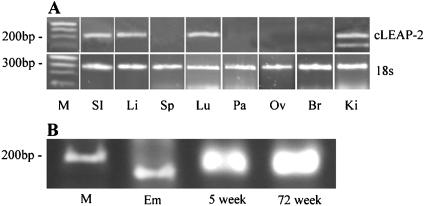
RT-PCR analyses with primers designed to amplify cLEAP-2 and sequencing of the cDNA products revealed cLEAP-2 expression in the small intestine, liver, lung, and kidney of 5-day-old chicks (Fig. 2A). Expression was also observed in the small intestinal tissues of chickens at 5 and 72 weeks of age. In contrast, no expression was detected in samples analyzed from the pancreas, brain, spleen, or ovary. Translation of the cDNA sequences isolated from each of the chick RNA samples into putative amino acid sequences revealed a peptide of 76 amino acids with significant identity to that encoded by the EST 497m18 clone (Fig. 3). Only three amino acid changes were observed, including the substitution of aspartic acids at positions 21 and 44 for tyrosine and valine residues, respectively, and the substitution of a proline at position 52 for a serine residue. Analyses of the encoded primary structure of the cLEAP-2 peptide indicated a putative signal sequence, suggesting that the peptide was secreted, and a unique proregion of 12 amino acids terminated by an endoproteinase cleavage site, RLKR, indicating that the propeptide was proteolytically cleaved to release the mature active peptide (14). The predicted molecular masses of the pro- and mature peptides were 6,074.2 and 4,593.3 Da, respectively (ExPASy Peptide Mass Program). Typical of cationic antimicrobial peptides, the mature peptide was composed of a high percentage of positively charged and hydrophobic amino acids, and within this region, four conserved cysteine residues were identified.
FIG. 2.
(A) Expression of cLEAP-2 in 5-day-old chick tissues. M, ladder standards; SI, small intestine; Li, liver; Sp, spleen; Lu, lung; Pa, pancreas; Ov, ovary; Br, brain; Ki, kidney. (B) Expression of cLEAP-2 in the small intestine of an 18-day chicken embryo (Em), a 5-week-old bird, and a 72-week-old bird.
FIG. 3.
The cDNA and peptide sequences of cLEAP-2. The prepropeptide contains 76 amino acids. The signal peptide is underlined, the propeptide is terminated by the motif RLKR (shown in bold), and the positions of introns 1 and 2 are marked by vertical arrows. The amino acids differing from those predicted from the chEST clone 497m18 and the four conserved cysteines are shown in italics. The nucleotide sequence of the 3′ and 5′ untranslated regions are presented, with locations of the TATA box (TATT), a putative NF-κB binding site (GGGCTCTTCC), and polyadenylation site (AATAAAA) indicated by underlining.
Analyses of small intestinal RNA from an 18-day-old chicken embryo revealed a unique 100-bp PCR product (Fig. 2B) which was also detected in the kidney tissues (Fig. 2A). This was identified as a putative cLEAP-2 splice variant in which the DNA sequence encoding amino acids 21 to 65 of cLEAP-2 had been excised.
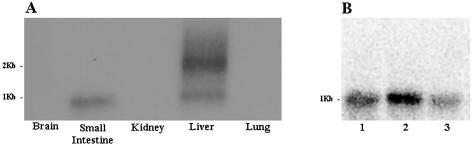
Hybridization of the 32P-labeled cLEAP-2 probe to poly(A+) RNA isolated from the chick brain, small intestine, kidney, liver, and lung verified the patterns of cLEAP-2 expression observed by RT-PCR in the liver, small intestine, and brain tissues but not in the kidney or lung (Fig. 4A). This lack of correlation in the latter tissues may have reflected the reduced sensitivity of the Northern analyses compared to the RT-PCR. Interestingly, the analysis of liver mRNA isolated from tissues of 4-day-old birds revealed two transcripts of approximately 1 and 2 kb, respectively, hybridizing to the cLEAP-2 probe, of which the latter appeared to be more highly expressed. In contrast, only the 1-kb transcript was identified in mRNA isolated from the small intestine. Surprisingly, Northern analyses of liver mRNAs isolated from 8-day-old birds did not reveal the larger 2-kb transcript (Fig. 4B).
