Abstract
Background
Presence of left ventricular hypertrophy on an electrocardiogram (ECG-LVH) is widely assessed clinically and provides prognostic information in some settings. There is evidence for significant heritability of ECG-LVH. We conducted a large-scale gene-centric association analysis of four commonly measured indices of ECG-LVH.
Methods and Results
We calculated Sokolow-Lyon Index, Cornell Product, 12-lead QRS Voltage Sum and 12-lead QRS Voltage Product in 10,256 individuals from 3 population based cohorts and typed their DNA using a customised gene array (the Illumina HumanCVD BeadChip 50K array) containing 49,094 genetic variants in ~2100 genes of cardiovascular relevance. We followed up promising associations in 11,777 additional individuals. We identified and replicated four loci associated with ECG-LVH indices - 3p22.2 (SCN5A, rs6797133, p=1.22×10−7) with Cornell Product and 12q13.3 (PTGES3, rs2290893, p=3.74×10−8), 15q25.2 (NMB, rs2292462, p=3.23×10−9) and 15q26.3 (IGF1R, rs4966014, p=1.26×10−7) with 12-lead QRS Voltage Sum. The odds ratio of being in the top decile for 12-lead QRS Voltage Sum for those carrying 6 trait-raising alleles at the 12q13.3, 15q25.2 and 15q26.3 loci versus those carrying 0–1 alleles was 1.60 (95% CI: 1.20 – 2.29). Lead SNPs at the 12q13.3 and 15q25.2 loci showed significant eQTL effects in monocytes.
Conclusions
These findings provide novel insights into the genetic determination of ECG-LVH. The findings could help to improve our understanding of the mechanisms determining this prognostically important trait.
Keywords: electrocardiography, left ventricular hypertrophy, genetics, genetic variation, association study
INTRODUCTION
Increased left ventricular (LV) mass (left ventricular hypertrophy, LVH) is a common manifestation of preclinical cardiovascular disease that predicts cardiovascular morbidity and mortality in some settings.1 LVH occurs in a wide range of disorders including hypertension, obesity and valvular heart disease. Its clinical relevance is further underscored by the finding that reduction of LVH with antihypertensive treatment is associated with decreased risk of cardiovascular events independent of treatment modality and degree of blood-pressure lowering.2 Several non-invasive measures of LVH have been developed including those based on echocardiography and magnetic resonance imaging. However, world-wide, the most commonly applied assessments clinically are based on the routinely recorded 12-lead electrocardiogram (ECG). These include calculations of the Sokolow-Lyon Index, the Cornell Product, the 12-lead QRS Voltage Sum and the 12-lead QRS Voltage Product (Table 1).3–6 ECG-indexed LVH and echocardiographically determined LVH (echo-LVH) predict cardiovascular mortality independently of each other7 suggesting that they capture somewhat different information on cardiac status.
TABLE 1.
Definitions of the ECG-LVH indices analysed:
|
Development of LVH in the context of pre-disposing conditions shows considerable inter-individual variability.1,2,7 This is at least partly due to genetic factors. ECG-indices of LVH have been shown to have significant heritability (Sokolow-Lyon Index ~40%, Cornell Product ~23%) that is at least similar if not higher than that for echo-LVH (~26%).8 A previous linkage analysis found suggestive evidence for loci on chromosomes 10q23.1 (LOD 2.21) for Sokolow-Lyon Index, on 17p13.3 (LOD 2.67) for Cornell Product and on 12q14.1 (LOD 2.19) for echo-LVH.9 Recently, a meta-analysis (n=12,612) of genome-wide association studies of echo-LV mass and LV wall thickness did not find any definite associations with either of these traits.10 To gain a better understanding into the genetic determinants of ECG-LVH, we undertook a large-scale cardiovascular gene-centric analysis of Sokolow-Lyon Index, Cornell Product, 12-lead QRS Voltage Sum and 12-lead QRS Voltage Product in three population-based cohorts followed by replication of promising signals in three further sample sets. For the discovery stage, we used a recently developed customised array, the Illumina HumanCVD BeadChip 50K array, which provides extensive coverage of ~2100 genes/loci of highest relevance to the cardiovascular system.11
MATERIALS AND METHODS
Study cohorts
For the discovery stage, we studied individuals from three population-based cohorts: The British Women’s Heart and Health Study (BWHHS, n= 3443)12, the Genetic Regulation of Arterial Pressure of Humans in the Community (GRAPHIC) Study (n= 2024)13 and the Whitehall II Study (WHII) (n=5059).14 The replication cohorts comprised individuals from the British Regional Heart Study (n=3519)15, the British Genetics of Hypertension (BRIGHT) Study (n=1198)16 and the Prevention of Renal and Vascular End stage Disease (PREVEND) Study (n=7060)17. Summary characteristics of the cohorts are given in Table 2 with further details on recruitment and assessment in Supplementary Materials.
TABLE 2.
