Abstract
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 subverts host cells through a type III secretion system encoded by the locus for enterocyte effacement (LEE). Genome sequencing of this pathotype revealed the existence of a gene cluster encoding components of a second cryptic type III secretion system, E. coli type III secretion system 2 (ETT2). Recently, we showed that the ETT2 gene cluster is present in whole or in part in the majority of E. coli strains but is unable to encode a functional secretion system in most strains, including EHEC O157:H7. However, here we show that mutational inhibition of two regulatory genes (ECs3720 or etrA and ECs3734 or eivF) from the ETT2 cluster in EHEC O157:H7 leads to greatly increased secretion of proteins encoded by the LEE and to increased adhesion to human intestinal cells. Studies in which transcriptional fusions and microarrays were used indicated that EtrA and EivF exert profound negative effects on gene transcription within the LEE. Consistent with these observations, expression of these regulators in an EHEC O26:H- strain led to suppression of protein secretion under LEE-inducing conditions. These findings provide fresh examples of the influence of mobile genetic elements on regulation of the LEE and of cross talk between type III secretion system gene clusters. In addition, they provide a cautionary tale because they show that the effects of regulatory genes can outlive widespread decay of other genes in a functionally coherent gene cluster, a phenomenon that we have named the “Cheshire cat effect.” It also seems likely that variations in the ETT2 regulator repertoire might account for strain-to-strain variation in secretion of LEE-encoded proteins.
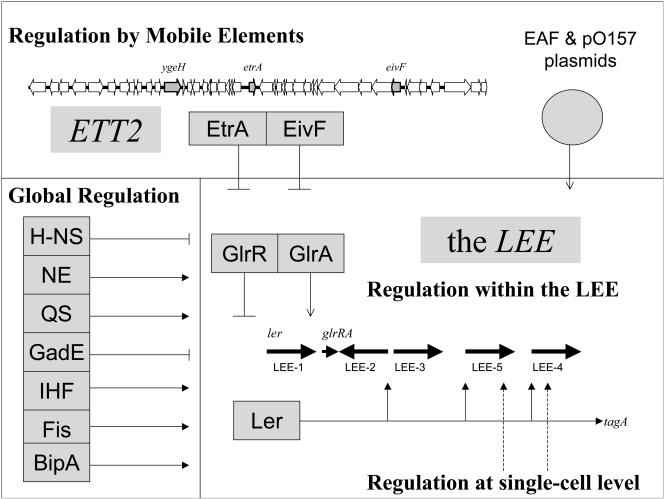
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC), like many other pathogenic gram-negative bacteria, utilize type III secretion to subvert eukaryotic signaling pathways by injecting bacterial effector proteins into the host cell cytoplasm (27, 30, 31). In these pathovars, a well-characterized type III secretion system (TTSS) is responsible for the development of the attaching and effacing lesions and for other effects on enterocyte function (31, 36, 37). Like almost all other TTSSs, this system is encoded by a pathogenicity island (in this case termed the locus of enterocyte effacement [LEE]), which contains virulence genes that are clustered on the chromosome and have been acquired en bloc by horizontal gene transfer (22, 37, 44). Regulation of gene expression within the LEE is known to be complex and governed by a large number of influences (Fig. 1).
FIG. 1.
Transcriptional regulation of the LEE: composite model based on evidence from three attaching and effacing strains (the EHEC-1 and EPEC-1 lineages and C. rodentium). The lines that end with arrowheads indicate positive influences; the lines that end with crossbars indicate negative influences. For references see the text. NE, norepinephrine and other catecholamines; QS, quorum sensing.
Analysis of complete genome sequences, initially of two strains of EHEC O157:H7 (24, 45) and later of the enteroaggregative strain E. coli 042 (47), revealed the existence of a gene cluster that might encode a second cryptic type III secretion system, which has been termed E. coli type III secretion system 2 (ETT2) (the term ETT1 is reserved for the LEE-encoded TTSS). ETT2 closely resembles the TTSS of Salmonella enterica, which is encoded by Salmonella pathogenicity island 1 (Spi-1) (33). Early reports suggested that the ETT2 gene cluster from EHEC O157:H7 was an insertion into the K-12 chromosome, that it might encode an intact secretion system, and that it might be linked to virulence in E. coli (23, 35). However, more recently, it has been shown that the ETT2 gene cluster is present in whole or in part in the majority of E. coli strains, irrespective of whether they are pathogens or commensals, and that there is a remnant of the cluster even in E. coli K-12; in other words, the difference between K-12 and O157 is a deletion in K-12 rather than an insertion in O157 (47). Furthermore, several decisive frameshift mutations in the secretion apparatus genes in the O157 ETT2 cluster have been identified, which means that ETT2 cannot function as a secretion system in EHEC O157, although a similar cluster in the enteroaggregative strain E. coli 042 may still be functional (47).
So far, there have been no reports ascribing a phenotype to this ETT2 gene cluster, and there are no homologues of known TTSS effectors encoded in the ETT2 gene cluster in EHEC O157:H7. However, from studies of other organisms, including S. enterica, Pseudomonas syringae, and Citrobacter rodentium (7, 12, 15, 19, 25, 41, 42, 46, 57-59), it is clear that TTSS effector genes do not have to be situated in a TTSS structural gene cluster. Thus, we reasoned that ETT2 effectors were probably encoded elsewhere on the chromosome in EHEC O157:H7, away from the ETT2 secretion gene cluster. Identification of genes that are outside TTSS islands but are still controlled by TTSS regulators has been used as an approach to find novel type III effectors and other virulence factors in several species, including genes regulated by SirA or SsrAB in S. enterica (1, 58) or by the TTSS regulators HrpL and HrpS in P. syringae (15, 59). Thus, in the hope of identifying ETT2 effector genes elsewhere on the chromosome (and before we realized that the secretion system itself was inactive), we created mutants with deletions in three regulatory genes in the ETT2 structural gene cluster in an EHEC strain. Surprisingly, instead of discovering novel ETT2 effectors, we obtained evidence that regulators from the ETT2 gene cluster influence expression and secretion of proteins by the LEE secretion system.
MATERIALS AND METHODS
Mutagenesis and complementation.
