Abstract
Leishmaniasis is endemic in southern Europe, and in other European countries cases are diagnosed in travellers who have visited affected areas both within the continent and beyond. Prompt and accurate diagnosis poses a challenge in clinical practice in Europe. Different methods exist for identification of the infecting Leishmania species. Sixteen clinical laboratories in 10 European countries, plus Israel and Turkey, conducted a study to assess their genotyping performance. DNA from 21 promastigote cultures of 13 species was analysed blindly by the routinely used typing method. Five different molecular targets were used, which were analysed with PCR-based methods. Different levels of identification were achieved, and either the Leishmania subgenus, species complex, or actual species were reported. The overall error rate of strains placed in the wrong complex or species was 8.5%. Various reasons for incorrect typing were identified. The study shows there is considerable room for improvement and standardisation of Leishmania typing. The use of well validated standard operating procedures is recommended, covering testing, interpretation, and reporting guidelines. Application of the internal transcribed spacer 1 of the rDNA array should be restricted to Old World samples, while the heat-shock protein 70 gene and the mini-exon can be applied globally.
Keywords: leishmaniasis, Typing, hsp70, rDNA ITS1, mini-exon, kinetoplast DNA
Introduction
Leishmaniasis is a vector-borne disease which is endemic in 98 countries worldwide [1]. It is caused by protozoan parasites of the genus Leishmania, which are transmitted by female sand flies of the genera Lutzomyia and Phlebotomus. Many infected individuals never develop symptoms, but those who do can exhibit various disease manifestations [2]. Visceral leishmaniasis (VL) or kala-azar is the severe form, whereby parasites infect internal organs and the bone marrow, a lethal condition if left untreated. Other disease types are restricted to the skin (cutaneous leishmaniasis, CL) or the mucosae of the nose and mouth (mucosal leishmaniasis, ML). Finally, a particular cutaneous disease sometimes develops in cured VL patients: post kala-azar dermal leishmaniasis (PKDL). Typically, VL is caused by two species: Leishmania donovani and Leishmania infantum. The latter can also cause CL, as can all other pathogenic species. Some particular species (e.g. L. braziliensis and L. aethiopica) can lead to overt ML.
As many as 20 different Leishmania species are able to infect humans, and globally there are over 1 million new disease cases per annum [1,3]. Leishmaniasis is endemic in southern Europe, and in other European countries cases are diagnosed in travellers who have visited affected areas both within the continent and beyond. Although treatment in practice is often guided only by clinical presentation and patient history, in some cases determination of the aetiological subgenus, species complex or species is recommended for providing optimal treatment [2,4,5]. For example, a patient returning from South America with CL might be infected with Leishmania braziliensis, which necessitates systemic drug therapy and counselling about the risk of developing mucosal leishmaniasis in the future. The same patient could also be infected with Leishmania mexicana, which is managed by less intensive treatment and which is not associated with mucosal disease [6]. Determining the infecting species and its probable source permits selection of the correct drug, route of administration (intralesional, oral systemic, or parenteral) and duration [7].
Unfortunately, for CL it is impossible to predict the species responsible for an ulcerating lesion clinically, and the morphology of amastigotes does not differ between species. When the geographical origin of infection is known, for instance when a patient in an endemic region is treated at a local hospital, the species can be guessed often from the known local epidemiology, as species distribution follows a geographical pattern [8]. However, especially in infectious disease clinics that treat patients who have stayed in various endemic countries, the geographic origin of infections may be unknown. For instance, people residing in Europe who have travelled outside Europe may come from, or have also visited, Leishmania-endemic areas within Europe, especially the Mediterranean basin. Even when the location of infection is known, several species can co-circulate in a given endemic area, in which case the species can only be determined by laboratory tests. Culture and subsequent isoenzyme analysis is time consuming and available in very few specialised centres, so it is impractical as a front-line diagnostic test in clinical laboratories. Hence, well-performed reliable molecular methods are necessary for species identification.
Several Leishmania typing methods have been published (reviewed in [9]), and as a result each laboratory uses its own preferred assay. The most popular assays nowadays are those that can be applied directly to clinical samples, thereby circumventing the need for parasite isolation and culture. However, few tests have been standardised, and no commercial kits are currently available. As a result, clinical and epidemiological studies make use of various techniques, and in patient management other methods are often deployed. In this study we compare the typing performance in 16 clinical laboratories across Europe, which use a variety of methods for species discrimination.
Methods
Participants and reference methods
Twenty one Leishmania isolates were typed by 16 laboratories in 12 countries in 2014. Table 1 lists the parasite strains that were used in this study, along with the reference method for species identification. Strains identified with a Laboratoire d'Ecologie Médicale (LEM) code were provided by the Centre National de Référence des Leishmanioses in Montpellier, France, which assigns LEM codes to each cryopreserved culture, while the remaining three strains were provided by the Institute of Tropical Medicine in Antwerp, Belgium.
