Abstract
Importance
Human episodic memory performance is linked to the function of specific brain regions, including the hippocampus, declines as a result of increasing age, and is markedly disturbed in Alzheimer’s disease (AD), an age-associated neurodegenerative disorder affecting primarily the hippocampus. Exploring the molecular underpinnings of human episodic memory is key to the understanding of hippocampus-dependent cognitive physiology and pathophysiology.
Objective
To determine whether biologically defined groups of genes are enriched in episodic memory performance across ages, in memory encoding-related brain activity and in AD.
Design, Setting, and Participants
In this multicenter collaborative study, gene set enrichment analysis was done by using primary and meta-analysis data from 57968 participants. The Swiss cohorts consisted of 3043 healthy young adults assessed for episodic memory performance. In a subgroup (1119 participants) of one of these cohorts, functional magnetic resonance imaging (fMRI) was used to identify gene set-dependent differences in brain activity related to episodic memory. The German AgeCoDe cohort consisted of 763 non-demented elderly participants assessed for episodic memory performance. The International Genomics of Alzheimer's Project (IGAP) case-control sample consisted of 54162 participants (17008 patients with sporadic AD, 37154 controls).
Main Outcomes and Measures
Gene set enrichment analysis in all samples was done by using genome-wide single nucleotide polymorphism (SNP) data. Episodic memory performance in the Swiss and AgeCoDe cohorts was quantified by picture and verbal delayed free recall tasks. In the fMRI experiment, activation of the hippocampus during encoding of pictures served as the phenotype of interest. In the IGAP sample, diagnosis of sporadic AD served as the phenotype of interest.
Results
We detected significant and consistent enrichment for genes constituting the Calcium Signaling Pathway (KEGG entry: hsa04020), especially those related to the elevation of cytosolic calcium (P=0.0002). This enrichment was observed in episodic memory performance in young and old, in hippocampal activation, and in the risk for sporadic AD.
Conclusion and Relevance
By detecting consistent significant enrichment in independent cohorts of young and elderly participants, this study identifies calcium signaling as a central player of hippocampus-dependent human memory processes, both in cognitive health and disease and contributes to the understanding -and hopefully treatment- of
Introduction
Episodic memory (EM), i.e. the ability to encode and retrieve a particular event along with its contextual information,1 is a polygenic cognitive trait, characterized by large interindividual variability and substantial heritability.2–5 (for review see5) As a consequence of physiological ageing processes in such brain regions as the hippocampus and the medial temporal lobe, performance in EM tasks declines with age.6–9 Pathological EM impairment is a behavioral hallmark of age-related neurodegenerative conditions, such as Alzheimer’s disease (AD).10,11
Genome-wide studies utilizing single-marker statistics have been successful in identifying single loci linked to intact and impaired EM.5,12–14 Triggered by statistical approaches for the analysis of gene expression, gene-set enrichment analysis (GSEA) has recently become available. By taking into account prior biological knowledge, GSEA examines whether test statistics for a group of related genes have consistent deviation from chance.15,16 As shown recently in studies on working memory,17 autism,18 bipolar disorder,19–21, ADHD22 and schizophrenia.21,23,24 GSEA can identify convergent molecular pathways relevant to neuropsychiatry.
Here we studied the enrichment of biologically defined gene sets in EM across ages, in EM-related brain activity, and in an EM-related neurodegenerative disorder (Fig. 1). Genome-wide GSEA of EM performance was performed in multiple independent data sets of young and aged cognitively healthy subjects (n=3806). In a large case-control sample (n=54162) we also performed GSEA for the risk of sporadic AD.
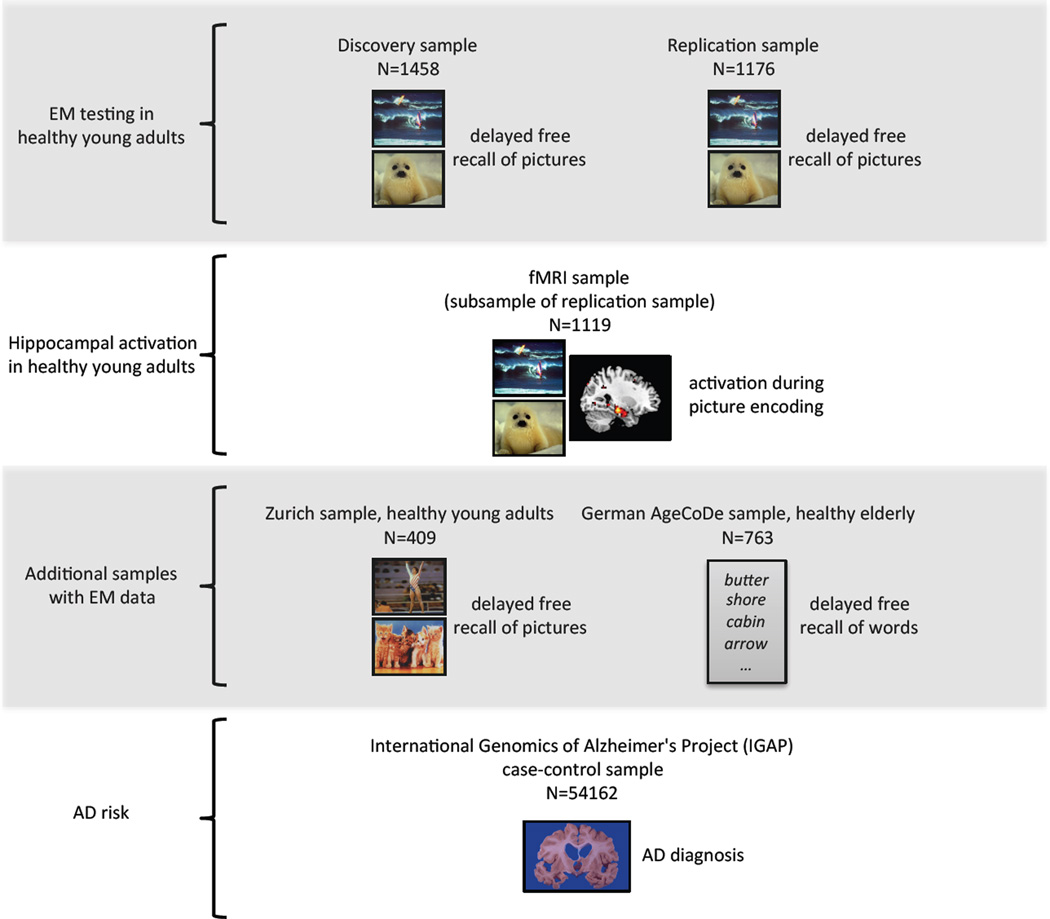
Fig. 1.
