Abstract
Genome editing allows for the versatile genetic modification of somatic cells, germ cells and embryos. In particular, CRISPR/Cas9 is worldwide used in biomedical research. Although the first report on Cas9-mediated gene modification in human embryos focused on the prevention of a genetic disease in offspring, it raised profound ethical and social concerns over the safety of subsequent generations and the potential misuse of genome editing for human enhancement. The present article considers germ line genome editing approaches from various clinical and ethical viewpoints and explores its objectives. The risks and benefits of the following three likely objectives are assessed: the prevention of monogenic diseases, personalized assisted reproductive technology (ART) and genetic enhancement. Although genetic enhancement should be avoided, the international regulatory landscape suggests the inevitability of this misuse at ART centers. Under these circumstances, possible regulatory responses and the potential roles of public dialogue are discussed.
Keywords: germ line genome editing, CRISPR/Cas9, disease prevention, assisted reproductive technology, genetic enhancement, global society
Introduction
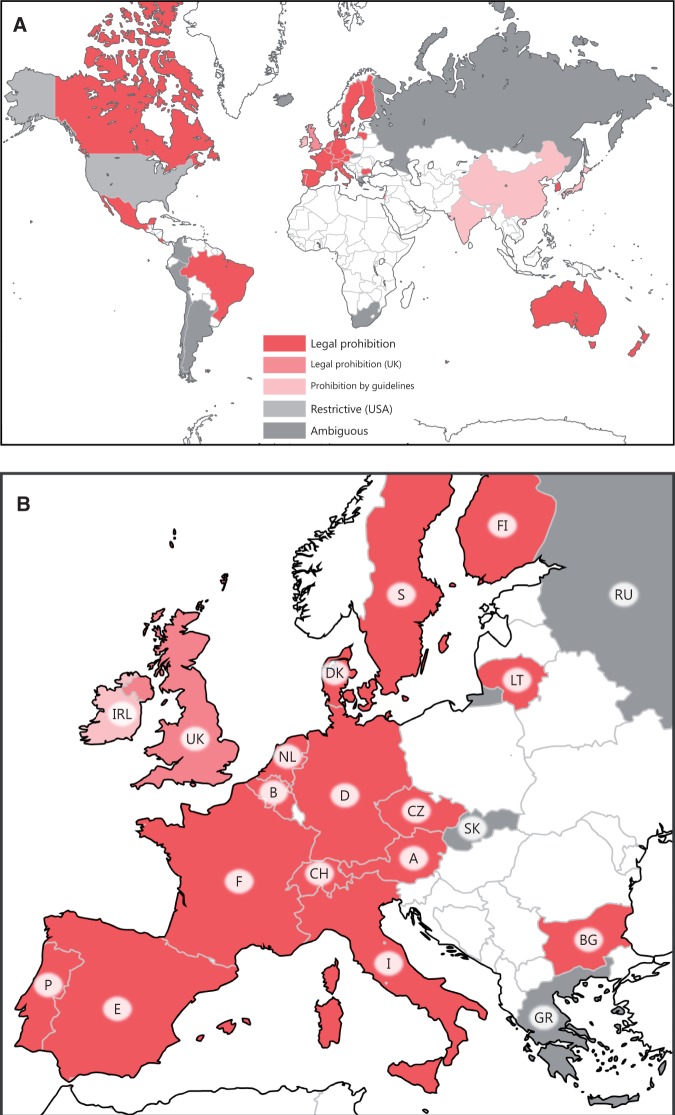
In 1969, Sinsheimer suggested that humans could be the agent of transition to a wholly new path of evolution via designed genetic changes [1]. Although germ line (oocytes, spermatozoa and embryos) gene modification can theoretically impact the entire body of an individual and subsequent generations, conventional genetic engineering has made this unrealistic owing to its inefficiency and imprecise nature [2, 3]. Furthermore, the reproductive use of germ line gene modification has been criticized for a wide array of biomedical and bioethical reasons, including the transgressions of the natural [4, 5] and divine laws [6, 7], irremediable risks to the offspring and future generations [2, 3] and the serious societal harms that eugenics and genetic enhancement (parents pursuing offspring with specific traits for social reasons) represents [2, 8, 9]. Thus, there appears to be a de facto global consensus that the genetic modification of the human germ line should not be permitted for reproductive purposes [10, 11] (Figure 1A). However, genome editing has increased the possibility that germ line gene modification will be practiced in the clinical setting [11–15]. Moreover, such prohibitive policies vary across regulatory systems. Some countries ban it under law on assisted reproduction, while others ban it under gene therapy or bioethics law. Some countries appear to be ill-prepared for germ line genome editing because their relevant regulations are based on conventional genetic engineering, or because the regulations are enforced by guidelines rather than legislation (Supplemental Table S1). Furthermore, the UK, the leading country for assisted reproductive technology (ART), has recently approved draft regulations on a form of germ line gene modification, mitochondrial donation, to prevent the onset of mitochondrial diseases in offspring [16] (Figure 1B). Such biomedical and regulatory situations urge us to reconsider the current policies on germ line gene modification.
Figure 1.
The international regulatory landscape of human germ line gene modification (permission to reuse and modify the figure was granted by the authors of [11]). (A) Thirty-nine countries were investigated and categorized with regard to their view on germ line gene modification. The different categories include ‘legal prohibition’ (24 countries, red), legal prohibition (the UK, pink), ‘prohibition by guidelines’ (four countries: China, India, Ireland and Japan; faint pink),‘ambiguous’ (nine countries: Argentina, Chile, Colombia, Greece, Iceland, Peru, Russia, Slovakia and South Africa; gray) and ‘restrictive’ (the USA, light gray). Note that the UK has recently legalized a form of germ line gene modification, mitochondrial donation (effective in October 2015). The noncolored countries were excluded from this survey. (B) An enlarged figure in Europe. See also the full list of the 39 countries shown in Supplementary Table S1 and Table 3 in the present article.
Genome editing is based on designable bacterial nucleases, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeat (CRISPR)/Cas systems such as Cas9 [17, 18]. Genome editing can efficiently add an exogenous gene, correct a mutation or disrupt an endogenous gene at target sites in mammalian genomes. CRISPR/Cas9 has an advantage over the other two systems in the user-friendliness of guide RNA (gRNA) preparation, hence its use in many laboratories throughout the world [18–20]. Cas9 also shows greater utility because it facilitates the simultaneous editing of several sites across the genome by using separate gRNAs [21, 22].
