Abstract
Disrupted in Schizophrenia-1 (DISC1) has been associated with a broad spectrum of mental disorders. DISC1 is a multi-compartmentalized protein found in the cytoplasm, centrosome, nuclei and mostly enriched in mitochondria. In order to shed light on DISC1 mitochondrial function, we have studied its topology within the organelle. We show in here that in mammals DISC1 resides in the ‘Mitochondrial contact site and Cristae Organizing system’ (MICOS) complex, involved in cristae organization. DISC1 knockdown in SH-SY5Y cells causes MICOS disassembly and fragmentation of the mitochondrial morphology network. Moreover, DISC1 depleted cells have decreased mitochondrial DNA (mtDNA) content and steady state levels of oxidative phosphorylation (OXPHOS) subunits. As a consequence, OXPHOS complexes and supercomplexes are partially disassembled in DISC1 knockdown cells, which suffer severe bioenergetic defects, evidenced by impaired oxygen consumption, adenosine triphosphate synthesis and mitochondrial membrane potential. Transfection of recombinant full-length human DISC1 restores MICOS complex assembly and rescues OXPHOS function, meanwhile overexpression of the DISC1 truncated form Δ597-854, known to be pathogenic, fails to rescue the bioenergetic impairment caused by DISC1 knockdown. These results should contribute to reveal DISC1 physiological function and potential pathogenic role in severe mental illnesses.
Introduction
Schizophrenia is a complex neuropsychiatric disorder whose pathophysiology is largely unknown. Growing evidences suggest that part of the pathological components of the disease can be attributable to mitochondrial function abnormalities, supported by recent findings suggesting mitochondrial roles to Disrupted in Schizophrenia-1 (DISC1). DISC1 was identified in a balanced reciprocal chromosomal translocation t(1;11) (q42;q14) that results in DISC1 truncated forms, such as the Δ597-854, co-segregating with schizophrenia and other major affective disorders in a large Scottish family (1–4). DISC1 is a multi-compartmentalized protein found in the cytoplasm, centrosome, nuclei and mostly enriched in mitochondria (5). The role of DISC1 in the mitochondria is under debate. It has been observed that modulation of DISC1 protein levels induces morphological abnormalities (6), causes deficiencies in important mitochondrial enzyme activities like NADH and L-Monoamine Oxidase (MAO)-A (7), and impairs mitochondrial trafficking throughout the axons (8,9). Moreover, overexpression of the resultant truncated proteins of the translocation identified in the Scottish family translates in aggregation of abnormal proteins in the mitochondria that induces clusterization and loss of mitochondrial membrane potential (mΔΨ) (4).
DISC1 acts as scaffold binding a number of other proteins. In mitochondria, it has been shown to interact with two main partners: Mic60/Mitofilin, with whom DISC1 collaborates in the maintenance of mitochondrial integrity (7), and the Miro-TRAK complex, with whom modulates mitochondrial motility (9,10). A third interacting partner of DISC1 in the mitochondria has also been identified, Mic25/CHCM1/Coiled-coil-Helix-Coiled-coil-Helix Domain Containing (CHCHD)6 (from now on, Mic25/CHCHD6), but the nature and the functional relevance of this interaction has not been described yet (11).
In order to shed light on the role of DISC1 in mitochondria, we aimed to disclose its sub-mitochondrial localization and determine whether DISC1 is recruited in a mitochondrial protein complex. We identified DISC1 associated to the mitochondrial contact site and cristae organizing system (MICOS), a protein complex that controls the inner mitochondrial membrane (IMM) structure and modulates mitochondrial dynamics and protein biogenesis (12). To further characterize the role of DISC1 at MICOS, we silenced DISC1 in human cells. DISC1 depletion not only affected MICOS assembly, but also impaired the oxidative phosphorylation system (OXPHOS) assembly and function, including ATP synthesis, respiration and mΔΨ. Interestingly, the expression of human recombinant full-length DISC1 partially rescued OXPHOS dysfunction meanwhile the overexpression of the aberrant DISC1 form found in the Scottish family failed to rescue DISC1 knockdown, suggesting that DISC1 modulation of OXPHOS through MICOS may play a role in the disease pathogenesis. Our results provide a detailed description of DISC1 topology in the mitochondria and shed light on its functional physiological role within the organelle and in the context of the pathophysiology of severe mental illnesses.
Results
Mitochondrial DISC1 anchors to the IMM facing the intermembrane space (IMS)
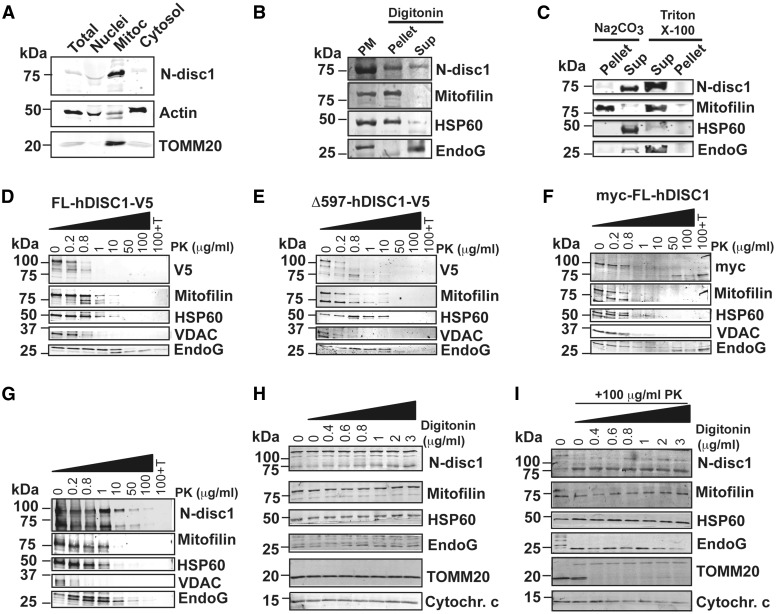
Several reports have pointed out that DISC1 is enriched at the mammalian mitochondria, but its mitochondrial topology remained to be investigated. By subcellular fractionation of adult mouse brain, we first confirmed that the 71 kDa isoform of mouse disc1 (mdisc1) is mostly located at the mitochondria. Total brain homogenates were differentially centrifuged and nuclei, mitochondrial and cytosolic protein extracts were separated by SDS-PAGE electrophoresis. In mouse brain, the 71 kDa mdisc1 isoform is mostly expressed in mitochondria, as evidenced by the immunoreactive band detected by an anti-disc1 antibody raised against the N-terminal domain (Fig. 1A). To describe the submitochondrial location of endogenous mdisc1, we performed mitoplast isolation from mouse brain mitochondria by digitonin solubilization (Fig. 1B) and sodium carbonate treatment (Fig. 1C). Immunoreactivity of mdisc1 was present in both pellet and supernatant, suggesting that part of the protein is associated with the inner membrane, and part is facing the intermembrane space (Fig. 1B–C). To further define the mitochondrial topology of mdisc1, we performed protease protection assays. Mitochondria isolated from human HEK293T cells transfected with full length recombinant human DISC1 (hDISC1) V5-tagged at the C-terminal domain (FL-hDISC1-V5) (Fig. 1D), with its truncated form Δ597-854 V5-tagged at the C-terminal domain (Δ597-854 hDISC1-V5) (Fig. 1E), and with full length hDISC1 myc-tagged at the N-terminal domain (myc-FL-hDISC1) (Fig. 1F), or mitochondria isolated from mouse brain (Fig. 1G) were incubated with increasing concentrations of proteinase K (PK) in presence or absence of Triton X-100. We investigated the accessibility of PK to selected proteins of the outer mitochondrial membrane (OMM), the intermembrane space (IMS), and the matrix. The patterns of digestion of the C-terminal domain of recombinant hDISC1, detected with an antibody against the V5 epitope, associated with the OMM protein voltage-dependent anion channel (VDAC) and the IMS protein Endonuclease G (EndoG) (Fig. 1D–E). Then, we studied the patterns of digestion of the N-terminal domain of recombinant myc-FL-hDISC1. The antibody against the myc epitope targeted two different regions, one that followed the patterns of digestion of Mic60/Mitofilin (inner mitochondrial membrane, IMM) and heat shock protein-60 (HSP60) (matrix) and another region that started being accessible to the antibody at high proteinase K concentrations (50 μg/ml) and remained being resistant to digestion and solubilization upon combined treatment of 100 μg/ml proteinase K and 0.1% Triton X-100 (Fig. 1F), suggestive of being protected by a protein-lipid interaction, a typical pattern of a transmembrane domain. We did not observe major differences between the patterns of PK digestion of FL-hDISC1-V5 and Δ597-854 hDISC1-V5. mDisc1 from mouse brain mitochondria, detected with an antibody raised against the N-terminal domain, showed a pattern of digestion paralleled to that of Mic60/Mitofilin, an IMM protein, and HSP60, a mitochondrial matrix protein (Fig. 1G). In order to discard that our observed results were masked by the fact that proteinase K penetrability to mitochondrial membranes is limited under isotonic conditions (13), we repeated the protease protection assay in crude mitochondria isolated from mouse brain, using increasing concentrations of digitonin combined with fixed treatment with 100 μg/ml of proteinase K (Fig. 1H–I). The immunoreactive band of ≈75kDa of m-disc1, detected with a polyclonal antibody raised at the N-terminal domain (Abbiotec), was protected to a comparable extent as Mic60/Mitofilin (IMM) and HSP60 (matrix) (Fig. 1I), further indicating that the N-terminal region of m-disc1 is associated with the IMM.