Schematic description of study workflow and included samples. First row, top: Testing for EM performance in the discovery sample (n=1458) and the replication sample (n=1176) of healthy young adults was done by quantifying the delayed free recall of IAPS pictures. The identical picture set was presented in both samples. Second row: This fMRI sample of healthy young adults (n=1119) was a subsample of the replication sample and was used for quantification of hippocampal activation during encoding of IAPS pictures (identical picture set as before). Third row: These additional samples of 409 healthy young and 763 healthy elderly adults were tested for EM performance by quantifying the delayed free recall of IAPS pictures (young sample, different picture set compared to the discovery and replication samples) or of words (elderly sample). Fourth row, bottom: This large case-control sample (n=54162) was used to test for enrichment of the episodic memory (EM) core gene set. Phenotype of interest was the diagnosis of Alzheimer’s disease.
Methods
Samples
Discovery sample
This sample is part of an ongoing, continuously recruiting behavioral genetics study in the city of Basel, Switzerland. For the purposes of this study (data lock August 2013), data from 1458 healthy young Swiss adults were available (eTable 1). Subjects were free of any neurological or psychiatric condition and did not take medication at the time of the experiment. All participants gave written informed consent before participation and completed a picture delayed free recall task, which reflects EM performance. For a detailed description of the procedure please refer to the online-only material.
Replication sample
This sample is part of an ongoing, continuously recruiting imaging genetics study in the city of Basel, Switzerland. For the purposes of this study (data lock August 2013), data from 1176 healthy young Swiss adults were available (eTable 1). Subjects were free of any neurological or psychiatric condition and did not take medication at the time of the experiment. All participants gave written informed consent before participation and, while undergoing fMRI acquisition, completed a similar picture delayed free recall task as in the discovery sample. For a detailed description of the procedure please refer to the online-only material.
The ethics committee of the Canton of Basel approved the study protocol.
Zurich sample
We recruited 409 healthy young Swiss adults for a behavioral genetics study in the city of Zurich, Switzerland (eTable 1). Subjects were free of any neurological or psychiatric condition and did not take medication at the time of the experiment. All participants gave written informed consent before participation and completed a picture delayed free recall task. For a detailed description of the procedure please refer to the online-only material.
The ethics committee of the Canton of Zurich approved the study protocol.
Healthy elderly sample
This sample consisted of 763 healthy elderly participants of the German Study on Ageing, Cognition and Dementia in primary care patients (AgeCoDe, total n=3327) (eTable 1). The AgeCoDe study is an ongoing primary care-based prospective longitudinal study on early detection of mild cognitive impairment and dementia established by the German Competence Network Dementia.25 Please also refer to the online-only material.
Genetic heterogeneity
For each of the four cognitively healthy samples, the genomic control inflation factor lambda (λGC) was calculated to assess admixture.26 λGC showed a range between 1.0046 and 1.0449, indicating the absence of noteworthy admixture in these samples (eFigures 2,3,4,5).
Array-based SNP genotyping
Samples were processed as described in the Genome-Wide Human SNP Nsp/Sty 6.0 User Guide (Affymetrix). SNP calls and array quality control were performed using the command line programs of the Affymetrix Power Tools package (version: apt-1.14.4.1). According to the manufacturer’s recommendation, contrast QC was chosen as QC metric, using the default value of greater or equal than 0.4. All samples passing QC criteria were subsequently genotyped using the Birdseed (v2) algorithm. Mean Call Rate for all samples averaged >98.5%. The sex-check in PLINK led to the exclusion of one individual in the discovery sample and three individuals in the replication sample. IBD analysis (PLINK) indicated the absence of duplicates within and between samples. For a detailed description of the procedure please refer to the online-only material.
Brain imaging
fMRI preprocessing and first level analyses
Preprocessing and data analysis was performed using SPM8 (Statistical Parametric Mapping, Wellcome Trust Centre for Neuroimaging; http://www.fil.ion.ucl.ac.uk/spm) implemented in MATLAB R2011b (MathWorks). The functional and structural images were spatially normalized by applying DARTEL, which leads to an improved registration between subjects.27,28 For a detailed description please refer to the online-only material.
fMRI group statistics
The first-level contrast parameters were used for behavioral analyses in a random effects model (second-level analysis). We used a regression model to analyze associations between brain activation differences (meaningful vs scrambled pictures) and the multi-allelic score. Age and sex were included as covariates. A one-sample t-test was computed to assess the significance of task-related activation (meaningful vs scrambled pictures) at the group level. The analysis was focused on the left and right hippocampi, defined using the template-based hippocampal ROIs (see below: Construction of a Population-Average Anatomical Probabilistic Atlas). For a detailed description please refer to the online-only material.