Demographic and clinical characteristics for discovery and replication studies
| Whitehall II | BWHHS | GRAPHIC | BRHS | BRIGHT | PREVEND | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Males (N=3721) |
Females (N=1338) |
Males (N=0) |
Females (N=3443) |
Males (N=1021) |
Females (N=1003) |
Males (N=3519) |
Females (N=0) |
Males (N=485) |
Females (N=713) |
Males (N=3298) |
Females (N=3762) |
|
| Age(yrs) | 60.8(5.9) | 61.2(6.1) | 68.9(5.5) | 39.4(15.1) | 39.2(13.9) | 68.8(5.5) | 56.7(10.7) | 57.7(10.1) | 49.6(12.8) | 47.9(12.1) | ||
| BMI(kg/m2) | 26.6(3.8) | 27.0(5.5) | 27.8(4.9) | 26.4(4.3) | 25.8(4.9) | 26.8(3.7) | 27.7(3.8) | 27.1(4.8) | 26.2(3.6) | 25.8(4.6) | ||
| DBP(mmHg) | 75.1(10.4) | 73.2(10.7) | 79.4(11.9) | 73.5(8.1) | 69.9(6.8) | 85.2(11.1) | 96.8(11.0) | 91.7(12.0) | 76.7(9.6) | 70.9(8.9) | ||
| SBP(mmHg) | 128.7(16.1) | 126.5(18.2) | 147.0(25.1) | 122.5(10.1) | 114.9(9.8) | 149.2(24.2) | 158.0(20.9) | 151.2(21.8) | 133.3(18.2) | 123.8(20.3) | ||
| % On BP-lowering drugs | 23.1 | 22.1 | 30 | 7.9 | 5.4 | 28.0 | 100 | 100 | 14.6 | 13.5 | ||
| Corrected SBP(mmHg) | 132.2(18.1) | 129.7(20.6) | 151.6(27.3) | 123.4(10.9) | 115.5(10.6) | 153.4(25.8) | 172.9(20.9) | 166.2(21.9) | 135.4(20.2) | 125.8(22.5) | ||
| Cornell Product(μV.s) | 147.5(58.8) | 163.8(54.1) | 183.9(53.3) | 137.0(48.6) | 143.5(40.0) | 143.4 (65.0) | 174.3(94.3) | 188.3(73.3) | 126.7(55.7) | 146.4(46.4) | ||
| QRS Voltage Product(μV.s) | 1437.5(398.9) | 1127.7(312.6) | 1197.6(340.4) | 1570.3(433.2) | 1130.4(265.8) | 1384.6(405.2) | 1471.5(413.6) | 1192.4(335.7) | 1576.6(365.8) | 1268.5(302.5) | ||
| QRS Voltage Sum(μV) | 14740.4(3056.7) | 12488.1(2629.2) | 13158.9(2812.2) | 16351.1(3830.4) | 12923.7(2636.3) | 13807.9(2887.8) | 14587(3407) | 12563(2990) | 15700.1(3349.6) | 13761.8(3020.5) | ||
| Sokolow-Lyon Index(μV) | 2156.2(718.1) | 1924.6(587.9) | 1850.3(541.0) | 2395.7(643.6) | 2125.1(539.2) | 2069.5(677.3) | 2326(743) | 2180(650) | 2522.5(695.6) | 2169.0(591.7) | ||
BMI=Body Mass Index; DBP=Diastolic Blood Pressure; SBP=Systolic Blood Pressure; μV=microvolts; μV.s=microvolt seconds.
Analysis in both the discovery and replication cohorts was restricted to subjects of European white descent. Ethics approval for all of studies was obtained from local research ethics committees and participants provided written informed consent.
Electrocardiographic measures of left ventricular hypertrophy
For the BWHHS, GRAPHIC, WII, BRHS and BRIGHT studies, standard 12-lead ECGs were recorded on either Siemens 460 electrocardiographs or Burdick Eclipse or Atria models. Digital ECG data was in all cases transferred to the University of Glasgow ECG Core Lab based in Glasgow Royal Infirmary and ECGs were analysed by the University of Glasgow ECG analysis program.18 This software meets all of the required specifications in terms of measurement accuracy and is used widely in various commercial products and clinical trials. All ECGs were reviewed manually and checked for technical problems which would have interfered with analysis and technically unsatisfactory ECGs were excluded. In PREVEND, standard 12-lead ECGs were recorded with CardioPerfect equipment (Cardio Control; currently Welch Allyn, Delft, The Netherland) and were electronically stored and digitally analysed using the computer program MEANS (Modular Electrocardiogram Analysis System).19
For each ECG-LVH index, outliers more than 3 standard deviations away from the mean were excluded from the analysis. Based on the observed distribution of the phenotypic measures (Supplementary Figure 1), analysis was done on untransformed data.
Genotyping and Quality Control (QC)
The discovery cohorts were all genotyped identically using the Illumina HumanCVD BeadChip 50K array.11 Genotypes were called using the Illumina BeadStudio (v3) Genotyping Module which uses the GenCall software application to automatically cluster, call genotypes and assign confidence scores. Samples with a genotype call rate less than 90% were removed from the analysis. SNPs were only included in the association analysis if they satisfied the following criteria: SNP call rate ≥ 0.95, minor allele frequency > 1% and Hardy-Weinberg Equilibrium p-value > 1 × 10−04. For replication of the SNPs taken forward from the discovery phase, genotypes were generated de novo for the PREVEND Study and BRHS using KASPAR assays (K Bioscience, UK) and extracted in silico from HumanCVD 50K array data for the BRIGHT Study.
Statistical Analysis
Discovery
For the discovery cohorts, separate within-cohort linear regression analyses were performed for each ECG-LVH trait using an additive genetic model adjusting for age, sex, BMI, and systolic blood pressure (SBP). For individuals on blood pressure lowering medication, SBP was adjusted by adding a constant of 15mmHg as recommended by Tobin et al 20. Covariates were selected based on prior knowledge and direct evidence of their correlation with ECG-LVH indices in the three cohorts (Supplementary Table 1). In GRAPHIC, additional adjustments for age2 (due to the two generational structure of the cohort) and familial correlation were taken into account using generalised estimating equations with an exchangeable correlation structure.
After verifying strand alignment, a meta-analysis of the results from the three studies was conducted. Because ~30% of SNPs passing our replication threshold of 1×10−4 showed moderate heterogeneity (I2 of between 40 and 70), using the DerSimonian-Laird estimator (I2) for heterogeneity21, we chose a random-effects model for our primary analysis. Use of a fixed effects model did not identify any additional loci passing this threshold. Bonferroni correction for 33,950 SNPs that passed QC in all 3 discovery cohorts is 1.47×10−6. However, given the fact that the prior odds of association in this cardiovascular gene-centric study are likely to be higher than for a whole-genome array, and the opportunity to eliminate false positives at the replication stage, the discovery threshold was relaxed to 1×10−4 to take SNPs forward for replication. Using the qvalue package in R (v1.26.0) (http://cran.r-project.org/web/packages/qvalue/index.html), this gave an estimated false discovery rate of (FDR) of 26.3%. Where multiple SNPs within the same locus reached significance, we selected the lead SNP from each locus to take forward for replication. At each locus conditional association analyses were performed, whereby the lead SNP was added to the model as a covariate to identify any additional independent signals.
Replication
Linear regression analysis was carried out in each of the three replication cohorts for the selected SNPs with the trait with which they were significantly associated in the discovery stage. Adjustments were again made for age, sex, BMI and corrected SBP. A meta-analysis of the results from the replication studies was done using the same method as in the discovery phase. A Bonferroni correction for the number of SNPs taken to replication (12) tested was applied to the standard 0.05 threshold, giving a significance threshold of p=4.17×10−3. Power of the replication study was calculated as detailed in Supplementary Materials.
Calculation of heritability of ECG-LVH traits
We took advantage of the family structure of the GRAPHIC Study13 to calculate heritability of the ECG-LVH traits. Heritability estimates were calculated using variance component analyses as implemented in SOLAR version 4.2.722 and adjusted for age and sex (model 1) and then for several other covariates (model 2) as described in Supplementary Materials.
