Abstract
1,4-Dihydroxy-2-naphthoic acid (1,4-DHNA) is a bacterial-derived metabolite that binds the aryl hydrocarbon receptor (AhR) and exhibits anti-inflammatory activity in the gut. The structure-dependent AhR activity of hydroxyl/carboxy-substituted naphthoic acids (NAs) was determined in young adult mouse colonic (YAMC) cells and human Caco2 colon cancer cells using CYP1A1/CYP1B1 mRNAs as Ah-responsive genes. Compounds used in this study include 1,4-, 3,5-, and 3,7-DHNA, 1,4-dimethoxy-2-naphthoic acid (1,4-DMNA), 1- and 4-hydroxy-2-naphthoic acid (1-HNA, 4-HNA), 1- and 2-naphthoic acid (1-NA, 2-NA), and 1- and 2-naphthol (1-NOH, 2-NOH). 1,4-DHNA was the most potent compound among hydroxyl/carboxy naphthalene derivatives, and the fold induction response for CYP1A1 and CYP1B1 was similar to that observed for 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in YAMC and Caco2 cells. 1- and 4-HNA were less potent than 1,4-DHNA but induced maximal (TCDD-like) response for CYP1B1 (both cell lines) and CYP1A1 (Caco2 cells). With the exception of 1- and 2-NA, all compounds significantly induced Cyp1b1 in YAMC cells and these responses were not observed in AhR-deficient YAMC cells generated using CRISPR/Cas9 technology. In addition, we also observed that 1- and 2-NOH (and 1,4-DHNA) were weak AhR agonists, and 1- and 2-NOH also exhibited partial AhR antagonist activity. Structure–activity relationship studies for CYP1A1 but not CYP1B1 were similar in both cell lines, and CYP1A1 induction required one or both 1,4-dihydroxy substituents and activity was significantly enhanced by the 2-carboxyl group. We also used computational analysis to show that 1,4-DHNA and TCDD share similar interactions within the AhR binding pocket and differ primarily due to the negatively charged group of 1,4-DHNA.
Keywords: 1,4-DHNA; structure-activity; Ah receptor; agonists; antagonists.
Bifidobacteria are prominent in the gastrointestinal tract, and these bacteria and their metabolites have been associated with promotion of good health and are used as probiotic agents (Cani and Delzenne, 2011; Picard et al., 2005). For example, bifidobacteria alone or in combination with other bacterial species (eg, Lactobacillus) result in decreased inflammation associated with Crohn’s disease and ulcerative colitis (Furrie et al., 2005; Ishikawa et al., 2011; Kato et al., 2004; Wildt et al., 2011). It was reported that cell-free filtrate from the bifidobacteria Propionibacterium freudenreichii stimulates bifodobacterial growth and this is attributed primarily to 2 microbial metabolites, namely 2-amino-3-carboxy-1,4-naphthoquinone (minor) and 1,4-dihydroxy-2-naphthoic acid (1,4-DHNA) (major) (Isawa et al., 2002; Mori et al., 1997). 1,4-DHNA is an intermediate in the biosynthesis of menaquinone (vitamin K2) (Bentley and Meganathan, 1982), and 1,4-DHNA has also been identified in lactic acid-producing bacterial Lactobacillus casei LP1 and in Korea traditional rice wine (Eom et al., 2012; Kang et al., 2015). Subsequent studies showed that 1,4-DHNA inhibits dextran sodium sulfate (DSS)-induced colitis in mice and also decreases induced inflammation and colitis in interleukin 10-deficient mice by suppressing macrophage-derived pro-inflammatory cytokines (Okada et al., 2006, 2013). These results suggest that 1,4-DHNA contributes to the health-promoting effects of bifidobacteria. It was also reported that 1,4-DHNA inhibits growth of Helicobacter pylori and induces apoptosis in human keratinocytes, indicating a potential application for treating psoriasis (Mok et al., 2013; Nagata et al., 2010).
Several studies show that ligands for aryl hydrocarbon receptor (AhR) including TCDD and 1,4-DHNA are inhibitors of colitis and development of colorectal cancer in rodent models (Benson and Shepherd, 2011; Furumatsu et al., 2011; Monteleone et al., 2011; Singh et al., 2011). In vitro studies in human Caco2 colon cancer cells showed that 1,4-DHNA induces CYP1A1 gene expression, a marker of Ah responsiveness, and similar results were observed in the small intestine of wild type (wt) but not AhR knockout mice (Fukumoto et al., 2014). 1,4-DHNA also inhibits DSS-induced colitis and this response is attenuated after cotreatment with 1,4-DHNA plus CH-223191, an AhR antagonist (Fukumoto, et al., 2014). Thus, the health-promoting effects of 1,4-DHNA in the gut are due, in part, to its activity as an AhR agonist.
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a prototypical and highly potent AhR agonist and environmental toxicant, and several halogenated aromatic industrial compounds and by-products and polynuclear aromatic hydrocarbons also act through the AhR (Van den Berg et al., 2006). In addition, other classes of AhR ligands include endogenous biochemicals such as indolo-2,3[b]-carbazole, kynurenine and microbiota-derived tryptophan metabolites, health promoting phytochemicals, and pharmaceuticals (Cheng et al., 2015; Denison et al., 2011; Hubbard et al., 2015; Jin et al., 2014). The structure-dependent effects of TCDD and related halogenated aromatics have been extensively investigated; however, less is known about other structural classes of AhR ligands. Therefore, in this study we further elucidated the structure–activity relationships (SARs) of 1,4-DHNA and related naphthalene analogs as AhR ligands and demonstrate the important roles of both the hydroxyl- and carboxyl substituents and their location in the naphthalene ring. Moreover, since the structures of TCDD and 1,4-DHNA are different, we have also used computational modeling approaches to investigate differences in their interactions with the AhR.