The strains and plasmids used are listed in Table 1. Mutants with deletions in the ygeH, etrA, and eivF genes of the Sakai 813 strain of enterohemorrhagic E. coli O157:H7 were obtained by using the one-step PCR-based method of Datsenko and Wanner (11). The pKD46 plasmid was transformed into the Sakai 813 strain by electroporation. Plasmid pKD3 was used as a template to amplify a chloramphenicol resistance marker flanked by gene-specific sequences. The 5′ ends of the primers (Table 2) contained homologous extension sequences, which consisted of the 40 nucleotides immediately before and after the coding sequences of the genes to be deleted. Mutants were created by transforming the Sakai 813 strain carrying pKD46 with linear PCR products and then plating the organisms on selective agar as described previously (11). Mutants were cured of the pKD46 plasmid by subculturing them on Luria-Bertani (LB) agar at a nonpermissive temperature (37°C). By using the helper plasmid pCP20, which encodes the FLP recombinase, the resistance genes were then eliminated. The helper plasmid was subsequently cured by growth at 37°C (11). Gene deletion mutants were confirmed by PCR and by sequencing with primers listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Sakai 813 | Isogenic stx1 and stx2-negative nontoxigenic derivative of the Sakai strain; contains insertion of kanamycin resistance gene into stxA2 gene and has deletion of 624-bp BsiWI fragment encompassing the start of the stxA1 gene and the 5′ flanking region | Chihiro Sasakawa |
| LZ1 | ygeH deletion mutant of Sakai 813 | This study |
| LZ2 | etrA deletion mutant of Sakai 813 | This study |
| LZ3 | eivF deletion mutant of Sakai 813 | This study |
| 193 Nalr | Spontaneous nalidixic acid-resistant mutant of recent bovine EHEC O26:H- isolate from the United Kingdom | This study |
| Plasmids | ||
| pKD46 | For deletion mutagenesis; encodes lambda Red genes | 11 |
| pKD3 | For deletion mutagenesis; template plasmid | 11 |
| pCP20 | For deletion mutagenesis; encodes FLP recombinase | 11 |
| pBAD30 | Low-copy-number expression vector with arabinose-inducible promoter | 20 |
| pBADetrA | etrA cloned into pBAD30 | This study |
| pBADeivF | eivF cloned into pBAD30 | This study |
| pAJR71 to pAJR75 | GFP fusion plasmids with promoters from LEE-1 to LEE-5 operons cloned in frame 5′ to the gene amplified from enhanced half-life GFP vector peGFP (Clontech, Ltd., Basingstoke, United Kingdom) | 48 |
TABLE 2.
Primers
| Primer | Sequence | Function |
|---|---|---|
| etrA-p1h1 | TAATATTTACCCTAAATGATTTTAGAGTACAGGTTGGCATGTGTAGGCTGGAGCTGCTTC | PCR for mutagenesis |
| etrA-p2h2 | GTTATTATGATTTTACTATTATATCATTACTAATGACATACATATGAATATCCTCCTTAG | PCR for mutagenesis |
| ygeH-p1h1 | GCTTATGCAGGATGCAAGAAACCAATTTTTTCATAGAGGTTAACTAGTGTAGGCTGGAGCTGCTTCGAA | PCR for mutagenesis |
| ygeH-p2h2 | AACAAGTTGCATACATCTAAATAATAAAATTGTGTCAATAAAAATTTACATATGAATATCCTCCTTAGTTCCT | PCR for mutagenesis |
| eivF-p1h1 | AGTTTATGGAGAATGAGAATATGGAATAAGGAGATGAAACGTGTAGGCTGGAGCTGCTTC | PCR for mutagenesis |
| eivF-p2h2 | AAGACTGAAATTAATATAATAGTAATGCGTAATTTTATTTCATATGAATATCCTCCTTAG | PCR for mutagenesis |
| YgeHfor1 | AGCGCAGGCATATATTGAACTCT | Confirmation of mutagenesis |
| YgeHrev1 | GAAAAGACGCCATCCATGTTGAA | Confirmation of mutagenesis |
| EtrAfor1 | TTCCCTCAACGTTAAGCAAATAT | Confirmation of mutagenesis |
| EtrArev1 | ACCTGTGCAAATTCGCGATAT | Confirmation of mutagenesis |
| EivFfor1 | ATGCGGGGAGTAACAAAACAC | Confirmation of mutagenesis |
| EivFrev1 | CAGGCGTTCAATCAAGGCAT | Confirmation of mutagenesis |
| Bad-etrA-for | CCCTTCTAGATTTTAGAGTACAGGTTGGCA | Complementation |
| Bad-etrA-rev | CTAATAAGCTTTCAACTTTCTCTTACGCAAGATTGG | Complementation |
| Bad-eivF-for | AGAATCTAGAATAAGGAGATGAAACATGA | Complementation |
| Bad-eivF-rev | GCATAAGCTTGAAATTAATATAATAGTAATGCGTAATTTTATTT | Complementation |
| Bad-for | CTGTTTCTCCATACCCGTT | Insert screening |
| Bad-rev | CTCATCCGCCAAAACAG | Insert screening |
Primers Bad-etrA-for and Bad-etrA-rev (Table 2) were used to PCR amplify etrA from genomic DNA of the Sakai 813 strain, with addition of an XbaI site to the 5′ end and a HindIII site to the 3′ end. Primers Bad-eivF-for and Bad-eivF-rev (Table 2) were used to PCR amplify eivF from genomic DNA of the Sakai 813 strain, with addition of an EcoRI site to the 5′ end and a SalI site to the 3′ end. Purified PCR products were digested with the relevant enzymes and then ligated into plasmid pBAD30 which had been digested with the same enzymes. Ligation products were transformed into Top10 competent cells (Invitrogen Ltd., Renfrew, Renfrewshire, United Kingdom) by using the manufacturer's protocol. Transformants were screened by PCR by using the primers flanking the insertion site (Bad-for and Bad-rev [Table 2]), and plasmids were purified from positive clones by using a QIAGEN mini-prep kit (QIAGEN Ltd., Crawley, Surrey, United Kingdom). The compositions of purified plasmids pBADetrA and pBADeivF were confirmed by sequencing by using primers BAD-for and BAD-rev.
Plasmid pBADetrA was electroporated into LZ2 to produce LZ2/pBADetrA, while plasmids pBADetrA and pBADeivF were electroporated into the O26:H- strain 193 Nalr to produce 193 Nalr/pBADetrA and 193 Nalr/pBADeivF, respectively. All strains were screened by PCR with primers BAD-for and BAD-rev to confirm that the plasmid insert had been retained.
Preparation and analysis of secreted protein profiles.
