Summary
Although specific microRNA (miRNA) signatures in classical Hodgkin lymphoma (cHL) have been proposed, their relationship with clinical outcome remains unclear. Despite treatment advances, a substantial subset of patients with advanced cHL are refractory to standard therapies based on adriamycin and its variants. Global miRNA expression data of 29 advanced cHL patients and five cHL-derived cell lines were used to identify profiles from Hodgkin-Reed-Sternberg (HRS) cells and their non-tumoural microenvironment. A cHL-miRNA signature was identified with 234 miRNAs differentially expressed. A subset of these miRNAs was associated with outcome and selected for study in an independent set of 168 cHL samples using quantitative reverse transcription polymerase chain reaction. Multivariate Cox regression analyses including cross-validation with failure-free survival (FFS) as clinical endpoint revealed a miRNA signature with MIR21, MIR30E, MIR30D and MIR92B* that identified two risk-groups with significant differences in 5-year FFS (81% vs. 35% 7%; P < 0·001). Additionally, functional silencing of MIR21 and MIR30D in L428 cells showed increased sensitivity to doxorubicin-induced apoptosis, pointing towards abnormalities of mitochondrial intrinsic and TP53-CDKN1A pathways as related to miRNA deregulation in cHL. These results suggest that clinical outcome in cHL is associated with a specific miRNA signature. Moreover, functional analyses suggest a role for MIR21 and MIR30D in cHL pathogenesis and therapeutic resistance.
Keywords: Hodgkin lymphoma, microRNA, RT-polymerase chain reaction, clinical outcome, molecular pathogenesis
MicroRNAs (miRNAs) are small, non-coding RNA molecules that regulate gene expression at the post-transcriptional level by targeting the 3′-untranslated regions of messenger RNA to promote their degradation or decrease their translation (Esquela-Kerscher & Slack, 2006). In humans, miRNA expression has been shown to be tissue-specific and temporally regulated. MiRNAs are important regulators of biological processes, such as proliferation and differentiation, and may act as oncogenes (‘oncomirs’) or tumour suppressor genes, suggesting an important role for miRNAs in cancerogenesis (Esquela-Kerscher & Slack, 2006). Indeed, most miRNAs are found in cancer-associated genomic regions or in chromosomal fragile sites (Calin et al, 2004) and their role in cancer pathogenesis has been demonstrated in several malignancies, including lymphomas (Arribas et al, 2012; Wang et al, 2012).
Classical Hodgkin lymphoma (cHL) is a lymphoid neoplasm characterized by the presence of relatively few tumour cells, the Hodgkin and Reed-Sternberg (HRS) cells, in a non-neoplastic and reactive cellular microenvironment. Despite progress in the treatment of patients with advanced cHL, a subset of patients fails to respond or suffers early relapse after complete remission. Therefore, identification of biological prognostic markers that can add value to the International Prognostic Score (IPS) would be useful to stratify cHL patients. Deregulation of specific molecular functions, such as apoptosis (Rassidakis et al, 2002; Garcia et al, 2003), cell cycle control,(Garcia et al, 2003; Sanchez-Aguilera et al, 2006) and factors related to the cellular composition of the reactive cellular background, such as the balance between cytotoxic and regulatory T cells (Alvaro et al, 2005, 2008; Tzankov et al, 2008) and the number of macrophages (Steidl et al, 2010; Kamper et al, 2011) have been associated with treatment response and prognosis in cHL patients. Gene expression signatures derived from non-neoplastic cells are correlated with response to therapy and models integrating specific gene signatures either from tumour cells and their reactive microenvironment have been described (Sanchez-Espiridion et al, 2010).
Within this context, several attempts have been made to elucidate the specific miRNA signatures of HRS cells and the cellular microenvironment of cHL. Specific deregulation of miRNAs has been previously described in HL-derived cell lines (Kluiver et al, 2005; Gibcus et al, 2009; Van Vlierberghe et al, 2009) and might be involved in oncogenic pathways targeting genes that affect survival, apoptosis, and B-cell function. Furthermore, preliminary data suggest that abnormal miRNA expression may be associated with clinical characteristics of cHL patients (Kluiver et al, 2005; Navarro et al, 2008, 2009; Gibcus et al, 2009; Van Vlierberghe et al, 2009) and studies of other lymphoma types (Zhao et al, 2010; Montes-Moreno et al, 2011) suggest that specific patterns of miRNA deregulation may participate in primary resistance to therapy, underscoring the potential relevance of miRNA expression analyses to outcome prediction (Chen et al, 2008). However, the possible prognostic role of miRNA signatures in cHL has yet to be well elucidated. In this study, we sought to further clarify the functional and practical relevance of miRNA expression in advanced cHL and identified a four-miRNA signature able to classify cHL patients according to treatment response. This signature included already described oncomirs, such as MIR21 and MIR30D (Medina et al, 2010; Kumar et al, 2011). Furthermore, functional studies with oligonucleotide anti-miRNAs revealed a reduction in cell viability and increased doxorubicin-induced apoptosis after MIR21 and MIR30D knockdown, probably mediated by the mitochondrial intrinsic apoptosis and TP53 pathways. These results point to a possible mechanism for chemoresistance in cHL patients.
Design and methods
Patients and samples
The initial dataset used for miRNA microarray hybridization included pretreatment frozen tissue samples from 29 patients with advanced cHL and five HL-derived cell lines (L428, L540, L1236, HDLM2 and HDMYZ). This dataset was used to define HRS cell and microenvironment miRNA signatures and to identify an initial set of miRNAs related to treatment outcome. The second part of the study enrolled a set of 168 formalin-fixed, paraffin-embedded (FFPE) cHL samples recruited from different institutions associated with the Spanish Hodgkin Lymphoma Study Group and from The University of Texas MD Anderson Cancer Center (Houston, TX). All tissue samples consisted of representative lymph node biopsy specimens obtained before treatment. Patients were eligible for this study if they fulfilled the following stringent criteria: age between 18 and 65 years, advanced cHL defined as Ann Arbor stage IV, III or IIB with a bulky mass, proven HIV-negative status, first line standard chemotherapy regimen that included doxorubicin, mainly ABVD [adriamycin® (doxorubicin), bleomycin, vinblastine, and dacarbazine] (Diehl et al, 2003) and had follow up of at least 12 months after initial treatment. Patients who had achieved complete remission (CR) for at least 12 months were considered to have had favourable outcomes, while those who failed to achieve CR or who relapsed within 12 months were considered to have had unfavourable outcomes. All study participants gave written informed consent, and the study was approved by the relevant Institutional Review Boards of the participant institutions.
Microarray hybridization
For the microarray hybridization study of freshly frozen tissue samples, RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA). After extraction, quality was assessed using a bioanalyser (Agilent Technologies Inc, Santa Clara, CA, USA), and 100 ng of total RNA were hybridized on an Agilent 8 × 15K human miRNA version 2 one-colour platform (Agilent Technologies Inc) containing probes for 723 human and 76 human viral miRNAs according to manufacturer guidelines (Agilent Technologies). The data were extracted by Feature Extraction software (Agilent Technologies) and between-array median normalization was then carried out.