It is hoped that genome editing can be widely used as a therapeutic tool because a clinical trial has demonstrated that infusions of autologous T cells in which CCR5 was disrupted by ZFNs are safe in HIV patients [23]. With regard to germ line genome editing, Cas9-mediated gene modification in human zygotes (one-cell-stage embryos) was recently reported from China [24]. Although the group intended to correct a mutation at HBB to prevent the onset of β-thalassemia in the progeny, ethical and social concerns regarding the potential risks to future generations and its potential use in genetic enhancement have been expressed worldwide [12–15]. Despite the current technical hurdles, further research will likely make germ line genome editing clinically feasible in the near future [11]. Recent discussions suggest the likelihood of the use of germ line editing to prevent the onset of genetic disease in offspring [11, 13–15]. However, some limitations may be necessary to increase the safety of germ line genome editing in the clinical setting. Moreover, germ line genome editing differs from somatic cell editing for existing patients because informed consent is given by the prospective parents. How should we weigh the parental benefit against the risk to future generations? Furthermore, how should we prepare for unwanted situations such as the misuse of germ line engineering?
In the present article, the author considers the appropriateness of some germ line genome editing approaches from clinical and ethical viewpoints and explores the objectives and the risk–benefit balance. The current regulatory landscape suggests that the germ line engineering will be inevitably misused in some countries with lax regulations. Based on this perspective, potential regulatory responses and the roles of public dialogue are discussed.
Appropriate modes of human germ line genome editing
The intracellular introduction of genome editing nucleases efficiently induces a double-strand break (DSB) at a specific site in the genome, which is directed by a designed targeting domain (in ZFNs or TALENs), or gRNA (in CRISPR/Cas9) [17, 18]. The induced DSBs are repaired via nonhomologous end joining (NHEJ) without exogenous DNA, or via homology-directed repair (HDR) with exogenous DNA. The NHEJ produces insertions or deletions (‘indels’) of various lengths, while the HDR can add an exogenous gene into, or functionally correct a mutation at a specific site. CRISPR/Cas9, which is based on the adaptive immune system of Streptococcus pyogenes SF370, is more useful than the other editing systems because the targeting molecule is separate gRNA, which facilitates the design and preparation of the editing system in most laboratories [18–20]. Furthermore, the incorporation of multiple gRNAs and/or multiple DNA templates in a CRISPR system allows for the simultaneous editing of several target sites across the genome [21, 22]. This form of genome editing is useful for generating animal models of multifactorial inheritance disorders such as heart disease, diabetes and cancer.
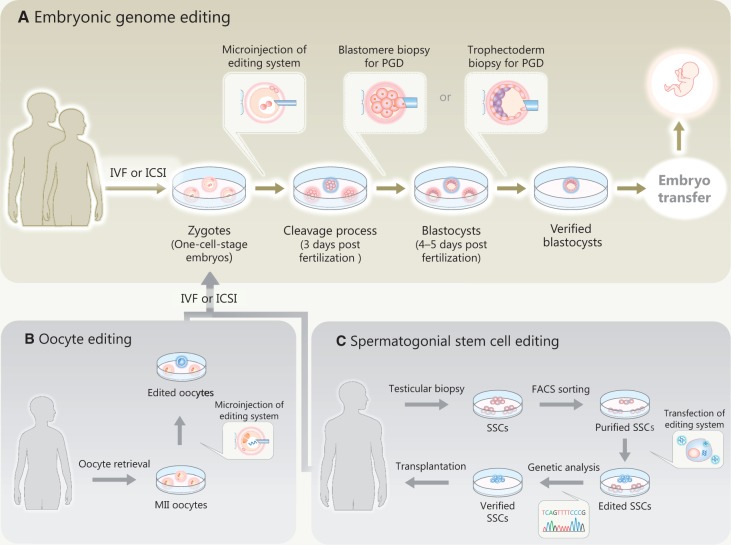
Mammalian germ line genome editing primarily begins with the microinjection of the nucleases into zygotes [11, 14] (Figure 2A), which is seemingly similar to a common ART technique, intracytoplasmic sperm injection (ICSI). In clinical practice, on-target gene modifications and off-target mutations would be investigated by using preimplantation genetic diagnosis (PGD), with either a blastomore or trophectoderm biopsy before fresh or frozen embryo transfer, respectively [25]. Other mammalian germ line genome editing approaches include oocyte editing (Figure 2B) and spermatogonial stem cell (SSC) editing (Figure 2C). Because there are currently a few reports on such germ cell editing approaches [14], the author primarily discusses embryonic genome editing (Figure 2A).
Figure 2.
An outline of human germ line genome editing. (A) Embryonic genome editing. The nucleases are microinjected into zygotes (one-cell-stage embryos). Subsequently, on-target gene modifications and off-target mutations are investigated by preimplantation genetic diagnosis (PGD) with either blastomore biopsy or trophectoderm biopsy before the embryo transfer. (B) Oocyte editing. (C) SSC editing. Genetically modified SSCs are transplanted for differentiation in vivo. The genetically modified oocytes or spermatozoa are used for in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI). The resultant embryos are also investigated by a PGD.
The safety of the unprecedented introduction of genome editing nucleases into human embryo deserves special scrutiny. Although embryonic genome editing can impact all of the cells of the resultant individual, the process may also be counterproductive. The designed nucleases could create off-target DSBs and thereby leave indels at nontarget sites [14]. The occurrence of an indel in a tumor suppressor gene (such as the TP53) could result in the child developing cancer [15]. In addition, off-target DSBs could lead to large-scale genomic alternations, including large deletions, inversions and translocations (by creating concurrent DSBs at two loci) [26, 27]. Such off-target effects could affect the entire body of the offspring, unless the designed gRNA in CRISPR/Cas9 display specificity. The use of the protein form of the nuclease [28, 29] or more sophisticated enzymes such as Cas9 nickase [30], and the genome-wide profiling of likely off-target effects [26, 27, 31] are expected to reduce such effects but the risk cannot be completely eliminated. To substantially reduce the off-target effects of introduced gRNAs, multiple gRNAs should not be used in embryonic genome editing. Therefore, Cas9 with a single type of gRNA should be the sole genome engineering tool, which is applicable to human zygotes for disease prevention or other reproductive purposes.