FIG. 4.
Northern blot analysis. (A) cLEAP-2 expression in 5-day-old chick tissues. (B) cLEAP-2 expression in the liver tissues of 8-day-old birds following infection with Salmonella or PBS. Lanes: 1, S. enterica serovar Typhimurium 1344; 2, S. enteritidis: 3, control birds given PBS.
Inducibility.
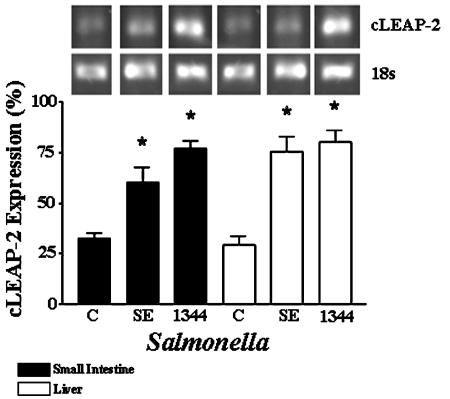
Antimicrobial peptides can be either constitutively expressed or induced in response to microbial infection, and regulation is often dependent on the site of synthesis. For example, bovine β-defensins are induced in some epithelial cells (20) but constitutively expressed in granulocytes (26). Similarly, gallinacin-3 has been shown to be constitutively expressed in the avian skin and tongue but inducible in the trachea (28). To determine whether the expression of cLEAP-2 in the small intestine and liver was constitutive or inducible in response to Salmonella infection, S. enteritidis, S. enterica serovar Typhimurium SL1344, or PBS was introduced to groups of 5-day-old chickens by gavage. After 4 days of infection, the liver and small intestines of the birds were extracted and analyzed for cLEAP-2 RNA expression by semiquantitative RT-PCR with the 1F-3R primer pair. This allowed amplification of cDNAs putatively encoding the cLEAP-2 prepropeptide. A significant increase (P < 0.01) in small intestinal cLEAP-2 expression was detected in the birds infected with Salmonella (Fig. 5), and a similar response was recorded in the liver (P < 0.01). These increases were observed regardless of the Salmonella strain and suggested that cLEAP-2 expression was induced in the avian tissues in response to either local or systemic infections.
FIG. 5.
Induction of cLEAP-2 expression in chick small intestinal and liver tissues following oral infection with Salmonella. RT-PCR analyses were performed on the chick tissues 3 days after infection. The number of experimental observations in each group was four, and values are means ± standard errors of the means. 1344, S. enterica serovar Typhimurium SL1344; SE, S. enteritidis: C, control birds given PBS; *, P < 0.01 compared to the appropriate control.
Interestingly, analyses of the Salmonella fecal numbers at day 3 postinfection were found to be one log lower in the S. enterica serovar Typhimurium SL1344-infected birds (3.5 × 104/gram of feces) than in those infected with S. enteritidis (4.0 ×105/gram of feces). These data suggested that enhanced S. enterica serovar Typhimurium SL1344 killing had occurred in the GI tracts of the chicks.
Antimicrobial activity.