Overnight bacterial cultures grown in LB broth were diluted 100-fold with fresh Dulbecco-modified Eagle medium (DMEM) (Invitrogen Ltd.) containing 25 μg of kanamycin per ml and then grown with shaking at 37°C to an optical density at 600 nm of 1.0. Cultures were centrifuged at 3,000 × g, and supernatants were collected. The supernatants were passed through a 0.22-μm-pore-size filter. A 50-ml portion of filtered supernatant was concentrated about 250-fold by using an Amicon Ultra-15 centrifugal filter device (Millipore, Watford, United Kingdom). A sample of the concentrated supernatant from each culture equivalent to 2.5 ml of the original supernatant was mixed with sodium dodecyl sulfate (SDS) gel loading buffer and boiled for 5 min. SDS-12% polyacrylamide gel electrophoresis (PAGE) gels were used to separate proteins in the samples. The gels were stained with Coomassie brilliant blue or blotted onto nitrocellulose membranes (Hybond ECL; Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). For immunoblotting we employed rabbit and mouse polyclonal antibodies against LEE-encoded secreted proteins, as described previously (8, 32). Horseradish peroxidase-linked anti-rabbit and anti-mouse antibodies (Sigma-Aldridge Ltd., Gillingham, Dorset, United Kingdom) were used as second antibodies. Bound immunoglobulin G was visualized by using 4-chloro-1-naphthol as the substrate. For protein identification by mass spectrometry, protein bands were excised from colloidal Coomassie brilliant blue-stained gels. Gel plugs were subjected to trypsin digestion, and peptides were extracted by using a QIAGEN Biorobot 3000 (QIAGEN Ltd.). The peptides were analyzed by using a Q-TOF Global mass spectrometer. The data were processed with the Bio Tool software, and processed data were searched against the Mascot database (http://www.matrixscience.com/cgi/index).
To determine whether complementation of the increased-protein-secretion phenotype occurred in LZ2/pBADetrA, three strains (the parent Sakai strain, LZ2, and LZ2/pBADetrA) were grown in LB broth overnight. The overnight cultures were diluted 100:1 with fresh DMEM containing 0.02% arabinose supplemented with 25 μg of kanamycin per ml (plus 100 μg of ampicillin per ml for LZ2/pBADetrA). Cells were harvested at an optical density at 600 nm of 1.0, and the supernatant was concentrated and analyzed by SDS-PAGE as described above. To determine whether expression of etrA and eivF could influence protein secretion in a heterologous host, protein secretion profiles were obtained by using the methods described above (except that trichloroacetic acid precipitation was used instead of a centrifugal filter device) for the high-secreting O26:H- strain 193 Nalr and for strains 193 Nalr/pBADetrA and 193 Nalr/pBADeivF.
Measurement of LEE promoter activity by using GFP plasmid reporter constructs.
To measure the activities of the promoters from operons LEE-1 to LEE-5, plasmids pAJR71 to pAJR75 (Table 1) were separately transformed into the Sakai 813 strain and the ygeH, etrA, and eivF mutant strains LZ1, LZ2, and LZ3 by electroporation by using a Bio-Rad Gene Pulser (Bio-Rad Ltd., Hemel Hempstead, United Kingdom) according to the manufacturer's instructions. Transformants were grown overnight in MEM-HEPES medium containing chloramphenicol and then subcultured 1:50 in fresh, prewarmed modified Eagle medium-HEPES medium. Typically, 15 ml was cultured in Erlenmeyer flasks shaken at 200 rpm and 37°C. The absorbance of the cultures was monitored by measurement with a spectrophotometer at 600 nm. At intervals, 200 μl of a culture was removed, and the green fluorescent protein (GFP) that had accumulated was quantified by using a Fluostar Optima fluorescence plate reader (BMG Labtechnologies Ltd., Aylesbury, Bucks, United Kingdom) at appropriate excitation and emission wavelengths. Control bacteria containing no plasmid were used to measure the background fluorescence, which, at the appropriate optical density, was subtracted from the other values.
Construction of E. coli whole-genome array.
We constructed a 70-mer oligonucleotide array containing 6,112 oligonucleotides that included samples of chromosomal genes identified in the genome sequences of E. coli K-12 strain MG1655, O157:H7 strain VT2 Sakai, and O157:H7 strain EDL933 and plasmid genes from the two O157:H7 strains whose genomes were sequenced (Operon Array Ready E. coli oligonucleotide set 1.0; QIAGEN-Operon, Hilden, Germany). Twelve positive control oligonucleotides and 12 negative control oligonucleotides were also included. The oligonucleotides were resuspended in 3× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and printed on Corning GAPS II slides (Corning B.V. Life Sciences, Schiphol-Rijk, Netherlands) by using a Biorobotics Microgrid TAS array printer. Amersham Lucidea array controls (Amersham Biosciences Ltd.) were printed in each subarray. After printing, slides were briefly rehydrated in steam, dried at 100°C, and UV cross-linked according to the manufacturer's instructions. The slides were stored desiccated and in the dark until they were used.
Preparation of mRNA, labeling, and hybridization.
Overnight cultures were diluted 100-fold with fresh DMEM (Invitrogen Ltd.) with 25 μg of kanamycin per ml and then grown with shaking at 37°C to an optical density at 600 nm of 1.0. Two-milliliter cultures were mixed with 4 ml of RNAprotect bacterial reagent (QIAGEN Ltd.) to stabilize the total RNA. An RNeasy minikit was used to prepare total RNA according to the manufacturer's instructions (QIAGEN Ltd.). Contaminating DNA was removed from the RNA preparation by using an RNase-free DNase set according to the manufacturer's protocol (QIAGEN Ltd.). The quality and quantity of the RNA preparations were determined with an Agilent 2100 Bioanalyzer by using the RNA 6000 nano assay Labchip (Agilent, Stockport, United Kingdom).
Aminoallyl-labeled cDNA was reverse transcribed from 10 μg of total bacterial RNA by using a CyScribe postlabeling kit (Amersham Biosciences Ltd.), reacted with Cy3 or Cy5 NHS esters (Amersham Biosciences Ltd.), and purified with a CyScribe GFX purification kit (Amersham) used according to the manufacturer's protocol. Lucidea (Amersham Biosciences Ltd.) test or reference RNA was added to the primer annealing step in the RNA labeling experiments to act as internal controls for both labeling and hybridization. The following slight modifications of the manufacturer's protocol for labeling were made: the length of the reverse transcription reaction was extended from 1.5 h to overnight, and the length of labeling of aminoallyl-modified cDNA with CyDye NHS esters was extended from 1 to 4 h. Five biological replicates of the microarray experiments were performed. In four of the experiments, aminoallyl-modified cDNA from the parent strain was labeled with Cy3, and cDNA samples from the mutants were labeled with Cy5, whereas in the dye swap experiment, the labels were reversed. DNA arrays were prehybridized by using a 25% formamide prehybridization solution at 42°C for 2 h. Forty picomoles of each CyDye-labeled cDNA was denatured at 95°C for 3 min and hybridized in the presence of Denhardt's solution and poly(A) oligonucleotide on the array for 16 h at 42°C. The slides were washed in 2× SSC-0.1% SDS at 42°C for 2 min, in 0.1× SSC for 2 min at room temperature, and finally in 0.01× SSC for 2 min at room temperature. The last two wash steps were repeated twice. The microarray slides were dried by centrifugation for 5 min at 1,500 rpm and were then scanned at 532 and 630 nm by using a Genepix 4000A scanner (Axon Instruments, Union City, Calif.).