Figure 1.
Mitochondrial mdisc1 is anchored to the Inner Mitochondrial Membrane facing the intermembrane space. (A) Subcellular fractionation of adult mouse brain followed by immunoblot with an antibody against the N-terminal domain of DISC1 (N-disc1) shows that mdisc1 concentrates in the mitochondria. (B, C) Mitoplast isolation from mouse brain mitochondria by digitonin solubilization (B), and alkaline extraction (C). Triton X-100 was used to completely solubilize the protein samples (C). Disc1 immunoreactivity was present in both pellet and supernatant (Sup), suggesting that part of the protein is associated with the inner membrane, and part is facing the intermembrane space. PM = Purified mitochondria. (D–I) Proteinase K (PK) protection assay performed with increasing concentrations of PK (D–G), or increasing concentrations of digitonin in the absence (H) or presence (I) of 100 μg/ml PK. Mitochondria isolated from HEK293T cells transfected with recombinant full length hDISC1 V5-tagged (FL-hDISC1-V5) (D), with its truncated form hDISC1-Δ597-854 V5 tagged (Δ597-hDISC1-V5) (E), or with full length hDISC1 myc-tagged (myc-FL-hDISC1) (F), and mitochondria isolated from mouse brain (G) were incubated with increased concentrations of PK, before analysis by immunoblotting. The combination of 100 μg/ml PK plus 0.1% Triton X-100 (100 + T) was used to fully solubilize the protein samples. hDISC1 immunoreactivity was lost in a Proteinase K-concentration-dependent manner, a pattern paralleled by that for Mic60/Mitofilin, an IMM protein. (H–I) Crude mitochondria isolated from mouse brain were permeabilized with increasing concentrations of digitonin (H–I), prior to be subject to digestion with 100 μg/ml of PK (I). Markers: Mic60/Mitofilin, inner mitochondrial membrane; HSP60, matrix; EndoG and Cytochrome c, intermembrane space; VDAC and TOMM20, outer mitochondrial membrane.
Overall, our submitochondrial localization studies strongly suggest that DISC1 is associated to the IMM facing the IMS, with the C-terminal domain exposed to the OMM and the N-terminal domain anchored to the IMM.
DISC1 associates with the MICOS complex
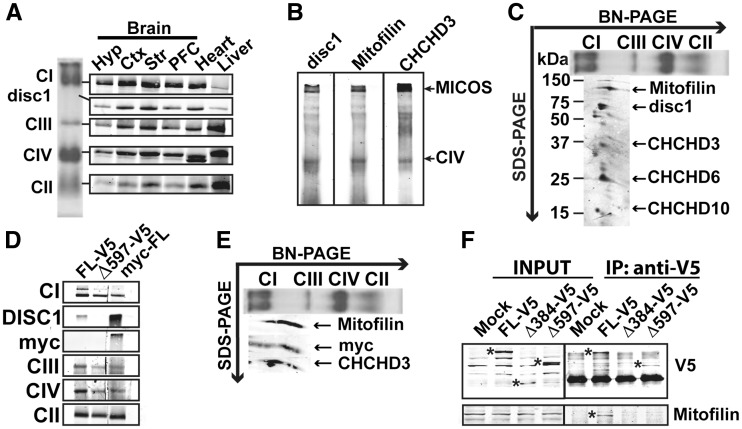
Our results on DISC1 mitochondrial topology strongly suggest an association of the N-terminal domain of DISC1 with the IMM. This domain is predicted to be composed by disordered residues (14). Disordered proteins can form very large intermolecular interfaces within complexes (15). Due to this reason and the fact that a previous report suggested that mitochondrial DISC1 interacts with Mic60/Mitofilin (7), the core component of the MICOS complex, we decided to investigate whether DISC1 could be associated to MICOS. Isolated mitochondria from mouse brain were solubilized with mild detergent (β-d-lauryl maltoside: β-d-LM) and the solubilized products were separated by Blue-Native poly-acrylamide gel electrophoresis (BN-PAGE) (Fig. 2A–B). By immunoblotting with the antibody against the N-terminal domain of mdisc1, we detected an immunoreactive band of approximately 700 kDa, below OXPHOS complex I, present in all analyzed tissues (brain, heart and liver), but enriched in cerebral cortex, striatum and heart (Fig. 2A). Disc1 immunoreactive complex was also detected with antibodies against the known subunits of MICOS Mic60/Mitofilin and Mic19/CHCHD3 (Fig. 2B). To verify that mdisc1-containing complex was MICOS, we performed two dimensional western-blotting (2D-WB) of the BN-PAGE, to separate the individual subunits of MICOS. By sequentially immunoblotting with the different MICOS subunits antibodies, we observed that mdisc1 immunoreactive band aligned with the known MICOS subunits Mic60/Mitofilin, Mic19/CHCHD3 and Mic25/CHCHD6 and with the recently reported novel MICOS subunit CHCHD10 (16) (Fig. 2C). We next checked whether recombinant hDISC1 could also assemble into MICOS. We transfected human HEK293T cells with FL-hDISC1-V5, Δ597-854 hDISC1-V5 and myc-FL-hDISC1 and we studied MICOS assembly by BN-PAGE electrophoresis (Fig. 2D) followed by 2D-WB (Fig. 2E). Both myc-N-ter tagged and V5-C-ter tagged hDISC1-FL were able to assemble into MICOS; by contrary, the truncated form Δ597-854-V5 failed to assemble (Fig. 2D). 2D-WB analysis confirmed that myc-hDISC1 fully aligned with Mic60/Mitofilin and Mic19/CHCHD3, further confirming that these three proteins are contained into the same multiprotein complex (Fig. 2C and E). To finally prove DISC1-Mic60/Mitofilin interaction, we performed a co-immunoprecipitation assay using the antibody against V5 in HEK293T cells overexpressing FL-hDISC1-V5 or its truncated forms Δ1-384 hDISC1-V5 and Δ597-854 hDISC1-V5, all tagged with the V5 epitope at the C-terminal domain. Only cells expressing FL-hDISC1-V5 were able to pull down endogenous Mic60/Mitofilin (Fig. 2F).
Figure 2.
hDISC1 associates with Mic60/Mitofilin and Mic19/CHCHD3 in the MICOS complex. (A) Blue-Native (BN) PAGE followed by immunoblotting of isolated mitochondria from mouse brain (Hyp: hypothalamus, Ctx: cortex, Str: striatum, PFC: prefrontal cortex), heart and liver. Complex I (CI) was detected with an antibody against the NDUFS9 subunit, complex II (CII) with the anti-SDHA subunit antibody, complex III (CIII) with the anti-Core I subunit antibody and complex IV (CIV) with the anti-COX1 subunit antibody. (A). Disc1 immunoreactive band was found to be associated with a protein complex of around 700 kDa. Disc1 protein complex is enriched in cerebral cortex, striatum and heart. (B) Disc1 immunoreactive band migrates at the same molecular weight than Mic60/Mitofilin and Mic19/CHCHD3. 200 μg of isolated mitochondria from mouse brain were loaded in three different lanes and electrophoresed in a BN-PAGE. Membrane was cut and every lane was blotted with the indicated antibodies. (C) Second dimension of the BN-PAGE showed that mdisc1 associates with Mic60/Mitofilin, Mic19/CHCHD3, Mic25/CHCHD6 and CHCHD10 in the MICOS complex. (D–E) Recombinant hDISC1 also associates with Mic60/Mitofilin in the MICOS complex. (D) BN-PAGE followed by immunoblotting of HEK293T cells transfected with full length recombinant human hDISC1 V5-tagged (FL-V5), with its truncated form Δ597-854 V5-tagged (Δ597-V5) or with full length hDISC1 myc-tagged (myc-FL), showed that both full lenght (FL)-hDISC1 proteins were able to assemble in the hDISC1/MICOS complex. By contrary, Δ597-854 did not assemble in MICOS. The same antibodies as in (A) were used to detect OXPHOS complexes. (E) Second dimension of the BN-PAGE showed that full length hDISC1 myc-tagged (myc-FL) associates with Mic60/Mitofilin and Mic19/CHCHD3 in the MICOS complex. (F) Co-immunonoprecipitation (Co-IP) performed with the anti-V5 antibody in HEK293T cells overexpressing either full length recombinant human hDISC1 V5-tagged (FL-V5), or its truncated forms Δ1-384 (Δ384-V5), or Δ597-854 (Δ597-V5). Only FL-V5 pulled down endogenous Mic60/Mitofilin. * Indicate the immunoreactive bands for V5 and Mitofilin antibodies. Mock: cells transfected with empty plasmid.