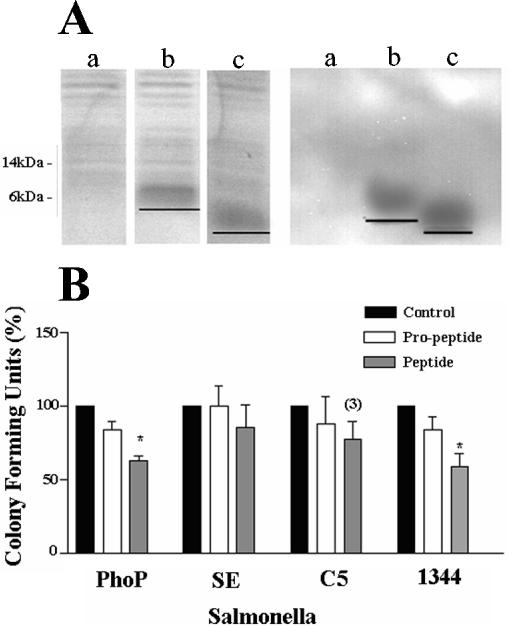
To investigate whether cLEAP-2 showed selective antimicrobial activity against Salmonella species, the cLEAP-2 DNA sequences encoding the propeptide and peptide were isolated by PCR and hyperexpressed in E. coli by using pPRSETA and the Origami B::pLysS strain. This strain contains the plasmid pLysS, which under the T7 promoter, prevents any expression of proteins until induced by IPTG and thus prevents any toxic effects caused by leakage of cLEAP-2 within the host strain. In addition, the cytoplasm of Origami B, in comparison with other E. coli strains, provides a higher oxidizing environment, facilitating the folding of proteins by promoting disulfide bond formation. SDS and Western analyses of culture extracts revealed peptide bands of approximately 6 and 4 kDa, respectively, in strains expressing the cLEAP-2 propeptide (amino acids 25 to 76) and mature peptide (amino acids 37 to 76) compared to the peptide-free control strain (Fig. 6A). Using a time-kill assay, the antimicrobial activities of the culture extracts (20 μg/ml) containing either no peptide, the cLEAP-2 propeptide, or cLEAP-2 peptide were tested against Salmonella species including the wild-type S. enteritidis and S. enterica serovar Typhimurium strains C5 and SL1344 and the mutant S. enterica serovar Typhimurium strain phoP. In comparison with the peptide-free control extract, reductions in phoP and SL1344 microbial numbers were observed using extracts containing the cLEAP-2 peptide but not the propeptide (Fig. 6B). The numbers of S. enterica serovar Typhimurium C5 and S. enteritidis were not affected following incubation with either of the recombinant cLEAP-2 peptides.
FIG. 6.
Antimicrobial activities of the cLEAP-2 propeptide (amino acids 25 to 76) and mature peptide (amino acids 37 to 76). (A) SDS-polyacrylamide gel electrophoresis and Western analyses of the peptide-free (a), 6-kDa cLEAP-2 propeptide (b), and 4.5-kDa cLEAP-2 peptide (c) E. coli preparations. (B) Time-kill assay analyses of cLEAP-2 activities. The number of CFU resulting from a 3-h incubation with either propeptide or mature peptide are presented relative to control incubations with a peptide-free E. coli extract. PhoP, S. enterica serovar Typhimurium PhoP mutant; SE, S. enteritidis; C5, S. enterica serovar Typhimurium C5; 1344, S. enterica serovar Typhimurium SL1344. Data are presented as means ± standard errors of the means (n = 3 for all S. enteritidis observations; n = 4 for all other observations, except where indicated). *, P < 0.05 compared to the appropriate negative control.
DISCUSSION
Antimicrobial peptides are important effectors of vertebrate innate defenses, and at each site of production, they provide an array of antimicrobial molecules, which work synergistically to prevent infection. Cells and tissues synthesizing antimicrobial peptides are generally those exposed to microbial pathogens. In the chicken, four β-defensins have been identified in heterophils (5, 8), and a fifth defensin-like antimicrobial peptide, gallinacin 3, is expressed in a number of avian tissues (28). To date, however, no cationic antimicrobial peptides have been identified in the chicken small intestine.
The novel cationic LEAP-2 peptide was first identified in human blood (14). The discovery of a homologue, cLEAP-2, in the avian species is important, especially given the remarkable sequence identity of cLEAP-2 and hLEAP-2 at the amino acid level. Amino acids 1 to 27 of the mature LEAP-2 peptide (MTPFWRGVSLRPVGASCRDNSECITMLCR) are encodedin exon 2 of the chicken gene. Of these amino acids, 25 of 27 (92.5%) are identical to those of hLEAP-2 and 26 of 27 (96.2%) are similar. Exon 2 also encodes 12 amino acids of the cLEAP-2 propeptide (LHQPQPLLRLKR). In contrast to the remarkably high conservation in the mature peptide domain, only 2 of 12 (16.6%) propeptide amino acids are identical in humans and chickens. Moreover, this limited homology is limited to the endoprotease processing site. The unusually high sequence identity of peptides from species whose last common ancestor existed over 100 million years ago suggests an important functional role.