Table 1. Strains used, study comparing Leishmania typing results in 16 European clinical laboratories, 2014.
| Strain (WHO code) | Culture name CNRLa | Speciesb | Reference typing methodc |
|---|---|---|---|
| MHOM/ET/83/130–83 | LEM1118 | Leishmania aethiopica | MLEE, MLSA |
| MHOM/GF/2002/LAV003 | LEM4351 | L. amazonensis | MLEE, MLSA |
| MHOM/VE/76/JAP78 | LEM0391 | L. amazonensis | MLEE, MLSA |
| MHOM/BR/75/M2903b | LEM0396 | L. braziliensis | MLEE, MLSA |
| MHOM/PE/83/STI139 | LEM0781 | L. braziliensis | MLEE, MLSA |
| MHOM/BO/2001/CUM555 | NA | L. braziliensis outlierd | AFLP [12], WGS, MLSA |
| MHOM/IN/--/LRC-L51 | LEM1070 | L. donovani | MLEE, MLSA |
| MHOM/KE/55/LRC-L53 | LEM0707 | L. donovani | MLEE, MLSA |
| MHOM/GF/86/LEM1034 | LEM1034 | L. guyanensis | MLEE, MLSA |
| MHOM/FR/78/LEM75 | LEM0075 | L. infantum | MLEE, MLSA |
| MCUN/BR/85/M9342 | LEM2229 | L. lainsoni | MLEE, MLSA |
| MHOM/IQ/86/CRE1 | LEM0858 | L. major | MLEE, MLSA |
| MHOM/BZ/82/BEL21 | LEM0695 | L. mexicana | MLEE, MLSA |
| MHOM/EC/87/EC103-CL8 | LEM1554 | L. mexicana | MLEE, MLSA |
| MDAS/BR/79/M5533 | LEM2204 | L. naiffi | MLEE, MLSA |
| MHOM/CO/86/UA126 | LEM1047 | L. panamensis | MLEE, MLSA |
| MHOM/CO/88/UA264 | LEM1492 | L. panamensis | MLEE, MLSA |
| MHOM/CO/88/UA316 | LEM1505 | L. panamensis / L. guyanensise | MLEE, MLSA |
| MHOM/PE/90/HB86 | NA | L. peruviana | AFLP [12], WGS, MLSA |
| MHOM/PE/90/LCA08 | NA | L. peruviana | AFLP [12], WGS, MLSA |
| MHOM/IL/80/SINGER | LEM0617 | L. tropica | MLEE, MLSA |
AFLP: amplified fragment length polymorphism; CNRL: Centre National de Référence des Leishmanioses (Montpellier, France); NA: not applicable; MLEE: multilocus enzyme electrophoresis; MLSA: multilocus sequence analysis; WGS: whole genome sequencing; WHO: World Health Organization.
a Identification in the Montpellier cryobank (Centre National de Référence des Leishmanioses).
b For the taxonomic position of each species (subgenus and species complex), please refer to Figure 2.
c Reference method used to determine the species of each isolate. MLEE [10]; MLSA based on seven genes [11]; AFLP analysis [12]; WGS (unpublished results).
d Group of distinct Leishmania braziliensis strains [9,12], also called L. braziliensis type 2 [15] or atypical L. braziliensis [18].
e This strain was typed as L. panamensis by MLSA, and as L. guyanensis by MLEE.
Four highly informative reference methods were used: multilocus enzyme electrophoresis (MLEE [10]), multilocus sequence analysis (MLSA [11], GenBank sequence accession numbers in Table 2), genome-wide amplified fragment length polymorphism (AFLP) analysis [12], and whole genome sequencing (unpublished results).
Table 2. GenBank sequence accession numbers from MLSA and hsp70, for sequences used in study comparing Leishmania typing results in 16 European clinical laboratories, 2014.
LEM: Laboratoire d'Ecologie Médicale; MLSA: multilocus sequence analysis; hsp70: heat-shock protein 70 gene; NA: not applicable; WHO: World Health Organization.
DNA was extracted from parasite cultures using either the DNeasy Blood and Tissue Kit or QIAamp DNA Mini Kit (Qiagen, www.qiagen.com), and the concentration was measured spectrophotometrically. The 21 DNAs were randomised at the United Kingdom (UK) National External Quality Assessment Service for Parasitology (UKNEQAS, London, UK), and every study participant received a blind panel containing 50 µl of a 10 ng/µl DNA solution. The participating laboratories are listed in Table 3.
Table 3. Participants in study comparing Leishmania typing results in 16 European clinical laboratories, 2014.
| Institute | City | Country |
|---|---|---|
| Institute of Tropical Medicine Antwerpa | Antwerp | Belgium |
| Centre National de Référence des Leishmaniosesa,b | Montpellier | France |
| Centre Hospitalier Universitaire de Rennes | Rennes | France |
| Charité-Universitätsmedizin Berlin | Berlin | Germany |
| Hebrew University-Hadassah Medical Centre | Jerusalem | Israel |
| Istituto Superiore di Sanità | Rome | Italy |
| National Institute for Infectious Diseases L. Spallanzani | Rome | Italy |
| Instituto de Higiene e Medicina Tropical | Lisbon | Portugal |
| Instituto de Salud Carlos III | Majadahonda | Spain |
| The Public Health Agency of Sweden | Stockholm | Sweden |
| Swiss Tropical and Public Health Institutea | Basel | Switzerland |
| Institute of Parasitology | Zürich | Switzerland |
| Academic Medical Center, University of Amsterdam | Amsterdam | The Netherlands |
| National Institute for Public Health and the Environment | Bilthoven | The Netherlands |
| St. Elisabeth Hospital | Tilburg | The Netherlands |
| Ege University Medical School | Izmir | Turkey |
| Hospital for Tropical Diseases | London | United Kingdom |
Institutes are listed in alphabetical order based on country and city.
a These laboratories provided parasite cultures and DNA.
b This laboratory applied one of the reference methods (MLSA) and did not participate in the comparative study of typing outcomes.
After performing the respective routine typing technology, each laboratory reported its results to UKNEQAS, who forwarded these along with the randomised code in one batch to the Institute of Tropical Medicine in Antwerp for analysis. Some participants used the term ‘L. braziliensis complex’ when referring to the L. (Viannia) subgenus, and where needed the reported results were adjusted. The results after these adjustments are presented in this analysis.