Calculation of variance explained
A univariate linear regression was used to determine the amount of trait variation explained by the SNP of interest within each study. In GRAPHIC only the parental generation was used to calculate the r2 value. To calculate the combined variance explained by each of the 4 SNPs across all 3 studies we used the metacor package (v1.0.1) (http://cran.r-project.org/web/packages/metacor/index.html) in R v2.11.1 (http://cran.r-project.org/) which implements the DerSimonian-Laird (DSL)21 random-effect meta-analytical approach with correlation coefficients as effect sizes.
Top decile analysis
We examined the effect of our four significant SNPs on the odds of being in the top decile of their respective trait, QRS Voltage Sum (rs2290893, rs2292462, rs4966014) and Cornell Product (rs6797133). This was performed in each discovery study individually using logistic regression and the results meta-analysed using random effects (fixed effects analysis provided similar results). In addition, for QRS Voltage Sum, a risk score was generated as the sum of risk alleles for each individual, assuming an additive effect, of the three SNPs significantly associated with this trait. Using the 0–2 risk allele carrying subjects as the reference group, the odds of being in the top decile of QRS Voltage Sum were calculated for subjects carrying 3, 4, 5 or 6 risk alleles.
Functional analysis
To identify potential functional variants in the 4 novel ECG-LVH loci, we used the “SNAP” tool of the Broad Institute (http://www.broadinstitute.org/mpg/snap/), to identify other SNPs correlated by either r2 (>0.5) or D′ (> 0.8) to the lead SNPs. The list of SNPs thus generated was then applied to the “SNP Function Portal” of Brain Array (http://brainarray.mbni.med.umich.edu/Brainarray/Database/SearchSNP/snpfunc.aspx) to assess for potential functionality.
We also analysed eQTLs related to the 4 novel ECG-LVH loci in monocyte transcriptomes from 395 healthy blood donors and 363 patients with premature MI assembled by the Cardiogenics consortium. Further details of this analysis are given in Supplementary Materials.
Analysis of ECG-LVH associated variants with Echo-LV mass
Association statistics for echo-LV mass were obtained from the genome-wide meta-analysis carried out by the ECHOGEN Consortium.10 The discovery analysis for this study combined data from 5 cohorts (Cardiovascular Health Study, Framingham Heart Study, Rotterdam Study, Multinational Monitoring of Trends and Determinants in Cardiovascular Disease study (MONICA-KORA), and Gutenberg Heart Study with total sample size of 12,612. Further details are given in Supplementary Materials.
RESULTS
Discovery Stage
The discovery sample comprised 10,526 individuals in total: 3443 from BWHHS, 2024 from GRAPHIC and 5059 from the WHII Study. Cohort characteristics are shown in Table 2. The distributions of the ECG measures were very similar in each cohort (Supplementary Figure 1). There were strong intra-individual correlations between 12-lead QRS Voltage Sum and 12-lead QRS Voltage Product, moderate correlations between either Cornell Product or Sokolow-Lyon index and the 12-lead QRS indices and no correlation between Cornell Product and Sokolow-Lyon index (Supplementary Table 1). Analysis of familial correlations in the GRAPHIC study confirmed the significant heritability of each of the traits, ranging between 31%–44% (Supplementary Table 2). The number of SNPs surviving QC and taken forward for association analysis in the discovery phase ranged between 34,321 and 36,381 for the three cohorts (Supplementary Table 3). The quantile-quantile plots for each cohort are shown in Supplementary Figure 2. The genomic inflation factors ranged between 1.000 and 1.079, suggesting no substantial population stratification.
A total of 47 SNPs in 12 loci passed the discovery meta-analysis p-value threshold of < 1×10−4. Conditional analysis of the lead SNPs in each locus did not identify any additional independent effects and these were therefore selected for replication. These comprised six SNPs selected on the basis of an association with 12-lead QRS Voltage Sum, one with 12-lead QRS Voltage Product, four with Cornell Product and one with Sokolow-Lyon Index (Table 3). Replication was undertaken in 3 additional cohorts (PREVEND, BRHS and BRIGHT) totalling 11,777 individuals. Details of the replication cohorts are given in Table 2.
TABLE 3.
Meta-analysis results for the 12 SNPs associated with ECG-LVH Indices
| Index of Left Ventricular Hypertrophy |
CHR | SNP | Coded allele |
Non- coded allele |
N | Mean MAF |
Gene Locus | Discovery Beta (95% CI) |
Discovery p-value |
Replication Beta (95% CI) |
Replicati on p- value |
Estimate of Power† |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cornell Product | 3p22.2 | rs6797133 | A | G | 9367 | 0.40 | SCN5A | −3.7(−5.3, −2.2) | 3.01×10−6 | −2.1(−3.6, −0.7) | 3.87×10−3 | 99.9% |
| 4q26 | rs9993110 | T | C | 9362 | 0.45 | MYOZ2/FABP2 /PDE5A | 3.3(1.8,4.9) | 1.88×10−5 | 1.1(−0.3,2.6) | 0.11 | 70.3% | |
| 11q12.1 | rs11603020 | C | T | 9375 | 0.27 | SERPING1 | 3.4(1.7,5.1) | 7.91×10−5 | 0.6(−1.7,2.9) | 0.59 | 3.3% | |
| 12q12 | rs860867 | T | C | 9387 | 0.10 | TMEM117 | −5.1(−7.7, −2.6) | 5.94×10−5 | 1.3(−1.0,3.7) | 0.27 | 4.4% | |
| QRS Voltage Product | 6p21.31 | rs4713854 | C | A | 9410 | 0.11 | PPARD | −33.3(−49.5, −17.1) | 5.84×10−5 | −0.4(−15.3,14.5) | 0.68 | 3.2% |
| QRS Voltage Sum | 4q26 | rs11724758 | A | G | 9413 | 0.47 | FABP2/MYOZ2 /PDE5A | 176.5(95.2,257.8) | 2.09×10−5 | 83.6(5.52,161.7) | 0.036 | 44.8% |
| 5p15.31 | rs3815743 | G | A | 9386 | 0.16 | MTRR | −302.8(−412.7, −192.8) | 6.76×10−8 | −22.6(−125.8,80.5) | 0.51 | 99.9% | |
| 11q12.1 | rs17597065 | G | A | 9427 | 0.04 | TCN1 | −396.6(−592.6, −200.6) | 7.32×10−5 | 110.0(−175.8,395.8) | 0.45 | 2.5% | |
| 12q13.3 | rs2290893 | A | G | 9427 | 0.35 | PTGES3/ATP5B | −201.4(−285.6, −117.1) | 2.84×10−6 | −126.7(−205.4, −48.1) | 1.59×10−3 | 99.8% | |
| 15q25.3 | rs2292462 | G | T | 9427 | 0.45 | NMB | −218.6(−315.7, −121.5) | 1.02×10−5 | −164.3(−249.8, −78.7) | 1.68×10−4 | 98.9% | |
| 15q26.3 | rs4966014 | C | T | 9437 | 0.30 | IGF1R | −181.8(−269.3, −94.4) | 4.75×10−5 | −143.5(−225.7, −61.3) | 6.23×10−4 | 3.0% | |
| Sokolow-Lyon | 2q31.2 | rs10497520 | T | C | 9321 | 0.13 | TTN/PRKRA | −68.9(−94.6, −43.1) | 1.58 ×10−7 | −24.3(−48.0, −0.55) | 0.045 | 99.9% |
The table shows the discovery statistics for the 12 SNPs take forward for replication. Results from the meta-analysis of the replication samples are shown in the penultimate two columns. Loci that were replicated (p<4.17×10−3) are shown in bold. CHR=chromosome; N=total number of subjects analysed in the discovery cohorts for the specific SNP; Mean MAF=Mean of minor allele frequency.