Analysis of microarray data.
The images were analyzed by using GenePix software (Axon Instruments), and the data were imported into GeneSpring, version 6.1 (Silicon Genetics, Redwood City, Calif.). To account for dye swapping, the signal channel and control channel measurements for the fifth experiment were reversed. A Lowess (locally weighted linear regression curve) curve was fitted to the plot of log intensity versus log ratio, and 20% of the data was used to calculate the Lowess fit at each point. The curve was used to adjust the control value for each measurement. If the control channel signal was below a threshold value of 10, then 10 was used instead. A whole-genome heat diagram of differentially expressed genes was produced (see Fig. 5). A cross-gene error model was constructed based on the five replicates, and genes that showed significantly different expression levels in the mutant strains (P < 0.05) were identified by using Student's t test. A second filter was then applied to significantly differentially expressed genes, which required a ≥2.5-fold change in gene expression.
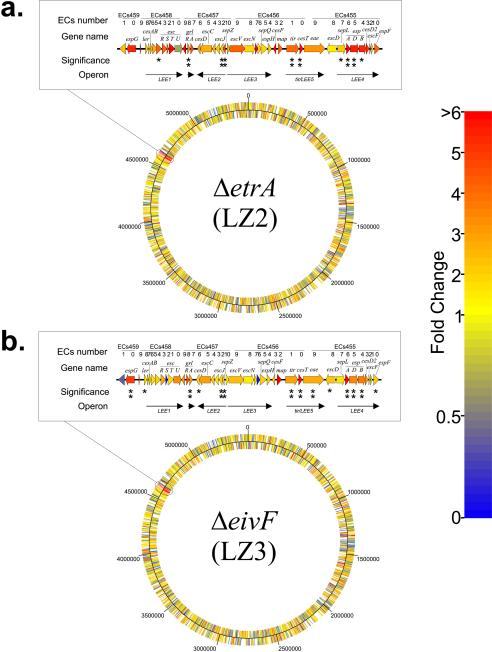
FIG. 5.
Up-regulation of LEE gene expression in ETT2 regulator mutants assayed by using microarrays: heat map of normalized microarray data showing changes relative to the parent strain for ΔetrA mutant LZ2 (a) and ΔeivF mutant LZ3 (b), colored on the basis of the fold changes. The circles represent the E. coli O157:H7 Sakai chromosome. The cutaway shows a magnification of the heat map for the LEE region. Asterisks in the LEE indicate the P values of the fold changes; one asterisk indicates that the P is <0.1, and two asterisks indicate that the P value is <0.05.
Hemolysis, immunofluorescence, and fluorescent actin staining.
The hemolysis assay was carried out by using the method of Shaw et al. (50). EspA filament detection and fluorescent actin staining were carried out as previously described (32).
Adherence to Int407 cells.
Adhesion of E. coli strains to Int407 cells (human embryonic intestine; ATCC CCL 6) was quantified essentially as described previously (38). Cells were seeded at a density of 2 × 105 cells per 35-mm dish on glass coverslips and grown for 18 h at 37°C in a humidified 5% CO2 atmosphere in modified Eagle medium buffered with 2 g of sodium bicarbonate per ml and supplemented with 10% (vol/vol) fetal calf serum (PAA Laboratories GmbH, Linz, Austria), nonessential amino acids, and 0.3 g of l-glutamine per liter. The cells were inoculated at a multiplicity of infection of approximately 50:1 with fresh stationary-phase LB cultures of wild-type or mutant strains adjusted to the same optical density. The cells were incubated for 3 h at 37°C in a humidified 5% CO2 atmosphere, washed three times with 2 ml of phosphate-buffered saline, and then incubated in fresh medium for an additional 2 h. The cells were washed five times with 2 ml of phosphate-buffered saline, fixed, and stained with Hemacolor rapid staining solutions (Merck, Darmstadt, Germany), and multiple images were captured at a magnification of ×400 by using a Leica DMLS microscope with a Polaroid digital microscope camera. For each independent assay 20 randomly selected fields containing 50 or more cells were examined, and the mean number of microcolonies (comprised of 10 or more bacteria) per cell was determined. The results are given below as the mean (± standard deviation) number of microcolonies per cell from three independent experiments.
RESULTS
Mutations in ETT2 regulators influence secreted protein profiles.
In the hope of identifying ETT2 effectors, we created Sakai 813 deletion mutants (Sakai 813 is an isogenic nontoxigenic derivative of the E. coli O157:H7 Sakai strain, whose genome has been sequenced) (Table 1). We created mutants with mutations in the following three ETT2 genes (Fig. 1) that had been identified as genes that code for regulators on the basis of homology searches: ygeH (Ecs3709), encoding a homologue of the Spi-1 master regulator HilA (2, 3); eivF (Ecs3734), encoding a homologue of the Spi-1 regulator InvF (9, 10); and a third putative regulator, Ecs3720, with no Spi-1 homologue and which we designated etrA (ETT2 regulator A) (47). There were dramatic differences in the secreted protein profiles between two of the mutants and the parent strain (Fig. 2). The parent strain was a poor secretor, while two of the mutant strains, the etrA deletion mutant LZ2 and the eivF deletion mutant LZ3, secreted abundant amounts of several proteins. These two high-secreting mutants had similar protein profiles, but greater quantities of secreted proteins were obtained from the ΔetrA mutant LZ2 than from the ΔeivF mutant LZ3. The third mutant strain, the ΔygeH mutant LZ1, had a secretion profile identical to that of the parent strain (data not shown).