We have shown, in agreement with Lynn et al. (15), that the cLEAP-2 gene encoding the novel antimicrobial peptide, cLEAP-2, is expressed in chicken epithelial tissues including the small intestine, liver, kidney, and lung. This suggests that cLEAP-2 is expressed as part of the epithelial innate defense system and functions to prevent potentially pathogenic microbes from interacting with epithelial surfaces and invading the tissues. The lack of cLEAP-2 expression in the pancreas, ovary, brain, or spleen may have reflected the sensitivity of the detection system or the reduced susceptibility of these tissues to microbial invasion. Interestingly, hLEAP-2 transcripts were detected in brain tissue, but only when using quantitative real-time PCR and at levels 104 lower than those of the GAPDH housekeeping gene (14).
The RT-PCR analyses of LEAP-2 expression in human tissues identified at least four splice variants. Employing a comparable primer pair, only two cLEAP-2 PCR products were identified in the avian tissues. The expression of a third transcript (2 kb) was identified by Northern blot analyses of liver mRNA isolated from 5-day-old birds and indicated the synthesis of an additional cLEAP-2 splice variant. The significance of these transcripts in avian tissue is not known. However, the 2-kb hLEAP-2 transcript has been attributed to the existence of an alternative LEAP-2 promoter site directing the synthesis of an mRNA species that includes intron 1 and which encodes a LEAP-2 peptide without a signal sequence. Interestingly, the comparable chick transcript was not detected in the birds challenged with Salmonella. These data suggest that cLEAP-2 peptides may have functions in addition to that of antimicrobial activity. This multifunctionality is not unique, for the angiogenins discovered nearly two decades ago and initially identified as having roles in vasculogenesis and tumor cell growth have recently been shown to exhibit bactericidal and fungicidal activity and have been proposed to play an important role in systemic innate immunity (11).
In certain epithelial cells, inducible antimicrobial peptide genes such as those encoding the bovine tracheal antimicrobial peptide and human β-defensin 2 become activated in response to bacteria and lipopolysaccharide (LPS) (1, 7, 23). Challenging 5-day-old chicks with Salmonella resulted in a significant increase in cLEAP-2 expression in both the small intestine and liver and provided evidence of cLEAP-2 induction in response to both local and systemic responses. Although we have no proof of increased peptide synthesis, these data suggest that cLEAP-2 is an inducible peptide. Whether induction occurred as a consequence of direct microbial contact or was mediated through bacterial products such as LPS or proinflammatory cytokines such as interleukin-1 and tumor necrosis factor is not known. However, the induction of antimicrobial peptides is known to follow the activation of a regulatory pathway involving Toll receptors and the transcription factor NF-κB (17). Toll-like receptors (TLRs) have been identified in the chicken, and the type 2 chTLR message has been shown to be ubiquitously expressed (9). Recent in silico studies have identified putative TLR pathways in the chicken (16), and a potential transcription factor binding site for NF-κB in the promoter region of the cLEAP-2 gene (Fig. 2) suggests that the increase in cLEAP-2 gene expression may occur through this signaling pathway.