Supervised analysis and identification of tumoural and microenvironment signatures
The analysis of the initial gene expression dataset was performed using the Pomelo II tool, available at http://pomelo2.bioinfo.cnio.es. To identify genes differentially expressed between cHL tumour samples and cell lines, we used a supervised method based on the Student’s T test with a correction for multiple testing. Unadjusted P values were obtained from 100 000 permutations of the dataset and false discovery rates (FDRs) were calculated by the method of Benjamini and Hochberg, considering genes with an FDR < 0·15 as differentially expressed. This comparison between miRNA expression profiles of cHL derived cell lines and tumour samples was performed in order to identify specific miRNA profiles from the tumour cells and their non-tumoural microenvironment without consideration of patient outcomes.
MiRNA expression by Quantitative-polymerase chain reaction (Q-PCR), using RNA from FFPE tissue
For FFPE samples, miRNA was extracted by using a phenolchlorophorm standard protocol after a deparaffinization step as reported previously (Sanchez-Espiridion et al, 2009). Q-PCR was performed for 12 of the 34 initially identified miRNAs associated with clinical outcome (Table II). This selection was based on their biological interest, classification strength and amplification efficiency to ensure high quality and fidelity of the assays (Table SI). Briefly, 250 ng of total miRNA was reverse-transcribed using a multiplex looped primer pool with miRNA probes selected for the study according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA). Each completed reaction was loaded onto the 384-well plate and Q-PCR was done on the ABI 7900HT Prism (Applied Biosystems) using SNORD44 and SNORD48 as endogenous controls for normalization. Ct values were exported using SDS software (SDS 2.2, Life Technologies, Carslbad, CA, USA), and data were analysed with Real-Time STATMINER software (INTEGROMICS TM; www.Integromics.com). All assays were performed in triplicate and results that did not meet methodological quality control criteria were omitted, considering a miRNA to be present if the Ct was less than 35 in all three biological replicates. Quantification of relative miRNA expression was performed using the delta Ct method, using -deltaCT values [–(Ct value of miRNA of interest–mean Ct value for SNORD44and SNORD48)] for further statistical analyses.
Table II.
cHL miRNA signature composition. Univariate regression analysis was done to identify the differentially expressed miRNAs between outcomes (P < 0·05 as cut-off value). Signatures expressed either from HRS cells or their reactive microenvironment were identified confronting the gene expression profiles of whole 29 frozen tissue samples to those of cHL derived cell lines. miRNA expressed by cHL cell lines were included in the HRS miRNA signature whereas those ones either not expressed or with lower expression levels by cHL cell lines were attributed to microenvironment.
| Microenvironment signature
|
HRS cells signature
|
||||
|---|---|---|---|---|---|
| miRNA | P value | Cell line expression | miRNA | P value | Cell line expression |
| MIR92B* | 0·005 | Low | MIR15B* | 0·036 | Low |
| MIR26A | 0·026 | Low | MIR17* | 0·006 | Low |
| MIR27B* | 0·045 | Low | MIR216A | 0·049 | Low |
| MIR132 | 0·006 | No | MIR23A | 0·048 | Low |
| MIR148B* | 0·001 | No | MIR25 | 0·024 | Low |
| MIR204 | 0·001 | No | MIR30D | 0·029 | Low |
| MIR300 | 0·030 | No | MIR30E | 0·037 | Low |
| MIR31* | 0·012 | No | MIR320 | 0·036 | Low |
| MIR423-5p | 0·032 | No | MIR93 | 0·049 | Low |
| MIR503 | 0·016 | No | MIR21 | 0·037 | Yes |
| MIR539 | 0·004 | No | MIR21* | 0·013 | Yes |
| MIR559 | 0·037 | No | MIR92A | 0·030 | Yes |
| MIR609 | 0·005 | No | MIR130B | 0·033 | Yes |
| MIR621 | 0·020 | No | MIR378 | 0·009 | Yes |
| MIR628-3p | 0·016 | No | MIR551A | 0·025 | Yes |
| MIR637 | 0·027 | No | MIR708* | 0·017 | Yes |
| MIR888 | 0·049 | No | – | – | – |
| MIR937 | 0·035 | No | – | – | – |
In vitro functional studies
The effect of MIR21 and MIR30D knock-down on cell growth and chemosensitivity was evaluated by transfecting cells with antisense oligonucleotides. Briefly, cHL-derived L428 cells were cultured in RPMI medium (Gibco-BRL, Burlington, Ontario, Canada) supplemented with antibiotics and 10% fetal bovine serum, and maintained at 37°C in 5% CO2. Exponentially growing cells were transfected with 150 nmol/l of anti-MIR21, anti-MIR30D or negative control (Ambion® miRNA Inhibitors, Applied Biosystems) using a capillary electroporation system (Digital Bio, Invitrogen Life Technologies, Barcelona, Spain) and the Neon transfection kit (Invitrogen), in 100 μl R Buffer. After transfection, relative levels of MIR30D and MIR21 after were checked by Q-PCR. Cells transfected with anti-MIR21, anti-MIR30D, or the negative control were treated with doxorubicin at a final concentration of 468 nmol/l (50% inhibitory concentration, IC50). After 24, 48 and 72 h, cells were harvested, washed in phosphate-buffered saline and centrifuged. The cells were then resuspended at 106 cells/100 μl and apoptosis was measured by allophycocyanin-conjugated annexin V (APC-AnnexinV) (BD Biosciences, Erembodegem-Aalst, Belgium) and 4′,6-diamidino-2-phenylindole (DAPI). The cells were immediately processed with afluorescence-activated cell sorting (FACS) scan flow cytometer (BD Biosciences). All experiments were done in quadruplicate.
For Western blot analyses, cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer. About 40 μg of total cellular protein were separated by 5–10% sodium dodecyl sulphate polyacrylamide gel electrophoresis. After electrophoresis, the separated proteins were transferred to Immobilon-P membranes (Millipore, ON, Canada). The primary antibodies used were BCL2 (clone 124, Dako, Glostrup, Denmark), BAX (polyclonal, Dako), BCL2L1 (BCL-XL; NC-1, Novocastra, Laboratories, Newcastle Upon Tyne, UK), TP53 (DO-7, Dako), CDKN1A (p21; Ab-1, Calbiochem, Darmstadt, Germany), and ACTB (Cell Signalling, Beverly, MA, USA). After incubation with secondary antibodies [goat anti-mouse IgG-horseradish peroxidase (HRP) or goat anti-rabbit IgG-HRP, Santa Cruz Technologies, Santa Cruz, CA, USA], the immunoblots were developed using the Amersham ECL™ detection kit (GE Healthcare, Buckinghamshire, UK) and chemiluminescence was detected using the Chemidoc XRS instrument (BioRad, Hercules, CA, USA) according to the manufacturer’s protocol. To measure relative proteins levels, experiments were done in quadruplicate.