Power estimate for replication calculated on the basis on an adjusted discovery beta corrected for possible Winner’s Curse as described by Zhong and Prentice.23
Replication Stage
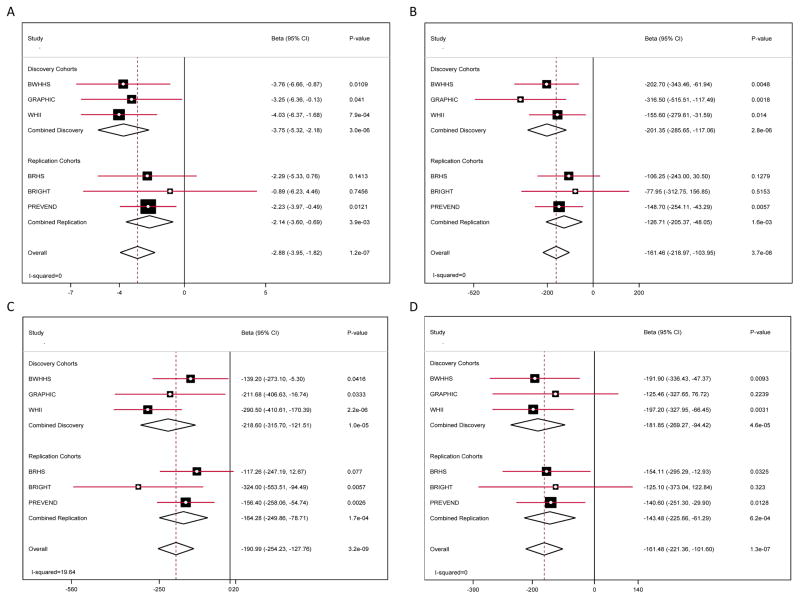
Of the 12 SNPs taken into replication, four showed evidence of significant association for their specific trait in the replication studies after allowing for the number of tests (p < 4.17×10−3) (Table 3). These were variants in the PTGES3 (12q13.3), NMB (15q25.2) and IGF1R (15q26.3) loci for 12-lead QRS Voltage Sum and the SCN5A (3p22.2) locus for Cornell Product. Meta-analysis of the combined discovery and replication data gave p-values of 3.74×10−8, 3.23×10−9, 1.26×10−7 and 1.22×10−7 for the lead SNPs in the PTGES3 (rs2290893), NMB (rs2292462), IGF1R (rs4966014) and SCN5A (rs6797133) loci, respectively (Figure 1). Summary findings in the replication samples for the SNPs that did not replicate are also shown in Table 3 and for individual studies in Supplementary Figure 3. The replication sample had > 95% power to replicate all the SNPs taken through for the effect sizes and MAF seen in the discovery cohorts. However, to formally assess the potential bias introduced by any Winner’s Curse, further power calculations were undertaken based on an adjusted discovery beta, corrected as described by Zhong and Prentice23 (see Supplementary Materials). This showed (Table 3) that for several SNPs, especially those near the p-value threshold used to select SNPs for replication, power for replication may have been very limited if the true effect sizes were actually much lower. Therefore, caution needs to be exercised in the interpretation of the non-replication of these variants.
Figure 1. Genetic association of four loci with ECG indices of left ventricular hypertrophy.
Forest plots for SNPs showing replicated association with a specific ECG index of LVH: (A) rs6797133 ((SCN5A locus) with Cornell Product; (B) rs2290893 (PTGES3) with QRS Sum; (C) rs2292462 (NMB) with QRS Sum; (D) rs4966014 (IGF1R) with QRS Sum. In each panel associations are reported by study separately for the discovery and replication cohorts, together with pooled estimates for discovery, replication and overall. Beta coefficients (with 95% confidence intervals) describe per allele effect of the minor allele of the SNP for the trait shown. A negative beta indicates that the trait had a lower value in those carrying the minor allele. The I2 value shown in the bottom left hand for each panel is the heterogeneity index for results in the meta-analysis of all studies.
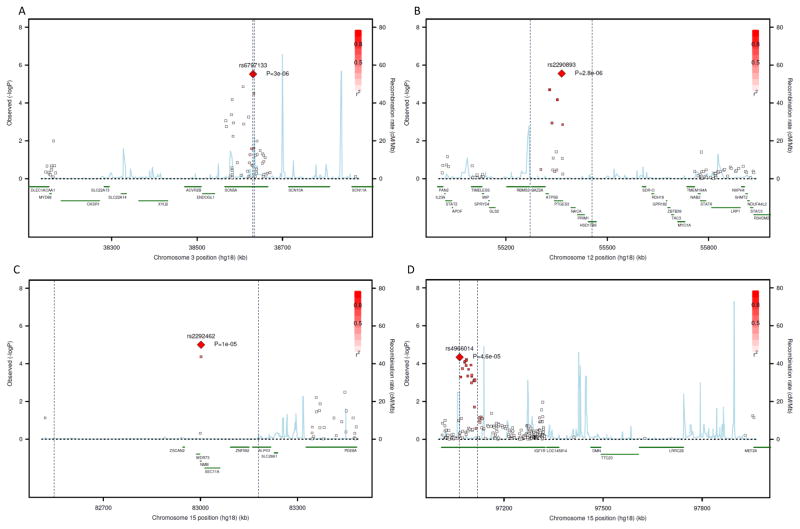
The regional association plots in the combined discovery samples for the four replicated loci, with the lead SNP highlighted are shown in Figure 2. For each locus, the chromosomal segment with SNPs showing a high (r2 > 0.8) linkage disequilibrium (LD) with the lead SNP is also marked (vertical lines). For the SCN5A and IGF1R loci, the signal was localised within the corresponding gene (Figure 2, panels A and D). However, for the PTGES3 and NMB loci, the region containing SNPs in strong LD with the lead SNP spanned several genes (Figure 2, panels B and C).
