Abstract
Inactivating mutations of the TSC1/TSC2 complex (TSC1/2) cause tuberous sclerosis (TSC), a hereditary syndrome with neurological symptoms and benign hamartoma tumours in the brain. Since TSC effectors are largely unknown in the human brain, TSC patient cortical tubers were used to uncover hyperphosphorylation unique to TSC primary astrocytes, the cell type affected in the brain. We found abnormal hyperphosphorylation of catenin delta-1 S268, which was reversible by mTOR-specific inhibitors. In contrast, in three metastatic astrocytoma cell lines, S268 was under phosphorylated, suggesting S268 phosphorylation controls metastasis. TSC astrocytes appeared epithelial (i.e. tightly adherent, less motile, and epithelial (E)-cadherin positive), whereas wild-type astrocytes were mesenchymal (i.e. E-cadherin negative and highly motile). Despite their epithelial phenotype, TSC astrocytes outgrew contact inhibition, and monolayers sporadically generated tuberous foci, a phenotype blocked by the mTOR inhibitor, Torin1. Also, mTOR-regulated phosphokinase C epsilon (PKCe) activity induced phosphorylation of catenin delta-1 S268, which in turn mediated cell-cell adhesion in astrocytes. The mTOR-dependent, epithelial phenotype of TSC astrocytes suggests TSC1/2 and mTOR tune the phosphorylation level of catenin delta-1 by controlling PKCe activity, thereby regulating the mesenchymal-epithelial-transition (MET). Thus, some forms of TSC could be treated with PKCe inhibitors, while metastasis of astrocytomas might be blocked by PKCe stimulators.
Introduction
Inactivating mutations of the tuberous sclerosis complex 1 and 2 (TSC1/2) cause an autosomal dominant hereditary tumour syndrome known as Tuberous Sclerosis Complex (TSC), which exhibits neurological symptoms and benign hamartoma in multiple tissues, including the brain, kidneys, skin, heart and lungs (1,2). In contrast to metastatic tumours, TSC hamartoma tumours are clinically benign, suggesting that elucidating TSC pathogenesis could unlock a set of fundamental mechanisms that underlie the transition from a benign to a metastatic lesion.
Mechanistically, the TSC1/2 complex accelerates the ras homolog enriched brain (Rheb) GTPase, which in turn downregulates mechanistic-target-of-rapamycin (mTOR) signalling (3). In astrocytes of wild-type mice, Rheb is predominantly GDP-bound and mTOR signalling is inactive (4,5). Constitutive stimulation of mTOR via Rheb likely has downstream effects on protein phosphorylation that should be evident in the TSC phosphoproteome. Most relevant proteomic studies, however, were conducted in fibroblasts from rodent models (6,7). In primary astrocytes from human patients, the TSC phosphoproteome was rarely examined. Thus, it is unclear whether the rodent studies are relevant to human TSC astrocytic tumours.
For patients with TSC, the recent FDA approval of a highly effective anti-epileptic drug (Vigabatrin) now makes removal of brain hamartomas rarely necessary. We nevertheless were able to obtain cortical tubers that were surgically removed from the brains of patients with TSC with a definitive diagnosis. These tissue samples are very valuable for the study of the human TSC hamartoma formation and analysis. We cultured primary astrocyte lines and profiled the phosphoproteome using stable isotope labelling by amino acids in cell culture (SILAC). Thus, we identified canonical signal transduction pathways affected in TSC brains and Torin1/rapamycin–sensitive events that implicate new downstream targets of mTOR in metastasis.
Results
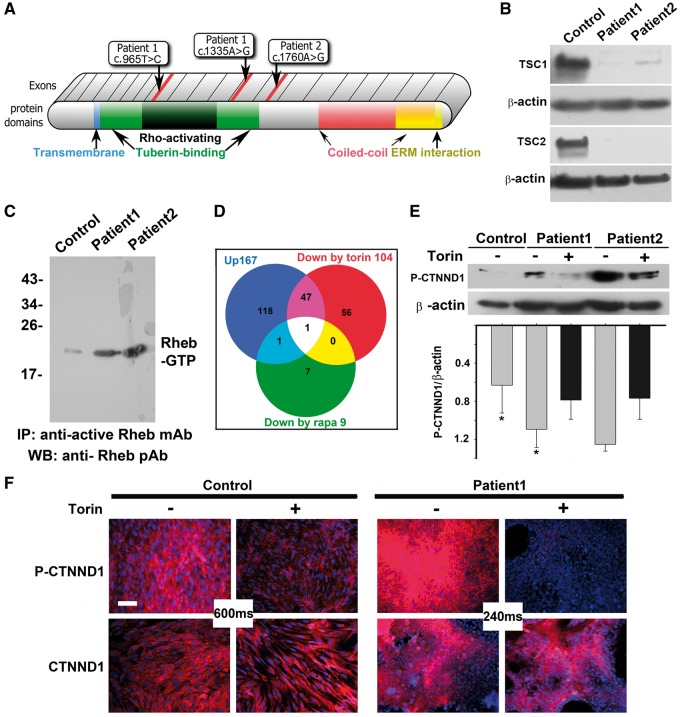
Genomic DNA was isolated from the cortical tubers, and the TSC1 and TSC2 genes were sequenced, identifying the disease mutations (Fig. 1A). Whole exome sequencing was performed by an external CLIA-certified lab, and no other clinically relevant mutations were identified. Astrocytic cell lines were derived from the tumours, and immunoblot analysis showed loss of TSC1 or TSC2 protein, in addition to confirming that the cell lines contained dramatically elevated levels of active, GTP-bound Rheb (Fig. 1B and C). In contrast, control astrocytes from a disease-free donor brain had a basal level of Rheb activation. Thus, we confirmed previous reports showing astrocytes derived from TSC cortical tubers are hyperactive for mTOR1/2 signalling (8).