Statistical analyses
Differences in the distributions of standard clinical parameters, including age, gender, stage, International Prognostic Score (IPS), the individual variables contained in IPS and outcome, in the miRNA microarray hybridization (discovery) and Q-PCR (FFPE) datasets were tested by Fisher’s exact test. We used response to standard first-line treatment for initial miRNA selection, considering favourable and unfavourable groups, as mentioned above, without considering data from second-line and salvage therapies and/or bone-marrow transplantation. Univariate regression analysis using the Pomelo II tool was used to identify the initial set of miRNAs differentially expressed in the discovery set (n = 29), and to select the most relevant miRNAs to set up a Q-PCR assay for additional study in the FFPE samples, considering genes with P < 0·05 as being differentially expressed. Final identification of miRNAs associated with treatment response was based on data from the FFPE series of cHL tumour samples (n = 168). Those miRNAs initially found to have a potential association with outcome were analysed in a backward stepwise multivariate Cox proportional hazards model with FFS as the endpoint, defined as the time interval between treatment initiation and treatment failure or last follow-up. Treatment failure was defined as either the failure to achieve complete remission, the occurrence of progressive disease, or relapse after initial complete remission. Multivariate models were trained on the data by inputting all miRNAs at once into a bidirectional, stepwise Cox regression algorithm using 10-fold cross-validation. The models were used to make predictions about patients in the testing folds using the MASS, survival, and survival receiver operating characteristic curve (ROC) packages in R 2.14.1(http://cran.r-project.org/bin/windows/base/old/2.14.1/). These patients were then classified into risk groups and plotted on a single, cross-validated Kaplan-Meier curve. Its performance was tested by ROC analysis and finally log-rank test and Kaplan-Meier analyses were used for survival comparison between risk groups, stratified into high- and low-risk Cox groups by the median of the miRNA score in the whole series. All statistical analyses were two-sided, taking P values less than 0·05 to be significant. Kaplan-Meier analyses were performed with SPSS version 19.0 (SPSS Inc., Chicago, IL, USA) and the already mentioned R 2.14.1 packages. Plots were generated using GraphPadPrism v.5 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Clinical parameters
There were no statistically significant differences in the distribution of most clinical parameters (gender, histology, stage, and IPS) between the groups of patients with favourable versus unfavourable outcomes, either in the initial discovery dataset (n = 29) or in the test dataset (n = 168). In addition, no significant differences or associations were found between miRNA expression and specific histological subtypes. Interestingly, confirming our previous observations (Guisado-Vasco et al, 2012) the number of patients older than 45 years of age was significantly higher in the unfavourable group in the FFPE cohort. These characteristics are summarized in Table I.
Table I.
Clinical characteristics of the cHL series. Summary of the clinical characteristics of the patients in the discovery and training sets that yielded suitable analysable data. All patients received anthracyclin-based therapy. Differences in the distribution of standard clinical parameters between favourable and unfavourable outcomes within each dataset were tested by Fisher’s exact test.
| Discovery set (n = 29)
|
P value | FFPE set (n = 168)
|
P value | |||
|---|---|---|---|---|---|---|
| Favourable (n = 23) | Unfavourable (n = 6) | Favourable (n = 104) | Unfavourable (n = 64) | |||
| Age | ||||||
| <45 years | 21 | 4 | 0·25 | 81 | 38 | 0·01 |
| >45 years | 3 | 2 | 22 | 26 | ||
| Missing | 0 | 0 | 1 | 0 | ||
| Gender | ||||||
| Female | 13 | 4 | 0·67 | 50 | 29 | 0·75 |
| Male | 11 | 2 | 54 | 35 | ||
| Missing | 0 | 0 | 0 | 0 | ||
| Histology | ||||||
| Mixed cellularity | 4 | 0 | 0·71 | 30 | 14 | 0·56 |
| Nodular sclerosis | 18 | 6 | 71 | 48 | ||
| Other | 2 | 0 | 2 | 2 | ||
| Missing | 0 | 0 | 1 | 0 | ||
| Stage | ||||||
| <IV (IIB & III) | 14 | 4 | 0·86 | 65 | 32 | 0·14 |
| IV | 8 | 2 | 38 | 31 | ||
| Missing | 1 | 0 | 1 | 1 | ||
| IPS | ||||||
| <3 | 16 | 5 | 0·64 | 36 | 26 | 0·24 |
| ≥3 | 8 | 1 | 63 | 30 | ||
| Missing | 0 | 0 | 5 | 8 | ||
IPS, International Prognostic score; FFPE, Formalin-fixed paraffin-embedded.
Recent reports describe that patients in older age groups do exceptionally worse (Evens et al, 2012). However, when using a different age cut-off (patients older than 60 years), we could not find significant association with unfavourable outcome, probably due to the small size of this subgroup (22 patients).
Supervised analysis and identification of miRNA signatures
Established cHL-derived cell lines undergo expression changes in order to survive in cell suspension. However, they retain the key features of primary HRS cells (Tiacci et al, 2012) and are commonly used as model systems for characterizing the biology of HRS cells, whereas tumoural samples represent a complex mixture of tumoural cells and reactive microenvironment. Thus, using cHL cell lines, we initially assessed their miRNA expression profile. Thereafter, we identified the miRNA profile of cHL tumour samples and compared the results with the data derived from cHL cell lines. The supervised T-test identified 234 miRNAs that were differentially expressed [false discovery rate (FDR) < 0·15] in cHL tumour samples and these miRNAs are attributable to both the HRS cells and the cellular microenvironment. The miRNAs overexpressed in cHL cell lines (134 of 234) were considered as the HRS miRNA signature, whereas miRNAs overexpressed in tumour samples (100 of 234) were considered to represent a microenvironment signature. Some of the miRNAs identified as part of this cHL signature have been previously described in this disease, such as MIR21, MIR31, MIR155 and MIR17HG (also known as MIR17-92) cluster members MIR17, MIR19A (Navarro et al, 2008; Gibcus et al, 2009). Other members included in the signatures, such as MIR30D, MIR30E, MIR590-5p, MIR143, MIR145 and MIR126, have been described in different tumour types (Michael et al, 2003; Yao et al, 2010).
Identification of candidate miRNAs associated with outcome
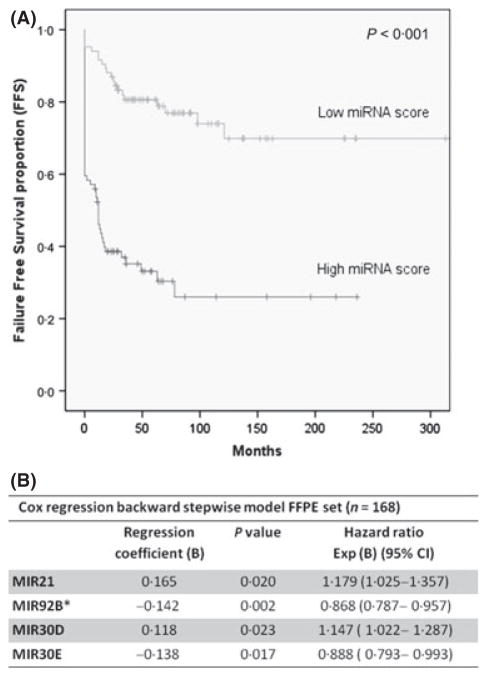
The study’s flowchart is depicted in Fig 1. Based on univariate regression analyses, when patient outcome was taken into account, a set of 34 miRNAs were identified as differentially expressed (P < 0·05) among patients with favourable and unfavourable outcomes, and considered for further evaluation in FFPE cHL tumour samples (Table II). From this set, some miRNAs showed remarkable expression in the cHL cell lines and thus probably reflect functional properties of the HRS cells, whereas other miRNAs not expressed in cHL cell lines probably reflect functional properties of the reactive cellular infiltrate (Figure S1). Using a backward stepwise selection algorithm, a multivariate Cox model for the entire series was constructed with FFS as the final endpoint, including a 1000 bootstrapping process. The final derived signature included four miRNAs that were significantly associated with FFS: MIR21, MIR92B*, MIR30D and MIR30E (Fig 2B). Classification accuracy of this miRNA signature was confirmed using the integrated area under the ROC curve (ROC value = 0·765; P = 0·000). When 10-fold cross-validation was applied, multivariate miRNA models were built for comparison. miRNAs most frequently found in the different trained models consisted of MIR21, MIR30D, MIR30E and MIR92B* (Table SII), in agreement with the miRNA signature identified initially. Details of the models, including cut-off values, trained on the data by use of the stepwise Cox regression algorithm using 10-fold cross-validation are shown in the Supporting Information. Moreover, this miRNA signature was able to stratify patients into low- and high-risk groups, respectively (cut-off point = 0·633) according to its median expression. Thus, Kaplan-Meier estimates of predicted risk groups showed significant differences in FFS probability between the low-risk and the high-risk groups with 5-year FFS of 81% vs. 35·7% (P < 0·001; Fig 2A). Moreover, the miRNA signature was also independent of the IPS and of age in multivariate Cox regression analysis (Table SIII).