Figure 2. Regional association plots of loci associated with indices of ECG-LVH.
Each plot shows a region of the relevant chromosome on the X-axis together with association p values from the discovery study meta-analysis for individual SNPs genotyped on the IBC array on the Y-axis. The lead SNP (one showing the strongest association) for each locus is shown as a large red diamond with its corresponding p value. The other SNPs are colour-coded according to their extent of LD (r2) with the lead SNP with bright red indicating high (> 0.9) LD (see scale). The two vertical dotted lines on each panel mark the segment containing SNPs with an LD > 0.5 with the lead SNP. The identity of genes within each region and their location are shown in green below the x-axis. The blue vertical lines identify locations of recombination hot spots with the height of the lines indicating the degree of recombination activity. Panel A: 3p22.2 (SCN5A) locus for Cornell Product; Panel B: 12q13.3 (PTGES3) locus for QRS Sum; Panel C: 15q25.2 (NMB) locus for QRS Sum; Panel D: 15q26.3 (IGF1R) locus for QRS Sum.
Functional Analysis
By cross-referencing to HapMap3 data, we examined whether there were any putative functional variants within each locus that were in LD with the lead SNPs. At the NMB locus, the lead SNP (rs2292462) was in LD (r2=0.476, D′=1.0) with a non-synonymous SNP (rs1051168, Proline to Threonine), 234 base pairs away that had an association signal (p=4.3×10−5) with QRS Voltage Sum in the discovery meta-analysis comparable with that of the lead SNP.
We also examined whether the lead SNP at each locus was independently associated with expression of genes in that locus in monocytes. Full findings are presented in Supplementary Table 4. There was a strong association of rs2958155, a perfect surrogate for rs2290893 in the PTGES3 locus, with expression level of RBMS2 (p=1.85×10−37) in a co-dominant fashion with the minor allele associated with higher level of RBMS2 expression and the variant explaining 17.9% of the total variance of RBMS2 transcript level. SNP rs2958155 was also more modestly associated with the expression of PRIM1 (p= 2.59×10−8). In the NMB locus, rs2292462 showed an association with expression of WDR73 (p=9.15×10−9) and with NMB itself (p=9.27×10−11), although it should be noted that while RBMS2, PRIM1 and WDR73 had abundant transcripts within monocytes, expression level of NMB was at the lower limit of detection on the gene expression array (Supplementary Table 4). Association of the minor allele of rs2290893 (PTGES3 locus) with higher RBMS2 expression was also seen in fibroblasts in two publicly-available eQTL datasets (see Supplementary Materials)
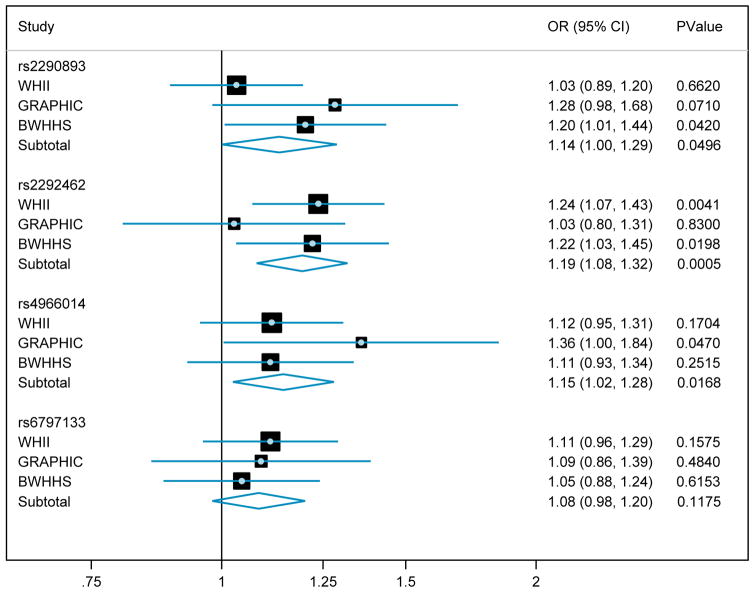
Variance Explained and Top Decile Analysis
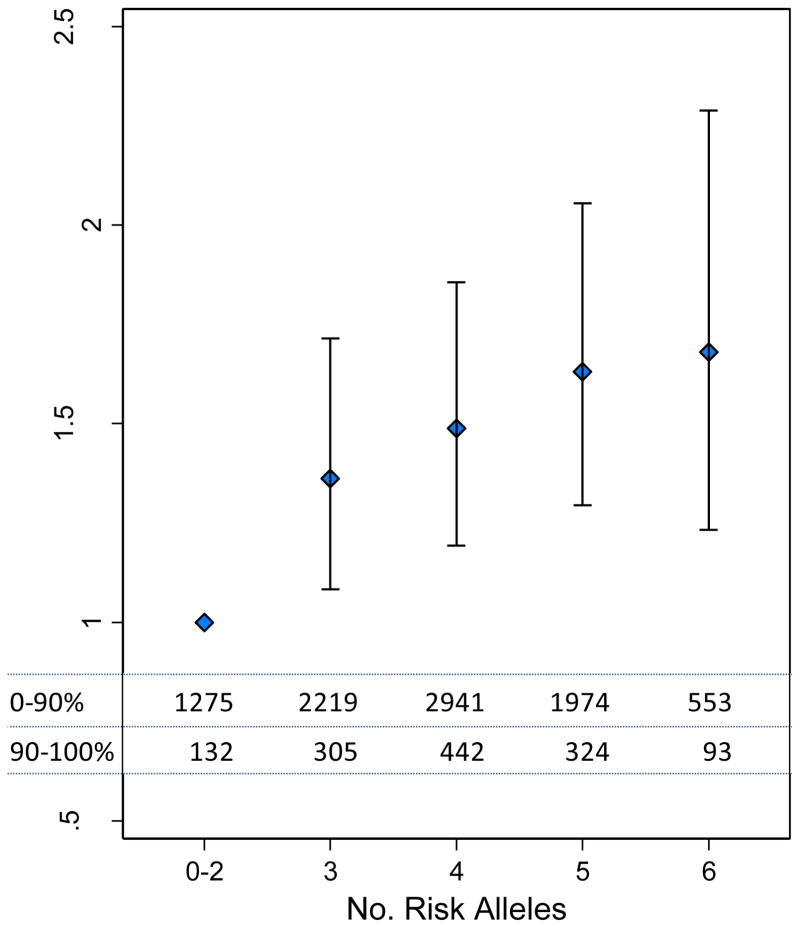
The amount of trait variance explained by each of the four novel loci was < 1% (Supplementary Table 5). To investigate the potential clinical relevance of our findings, we examined the extent to which carriage of a trait-raising allele increased the chance of being in the top 10% of individuals for that trait. This showed that this risk was increased by between 8% (rs6797133, SCN5A) and 19% (rs2292462, NMB) per allele copy (Figure 3). To assess the combined effects of the three loci (PTGES3, NMB and IGF1R) affecting QRS Voltage Sum, we compared the odds ratio of being in the top decile for the trait for those carrying 6 trait-raising alleles versus those carrying 0–2 alleles. This showed a 1.60-fold (95% CI: 1.20 – 2.29) increased likelihood of being in the top decile (Figure 4).