The fecal counts from the in vivo studies suggested that the S. enterica serovar Typhimurium SL1344 enteropathogen was more sensitive to the bactericidal activity of the chick gut than S. enteritidis. The MICs of the more effective antimicrobial peptides are in the range of 0.1 to 10 μg/ml (13), and sufficient recombinant extract (20 μg/ml) was isolated to investigate cLEAP-2 antimicrobial activity against Salmonella sp. in vitro. Microgram amounts of peptide were found to be effective against the mutant Salmonella phoP and wild-type S. enterica serovar Typhimurium SL1344 strains but ineffective against S. enteritidis and S. enterica serovar Typhimurium C5. Cationic antimicrobial peptides function by disrupting the structure of the outer membrane of gram-negative bacteria, but their bactericidal activity is also dependent on their disrupting the cytoplasmic membrane. Most antimicrobial peptides are too large to use porin channels, so the surface charge and permeability of the outer microbial membrane determines the ability of each peptide to reach its target. The susceptibility of S. enterica serovar Typhimurium SL1344 and the resistance of S. enteritidis and S. enterica serovar Typhimurium C5 to cLEAP-2 activity were probably a reflection of the different compositions of the bacterial membranes. Salmonella spp. are known to resist cationic antimicrobial peptide activity by reducing the negative charge of their outer membranes. This is known to occur through modification of the anionic membrane molecules, including teichoic acids, LPS, or lipid A, with positively charged substituents (18). Several modifications of lipid A are responsible for cationic antimicrobial resistance in Salmonella, and the enzymes responsible for these are linked to the expression of phoP and phoQ (2). Indeed, the S. enterica serovar Typhimurium phoP mutant strain, which has lost the ability to modify its membranes, was more sensitive to cLEAP-2, as reflected by the 40% reduction in numbers (Fig. 6B). It was interesting that the in vivo reduction in S. enterica serovar Typhimurium SL1344 numbers was paralleled in vitro. However, the in vivo observation probably reflected the synergistic activity of an array of cationic antimicrobial peptides rather than cLEAP-2 per se. Furthermore, it is also unlikely that cLEAP-2 is only active against gram-negative bacteria, as synthetic hLEAP-2 has been shown to demonstrate activity against gram-positive bacteria and yeasts.
These data suggest that the cLEAP-2 gene encodes a novel cationic antimicrobial peptide, which is induced in chick tissues in response to Salmonella infection and which forms an integral part of the avian innate host defense system.
Acknowledgments
The Salmonella phoP strain was a gift from S. I. Miller.
C.L.T. and C.J.N. are supported by BBSRC studentships. G.M. is supported by the Greek State Scholarships Foundation. We acknowledge the support of the Yorkshire Agricultural Society.
Editor: F. C. Fang
REFERENCES
- 1.Becker, M. N., G. Diamond, M. W. Verghese, and S. H. Randall. 2000. CD14-dependent lipopolysaccharide-induced beta-defensin-2 expression in human tracheobronchial epithelium. J. Biol. Chem. 275:29731-29736. [DOI] [PubMed] [Google Scholar]
- 2.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boardman, P. E., J. Sanz-Ezquerro, I. M. Overton, D. W. Burt, E. Bosch, W. T. Fong, C. Tickle, W. R. Brown, S. A. Wilson, and S. J. Hubbard. 2002. A comprehensive collection of chicken cDNAs. Curr. Biol. 12:1965-1969. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G., B. Agerberth, and A. Boman. 1993. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 61:2978-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockus, C. W., M. W. Jackwood, and B. G. Harmon. 1998. Characterisation of beta-defensin prepropeptide mRNA from chicken and turkey bone marrow. Anim. Genet. 29:283-289. [DOI] [PubMed] [Google Scholar]
- 6.Chan, Y. R., and R. I. Gallo. 1998. PR-39, a syndecan-inducing antimicrobial peptide, binds and effects p130(Cas). J. Biol. Chem. 273:28978-28985. [DOI] [PubMed] [Google Scholar]