Embryonic genome editing may affect embryonic or subsequent development owing to its physical and biological activities [11]. The different types of genome editing approaches also require due consideration. As suggested by alipogene tiparvovec (the first somatic gene therapy product approved in Western countries) [32], the introduction of an additional gene copy via HDR seems attractive. However, this approach should not be adopted in human embryos even if the exogenous gene is introduced in a genomic safe harbor (GSH; a chromosomal site where an introduced gene can be stably expressed in all tissues without affecting endogenous gene structure or expression) [33]. The three loci that have been proposed as GSHs (AAVS1, CCR5 and ROSA26) are all located in gene-rich regions [33], which likely has an adverse effect on zygotic genome activation, which is initiated immediately after fertilization [34]. The three have been implicated in cancer [33]. Embryonic gene disruption via NHEJ sometimes sounds plausible, as demonstrated by the clinical use of somatic CCR5 disruption, which reproduced a naturally occurring variant (CCR5 delta32), which confers resistance to HIV infection [23, 35]. However, this type of genetic modification should not be attempted in the human embryo. To begin with, there is an inherent problem with the premise that such children may be exposed to a high risk of HIV infection. Moreover, intentional CCR5 disruption to protect the offspring against HIV infection may have other biological effects. Although CCR5–/– mice display no serious abnormalities, the loss of CCR5 results in glucose intolerance in mice with diet-induced obesity [36]. In humans, CCR5 mutations increase the risk of West Nile virus infection [37]. The systemic effect of naturally occurring variants remains to be fully characterized in humans [15].
In contrast, the functional correction of a small mutation in the embryo via HDR along with a short DNA template appears to be acceptable because this form of genetic modification can leave a wild-type gene, which is in a natural genetic state, and would fall outside of one of the ethical objections against germ line gene modification: transgression of the natural laws [4, 5]. The copying of a naturally occurring variant via HDR along with a short DNA template might be considered to be natural. Both gene addition via HDR and gene disruption by the introduction of indels of various lengths via NHEJ would largely be considered unnatural. However, there is still room for careful consideration. Embryonic editing to correct a mutation or copy a naturally occurring variant might be seen as contrary to the natural course of events, and therefore contrary to human nature [4, 5]. Health care providers who intentionally edit the human germ line genome could be considered to be transgressing the divine laws, or ‘playing God’ [5, 6]. However, such concerns might be dismissed if the role of embryonic genome editing is well-defined and if the ethical and social implications are given due consideration.
Based on these considerations, if embryonic editing by CRISPR/Cas9 is permitted to correct a mutation or copy a naturally occurring variant, it should be limited to gene modification via HDR using a short DNA template along with a single type of gRNA. Such limitations increase the safety and help to clarify the objectives of embryonic editing. Without any limitations on CRISPR/Cas9, its clinical use will be more likely to cause adverse effects and/or incur public blame for ‘the creation of GM humans’ [38].
Objectives
Recent arguments suggest that a likely use of germ line genome editing is to prevent the onset of genetic diseases in offspring [11, 13–15]. Hereafter, in addition to the disease prevention, another medical purpose and two social purposes are discussed.
Disease prevention
In the first study on human embryonic genome editing [24], zygotes were treated with CRISPR/Cas9 to repair an HBB mutation that is responsible for β-thalassemia (an autosomal recessive disease), via HDR. The prevention of such monogenic diseases before childbirth appears to be a reasonable application of embryonic genome editing [11, 14, 15]. This possibility is supported by some examples of HDR-mediated gene modification in mammalian zygotes (Table 1) [24, 39–43]. The efficiency of genetic modification in neonates is, for the most part, low (<10%); however, some cases show higher efficiency using Cas9 (Crb1: 27%, Asip: 18.2%) (Table 1) [42, 43]. Notably, the microinjection of Cas9, along with a repair DNA, into mouse zygotes with a dominant mutation in Crygc prevented the onset of cataracts in offspring [40]. Owing to rapid advances in genome editing, the correction of mutations in human embryos by genome editing might reach the clinical stage in the near future [11, 14].
Table 1.
Examples of HDR-mediated gene modification in mammalian zygotes
| Subject | Gene modification | Efficiency in neonates (embryos) | Off-target mutation | Mosaicism | Genome editing | Remarks | Reference |
|---|---|---|---|---|---|---|---|
| Mouse zygotes | Introduction of V5 tag (42bp) into Sox2, two loxP (34 bp) sites into Mecp2 | Sox: 6.0%, Mecp2: 0.8% | Yes (Mecp2) | Yes | Cas9 | Cytoplasmic or pronuclear injection | [39] |
| Mouse zygotes | Correction of Crytg with 1bp deletion in exon3 | 4.4∼5.7% | Yes | N.D. | Cas9 | Cytoplasmic injection | [40] |
| Mouse zygotes | Correction of Dmdmdx | 9.1% | (No)a | Yes | Cas9 | Pronuclear injection only, or pronuclear and cytoplasmic injections | [41] |
| Mouse zygotes | Correction of Crb1rd8 | 27% | Yes | Yes | TALEN | Pronuclear injection | [42] |
| Rat zygotes | Correction of Tyrc, Asipa, Kit h | Tyr: 7.7 %, Asip:18.2%, Kit: 4.0% | (No)a | Yes | Cas9 | Pronuclear injection | [43] |
| Human zygotes | Introduction of silent mutations into HBB | (4.7%) | Yes | Yes | Cas9 | Cytoplasmic injection | [24] |
(No) shows that no off-target mutations were identified at potential off-target sites.
PGD, which entails an embryo-invasive procedure, is practiced in many countries to identify embryos that are free from specific genetic conditions (http://www.coe.int/t/dg3/healthbioethic/Source/INF_2010_6_dpidpn_en.pdf). Prospective parents who use PGD wish to have healthy, genetically related children; thus, gamete donation would be unacceptable. Similarly, the clinical use of embryonic genome editing is conceivable in some cases where there is a high probability that a genetic condition will be inherited.
Personalized ART
A more straightforward objective may be to enhance the ART success rate by genetically editing embryos or germ cells. The efficacy of present-day ART treatment is limited. An international ART analysis indicates that the average delivery rate was 20.5% per aspiration and that the cumulative delivery rate in a single treatment cycle was 25.2% (http://www.eshre.eu/Guidelines-and-Legal/ART-fact-sheet.aspx). In some cases, infertility is associated with a genetic condition within the patients [44, 45]. Germ line genome editing can be considered as a form of ‘personalized ARTs’ [46].