Statistical genetic analysis
Genome-wide association analyses
For each genome-wide analysis (Genome Reference Consortium GRCh37, hg19), P values of effectively genotyped (i.e. not imputed) variants were obtained using linear regression analyses as implemented in PLINK (number of SNPs: discovery sample: 730540 SNPs, replication sample: 736286 SNPs, Zurich sample: 737533 SNPs, AgeCoDe sample: 683072 SNPs).29 Sex and age were included as covariates. We applied the following quality control criteria: Non-significant deviation from Hardy-Weinberg equilibrium (HWE; P(HWE>0.0001)) and a minor allele frequency (MAF)>0.01. Mean per SNP call rate was >99%.
Pathway analysis
GSEA was performed using MAGENTA.30 Briefly, the method first maps SNPs onto genes. In this study, only intragenic SNPs were used for SNP-to-gene mapping (i.e., ±0kb of the annotated gene according to Genome Reference Consortium GRCh37, hg19). We chose the ±0kb threshold to avoid potentially significant biases, which are introduced by overlapping signals especially in gene-rich regions. After SNP-to-gene mapping MAGENTA assigns each gene a SNP association score (i.e. the maximum SNP P value). The analysis is corrected for gene size, number of SNPs, number of independent SNPs, number of recombination hotspots, linkage disequilibrium (LD) and genetic distance. Lastly, a gene-set enrichment-like statistical test is applied to determine if a gene set is enriched for highly ranked P values compared to a gene-set of identical size, randomly drawn from the genome. False-discovery rate (FDR) based on the 75th percentile of association P values from all genes was used for multiple testing correction. As recommended, we used the 75th percentile cutoff because it yields optimal power for weak genetic effects that are expected for highly polygenic traits (e.g., EM performance).30 The utilized gene sets are extracted and curated from the MSigDB v3.1 database (http://www.broadinstitute.org/gsea/msigdb, downloaded in February 2013), including gene-sets from different online databases (KEGG: http://www.genome.jp/kegg/, Gene Ontology GO: http://geneontology.org/, BioCarta: http://www.biocarta.com/ and Reactome: http://www.reactome.org/).31,32 We used a gene set size ranging between 20 and 200 genes to avoid both overly narrow and broad functional gene-set categories, resulting in 1’411 to be analyzed gene-sets.
We also applied INRICH (http://atgu.mgh.harvard.edu/inrich/), a software tool that examines enrichment of association signals for genetic gene-sets.33 Unlike MAGENTA, which uses each gene’s lowest P value of association with the phenotype to quantify gene set enrichment, INRICH defines LD-independent genomic regions (i.e. intervals) that contain the top GWAS hits. These intervals are then tested for overlaps with gene targets. A permutation-based resampling method is used to obtain the null distribution by random data shuffling and calculating the overlap between random test intervals of equal size as the observed ones and the gene target sets.
Multilocus genetic score calculation
To capture the multi-allelic effect of the Calcium Signaling Pathway gene set, we generated an individual multilocus genetic score.17 The score comprises variants of the Calcium Signaling Pathway gene-set (one SNP per gene) of the discovery sample that remained significant (P < 0.05) after correction for number of independent SNPs per gene, gene size, genetic distance and recombination hotspots (Table e3). The algorithm calculates the score by summing up the individual number of reference alleles over all SNPs, and finally averages the score by the number of non-missing SNPs. We weighted each SNP by the direction of effect on EM performance (i.e., with “1” (the reference allele enhances performance) or “−1” (the reference allele decreases performance)). This procedure harmonizes the single variant effects and avoids bias due to overestimation of accidentally large SNP effects.
Results
GSEA of EM in young healthy adults
Discovery sample (n=1458)
GSEA was performed with MAGENTA.30 Among the 1411 database-derived gene-sets, MAGENTA identified significant enrichment (FDR < 0.05; multiple testing-corrected) for one gene set, the Calcium Signaling Pathway gene set (KEGG entry: hsa04020) (Table 1). The Calcium Signaling Pathway gene set was also significant when applying INRICH,33 an alternative GSEA method. Of 864 independent intervals that contained the best genome-wide association signals, INRICH identified 26 intervals overlapping with the target genes of the Calcium Signaling Pathway gene set. Subsequent permutation analysis was done by generating a null distribution through calculation of multiple random intervals of the same size as the observed ones and computation of the overlap with the target gene-sets. This analysis revealed significant enrichment for the Calcium Signaling Pathway gene set (Pempirical=0.021).
Table 1.
GSEA results (FDR q < 0.25) in the discovery sample.
| Database | Gene-set ID | Gene set name | Number of genes |
Number of significant genes |
P-value | FDR q value |
|---|---|---|---|---|---|---|
| KEGG | hsa4020 | calcium signaling pathway | 178 | 56 | 0.0002 | 0.024 |
| Gene Ontology | GO:0015837 | amine transport | 38 | 17 | 0.0013 | 0.110 |
| Gene Ontology | GO:0015171 | amine transmembrane transporter activity | 41 | 18 | 0.0006 | 0.122 |
| Gene Ontology | GO:0043566 | structure specific DNA binding | 56 | 21 | 0.0024 | 0.140 |
| Gene Ontology | GO:0008271 | active transmembrane transporter activity | 122 | 48 | 0.0007 | 0.142 |
| Gene Ontology | GO:0045935 | positive regulation of nucleobase-nucleoside- nucleotide and nucleic acid metabolic process |
154 | 38 | 0.0017 | 0.148 |
| Gene Ontology | GO:0045893 | positive regulation of transcription-DNA dependent |
118 | 24 | 0.0025 | 0.175 |
| Gene Ontology | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter |
65 | 38 | 0.001 | 0.184 |
| Gene Ontology | GO:0051252 | positive regulation of RNA metabolic process | 120 | 48 | 0.0039 | 0.187 |
| Gene Ontology | GO:0004930 | G protein coupled receptor activity | 191 | 16 | 0.004 | 0.193 |
| Gene Ontology | GO:0017171 | serine hydrolase activity | 46 | 16 | 0.0059 | 0.195 |
| Gene Ontology | GO:0008236 | serine type peptidase activity | 45 | 52 | 0.0064 | 0.195 |
| Gene Ontology | GO:0008233 | peptidase activity | 176 | 10 | 0.0027 | 0.197 |
| Gene Ontology | GO:0015082 | di_tri_valent inorganic cation transmembrane transporter activity |
22 | 17 | 0.0098 | 0.199 |
| Gene Ontology | GO:0008081 | phosphoric diester hydrolase activity | 40 | 17 | 0.0047 | 0.207 |
| Gene Ontology | GO:0005792 | microsome | 42 | 17 | 0.0049 | 0.209 |
| Gene Ontology | GO:0042598 | vesicular fraction | 44 | 16 | 0.0095 | 0.248 |
Replication sample (n=1176)
Next, GSEA of the identical task (picture free recall task, see Methods) was performed in an independently recruited replication sample. The Calcium Signaling Pathway gene set was enriched significantly (P=0.015).