DISC1 knockdown partially impairs MICOS assembly
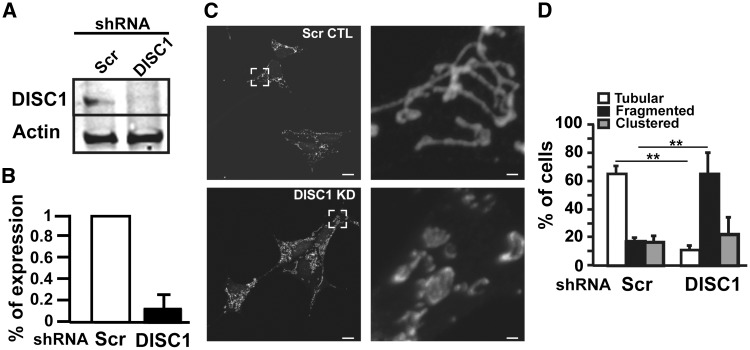
To further investigate the role of DISC1 in MICOS, we generated stable human neuroblastoma cell lines SH-SY5Y depleted of hDISC1 by lentiviral infection of shRNA against hDISC1. The lentiviral construct, aside of the hDISC1 shRNA, also contained a resistance to the antibiotic puromycin. Infected cells were grown in puromycin supplemented media culture and 15 days post-infection, clones were selected according to their DISC1 expression levels, measured by WB analysis. The studied clone was severely depleted of DISC1 expression (77% ± 8 of depletion as compared to the mass culture of shRNA scrambled infected control cells) (Fig. 3A–B). Since MICOS has been shown to be crucial to maintain the mitochondrial architecture of cristae morphology (11,17–23), we first checked how DISC1 knockdown (KD) affected the mitochondrial network and distribution by confocal microscopy analysis (Fig. 3C–D). We labeled mitochondria with an antibody against the translocator of the outer membrane 20 (TOMM20) and classified DISC1 depleted cells based on mitochondrial morphology criteria. Under normal conditions, SH-SY5Y cells have variable shape and distribution of the mitochondrial network. Cells normally display a tubular mitochondrial network but can also show clustering of mitochondria around the nucleus or fragmented network with round mitochondria. An excessive clustering or fragmentation is indicative of mitochondrial dysfunction (24). DISC1 KD resulted in decreased tubular morphology and increased fragmentation of the mitochondrial network (control cells: 17.6% ± 2.5 of fragmented mitochondria, DISC1 KD cells: 65% ± 15 of fragmented mitochondria, P < 0.01; Fig. 3C–D).
Figure 3.
hDISC1 knockdown impairs the mitochondrial morphology network. (A) Western Blot showing that stable SH-SY5Y expressing shRNA against hDISC1 were severely depleted of hDISC1 protein expression. (B) hDISC1 silencing efficiency, measured from three independent WB experiments, showed that stable SH-SY5Y hDISC1 knockdown (KD) cells only express 13 ± 6% of hDISC1, compared with scrambled (Scr) shRNA expressing control cells. (C–D) Mitochondrial morphology changes in hDISC1 KD cells. The mitochondrial network was resolved by immunofluorescence microscopy using an antibody anti-TOMM20. (C) Representative images show that hDISC1 knockdown caused increased mitochondrial fragmentation. The right panel corresponds to the 10x zoomed area framed in the left panel, Control cells (Scr CTL) and hDISC1 knockdown cells (DISC1 KD). Scale bars: 10 μm and 1 μm, for the left and right panel, respectively. (D) Quantification of mitochondrial morphology showing that control cells display a high proportion of tubular mitochondria, whereas hDISC1 KD cells have significantly increased fragmented mitochondria. Error bars represent ± SD of three independent experiments. In each experiment, at least 100 cells were analyzed. **P < 0.01 compared with scrambled controls.
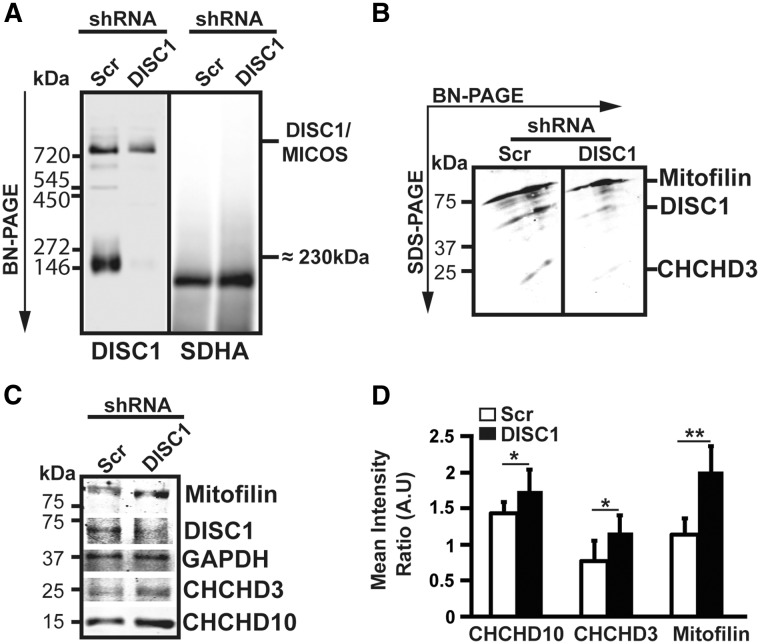
In order to see whether this aberrant mitochondrial morphology could be caused by MICOS disassembly, we performed BN-PAGE analysis on DISC1 KD cells followed by immunoblotting with an antibody against the N-terminal domain of DISC1. We found a partial disassembly of the DISC1/MICOS complex in SH-SY5Y DISC1 KD cells (Fig. 4A). 2D-WB analysis of the BN-PAGE demonstrated that the assembled steady-state levels of DISC1 were severely depleted, as well as the ones of Mic19/CHCHD3. However, Mic60/Mitofilin assembled levels did not show significant differences between control cells and DISC1 KD, suggesting that hDISC1 and Mic19/CHCHD3 are located in a more peripheral position within the complex (Fig. 4B). By contrary, total steady-state levels of MICOS proteins, analyzed by WB, showed that DISC1 KD cells tend to compensate the loss of DISC1 by up-regulating MICOS subunits, as it is the case for Mic60/Mitofilin, Mic19/CHCHD3 or CHCHD10 (Fig. 4C–D).
Figure 4.
hDISC1 knockdown induces a partial disassembly of the MICOS complex. (A) BN-PAGE analysis on SH-SY5Y DISC1 KD cells followed by immunoblotting with an antibody against the N-terminal domain of DISC1 showing a partial disassembly of the hDISC1/Mitofilin/CHCHD3 containing complex, and almost a complete disassembly of the ≈230 kDa complex which nature remains to be determined. N-DISC1 anti-rabbit antibody was used to detect hDISC1/MICOS complex. The mouse raised antibody anti-SDHA subunit OXPHOS complex II was used as loading control. (B) Second dimension of the BN-PAGE followed by immunoblotting with the indicated antibodies showed severe reductions of assembled hDISC1 and Mic19/CHCHD3 steady-state levels. Mic60/mitofilin steady-state levels were not affected. (C–D) WB analysis showed that the steady-state levels of MICOS subunits Mic60/Mitofilin, Mic19/CHCHD3 and CHCHD10 were upregulated in the hDISC1 KD cells. (D) Quantification of WB analysis shown in (C). Column bars indicate the immunoreactivity of each protein as determined by densitometric analysis (integrated optical density of each protein band vs. that of GAPDH band), and are the mean ± SD of three independent experiments. A.U.=Arbitrary Units. *P < 0.05 and **P < 0.01, compared with scrambled controls.
DISC1 knockdown impairs OXPHOS structure and function
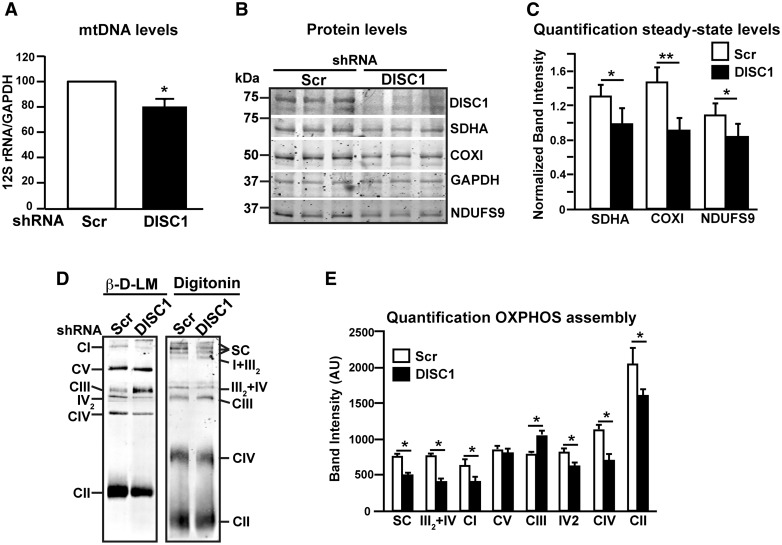
Since MICOS alterations have been associated with deficits in mitochondrial biogenesis and mitochondrial DNA (mtDNA) instability (16,17,25,26), we investigated whether hDISC1 depletion in SH-SY5Y cells affects mtDNA levels and OXPHOS proteins steady-state levels. Total mtDNA content was decreased by 20% ± 6.8 (P < 0.05) in hDISC1 KD cells (Fig. 5A). Next, we studied if this decrease in mtDNA levels found in hDISC1 depleted cells could result in a decreased expression of its encoded proteins (Fig. 5B). In this sense, we observed that the steady-state levels of the mtDNA encoded cytochrome c oxidase subunit 1 (COX1) of Complex IV (CIV) were down-regulated by 38% ± 11.6 in DISC1 KD cells as compared to scrambled control cells (P < 0.01). However, the nuclear encoded subunits NADH dehydrogenase ubiquinone 1 alpha subcomplex subunit 9 (NDUFS9) of OXPHOS Complex I (CI) and the succinate dehydrogenase complex, subunit A (SDHA) of Complex II (CII) were also down-regulated (22% ± 13 and 24.6% ± 10, respectively, P < 0.05) (Fig. 5B–C), suggesting that in these cells the mitochondrial protein import machinery may also be impaired. In this regard, we also found that the assembly of OXPHOS complexes I, II and IV, resolved by β-D-LM, were significantly decreased in DISC1 KD cells as compared to scrambled control cells (35% ± 19, 21% ± 18, and 37% ± 17, respectively, P < 0.05), Paradoxically, CIII was over-represented compared to scrambled controls (29% ± 6, P < 0.05) (Fig. 5D–E). OXPHOS supercomplexes, resolved by digitonin solubilization, were also downregulated in DISC1 KD cells (34% ± 5, P < 0.05; Fig. 5D–E).