Chromosomal rearrangements in prospective parents are associated with recurrent pregnancy loss and the birth of offspring with congenital anomalies. The most common chromosomal rearrangements are reciprocal translocations (1 in 500 individuals) that are an exchange between two different chromosomal terminals without a loss of genetic material (balanced) [45]. Genome editing might be able to correct such chromosomal abnormalities. Indeed, a recent report demonstrated that Cas9 treatment functionally corrected large Factor VIII gene chromosomal inversions in hemophilia patient-derived induced pluripotent stem cells [47]. However, such chromosomal corrections require at least two types of gRNA, which would make them unadvisable. In contrast, a recent study reported that a Cdk2 allele, which mimicked a human single nucleotide polymorphism (SNP; rs3087335) using CRISPR/Cas9 caused infertility in male mice owing to an SSC maintenance dysfunction [48]. Thus, genome editing has the potential to correct such infertility-related SNPs in the patient’s SSCs via HDR. However, this strategy requires SSC transplantation in patients to allow the corrected SSCs into spermatozoa; at present, this is only at experimental stage (Figure 2C) [49].
As the current technical hurdles decrease, genome editing-mediated personalized ART is likely to become an option for infertile patients who do not wish to use donor gametes or adopt. The objective will increase the motive to perform genome editing at ART centers. This possibility is supported by two case reports on the use of germ line gene modification for reproductive purposes, in which ooplasmic transfer and pronuclear transfer were used to treat for females with quality-compromised oocytes [14, 50–52].
Genetic enhancement
The potential use of genome editing for genetic enhancement may be found in a demand when people choose a sperm donor. A recent study showed that 1597 women who formed their families using donor spermatozoa indicated that, other than the donor’s health, 50.0% of them viewed the donor’s intelligence as important, while 42.7% and 40.7%, respectively, indicated the importance of the donor’s height and ethnicity [53]. It would be difficult to enhance the future child’s intelligence by embryonic editing. Although three SNPs were found to be associated with educational attainment in a genome-wide association study (GWAS) of 126 559 individuals, the effects of the SNPs were all small [54]. Moreover, introducing three SNPs entails risky multiplex editing. The same is true with regard to the height [15].
With regard to ethnicity, externally visible traits, such as eye, hair and skin color, may be considered as a target phenotype particularly in countries with ethnically diverse populations [15]. In a recent study using rat zygotes, Cas9 treatment corrected Tyrc, Asipa, Kith via HDR and thereby led to recovery from albinism via an SNP exchange, in the non-agouti and hooded phenotypes through integration of a 19 bp DNA sequence and the elimination of a 7098 bp insertional DNA fragment, respectively (Table 1) [43]. The successful changes of rat coat color suggest the possibility that a naturally occurring variant could be copied to induce a pigmentation change in humans via embryonic editing. A GWAS identified some variants associated with hair and eye pigmentation in European individuals [55]. Of note, a variant in OCA2 (rs1667394 A) is associated with blue eyes and blond hair. Although some prospective parents might wish to introduce the OCR2 variant in their embryos via HDR, it is unlikely that their desired iris and hair phenotypes will be achieved in a simplistic manner. Because the human iris color is regulated by at least 16 genes [56], there would be a high degree of uncertainty in the appearance of the resultant offspring owing to the genetic backgrounds of the parents. Although pink eyes, which are a common trait of albinism would be attainable using an OCA2, TYR,TYRP1 or SLC45A2 variant [57], children with this genetic condition would likely experience vision defects and be more sensitive to sunburn and skin cancer. Likewise, introducing a variant in MC1R (rs1805008 T) might result in the birth of offspring with red hair, but would make his/her skin sensitive to the sun [55].
Thus, while it may be possible to control externally visible traits by introducing a naturally occurring variant via HDR, this would likely cause problems with child’s appearance and/or health.
Eugenic applications
Another possible application of germ line genome editing for a social purpose is eugenics [58]. The goal of eugenics is to improve human genetic traits by reducing the reproduction of people with socially undesired traits (negative eugenics) and/or promoting the reproduction of people with socially desired traits (positive eugenics). Notably, Nazi Germany attempted to eliminate socially undesired people, while creating so-called ‘perfect’ and ‘higher’ humans, thus resulting in the Holocaust and numerous forced sterilizations. The use of a sex ratio distortion system (selective induction of DSBs in the X chromosome during spermatogenesis by engineered nucleases to distort the sex ratio of the progeny) [59] for negative eugenics might be developed based on a genome editing system. However, its large-scale use for negative eugenics would amount to a crime against humanity [60].
Embryonic genome editing could, under unusual circumstances, be considered for positive eugenics. In a society where people require excellent athletic abilities, the copying of a naturally occurring MSTN variant [61], via HDR, could be considered to increase the number of people with muscle hypertrophy. Although MSTN disruption in livestock suggests that this may be feasible [62], such a social goal would be difficult to attain. First, genome editing requires the design of gRNA to be meticulously optimized in each case. Moreover, PGD entails the time-consuming and inefficient process of embryonic cell biopsy. Second, the implementation of this procedure on a large scale would increase social costs, even in countries that permit the diffusion of ART, as it would add to the expenses of ART, which are generally high [63]. Third, the effect of this movement would be difficult to assess owing to the length of the human generation period. Thus, in most countries, embryonic genome editing is unlikely to be considered for eugenics.
Risks and benefits
The above-mentioned considerations confirmed three likely objectives: the prevention of monogenic diseases, personalized ART and genetic enhancement. However, unlike PGD and mitochondrial donation, the designed nucleases could introduce off-target effects into human zygotes. Embryonic genome editing may therefore pose substantial risks to the embryo, fetus or resultant child. The occurrence of genetic mosaicism in which wild-type and modified cells coexist likely indicates the failure to prevent the onset of genetic disease in offspring, although the injection of Cas9 into mouse embryos of a muscular dystrophy model demonstrated that the degree of muscle phenotypic rescue in mosaic mice exceeded the efficiency of gene correction [41]. In addition to rodent experiments (Table 1), four recent nonhuman primate experiments have underscored the risk of mosaicism [64–67]. Mosaicism was frequently observed in genetically modified monkey neonates (3/4; Table 2). Additionally, the first report on human embryonic genome editing showed mosaicism in the resultant embryos [24]. Furthermore, despite the absence of off-target mutations at the potential off-target sites in the nonhuman primate study (3/4; Table 2), Cas9 treatment has been demonstrated to create off-target mutations in the human genome [24]. In ARTs such as in vitro fertilization (IVF), ICSI and PGD, informed consent is obtained from the prospective parent(s) [10]. In embryonic genome editing, which is more invasive than the common ARTs, parental consent is justified when the benefits for the parent(s) and/or resultant child exceed the risks to the child.