Figure 1.
mTOR inhibitors block hyperphosphorylation of CTNND1 serine 268 in TSC patient astrocytes. (A) Diagram of TSC1 protein, showing known motifs and the location of patient mutations. Patient 1 had compound, heterozygous base substitutions in both TSC1 alleles: c.965T > C in the Rho-activating domain of one allele, and C.1335A > G in the tuberin-binding domain of the other allele. Patient 2 exhibited a single c.1760A > G mutation in one of their TSC1 alleles. No deletions were found in microarray analyses. (B) Immunoblot analysis of astrocytic cell lines showed loss of TSC1 or TSC2 protein. Control human astrocytes were from a commercial source (ScienCell Research Laboratories #1800, Carlsbad, CA). (C) Active GTP-bound Rheb is upregulated in TSC patient astrocytes. Lysates from control human astrocytes and patient astrocytes, cultured to 80 to 90% confluence, were immunoprecipitated with an antibody that specifically recognizes GTP-bound Rheb. Immunoprecipitated protein was compared via immunoblot. (D) Stable Isotope Labelling by Amino acids in Cell culture (SILAC) in the absence and presence of Torin1 or rapamycin identified hyperphosphorylation of catenin delta-1 (CTNND1) alone as driving TSC1 mTOR-opathy. Venn diagram depicts proteins hyperphosphorylated in TSC patient astrocyte cultures grown in the presence of vehicle (blue circle; 167 phospo-peptides), rapamycin (green circle; 9 phospo-peptides) or Torin1 (red circle; 104 phospo-peptides). In lysates from TSC astrocytes, both mTOR inhibitors prevented hyperphosphorylation of the mTOR pathway component, CTNND1, on serine 268. (E,F) CTNND1 serine 268 is hyperphosphorylated via the overactive mTOR pathway in TSC patient astrocytes. Confluent cultures of control and patient astrocytes were incubated for 1 h in the absence or presence of the mTOR inhibitor, Torin1. E,Immunoblots of lysates derived from two patient cell lines (normalized to beta actin expression) show hyperphosphorylation of CTNND1 S268 was blocked by Torin1. Band intensity was measured via densitometry and normalized to β-actin levels. The relative value of normal was 0.52 ±0.24, whereas patient values were 1.17 ± 0.3131 (n = 3 immunoblots; *P = 0.048). F, Immunostaining for CTNND1 P-S268 shows hyperphosphorylation (but not expression) of CTNND1 in TSC patient astrocytes is downregulated by Torin1. Image acquisition times were dramatically reduced for mutant cultures (240 ms vs. 600 ms). Scale bar, 100 µm.
To uncover how constitutive mTOR stimulation changes protein phosphorylation, we analysed the TSC astrocyte phosphoproteome using SILAC. Patient-derived astrocytes were compared to normal astrocytes and showed hyperphosphorylation of 167 proteins by at least two-fold (Fig. 1D). After determining the timing and dose of rapamycin and Torin1 which would induce the most divergent kinomic change (Supplementary Material, Figs S2 and S3), each was administered and observed to inhibit phosphorylation of 48 and 2 of the SILAC-identified proteins, respectively, (Fig. 1D). Remarkably, of the 167 proteins hyperphosphrylated in our TSC sample, only one phosphorylation event was inhibited by both Torin1 and rapamycin—phosphorylation of catenin delta-1 at S268 (CTNND1 P-S268).
In cultured, primary TSC astrocytes, immunoblots confirmed that Torin1 suppressed CTNND1 S268 hyperphosphorylation (Fig. 1E). In the absence of Torin1, TSC cells immunolabelled for CTNND1 P-S268 emitted a signal that was so intense that image-acquisition times needed to be reduced to compare TSC to WT (240 milliseconds for the TSC samples vs. 600 milliseconds for the WT; Fig. 1F). As positive controls, we also immunolabelled astrocytes for phosphorylation of two well-known mTOR downstream targets: ribosomal S6 protein kinase p70 (S6) and eukaryotic translation-initiation factor 4E-binding protein (4EBP1; an effector of mTOR) (9). Like CTNND1 S268, S6 and 4EBP1 phosphorylation was upregulated and suppressible by Torin1 (Supplementary Material, Figs S1). In summary, these SILAC, pharmacological, biochemical and immunolabelling data identify CTNND1 as the mTOR effector specifically hyperphosphorylated in TSC astrocytes.
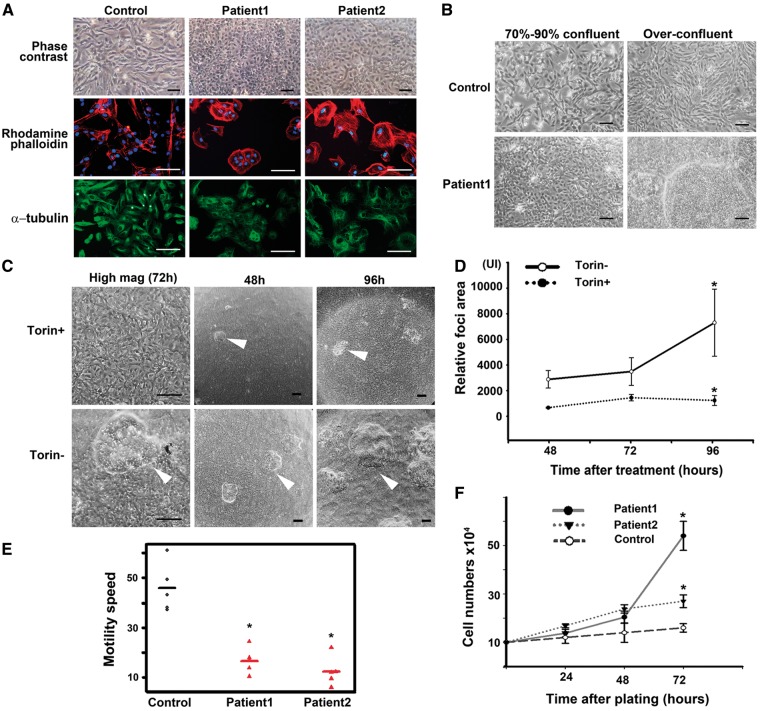
CTNND1 S268 phosphorylation controls homodimerization of E-cadherin extracellular domains on abutting cells, allowing cells to bind tightly to one another (10). Epithelial cells are rich in CTNND1 P-S268 and E-cadherin, and so tightly adhere to one another, assuming a cuboidal, cobblestone morphology in culture (11). Consistent with S268 hyperphosphorylation, TSC astrocytes appeared cuboidal under phase contrast microscopy, and immunostaining for actin and tubulin confirmed an epithelial morphology. In contrast, control astrocytes had a mesenchymal morphology (Fig. 2A). When cultured astrocytes were grown beyond confluency, TSC monolayers gave rise to tuber-like foci that grew vertically. Such foci never grew from control astrocyte monolayers, at any density (see Fig. 2B). Torin1 blocked both the tuber-like foci and epithelial fate of TSC astrocytes (Fig. 2C and D), suggesting both were under mTOR control.