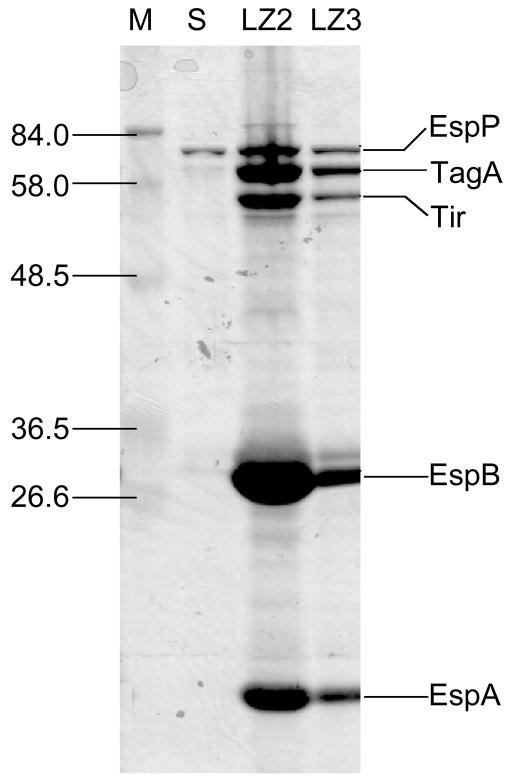
FIG. 2.
SDS-PAGE gel showing secreted protein profiles for ETT2 regulator mutants and the parent strain. Lane M, molecular weight markers; lane S, parent strain Sakai 813; lane LZ2, ΔetrA ETT2 regulator mutant LZ2; lane LZ3, ΔeivF ETT2 regulator mutant LZ3. Bands identified by mass spectrometry are indicated on the right. Molecular sizes in kilodaltons are indicated on the left.
Five secreted proteins from LZ2 were gel purified and identified by using mass spectrometry as EspP, TagA, Tir, EspB, and EspA (Fig. 2). The last four of these proteins are known to be coregulated at the transcriptional level by the LEE-encoded regulator, Ler (14). Thus, we speculated that the effects of EtrA and EivF on protein secretion profiles might be mediated at the transcriptional level. Western blotting with polyclonal antibodies against the LEE-encoded secreted proteins Tir, EspA, EspB, and EspD confirmed that secretion of these proteins was increased in LZ2 and LZ3 compared to the parent strain (Fig. 3a). The etrA deletion in LZ2 was complemented in trans by expressing the gene on a low-copy-number plasmid. As expected, decreased levels of protein secretion were seen in the complemented mutant compared to the noncomplemented mutant (Fig. 3b).
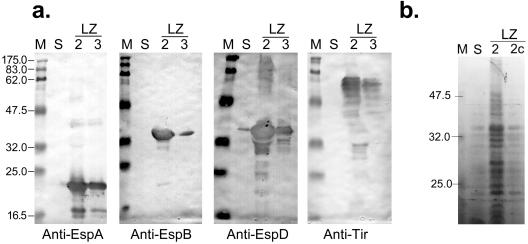
FIG. 3.
(a) Immunoblots showing changes in secretion of specific proteins in ETT2 regulator mutants. The antibodies used are indicated at the bottom. Lane M, molecular weight markers; lane S, parent strain Sakai 813; lane LZ2, ΔetrA ETT2 regulator mutant LZ2; lane LZ3, ΔeivF ETT2 regulator mutant LZ3. (b) SDS-PAGE gel showing complementation of secretion phenotype of the ΔetrA ETT2 regulator mutant. Lane M, molecular weight markers; lane S, parent strain Sakai 813; lane LZ2, ΔetrA ETT2 regulator mutant LZ2; lane LZ2c, complemented mutant strain LZ2/pBADetrA. Molecular sizes in kilodaltons are indicated on the left.
ETT2 regulators control expression of LEE-encoded genes at the transcriptional level.
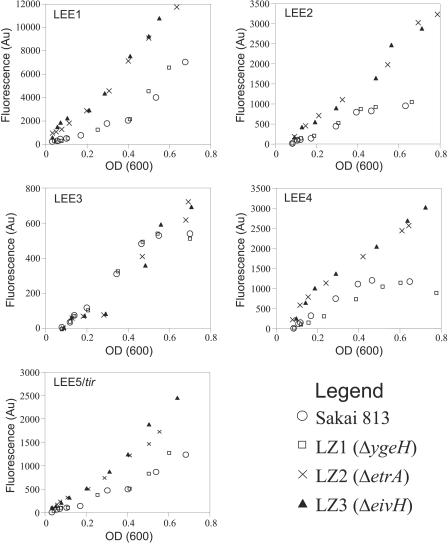
We wished to confirm our hypothesis that the effects on secretion of LEE-encoded proteins in the ΔetrA mutant LZ2 and the ΔeivF mutant LZ3 were the result of increased transcription of the LEE, especially as it is known that secretion of LEE-encoded effectors can be influenced at the posttranscriptional level, e.g., by toxB and other factors (48, 55). Thus, we examined expression of the LEE using transcriptional fusions to the gene for green fluorescent protein. Expression of the LEE-1 (which encodes Ler), LEE-2, LEE-4 (which encodes the secreted proteins EspA, EspB, and EspD) and LEE-5 operons was up-regulated under LEE secretion-inducing conditions around two- to threefold more strongly in LZ2 and LZ3 than in the parent strain or in LZ1 (Fig. 4). LEE-3, which encodes much of the secretion apparatus, showed similar weak patterns of induction in all strains (Fig. 4).
FIG. 4.
Up-regulation of LEE gene expression in ETT2 regulator mutants assayed by using GFP fusions. LEE-1 to LEE-5 promoter-egfp fusions were transformed into the appropriate strain, and fluorescence was determined during the growth cycle. Note the different scales used in the graphs, illustrating the high level of expression for LEE-1 (changing in LZ2 and LZ3), the intermediate levels of expression for LEE-2, LEE-4, and LEE-5 (changing in LZ2 and LZ3), and the low level of expression for LEE-3 (unchanged in LZ2 and LZ3). OD (600), optical density at 600 nm; Au, arbitrary units.
We then took an alternative look at changes in gene expression through hybridization to a whole-genome microarray based on the E. coli O157 genome sequences. Genome-wide transcriptional profiles obtained from mid- to late-log-phase cells of mutant strains LZ1 and LZ2 grown in DMEM (conditions which induce LEE-mediated secretion) were compared with profiles obtained under identical conditions for the parent strain. In the unfiltered data, several hundred genes apparently showed differential expression in the mutant strains (data not shown), and genome-wide heat maps showed clear clustering of these differentially expressed genes within the LEE in both mutants (Fig. 5). However, as expected, when the statistical filter (Student's t test for five replicates; P < 0.05) and the fold change filter (≥2.5-fold change) were applied to each mutant-parent comparison, for the vast majority of genes there was not a significant and substantial difference in expression between the parent and mutant strains (and none of the genes was significantly down-regulated in either mutant).