Table 2.
Examples of targeted gene disruption in nonhuman primate zygotes via NHEJ
| Subject | Gene disruption | Efficiency in neonates (embryos)a | Off-target mutationb | Mosaicism | Genome editing | Remarks | Reference |
|---|---|---|---|---|---|---|---|
| Cynomolgus zygotes | NR0B1, PPARG, RAG1 | (Single gene: 18.2–40.7%) (PPARG&RAG1: 9.1–27.3%) | No | Yes | Cas9 | Cytoplasmic injection | [64] |
| Rhesus and cynomolgus zygotes | MECP2 | Rhesus: 9.5% Cynomolgus: 3.7% | No | N.D. | TALEN | Cytoplasmic injection | [65] |
| Cynomolgus zygotes | MECP2 | 2.0% | N.D. | Yes | TALEN | – | [66] |
| Rhesus zygotes | DMD | 6.1% (46.47%) | No | Yes | Cas9 | Cytoplasmic injection | [67] |
Denotes the result of genetically modified neonates (including fetus or stillborn) per transferred embryo (%) or genetically modified embryos per injected zygote (%).
‘No’ shows that no off-target mutations were identified at potential off-target sites.
With regard to the prevention of severe types of monogenic disease in offspring, the authors of the first study on human embryonic editing did not address the possible use of PGD for selecting an embryo with none of the homozygous mutations that are responsible for β-thalassemia [24]. Likewise, in mitochondrial donation, PGD could prevent the birth of children affected by mitochondrial diseases. Oocytes with a low load of mitochondrial DNA (mtDNA) mutations can be identified among the patient’s (or carrier’s) oocytes. However, mitochondrial donation might be the only option for the female patients who wish to have healthy genetically related children, if all of their oocytes have a high load of mtDNA mutations [10, 16, 68]. Mitochondrial donation appears to justify disease prevention by embryonic genome editing in cases where PGD is not clinically applicable. Embryonic genome editing seems justifiable in specific cases where both parents are homozygous, even in cases of autosomal recessive disease [11, 15]. Another presumable use for germ line editing is in the cases of autosomal dominant diseases in which a parent(s) is homozygous [11, 15]. Although such cases are extremely rare [15], the rarity is not a sufficient reason for ethical rejection. Procedures may be carefully considered in health care for minority populations [10]. The benefits for the parents and the child would largely exceed the risk to the child if the safety is sufficiently maximized by preclinical research [69]. In the last two cases, children will develop genetic disease unless the pathogenic mutation is corrected by genome editing.
It is likely that embryonic genome editing will be an option for preventing severe types of monogenic disease in cases in which there is a high probability that the child will inherit a disease-causing mutation. Moreover, the disease prevention aspect may be positively considered for early-onset diseases, such as cystic fibrosis and familial adenomatous polyposis [11] because the procedure is a preventive intervention that can be performed at the embryonic stage. Such genetic conditions can be found in authorized PGD conditions, which can be licensed by the UK’s Human Fertilisation and Embryology Authority (HFEA) (http://guide.hfea.gov.uk/pgd/). Furthermore, the requirements for the clinical use of embryonic genome editing may include a parental history of childbirth with such genetic conditions because the need for the preventive intervention is suggested by their clinical history. Conversely, in genome editing for enhancement, which is probably performed in the pursuit of a specific appearance, parental consent is not justifiable and the benefits for the parents and/or child seem unclear. Although the use of genome editing for social reasons could enhance the child’s life by providing a socially favored appearance, it also could affect the child’s health owing to the introduction of off-target mutations. Plastic surgery is, despite the inherent health risks, allowed for patients to pursue mental and social well-being [70]. In this regard, patients who undergo the surgery consent to the procedure. Unborn children cannot express worry about their appearance or consent to genome editing. Moreover, as discussed above, genome editing-mediated genetic enhancement to achieve a specific appearance does not necessarily lead to the attainment of the desired phenotype in the resultant child. It is likely that the parents would value a designed appearance characteristic in their child; however, serious discord may occur within a family if the child does not exhibit that characteristic immediately after birth or in adulthood. Even if a naturally occurring variant was successfully introduced and no adverse effects were observed in the children, the children may suffer distress. Parents whose children were conceived with donor gametes frequently confront challenges because the genetic links are commonly reinforced through observations about a child’s physical similarity to the parents [71]. In the genetic enhancement of a specific appearance, children may be left in an unsettled state of mind or become disillusioned with their appearance because it was imposed through means other than a blood relationship. The benefits for parents and the child are unclear, whereas the risks to physical and mental health, primarily the child’s, are substantial.
Personalized ART appears to be similar to ARTs, which use the patient’s gametes with regard to informed consent. Although the prospective parent is the primarily beneficiary of embryonic editing in personalized ART, the risks are currently greater than in ARTs such as IVF owing to the involvement of genome editing. However, the risks will be reduced. Recent mammalian genome editing research (Tables 1 and 2) and the development of mitochondrial donation suggest that additional works including gene correction via HDR in nonhuman primates and in vitro human embryo research will be useful [14, 68]. An intergenerational risk assessment using rodents would also be valuable to assess the long-term safety. Furthermore, the rapid advances in PGD [72] will enable us to investigate off-target mutations in embryos. However, it should be noted that PGD-based whole-genome prediction is limited in that it can only analyze a specific locus, even if it is compared with the parent’s genome information, owing to stochastic biases during the amplification of genomic DNA derived from a few biopsied cells [73]. Ultimately, parental consent for this procedure may be justified if the safety can be considered to equal that of the common ARTs.
With the increasing safety of genome editing [11, 14], embryonic genome editing will be positively considered for two medical purposes: disease prevention and personalized ART.