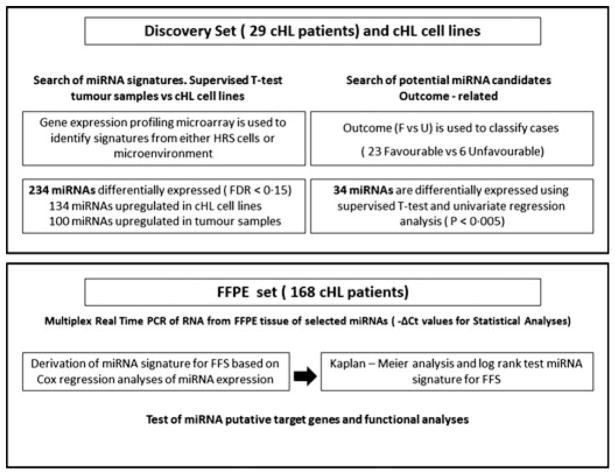
Fig 1.
Experimental design. The whole series of patients was divided in two major groups, a discovery group of 29 patients with available frozen tissue for miRNA profiling using array technologies and an additionally independent group of 168 patients with available formalin-fixed paraffin-embedded (FFPE) tissue from the diagnostic pathological sample that was further evaluated by ten-fold cross-validation. The clinical characteristics of both datasets are summarized in Table I. cHL, classical Hodgkin lymphoma; FDR, false discovery rate; F, favourable; U, unfavourable; PCR, polymerase chain reaction; FFS, failure-free survival.
Fig 2.
Survival analyses of the miRNA signature. (A) Kaplan-Meier estimates of the miRNA signature identified by Cox regression model. MiRNA-based Cox survival scores were calculated for the whole patient series (N = 168). The miRNA score was defined as the continuous survival function derived from the Cox regression model, being afterwards validated by leave one out cross-validation. After median stratification patients were classified into high and low risk groups with significant differences in failure-free survival (FFS; log rank test. P < 0·001). (B) Cox regression backward stepwise analysis for FFS in the formalin-fixed paraffin-embedded (FFPE) dataset (n = 168). 95% CI, 95% confidence interval; Exp (B), exponential B.
Given that age has been demonstrated as a major factor associated with outcome in cHL (Evens et al, 2012; Guisado-Vasco et al, 2012), we analysed the predictive power of the model for the selection of patients older than 45 years (48 cases) and older than 60 years (22 cases), finding significant results in both subgroups of age.
MIR21 and MIR30D sensitizes L428 cells to doxorubicin-induced apoptosis
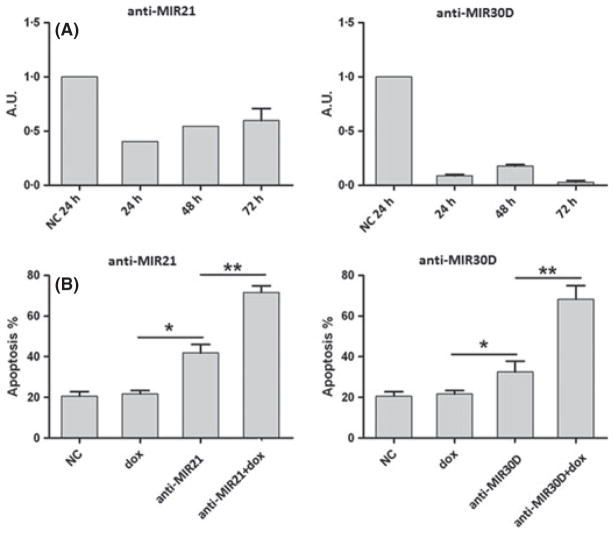
We selected the L428 cell line for functional experiments because expression levels of MIR21 and MIR30D were higher in L428 cells, in terms of fold change, compared with the other cHL-derived cell lines. Additionally, L428 cells represent a well-known model of drug resistance (Abu-Ghanem et al, 2007). As expected, doxorubicin treatment resulted in low apoptosis levels, with an IC50 value of 468·3 mmol/l in basal conditions. To further explore the potential association of MIR21 and MIR30D with chemoresistance, cells were transfected with MIR21- and/or MIR30D-specific antisense inhibitors. The transfection efficiency was assessed by MIR21 and MIR30D Q-PCR, showing a median decrease of 48·5% and 90·3% for MIR21 and MIR30D expression levels, respectively (Fig 3A). Furthermore, silencing of both miRNAs induced a moderate increase in apoptosis (Fig 3B). Given that overexpression of both MIR21 and MIR30D was found to be associated with the therapy-resistant group of cHL patients, we studied the effects on doxorubicin sensitivity after miRNA knockdown. In agreement with previous observations (Mei et al, 2010; Yao et al, 2010), we observed increased apoptosis after transfection with the antisense inhibitors. As shown in Fig 3B, transfection with either anti-MIR21 or anti-MIR30D resulted in substantially increased apoptosis when combined with doxorubicin treatment. Combined co-transfection with anti-MIR21 and anti-MIR30D did not result in additive effects or synergy (data not shown).
Fig 3.
Oligonucleotide anti-miRNA in vitro studies. The L428 cell line was pretreated with oligonucleotide anti-MIR21 and anti-MIR30D at 150 nmol/l concentration for 24 h, followed by doxorubicin treatment for an additional 24, 48 and 72 h. (A) Q-PCR analyses of MIR21 and MIR30D after miRNA inhibition showing miRNA downregulation expression levels. (B) Increased doxorubicin-mediated apoptosis was observed after miRNA transfection at all-time points with significant differences at 72 h. NC, negative control; dox, doxorubicin; *P < 0·05; **P < 0·01.