Figure 5.
hDISC1 knockdown impairs OXPHOS proteins translation and supercomplexes formation. (A) Transcription levels of mtDNA. Real-time PCR analysis of the mtDNA transcript 12S rRNA from SH-SY5Y cells stably expressing either scrambled shRNA (Scr) or hDISC1 shRNA (hDISC1). Data expressed relative to GAPDH of three independent determinations ± SD. *P < 0.05. (B) WB analysis of OXPHOS proteins showed that SH-SY5Y hDISC1 KD cells are depleted of both mtDNA and nuclear DNA encoded subunits. (C) Quantification of the steady-state levels of OXPHOS proteins. Bars represent the mean ± SD of 5 independent western blot experiments relative to GAPDH; * P < 0.05; ** P < 0.01. (D) hDISC1 KD cells present partial disassembly of OXPHOS complexes and supercomplexes. BN-PAGE analysis of OXPHOS complexes and supercomplexes. Cells were solubilized either with β-D-lauryl maltoside (β-D-LM) to resolve individualized OXPHOS complexes or with digitonin to resolve OXPHOS supercomplexes. Complexes I to IV of OXPHOS were detected with an anti-NDUFS9 subunit antibody (CI), SDHA subunit antibody (CII), Core1 antibody (CIII) and COXI antibody (CIV). ATP synthase (CV) was detected with the anti-subunit β antibody. IV2, dimer of CIV detected with the COX1 antibody; SC, supercomplexes I + III2 + IVn. (E) Quantification of OXPHOS complexes and supercomplexes assembly, comparing the immunoreactive band intensities of the antibodies used in (D) between DISC1 KD cells and its Scr control cells. Bars represent the mean ± SD of 5 independent BN-PAGE experiments; * P < 0.05.
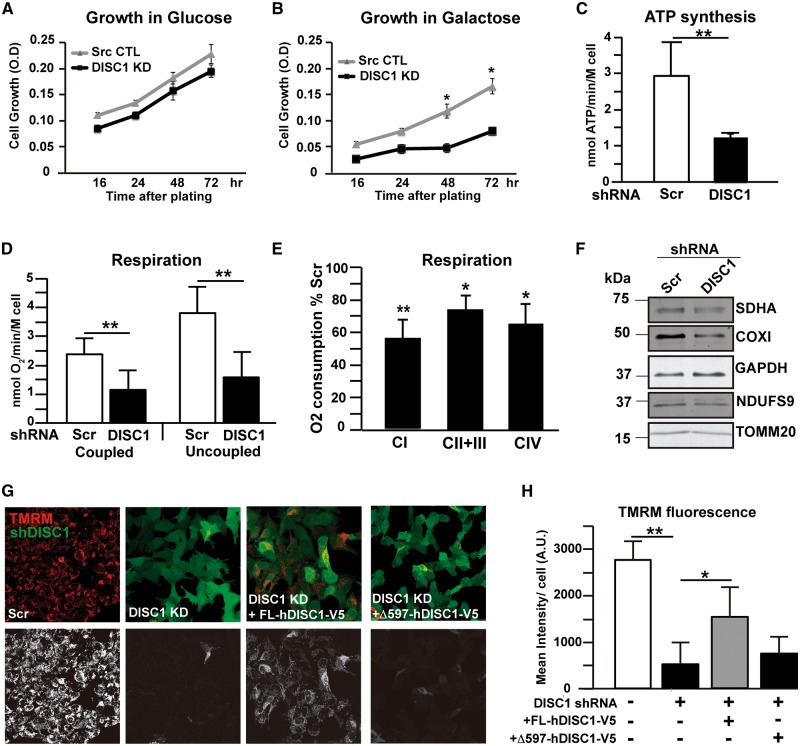
We then checked whether OXPHOS structural abnormalities translated into bioenergetics dysfunction in DISC1 KD cells. First, we measured cell replication in medium containing either glucose or galactose as energetic substrate. No differences were observed between control and DISC1 KD cells once grown in glucose (Fig. 6A). However, in galactose medium, where cells have to rely primarily on OXPHOS for ATP production, the replication rate of DISC1 KD cells was severely reduced (Fig. 6B). To characterize OXPHOS function, we measured mitochondrial ATP synthesis in digitonin permeabilized SH-SY5Y cells using malate and pyruvate as substrates. DISC1 KD cells displayed a 48% ± 14 (p < 0.01) decrease in ATP production compared to control cells (Fig. 6C). Since ATP synthase activity is coupled to the mitochondrial electron transfer and proton translocation, it directly reflects the efficiency of the respiratory chain. Consequently, differences in ATP synthesis may depend upon changes in respiratory chain function upstream of the ATP synthase. We therefore measured mitochondrial respiration, as oxygen consumption, in intact SH-SY5Y cells using pyruvate as substrate and, as expected, the results correlated with the ones obtained for ATP synthesis. Coupled respiration in DISC1 KD cells was reduced by 51% ± 16.2 (P < 0.05) as compared to control cells (Fig. 6D). To achieve the maximum respiratory capacity, independent of ATP synthase, cells were incubated with 1 μM of the uncoupling agent carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP). Uncoupled respiration was severely decreased in DISC1 KD cells (57% ± 14.3, P < 0.05; Fig. 6D) compared to control cells, evidencing the severe deficit of the respiratory chain electron transfer caused by DISC1 loss. To further characterize the respiratory deficit, we measured the contribution of each complex to oxygen consumption in digitonin-permeabilized cells. DISC1 KD cells incubated with glutamate plus malate, substrates that allow the generation of NADH to deliver electrons to complex I, exhibited a 43% ± 10 reduction in O2 consumption compared to scrambled controls (P < 0.01, Fig. 6E). Complex II + III-dependent respiration, measured with succinate after inhibition of complex I by rotenone, was also impaired in DISC1 KD cells, although to a comparable extent than complex IV-driven respiration (26% ± 8 versus 35% ± 12 decrease compared to scrambled controls, respectively, P < 0.05; Fig. 6E). Together, all these results suggest that complexes I and IV are the major contributors to the respiration deficit exhibited by DISC1 KD cells. With the aim to correlate the mitochondrial protein load with the biochemical findings, proteins were extracted from the cells used to measure respiration and were analyzed by western blotting. As already shown, all OXPHOS proteins analyzed (SDHA, COXI and NDUFS9) were downregulated in DISC1 KD cells, as well as TOMM20 (Fig. 6F), supporting a global mitochondrial protein import impairment. These results also suggest that the bioenergetics defects of DISC1 KD cells were due to OXPHOS protein loss. To finally characterize OXPHOS function, we measured mΔΨ by tetramethylrhodamine methyl ester (TMRM) staining followed by live-imaging. TMRM is a fluorescent cationic dye that gets into mitochondria depending on mΔΨ. The fluorescence intensity of the dye is directly proportional to the mΔΨ. DISC1 KD cells showed decreased capacity to retain the mitochondrial TMRM fluorescence compared to control cells (82% ± 12.2, P < 0.01; Fig. 6G–H). Interestingly, overexpression of recombinant hDISC1 on DISC1 KD cells could partially rescue the mΔΨ deficiency. DISC1 KD cells had only 18% ± 12.2 mΔΨ compared to control cells; however, once they were transfected with hDISC1-V5, their mΔΨ rate increased up to 56.4% ± 12.4 (P < 0.05). In contrast, the overexpression of the pathogenic form Δ597-854 failed to rescue the mΔΨ impairment (Fig. 6G–H).