Global society
Although the whole-life safety of PGD has not been confirmed [72], genetic testing has been used for sex selection by parental preference (nonmedical reason) in some European countries (this is illegal in many European countries) (http://www.coe.int/t/dg3/healthbioethic/Source/INF_2010_6_dpidpn_en.pdf), and in the USA [74]. In addition, PGD to select a disability has been offered at some US ART centers [74]. PGD for nonmedical sex selection has already become a lucrative global trade [75]. When ART centers initiate the use of embryonic genome editing in countries with lax regulations, genome editing-mediated enhancement will likely be offered. Baylis and Robert also predict the inevitability of genetic enhancement based on the worldview of that humans are the masters of the human evolutionary future [8].
Table 3 shows the number of ART centers and relevant legislation in the G8 nations and China in which the first human embryonic editing was performed [11, 13, 76–79]. Japan, the USA and China, where few or no ART-specific national laws have been established, have more ART centers than Canada and the four European countries with ART-specific national laws (excluding Russia). Remarkably, Japan has 606 ART centers (4.74 per million). Although the UK is the first nation to permit a form of germ line gene modification, mitochondrial donation will only be offered at ART centers with an HFEA license [16]. Meanwhile, the USA has no federal legislation regarding germ line gene modification for reproductive purposes and restricts this procedure via FDA review of its clinical trial and the NIH Guidelines for Research Involving Recombinant or Synthetic Nucleic Acid Molecules (2013). Japan and China prohibit this procedure under guidelines that involve less enforcement than law, and which are subject to amendment. Russia is legally ambiguous about this subject. Under such circumstances, Japan, the USA, China and Russia might confront more serious social issues owing to the misuse of embryonic genome editing than those that have been encountered with the nonmedical use of PGD. The current regulatory situation is incommensurate with the proposed policy of clinical use of CRISPR/Cas9 with one gRNA (Figures 1A and B, Table 3).
Table 3.
ART center and relevant national law in the G8 and China
| ART centers 2007a | Population 2007 (millions) | Centers/ million | ART-specific national law | Prohibition of reproductive cloning by national law | Prohibition of GGMb by national law | Relevant national laws and guidelines | |
|---|---|---|---|---|---|---|---|
| Canada | 26 | 33 | 0.79 | Yes | Yes | Yes | Assisted Human Reproduction Act (2004, amended 2006, 2007, 2012). |
| Russia | 69 | 143 | 0.48 | Noc | Yes | Ambiguousc | Law No. 323 on Fundamentals of Citizens’ Health Protection in the Russian Federation (2011). Order of the Ministry of Health No. 107n on the Use of Assisted |
| Reproductive Technologies, Contraindications and Limitations to their Use (2012). | |||||||
| Law No. 54 on the Temporary Ban on Human Cloning (2002, amended 2010) | |||||||
| UK | 70 | 61 | 1.15 | Yes | Yes | Yesd | Surrogacy Arrangement Act (1985). Human Embryology and Fertilisation Act (1990, amended 2008). Human Fertilisation and Embryology (Mitochondrial Donation) Regulations (2015). |
| France | 104 | 62 | 1.68 | Yes | Yes | Yes | Law on the Donation and Use of Elements and Products of the Human Body, Medically Assisted Procreation, and Prenatal Diagnosis, No. 654 (1994). Bioethics Law No. 800 (2004, amended 2009, 2011). |
| Germany | 118 | 82 | 1.44 | Yes | Yes | Yes | Embryo Protection Law (1990, amended 2001, 2011). Adoption Brokerage Law (2006). Guideline of the German Federal Medical Chamber (2006) |
| Italy | 202 | 58 | 3.47 | Yes | Yes | Yes | Law No. 40 Rules in the Field of Medically Assisted Reproduction (2004) |
| China | 358e | 1,354 | 2.65 | Nof | Nof | Nof | Ministry of Health: Technical Standards and Ethical Principles of Assisted Reproductive Technologies and Sperm Banks (2003). |
| USA | 483 | 302 | 1.60 | Yesg | Nog | No | Fertility Clinic Success Rate and Certification Act (1992). |
| Japan | 606 | 128 | 4.74 | No | Yes | Noh | Law Concerning Regulation Relating to Human Cloning Techniques and Other Similar Techniques (2001). Ministry of Health, Labour and Welfare: Guidelines for Clinical Research Such as Gene Therapy (2015). |
Based on Ishihara et al. Fertil Steril. 2015;103:402–13.
GGM: human germ line gene modification for reproduction, other than reproductive cloning
No ART-specific federal law. ART is stipulated in the chapter 6, article 55 of the law No. 323. The details of ART operation is indicated by the order No.107n. Although human embryo research is legal, germ line gene modification for reproduction is not considered in relevant legislation or the Order.
A form of germ line gene modification, mitochondrial donation will be allowed under the regulations (effective on 29 October 2015).
Based on Qiao J, Feng H.L. Transl. Pediatr 3, 91–7 (2014). The numbers of center and population as of 2012.
ART is regulated under guidelines. Reproductive cloning and GGM are also prohibited under guidelines, not legislation.
The ART-specific federal law only requires ART centers to report pregnancy success rates. Regulation of ART activities varies at the state level (https://www.asrm.org/Oversight_of_ART/). Some state laws prohibit human cloning (http://www.ncsl.org/research/health/human-cloning-laws.aspx).
GGM is prohibited by guidelines, not lelgislation.
One might propose the global prohibition of germ line genome editing for enhancement or for all reproductive purposes. The proposal could be brought to the attention of the United Nations (UN) because the UN Declaration on Human Cloning resulted in the establishment of relevant legislation in many countries [80]. Nonetheless, it would likely be difficult to establish legislation regarding germ line editing in some countries in which ART and/or reproductive cloning are not legislated (Table 3). Eventually, differences in national policy might start to reemerge throughout the world. Although it might be difficult to impose worldwide legislation standards, we can seek to improve the situation in each country. It would be crucial to informs the public of the pros and cons of germ line genome editing in the ARTs [14] (http://costep.hucc.hokudai.ac.jp/costep/contents/article/1357/). Such public dialogue is also important in countries with legislation on ART and germ line gene modification because the people could be involved in the misuse of germ line editing in countries with lax regulations. This article highlighted the prevention of inheritance of monogenic diseases and personalized ART as the likely objectives of germ line editing applications, which may have clear benefits. Prospective parents with a genetic condition may have healthy, genetically related children without using donor gametes. However, the clinical use of germ line genome editing for such purposes is not the first option to build family. Prospective parents should also consider adoption, which can offer valuable opportunities for children who are in need of a loving family. In 2001, there were 127 407, 45 844 and 23 108 adoptions in the USA, China and Russia, respectively. In contrast, there were only 1931 adoptions in Japan in 1995 [81]. Moreover, it should be noted that the clinical uses of germ line editing would be expensive and might result in adverse societal effects, such as reproductive disparity [11]. Philosophical questions, such as whether germ line genome editing runs contrary to the evolutionary course of events (a species corrects a mutation in the germ line or creates a variant) [11] should be included in such discussions.