Figure 2.
Cultured TSC astrocytes assume an adherent, mitotic phenotype that is mTOR-dependent. (A) TSC patient astrocytes were cultured and cell morphology visualized via phase contrast, rhodamine-phalloidin (β-actin marker, red), or α-tubulin immunolabelling (green). Images were merged with DAPI (blue) to show nuclei. Scale bars, 100 µm. (B) Control and TSC astrocytes were cultured to confluency or over-confluency and imaged with phase contrast. Scale bar, 100 µm. (C,D) Epithelial morphology and tuberous foci of TSC1 astrocytes are inhibited by the mTOR inhibitor, Torin1. Cells were seeded in 35-mm wells (∼100 x 104 cells/well), cultured to confluence, and grown in the presence or absence of 250 nM Torin1 for 48, 72 and 96 h. (C) Cells exposed to Torin1 assume a mesenchymal morphology. Scale bar, 100 µm. (D) At terminal time points, the area occupied by foci was quantified (3 areas/well, 3 wells). After 96 h, the mean sizes of foci were 873.33 ± 429 pixels in the presence of Torin1, and 6993.25 ± 2993 in the absence of Torin1 (n = 9 areas). *P = 0.026. Scale bar, 100 µm. (E) TSC astrocytes are less motile in an in vitro scratch-wound assay. A confluent monolayer of TSC astrocytes was ‘scratched’ to introduce an area devoid of cells. Cultures were imaged to measure cell migration into the void. Five cells of each sample were tracked to estimate the mean speed for each. The mean difference between patients and normal was 15.7 h (SE = 2). P <0.0001. (F) Proliferation of TSC astrocytes is resistant to contact inhibition. Cells were counted at 24, 48 and 72 h after plating. The mean difference in cell number between patients and control is 13.1 × 104 (SE = 2.6) at 72 h. The P-value is 0.002.
Epithelial cells are characterized by limited motility, so we tested the TSC patient astrocytes in an in vitro scratch-motility assay (Fig. 2E), in which cells were tested for traversal of a ‘wound’ scratched into a confluent monolayer. As would be expected of epithelialized cells, TSC astrocytes were less motile than control astrocytes. Although TSC primary astrocytes proliferated faster than controls (Fig. 2F), they did not migrate into a scratched void (Fig. 2E). Thus, the TSC astrocytes comprising the tuber-foci appeared trapped in the late MET: despite showing hallmarks of epithelialized cells, these astrocytes did not respond to contact inhibition and continued to divide (Fig. 2F).
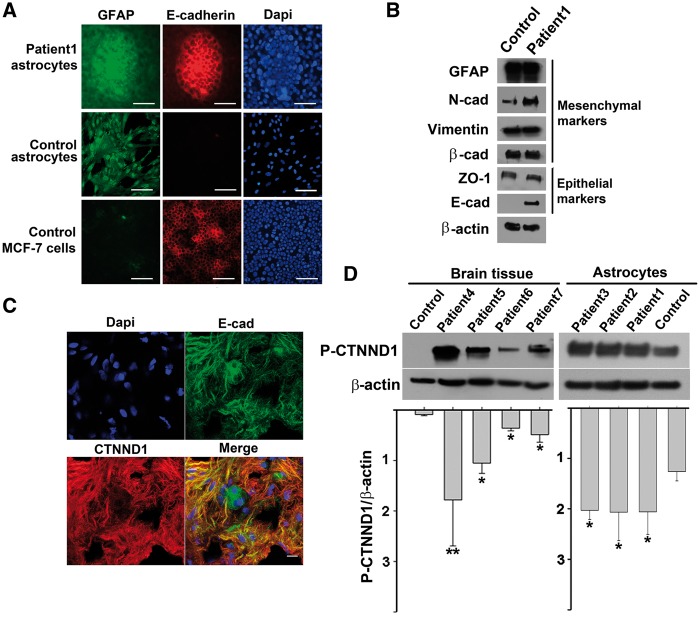
CTNND1 binds E-cadherin at points of adhesion between epithelial cells (12) and less-adherent, mesenchyme-like cells are E-cadherin negative (13). Accordingly, control astrocytes did not express detectable levels of E-cadherin (Fig. 3A and B). In contrast, both the foci from over-confluent TSC astrocytes and MCF-7 breast cancer cells (which also have hyperstimulated mTOR signalling (14)) robustly expressed E-cadherin.
Figure 3.
TSC astrocytes express CTNND1 P-S268, E-cadherin, and markers of both epithelial and mesenchymal cells A, B, TSC astrocyte foci and epithelial MCF-7 breast cancer cells (but not control astrocytes) express the epithelial marker, E-cadherin, whereas TSC and control astrocytes (but not MCF-7 cells) express the mesenchymal marker GFAP. (A) Patient TSC astrocytes and control cells (normal astrocytes and epithelial MCF-7 breast cancer cells) were cultured beyond confluency, and triple labelled: GFAP immunolabelling (green), E-cadherin immunolabelling (red) and DAPI staining (blue). Scale bar, 100 µm. (B) Two days after reaching confluency, control and patient cells were harvested and lysates were immunoblotted for mesenchymal and epithelial markers. β-actin was used as a loading control. (C) CTNND1 P-S268 and E-cadherin are localized to the membrane of astrocyte processes in a TSC surgical specimen. Merged image shows staining for CTNND1 P-S268 (red), E-cadherin (green), and DAPI (blue) overlayed on the same image. Scale bar, 10 µm. (D) CTNND1 is hyperphosphorylated in surgical specimens from four additional TSC brains and a third TSC cell line. Patient-3, TSC2 c.488T > C. p.F163S; Patient-4, TSC1 c.901C > T, p.Q301X; Patient-5, TSC1, c.1525C > T, p.R509Ter; Patient-6, c.1821_1834del14bp.; and Patient-7, TSC2, c.2251C > T, p.Arg751Ter. TSC astrocyte line with higher cellular homogeneity also showed higher levels of CTNND1 P-S268 than surgical specimens. (* = P < 0.05; ** = P < 0.005).