Figure 3. Odds Ratios for being in the top decile for the associated trait per carriage of a copy of a trait-raising allele for each of the four novel ECG-LVH loci.
Data from meta-analysis of all three discovery cohorts are shown and represented as odds ratio (95% CI) and association p-value.
Figure 4. Meta analysis of the Odds Ratio for being in the top decile for QRS Voltage Sum decile for increasing carriage of trait raising alleles at the three loci associated with QRS Voltage Sum.
The group carrying 0–2 alleles was used as the reference group. Data shows the odds ratio and 95% CI of per allele count of the three SNPs associated with this trait (rs2290893, rs2292462, rs4966014). The combined discovery cohort frequencies by allele group are shown in the table within the figure.
ECG-LVH variants and Echo-LV mass
To assess if any of our replicated variants associated with echo-LV mass, we used data from the GWA meta-analysis of this trait reported by the ECHOGEN Consortium. We found no evidence for association of any of the variants with echo-LV mass (Supplementary Table 6).
Previous candidate genes and ECG-LVH
Previous studies have suggested an association of specific variants in genes of the renin-angiotensin system cascade and LVH, notably the A1166C variant in the angiotensin 1 receptor gene, the M235T polymorphism in angiotensinogen, the insertion/deletion (I/D) polymorphism in the angiotensin converting enzyme gene and the −344 C/T polymorphism in the CYP11B2 gene. We found no evidence that these variants were associated with ECG-LVH traits in our study (Supplementary Table 7). We also analysed whether variants in other genes and pathways linked to development of LVH showed any association with ECG-LVH traits. Here we found nominal associations with variants in several genes (Supplementary Table 8). Although the level of significance achieved for these variants in the context of the large number of SNPs examined cannot exclude the possibility that many of these associations are false positives, their location within genes known to be involved in LVH, indicates that further analysis may be warranted.
DISCUSSION
In this study we confirm the significant heritability of ECG indices of left ventricular hypertrophy and report the first genetic variants robustly associated with some of these indices, providing novel insights into the determinants of this widely assessed cardiovascular trait. We further show that these variants do not associate with echo-LV mass suggesting that these phenotypes measure somewhat distinct aspects of cardiac biology.
The association of variants in the insulin growth factor 1 receptor gene IGF1R with ECG-LVH is consistent with current understanding of the role of IGF pathway in the heart where it has been linked to inhibition of cardiac muscle growth through myostatin.24 Mutations in SCN5A which encodes the sodium channel, voltage-gated, type V, alpha subunit cause long QT syndrome and common variants in SCN5A and the adjacent and functionally related gene SCN10A (Figure 2, panel A) have been associated with two other ECG parameters, PR Interval and QRS Duration.25,26 Our finding of an association of variants in SCN5A with an ECG marker of LVH suggests that the SCN family of genes may influence several aspects of cardiac electrophysiology possibly through an effect on myocyte size. The likely causal gene at the other two loci, PTGES3 and NMB, is less clear. PTGES3 (also known as p23 or TERT binding protein) interacts with the telomerase reverse transcriptase (TERT) protein which has been shown to promote hypertrophy in cultured cardiomyocytes.27 However, at this locus we also observed a strong eQTL in monocytes and fibroblasts of the lead SNP with expression of RBMS2 (RNA-binding motif protein, single stranded-interacting, 2) and a modest association with expression of PRIM1 (Primase polypeptide 1) in monocytes. RBMS2 belongs to a family of proteins which bind single stranded DNA and RNA and which have been implicated in diverse functions such as DNA replication, gene transcription, cell cycle progression and apoptosis28 while PRIM1 is involved in DNA replication.29 At the NMB locus, we also observed modest eQTL associations in monocytes of the lead SNP with expression of NMB and WDR73. Furthermore, the lead SNP is in LD (r2=0.476, D′=1.0) with a non-synonymous coding SNP in NMB, which codes for neuromedin B, the mammalian homologue of bombesin-like peptide. This variant has previously been associated with eating disorders and obesity.30 Although, there are no previous data directly linking neuromedin B to cardiac structure, it is involved in angiogenesis.31 There are also data linking neuromedin B with PTK2 (Protein Tyrosine Kinase 2) and Ca2+ levels.32 Interference of PTK2 mRNA by siRNA reverses load-induced ventricular hypertrophy in mice. As with any study of this type, the association and eQTL findings provide a first step towards identifying and studying the functionality of candidate genes in the associated region. Also, although the SNPs in MYOZ2 (Myozenin 2) and TTN (Titin), did not pass the criteria for replication (Table 2), they showed suggestive evidence for association in the replication studies, and given that both genes associated with hypertrophic cardiomyopathy33,34 warrant follow-up.
Interestingly, the loci that we discovered here to be associated with ECG indices of LVH did not show evidence of association with echo-derived LV mass in the large meta-analysis of by the ECHOGEN Consortium10 (Supplementary Table 6). This may seem paradoxical and the lack of association of our loci with echo-derived LV mass may be due to chance. However, several observations indicate that the two may reflect different biological processes. The electrocardiogram measures the algebraic sum of the action potentials of myocardial fibres. Therefore, the ECG changes in cardiac hypertrophy reflect the electrical remodelling of the action potential of the cardiac myofibres, which is measured in voltage and time.35 By contrast, the echocardiogram captures anatomical remodelling of the myofibres, fibroblasts, other interstitial changes (such as inflammation) and the cardiac chambers of the heart. This is reflected in the poor correlation between ECG and echo measures of LVH in several clinical contexts.36,–37 More direct evidence that they may be genetically different comes from assessment of ECG-LVH indices and Echo-LV mass in the same families showing greater heritability for the ECG indices8 and distinct putative loci on linkage mapping.9 Finally, ECG and echo measures of LVH provide independent prognostic information in population studies.7 This observation underscores the importance and relevance of identifying genetic determinants of both traits.