Genome targets for typing
The 16 laboratories used a total of five genome targets for typing (Table 4): the internal transcribed spacer 1 of the rDNA array (ITS1), the mini-exon, kinetoplast minicircle DNA (kDNA), the heat-shock protein 70 gene (hsp70), and a repetitive DNA sequence. One laboratory reported two sets of result from two different targets, which are treated in the analysis as if they were from separate laboratories, which is why the results section describes 17 instead of 16 outcomes. The targets were analysed with PCR, generally followed by sequencing or restriction fragment length polymorphism (RFLP) analysis, as shown in Table 4 and Figure 1. Four laboratories used in-house sequencing, while five others used the service of an external sequencing facility. PCRs based on kDNA did not require post-PCR manipulations other than gel analysis.
Table 4. Typing methods used in study comparing Leishmania typing results in 16 European clinical laboratories, 2014.
| Genomic locus / gene | Analysis method | Number of laboratoriesa |
|---|---|---|
| ITS1 | RFLP [13,14] | 7 |
| Sequencing [15] | 4 | |
| hsp70 | Sequencing [15,16] | 5 |
| RFLP [17] | 2 | |
| Mini-exon | Sequencing [19-21] | 3 |
| kDNA minicircles | RFLP [24,25] | 1 |
| Specific PCR [23] | 1 | |
| Repetitive DNA | RFLP [26] | 1 |
ITS: internal transcribed spacer; hsp70: heat-shock protein 70 gene; kDNA: kinetoplast minicircle DNA; RFLP: restriction fragment length polymorphism.
a The total number is higher than the 16 participating laboratories, because several laboratories used different methods in parallel.
Figure 1 indicates for each laboratory individually which method or methods were used, but not all samples were necessarily analysed with each method. Of the 16 laboratories, 11 used the ITS1 target, either applying RFLP (n=7) or sequencing (n=4). All of them based their analysis on the fragment described in [13], except for laboratory L which used a larger region also including ITS2 [14]. Five laboratories based typing on hsp70: four (A-D) used sequencing of the F fragment described in [15-17], while one (E) used the N fragment. Two laboratories (F and G) analysed this gene with RFLP [17,18]. Three laboratories used sequence analysis of the mini-exon gene: laboratory O [19,20], laboratory P [21], and laboratory Q [22]. Two laboratories based typing partly on kDNA: laboratory K [23], and laboratory L [24,25]. Finally, laboratory J complemented ITS1-RFLP with RFLP analysis of a repetitive DNA sequence [26].
Grading of results
Each individual result was graded as follows. The best ranking was given to reported species agreeing with the reference methods, whereby L. garnhami was considered a synonym of L. amazonensis [27]. Results reporting MHOM/BO/2001/CUM555 as L. braziliensis were considered correct. Although this strain belongs to a group of clearly distinguishable outliers (Table 1), it has so far not been described as a separate species. Next were identifications that reported the species complex rather than the actual species (see Figure 2), and were in agreement with the reference methods. The lowest ranking of correct results was given to those identifying the subgenus, i.e. L. (Viannia) or L. (Leishmania), without specification of species or species complex. Identification errors were graded at two levels. First, some laboratories reported a species within the correct complex, but identified the wrong species within that complex. Second, some isolates were placed in an erroneous species complex altogether. A peculiar case was presented by strain MHOM/CO/88/UA316, which was L. guyanensis based on MLEE, but L. panamensis based on MLSA (Table 1). For this strain, all results reporting either L. guyanensis or L. panamensis were considered to have identified the correct species complex.
In a next level of the analysis, the cause of erroneous typings was sought by means of in-depth assessment of the methods. The reasons for different identification outcomes of laboratories using the same methods were also identified. Sequences from laboratories that based their typing on the same genes were compared by alignment in the software package MEGA5 [28].
Results
Results from all analyses are summarised in Figure 1, details are available from [29]. One laboratory reported two sets of results because identification based on hsp70 sequences was sometimes in conflict with those of ITS1 sequencing. These results are listed separately from laboratories E and M respectively, which brings the number of reported result sets from the 16 laboratories to 17. Most laboratories succeeded in typing all 21 samples, but in some cases results were reported for 20/21 isolates only (laboratories D, K, L, M). The total number of erroneous identifications amounted to 30, with 23 of these being classified in an incorrect species complex. On a total of 353 results, these represent 8.5% and 6.5% respectively. The correct species was identified in 211 typing results (60%), while 58 (16%) identified the correct species complex, and 54 (15%) the correct subgenus. Eight laboratories made no incorrect assignments, while the laboratory with most errors (laboratory J) misidentified 10 out of 21 samples, seven of which were placed in the wrong species complex. Laboratories relying only on kDNA and ITS1 more frequently reported results to the subgenus level, while laboratories using the mini-exon or hsp70 often succeeded in obtaining identification either to the species or complex level.
Figure 1.
Typing results obtained in study comparing Leishmania typing results in 16 European clinical laboratories, 2014
ITS: internal transcribed spacer; hsp70: heat-shock protein 70 gene; kDNA: kinetoplast minicircle DNA; RFLP: restriction fragment length polymorphism.
a RFLP was performed on a fragment covering both ITS1 and ITS2 [14].
b One laboratory reported the use of two separate methods. Results E and M.