Only 10 genes in the ΔetrA mutant LZ2 and 12 genes in the ΔeivF mutant LZ3 survived the filtering process (Table 3). Six of these genes in LZ2 and nine of these genes in LZ3 were genes in the LEE, supporting our conclusion that EtrA and EivF influence transcription of this locus. Microarray-determined patterns of gene expression in the LEE mirrored the GFP fusion data in that in both mutants, genes showing significant and substantial changes populated the LEE-2, LEE-4, and LEE-5 operons but not LEE-3, and some LEE-1 genes narrowly missed the significance cutoff (Fig. 5). Furthermore, in both mutants, even among the genes in the LEE that missed the significance cutoff, there was a tendency toward up-regulation (Fig. 5). Interestingly, in both mutants there was up-regulation of the pair of genes encoding the recently characterized high-level regulators of the LEE, GrlA and GrlR (12), although only one of the pair passed the significance cutoff in each mutant (grlR in the etrA mutant and grlA in the eivF mutant) (Fig. 5 and Table 3). This suggests that the ETT2 regulators might influence the LEE indirectly through GrlA and GrlR.
TABLE 3.
Up-regulated genesa
| Strain | Sakai nomenclature | Gene name | Fold change | P value | Protein annotation or function |
|---|---|---|---|---|---|
| LZ2 | ECs4556 | espA | 9.18 | 0.018 | TTSS secreted protein EspA |
| ECs4560 | cesT | 8.62 | 0.037 | TTSS chaperone CesT | |
| ECs4555 | espD | 5.44 | 0.034 | TTSS secreted protein EspD | |
| ECs4571 | sepZ | 4.69 | 0.001 | TTSS SepZ protein | |
| ECs3948 | rpsU | 4.38 | 0.037 | 30S ribosomal subunit protein S21 | |
| ECs0814 | 4.37 | 0.008 | Unknown protein encoded by prophage CP-933K | ||
| ECs4561 | tir | 3.95 | 0.033 | Translocated intimin receptor | |
| ECs4578 | grlR | 3.51 | 0.027 | TTSS repressor GrlR | |
| On plasmid pO157 | tagA | 3.13 | 0.042 | Plasmid-encoded lipoprotein TagA (pO157) | |
| ECs1192 | 2.87 | 0.023 | Unknown protein encoded by prophage BP-933W | ||
| LZ3 | ECs4556 | espA | 6.02 | 0.009 | TTSS secreted protein EspA |
| ECs4560 | cesT | 5.86 | 0.00004 | TTSS chaperone CesT | |
| ECs2711 | yodA | 4.93 | 0.045 | Metal-binding, lipocalin-like protein YodA | |
| ECs4590 | espG | 4.78 | 0.003 | TTSS secreted protein EspG | |
| ECs4554 | espB | 3.61 | 0.007 | TTSS secreted protein EspB | |
| ECs4555 | espD | 3.33 | 0.017 | TTSS secreted protein EspD | |
| ECs4553 | cesd2 | 2.96 | 0.038 | TTSS chaperone CesD2 | |
| ECs4577 | grlA | 2.95 | 0.047 | TTSS activator GrlA | |
| ECs2819 | hisL | 2.87 | 0.031 | His operon leader peptide | |
| ECs4561 | tir | 2.80 | 0.002 | Translocated intimin receptor | |
| ECs0814 | 2.70 | 0.003 | Unknown protein encoded by prophage CP-933K | ||
| ECs4559 | eae | 2.66 | 0.003 | Intimin |
Summary of normalized microarray data for ΔetrA strain LZ2 and ΔeivF strain LZ3. Data are shown for genes that were up-regulated ≥2.5 fold, with a P value of <0.05. Genes located within the LEE are indicated by boldface type. espB showed a 5.24-fold increase in gene expression in LZ2, which fell just outside the significance cutoff (P = 0.058).
Six genes outside the LEE showed significant and substantial up-regulation in one or both mutants (Table 3). As expected from the secretion data and from the previously known link to Ler (14), the plasmid-encoded tagA gene was up-regulated threefold and with significance (P = 0.042) in the ΔetrA mutant LZ2 and just missed our fold change cutoff in the ΔeivF mutant LZ3 (up-regulated 2.18-fold; P = 0.019). Expression of the other plasmid-borne gene encoding a protein detected in the secretion experiments, espP, was weakly up-regulated in both mutants, but the up-regulation was significant in only one mutant (1.57-fold up-regulation in LZ3 [P = 0.048]; 1.3-fold up-regulation in LZ2 [P > 0.1]). Two prophage-encoded genes with unknown functions (ECs1192 and ECs0814) were up-regulated, suggesting that they might encode novel virulence determinants. Up-regulation of the gene encoding YodA, a metal-binding, lipocalin-like protein, in LZ3 is hard to explain, as is the apparent up-regulation of two housekeeping genes (hisL and rpsU); these three genes may represent statistical artifacts.
ETT2 regulators influence interactions with human cells.
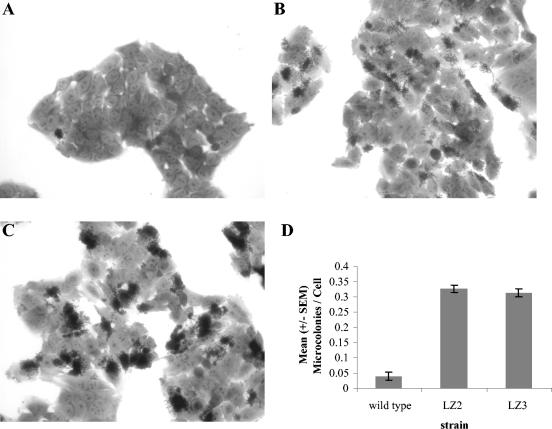
We could not detect any quantifiable effects of mutations in eivF and etrA on the ability of cells to form the EspA filament (as measured by red cell lysis and immunofluorescence) or to trigger actin polymerization (data not shown). However, clear, quantifiable differences from the parent strain were seen when the ΔetrA and ΔeivF mutants were applied to cultured intestinal epithelial (Int407) cells. Both mutants were considerably more adherent than the parent, forming six times as many microcolonies (Fig. 6). Although the LZ2 and LZ3 strains produced similar numbers of microcolonies, there was a clear difference in colonial morphology between the two; the colonies formed by ΔeivF mutant LZ3 tended to be dense, whereas those formed by the ΔetrA mutant LZ2 were more diffuse.
FIG. 6.
Adhesion to human epithelial cells. (A to C) Int407 cells infected with the parent strain (A), ΔetrA mutant LZ2 (B), or ΔeivF mutant LZ3 (C) and subjected to Hemacolor staining. Magnification, ×400. (D) Semiquantitative analysis of adherence. The number of microcolonies (a microcolony consisted of >10 adherent bacteria) per cell was calculated from 20 fields for each experiment, and the means were determined from three independent experiments. An average of 140 cells were examined in each field.