We characterized the expression of several other markers of the epithelial vs. mesenchymal fate. When immunolabelling for GFAP (a mesenchymal marker), TSC astrocyte cultures and their foci stained brightly (Fig. 3A), indicating that epithelial-like TSC astrocytes retained mesenchymal qualities. Additionally, imunnoblotting showed lysates of over-confluent TSC astrocytes (i.e. foci-containing) expressed mesenchymal markers (β-cadherin, N-cadherin and vimentin (15–17)) and epithelial markers (E-cadherin, and ZO-1; Fig. 3B). In surgical specimens, TSC astrocytic processes also expressed both E-cadherin and high levels of CTNND1 P-S268 (Fig. 3C).
In addition to the discovery cohort, we verified CTNND1 S268 hyperphosphorylation in five additional patients with TSC, whose ages varied from 1 to 60 years old, sampling lysates of naïve surgical specimens, astrocytomas, or cultured primary astrocytes (Fig. 3D). Although the level of S268 phosphorylation was more uniform in cultured TSC astrocytes compared to surgical specimens, hyperphosphorylation was a feature of both. In surgical specimens, both CTNND1 P-S268 and E-cadherin co-localized to the membrane of astrocytic processes (Fig. 3C). CTNND1 S268 was also hyperphosphorylated in TSC brain and astrocyte specimens from the other patients (Fig. 3D).
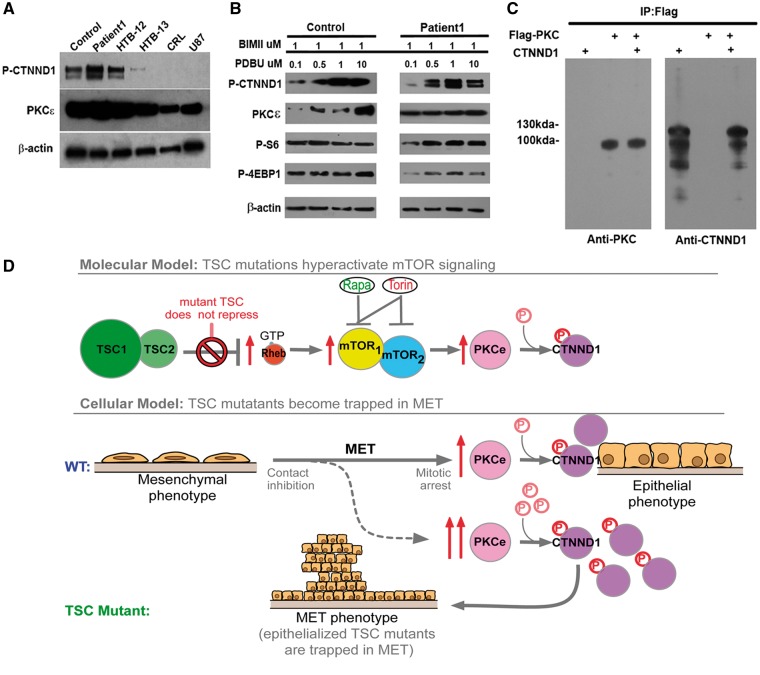
In astrocytes, an upstream kinase for CTNND1 is protein kinase C epsilon (PKCe). PKCe is also an mTOR substrate that regulates the epithelial-mesenchymal transition (EMT) and its reverse process, the mesenchyme to epithelial transition (MET) (18,19). We found levels of CTNND1 S268 phosphorylation and PKCe (18,19) activity were inversely correlated with metastatic potential, in that highly metastatic astrocytoma cell lines contained less PKCe and less CTNND1 P-S268 (Fig. 4A). In contrast to malignant astrocytomas, TSC surgical samples contained hyperphosphorylated CTNND1 S268 and more active PKCe (Fig. 4A).
Figure 4.
PKCe drives hyperphosphorylation of CTNND1 P-S268 and traps TSC astrocytes in the MET. (A) CTNND1 P-S268 and PKCe levels were higher in TSC patient samples than in malignant astrocytomas. HTB-12, -13, CRL-1718 and U87 are established malignant astrocytoma lines carrying mutations in tumour suppressor genes: PTEN, homozygous deletion (c.165_1212del1048) in HTB-12; c.209 + 1G > T in HTB-13; c.335T > G in CRL-1718; c.209 + 1G > T in U87. Cyclin-dependent kinase inhibitor 2A (CDKN2A) mutations: homozygote c.1_471del471 in HTB-12. TP53 homozygote c.817C > T in HTB-12; RB1 mutations: homozygote c.817C > T in HTB-12; compound heterozygote c.817C >T and c818G > A in HTB-13. (B) CTNND1 S268 is fully phosphorylated in patient but not control astrocytes. Control and TSC1-patient astrocyte lines were plated in 35-mm wells (100 x 104 cells/well) and grown to confluency (i.e. for 2 days); then the PKC inhibitor BIMII was added to eliminate basal activity of all PKC isoforms. PKCe was then specifically activated in a does-dependent manner, with the PKCe agonist, PDBU. (C) Co-immunoprecipitation of PKCe and CTNND1. HEK293 cells were transfected with FLAG-tagged PKCe and CTNND1. 24 hr after transfection, whole-cell extracts were immunoprecipitated with FLAG antibodies, and immunoblotted for PKCe and CTNND1. IgG antibody was used as a negative control for immunoprecipitation and 25–50μg of whole-cell protein lysate was used as input. (D) Inactivating mutations of the TSC complex epithelialize astrocytes, and make them prone to becoming trapped in the late mesenchymal-epithelial transition. In TSC astrocytes, constitutive mTOR signaling drives PKCe to fully phosphorylate CTNND1 and causes an epithelial phenotype. Such cells can get trapped in the late MET and continue to proliferate, despite their otherwise epithelialized phenotype.