MIR21 and MIR30D target analyses by Western-blot
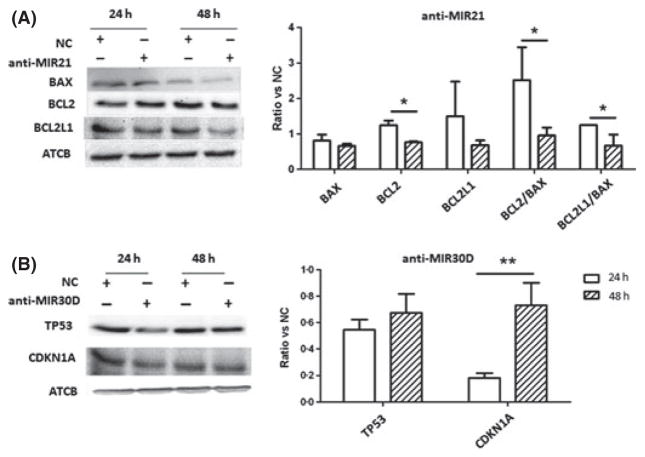
We performed Western blot analyses for selected genes to analyse potential pathways involved in the biological effect of MIR21 and MIR30D. We selected these putative targets based on prediction algorithms (TargetScan, http://www.targetscan.org, and miRanda http://www.microrna.org), considering also putative targets previously reported in different cell models (Shi et al, 2010; Kumar et al, 2011). Inhibition of MIR21 expression was associated with decreased BCL2/BAX and BCL2L1/BAX ratios. On the other hand, downregulation of MIR30D expression was associated with increased CDKN1A levels, associated with a lower increase of TP53 protein levels, consistent with the activation of a functional TP53 pathway (Fig 4).
Fig 4.
Western blot analyses after MIR21 and MIR30d inhibition. (A) Inhibition of MIR21 expression is associated with decreasing of BCL2/BAX and BCL2L1/BAX ratios. (B) Downregulation of MIR30D expression is associated with an increase of TP53 and CDKN1A proteins. Histograms illustrate protein levels normalized to ACTB; each bar represents mean ± standard error of quadruplicate cultures. *P < 0·05; **P < 0·01.
Discussion
Classical HL, at one time an aggressive B-cell lymphoma with fatal outcome because adequate therapy was not available, is now often cured with current chemotherapy regimens, particularly in patients with localized disease. However, a fraction of patients with advanced cHL present with primary refractory tumours that do not completely respond to therapy, or who relapse after short periods of remission (Diehl et al, 2003; Duggan et al, 2003). For this reason, current research is focused on identifying patients with a low probability of cure and who may benefit from novel therapies.
In recent years, advances in our understanding of cHL biology and interactions between HRS cells and the tumour microenvironment have led to the discovery of new potentially prognostic factors associated with treatment failure. We (Garcia et al, 2003; Montalban et al, 2004) and others (Smolewski et al, 2000; Rassidakis et al, 2002; Bai et al, 2007) previously showed the prognostic value of markers of cell cycle deregulation and apoptosis, such as BCL2, BCL2L1, or TP53. Recent evidence also suggests that the cellular composition of the tumour microenvironment can modify clinical outcome in cHL patients, with suggested factors including the reactive T-cell phenotype (Alvaro et al, 2005, 2008; Tzankov et al, 2008) or monocyte/macrophage activation (Steidl et al, 2010; Kamper et al, 2011). In the last decade, new molecular profiling technologies have identified biological alterations and new prognostic factors that may be useful for risk stratification and predicting response to chemotherapy (Devilard et al, 2002; Chetaille et al, 2009; Steidl et al, 2010). Our group has recently described a gene expression model that included 11 genes belonging to four functional pathways: apoptosis, cell-cycle regulation, macrophage activation, and interferon regulatory factor 4. This model is associated with risk of cHL relapse (Sanchez-Espiridion et al, 2010). Additionally, data from gene expression studies have identified a gene signature of tumour-associated macrophages that was associated with primary treatment failure in cHL patients (Steidl et al, 2010). The results of these studies suggest that specific gene signatures are useful for predicting outcome, and that factors related to HRS cells as well as the reactive microenvironment, may be associated with therapy response.
Preliminary studies have shown that miRNA losses and gains may explain some of the biological and clinical features of different lymphoma types, and recent analyses of miRNA expression in cHL cell lines and tumours have demonstrated deregulated expression of some miRNAs. Overexpression of MIR155 has been found in cHL cell lines by Q-PCR analysis (Kluiver et al, 2005). MiRNA profiling of cHL-derived cell lines also has revealed specific miRNAs including MIR17HG cluster members, MIR16, MIR21, MIR24 and MIR155 (Gibcus et al, 2009). In primary cHL tumour samples, comparison of miRNA profiles of microdissected HRS cells, and analysis of four common cHL cell lines (HDLM2, L540, KMH2 and L1236) and CD77+ normal germinal centre B-cells yielded a distinct cHL signature of 12 over- and three under-expressed miRNAs (Van Vlierberghe et al, 2009). Some preliminary observations suggest that expression of selected miRNAs can be associated with clinical characteristics in cHL patients. Thus, overexpression of MIR328 has been found in advanced cHL stages (III–IV stages; Navarro et al, 2008), and low MIR135 levels are associated with higher relapse rates and shorter disease-free survival (Navarro et al, 2009). However, the potential prognostic role of miRNA signatures in cHL has not been yet fully investigated in larger and more homogeneous cHL series.
The present study was designed for hypothesis generation rather than a model for outcome prediction. The main goal was to propose a miRNA signature that could be associated with clinical outcome in advanced cHL patients treated with ABVD or similar therapies, and technical validation using FFPE tissue samples. MiRNA expression and the supervised T-test identified specific miRNA profiles from HRS cells and their non-tumoural cellular microenvironment, with 234 miRNAS differentially expressed at significant levels. Recent studies have shown that cHL cell lines, although differing from primary HRS cells, retain their key features (Tiacci et al, 2012). Thus, within the identified profiles, a set of 34 miRNAs was differentially expressed between patients with favourable and unfavourable outcome with some miRNAs showing remarkable expression in the cHL cell lines and thus probably reflecting functional properties of the HRS cells, whereas the other miRNAs probably reflecting the reactive cellular microenvironment. Using Q-PCR in an independent series of 168 patients, a final Cox model identified four miRNAs associated with FFS that, interestingly, have been described previously as being implicated in tumourigenesis: MIR21, MIR30D, MIR30E, and MIR92B. Here we further analysed the functional consequences of MIR21 and MIR30D overexpression because both miRNAs were highly expressed in the cHL-derived cell lines and upregulated in the unfavourable group of cHL patients.
Overexpression of MIR21 has been reported in cHL and other lymphoma types (Navarro et al, 2008; Gibcus et al, 2009; Rossi et al, 2010). Additionally, MIR21 overexpression has been associated with poorer prognosis in many tumour types including squamous cell carcinoma (Gao et al, 2011), oesophageal cancer (Hummel et al, 2011), colorectal adenocarcinoma (Kulda et al, 2010), pancreatic cancer (Hwang et al, 2010), and astrocytoma (Zhi et al, 2010). Different mouse models also have yielded additional evidence for a specific role for MIR21 in tumourigenesis. Anti-MIR21 treatment inhibited cell growth in vitro, and inhibited tumour growth in a breast cancer mouse model (Si et al, 2007). A mouse model conditionally expressing MIR21 led to a pre-B-cell lymphoma (Medina et al, 2010), and when MIR21 was inactivated, the tumours regressed completely. This study additionally identified MIR30D as related to outcome. This miRNA has been reported to promote tumour invasion and metastasis in hepatocellular carcinoma, and a prognostic signature for this tumour also included this miRNA (Yao et al, 2010). MIR30D is co-amplified with MIR30B (8q24.22-23) in medulloblastoma, independently of MYC amplification (Lu et al, 2009), and has been shown to negatively regulate TP53 expression, inhibiting cellular apoptosis in several cell lines, including one from multiple myeloma without TP53 mutations (Kumar et al, 2011). Finally, the other two members of the identified signature, MIR30E and MIR92B*, have also been related to several cancer types. Thus, MIR30E has been related to metastasis-related hepatocellular carcinoma (Budhu et al, 2008) whereas MIR92B* has been related to colorectal and cervical carcinomas.