ETT2 regulators can down-regulate protein secretion in a high-secreting O26:H- EHEC strain.
There is considerable variation in LEE-mediated protein secretion among attaching and effacing strains (13, 39). From previous surveys of ETT2 gene clusters, it is also clear that the repertoire of ETT2 regulators varies from one lineage to the next; for example, the EHEC O157:H7 strains possess both etrA and eivF, while the EPEC-2 lineage (which includes strain B171-8) possesses etrA but lacks eivF (23, 35, 47). We speculated that variations in the ETT2 regulator repertoire might account for some of the known variation in protein secretion, particularly as Sakai 813, which possesses both regulators, is a low-secreting strain of EHEC. We also reasoned that expression of the ETT2 regulators in a host lacking one of the two regulators might also allow us to investigate the hierarchical relationship between them. We therefore expressed the etrA and eivF genes from Sakai 813 in a high-secreting EHEC O26:H- strain, 193 Nalr, which was shown by tiling path PCR to possess the ETT2 genotype common to the EPEC-2 lineage (i.e., it lacks eivF) (47). Expression of either etrA or eivF in this strain led to decreased protein secretion (Fig. 7). Interestingly, the relative magnitude of the change, even though it was in the opposite direction, mirrored the relative magnitude seen in the mutants; i.e., expression of etrA produced a greater effect than expression of eivF.
FIG. 7.
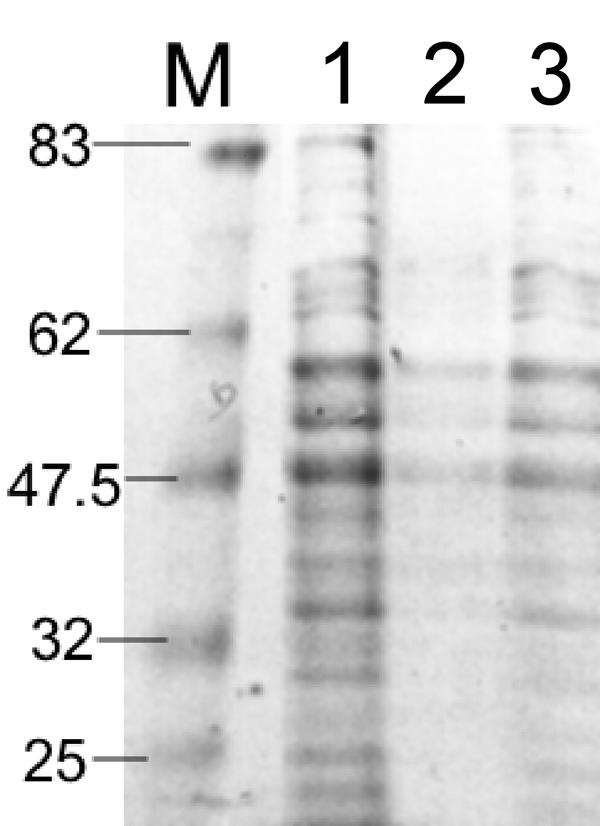
SDS-PAGE gel showing that ETT2 regulators suppress protein secretion under LEE-inducing conditions in an EHEC O26:H- strain. Lane M, molecular mass markers; lane 1, EHEC O26:H- strain 193 Nalr bearing empty plasmid pBAD30; lane 2, strain 193 Nalr bearing plasmid pBADetrA; lane 3, strain 193 Nalr bearing plasmid pBADeivF. Molecular sizes in kilodaltons are indicated on the left.
DISCUSSION
ETT2: an additional influence on expression of the LEE.
Regulation of the LEE is complex (Fig. 1) and is governed by at least four different kinds of regulatory influences, including (i) a regulatory hierarchy encoded within the LEE that includes the positive regulator Ler (40) and at least two additional regulators, GlrR and GlrA (12); (ii) unidentified non-LEE-encoded factors that regulate LEE-4 and LEE-5 expression at the single-cell level and govern type III-secreted protein levels (48; D. Gally, personal communication); and (iii) global regulatory systems that influence LEE expression, including heat-stable nucleoid-structuring protein (H-NS) (4, 5, 21, 49), integration host factor (IHF) (16), factor for inversion stimulation (FIS) (17), quorum sensing and catechol amines (28, 29, 51-54), BipA (18), and YhiE/GadE, YhiF, and YhiX/GadX (global regulators involved in acid resistance) (26, 34, 56).
This study provides a fresh example of a fourth class of regulatory influence: regulation of the LEE by regulators encoded in mobile genetic elements. The first example of this phenomenon was regulation of the LEE by the plasmid-encoded regulator Per in EPEC strain E2348/69 (40). Here we show that two regulators encoded in the chromosomal ETT2 gene cluster (like the LEE, a pathogenicity island, as shown by comparative genomics and the G+C content [47]) have a profound effect on the LEE. Mutational inhibition of these regulators results in a marked increase in the secretion of proteins by the LEE-encoded TTSS through an effect on LEE gene expression in the EHEC O157:H7 Sakai strain. This provides an additional level of complexity for what is already a highly complex regulatory network governing expression of the LEE genes (Fig. 1) and shows that other chromosomal pathogenicity islands affect LEE gene expression, in addition to plasmid-encoded factors.
The fact that two regulators encoded in different parts of the ETT2 gene cluster have similar effects on LEE-encoded protein secretion suggests that they work together, perhaps in some kind of hierarchy. When we introduced these regulators into a strain that lacked eivF, we saw suppression of protein secretion under LEE-inducing conditions with both the etrA- and eivF-containing plasmids. This rules out the hypothesis that EtrA acts via EivF because we saw the suppressant effect in the O26:H- strain carrying the plasmid-encoded EtrA, even though there was no eivF gene in the strain. However, the precise relationship between these two regulators remains unclear, as does the issue of whether the regulators influence LEE gene expression directly or through other regulators (the newly described GlrA and GlrR regulators are obvious candidates). Planned future experiments include chromatin immunoprecipitation studies to identify chromosomal binding sites for these regulators, which should clarify these points.
Cross talk between TTSSs.
The ETT2-LEE interaction provides a cogent example of the potential for interaction between different TTSSs within the same cell, particularly as EHEC joins a growing list of bacteria that possess more than one virulence-associated TTSS, including S. enterica, Yersinia spp., Vibrio parahaemolyticus, Burkholderia spp., and Chromobacterium violaceum (43). We assume that cross talk between different TTSSs results from a need to avoid expression of two highly complex multiprotein complexes at the cell surface at the same time in order to prevent structural or functional clashes and/or excessive demands for energy.