CTNND1 was tracked by an antibody specific for phospho-S268, showing it was phosphorylated in a strictly PKCe-dependent manner (Figs 1E and F, 3C and D, 4A and B). To test whether levels of S268 phosphorylation were directly proportional to PKC activity, we used the PKC agonist, phorbol-12,13-dibutyrate (PDBu) (Fig. 4B). In cells where the basal activities of all PKC isoforms were blocked by BIMII, administration of PDBu increased S268 phosphorylation in a dose-dependent manner in wild-type control astrocytes (Fig. 4B). In contrast, CTNND1 phosphorylation in TSC astrocytes was unaffected, suggesting both S268 and PKCe were already in a fully activated state (Fig. 4B) as a result of mTOR activation due to loss of TSC function. Meanwhile, other mTOR targets (i.e. S6 and 4EBP1) were not affected by PDBu in either astrocyte line, suggesting they act independently of the mTOR-PKCe signalling pathway. CTNND1 and PKCe also co-immunoprecipitated (Fig. 4C), which strongly suggests PKCe directly controlled the phosphorylation status of CTNND1 P-S268 in TSC astrocytes (18). Thus, we identified both the substrate uniquely phosphorylated in TSC astrocytes and the upstream kinase regulating the level of this phosphorylation (Fig. 4C).
Discussion
In this study, a SILAC analysis of rare, primary TSC astrocytes found they contain abnormally hyperphosphorylated CTNND-1 S268. Accordingly, they were less motile, consistent with CTNNSD1 PS268 and tightly adherent cells. In contrast, metastatic cancer cells were underphoshporylated on CTNND-1 S268 (Fig. 4A), suggesting pCTN augmented the MET in naïve TSC patient surgical specimens, and decreased pCTN allowed malignant astrocytomas (i.e. HTB-13. CRL-1718, UB7) to traverse from MET to EMT. In cancer, reduced CTNND-1 S268 expression correlates with reduced E-cadherin function with tumour progression (20), while increased CTNND-1 S268 may reduce tumour’s progression. Therefore, we expected that the non-metastatic character of hamartomas from brain samples could be related to the hyperphosphorylation of CTNND-1 S268.
Previous studies showed that elevated mTORC1 and mTORC2 activity played a role in regulating the epithelial-mesenchymal transition (EMT), motility and metastasis of colorectal cancer (21). It has also been shown that CTNND1 P-S268 is an E-cadherin-associated protein that modulates E-cadherin function and stability (20). In our study, the expression of E-cadherin was detected in patients’ samples when the ‘tubular foci’ was formed, whereas TSC astrocytes did not overtly express E-cadherin before the ‘foci’ formation. Moreover, mTOR inhibitors appeared capable of blocking the ‘foci’ in this study. Hence, CTNND1 S268 phosphorylation and E-cadherin expression may be controlled by the mTOR pathway in TSC hamartoma. Elevated mTORC1, mTORC2 activity in patients with TSC with phosphorylation of CTNND1 S268 may therefore play a role in regulating MET.
Our data imply that the TSC/mTOR complex controls cellular traversal through the MET and EMT in human astrocytes. Cultured TSC astrocytes showed epithelial-like qualities, such as low motility, tight cell–cell connections and cuboidal morphology. Over confluent TSC astrocyte monolayers were resistant to contact inhibition and generated foci that continued to grow vertically. This phenotype—epithelialized astrocytes that continue to divide—indicates that the cells may have been trapped in late MET (Fig. 4D). mTOR inhibitors, specifically Torin1, blocked this phenotype (Fig. 2C and D), allowing us to isolate a molecular event that may be responsible for traversal through the late MET: the phosphorylation of CTNND1 S268.
Hyper-phosphorylated CTNND1 S268 was a feature of low-motility TSC astrocytes. In contrast, under-phosphorylated CTNND1 S268 was a feature of highly motile, metastatic cancer cells from malignant astrocytomas (i.e. HTB-13. CRL-1718, UB7; Fig. 4A). These data suggest CTNND1 S268 phosphorylation may be a feature of TSC surgical specimens, which consist of epithelialized, self-adhering, proliferative cells. Since PKCe co-immunopreciptated with CTNND1 in TSC astrocytes, the mTOR pathway may regulate S268 phosphorylation (and cell–cell adhesion) by controlling PKCe (Fig. 4C). Phospho-proteome analyses of TSC surgical specimens allowed mapping a pathway through which TSC mutations trigger growth of astrocytic cells in hamartoma (Fig. 2A).
An upstream kinase for CTNND1 is Protein Kinase C epsilon (PKCe), which has been extensively researched in relation to breast cancer (18). PKCe is also an mTOR substrate that regulates the EMT and the reverse process, the MET (18,19). Co-immunoprecipitation of PKCe and CTNND-1 in this study suggests that PKCe is plausibly the mTOR-sensitive kinase controlling CTNND1-S268 phosphorylation (Fig. 4C). Through these results, we have mapped a potential pathway that demonstrates how TSC mutations may trigger growth of astrocytic hammartoma tumours (Fig. 4D). Moreover, since PKCe is among the most important classes of pharmaceutical drug targets today, using already available compounds to target CTNND-1 signalling provides an alternative pharmacological target for the treatment of TSC and mTOR-opathies.
Materials and Methods
Patients with TSC
Patient 1, the proband, was a 2-year-old girl with a history of tuberous sclerosis who had failed multiple pharmacological seizure therapies. Resection of residual epileptogenic frontal and parieto-occipital tubers and peri-tuberal tissue was preformed to control seizures. Pathologist reported seeing enlarged astrocytes with a balloon-like cytoarchitecture that stained for glial fibrillary acidic protein (GFAP). GFAP staining also revealed abundant, thick, glial fibers throughout the specimen and a marked subpial gliosis. Patient 2 was a 13-year-old boy with a history of infantile spams and focal epilepsy related to tuberous sclerosis. Enlarged cells with a balloon-like cytoarchietcture were also seen, some of which showed basophilic material around the periphery of the cell. A stain for GFAP revealed abundant, thick, glial fibers throughout the specimen and a marked subpial gliosis. During surgery, astrocytic hamartoma tissue was sampled to establish TSC cell lines from these two patients. For both patients, whole exome sequencing was performed and identified two disruptive variants in the EYS gene. No other variants of known clinical significance were identified. All procedures adhered to the tenets of the Declaration of Helsinki, and written informed consent was obtained from all subjects after a full explanation of the procedures was provided. The protocol was approved by the Institutional Review Boards of University of Iowa (ID#200112047) and Columbia University (ID#AAAB6560).