Calcium Signaling Pathway allelic load correlates with hippocampal activation
In an additional experiment, conducted in a subgroup (n=1119) of the replication sample, functional magnetic resonance imaging (fMRI) was used to identify gene set-dependent differences in brain activity related to EM (see Methods). We focused our search on the hippocampus because i) the Calcium Signaling Pathway gene set was associated with EM, which depends on the hippocampus,34–37 and ii) genes of this gene set are part of the signaling cascade involved in the formation of hippocampus-dependent memory in vertebrates.38–42 Thus, the left and right hippocampi served as regions of interest (ROI). Independently of allelic load, we detected highly robust picture encoding-related activation (contrast: meaningful vs scrambled pictures) in the hippocampus (Fig. e1). To capture the multi-allelic effect of the Calcium Signaling Pathway gene set on hippocampal activity, we generated an individual multilocus genetic score17 (see Methods). Of note, the multiallelic score for this sample was calculated by using only those SNPs (including the respective directions of effect) that proved significant in the independent discovery sample (see Methods). This procedure prevented model overfitting.
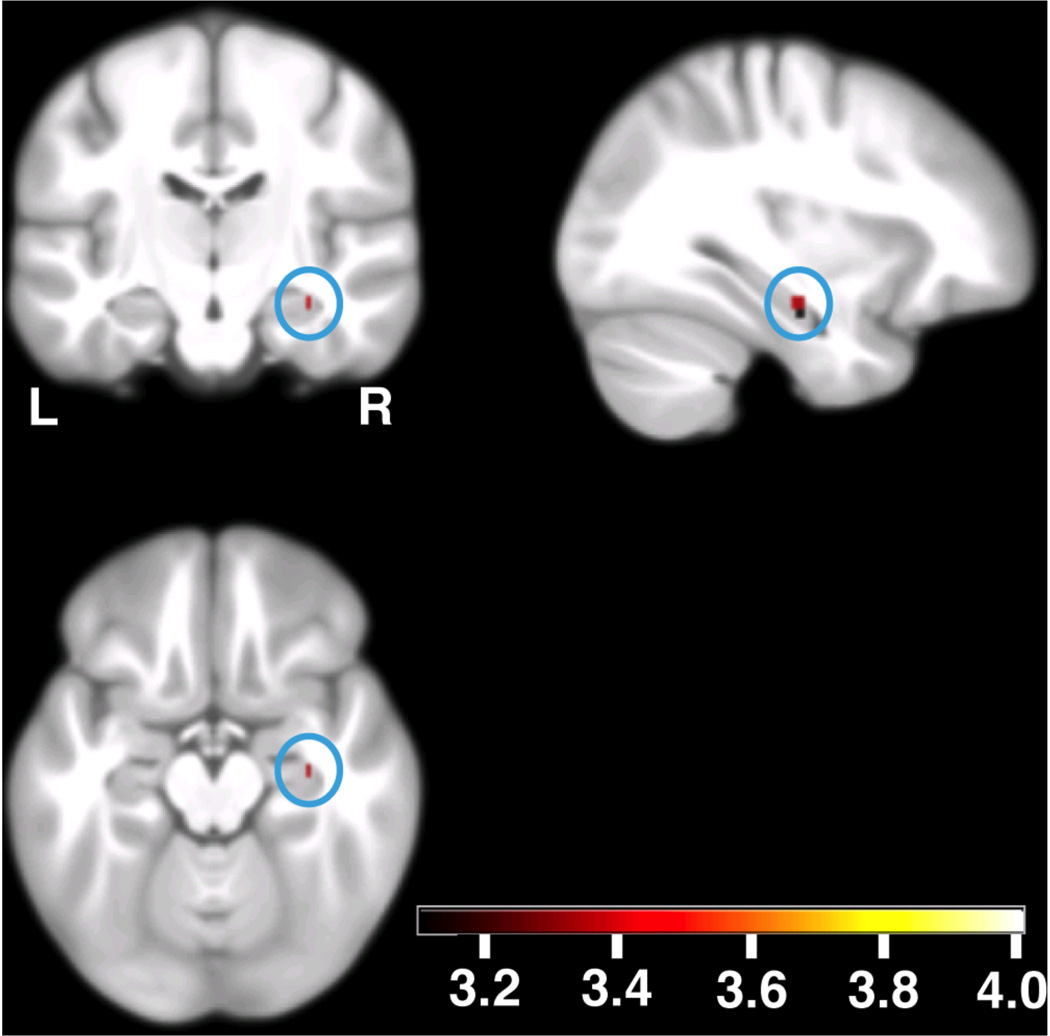
Genetic score-dependent analysis revealed a significant positive correlation between genetic score values and activation in the right hippocampus (peak at [33 −16.5 −16]; t=3.35; Puncorrected=0.0004, Psmall volume correction (SVC)<0.05, Fig. 2; genotype-independent task-related activation at this coordinate t=42.51). The peak association in the left hippocampus [−24.75 −13.75 −28] did not survive small volume correction (t=2.36; Puncorrected=0.009, PSVC> 0.05). No significant effect of the multi-allelic score on gray matter volume was found within the hippocampal ROI.