Analyses using the L428 cell line demonstrated a link between upregulation of MIR21 and MIR30D and doxorubicin-induced apoptosis, consistent with the observed overexpression of these two miRNA in the treatment-resistant group of patients. These results are in agreement with previous observations in other tumour cell types, such as in cell lines of leukaemia (Li et al, 2010; Bai et al, 2011), pancreatic adenocarcinoma (Hwang et al, 2010), breast cancer (Mei et al, 2010), or glioblastoma (Shi et al, 2010), and can provide additional rationale for targeting miRNAs in cHL and other cancers (Garzon et al, 2010). As could be expected, observed modifications at the protein level after MIR21 (BAX, BCL2, and BCL2L1), and MIR30D knockdown (TP53 and CDKN1A) are subtle, probably indicating indirect and complex modulation of the intrinsic mitochondrial pathway and CDKN1A-mediated apoptosis respectively. Compared with control cells, cHL cells after MIR21 inhibition showed a decreased BCL2/BAX ratio and, more slightly, the BCL2L1/BAX ratio, in agreement with observations in other tumour models (Shi et al, 2010). These alterations suggest that overexpression of MIR21 could inhibit apoptosis, at least in part, by increasing this ratio, highlighting the potential role of MIR21 in clinical resistance to therapy in cHL patients. Additionally, when the MIR30D inhibitor was introduced we observed increased CDKN1A protein levels, with a slight (although not significant) increase in TP53 protein levels, consistent with negative regulation of the TP53 pathway by MIR30D. Recent studies using multiple myeloma cell lines with a wild-type TP53 gene showed that MIR30D directly targets the 3′ untranslated region (UTR) of TTP53 to down-regulate TP53 protein levels and reduce the expression of genes that are transcriptionally activated by TP53 (Kumar et al, 2011), as a mechanism to negatively regulate apoptosis.
In conclusion, we have identified a miRNA signature related to outcome in patients with advanced cHL. This miRNA signature represents an integration of tumour (HRS) cells and microenvironment factors. In addition, functional analyses for MIR30D and MIR21 revealed their potential value as modulators of the therapeutic response in cHL patients. Proper validation of the current data is needed in an independent cohort and future studies should address the specific prognostic value of these miRNAs in the context of other biological and clinical variables. However, our results suggest that overexpression of these miRNAs can be associated with inhibition of drug-induced apoptosis, and could be linked with the clinical resistance to chemotherapy in some cHL patients.
Supplementary Material
Fig S1. Unsupervised clustering and heatmap image from the initially identified 34 miRNAs outcome related and cHL cell lines.
Table SI. Assay details miRNAS used for quantitative PCR (Q-PCR) analyses of the FFPE dataset (n = 168).
Table SII. miRNAs retained in the different Cox backward stepwise models in Crossvalidation Runs.
Table SIII. Multivariate analysis using Cox proportional hazards regression model in the FFPE cohort.
Appendix S1. Members of the Spanish Hodgkin Lymphoma Study Group.
Data S1. miRNA selection, cross-validation runs details and R scripts.
Acknowledgments
The authors thank María E. Castillo and the CNIO Tumour Bank Unit (María J. Artiga, Laura Cereceda, and Manuel Morente) for their skillful retrieval and handling of clinical data from different clinical institutions. Special thanks also to Douglas A. Mata, from Baylor College of Medicine (Houston), for his invaluable contributions to the biostatistics analyses and for his perceptive discussions of the manuscript. We also acknowledge all of the clinical colleagues who kindly completed the clinical data form and Wesley O. Greaves for his kind help when selecting cases from MD Anderson Cancer Center (Houston). Members of the Spanish Lymphoma Study Group are cited in the Appendix S1. This work was supported by grants from the Fondo de Investigaciones Sanitarias (FIS, PI08/1985 and PI12/1832, RTICC RD06/0020/0107), Ministerio de Educación y Ciencia, Spain (SAF2008-03871), the Association for Cancer Research Spanish (AECC), and from the U.T M.D. Anderson Cancer Center Funds.
Footnotes
Members of the Spanish Hodgkin Lymphoma Study Group are listed in Appendix S1.
Conflict of interest
The authors have no competing interest.
Author’s contributions
Conception and design: C. Montalban, M.A. Piris, J.F. Garcia. Development of methodology: B. Sanchez-Espiridion, A.M. Martin-Moreno, M.E. Rodriguez, V. Figueroa. Acquisition of data (provided animals or samples, acquired and managed patients, provided facilities, etc.): F. Vega, A. Younes, L.J. Medeiros, F.J. Alves, M. Canales, M. Estevez, J. Menarguez, P. Sabin, M.C. Ruiz-Marcellan, A. Lopez, P. Sanchez-Godoy, F. Burgos, C. Santonja, M. M. Morente, J.L. Lopez. Analysis and interpretation of the data (e.g. Statistical analysis, biostatistics, computational analysis): B. Sanchez-Espiridion, A.M. Martin-Moreno, V. Figueroa. Writing, review, and/or revision of the manuscript: B. Sanchez-Espiridion, A. M. Martin- Moreno, F. Vega, A. Younes, L.J. Medeiros, F.J. Alves, M. Canales, M. Estevez, J. Menarguez, P. Sabin, M.C. Ruiz-Marcellan, A. Lopez, P. Snachez-Godoy, F. Burgos, C. Santonja,M. M. Morente, J.L. Lopez. Administrative,technical, or material support (i.e., reporting or organizing data, constructing databases): B. Sanchez-Espiridion, A.M. Martin-Moreno, J.F. Garcia. Study supervision: C. Montalban, M.A. Piris and J.F. Garcia.