Consistent with variants identified for other complex quantitative traits, the amount of trait variance by each of the identified loci was low (<1%). Carriage of the trait-raising allele at each of the locus was associated with a between 8–19% higher probability of lying in the top 10% of the population for that trait. The effect of the three loci affecting QRS Voltage Sum was additive. Individuals carrying all six trait raising alleles for these loci (~ 6.1% of the population) had a 1.60 (95% CI = 1.23 – 2.29) fold increased probability of lying in the top decile for QRS Voltage Sum compared with those carrying 0–2 alleles. Whether these differences impact on the cardiovascular risk associated with ECG-LVH will require further evaluation in large-scale population samples.
LIMITATIONS
Our findings need to be considered in the context of some limitations of the study. The HumanCVD BeadChip 50K array contains only about 10% of all genes in the human genome with a known or suspected cardiovascular function.11 While providing a cost-effective analysis of variants in these genes, a significant limitation is that it does not provide full genome coverage and we may have missed several other variants with similar or indeed greater effects than those identified. Indeed, even within the genes studied we may have missed significant associations (for example in those genes shown in Supplementary Table 8) given the threshold used for taking variants forward for replication and the power of our study. Larger and more comprehensive studies will be required to identify more loci associated with ECG-LVH traits and explain a larger proportion of their variances. Furthermore, although e-QTL analysis can help to prioritise genes for further study in a locus, e-QTLs can be tissue-specific and especially since we examined peripheral rather than cardiac cells, caution needs to be exercised in the interpretation of these findings and their relevance to the observed associations.
Notwithstanding, these limitations, we report the identification and replication of four novel loci associated with ECG indices of LVH. The findings provide new insights into the genetic influences on a routinely recorded clinically-relevant cardiovascular trait with important prognostic consequences which may help to direct research into possible therapeutic targets.
Supplementary Material
Acknowledgments
We acknowledge the work of Shahid Latif, Louise Inglis, Kathryn McLaren and Jean Watts at the Glasgow ECG Centre for the processing and analysis of ECGs. We are grateful to the ECHOGEN Study, Cardiogenics and the BRIGHT Study for access to their datasets. Members of these studies are listed in the Supplementary Materials. J.F.F. is member of the Netherlands Consortium on Healthy Ageing.
Footnotes
DISCLOSURES
None
FUNDING SOURCES
Details are provided in Supplementary Materials.
References
- 1.Kannel WB, Gordon T, Offutt D. Left ventricular hypertrophy by electrocardiogram. Prevalence, incidence, and mortality in the Framingham study. Ann Int Med. 1969;71:89–105. doi: 10.7326/0003-4819-71-1-89. [DOI] [PubMed] [Google Scholar]
- 2.Okin PM, Devereux RB, Jern S, Kjeldsen SE, Julius S, Nieminen MS, Snapinn S, Harris KE, Aurup P, Edelman JM, Wedel H, Lindholm LH, Dahlöf B LIFE Study Investigators. Regression of electrocardiographic left ventricular hypertrophy during antihypertensive treatment and the prediction of major cardiovascular events. JAMA. 2004;292:2343–2350. doi: 10.1001/jama.292.19.2343. [DOI] [PubMed] [Google Scholar]
- 3.Sokolow M, Lyon TP. The ventricular complex in left ventricular hypertrophy as obtained by unipolar precordial and limb leads. Am Heart J. 1949;37:161–186. doi: 10.1016/0002-8703(49)90562-1. [DOI] [PubMed] [Google Scholar]
- 4.Casale PN, Devereux RB, Alonso DR, Campo E, Kligfield P. Improved sex-specific criteria of left ventricular hypertrophy for clinical and computer interpretation of electrocardiograms: validation with autopsy findings. Circulation. 1987;75:565–572. doi: 10.1161/01.cir.75.3.565. [DOI] [PubMed] [Google Scholar]
- 5.Okin PM, Roman MJ, Devereux RB, Kligfield P. Electrocardiographic identification of increased left ventricular mass by simple voltage-duration products. J Am Coll Cardiol. 1995;25:417–423. doi: 10.1016/0735-1097(94)00371-v. [DOI] [PubMed] [Google Scholar]
- 6.Molloy TJ, Okin PM, Devereux RB, Kligfield P. Electrocardiographic detection of left ventricular hypertrophy by the simple QRS voltage-duration product. J Am Coll Cardiol. 1992;20:1180–1186. doi: 10.1016/0735-1097(92)90376-x. [DOI] [PubMed] [Google Scholar]
- 7.Sundström J, Lind L, Arnlöv J, Zethelius B, Andrén B, Lithell HO. Echocardiographic and electrocardiographic diagnoses of left ventricular hypertrophy predict mortality independently of each other in a population of elderly men. Circulation. 2001;103:2346–2351. doi: 10.1161/01.cir.103.19.2346. [DOI] [PubMed] [Google Scholar]
- 8.Mayosi BM, Keavney B, Kardos A, Davies CH, Ratcliffe PJ, Farrall M, Watkins H. Electrocardiographic measures of left ventricular hypertrophy show greater heritability than echocardiographic left ventricular mass. Eur Heart J. 2002;23:1963–1971. doi: 10.1053/euhj.2002.3288. [DOI] [PubMed] [Google Scholar]
- 9.Mayosi BM, Avery PJ, Farrall M, Keavney B, Watkins H. Genome-wide linkage analysis of electrocardiographic and echocardiographic left ventricular hypertrophy in families with hypertension. Eur Heart J. 2008;29:525–530. doi: 10.1093/eurheartj/ehn028. [DOI] [PubMed] [Google Scholar]