Figure 6.
hDISC1 knockdown impairs OXPHOS function. (A–B) Cell growth in glucose (A) and galactose medium (B) measured in hDISC1 KD (DISC1-KD) and scrambled control cells (Src-CTL). Equal number of cells were plated and cell proliferation was measured by the MTT assay. (C) ATP synthesis by using a luciferin–luciferase method with malate plus pyruvate (30 μM each) as substrates, measured in digitonin-permeabilized hDISC1 KD or control (Scr) SH-SY5Y cells, as indicated. (D) Oxygen consumption measured in intact cells using 1 mM pyruvate as substrate, with or without 1 µM of the uncoupler FCCP, and 1.5 mM KCN as the terminal inhibitor (E) Complex-driven oxygen consumption in digitonin permeabilized cells. Complex I-driven respiration (CI) was achieved using glutamate (5 mM) plus malate (5 mM) as substrates; complex II + III (CII + III) was performed using succinate (5 mM) and glycerol-3-phosphate (5 mM) as substrates; and complex IV-driven respiration (CIV) was done by addition of ascorbate (5 mM) plus TMPD (2mM) as electron donor. Rotenone (100 mM), antimycin A (20 nM) and KCN (1 mM) were used to inhibit CI, CIII and CIV activities, respectively. Bars represent the mean ± SD of the percentage of oxygen consumed referred to control cells of 3 independent experiments. * P < 0.05; ** P < 0.01. (F) Western blotting analysis of proteins extracts from cells used to measure respiration. Mitochondrial proteins levels were decreased in DISC1 KD cells. (G–H) ΔΨm measured by TMRM fluorescence. (G) Representative images evidence the lower fluorescence of TMRM in hDISC1 KD cells compared to control cells (Scr). TMRM fluorescence was partially rescued by overexpressing recombinant hDISC1 full-length (DISC1 KD + FL-hDISC1-V5), but not by overexpressing the truncated hDISC1 form Δ597-854 (DISC1 KD + Δ597-hDISC1-V5). Upper panels show GFP fluorescence expression of hDISC1-KD in green and TMRM fluorescence in red. Lower panels show only the TMRM signal. Scale bars: 10 μm. (H) Quantification of TMRM fluorescence in the above experimental paradigms. Error bars represent ± SD of three independent experiments. * P < 0.05; ** P < 0.01.
Taken together, our results indicate that mitochondrial DISC1 associates to the MICOS complex and modulates OXPHOS function in human cells. Truncated DISC1 protein identified in schizophrenic patients compromises its assembly to MICOS and fails to rescue OXPHOS impairment.
Discussion
In this study we have identified DISC1 as a novel component of the MICOS complex in human cells and mouse brain mitochondria. MICOS is conserved from yeast to humans. In yeast, it is comprised of six core subunits, Mic60, Mic27, Mic26, Mic19, Mic12 and Mic10 (17,27). To date, six mammalian homologues of five of the yeast MICOS proteins have been identified: Mic60/Mitofilin (18,28), Mic27/APOOL (21,29), Mic26/APOO (19), Mic10/MINOS1 (23,29) and Mic19/CHCHD3 and Mic25/CHCHD6 (11,26), both homologues of yeast Mic19. The combined approximate molecular weight of these subunits is ∼200 kDa; however, others and we have identified the mammalian MICOS at ∼700 kDa migration band in native gels (16,30), suggesting that there are novel subunits that remain to be identified. Among these ones, QIL1/C19orf70 (30) has been recently identified, shown to be the mammalian ortholog of Mic12 (31), and we recently identified CHCHD10 as a novel subunit of MICOS (16).
Our results disclose DISC1 mitochondrial topology. DISC1 appears to be anchored at the IMM through its N-terminal domain, facing the IMS (Fig. 1). This DISC1 topology assembles the ones previously described for the MICOS subunits Mic19/CHCHD3 and Mic60/Mitofilin. Both proteins were found to be anchored at the IMM through an amino-terminal transmembrane domain while the rest of the proteins is extruding into the IMS (26,32,33). Contrarily to Mic60/Mitofilin and Mic19/CHCHD3, DISC1 does not have a canonical amino-terminal transmembrane domain. Its N-terminal domain is predicted to be disordered, meaning that it lacks a fixed tertiary structure. Intrinsically disordered proteins (IDP) are depleted in hydrophobic residues that usually drive protein folding, and are enriched in charged and polar residues (34), as is the case for the 60% out of the 350 amino acids that form DISC1 N-terminal domain (14). IDPs are important constituents of protein complexes, playing crucial roles in assembly, as the molecular stick that strengthens complexes or as highly flexible scaffolds that mediate interactions with other complexes (35). This suggests that the association of DISC1 to the IMM is mediated by its binding to another MICOS subunit. Prior to this study, Park and co-workers described the functional interaction between hDISC1 and Mic60/Mitofilin by interaction analysis and identified two potential DISC1 binding regions, one contained in the N-terminal domain (1–347 amino acids) and one at the C-terminal domain, residues 600–854 (7). We could confirm FL-hDISC1 interaction with Mic60/Mitofilin by co-immunoprecipitation, but with our experimental settings we failed to confirm Mic60/Mitofilin pull-down with the DISC1 truncated construct containing the first 384 amino acids of the protein (1–384), including the N-terminal domain, suggesting that the N-terminal domain of DISC1 per se it is not sufficient to promote the interaction of DISC1 with Mic60/Mitofilin in human cells (Fig. 2F). Another potential interacting partner of DISC1 within MICOS is CHCHD3, since its MICOS assembled steady-state levels decrease in DISC1 KD cells and, paradoxically, the total protein levels, measured by WB, are increased (Fig. 4B–C). Now, by using BN-PAGE analysis followed by 2D-WB, we demonstrate that DISC1, both recombinant hDISC1 and endogenous mdisc1, assemble into MICOS (Fig. 2). The interaction of DISC1 with known MICOS subunits is also reflected in the significant alteration of the expression levels of the MICOS subunits in DISC1 depleted cells, most of them being up-regulated, suggesting that the transcription-translation machinery tries to compensate an abnormal situation (Fig. 4C). This may seem contradictory compared to previous observations in which the knockdown of Mic60/Mitofilin or Mic19/CHCHD3 for 7 days induced depletion of the other MICOS subunits analyzed, whereas the knockdown of Mic25/CHCHD6, Mic27/ApoOL and Mic23/ApoO had no effect on other MICOS components (36). These conflicting results may be due to the fact that our cells were already stably depleted of hDISC1 once MICOS subunits expression was tested. We cannot discard that MICOS subunits overexpression in DISC1 knockdown cells may also be related with the organizing role of DISC1 within MICOS, as it has been recently shown in yeast that non-core MICOS subunits have distinct roles within the complex (37). The fact that MICOS does not suffer a major disassembly upon hDISC1 silencing, together with its described topology, suggests that DISC1 may be a peripheral protein within the complex (Fig. 4A).
DISC1 knockdown also affected the mitochondrial tubular network of the SH-SY5Y cells. Cells depleted of hDISC1 displayed a significant increase of fragmented mitochondria with onion-ring structures (Fig. 3). Together with aberrant mitochondria, hDISC1 loss also resulted in partial disassembly of MICOS (Fig. 4). MICOS is thought to reside at the cristae junctions (CJs) and contact sites between the IMM and the OMM, and controls IMM architecture. The mitochondrial morphology of DISC1 KD cells resembles the ones observed upon genetic depletion of several MICOS subunits, in which mitochondrial fragmentation has been associated to CJs loss with aberrant concentric stacks of IMM dissociated from the inner boundary membranes (17,18,20,21,23,26,27). The loss of CJs is thought to separate the respiratory chain complexes from the protein import machinery, affecting the supply with substitute proteins required for the replacement of damaged subunits (22). In fact, DISC1 KD cells evidenced a significant impaired assembly of OXPHOS complexes and supercomplexes and decreased steady-state levels of OXPHOS subunits encoded by both genomes, the mtDNA, as shown for COXI CIV (Fig. 5B–C) and the nuclear DNA, as shown for NDUFS9 subunit of CI or SDHA of CII (Fig. 5B–C). These results suggest that DISC1 loss causes a general mitochondrial protein import defect. In fact, it is known that MICOS promotes protein import by interacting with the translocase of the outer mitochondrial membrane (TOMM) complex, the main entry gate for the majority of mitochondrial precursor proteins, via the mitochondrial intermembrane space assembly machinery (MIA) (38). MICOS controls protein biogenesis not only interacting with TOMM and MIA, but also through its interaction with components of the sorting and assembly machinery (SAM) complex of the outer membrane to form the mitochondrial intermembrane space bridging (MIB) complex (39). Aligning with our observed results, long-term depletion of SAM50, a core component of the SAM complex, or Mic60/Mitofilin translate into down-regulation of OXPHOS proteins and impacts on the assembly of the respiratory complexes and the maintenance of the mtDNA (39). The MIB/MICOS complex also plays an important role in nucleoid organization (40), and defects in MICOS subunits translate in nucleoid disorganization, leading to mtDNA damage after oxidative stress (16). DISC1 KD cells displayed a significant depletion of mtDNA that could also account for the decreased steady-state levels of COXI, observed in DISC1 KD cells. This mtDNA encoded subunit depletion could also be caused by impairment of the mtDNA transcription. Yang and co-workers have recently published that Mic60/Mitofilin deficiency impairs mtDNA transcription by interacting with the mitochondrial transcription factors TFAM, TFB2M and TFB1M (25). Mic60/Mitofilin depletion decreased TFAM binding and mitochondrial RNA polymerase recruitment to mtDNA promoters, contributing to downregulation of mtDNA transcription and OXPHOS activities (25). Whether or not hDISC1 depletion exerts its effects on mtDNA transcription modulating Mic60/Mitofilin-TFAM binding will have to be studied in further investigations. It is intriguingly interesting that patients with bipolar disorder have also decreased mtDNA plasma levels (41).