Supporting Information
Additional Supporting Information may be found in the online version of this article
References
- Abu-Ghanem S, Oberkovitz G, Benharroch D, Gopas J, Livneh E. PKCeta expression contributes to the resistance of Hodgkin’s lymphoma cell lines to apoptosis. Cancer Biology & Therapy. 2007;6:1375–1380. doi: 10.4161/cbt.6.9.4527. [DOI] [PubMed] [Google Scholar]
- Alvaro T, Lejeune M, Salvado MT, Bosch R, Garcia JF, Jaen J, Banham AH, Roncador G, Montalban C, Piris MA. Outcome in Hodgkin’s lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clinical Cancer Research. 2005;11:1467–1473. doi: 10.1158/1078-0432.CCR-04-1869. [DOI] [PubMed] [Google Scholar]
- Alvaro T, Lejeune M, Garcia JF, Salvado MT, Lopez C, Bosch R, Jaen J, Escriva P, Pons LE. Tumor-infiltrated immune response correlates with alterations in the apoptotic and cell cycle pathways in Hodgkin and Reed-Sternberg cells. Clinical Cancer Research. 2008;14:685–691. doi: 10.1158/1078-0432.CCR-07-1246. [DOI] [PubMed] [Google Scholar]
- Arribas AJ, Campos-Martin Y, Gomez-Abad C, Algara P, Sanchez-Beato M, Rodriguez-Pinilla MS, Montes-Moreno S, Martinez N, Alves-Ferreira J, Piris MA, Mollejo M. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119:e9–e21. doi: 10.1182/blood-2011-02-339556. [DOI] [PubMed] [Google Scholar]
- Bai M, Papoudou-Bai A, Horianopoulos N, Grepi C, Agnantis NJ, Kanavaros P. Expression of bcl2 family proteins and active caspase 3 in classical Hodgkin’s lymphomas. Human Pathology. 2007;38:103–113. doi: 10.1016/j.humpath.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Bai H, Xu R, Cao Z, Wei D, Wang C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Letters. 2011;585:402–408. doi: 10.1016/j.febslet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, Tang ZY, Wang XW. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Kitabayashi N, Zhou XK, Fahey TJ, 3rd, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Modern Pathology. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- Chetaille B, Bertucci F, Finetti P, Esterni B, Stamatoullas A, Picquenot JM, Copin MC, Morschhauser F, Casasnovas O, Petrella T, Molina T, Vekhoff A, Feugier P, Bouabdallah R, Birnbaum D, Olive D, Xerri L. Molecular profiling of classical Hodgkin lymphoma tissues uncovers variations in the tumor microenvironment and correlations with EBV infection and outcome. Blood. 2009;113:2765–3775. doi: 10.1182/blood-2008-07-168096. [DOI] [PubMed] [Google Scholar]
- Devilard E, Bertucci F, Trempat P, Bouabdallah R, Loriod B, Giaconia A, Brousset P, Granjeaud S, Nguyen C, Birnbaum D, Birg F, Houlgatte R, Xerri L. Gene expression profiling defines molecular subtypes of classical Hodgkin’s disease. Oncogene. 2002;21:3095–3102. doi: 10.1038/sj.onc.1205418. [DOI] [PubMed] [Google Scholar]
- Diehl V, Franklin J, Pfreundschuh M, Lathan B, Paulus U, Hasenclever D, Tesch H, Herrmann R, Dorken B, Muller-Hermelink HK, Duhmke E, Loeffler M. Standard and increased-dose BEACOPP chemotherapy compared with COPP-ABVD for advanced Hodgkin’s disease. New England Journal of Medicine. 2003;348:2386–2395. doi: 10.1056/NEJMoa022473. [DOI] [PubMed] [Google Scholar]
- Duggan DB, Petroni GR, Johnson JL, Glick JH, Fisher RI, Connors JM, Canellos GP, Peterson BA. Randomized comparison of ABVD and MOPP/ABV hybrid for the treatment of advanced Hodgkin’s disease: report of an intergroup trial. Journal of Clinical Oncology. 2003;21:607–614. doi: 10.1200/JCO.2003.12.086. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Evens AM, Helenowski I, Ramsdale E, Nabhan C, Karmali R, Hanson B, Parsons B, Smith S, Larsen A, McKoy JM, Jovanovic B, Gregory S, Gordon LI, Smith SM. A retrospective multicenter analysis of elderly Hodgkin lymphoma: outcomes and prognostic factors in the modern era. Blood. 2012;119:692–695. doi: 10.1182/blood-2011-09-378414. [DOI] [PubMed] [Google Scholar]
- Gao W, Shen H, Liu L, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. Journal of Cancer Research and Clinical Oncology. 2011;137:557–566. doi: 10.1007/s00432-010-0918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Camacho FI, Morente M, Fraga M, Montalban C, Alvaro T, Bellas C, Castano A, Diez A, Flores T, Martin C, Martinez MA, Mazorra F, Menarguez J, Mestre MJ, Mollejo M, Saez AI, Sanchez L, Piris MA. Hodgkin and Reed-Sternberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue microarrays. Blood. 2003;101:681–689. doi: 10.1182/blood-2002-04-1128. [DOI] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature Reviews Drug Discovery. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Tan LP, Harms G, Schakel RN, de Jong D, Blokzijl T, Moller P, Poppema S, Kroesen BJ, van den Berg A. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–176. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisado-Vasco P, Arranz-Saez R, Canales M, Canovas A, Garcia-Larana J, Garcia-Sanz R, Lopez A, Lopez JL, Llanos M, Moraleda JM, Rodriguez J, Rayon C, Sabin P, Salar A, Marin-Niebla A, Morente M, Sanchez-Godoy P, Tomas JF, Muriel A, Abraira V, Piris MA, Garcia JF, Montalban C. Stage IV and age over 45 years are the only prognostic factors of the International Prognostic Score for the outcome of advanced Hodgkin lymphoma in the Spanish Hodgkin Lymphoma Study Group series. Leukaemia & Lymphoma. 2012;53:812–819. doi: 10.3109/10428194.2011.635861. [DOI] [PubMed] [Google Scholar]
- Hummel R, Hussey DJ, Michael MZ, Haier J, Bruewer M, Senninger N, Watson DI. MiRNAs and their association with loco-regional staging and survival following surgery for esophageal carcinoma. Annals of Surgical Oncology. 2011;18:253–260. doi: 10.1245/s10434-010-1213-y. [DOI] [PubMed] [Google Scholar]
- Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, Kim SW, Del Chiaro M, Peters GJ, Giaccone G. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamper P, Bendix K, Hamilton-Dutoit S, Honore B, Nyengaard JR, d’Amore F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin’s lymphoma. Haematologica. 2011;96:269–276. doi: 10.3324/haematol.2010.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. Journal of Pathology. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Kulda V, Pesta M, Topolcan O, Liska V, Treska V, Sutnar A, Rupert K, Ludvikova M, Babuska V, Holubec L, Jr, Cerny R. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genetics and Cytogenetics. 2010;200:154–160. doi: 10.1016/j.cancergencyto.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS, Young KH, Li Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30:843–853. doi: 10.1038/onc.2010.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu X, Gu J, Dong D, Yao J, Lin C, Huang K, Fei J. Anti-miR-21 oligonucleotide sensitizes leukemic K562 cells to arsenic trioxide by inducing apoptosis. Cancer Science. 2010;101:948–954. doi: 10.1111/j.1349-7006.2010.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ryan SL, Elliott DJ, Bignell GR, Futreal PA, Ellison DW, Bailey S, Clifford SC. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS ONE. 2009;4:e6159. doi: 10.1371/journal.pone.0006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Mei M, Ren Y, Zhou X, Yuan XB, Han L, Wang GX, Jia Z, Pu PY, Kang CS, Yao Z. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technology in Cancer Research & Treatment. 2010;9:77–86. doi: 10.1177/153303461000900109. [DOI] [PubMed] [Google Scholar]
- Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Molecular Cancer Research. 2003;1:882–891. [PubMed] [Google Scholar]
- Montalban C, Garcia JF, Abraira V, Gonzalez-Camacho L, Morente MM, Bello JL, Conde E, Cruz MA, Garcia-Sanz R, Garcia-Larana J, Grande C, Llanos M, Martinez R, Flores E, Mendez M, Ponderos C, Rayon C, Sanchez-Godoy P, Zamora J, Piris MA. Influence of biologic markers on the outcome of Hodgkin’s lymphoma: a study by the Spanish Hodgkin’s Lymphoma Study Group. Journal of Clinical Oncology. 2004;22:1664–1673. doi: 10.1200/JCO.2004.06.105. [DOI] [PubMed] [Google Scholar]
- Montes-Moreno S, Martinez N, Sanchez-Espiridion B, Diaz Uriarte R, Rodriguez ME, Saez A, Montalban C, Gomez G, Pisano DG, Garcia JF, Conde E, Gonzalez-Barca E, Lopez A, Mollejo M, Grande C, Martinez MA, Dunphy C, Hsi ED, Rocque GB, Chang J, Go RS, Visco C, Xu-Monette Z, Young KH, Piris MA. miRNA expression in diffuse large B-cell lymphoma treated with chemoimmunotherapy. Blood. 2011;118:1034–1040. doi: 10.1182/blood-2010-11-321554. [DOI] [PubMed] [Google Scholar]
- Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, Gel B, Abrisqueta P, Lopez-Guillermo A, Artells R, Montserrat E, Monzo M. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- Navarro A, Diaz T, Martinez A, Gaya A, Pons A, Gel B, Codony C, Ferrer G, Martinez C, Montserrat E, Monzo M. Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood. 2009;114:2945–2951. doi: 10.1182/blood-2009-02-204842. [DOI] [PubMed] [Google Scholar]
- Rassidakis GZ, Medeiros LJ, Vassilakopoulos TP, Viviani S, Bonfante V, Nadali G, Herling M, Angelopoulou MK, Giardini R, Chilosi M, Kittas C, McDonnell TJ, Bonadonna G, Gianni AM, Pizzolo G, Pangalis GA, Cabanillas F, Sarris AH. BCL-2 expression in Hodgkin and Reed-Sternberg cells of classical Hodgkin disease predicts a poorer prognosis in patients treated with ABVD or equivalent regimens. Blood. 2002;100:3935–3941. doi: 10.1182/blood.V100.12.3935. [DOI] [PubMed] [Google Scholar]
- Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, Fabbri M, Lerner S, Barron LL, Rassenti LZ, Jiang L, Xiao L, Hu J, Secchiero P, Zauli G, Volinia S, Negrini M, Wierda W, Kipps TJ, Plunkett W, Coombes KR, Abruzzo LV, Keating MJ, Calin GA. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Aguilera A, Montalban C, de la Cueva P, Sanchez-Verde L, Morente MM, Garcia-Cosio M, Garcia-Larana J, Bellas C, Provencio M, Romagosa V, de Sevilla AF, Menarguez J, Sabin P, Mestre MJ, Mendez M, Fresno MF, Nicolas C, Piris MA, Garcia JF. Tumor microenvironment and mitotic checkpoint are key factors in the outcome of classic Hodgkin lymphoma. Blood. 2006;108:662–668. doi: 10.1182/blood-2005-12-5125. [DOI] [PubMed] [Google Scholar]
- Sanchez-Espiridion B, Sanchez-Aguilera A, Montalban C, Martin C, Martinez R, Gonzalez-Carrero J, Poderos C, Bellas C, Fresno MF, Morante C, Mestre MJ, Mendez M, Mazorra F, Conde E, Castano A, Sanchez-Godoy P, Tomas JF, Morente MM, Piris MA, Garcia JF. A TaqMan low-density array to predict outcome in advanced Hodgkin’s lymphoma using paraffin-embedded samples. Clinical Cancer Research. 2009;15:1367–1375. doi: 10.1158/1078-0432.CCR-08-1119. [DOI] [PubMed] [Google Scholar]
- Sanchez-Espiridion B, Montalban C, Lopez A, Menarguez J, Sabin P, Ruiz-Marcellan C, Ramos R, Rodriguez J, Canovas A, Camarero C, Canales M, Alves J, Arranz R, Acevedo A, Salar A, Serrano S, Bas A, Moraleda JM, Sanchez-Godoy P, Burgos F, Rayon C, Fresno MF, Larana JG, Garcia-Cosio M, Santonja C, Lopez JL, Llanos M, Mollejo M, Gonzalez-Carrero J, Marin A, Forteza J, Garcia-Sanz R, Tomas JF, Morente MM, Piris MA, Garcia JF. A molecular risk score based on 4 functional pathways for advanced classical Hodgkin lymphoma. Blood. 2010;116:e12–e17. doi: 10.1182/blood-2010-02-270009. [DOI] [PubMed] [Google Scholar]
- Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Research. 2010;1352:255–264. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Smolewski P, Robak T, Krykowski E, Blasinska-Morawiec M, Niewiadomska H, Pluzanska A, Chmielowska E, Zambrano O. Prognostic factors in Hodgkin’s disease: multivariate analysis of 327 patients from a single institution. Clinical Cancer Research. 2000;6:1150–1160. [PubMed] [Google Scholar]
- Steidl C, Lee T, Shah SP, Farinha P, Han G, Nayar T, Delaney A, Jones SJ, Iqbal J, Weisenburger DD, Bast MA, Rosenwald A, Muller-Hermelink HK, Rimsza LM, Campo E, Delabie J, Braziel RM, Cook JR, Tubbs RR, Jaffe ES, Lenz G, Connors JM, Staudt LM, Chan WC, Gascoyne RD. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. New England Journal of Medicine. 2010;362:875–885. doi: 10.1056/NEJMoa0905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiacci E, Doring C, Brune V, van Noesel CJ, Klapper W, Mechtersheimer G, Falini B, Kuppers R, Hansmann ML. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood. 2012;120:4609–4620. doi: 10.1182/blood-2012-05-428896. [DOI] [PubMed] [Google Scholar]
- Tzankov A, Meier C, Hirschmann P, Went P, Pileri SA, Dirnhofer S. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin’s lymphoma. Haematologica. 2008;93:193–200. doi: 10.3324/haematol.11702. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, De Weer A, Mestdagh P, Feys T, De Preter K, De Paepe P, Lambein K, Vandesompele J, Van Roy N, Verhasselt B, Poppe B, Speleman F. Comparison of miRNA profiles of microdissected Hodgkin/Reed-Sternberg cells and Hodgkin cell lines versus CD77+ B-cells reveals a distinct subset of differentially expressed miRNAs. British Journal of Haematology. 2009;147:686–690. doi: 10.1111/j.1365-2141.2009.07909.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Corrigan-Cummins M, Hudson J, Maric I, Simakova O, Neelapu SS, Kwak LW, Janik JE, Gause B, Jaffe ES, Calvo KR. MicroRNA profiling of follicular lymphoma identifies microRNAs related to cell proliferation and tumor response. Haematologica. 2012;97:586–594. doi: 10.3324/haematol.2011.048132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, Wan D, Yao M, Li J, Gu J, He X. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, Sotomayor E, Tao J, Cheng JQ. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, Shao N, Qu H, Yang C, Zhang Y, Wang Q, Wang R, Zen K, Zhang CY, Zhang J, Yang Y. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. European Journal of Cancer. 2010;46:1640–1649. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Unsupervised clustering and heatmap image from the initially identified 34 miRNAs outcome related and cHL cell lines.
Table SI. Assay details miRNAS used for quantitative PCR (Q-PCR) analyses of the FFPE dataset (n = 168).
Table SII. miRNAs retained in the different Cox backward stepwise models in Crossvalidation Runs.
Table SIII. Multivariate analysis using Cox proportional hazards regression model in the FFPE cohort.
Appendix S1. Members of the Spanish Hodgkin Lymphoma Study Group.
Data S1. miRNA selection, cross-validation runs details and R scripts.