For each method, the number of correct typings to species, species complex, and subgenus level are shown in different colours. In addition, the incorrect species designations are indicated, some of which identified the wrong species in the correct complex (purple bars), others placing a strain in the wrong complex (red bars). The methods or combination of methods that were used to obtain the given results are shown on top.
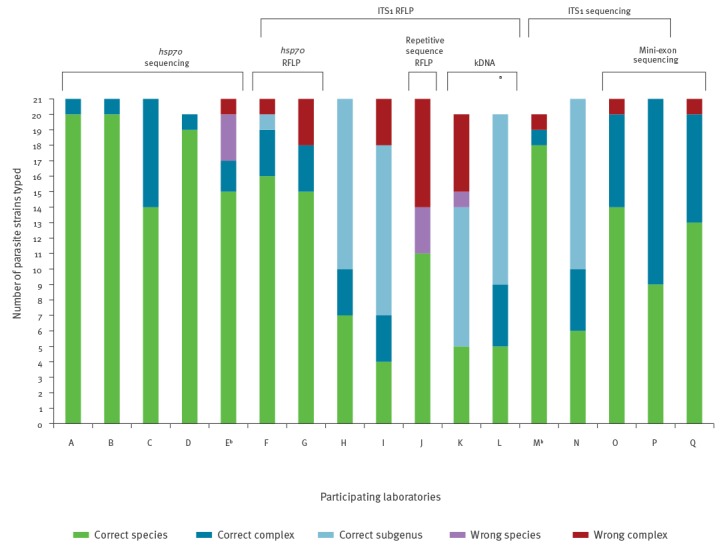
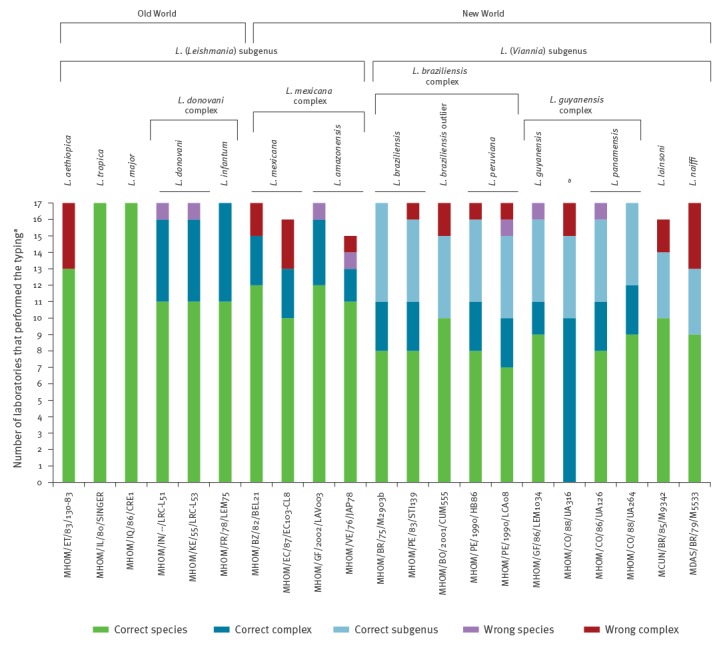
Figure 2 depicts the typing results for each strain, irrespective of the methods used. The only two species that were correctly identified with all methods were L. tropica and L. major. Strains from the L. (Leishmania) subgenus were identified to either the species or complex level by all laboratories. This was in contrast to the 11 strains from the L. (Viannia) subgenus, each of which was typed by four to six laboratories only to the subgenus level. The error rate for both subgenera was comparable: 8.4% (14/167) for L. (Leishmania) and 8.6% (16/186) for L. (Viannia). The error rate in Old World strains was lower than for strains of the New World: 5.9% (6/102) and 9.6% (24/251) respectively.
Figure 2.
Typing results for each of the 21 strains included in study comparing Leishmania typing results in 16 European clinical laboratories, 2014
MLSA: multilocus sequence analysis; WHO: World Health Organization.
a One laboratory reported the use of two separate methods.
b Strain MHOM/CO/88/UA316 is L. guyanensis based on MLEE, but L. panamensis based on MLSA (Table 1).
For each strain, the number of correct typings at species, species complex, and subgenus level are reported. In addition, the incorrect species designations are indicated, some of which identified the wrong species in the correct complex (purple bars), others placing an isolate in the wrong complex (red bars). The strain identification by WHO code (Table 1) is given with the abscissa. Species, complexes, and subgenera are represented on top, with an indication of the New or Old World strain origin.
When comparing the hsp70 sequences provided by four laboratories (A-D), there were marked differences in sequence quality. Three laboratories (A, B, C) succeeded in sequencing the entire or nearly entire fragment F [17], with few or no sequence ambiguities. The sequence sets of two laboratories (A and C) contained one insertion and one deletion relative to the other data, indicating sequence mistakes as the gene shows no size variation [15,16]. In contrast, the quality of the fragment F sequences from one laboratory (D) was considerably lower. Sequences were largely incomplete at their 5’ end and to a lesser extent at their 3’ terminus, and numerous insertions, deletions, and unresolved nucleotides (nt) were present. One laboratory (E) sequenced only the N fragment [17], but base calling quality was poor in the 40 terminal 3’ nt. The consensus hsp70 sequences were deposited in GenBank (Table 2).
Three laboratories (M, N, O) determined the ITS1 sequence of all isolates, while one laboratory (P) sequenced only MHOM/GF/2002/LAV003. The sequences of two laboratories (N and P) covered the entire amplified PCR product, while some of two others (O and M) were incomplete at the termini. Apart from some insertions in the sequences of one laboratory (N) and occasional unresolved nt in those of another (O), the sequences were identical, except for isolate MHOM/CO/88/UA316. Here, up to 9 nt differences were present in a 120 nt stretch.