Tissue, cells and cultures
TSC tissue and cell lines
Astrocytic hamartoma tissue isolated from patient surgical specimens was cut into 0.5mm thick slices and digested with 2 mg/ml papain for 30 min at 30°C (in Hibernate A solution, BrainBits LLC) without calcium, followed by gentle trituration. Cell suspensions settled by gravity for 2 min, to partition large debris, and the supernatant was collected. The sediment from the first tube was resuspended in 2 ml medium, and the procedure repeated twice or more. Cells were centrifuged at 1000g for 5 to 8 min. Cell pellets were gently resuspended in astrocyte media supplemented with 2% B27 (Gibco, Grand Island, NY), 500 μM l-glutamine, 10 units/ml penicillin, and 10 μg/ml streptomycin, and the suspension was dispensed into poly-D-lysine-coated plates at 1 × 105 cells/cm2, and incubated at 37°C with 5% CO2. Eighteen to twenty-four hours after plating, the media was changed. On subsequent days, half of the culture volume was replaced. Cells were routinely used at 12–14 days after plating,
Control tissue and cell lines. Wild-type human astrocytes cells were purchased from a commercial source (ScienCell Research Laboratories, Carlsbad, CA). All malignant astrocytoma cell lines HTB-12, HTB-13, CRL-1718, U87 and the MCF-7 epithelial cell line were also from a commercial source (Coriell Institute for Medical Research Camden, NJ. USA). The hamartoma TSC samples originated from the Universities of Washington, Iowa and Texas.
DNA sequencing
Next generation sequencing (NGS) of TSC surgical specimens’ determined TSC gene variants. Mutants chosen for further analysis met three criteria: i) present at < 0.5% in ESP6500 (http://eversusgs.washington.edu/EVS/) and 1000 Genomes; ii) present in protein-coding regions and/or canonical-splice sites; iii) identification as missense, nonsense, frameshift, or splice-site.
Rheb assay
Lysates were prepared from astrocytes cultured to 80%-90% confluence. GTP-bound, active Rhe was pulled down by protein A/G agarose and the then detected by immunoblot using anti- Rheb rabbit polyclonal antibody (Rheb Activation Assay Kit ab173243).
Pharmacological treatments
Cells were serum-deprived for 17 h before any pharmacological treatment. Rapamycin, Torin1, PDBU and bisindolylmaleimide (BIMII) were dissolved in DMSO. For SILAC 20nM rapamycin was treated on patient1 and control. Torin1 treatment: Various time points and dosages were all tried in Torin1 group. 250nm and 30 min Torin1 treatment were chosen for SILAC test. 1 um BIMII pretreated on the control and patient 1 cell line half an hour, then treated 0.1 to 20um PDBu for PKCe hyperphosphorylation analysis.
Cell culture for SILAC
Mass spectrometry data were obtained using the SILAC method on diseased or wild-type cells grown in light and heavy DMEM ([12C614N2] L-Lysine-HCl, [12C614N4]L-Arginine-HCl and [13C615N2]L-Lysine-HCl, [13C615N4] L-Arginine-HCl, respectively (Thermo Fisher Scientific, USA), for more than five passages (22,23). All DMEM was supplemented with 10% dialyzed FBS (Thermo Fisher Scientific, USA,). The log base 2 ratio between patient and normal was taken.
Sample preparation for mass spectrometric analysis
Cells for SILAC test were lysed in urea buffer (8 M urea, 20 mM HEPES pH 7.0, 75 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 mM DTT and 1.5 mM EGTA) and the lysates were combined at a 1:1 ratio. Lysates were reduced by adding DTT (3 mM final concentration) and incubating them at room temperature for 20 min.
Mass spectrometry analysis and data processing
FASP (filter-aided sample preparation)
To identify phospho-peptides enriched in TSC patient astrocytes (compared to control), 200 µg of protein lysate from DMSO-treated ‘light’ (Lys0/Arg0) patient-1 astrocytes was mixed with 200 µg of protein lysate from DMSO-treated ‘heavy’ (Lys8/Arg10) control astrocytes. To identify phospho-peptides that are less abundant in the TSC samples after rapamycin treatment, 200 µg of cell lysate from DMSO-treated heavy (Lys8/Arg10) patient-1 astrocytes was mixed with rapamycin-treated light (Lys0/Arg0) patient-1 astrocytes. To identify phospho-peptides that are less abundant in the patient samples after Torin1 treatment, 200 µg of protein in the lysate from DMSO-treated light (Lys0/Arg0) patient astrocytes was mixed with 200 µg protein from Torin1-treated heavy (Lys8/Arg10) patient astrocyte lysates. For each of these three combinations, the resulting 400 µg proteins were digested with a modified FASP procedure (24) Briefly, cysteine residues were reduced with dithiothreitol and alkylated with 2-iodoacetamide; samples were centrifuged on a 30 kDa cut-off filter (PALL), which was washed thoroughly, and retained proteins digested with Lys-C (Wako) and trypsin (Promega). The resulting peptides were eluted using 50 mM ammoniumbicarbonate/2% acetonitrile (ACN) and concentrated to 10 µg/µl, in a total of 40 µl.
Enrichment of Phospho-peptides using TiO2 matrix
Phospho-peptides in digested cell lysates were enriched using TiO2 beads (GL Science, Titansphere 5µm bulk). 2.5 mg of beads were resuspended in water, washed once with ACN and then with 60% ACN/4% trifluoroacetic acid (TFA). Peptides from eluted FASP digests were diluted 5-fold with 80% ACN/5% TFA and added to the prepared TiO2 beads. After a 2 h incubation, shaking, at room temperature, beads were centrifuged and washed three times with 50% ACN/0.1% TFA. Bound phospho-peptides were eluted in two steps with 10% ammonium hydroxide. Formic acid was added to pH 2 for subsequent direct LC-MSMS.