Fig. 2.
Allelic load-dependent increases in EM-related brain activity (n=1119). The blue circles show the activation in the right hippocampus. The local maximum is located at [33 −16.5 −16]; t=3.35, Puncorrected=0.0004, Psmall volume correction (SVC) < 0.05. Activations are overlaid on coronal (upper left), sagittal (upper right) and axial (lower left) sections of the study specific group template (see Methods); displayed at Puncorrected=0.001, using color-coded t values. L, left side of the brain; R, right side of the brain.
EM core gene set
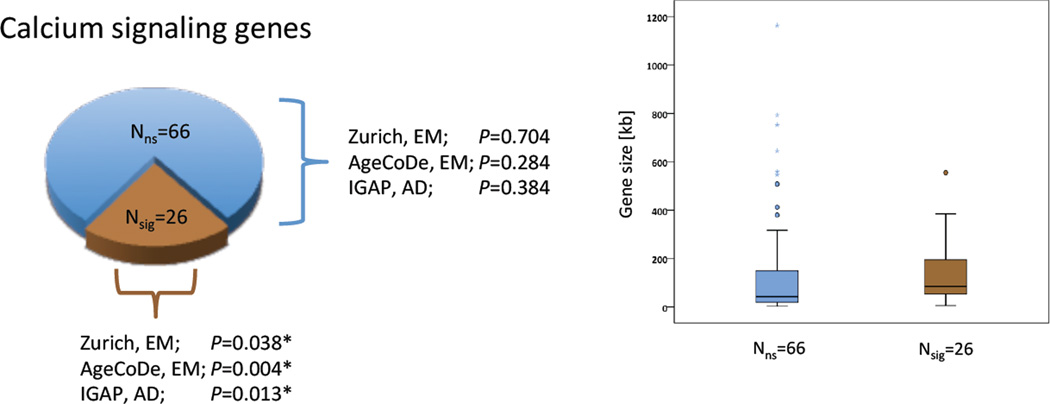
GSEA tests for statistical enrichment at gene set level. Thus, a certain gene set might prove significant in two different samples without any overlap of the gene set components, which gave rise to the significant enrichment (i.e. significantly associated genes), between samples. We tested this possibility by comparing the significant components of the Calcium Signaling Pathway gene set between the discovery and the replication sample. The overlap was significant (P=0.007; exact hypergeometric probability). Of the 144 Calcium Signaling Pathway genes, 26 genes contributed to gene set significance in both samples, whereas 66 genes did not contribute to gene set significance in either sample. The remaining 52 genes contributed to gene set significance in one of the two samples. Thus, the former group of 26 genes was defined as the replicated EM core gene set (Table 2, Fig. 3, Table e2, Table e4). Further exploratory analysis (see Methods) revealed that the EM core gene set was highly significantly enriched with genes involved in the elevation of cytosolic calcium (42.3% of the genes, P=8.9 × 10−18). In comparison, the enrichment of the group of 66 non-contributing genes with molecules involved in the elevation of cytosolic calcium (10.6% of the genes, P=2.7 × 10−7) was 10 orders of magnitude weaker.
Table 2.
GSEA results in the discovery and replication samples. Genes shown in this table were identified by GSEA as significant constituents of the Calcium Signaling Pathway in the respective sample. Genes highlighted bold are members of the EM core gene set (i.e. significant in the discovery and the replication sample). All gene symbols according to HGNC nomenclature.
| Discovery sample (Basel 1, n=1458) | ||||||||
| ADCY2 | ADCY4 | ADCY8 | ADCY9 | ADRA1A | ADRA1B | ADRA1D | ATP2A2 | ATP2B2 |
| ATP2B4 | AVPR1A | BST1 | CACNA1A | CACNA1B | CACNA1E | CACNA1G | CACNA1S | CAMK2G |
| CCKBR | CHRM1 | CHRM3 | CHRM5 | EGFR | GNA15 | GNAQ | GRIN2A | GRM1 |
| HTR2A | ITPKB | ITPR1 | ITPR2 | ITPR3 | LHCGR | MYLK | MYLK2 | NOS3 |
| NTSR1 | OXTR | P2RX5 | P2RX7 | PDE1B | PDGFRA | PDGFRB | PLCB2 | PLCD4 |
| PLCG2 | PPP3CA | PPP3R1 | PRKCB | PTAFR | PTGER3 | RYR3 | SLC8A1 | SLC8A3 |
| TACR1 | TRPC1 | |||||||
| Replication sample (Basel 2, n=1176) | ||||||||
| ADCY3 | ADCY8 | ADRA1A | ATP2B4 | AVPR1A | CACNA1E | CACNA1G | CACNA1I | CACNA1S |
| CAMK2A | CAMK2B | CAMK2G | CCKAR | CCKBR | CHP2 | CHRM5 | CHRNA7 | EDNRA |
| EDNRB | GNA14 | GNA15 | GNAL | HRH2 | HTR2A | HTR5A | ITPKB | ITPR1 |
| NOS1 | NOS2 | P2RX4 | P2RX7 | PDGFRA | PDGFRB | PLCB2 | PLCD4 | PLCE1 |
| PLCG2 | PLCZ1 | PLN | PPP3CA | PPP3R1 | PRKCB | PTGER3 | PTK2B | RYR3 |
| TACR1 | TNNC2 | VDAC2 | ||||||
Fig. 3.