Three laboratories (O, P, Q) determined the mini-exon sequences. For some strains the sequences of these laboratories were nearly identical, but for others large size differences of the determined fragment were seen, and deletions and nt identity discrepancies were observed. Also, many nt were not fully resolved.
Discussion
As a general observation, eight laboratories who participated in this comparison typing performance made no errors, and often laboratories using the same typing marker reported different results (Figure 1). Two of the ‘error-free’ laboratories obtained the highest typing accuracy, with 20 out of 21 strains typed to the species level, and strain MHOM/CO/88/UA316 at the complex level. Using our reference methods MLSA and MLEE (Table 1), the latter species could not be classified unequivocally, and hence results placing it in the L. guyanensis complex were regarded as correct. These two laboratories (A and B) based their typing on hsp70 gene sequencing, which was identified as one of the typing methods with the highest resolution in other comparative studies [9,15]. One other laboratory (C) also made use of this method, but typed several strains only to the complex level. Even though the hsp70 gene often permits distinction between closely related species, separating them is not always straight-forward. For instance, some MLEE-defined L. guyanensis have the same sequence as L. panamensis [16]. Because identifying the exact species within a given complex can therefore be difficult, one laboratory (C) decided to identify the species complex rather than the exact species in case of doubt. Apparently the low sequence quality obtained by one of the participants (D) had no adverse effects on the results, probably because species-specific nt identities were not affected. The sequence quality was not influenced by the use of in-house vs external sequencing services.
One laboratory (E) reported four mistakes based on hsp70 sequences. As opposed to laboratories A-D, the analysis was based on a smaller part of the gene, fragment N [17], which is not suited for typing all species [15]. Nevertheless, several of these species were called based on a BLAST search in GenBank [https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch], from which the first listed species was regarded as the final result, regardless of identical similarity scores obtained from other species. In this process some species were by chance determined correctly, while others were erroneously identified. This stresses the importance of correctly interpreting output lists generated by BLAST, because different species can have the same similarity score when the marker is too conservative for discriminating between them. To avoid such errors the species complex rather than the species itself should have been reported. On one occasion, the applied methodology even identified an erroneous complex, i.e. MHOM/ET/83/130–83 was typed as L. donovani instead of L. aethiopica, based on an erroneous annotation in GenBank. Indeed, several GenBank entries of [30] were wrongfully submitted as L. donovani, while they derived in fact from other species [16]. This illustrates the importance of critically evaluating BLAST results, and underscores the importance of an agreed reference panel of sequences from trustworthy laboratories and knowledge of the limitations of a typing marker.
The same laboratory E reported a second results set based on ITS1 sequence analysis, listed under laboratory M in Figure 1. Again, BLAST analysis was applied, and even though ITS1 is not suitable for discriminating L. braziliensis and L. guyanensis complex species [15], several species were reported. Except for one misclassified L. braziliensis outlier strain (Figure 2), species were correctly assigned by laboratory M. However, in several cases also other species showed the same similarity scores, and hence there was no ground for naming the exact species. In contrast, another laboratory (N), which also used ITS1 sequence analysis, reported L. (Viannia) strains at subgenus level with no further attempt to determine the complex or species. Thereby they respected the limitations of ITS1, although some L. (Viannia) complexes could have been identified based on their data.
The majority of study participants that used ITS1 did not sequence the target, but relied on RFLP analysis. Laboratories basing their results on this method reported some typical errors: L. tropica was mixed up with L. aethiopica; the L. donovani complex was confused with L. mexicana; unsuccessful attempts were made to separate L. infantum from L. donovani; and on one occasion L. amazonensis was identified as L. major. When digesting the PCR products with the popular enzyme HaeIII, sufficient gel resolution is needed in order not to mix up the aforementioned species, as their RFLP fragments are similar in size. In addition, contrary to what was originally published [13], L. infantum cannot be distinguished from L. donovani [9] and therefore ITS1 can only type to the L. donovani complex, without further specification.
Two laboratories (F and G) complemented ITS-RFLP with hsp70-RFLP, and both mistook L. naiffi for L. braziliensis. This is a result of identical patterns generated from L. naiffi and many L. braziliensis strains with restriction endonucleases HaeIII and RsaI. The mistake could have been avoided by using the appropriate enzyme SduI [18].
Only one laboratory (J) made use of a repetitive DNA sequence originally described in [31]. In combination with ITS1, 10 out of the 21 typings were incorrect, whereby seven strains were assigned to the wrong complex. Of the 10 mistakes, nine were made in the L. (Viannia) subgenus, while the remaining error was due to the unsuccessful separation of L. infantum from L. donovani. ITS1-RFLP is not suitable for discriminating these species, and the repetitive sequence RFLP was designed for typing Old World strains, where only the L. (Leishmania) subgenus is encountered. Such mistakes once more underline the importance of knowing the limits of the typing marker chosen.
Kinetoplast DNA is primarily a useful marker to discriminate the two Leishmania subgenera, but is less suited for typing to the actual species level (reviewed in [9]). In combination with the fact that also ITS1-RFLP does not discriminate many L. (Viannia) species, the two laboratories (K and L) using these methods reported typing mostly to the subgenus or species complex level. One of them (K) had a particularly high error rate (6/20) using these markers, probably related to the previously mentioned gel resolution problems and separation of L. infantum from L. donovani with ITS1-RFLP. In addition the laboratory used ‘L. braziliensis complex / L. guyanensis complex’ as a synonym for L. (Viannia), while two strains were L. naiffi and L. lainsoni.