Mass spectrometric measurements
Samples were analysed online by LC-MSMS on an Ultimate3000 nano RSLC system (Dionex, Sunnyvale, CA) coupled to a LTQ OrbitrapXL mass spectrometer (Thermo Fisher Scientific) with a nano spray ion source, as described previously (Hauck et al., 2010; Merl et al., 2012). Samples were automatically injected and loaded onto a trap column (100µm inner diameter x 2 cm, packed with Acclaim PepMap100 C18, 5 µm, 100 Å, Dionex) at a flow rate of 30 l/min in 5% buffer B (75% ACN/0.1% formic acid in HPLC-grade water) and 95% buffer A (2% ACN/0.1% FA in HPLC-grade water). After 5 min, peptides were eluted from the trap column and separated on an analytical column (Acclaim PepMap C18, 25 cm, 75µm inner diameter, 2 µm/100 Å pore size, LC Packings) by a 140 min gradient (5–50%) of buffer B at a 300 nl/min flow rate, followed by a short gradient from 50 to 95% buffer B for 5 min. Between each sample, the gradient was set back to 5% buffer B and left to equilibrate for 20 min. From the MS pre-scan, the 10 most abundant peptide ions were selected for fragmentation in the linear ion trap if they exceeded an intensity of at least 200 counts and if they were at least doubly charged. Every ion selected for fragmentation was excluded for the next 60 seconds, by dynamic exclusion. During fragment analysis a high-resolution (60,000 full-width half maximum) MS spectrum was acquired in the Orbitrap with a mass range from 300 to 1500 Da.
MaxQuant (MQ)
Sample compositions were analysed using MQ software (version 1.4.0.5, Max Planck Institute of Biochemistry, Martinsried)(25) with its internal Andromeda search engine (26). Except for the precursor tolerance of 10 ppm and the fragment tolerance of 0.6 Da, all settings were at default. Labelling was set to doublets (0/0 and 8/10). Carbamidomethylation of cysteine residues was set as a fixed modification and methionine oxidation and phosphorylation of serine, threonine and tyrosine set as variable modifications. The Andromeda search engine was configured for the Ensembl Human protein database (release 72, 40047703 residues, 105287 sequences). The software includes a decoy database and a ‘common contaminants’ database, to determine the false discovery rate and to exclude false positive hits. The lists of identified phospho-peptides in the 3 datasets, with their respective up- or down-regulation values, were exported (Supplementary Material, Table S1).
Immunoblots
Cells were lysed in 8 M urea, 20 mM HEPES pH7.0, 75 mM beta-glycerol phosphate, 1 mM sodium vanadate, 1 mM DTT, 1.5 mM EGTA. Protein concentration was measured by Bradford assay. Band densitometry was determined using image J software.
Immunofluorescence
Normal/control cells, patient-1 and -2 cells, and four human astrocytoma cell lines were plated in 35-mm wells at 70% confluence or at over confluence for foci staining. All images were obtained with a fluorescence microscope (Leica DM 5000 B), using the same settings.
Co-immunoprecipitation of PKCe and CTN
Extracts were probed for PKCe and CTNND1 by immunoblotting. 293 cells were transfected with FLAG-tagged PKCe and FLAG-tagged CTN. 24 hrs later, whole-cell extracts were immunoprecipitated using FLAG antibodies, followed by immunoblot analysis using anti-PKCe and anti-CTN antibodies, respectively. IgG antibody was used as a negative control for immunoprecipitation. 25–50μg of whole-cell protein lysate was used as input. Arrowheads denote the position of the ∼100-kDa FLAG -tagged PKCe immunoreactivity on each blot.
Antibodies
The EMT Epithelial-Mesenchymal Transition Kit to detect specific EMT protein markers (E-caherin, Claudin-1, ZO-1, Vimentin, Snail, Slug, TCF8/ZEB1 and beta catenin) was purchased from Cell Signalling (Catalog number 9782S; Fig. 3D). Antibodies used to characterize the cells or proteins are shown in Supplementary Material, Table S2. Secondary antibodies were either Alexa Fluor 488-conjugated goat anti-rabbit antibody or Alexa Fluor 555-conjugated goat anti-mouse IgG (1:1,000, Invitrogen, Life Technologies). DAPI was used to stain cell nuclei.
Motility assay
Astrocytes were cultured to confluence in 6-well plates; and then a ‘wound’ was introduced by drawing a tip across the monolayer. Cultures were imaged by using Microscope (Leica DMi8) every 2 min from 0 to 12 h, to measure cell migration. Five cells of each sample were tracked as subjects by ‘R’ version 3.2.2 to estimate the mean speed for each.
Cell proliferation assay
Patient-1, patient-2, and control astrocytes were cultured to 80–90% confluence, resuspended, and 104 cells were replated in each well of a 12-well plate. Nine wells were prepared for each sample. Three wells containing the different samples were monitored at 24, 48 and 72 h intervals. Astrocytes were resuspended, stained with trypan blue, and counted using a hemocytometer.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We would like to thank members of the Jonas Stem Cell Laboratory and the Brown Glaucoma Laboratory for sharing ideas and equipment and, especially Wei-Pu Wu for data analysis of cell motility, Yi-Ting Tsai and Wen-Hsuan Wu for genotyping instruction, and Rebecca Tuttle, Sally Justus for critical reviews of the manuscript. We also thank Jimmy Duong for comments on the statistical analyses. We would also like to thank the Columbia University Pathology Laboratory for their extensive assistance in exome sequencing for patient samples.
Conflict of Interest statement. None declared.
Funding
This work was conducted in the Barbara & Donald Jonas Laboratory of Regenerative Medicine and Bernard & Shirlee Brown Glaucoma Laboratory, which are supported by the Department of Defense CDMRP TSCRP:TS080017, National Institute of Health [5P30EY019007, R01EY018213, R01EY024698, 1R01EY026682, R21AG050437]; National Cancer Institute Core [5P30CA013696]; the Research to Prevent Blindness (RPB) Physician-Scientist Award; unrestricted funds from RPB, New York, NY, USA; the Tistou and Charlotte Kerstan Foundation; the Crowley Family Fund; the Schneeweiss Stem Cell Fund; New York State [C029572]; the Foundation Fighting Blindness New York Regional Research Center Grant [C-NY05-0705-0312]; and the Gebroe Family Foundation. JY is supported by National Natural Science Funds [81400412], the US Army Medical Research and Material Command [TS080017], China and the Key Program of Tianjin Natural Science Foundation [15JCZDJC34500] Tianjin, China. VBM is supported by NIH grants [K08EY020530, R01EY024665, R01EY025225, R01EY024698 and R21AG050437] and RPB. AGB is supported by 1R01NS098590-01.