- 10.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, Watzinger N, Larson MG, Smith NL, Dehghan A, Grosshennig A, Schillert A, Teumer A, Schmidt R, Kathiresan S, Lumley T, Aulchenko YS, König IR, Zeller T, Homuth G, Struchalin M, Aragam J, Bis JC, Rivadeneira F, Erdmann J, Schnabel RB, Dörr M, Zweiker R, Lind L, Rodeheffer RJ, Greiser KH, Levy D, Haritunians T, Deckers JW, Stritzke J, Lackner KJ, Völker U, Ingelsson E, Kullo I, Haerting J, O’Donnell CJ, Heckbert SR, Stricker BH, Ziegler A, Reffelmann T, Redfield MM, Werdan K, Mitchell GF, Rice K, Arnett DK, Hofman A, Gottdiener JS, Uitterlinden AG, Meitinger T, Blettner M, Friedrich N, Wang TJ, Psaty BM, van Duijn CM, Wichmann HE, Munzel TF, Kroemer HK, Benjamin EJ, Rotter JI, Witteman JC, Schunkert H, Schmidt H, Völzke H, Blankenberg S. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SFA, Farlow DN. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PloS one. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebrahim S, Lawlor DA, Shlomo YB, Timpson N, Harbord R, Christensen M, Baban J, Kiessling M, Day I, Gaunt T, Davey-Smith G. Alcohol dehydrogenase type 1C (ADH1C) variants, alcohol consumption traits, HDL-cholesterol and risk of coronary heart disease in women and men: British Women’s Heart and Health Study and Caerphilly cohorts. Artherosclerosis. 2008;196:871–878. doi: 10.1016/j.atherosclerosis.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 14.Marmot M, Brunner E. Cohort Profile: the Whitehall II study. Int J Epidemiol. 2005;34:251–256. doi: 10.1093/ije/dyh372. [DOI] [PubMed] [Google Scholar]
- 15.Walker M, Whincup PH, Shaper AG. The British Regional Heart Study 1975–2004. Int J Epidemiol. 2004;33:1185–1192. doi: 10.1093/ije/dyh295. [DOI] [PubMed] [Google Scholar]
- 16.Caulfield M, Munroe P, Pembroke J, Samani N, Dominiczak A, Brown M, Benjamin N, Webster J, Ratcliffe P, O’Shea S, Papp J, Taylor E, Dobson R, Knight J, Newhouse S, Hooper J, Lee W, Brain N, Clayton D, Lathrop GM, Farrall M, Connell J. Genome-wide mapping of human loci for essential hypertension. Lancet. 2003;361:2118–2123. doi: 10.1016/S0140-6736(03)13722-1. [DOI] [PubMed] [Google Scholar]
- 17.Smilde TDJ, Asselbergs FW, Hillege HL, Voors AA, Kors JA, Gansevoort RT, Gilst WH, Jong PE, Van Veldhuisen DJ. Mild renal dysfunction is associated with electrocardiographic left ventricular hypertrophy. Am J Hypertens. 2005;18:342–347. doi: 10.1016/j.amjhyper.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Macfarlane P, Devine B, Clark E. The University of Glasgow (Uni-G) ECG Analysis Program. Comput Cardiol. 2005;32:451–454. [Google Scholar]
- 19.Bemmel JH, Kors JA, Herpen G. Methodology of the modular ECG analysis system MEANS. Method Inform Med. 1990;29:346–353. [PubMed] [Google Scholar]
- 20.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 21.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong H, Prentice RL. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics. 2008;9:621–634. doi: 10.1093/biostatistics/kxn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaussin V, Depre C. Myostatin, the cardiac chalone of insulin-like growth factor-1. Cardiovasc Res. 2005;68:347–349. doi: 10.1016/j.cardiores.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Chambers JC, Zhao J, Terracciano CM, Bezzina CR, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi JS, Kooner MK, Deng G, Siedlecka U, Parasramka S, El-Hamamsy I, Wass MN, Dekker LR, de Jong JS, Sternberg MJ, McKenna W, Severs NJ, de Silva R, Wilde AA, Anand P, Yacoub M, Scott J, Elliott P, Wood JN, Kooner JS. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 26.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen AB. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 27.Oh H, Taffet GE, Youker KA, Entman ML, Overbeek PA, Michael LH, Schneider MD. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. P Natl Acad Sci USA. 2001;98:10308–10313. doi: 10.1073/pnas.191169098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanaoka Y, Nojima H. SCR: novel human suppressors of cdc2/cdc13 mutants of Schizosaccharomyces pombe harbour motifs for RNA binding proteins. Nucleic Acids Res. 1994;22:2687–2693. doi: 10.1093/nar/22.13.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadlbauer F, Brueckner A, Rehfuess C, Eckerskorn C, Lottspeich F, Forster V, Tseng BY, Nasheuer HP. DNA replication in vitro by recombinant DNA-polymerase-alpha-primase. Eur J Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- 30.Bouchard L, Drapeau V, Provencher V, Lemieux S, Chagnon Y, Rice T, Rao DC, Vohl MC, Tremblay A, Bouchard C, Pérusse L. Neuromedin beta: a strong candidate gene linking eating behaviors and susceptibility to obesity. Am J Clin Nutr. 2004;80:1478–1486. doi: 10.1093/ajcn/80.6.1478. [DOI] [PubMed] [Google Scholar]
- 31.Park H, Kim S, Bae S, Choi YK, Bae Y, Kim EC, Kim WJ, Jang H, Yun I, Kim Y, Bae M. Neuromedin B induces angiogenesis via activation of ERK and Akt in endothelial cells. Exp Cell Res. 2009;315:3359–3369. doi: 10.1016/j.yexcr.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Tsuda T, Kusui T, Jensen RT. Neuromedin B receptor activation causes tyrosine phosphorylation of p125FAK by a phospholipase C independent mechanism which requires p21rho and integrity of the actin cytoskeleton. Biochemistry. 1997;36:16328–16337. doi: 10.1021/bi971448o. [DOI] [PubMed] [Google Scholar]
- 33.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, Roberts R, Willerson JT, Marian AJ. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circulation. 2007;100:766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem Bioph Res Comm. 1999;262:411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 35.Hill JA. Electrical remodeling in cardiac hypertrophy. Trends Cardiovas Med. 2003;13:316–322. doi: 10.1016/j.tcm.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Rosenzweig A, Watkins H, Hwang DS, Miri M, McKenna W, Traill TA, Seidman JG, Seidman CE. Preclinical diagnosis of familial hypertrophic cardiomyopathy by genetic analysis of blood lymphocytes. New Engl J Med. 1991;325:1753–1760. doi: 10.1056/NEJM199112193252501. [DOI] [PubMed] [Google Scholar]
- 37.Epstein ND, Lin HJ, Fananapazir L. Genetic evidence of dissociation (generational skips) of electrical from morphologic forms of hypertrophic cardiomyopathy. Am J Cardiol. 1990;66:627–631. doi: 10.1016/0002-9149(90)90492-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.