Careful public dialogue will foster a constructive atmosphere for shaping the appropriate roles of germ line genome editing in the ARTs (if any), and may lead to the amendment or establishment of relevant regulations.
Key points
Germ line genome editing is likely used for the prevention of monogenic diseases, personalized assisted reproductive technology and genetic enhancement.
Germ line genome editing-mediated genetic enhancement should be avoided in global society.
Careful public dialogue will shape the appropriate roles of germ line genome editing in the ARTs, and may lead to the amendment or establishment of relevant regulations.
Supplementary Material
Acknowledgements
The author thanks Motoko Araki for supporting the preparation of figures.
Funding
This work was supported by JSPS KAKENHI Grant Number 26460586 (T.I.).
Biography
Tetsuya Ishii is a Professor at the Office of Health and Safety, Hokkaido University. His interests are the ethics of genetic engineering, stem cells and assisted reproductive technology.
References
- 1. Sinsheimer RL. The prospect for designed genetic change. Am Sci 1969;57:134−42. [PubMed] [Google Scholar]
- 2. Frankel MS, Chapman AS. Human Inheritable Genetic Modifications. Assessing Scientific, Ethical, Religious and Policy Issues. Washington, DC: American Association for the Advancement of Sciences, 2000. [Google Scholar]
- 3. Billings PR, Hubbard R, Newman SA. Human germline gene modification: a dissent. Lancet 1999;353:1873−75. [DOI] [PubMed] [Google Scholar]
- 4. Leiter B. The Future for Philosophy. Oxford University Press, 2004. [Google Scholar]
- 5. Trigg R. Ideas of Human Nature: An Historical Introduction. Oxford: Basil Blackwell, 1988. [Google Scholar]
- 6. Messer N. Human cloning and genetic manipulation: some theological and ethical issues. Studies in Christian Ethics 1999;12:1–16. [DOI] [PubMed] [Google Scholar]
- 7. Glover J. What Sort of People Should There Be? London: Penguin Book, 1984. [Google Scholar]
- 8. Baylis F, Robert JS. The inevitability of genetic enhancement technologies. Bioethics 2004;18:1–26. [DOI] [PubMed] [Google Scholar]
- 9. Sandel M. The case against perfection. Atl Mon 2004;293:51–62. [PubMed] [Google Scholar]
- 10. Ishii T. Potential impact of human mitochondria: replacement on global policy regarding germline gene modification. Reprod Biomed Online 2014;29:150–5. [DOI] [PubMed] [Google Scholar]
- 11. Araki M, Ishii T. International regulatory landscape and integration of corrective genome editing into in vitro fertilization. Reprod Biol Endocrinol 2014;12:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lanphier E, Urnov F, Haecker SE, et al. Don't edit the human germ line. Nature 2015;519:410–1. [DOI] [PubMed] [Google Scholar]
- 13. Baltimore D, Berg P, Botchan M, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 2015;348:36–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishii T. Germline genome-editing research and its socioethical implications. Trends Mol Med 2015;21:473–81. [DOI] [PubMed] [Google Scholar]
- 15. Lander ES. Brave new genome. N Engl J Med 2015;373:5–8. [DOI] [PubMed] [Google Scholar]
- 16. The Human Fertilisation and Embryology (Mitochondrial Donation) Regulations 2015 No. 572. http://www.legislation.gov.uk/ukdsi/2015/9780111125816/contents (16 October 2015, date last accessed).
- 17. Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet 2014;15:321–34. [DOI] [PubMed] [Google Scholar]
- 19. Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Findlay GM, Boyle EA, Hause RJ, et al. Saturation editing of genomic regions by multiplex homology-directed repair. Nature 2014;513:120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 2014;370:901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang P, Xu Y, Zhang X, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015;6:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bolton VN. Embryology and PGD. In: El-Toukhy T, Braude P. (eds). Preimplantation Genetic Diagnosis in Clinical Practice. London: Springer-Verlag, 2014, 55–64. [Google Scholar]
- 26. Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frock RL, Hu J, Meyers RM, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 2015;33:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaj T, Guo J, Kato Y, et al. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 2012;9:805–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014;24:1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154:1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim D, Bae S, Park J. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 2015;12:237–43, 1 p following 243. [DOI] [PubMed] [Google Scholar]
- 32. Wierzbicki AS, Viljoen A. Alipogene tiparvovec: gene therapy for lipoprotein lipase deficiency. Expert Opin Biol Ther 2013;13:7–10. [DOI] [PubMed] [Google Scholar]
- 33. Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer 2012;12:51–8. [DOI] [PubMed] [Google Scholar]
- 34. Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 2014;30:581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996;86:367–77. [DOI] [PubMed] [Google Scholar]
- 36. Kennedy A, Webb CD, Hill AA, et al. Loss of CCR5 results in glucose intolerance in diet-induced obese mice. Am J Physiol Endocrinol Metab 2013;305:E897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim JK, Glass WG, McDermott DH, et al. CCR5: no longer a "good for nothing" gene--chemokine control of West Nile virus infection. Trends Immunol 2006;27:308–12. [DOI] [PubMed] [Google Scholar]
- 38. Melo-Martín Id. Genetically modified organisms (GMOs): human beings. In ten Have H. (ed). Encyclopedia of Global Bioethics. Springer International Publishing, 2015. http://link.springer.com/referenceworkentry/10.1007/978-3-319-05544-2_210-1. [Google Scholar]
- 39. Yang H, Wang H, Shivalila CS, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013;154:1370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Y, Liang D, Wang Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013;13:659–62. [DOI] [PubMed] [Google Scholar]
- 41. Long C, McAnally JR, Shelton JM, et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014;345:1184–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Low BE, Krebs MP, Joung JK, et al. Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6N mice via TALEN-mediated homology-directed repair. Invest Ophthalmol Vis Sci 2014;55:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoshimi K, Kaneko T, Voigt B, et al. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun 2014;5:4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hotaling J, Carrell DT. Clinical genetic testing for male factor infertility: current applications and future directions. Andrology 2014;2:339–50. [DOI] [PubMed] [Google Scholar]