References
- 1.Xu L., Burke T.R., Greenberg J.P., Mahajan V.B., Tsang S.H. (2012) Infrared imaging and optical coherence tomography reveal early-stage astrocytic hamartomas not detectable by fundoscopy. Am. J. Ophthalmol., 153, 883–889 e882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tee A.R., Sampson J.R., Pal D.K., Bateman J.M. (2016) The role of mTOR signalling in neurogenesis, insights from tuberous sclerosis complex. Semin. Cell Dev. Biol., 52, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon B.S., Kazi A.A., Coleman C.S., Dennis M.D., Chau V., Jefferson L.S., Kimball S.R. (2014) RhoA modulates signaling through the mechanistic target of rapamycin complex 1 (mTORC1) in mammalian cells. Cell Signal, 26, 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uhlmann E.J., Li W., Scheidenhelm D.K., Gau C.L., Tamanoi F., Gutmann D.H. (2004) Loss of tuberous sclerosis complex 1 (Tsc1) expression results in increased Rheb/S6K pathway signaling important for astrocyte cell size regulation. Glia, 47, 180–188. [DOI] [PubMed] [Google Scholar]
- 5.Martin T.D., Chen X.W., Kaplan R.E., Saltiel A.R., Walker C.L., Reiner D.J., Der C.J. (2014) Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol. Cell, 53, 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu P.P., Kang S.A., Rameseder J., Zhang Y., Ottina K.A., Lim D., Peterson T.R., Choi Y., Gray N.S., Yaffe M.B., et al. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science, 332, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villen J., Kubica N., Hoffman G.R., Cantley L.C., Gygi S.P., et al. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science, 332, 1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutt-Cabezas M., Karajannis M.A., Zagzag D., Shah S., Horkayne-Szakaly I., Rushing E.J., Cameron J.D., Jain D., Eberhart C.G., Raabe E.H., et al. (2013) Activation of mTORC1/mTORC2 signaling in pediatric low-grade glioma and pilocytic astrocytoma reveals mTOR as a therapeutic target. Neuro. Oncol., 15, 1604–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alain T., Sonenberg N., Topisirovic I. (2012) mTOR inhibitor efficacy is determined by the eIF4E/4E-BP ratio. Oncotarget, 3, 1491–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H., Paradies N.E., Fedor-Chaiken M., Brackenbury R. (1997) E-cadherin mediates adhesion and suppresses cell motility via distinct mechanisms. J. Cell Sci., 110 (Pt 3), 345–356. [DOI] [PubMed] [Google Scholar]
- 11.Lopes M.B., Altermatt H.J., Scheithauer B.W., Shepherd C.W., VandenBerg S.R. (1996) Immunohistochemical characterization of subependymal giant cell astrocytomas. Acta Neuropathol., 91, 368–375. [DOI] [PubMed] [Google Scholar]
- 12.Lu Q. (2010) delta-Catenin dysregulation in cancer: interactions with E-cadherin and beyond. J. Pathol., 222, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Roy F. (2014) Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat. Rev. Cancer, 14, 121–134. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J., Wulfkuhle J., Zhang H., Gu P., Yang Y., Deng J., Margolick J.B., Liotta L.A., Petricoin E, 3rd, Zhang Y. (2007) Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl Acad. Sci. U S A, 104, 16158–16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neugebauer V., Schaible H.G. (1988) Peripheral and spinal components of the sensitization of spinal neurons during an acute experimental arthritis. Agents Actions, 25, 234–236. [DOI] [PubMed] [Google Scholar]
- 16.Menet V., Gimenez y., Ribotta M., Chauvet N., Drian M.J., Lannoy J., Colucci-Guyon E., Privat A. (2001) Inactivation of the glial fibrillary acidic protein gene, but not that of vimentin, improves neuronal survival and neurite growth by modifying adhesion molecule expression. J. Neurosci., 21, 6147–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanemaru K., Nishiyama Y., Yoshioka H., Satoh K., Hashimoto K., Hanihara M., Horikoshi T., Ozaki Y., Kinouchi H. (2013) In-stent thrombosis after carotid artery stenting despite sufficient antiplatelet therapy in a bladder cancer patient. J. Stroke Cerebrovasc. Dis., 22, 1196–1200. [DOI] [PubMed] [Google Scholar]
- 18.Dann S.G., Golas J., Miranda M., Shi C., Wu J., Jin G., Rosfjord E., Upeslacis E., Klippel A. (2014) p120 catenin is a key effector of a Ras-PKCvarepsilon oncogenic signaling axis. Oncogene, 33, 1385–1394. [DOI] [PubMed] [Google Scholar]
- 19.Jain K., Basu A. (2014) Protein Kinase C-ϵ Promotes EMT in Breast Cancer. Breast Cancer (Auckl), 8, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kourtidis A., Ngok S.P., Anastasiadis P.Z. (2013) p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl Sci., 116, 409–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulhati P., Bowen K.A., Liu J., Stevens P.D., Rychahou P.G., Chen M., Lee E.Y., Weiss H.L., O'Connor K.L., Gao T., et al. (2011) mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res., 71, 3246–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liffers S.-T., Ozlu N., Bensaddek D., Steen J., Steen H. Biomolecular MS-Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC): A Primer. https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Aldrich/General_Inf ormation/silac_primer.pdf. Updated Jan 16, 2008. Accessed Jan 29, 2016.
- 23.Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol., 7, 952–958. [DOI] [PubMed] [Google Scholar]
- 24.Wiśniewski J.R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res., 8, 5674–5678. [DOI] [PubMed] [Google Scholar]
- 25.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc., 4, 698–705. [DOI] [PubMed] [Google Scholar]
- 26.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res., 10, 1794–1805. [DOI] [PubMed] [Google Scholar]