GSEA results of two different gene sets: EM core gene set (brown color), group of 66 non-significant genes (blue color). Left panel: enrichment P values of the respective gene sets in three samples (Zurich sample, AgeCoDe sample, IGAP sample). Right panel: Box-plot of gene size distribution stratified by gene set. Filled circles indicate outliers, stars indicate extreme values. The average length (± S.D.) of the 26 significant genes is 144.6 kb ± 139.7 kb. The average length of the 66 non-significant genes is 143.8 kb ± 226.9 kb. The difference is not significant (P=0.9).
GSEA in additional samples
Zurich sample (n=409)
Participants performed a picture free recall task similar to the task used in the discovery and replication samples (see Methods). The EM core gene set was significantly enriched (P=0.038, Fig. 3). No significant enrichment (P=0.704, Fig. 3) was found for the set of 66 genes, which did not contribute to the significance of the Calcium Signaling Pathway gene set in any of the discovery and the replication samples.
GSEA in non-demented elderly subjects, AgeCoDe sample (n=763)
This sample of cognitively healthy elderly individuals was included to investigate whether the observed association of the EM core gene set with EM performance can be also observed in older adults. In analogy to the discovery, replication, and the Zurich sample, genome-wide P values for association with EM performance (delayed verbal free recall, see Methods) under the additive genetic model were used for GSEA. MAGENTA revealed significant enrichment (P=0.004, Fig. 3) of the EM core gene set. Also in this sample, no significant enrichment (P=0.284, Fig. 3) was found for the set of 66 genes, which did not contribute to the significance of the Calcium Signaling Pathway gene set.
GSEA in Alzheimer’s disease
EM deficits represent a behavioral hallmark of AD11 and are observed early in the course of the disease. We investigated the enrichment of the EM core gene set in a large AD case-control sample (for a detailed study description, please refer to the online-only material).
International Genomics of Alzheimer's Project (IGAP) case-control sample43 (n=54162; ncases=17008, ncontrols=37154)
7036050 autosomal SNP P values of association with sporadic AD served as input for MAGENTA. MAGENTA was run with the identical parameters as in the studies of cognitively healthy subjects. The EM core gene set was significantly enriched (P=0.013, Fig. 3). Also in this sample, no significant enrichment (P=0.384, Fig. 3) was found for the set of 66 genes, which did not contribute to the significance of the Calcium Signaling Pathway gene set in the EM samples. No significant enrichment was observed for the entire Calcium Signaling pathway gene set (P=0.155).
Discussion
We detected consistent and robust associations between Calcium Signaling Pathway genes and human EM performance. In particular, a core gene set comprising 26 genes was significantly enriched in 4 independent cohorts of young and elderly cognitively healthy individuals (n=3806). This finding is compatible with the critical role of calcium signaling in molecular processes underlying memory, as shown in model organisms and in vitro studies:44 For example, increases in intracellular calcium are causally related to the induction of long-term potentiation (LTP) and long-term depression (LTD),45–47 two cellular correlates of learning and memory.48,49 The results of the present study suggest that genes involved in calcium signaling are also related to human episodic memory throughout adulthood.
Moreover, fMRI data revealed that the individual Calcium Signaling-related allelic load correlated with hippocampal activity measured during memory encoding. Animal studies have amply demonstrated that calcium signaling genes are crucial for the formation of hippocampus-dependent memory.38–42 Our findings suggest that calcium signaling genes are related to the formation of hippocampus-dependent memory also in humans.
Interestingly, the EM core gene set was also significantly enriched in a large case-control study of sporadic AD. Calcium signaling dysregulation has been repeatedly observed in cell culture and animal models of AD.50,51 Our results argue in favor of a role for calcium signaling genes in AD and support recently published studies, which identified significant enrichment of the Calcium Signaling Pathway in AD.52,53
Interestingly, the EM core gene set was significantly enriched with genes involved in the elevation of cytosolic calcium. A local increase in calcium concentrations results in a number of short-term and long-term synapse-specific alterations that are essential for dendritic development, neuronal survival, synaptic plasticity, learning, and memory.54,55 A genetic profile favoring the local increase in calcium concentrations might therefore be related to better cognitive capacities and consequently to a delayed clinical manifestation of cognitive decline in AD patients. On the other hand, a sustained increase in intracellular calcium can, in the long run, lead to neurodegeneration and cell death.56–58 In this case, a genetic profile favoring the local increase in calcium concentrations could also increase AD risk. Thus, given the complexity of the phenotypic, biological, and temporal relationship between cognition, physiological brain aging, and neurodegeneration,59,60 it is not possible to make definite inferences regarding the direction of effect of the results presented herein.
We stress that genes not contributing significantly to pathway enrichment cannot be excluded from being related to human EM. This is due to a number of possible reasons. For example, some genes might be associated with other forms or stages of EM as those investigated in the present study. Some genes may not be related to variability of memory performance because their expression in the brain might be too tightly regulated and independent of common genetic variability. Further, the results of the present study must not lead to the erroneous assumption of an exclusive role of calcium signaling genes in human EM and AD. Given the ubiquity and versatility of calcium signaling,61 it is apparent that it must play a role in a variety of neurocognitive traits.17 Nonetheless, by showing robust and consistently significant enrichment in independent cohorts of young and elderly participants, our study identifies calcium signaling as a central player of hippocampus-dependent human memory processes, both in cognitive health and disease and thereby contributes to the understanding -and hopefully treatment- of hippocampus-dependent cognitive pathology.
Supplementary Material
Acknowledgments
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
We thank Elmar Merkle, MD, Christoph Stippich, MD, and Oliver Bieri, MD for granting access to the fMRI facilities of the University Hospital Basel.