With the mini-exon sequences, only two mistakes were reported. One laboratory (O) identified L. mexicana strain MHOM/EC/87/EC103-CL8 as L. donovani, but after disclosing the results realised a mistake in reporting, as their analysis actually did show the correct species. In a comparative analysis of four markers [15], the mini-exon together with hsp70 were identified as the most discriminative markers worldwide, which is confirmed by the results presented here. Some species within the complexes can, however, not be resolved based on the mini-exon, as also reflected in the current analysis, where often complexes rather than species were identified.
When looking at the typing results for each of the 21 strains (Figure 2), it is apparent that strains of the L. (Viannia) subgenus were more often typed to the subgenus level, while those of the L. (Leishmania) subgenus were more often reported at the species level. Given that ITS1 was the most popular marker, this is a logical result in view of the poor discrimination of L. (Viannia) species by ITS1. Also the fact that for Old World strains 5.9% of typings were erroneous, in comparison to 9.6% New World strains, relates to the use of methods that are tailored to Old World strains. Only two strains were identified to the species level by all laboratories and all methods: MHOM/IL/80/SINGER (L. tropica) and MHOM/IQ/86/CRE1 (L. major). The results show that several laboratories are currently unable to discriminate L. (Viannia) species, which is partly explained by the participation in the study of six groups that are situated in a European country where Leishmania is actively transmitted. Hence, they mainly diagnose patients infected by endemic species, and use methods primarily tailored to species in the Old World. On the contrary, the remaining laboratories are dealing only with imported leishmaniasis cases, which can originate from anywhere in the world, and for which the origin of infection is sometimes unknown. This forces them to apply assays that are able to identify species from everywhere around the globe.
With regard to nomenclature, there is an evident need for standardisation. When the first results were reported, several laboratories used the term ‘L. braziliensis complex’ to refer to L. (Viannia). For many years these have been synonyms, but current literature restricts this term to L. braziliensis and L. peruviana [27]. Another confusion can arise from the fact that each complex bears the name of one of its constituent species. For instance, a typing outcome reported as ‘L. guyanensis’ has to be clearly distinguished from ‘L. guyanensis complex’. Even though this particular problem did not seem to occur in our analysis, one could easily envision such occurrence. One laboratory (K) reported several results as ‘L. braziliensis complex / L. guyanensis complex’ for referring to L. (Viannia), but with this term L. naiffi and L. lainsoni were excluded.
Finally, the particular case of strain MHOM/CO/88/UA316 draws attention to problems in species definitions, as this strain was typed as L. guyanensis with MLEE, but as L. panamensis with MLSA (Table 1). Reported correct results for this strain were either L. guyanensis complex, L. guyanensis, or L. panamensis, but this was irrespective of the method or target used [29]. Such occasional dubious results are unavoidable when dealing with closely related species, in particular L. guyanensis-L. panamensis; L. braziliensis-L. peruviana; L. mexicana-L. amazonensis; and L. donovani-L. infantum [9]. Also newly documented parasite species such as L. martiniquensis [32] and L. waltoni [33], and variants as the L. braziliensis outlier [9,12,15,18] further complicate the interpretation of typing results. It is therefore of utmost importance that species identification is performed with a well-documented standard operating procedure (SOP), clearly describing not only experimental procedures, but also in detail how results should be analysed, interpreted, and reported.
The current study was performed on cultured parasite isolates, so all participants received a high amount of pure parasite DNA. Yet, 8.5% errors were seen, and in four cases no result was obtained. When dealing with patient material, the amount of parasite DNA is much lower, and vastly exceeded by human DNA. As the current study did not assess the sensitivity of the methods used, it is expected that typing success based on clinical samples will be considerably lower. In view of the fact that only recognised reference laboratories participated in this study, there is a clear need for optimisation. On the other hand, in many clinical settings the suspected origin of infection can help in interpretation of typing outcomes, thereby possibly lowering the error rate.
Conclusions
There is considerable room for improvement of current Leishmania typing strategies, and inter-laboratory comparisons such as the one we conducted can contribute to enhance typing quality. Whichever the clinical need for determining the subgenus, complex, or species, and whichever the technology used in a particular setting, typing should be based on a well-defined and validated SOP designed by an expert in Leishmania taxonomy. This SOP should cover not only testing, but also analysis and interpretation procedures, and a clear description of how species should be named and reported, taking into account the limitations of each marker and technique, and the problem of resolving closely related species or occasional inter-species hybrids. Validation should be performed on a sufficient amount of reference isolates from various geographic origins to cover each species’ variability. When using sequencing, sequence errors should be avoided, and a well-validated sequence reference set is recommended over BLAST analysis using GenBank, which lacks quality control. In cases where treatment is species- or complex-dependent, clinicians should be made aware of the limitations of the technology used whenever results are reported, especially when closely related species are involved. The use of real-time PCR assays developed for specific complexes or species could speed up typing and facilitate interpretation of results, but currently no globally applicable methods are available. As previously recommended [15] and also apparent from this analysis, hsp70 and the mini-exon currently offer the best Leishmania typing tools world-wide, and the use of ITS1 should be restricted to the Old World. Setting up similar evaluations outside Europe, in institutes in endemic as well as non-endemic countries, would shed additional light on the quality of Leishmania typing across the globe.