Cristae increase the membrane area available for OXPHOS and play an important role in the organization of ATP synthases dimers, which have been linked to more proficient ATP synthesis (42–45). Destructured MICOS caused by DISC1 knockdown ends up causing OXPHOS disassembly and dysfunction, with the consequent bioenergetics deficit (Fig. 6). Indeed, OXPHOS dysfunction is a common phenotype observed in cells depleted of MICOS subunits (19,25,26,46). In this sense, it is worth to mention that recent studies based on magnetic resonance brain imaging spectrometry strongly suggest that OXPHOS function is also impaired in psychiatric patients (47,48).
Mitochondrial dysfunction has been widely associated with psychiatric disorders (reviewed in (49)) and hDISC1 is one of the few susceptibility genes found till date that reinforces this association. Although hDISC1 is enriched in the mitochondria (5), the relationship of its mitochondrial function with the pathogenesis of the disease is still under debate. Overexpression of truncated hDISC1 isoforms induced abnormal mitochondrial morphologies, indicating changes in mitochondrial dynamics such as fission and fusion (4,6,9). Our hDISC1 depleted cells exhibited an overload of fragmented mitochondria, indicative of excess of fission or loss of fusion. Mitochondrial fission is regulated by Drp1 (dynamin-related protein 1), which is recruited to the mitochondria by anchors such as Fis1 (mitochondrial fission protein 1) and fusion is coordinated by the GTPases Mitofusin1 and 2 at the outer OMM, which tether two mitochondria together, and OPA1 at the IMM (24). OPA1 cooperates with the MICOS subunits Mic60/Mitofilin and Mic19/CHCHD3 to maintain cristae morphology (26), which is crucial to adapt cells to physiological changes. It has recently been demonstrated that MICOS-OPA1 synergy regulates the response to hypoxia by modulating cristae outlets and decreasing the number of ATP synthases dimers (50). Moreover, hDISC1 has been reported previously to inhibit glycogen synthase kinase 3β (GSK3β) (51), which in turn phosphorylates Drp1, increasing its GTPase activity and mitochondrial recruitment, with the resulting trigger of mitochondrial fragmentation (52). Whether DISC1 modulates mitochondrial dynamics through its function at MICOS or through its control on GSK3β-Drp1 will have to be explored in further investigations. However, our data supports a pathogenic role of DISC1 in the mitochondria mediated by its function within MICOS, since the recombinant truncated hDISC1 form identified in the Scottish family does not assemble into MICOS and therefore, fails to rescue the mΔΨ deficit of hDISC1 depleted cells. Investigating the role of hDISC1/MICOS throughout the neurodevelopment will allow deciphering its role with the disease.
Materials and Methods
Cell culture, transfection and infection
HEK293 and SH-SY5Y cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen) containing 10% fetal bovine serum at 37 °C in a 5% CO2 humidified atmosphere. Transfection of cultured cells was conducted using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Twenty-four hour post-transfection, cells were harvested for mitochondria isolation, immunofluorescence, BN-PAGE or WB experiments.
Stable SH-SY5Y cells silenced of hDISC1 were made by infection of lentiviral shRNA particles specifically targeting hDISC1 (a combination of two different shRNAs at an equimolar concentration: TAATCTTTGTTGTAACGTCTC and ATAATCATCAT TC TC AACTGC; SMARTchoice®, Thermo Scientific Dharmacon) followed by growth in selection media (regular media supplemented with 5 μg/ml Puromycin).
Plasmids and constructs
Human hDISC1 myc-tagged at the N-terminal domain was a kind gift of Dr. BK. Lipska (Clinical Brain Disorders Branch, National Institutes of Mental Health, National Institute of Health, Bethesda, MD, USA; for cloning details see (53)). C-terminal V5 full-length human hDISC1 and the Δ597-854 and the Δ1-384 truncated forms were kind gifts of Dr. JK Millar (University of Edinburgh Centre for Genomics and Experimental Medicine, MRC Institute of Genetics and Molecular Medicine, Edinburgh, UK; for cloning details see (6)).
Mitochondrial and mitoplast isolation, alkaline extraction and proteinase K protection assay
All of the procedures were approved by the Animal Care and Use Committee of the Universitat de les Illes Balears (UIB). Mitochondria were purified as described previously (54), with some modifications. Briefly, tissues were excised and homogenized (glass-Teflon Potter-Elvehjem homogenizer) in buffer H (10 mM Tris, pH 7.4, 320 mM sucrose, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM DTT, and 1 mg/ml fatty-acid-free bovine serum albumin (BSA) (Sigma-Aldrich). The homogenate was centrifuged at 1500g for 5 min at 4 °C. The pellet was resuspended in one-half of the original volume of buffer H and centrifuged again under the same conditions. The supernatants were combined and centrifuged at 15 000g for 20 min at 4 °C. The crude mitochondrial pellet was resuspended in 1 ml of buffer H without BSA and then layered on top of a chilled, 7.5/10% (w/v) discontinuous Ficoll gradient (made in buffer H without BSA), using 5 ml for each layer. The samples were centrifuged at 100 000g for 24 min at 4 °C in a Beckman Coulter Optima LE-80K ultracentrifuge. The purified mitochondrial pellet was resuspended in 200 μl of chilled buffer H. Sample aliquots were snap frozen in liquid nitrogen and kept at −80 °C.
To obtain mitoplasts, i.e. mitochondria whose inner membrane had been exposed, the outer membrane was disrupted with 0.11 mg digitonin/mg protein and incubation for 5–10 min on ice. Mitoplasts were collected by centrifugation at 8000 g and suspended in mannitol-HEPES containing 1 mM EDTA and 0.01 mg/ml proteolytic inhibitors: aprotinin, leupeptin, pepstatin A and phenylmethylsulfonylfluoride (PMSF). Final solubilization of this fraction was accomplished with 2% Triton X-100 in mannitol-HEPES buffer and clarification at high speed (100 000 g).
For alkaline extraction, purified whole mitochondria were incubated for 30 min on ice in the presence of 20 volumes of freshly prepared buffer H plus 0.1 M sodium carbonate, pH 11.5, or buffer H plus 0.5% Triton X-100 (Sigma-Aldrich). The samples were then centrifuged at 91 000g for 25 min at 4 °C. The supernatants were precipitated with ice-cold 12% trichloroacetic acid (TCA) and centrifuged at 18 000g for 15 min at 4 °C, followed by an ice-cold acetone wash using the same centrifugation conditions.
For protease protection assay, 25 μg of mouse brain mitochondria or 50 μg of mitochondrial protein from HEK293T transfected cells were treated with 0.2, 0.8, 1, 10, 50 or 100 μg/mL proteinase K for 20 min on ice. To fully solubilize the protein sample, 100 μg/mL proteinase K treatment was combined with 0.1% of Triton X-100 detergent. Then, the proteinase K was inactivated with 100 mM PMSF for 10 min on ice. Alternatively, 60 μg of crude mitochondria extracted from mouse brain were resuspended in a sucrose buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7, 2) and treated with digitonin (0–3%, Sigma‐Aldrich) for 1 min on ice. 30 μg of the permeabilized crude mitochondria were further subject to digestion with 100 μg/ml of Proteinase K for 20 min on ice, in a 50 μl final volume. Proteinase K was then inhibited by addition of 4 mM PMSF. 20 μg of protein sample was loaded to 5–20% Tris-HCl gels for Western blotting. Protein concentration was determined by a colorimetric assay kit (Bio-Rad).
Blue native electrophoresis (BN-PAGE) and SDS-PAGE
Mitochondrial membranes were isolated from 2.5 × 106 cells or from 200 μg of pure mitochondria (PM) as described previously (55). Cells were solubilized with 3% digitonin (wt/vol) (Sigma-Aldrich) and 0.4% (wt/vol) lauryl maltoside (Sigma-Aldrich) or 1% digitonin (wt/vol). PM were solubilized with two rounds of digitonin (6g/g and 4g/g). Twenty μl of samples were electrophoresed on a 5–13% gradient polyacrylamide gel as described previously (55). Transfer of proteins onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) was carried out overnight at 30 V at 4 °C. For second-dimension gel electrophoresis, a lane excised from the first dimension native gel was first treated for 30 min with denaturing buffer containing 1% β-mercaptoethanol and 1% SDS and then washed in 1% SDS for 1 h. The gel strip was electrophoresed on a tricine-SDS-polyacrylamide gel as described previously (56).
For WB analysis, fifty μg of protein were separated by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto PVDF filters (Bio-Rad).