This work was funded by the Swiss National Science Foundation (Sinergia grant CRSII1_136227 to D.Q. and A.P.), by the 7th framework programme of the European Union (ADAMS project, HEALTH-F4-2009-242257), and by the German Research Network on Dementia (KND) and the German Research Network on Degenerative Dementia (KNDD), German Federal Ministry of Education and Research grants 01GI0420 and 01GI0711.
We also thank the International Genomics of Alzheimer's Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer's disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer's Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer's Association grant ADGC–10–196728.
Angela Heck had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Author contributions: A.H., M.F., H.B., F.J., H.K., W.M., S.R-H, M.W., S.We., D. J.-F. d. Q. & A.P. designed and conceptualized the study. A.H., M.F., B.A., D.C., L.G., V.F., F.J., H.K., A.M., M.P., S.R-H, C.V., M.W., S. We., S. Wo. & A.P. did the data analysis. F.H., H.B., W.M., M.P., K.S., M.W., S.We. & S.Wo. were responsible for data collection and preparation. D. J.-F. d. Q. & A.P. supervised the project. All authors contributed to the writing of the manuscript.
Conflicts of interest: There are no conflicts of interest to declare.
References
- 1.Tulving E. Episodic memory: from mind to brain. Annu Rev Psychol. 2002;53:1–25. doi: 10.1146/annurev.psych.53.100901.135114. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon M, Plomin R, Fulker DW, Corley R, DeFries JC. Multivariate path analysis of specific cognitive abilities data at 12 years of age in the Colorado Adoption Project. Behav Genet. 1998 Jul;28(4):255–264. doi: 10.1023/a:1021667213066. [DOI] [PubMed] [Google Scholar]
- 3.Bouchard TJ, Jr, Lykken DT, McGue M, Segal NL, Tellegen A. Sources of human psychological differences: the Minnesota Study of Twins Reared Apart. Science. 1990 Oct 12;250(4978):223–228. doi: 10.1126/science.2218526. [DOI] [PubMed] [Google Scholar]
- 4.McClearn GE, Johansson B, Berg S, et al. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 1997 Jun 6;276(5318):1560–1563. doi: 10.1126/science.276.5318.1560. [DOI] [PubMed] [Google Scholar]
- 5.Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: dealing with complexity. Trends Cogn Sci. 2011 Sep;15(9):381–387. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Balota DA, Dolan PO, Duchek JM. Memory changes in healthy young and older adults. In: Tulving E, Craik FIM, editors. Handbook of memory. Oxford: Oxford University Press; 2000. pp. 395–409. [Google Scholar]
- 7.Bernard FA, Desgranges B, Eustache F, Baron JC. Neural correlates of age-related verbal episodic memory decline: a PET study with combined subtraction/correlation analysis. Neurobiol Aging. 2007 Oct;28(10):1568–1576. doi: 10.1016/j.neurobiolaging.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Grady CL, Craik FI. Changes in memory processing with age. Curr Opin Neurobiol. 2000 Apr;10(2):224–231. doi: 10.1016/s0959-4388(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 9.Light LL. Memory and aging: four hypotheses in search of data. Annu Rev Psychol. 1991;42:333–376. doi: 10.1146/annurev.ps.42.020191.002001. [DOI] [PubMed] [Google Scholar]
- 10.Langbaum JB, Fleisher AS, Chen K, et al. Ushering in the study and treatment of preclinical Alzheimer disease. Nature reviews. Neurology. 2013 Jul;9(7):371–381. doi: 10.1038/nrneurol.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & dementia : the journal of the Alzheimer's Association. 2011 May;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer's disease. Lancet neurology. 2013 Jan;12(1):92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- 13.Debette S, Ibrahim Verbaas CA, Bressler J, et al. Genome-wide studies of verbal declarative memory in non-demented older people: the CHARGE consortium. Biological Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez A, van der Flier WM, Herold C, et al. SUCLG2 identified as both a determinator of CSF Abeta1-42 levels and an attenuator of cognitive decline in Alzheimer's disease. Hum Mol Genet. 2014 Dec 15;23(24):6644–6658. doi: 10.1093/hmg/ddu372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends in genetics : TIG. 2012 Jul;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011 Jul;98(1):1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heck A, Fastenrath M, Ackermann S, et al. Converging genetic and functional brain imaging evidence links neuronal excitability to working memory, psychiatric disease, and brain activity. Neuron. 2014 Mar 5;81(5):1203–1213. doi: 10.1016/j.neuron.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011 Jun 16;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmans P, Green EK, Pahwa JS, et al. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009 Jul;85(1):13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 2011 Oct;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The N, Pathway Analysis Subgroup of the Psychiatric Genomics C. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18(2):199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stergiakouli E, Hamshere M, Holmans P, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012 Feb;169(2):186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Dushlaine C, Kenny E, Heron E, et al. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011 Mar;16(3):286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 24.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014 Jul 24;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luck T, Riedel-Heller SG, Kaduszkiewicz H, et al. Mild cognitive impairment in general practice: age-specific prevalence and correlate results from the German study on ageing, cognition and dementia in primary care patients (AgeCoDe) Dement Geriatr Cogn Disord. 2007;24(4):307–316. doi: 10.1159/000108099. [DOI] [PubMed] [Google Scholar]