Acknowledgements
The authors would like to thank the ESCMID study group for Clinical Parasitology (ESGCP, headed by Titia Kortbeek, National Institute for Public Health and the Environment, RIVM, Bilthoven, The Netherlands) for financial support (ESCMID Study Group Research Grant 2013 to Peter L. Chiodini). We are grateful to Jean-Pierre Gangneux (ESGCP coordinator for leishmaniasis, Centre Hospitalier Universitaire de Rennes, Rennes, France) and the LeishMan consortium [www.leishman.eu] for conceptual support and promoting the study. We acknowledge the valuable comments of Titia Kortbeek, and the technical assistance of Sofia Andersson (The Public Health Agency of Sweden, Stockholm, Sweden), José M. Cristóvão (Global Health and Tropical Medicine, GHTM, Instituto de Higiene e Medicina Tropical, UNL, Lisbon, Portugal), Mehmet Karakus (Ege University, Faculty of Medicine, Department of Parasitology, Izmir, Turkey), Ilse Maes (Institute of Tropical Medicine, Antwerp, Belgium), Abed Nasereddin (Hebrew University, Hadassah Medical Centre, Jerusalem, Israel), Chris Stalder (Swiss Tropical and Public Health Institute, Basel, Switzerland), Antonietta Toffoletti and Antonella Vulcano (National Institute for Infectious Diseases (INMI) Lazzaro Spallanzani, Rome, Italy), Carla Wassenaar (Academic Medical Center, Amsterdam, The Netherlands), Julie Watson and Spencer Polley (Hospital for Tropical Diseases, London, United Kingdom). Gert Van der Auwera is supported by the Third Framework program between ITM and the Belgian Directorate General for Development. Peter L. Chiodini is supported by the Biomedical Research Centre of the University College London Hospitals and National Institute for Health Research.
Conflict of interest: None declared.
Authors’ contributions: Gert Van der Auwera, Aldert Bart, Ingrid Felger, Christophe Ravel, Jean-Claude Dujardin, and Peter L. Chiodini conceptualised the study. Gert Van der Auwera coordinated the study, analysed the data, and drafted the publication. Christophe Ravel and Gert Van der Auwera provided the cultures, from which DNA was extracted by Ingrid Felger and Gert Van der Auwera. Monika Manser was responsible for blinding the samples and collecting the results. Gert Van der Auwera, Aldert Bart, Carmen Chicharro, Sofia Cortes, Leigh Davidsson, Trentina Di Muccio, Ingrid Felger, Maria Grazia Paglia, Felix Grimm, Gundel Harms, Charles L. Jaffe, Christophe Ravel, Florence Robert-Gangneux, Jeroen Roelfsema, Seray Töz, and Jaco J. Verweij supervised or carried out the assays. All authors gave their input on the manuscript draft.
References
- 1. Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. WHO Leishmaniasis Control Team . Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671. 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Control of the leishmaniasis: report of a meeting of the WHO Expert Committee on the Control of Leishmaniases, Geneva, 22-26 March 2010. Geneva, Switzerland: World Health Organization; 2010. [Google Scholar]
- 3.Boelaert M, Sundar S. Leishmaniasis. In: Farrar J, Hotez P, Junghanss T, Kang G, Lalloo D, White N, eds. Manson's Tropical Diseases. 23 ed. Philadelphia: Elsevier Saunders, 2013. p. 631-51. [Google Scholar]
- 4. Blum J, Buffet P, Visser L, Harms G, Bailey MS, Caumes E, et al. LeishMan recommendations for treatment of cutaneous and mucosal leishmaniasis in travelers, 2014. J Travel Med. 2014;21(2):116-29. 10.1111/jtm.12089 [DOI] [PubMed] [Google Scholar]
- 5. Blum J, Lockwood DN, Visser L, Harms G, Bailey MS, Caumes E, et al. Local or systemic treatment for New World cutaneous leishmaniasis? Re-evaluating the evidence for the risk of mucosal leishmaniasis. Int Health. 2012;4(3):153-63. 10.1016/j.inhe.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 6. Bailey MS, Green AD, Ellis CJ, O’Dempsey TJ, Beeching NJ, Lockwood DN, et al. Clinical guidelines for the management of cutaneous leishmaniasis in British military personnel. J R Army Med Corps. 2005;151(2):73-80. 10.1136/jramc-151-02-03 [DOI] [PubMed] [Google Scholar]
- 7. Lawn SD, Whetham J, Chiodini PL, Kanagalingam J, Watson J, Behrens RH, et al. New world mucosal and cutaneous leishmaniasis: an emerging health problem among British travellers. QJM. 2004;97(12):781-8. 10.1093/qjmed/hch127 [DOI] [PubMed] [Google Scholar]
- 8. Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, et al. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis. 2016;10(3):e0004349. 10.1371/journal.pntd.0004349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van der Auwera G, Dujardin JC. Species typing in dermal leishmaniasis. Clin Microbiol Rev. 2015;28(2):265-94. 10.1128/CMR.00104-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp. 1990;65(3):111-25. 10.1051/parasite/1990653111 [DOI] [PubMed] [Google Scholar]
- 11. El Baidouri F, Diancourt L, Berry V, Chevenet F, Pratlong F, Marty P, et al. Genetic structure and evolution of the Leishmania genus in Africa and Eurasia: what does MLSA tell us. PLoS Negl Trop Dis. 2013;7(6):e2255. 10.1371/journal.pntd.0002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Odiwuor S, Veland N, Maes I, Arévalo J, Dujardin JC, Van der Auwera G. Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol. 2012;12(8):1994-2002. 10.1016/j.meegid.2012.03.028 [DOI] [PubMed] [Google Scholar]