Antibodies
Primary antibodies were used as follows: anti-C-ter hDISC1 (Santa Cruz; 1:2000), anti-mouse N-terminal mdisc1 polyclonal antibody (pAb) (Abbiotec; 1:500), anti-V5 pAb (Invitrogen; 1:500), anti-Mitofilin pAb (Abcam; 1:2000), anti-CHCHD3 pAb (Abcam; 1:1000), anti-CHCHD10 pAb (Sigma-Aldrich; 1:200), anti-CHCHD6 pAb (ProSci; 1:1000), anti-Myc pAb (Abcam; 1:5000), anti-HSP60 pAb (Abcam; 1:20000), anti-EndoG pAb (Abcam; 1:2000), anti-cytochrome c monoclonal antibody (mAb) (Abcam; 1:1000), anti-TOMM20 mAb (Santa Cruz; 1:500), anti-VDAC pAb (Cell Signaling; 1:1000), anti-GAPDH (Abcam; 1:2000), anti-actin pAb (Sigma-Aldrich; 1:200), and the mAb anti-OXPHOS subunits NDUFS9 CI, SDH CII, Core I CIII, COXI CIV, subunit β CV (Thermo; 1:2000). Protein bands were detected using the Odyssey Imaging System (LI-COR). Protein band intensities were evaluated with NIH ImageJ version 1.47 software.
Real-time PCR to quantify mtDNA levels
Quantification of mtDNA relative to nuclear DNA (nDNA) was performed by real-time PCR in a CFX96 Touch Real Time PCR Detection System (BioRad), using 12S rRNA and GAPDH primers, respectively, and the Sso Advanced Universal SYBR Green Supermix Kit (BioRad), according to the manufacturer’s instructions. Relative amounts of mtDNA and nDNA were calculated using linear amplification standard curves of serially diluted DNA samples.
Immunofluorescence microscopy
Cells were grown on coverslips, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 10% goat serum (GS), 1% albumin (BSA). Anti-TOMM20 primary antibody was added for 1 h at 37 °C in phosphate-buffered saline (PBS) containing 10% goat serum, 1% bovine serum albumin and 0.1% Triton X-100. After washing, cells were incubated with cyanine (Cy)3 conjugated secondary antibody (Jackson ImmunoRe search Laboratories), and mounted in immunofluorescence mounting medium (Fluoroshield with DAPI, Sigma-Aldrich). Samples were analyzed using an LSM710 laser scanning confocal microscope (Carl Zeiss Microimaging) equipped with a 63 × /1.25 numerical aperture oil lens. A series of z-sections were taken spanning the thickness of the cell, with intervals between sections set at 0.7 μm. The z-stack images were projected onto a single plane using Zen 2008 LSM 710, V5.0 SP1.1 (Carl Zeiss MicroImaging). To study mitochondrial morphology and distribution, images from at least 20 random fields from three independent experiments were taken and analyzed.
Live cell ΔΨm microscopy and TMRM fluorescence quantification
For live cell imaging, cells were grown on four-well chamber slides (Nalge Nunc International) and monitored using the Zeiss Cell Observer Microscope equipped with the Zeiss incubation modules CO2 Module S, TempModule S and Heating Unit XL S. To measure ΔΨm, cells first were incubated for 1 h in medium containing 1 mM pyruvate as sole carbon energy source, followed by incubation in 10 nM TMRM (Invitrogen) in the same medium for 20 min. Live imaging was performed as described previously (57). The uncoupling agent carbonyl cyanide p-trifluoromethoxyphenylhydrazone (3 mM) was added to the cells to depolarize mitochondria and measure residual, non–ΔΨm-dependent fluorescence, which was subtracted from the initial fluorescence values. All imaging settings were kept constant among all experiments to allow a direct comparison between control and treated cells. At least 100 cells/sample were analyzed in three independent experiments.
Biochemical assays
ATP synthesis was measured in digitonin-permeabilized SH-SY5Y cells by using a luciferin–luciferase method with malate plus pyruvate as substrates, as described previously (58). Oxygen consumption was measured in intact cells using 1 mM pyruvate as substrate, with or without 1 µM of the uncoupler FCCP, and 1.5 mM KCN as the terminal inhibitor, in an Oxygraph chamber equipped with a Clark-type electrode (Hansatech) at 37 °C, as described in (57). For complex-driven respiration we proceeded as described (59), with minor modifications. 5.0 × 106 cells per ml were resuspended in 1 ml of a buffer containing 20 mM HEPES pH 7.1, 250 mM sucrose, 10 mM MgCl2, 1 mM ADP and 2 mM inorganic phosphate. Plasma membranes were permeabilized with digitonine (8 µg per million cells) and oxygen consumption was measured as described above, after addition of complex I substrates glutamate and malate (5mM of each, Sigma‐Aldrich) for the generation of intramitochondrial NADH. Electrons to complex II were provided by succinate added with glycerol-3-phosphate (5mM of each, Sigma‐Aldrich). Complexes I and III were inhibited by addition of 100 nM rotenone (Sigma‐Aldrich) and 20 nM antimycin A (Sigma‐Aldrich), respectively. Complex IV-driven respi ration was then achieved by addition of 5 mM ascorbate and 2 mM TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine, Sigma‐Aldrich); as a direct electron donor to complex IV. Complex IV was inhibited by addition of 1 mM KCN.
Statistical analyses
Data are expressed as mean ± SD. Comparisons between experimental groups were made using unpaired Student’s t-test. Differences were considered statistically significant at P < 0.05. Data analyses were performed using the statistical program IBM SPSS Statistics.
Conflict of Interest statement. None declared.
Funding
This work was supported by grants of the Spanish National Institute of Health, Instituto de Salud Carlos III [CP11/00046, PI15/00809 and MS11/00046 to C.V.B]. E.P.M. and L.R.G. were respectively supported by fellowships from ‘Govern de les Illes Balears’, ‘Conselleria d’Educació, Cultura i Universitats’ and ‘Conselleria d’Innovació, Recerca i Turisme’, cofinanced with European Social Funds. J.P.F. was supported with a fellowship from the Spanish Ministry of Education.
References
- 1.Millar J.K., Wilson-Annan J.C., Anderson S., Christie S., Taylor M.S., Semple C.A., Devon R.S., St Clair D.M., Muir W.J., Blackwood D.H. et al. (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet., 9, 1415–1423. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood D.H., Fordyce A., Walker M.T., St Clair D.M., Porteous D.J., Muir W.J. (2001) Schizophrenia and affective disorders–cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet., 69, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.St Clair D., Blackwood D., Muir W., Carothers A., Walker M., Spowart G., Gosden C., Evans H.J. (1990) Association within a family of a balanced autosomal translocation with major mental illness. Lancet, 336, 13–16. [DOI] [PubMed] [Google Scholar]
- 4.Eykelenboom J.E., Briggs G.J., Bradshaw N.J., Soares D.C., Ogawa F., Christie S., Malavasi E.L., Makedonopoulou P., Mackie S., Malloy M.P. et al. (2012) A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum. Mol. Genet., 21, 3374–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James R., Adams R.R., Christie S., Buchanan S.R., Porteous D.J., Millar J.K. (2004) Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell. Neurosci., 26, 112–122. [DOI] [PubMed] [Google Scholar]
- 6.Millar J.K., James R., Christie S., Porteous D.J. (2005) Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol. Cell. Neurosci., 30, 477–484. [DOI] [PubMed] [Google Scholar]
- 7.Park Y.U., Jeong J., Lee H., Mun J.Y., Kim J.H., Lee J.S., Nguyen M.D., Han S.S., Suh P.G., Park S.K. (2010) Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc. Natl. Acad. Sci. U. S. A., 107, 17785–17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkin T.A., MacAskill A.F., Brandon N.J., Kittler J.T. (2011) Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons. Mol. Psychiatry, 16, 122–121. [DOI] [PubMed] [Google Scholar]
- 9.Norkett R., Modi S., Birsa N., Atkin T.A., Ivankovic D., Pathania M., Trossbach S.V., Korth C., Hirst W.D., Kittler J.T. (2016) DISC1-dependent Regulation of Mitochondrial Dynamics Controls the Morphogenesis of Complex Neuronal Dendrites. J. Biol. Chem., 291, 613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa F., Malavasi E.L., Crummie D.K., Eykelenboom J.E., Soares D.C., Mackie S., Porteous D.J., Millar J.K. (2013) DISC1 complexes with TRAK1 and Miro1 to modulate anterograde axonal mitochondrial trafficking. Human Mol. Genet., 23, 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An J., Shi J., He Q., Lui K., Liu Y., Huang Y., Sheikh M.S. (2012) CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J. Biol. Chem., 287, 7411–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfanner N., van der Laan M., Amati P., Capaldi R.A., Caudy A.A., Chacinska A., Darshi M., Deckers M., Hoppins S., Icho T. et al. (2014) Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol., 204, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Vlies N., Ofman R., Wanders R.J., Vaz F.M. (2007) Submitochondrial localization of 6-N-trimethyllysine dioxygenase - implications for carnitine biosynthesis. FEBS J., 274, 5845–5851. [DOI] [PubMed] [Google Scholar]
- 14.Soares D.C., Carlyle B.C., Bradshaw N.J., Porteous D.J. (2011) DISC1: structure, function, and therapeutic potential for major mental illness. ACS Chem. Neurosci., 2, 609–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fink A.L. (2005) Natively unfolded proteins. Curr. Opin. Struct. Biol., 15, 35–41. [DOI] [PubMed] [Google Scholar]