- 45. Scriven PM, Ogilvie CM. PGD for sex determination and chromosome rearrangements: FISH and emerging technologies. In El-Toukhy T, Braude P. (ed). Preimplantation Genetic Diagnosis in Clinical Practice. London: Springer-Verlag, 2014, 65–82. [Google Scholar]
- 46. Simon C. Personalized assisted reproductive technology. Fertil Steril 2013;100:922–23. [DOI] [PubMed] [Google Scholar]
- 47. Park CY, Kim DH, Son JS, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs Using CRISPR-Cas9. Cell Stem Cell 2015;17:213–20. [DOI] [PubMed] [Google Scholar]
- 48. Singh P, Schimenti JC. The genetics of human infertility by functional interrogation of SNPs in mice. Proc Natl Acad Sci USA 2015;112:10431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tournaye H, Dohle GR, Barratt CL. Fertility preservation in men with cancer. Lancet 2014;384:1295–301. [DOI] [PubMed] [Google Scholar]
- 50. Cohen J, Scott R, Schimmel T, et al. Birth of infant after transfer of anucleate donor oocyte cytoplasm into recipient eggs. Lancet 1997;350:186–7. [DOI] [PubMed] [Google Scholar]
- 51. Barritt JA, Brenner CA, Malter HE, et al. Mitochondria in human offspring derived from ooplasmic transplantation. Hum Reprod 2001;16:513–6. [DOI] [PubMed] [Google Scholar]
- 52. Zhang J, Zhuang G, Zeng Y, et al. Pregnancy derived from human nuclear transfer. Fertil Steril 2003;80:56. [Google Scholar]
- 53. Sawyer N, Blyth E, Kramer W, et al. A survey of 1700 women who formed their families using donor spermatozoa. Reprod Biomed Online 2013;27:436–47. [DOI] [PubMed] [Google Scholar]
- 54. Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 2013;340:1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sulem P, Gudbjartsson DF, Stacey SN, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet 2007;39:1443–52. [DOI] [PubMed] [Google Scholar]
- 56. White D, Rabago-Smith M. Genotype-phenotype associations and human eye color. J Hum Genet 2011;56:5–7. [DOI] [PubMed] [Google Scholar]
- 57. Kamaraj B, Purohit R. Mutational analysis of oculocutaneous albinism: a compact review. Biomed Res Int 2014;2014:905472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guvercin CH, Arda B. Eugenics concept: from Plato to present. Hum Reprod Genet Ethics 2008;14:20–6. [DOI] [PubMed] [Google Scholar]
- 59. Galizi R, Doyle LA, Menichelli M, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun 2014;5:3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. United Nations. Article 7 of the Rome Statute of the International Criminal Court, United Nations, 1998. https://childrenandarmedconflict.un.org/keydocuments/english/romestatuteofthe7.html.
- 61. Schuelke M, Wagner KR, Stolz LE, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 2004;350:2682–8. [DOI] [PubMed] [Google Scholar]
- 62. Proudfoot C, Carlson DF, Huddart R, et al. Genome edited sheep and cattle. Transgenic Res 2015;24:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chambers GM, Adamson GD, Eijkemans MJ. Acceptable cost for the patient and society. Fertil Steril 2013;100:319–27. [DOI] [PubMed] [Google Scholar]
- 64. Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014;156:836–43. [DOI] [PubMed] [Google Scholar]
- 65. Liu H, Chen Y, Niu Y, et al. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 2014;14:323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu Z, Zhou X, Zhu Y, et al. Generation of a monkey with MECP2 mutations by TALEN-based gene targeting. Neurosci Bull 2014;30:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen Y, Zheng Y, Kang Y, et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet 2015;24:3764–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Human Fertilisation and Embryology Authority. Third scientific review of the safety and efficacy of methods to avoid mitochondrial disease through assisted conception: update, 2014. http://www.hfea.gov.uk/docs/Third_Mitochondrial_replacement_scientific_review.pdf (16 October 2015, date last accessed).
- 69. Bredenoord AL, Braude P. Ethics of mitochondrial gene replacement: from bench to bedside. BMJ 2010;341:c6021. [DOI] [PubMed] [Google Scholar]
- 70. Sethi N. Ethics and the facial plastic surgeon. Eur Arch Otorhinolaryngol 2015. doi: 10.1007/s00405-015-3745-z, http://link.springer.com/article/10.1007%2Fs00405-015-3745-z. [DOI] [PubMed] [Google Scholar]
- 71. Becker G, Butler A, Nachtigall RD. Resemblance talk: a challenge for parents whose children were conceived with donor gametes in the US. Soc Sci Med 2005;61:1300–9. [DOI] [PubMed] [Google Scholar]
- 72. Stern HJ. Preimplantation genetic diagnosis: prenatal testing for embryos finally achieving its potential. J Clin Med 2014;3:280–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kumar A, Ryan A, Kitzman JO, et al. Whole genome prediction for preimplantation genetic diagnosis. Genome Med 2015;7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Baruch S, Kaufman D, Hudson KL. Genetic testing of embryos: practices and perspectives of US in vitro fertilization clinics. Fertil Steril 2008;89:1053–8. [DOI] [PubMed] [Google Scholar]
- 75. Whittaker AM. Reproduction opportunists in the new global sex trade: PGD and non-medical sex selection. Reprod Biomed Online 2011;23:609–17. [DOI] [PubMed] [Google Scholar]
- 76. Ishihara O, Adamson GD, Dyer S, et al. International committee for monitoring assisted reproductive technologies: world report on assisted reproductive technologies, 2007. Fertil Steril 2015;103:402–413.e411. [DOI] [PubMed] [Google Scholar]
- 77. Qiao J, Feng HL. Assisted reproductive technology in China: compliance and non-compliance. Transl Pediatr 2014;3:91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kirillova YA, Bogdan VV. The role and significance of assisted reproductive technology in Russia's inheritance law. Middle East J Sci Res 2013;17:1641–45. [Google Scholar]
- 79. Svitnev K. New Russian Legislation on assisted reproduction. Open Access Sci Rep 2012;1:27. [Google Scholar]
- 80. Isasi R, Knoppers BM. Oversight of human inheritable genome modification. Nat Biotechnol 2015;33:454–5. [DOI] [PubMed] [Google Scholar]
- 81. United Nations. Child Adoption: Trends and Policies. United Nations; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.