- 13. Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47(1):349-58. 10.1016/S0732-8893(03)00093-2 [DOI] [PubMed] [Google Scholar]
- 14. Mauricio IL, Stothard JR, Miles MA. Leishmania donovani complex: genotyping with the ribosomal internal transcribed spacer and the mini-exon. Parasitology. 2004;128(Pt 3):263-7. 10.1017/S0031182003004578 [DOI] [PubMed] [Google Scholar]
- 15. Van der Auwera G, Ravel C, Verweij JJ, Bart A, Schönian G, Felger I. Evaluation of four single-locus markers for Leishmania species discrimination by sequencing. J Clin Microbiol. 2014;52(4):1098-104. 10.1128/JCM.02936-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van der Auwera G, Maes I, De Doncker S, Ravel C, Cnops L, Van Esbroeck M, et al. Heat-shock protein 70 gene sequencing for Leishmania species typing in European tropical infectious disease clinics. Euro Surveill. 2013;18(30):20543. 10.2807/1560-7917.ES2013.18.30.20543 [DOI] [PubMed] [Google Scholar]
- 17. Montalvo AM, Fraga J, Maes I, Dujardin JC, Van der Auwera G. Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis. 2012;31(7):1453-61. 10.1007/s10096-011-1463-z [DOI] [PubMed] [Google Scholar]
- 18. Fraga J, Montalvo AM, Maes L, Dujardin JC, Van der Auwera G. HindII and SduI digests of heat-shock protein 70 PCR for Leishmania typing. Diagn Microbiol Infect Dis. 2013;77(3):245-7. 10.1016/j.diagmicrobio.2013.07.023 [DOI] [PubMed] [Google Scholar]
- 19. Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J Clin Microbiol. 2003;41(7):3147-53. 10.1128/JCM.41.7.3147-3153.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roelfsema JH, Nozari N, Herremans T, Kortbeek LM, Pinelli E. Evaluation and improvement of two PCR targets in molecular typing of clinical samples of Leishmania patients. Exp Parasitol. 2011;127(1):36-41. 10.1016/j.exppara.2010.06.024 [DOI] [PubMed] [Google Scholar]
- 21. Marfurt J, Niederwieser I, Makia ND, Beck HP, Felger I. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn Microbiol Infect Dis. 2003;46(2):115-24. 10.1016/S0732-8893(03)00040-3 [DOI] [PubMed] [Google Scholar]
- 22. Bart A, van Thiel PP, de Vries HJ, Hodiamont CJ, Van Gool T. Imported leishmaniasis in the Netherlands from 2005 to 2012: epidemiology, diagnostic techniques and sequence-based species typing from 195 patients. Euro Surveill. 2013;18(30):20544. 10.2807/1560-7917.ES2013.18.30.20544 [DOI] [PubMed] [Google Scholar]
- 23. Cortes S, Rolão N, Ramada J, Campino L. PCR as a rapid and sensitive tool in the diagnosis of human and canine leishmaniasis using Leishmania donovani s.l.-specific kinetoplastid primers. Trans R Soc Trop Med Hyg. 2004;98(1):12-7. 10.1016/S0035-9203(03)00002-6 [DOI] [PubMed] [Google Scholar]
- 24. de Bruijn MH, Barker DC. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop. 1992;52(1):45-58. 10.1016/0001-706X(92)90006-J [DOI] [PubMed] [Google Scholar]
- 25. Noyes HA, Reyburn H, Bailey JW, Smith D. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J Clin Microbiol. 1998;36(10):2877-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minodier P, Piarroux R, Gambarelli F, Joblet C, Dumon H. Rapid identification of causative species in patients with Old World leishmaniasis. J Clin Microbiol. 1997;35(10):2551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schönian G, Mauricio I, Cupolillo E. Is it time to revise the nomenclature of Leishmania? Trends Parasitol. 2010;26(10):466-9. 10.1016/j.pt.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Institute of Tropical Medicine (ITG). Leishmania Typing Results, 2014. Table S1. Antwerp: ITG. [Accessed 29 Nov 2016]. Available from: www.itg.be/leishmaniatyping
- 30. Zhang CY, Lu XJ, Du XQ, Jian J, Shu L, Ma Y. Phylogenetic and evolutionary analysis of Chinese Leishmania isolates based on multilocus sequence typing. PLoS One. 2013;8(4):e63124. 10.1371/journal.pone.0063124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Piarroux R, Fontes M, Perasso R, Gambarelli F, Joblet C, Dumon H, et al. Phylogenetic relationships between Old World Leishmania strains revealed by analysis of a repetitive DNA sequence. Mol Biochem Parasitol. 1995;73(1-2):249-52. 10.1016/0166-6851(95)00097-K [DOI] [PubMed] [Google Scholar]
- 32. Desbois N, Pratlong F, Quist D, Dedet JP. Leishmania (Leishmania) martiniquensis n. sp. (Kinetoplastida: Trypanosomatidae), description of the parasite responsible for cutaneous leishmaniasis in Martinique Island (French West Indies). Parasite. 2014;21:12. 10.1051/parasite/2014011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shaw J, Pratlong F, Floeter-Winter L, Ishikawa E, El Baidouri F, Ravel C, et al. Characterization of Leishmania (Leishmania) waltoni n.sp. (Kinetoplastida: Trypanosomatidae), the Parasite Responsible for Diffuse Cutaneous Leishmaniasis in the Dominican Republic. Am J Trop Med Hyg. 2015;93(3):552-8. 10.4269/ajtmh.14-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]