- 7.Diamond, G., V. Kaiser, J. Rhodes, J. P. Russell, and C. L. Bevins. 2000. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect. Immun. 68:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, E. W., F. G. Beach, K. M. Moore, M. W. Jackwood, J. R. Glisson, and B. G. Harmon. 1995. Antimicrobial activity of chicken and turkey heterophils CHP1, CHP2, THP1 and THP3. Vet. Microbiol. 47:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukui, A., N. Inoue, M. Matsumoto, M. Nomura, K. Yamada, Y. Matsuda, K. Toyoshima, and T. Seya. 2001. Molecular cloning and functional characterisation of chicken toll-like receptors. J. Biol. Chem. 276:47143-47149. [DOI] [PubMed] [Google Scholar]
- 10.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 11.Hooper, L. V., T. S. Stappenbeck, C. V. Hong, and J. I. Gordon. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4:269-273. [DOI] [PubMed] [Google Scholar]
- 12.Hormaeche, C. E. 1979. Natural resistance to Salmonella typhimurium in different inbred mouse strains. Immunology 37:311-318. [PMC free article] [PubMed] [Google Scholar]
- 13.Koczulla, A. R., and R. Bals. 2003. Antimicrobial peptides. Current status and therapeutic potential. Drugs 63:389-406. [DOI] [PubMed] [Google Scholar]
- 14.Krause, A., R. Sillard, B. Kleemeier, E. Kluver, E. Maronde, J. R. Conejo-Garcia, W. G. Forssmann, P. Schulz-Knappe, M. C. Nehls, F. Wattler, S. Wattler, and K. Adermann. 2003. Isolation and biochemical characterisation of LEAP-2 a novel blood peptide expressed in the liver. Protein Sci. 12:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynn, D. J., R. Higgs, T. Gaines, J. Tierney, T. James, A. T. Lloyd, M. A. Fares, G. Mulcahy, and C. O. O'Farrelly. 2004. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 56:170-177. [DOI] [PubMed] [Google Scholar]
- 16.Lynn, D. J., A. T. Lloyd, and C. O. O'Farrelly. 2003. In silico identification of components of the Toll-like receptor (TLR) signalling pathway in clustered chicken expressed sequence tags (ESTs). Vet. Immunol. Immunopathol. 93:177-184. [DOI] [PubMed] [Google Scholar]
- 17.Meng, X., B. S. Khanuja, and Y. T. Ip. 1999. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 13:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 19.Salzman, N. H., D. Ghosh, K. M. Huttner, Y. Paterson, and C. L. Bevins. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522-526. [DOI] [PubMed] [Google Scholar]
- 20.Tarver, A. P., D. P. Clark, G. Diamond, J. P. Russell, P. Erdjument-Bromage Tempst, K. S. Cohen, D. E. Jones, R. W. Sweeney, M. Wines, S. Hwang, and C. L. Bevins. 1998. Enteric β-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect. Immun. 66:1045-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells, M., B. Brown, and J. Hall. 2003. Pepsinogen C expression in intestinal IEC-6 cells. Cell. Physiol. Biochem. 13:249-328. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, C. L., A. J. Ouellette, D. P. Satchel, T. Ayabe, Y. S. Lopez-Boado, J. L. Stratman, S. J. Hultgren, L. M. Matrisian, and W. C. Parks. 1998. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defence. Science 286:113-117. [DOI] [PubMed] [Google Scholar]
- 23.Wu, H., G. Zhang, J. E. Minton, C. R. Ross, and F. Blecha. 2000. Regulation of cathelicidin gene expression: induction by lipopolysaccharide, interleukin-6, retinoic acid, and Salmonella enterica serovar Typhimurium infection. Infect. Immun. 68:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, D., A. Biragyn, L. W. Kwak, and J. J. Oppenheim. 2002. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 23:291-296. [DOI] [PubMed] [Google Scholar]
- 25.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. β-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 26.Yount, N. Y., J. Yuan, A. Tarver, T. Castro, G. Diamond, P. A. Tran, L. C. McCullough, J. S. Cullor, C. L. Bevins, and M. E. Selsted. 1999. Cloning and expression of bovine neutrophil beta-defensins. Biosynthetic profile during neutrophilic maturation and localisation of mature peptide to novel cytoplasmic dense granules. J. Biol. Chem. 274:26249-26258. [DOI] [PubMed] [Google Scholar]
- 27.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 28.Zhao, C., T. Nguyen, L. Liu, R. E. Sacco, K. A. Brogden, and R. I. Lehrer. 2001. Gallinacin-3, an inducible epithelial β-defensin in the chicken. Infect. Immun. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]