- 26.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999 Dec;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007 Oct 15;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Klein A, Andersson J, Ardekani BA, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009 Jul 1;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segre AV, Consortium D, et al. investigators M, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010 Aug;6(8) doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000 May;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research. 1999 Jan 1;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee PH, O'Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012 Jul 1;28(13):1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habib R, Nyberg L. Neural correlates of availability and accessibility in memory. Cereb Cortex. 2008 Jul;18(7):1720–1726. doi: 10.1093/cercor/bhm201. [DOI] [PubMed] [Google Scholar]
- 35.Milner B. Disorders of learning and memory after temporal lobe lesions in man. Clinical neurosurgery. 1972;19:421–446. doi: 10.1093/neurosurgery/19.cn_suppl_1.421. [DOI] [PubMed] [Google Scholar]
- 36.Schacter DL, Tulving E. Memory Systems. Cambridge, MA: MIT Press; 1994. [Google Scholar]
- 37.Squire LR, Alvarez P. Retrograde amnesia and memory consolidation: a neurobiological perspective. Curr Opin Neurobiol. 1995 Apr;5(2):169–177. doi: 10.1016/0959-4388(95)80023-9. [DOI] [PubMed] [Google Scholar]
- 38.Cummings JA, Mulkey RM, Nicoll RA, Malenka RC. Ca2+ signaling requirements for long-term depression in the hippocampus. Neuron. 1996 Apr;16(4):825–833. doi: 10.1016/s0896-6273(00)80102-6. [DOI] [PubMed] [Google Scholar]
- 39.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001 Nov 2;294(5544):1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 40.Nicoll RA, Malenka RC. Contrasting properties of two forms of long-term potentiation in the hippocampus. Nature. 1995 Sep 14;377(6545):115–118. doi: 10.1038/377115a0. [DOI] [PubMed] [Google Scholar]
- 41.Shobe J. The role of PKA, CaMKII, and PKC in avoidance conditioning: permissive or instructive? Neurobiol Learn Mem. 2002 May;77(3):291–312. doi: 10.1006/nlme.2001.4022. [DOI] [PubMed] [Google Scholar]
- 42.Tonegawa S, Nakazawa K, Wilson MA. Genetic neuroscience of mammalian learning and memory. Philos Trans R Soc Lond B Biol Sci. 2003 Apr 29;358(1432):787–795. doi: 10.1098/rstb.2002.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013 Dec;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker KD, Edwards TM, Rickard NS. The role of intracellular calcium stores in synaptic plasticity and memory consolidation. Neurosci Biobehav Rev. 2013 Aug;37(7):1211–1239. doi: 10.1016/j.neubiorev.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004 Sep 30;44(1):5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Malenka RC, Lancaster B, Zucker RS. Temporal limits on the rise in postsynaptic calcium required for the induction of long-term potentiation. Neuron. 1992 Jul;9(1):121–128. doi: 10.1016/0896-6273(92)90227-5. [DOI] [PubMed] [Google Scholar]
- 47.Mulkey RM, Malenka RC. Mechanisms underlying induction of homosynaptic long-term depression in area CA1 of the hippocampus. Neuron. 1992 Nov;9(5):967–975. doi: 10.1016/0896-6273(92)90248-c. [DOI] [PubMed] [Google Scholar]
- 48.Artola A, Singer W. Long-term depression of excitatory synaptic transmission and its relationship to long-term potentiation. Trends Neurosci. 1993 Nov;16(11):480–487. doi: 10.1016/0166-2236(93)90081-v. [DOI] [PubMed] [Google Scholar]
- 49.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993 Jan 7;361(6407):31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 50.Missiaen L, Robberecht W, van den Bosch L, et al. Abnormal intracellular ca(2+)homeostasis and disease. Cell calcium. 2000 Jul;28(1):1–21. doi: 10.1054/ceca.2000.0131. [DOI] [PubMed] [Google Scholar]
- 51.Thibault O, Porter NM, Chen KC, et al. Calcium dysregulation in neuronal aging and Alzheimer's disease: history and new directions. Cell calcium. 1998 Nov-Dec;24(5–6):417–433. doi: 10.1016/s0143-4160(98)90064-1. [DOI] [PubMed] [Google Scholar]
- 52.Perez-Palma E, Bustos BI, Villaman CF, et al. Overrepresentation of glutamate signaling in Alzheimer's disease: network-based pathway enrichment using meta-analysis of genome-wide association studies. PloS one. 2014;9(4):e95413. doi: 10.1371/journal.pone.0095413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramanan VK, Kim S, Holohan K, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain imaging and behavior. 2012 Dec;6(4):634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson RS, Barral S, Lee JH, et al. Heritability of different forms of memory in the Late Onset Alzheimer's Disease Family Study. J Alzheimers Dis. 2011;23(2):249–255. doi: 10.3233/JAD-2010-101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng Y, Chang W, Shu C, et al. Decreased cognitive function in extended family members from the single late-onset-Alzheimer's-disease pedigree. Journal of the International Neuropsychological Society : JINS. 2013 Aug;19(7):809–819. doi: 10.1017/S1355617713000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amieva H, Mokri H, Le Goff M, et al. Compensatory mechanisms in higher-educated subjects with Alzheimer's disease: a study of 20 years of cognitive decline. Brain. 2014 Apr;137(Pt 4):1167–1175. doi: 10.1093/brain/awu035. [DOI] [PubMed] [Google Scholar]
- 57.Barral S, Bird T, Goate A, et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012 May 8;78(19):1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stern Y. Cognitive reserve. Neuropsychologia. 2009 Aug;47(10):2015–2028. doi: 10.1016/j.neuropsychologia.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redmond L, Ghosh A. Regulation of dendritic development by calcium signaling. Cell calcium. 2005 May;37(5):411–416. doi: 10.1016/j.ceca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 60.Rogaev EI. Genomics of behavioral diseases. Frontiers in genetics. 2012;3:45. doi: 10.3389/fgene.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marambaud P, Dreses-Werringloer U, Vingtdeux V. Calcium signaling in neurodegeneration. Mol Neurodegener. 2009;4:20. doi: 10.1186/1750-1326-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.