- 16.Genin E.C., Plutino M., Bannwarth S., Villa E., Cisneros-Barroso E., Roy M., Ortega-Vila B., Fragaki K., Lespinasse F., Pinero-Martos E. et al. (2015) CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol. Med., 8, 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harner M., Korner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W. (2011) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J., 30, 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John G.B., Shang Y., Li L., Renken C., Mannella C.A., Selker J.M., Rangell L., Bennett M.J., Zha J. (2005) The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol.Biol. Cell, 16, 1543–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koob S., Barrera M., Anand R., Reichert A.S. (2015) The non-glycosylated isoform of MIC26 is a constituent of the mammalian MICOS complex and promotes formation of crista junctions. Biochim. Biophys. Acta, 1853, 1551–1563. [DOI] [PubMed] [Google Scholar]
- 20.van der Laan M., Bohnert M., Wiedemann N., Pfanner N. (2012) Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol., 22, 185–192. [DOI] [PubMed] [Google Scholar]
- 21.Weber T.A., Koob S., Heide H., Wittig I., Head B., van der Bliek A., Brandt U., Mittelbronn M., Reichert A.S. (2013) APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PloS One, 8, e63683.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerbes R.M., van der Klei I.J., Veenhuis M., Pfanner N., van der Laan M., Bohnert M. (2012) Mitofilin complexes: conserved organizers of mitochondrial membrane architecture. Biol. Chem., 393, 1247–1261. [DOI] [PubMed] [Google Scholar]
- 23.Alkhaja A.K., Jans D.C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S. et al. (2012) MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell, 23, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wai T., Langer T. (2016) Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab., 27, 105–117. [DOI] [PubMed] [Google Scholar]
- 25.Yang R.F., Sun L.H., Zhang R., Zhang Y., Luo Y.X., Zheng W., Zhang Z.Q., Chen H.Z., Liu D.P. (2015) Suppression of Mic60 compromises mitochondrial transcription and oxidative phosphorylation. Sci. Rep., 5, 7990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darshi M., Mendiola V.L., Mackey M.R., Murphy A.N., Koller A., Perkins G.A., Ellisman M.H., Taylor S.S. (2011) ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem., 286, 2918–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoppins S., Collins S.R., Cassidy-Stone A., Hummel E., Devay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., Nunnari J. (2011) A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol., 195, 323–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie J., Marusich M.F., Souda P., Whitelegge J., Capaldi R.A. (2007) The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett., 581, 3545–3549. [DOI] [PubMed] [Google Scholar]
- 29.Koob S., Reichert A.S. (2014) Novel intracellular functions of apolipoproteins: the ApoO protein family as constituents of the Mitofilin/MINOS complex determines cristae morphology in mitochondria. Biol. Chem., 395, 285–296. [DOI] [PubMed] [Google Scholar]
- 30.Guarani V., McNeill E.M., Paulo J.A., Huttlin E.L., Frohlich F., Gygi S.P., Van Vactor D., Harper J.W. (2015) QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. ELife, 4, doi: 10.7554/eLife.06265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynen M.A., Muhlmeister M., Gotthardt K., Guerrero-Castillo S., Brandt U. (2016) Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim. Biophys. Acta, 1863, 91–101. [DOI] [PubMed] [Google Scholar]
- 32.Odgren P.R., Toukatly G., Bangs P.L., Gilmore R., Fey E.G. (1996) Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J. Cell Sci., 109 (Pt 9), 2253–2264. [DOI] [PubMed] [Google Scholar]
- 33.Gieffers C., Korioth F., Heimann P., Ungermann C., Frey J. (1997) Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp. Cell Res., 232, 395–399. [DOI] [PubMed] [Google Scholar]
- 34.Tusnady G.E., Dobson L., Tompa P. (2015) Disordered regions in transmembrane proteins. Biochim. Biophys. Acta, 1848, 2839–2848. [DOI] [PubMed] [Google Scholar]
- 35.Uversky V.N. (2015) The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett., 589, 2498–2506. [DOI] [PubMed] [Google Scholar]
- 36.Ott C., Dorsch E., Fraunholz M., Straub S., Kozjak-Pavlovic V. (2015) Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PloS One, 10, e0120213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerbes R.M., Hoss P., Pfanner N., van der Laan M., Bohnert M. (2016) Distinct roles of Mic12 and Mic27 in the mitochondrial contact site and cristae organizing system. J. Mol. Biol., 428, 1485–1492. [DOI] [PubMed] [Google Scholar]
- 38.von der Malsburg K., Muller J.M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D. et al. (2011) Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell, 21, 694–707. [DOI] [PubMed] [Google Scholar]
- 39.Ott C., Ross K., Straub S., Thiede B., Gotz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T. et al. (2012) Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol., 32, 1173–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Ruan Y., Zhang K., Jian F., Hu C., Miao L., Gong L., Sun L., Zhang X., Chen S., et al. (2016) Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ., 23, 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang C.C., Jou S.H., Lin T.T., Liu C.S. (2014) Mitochondrial DNA variation and increased oxidative damage in euthymic patients with bipolar disorder. Psychiatry Clin. Neurosci., 68, 551–557. [DOI] [PubMed] [Google Scholar]
- 42.Bornhovd C., Vogel F., Neupert W., Reichert A.S. (2006) Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J. Biol. Chem. 281, 13990–13998. [DOI] [PubMed] [Google Scholar]
- 43.Davies K.M., Strauss M., Daum B., Kief J.H., Osiewacz H.D., Rycovska A., Zickermann V., Kuhlbrandt W. (2011) Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. U. S. A., 108, 14121–14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss M., Hofhaus G., Schroder R.R., Kuhlbrandt W. (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J., 27, 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jimenez L., Laporte D., Duvezin-Caubet S., Courtout F., Sagot I. (2014) Mitochondrial ATP synthases cluster as discrete domains that reorganize with the cellular demand for oxidative phosphorylation. J. Cell Sci., 127, 719–726. [DOI] [PubMed] [Google Scholar]
- 46.Ding C., Wu Z., Huang L., Wang Y., Xue J., Chen S., Deng Z., Wang L., Song Z., Chen S. (2015) Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep., 5, 16064.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi X.F., Kondo D.G., Sung Y.H., Hellem T.L., Fiedler K.K., Jeong E.K., Huber R.S., Renshaw P.F. (2012) Frontal lobe bioenergetic metabolism in depressed adolescents with bipolar disorder: a phosphorus-31 magnetic resonance spectroscopy study. Bipolar Disord., 14, 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goh S., Dong Z., Zhang Y., DiMauro S., Peterson B.S. (2014) Mitochondrial dysfunction as a neurobiological subtype of autism spectrum disorder: evidence from brain imaging. JAMA Psychiatry, 71, 665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clay H.B., Sillivan S., Konradi C. (2011) Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int. J. Dev. Neurosci., 29, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plecita-Hlavata L., Engstova H., Alan L., Spacek T., Dlaskova A., Smolkova K., Spackova J., Tauber J., Stradalova V., Malinsky J. et al. (2016) Hypoxic HepG2 cell adaptation decreases ATP synthase dimers and ATP production in inflated cristae by mitofilin down-regulation concomitant to MICOS clustering. FASEB J., 30, 1941–1957. [DOI] [PubMed] [Google Scholar]
- 51.Mao Y., Ge X., Frank C.L., Madison J.M., Koehler A.N., Doud M.K., Tassa C., Berry E.M., Soda T., Singh K.K., et al. (2009) Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell, 136, 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan J., Liu X.H., Han M.Z., Wang Y.M., Sun X.L., Yu N., Li T., Su B., Chen Z.Y. (2015) Blockage of GSK3beta-mediated Drp1 phosphorylation provides neuroprotection in neuronal and mouse models of Alzheimer’s disease. Neurobiol. Aging, 36, 211–227. [DOI] [PubMed] [Google Scholar]
- 53.Newburn E.N., Hyde T.M., Ye T., Morita Y., Weinberger D.R., Kleinman J.E., Lipska B.K. (2011) Interactions of human truncated DISC1 proteins: implications for schizophrenia. Transl. Psychiatry, 1, e30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pallotti F., Lenaz G. (2001) Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol., 65, 1–35. [DOI] [PubMed] [Google Scholar]
- 55.Nijtmans L.G., Henderson N.S., Holt I.J. (2002) Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods, 26, 327–334. [DOI] [PubMed] [Google Scholar]
- 56.Brookes P.S., Pinner A., Ramachandran A., Coward L., Barnes S., Kim H., Darley-Usmar V.M. (2002) High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics, 2, 969–977. [DOI] [PubMed] [Google Scholar]
- 57.Vives-Bauza C., Anand M., Shiraz A.K., Magrane J., Gao J., Vollmer-Snarr H.R., Manfredi G., Finnemann S.C. (2008) The age lipid A2E and mitochondrial dysfunction synergistically impair phagocytosis by retinal pigment epithelial cells. J. Biol. Chem., 283, 24770–24780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vives-Bauza C., Yang L., Manfredi G. (2007) Assay of mitochondrial ATP synthesis in animal cells and tissues. Methods Cell Biol., 80, 155–171. [DOI] [PubMed] [Google Scholar]
- 59.Hofhaus G., Attardi G. (1993) Lack of assembly of mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase and loss of enzyme activity in a human cell mutant lacking the mitochondrial ND4 gene product. EMBO J., 12, 3043–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]