ETT2, the “Cheshire cat effect,” and strain-to-strain variation.
In a previous study it was shown that the primary ETT2 gene cluster was present in whole or in part in the majority of E. coli strains but that it had undergone mutational attrition in almost all cases (47). It was concluded that ETT2 could not function as a secretion system in either of the two EHEC strains whose genomes had been sequenced. However, this study showed that in EHEC, although the ETT2 secretion system may be inhibited, the ETT2 pathogenicity island is still capable of exerting powerful phenotypic effects through gene regulation. In other words, the effects of regulatory genes can outlive widespread decay of structural genes in a functionally coherent gene cluster. A name has been proposed for this phenomenon, the Cheshire cat effect, after Alice in Wonderland, in which a grin outlives the disappearance of its owner, the cat (6, 47). We suspect that the Cheshire cat effect is widespread in nature; indeed, ETT2 might even provide a second example in that a degenerate ETT2 cluster influences virulence in an avian pathogenic strain of E. coli that lacks the LEE (E. Ron, personal communication). Thus, it is not safe to dismiss an apparently degenerate gene cluster as nonfunctional without a full gene-by-gene dissection of the kind recently applied to the LEE (12).
Expression of the etrA and eivF genes obtained from the low-secreting EHEC strain Sakai 813 in a high-secreting EHEC O26:H- strain that lacks eivF led to suppression of protein secretion under LEE-inducing conditions. This suggests that variations in the ETT2 regulator repertoire might account for some of the known variation in LEE-mediated protein secretion among attaching and effacing strains (13, 39) and might provide a new target for comparative studies of strain-to-strain variation in regulation of the LEE.
Acknowledgments
We thank the BBSRC for funding this work through grant EGA16107 under the Exploiting Genomics Initiative.
We are grateful to Steve Minchin and others in the University of Birmingham E. coli consortium for assistance in designing and setting up a microarray. We thank Barry Wanner for kindly supplying plasmids and cells required for one-step PCR-based mutagenesis. We thank Chihiro Sasakawa for the kind gift of the Sakai 813 strain. We are grateful to Michael Russell and Chengjie Liu for medium preparation.
Editor: A. D. O'Brien
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Beltrametti, F., A. U. Kresse, and C. A. Guzman. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, L., and J. Tenniel. 2001. Alice in Wonderland. Dover Publications, Mineola, N.Y.
- 7.Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. J. Schneider, and J. R. Alfano. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10:462-469. [DOI] [PubMed] [Google Scholar]
- 8.Daniell, S. J., R. M. Delahay, R. K. Shaw, E. L. Hartland, M. J. Pallen, F. Booy, F. Ebel, S. Knutton, and G. Frankel. 2001. Coiled-coil domain of enteropathogenic Escherichia coli type III secreted protein EspD is involved in EspA filament-mediated cell attachment and hemolysis. Infect. Immun. 69:4055-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebel, F., C. Deibel, A. U. Kresse, C. A. Guzman, and T. Chakraborty. 1996. Temperature- and medium-dependent secretion of proteins by Shiga toxin-producing Escherichia coli. Infect. Immun. 64:4472-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. L. Gwinn, S. G. Lazarowitz, N. C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 18.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol 48:507-521. [DOI] [PubMed] [Google Scholar]
- 19.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 23.Hartleib, S., R. Prager, I. Hedenstrom, S. Lofdahl, and H. Tschape. 2003. Prevalence of the new, SPI1-like, pathogenicity island ETT2 among Escherichia coli. Int. J. Med. Microbiol. 292:487-493. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hommais, F., E. Krin, J. Y. Coppee, C. Lacroix, E. Yeramian, A. Danchin, and P. Bertin. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61-72. [DOI] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanamaru, K., I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. Regulation of virulence factors of enterohemorrhagic Escherichia coli O157:H7 by self-produced extracellular factors. Biosci. Biotechnol. Biochem. 64:2508-2511. [DOI] [PubMed] [Google Scholar]
- 29.Kanamaru, K., I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805-816. [DOI] [PubMed] [Google Scholar]
- 30.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 31.Kenny, B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469-477. [DOI] [PubMed] [Google Scholar]
- 32.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 34.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 35.Makino, S., T. Tobe, H. Asakura, M. Watarai, T. Ikeda, K. Takeshi, and C. Sasakawa. 2003. Distribution of the secondary type III secretion system locus found in enterohemorrhagic Escherichia coli O157:H7 isolates among Shiga toxin-producing E. coli strains. J. Clin. Microbiol. 41:2341-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 38.McKee, M. L., and A. D. O'Brien. 1996. Truncated enterohemorrhagic Escherichia coli (EHEC) O157:H7 intimin (EaeA) fusion proteins promote adherence of EHEC strains to HEp-2 cells. Infect. Immun 64:2225-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNally, A., A. J. Roe, S. Simpson, F. M. Thomson-Carter, D. E. Hoey, C. Currie, T. Chakraborty, D. G. Smith, and D. L. Gally. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect. Immun. 69:5107-5114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 41.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirold, S., W. Rabsch, H. Tschape, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector sopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 43.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 44.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 46.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan, Y. Jamir, L. M. Schechter, M. D. Janes, C. R. Buell, X. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren, C. P., R. R. Chaudhuri, A. Fivian, C. M. Bailey, M. Antonio, W. M. Barnes, and M. J. Pallen. 2004. The ETT2 gene cluster encoding a second type III secretion system from Escherichia coli is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186:3547-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roe, A. J., H. Yull, S. W. Naylor, M. J. Woodward, D. G. Smith, and D. L. Gally. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71:5900-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw, R. K., S. Daniell, F. Ebel, G. Frankel, and S. Knutton. 2001. EspA filament-mediated protein translocation into red blood cells. Cell. Microbiol. 3:213-222. [DOI] [PubMed] [Google Scholar]
- 51.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatsuno, I., K. Nagano, K. Taguchi, L. Rong, H. Mori, and C. Sasakawa. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood, M. W., M. A. Jones, P. R. Watson, S. Hedges, T. S. Wallis, and E. E. Galyov. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883-891. [DOI] [PubMed] [Google Scholar]
- 58.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 59.Zwiesler-Vollick, J., A. E. Plovanich-Jones, K. Nomura, S. Bandyopadhyay, V. Joardar, B. N. Kunkel, and S. Y. He. 2002. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 45:1207-1218. [DOI] [PubMed] [Google Scholar]