MATERIALS AND METHODS
Cell lines, antibodies, and reagents
The young adult mouse colonic (YAMC) cell line was initially generated from the Immorto mouse (Whitehead et al., 1993) and has been previously used in our studies (Turk et al., 2011; Weige et al., 2009). Cells were maintained in RPMI 1640 medium with 5% fetal bovine serum (FBS), 5 U/ml mouse interferon-γ (IF005) (EMD Millipore, Massachusetts), 1% ITS “−” minus (insulin, transferrin, selenium) (41-400-045) (Life Technologies, Grand Island, New York) at 33 °C (permissive conditions). In preparation for experiments, cells were transferred to 37 °C (nonpermissive conditions). Caco2 human colon cancer cell line was obtained from the American Type Culture Collection (ATCC, Manassas, Virginia). Caco2 cells were maintained in Dulbecco’s modified Eagle’s medium nutrient mixture supplemented with 20% FBS, 10 ml/l 100× MEM nonessential amino acid solution (Gibco), and 10 ml/l 100× antibiotic/antimycotic solution (Sigma-Aldrich). Caco2 cells were maintained at 37 °C in the presence of 5% CO2, and the solvent (dimethyl sulfoxide, DMSO) used in the experiments was ≤ 0.2%. Mouse AhR antibody (BML-SA210) was purchased by Enzo (Enzo Life Sciences Inc., Farmingdale, New York). β-Actin (A1978) was purchased from Sigma-Aldrich (St Louis, Missouri), and mouse Cyp1a1 antibody was kindly provided by the late Dr Paul Thomas (Rutgers University) and Dr B. Moorthy (Baylor College of Medicine, Houston). Human CYP1A1, AHR, and GAPDH antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, California). 1,4-DHNA, 3,5-DHNA, 3,7-DHNA, 1,4-dimethoxy-2-naphthoic acid (1,4-DMNA), 1-naphthoic acid (1-NA), 2-naphthoic acid (2-NA), 1-naphthol (1-NOH), and 2-naphthol (2-NOH) used in this study were purchased from Sigma-Aldrich (St Louis, Missouri). 1-Hydroxy-2-naphthoic acid (1-HNA) was purchased from Alfa Aesar (Ward Hill, Massachusetts, USA) and 4-HNA was purchased from Chem Scene (www.chemscene.com).
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed using the ChIP-IT Express Magnetic Chromatin Immunoprecipitation kit (Active Motif, Carlsbad, California) according to the manufacturer’s protocol. YAMC cells (1.2 × 107 cells) were treated with TCDD and/or compounds for 2 or 24 h. Caco2 cells (5 × 106 cells) were treated with TCDD and/or compounds for 2 h. The cells were then fixed with 1% formaldehyde, and the cross-linking reaction was stopped by addition of 0.125 M glycine. After washing with phosphate-buffered saline, cells were scraped and pelleted. The collected cells were hypotonically lysed, and nuclei were collected and then sonicated to the desired chromatin length (200–1500 bp). The sonicated chromatin was immunoprecipitated with normal rabbit IgG or AhR antibodies and protein A-conjugated magnetic beads at 4 °C for overnight. After the magnetic beads were extensively washed, protein-DNA crosslinks were reversed and eluted. DNA was prepared by proteinase K digestion followed by polymerase chain reaction (PCR) amplification. The mouse Cyp1a1 primers were 5′-CAG GAG AGC TGG CCC TTT A-3′ (sense) and 5′-TAA GCC TGC TC ATC CTG TG-3′ (antisense), and subsequently amplified by targeting a 215-bp region of mouse Cyp1a1 promoter, which contained the AhR-binding sequences. The human CYP1A1 primers were 5′-TCA GGG CTG GGG TCG CAG CGC TTC T-3′ (sense) and 5′-GCT ACA GCC TAC CAG GAC TCG GCA G-3′ (antisense) which amplified a 112-bp region of the human Cyp1A1 promoter which containing the AhR-binding sequences. PCR products were resolved on a 2% agarose gel in the presence of ETBR.
Quantitative real-time PCR
Total RNA was isolated using Zymo Quick RNA MiniPrep Kit (Zymo Research, Irvine, California) according to the manufacturer’s protocol. RNA was eluted with RNase-free water and stored at −80 °C. Real-time (RT)-PCR was carried out using iTaq Universal SYBR Green One-step Kit (Bio-Rad, Hercules, California). The following primers were used.
Mouse
| Name | Forward Primer | Reverse Primer |
|---|---|---|
| TBP | GAACAATCCAGACTAGCAGCA | GGGAACTTCACATCACAGCTC |
| Cyp1a1 | ATCCAAGGCAGAATACGGTG | TCCACTCCATCTTCCGACTT |
| Cyp1b1 | GGATATCAGCCACGACGAAT | ATTATCTGGGCAAAGCAACG |
Human
| Name | Forward Primer | Reverse Primer |
|---|---|---|
| TBP | GATCAGAACAACAGCCTGCC | TTCTGAATAGGCTGTGGGGT |
| CYP1A1 | GACCACAACCACCAAGAAC | AGCGAAGAATAGGGATGAAG |
| CYP1B1 | CACTGACATCTTCGGCG | ACCTGATCCAATTCTGCCTG |
Western blot analysis
Cells were treated with different concentrations of the compounds for 18 h and then collected using high-salt buffer (50 mM HEPES, 0.5 M NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, and 1% Triton-X-100) and 10 mM Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO). Protein lysates were incubated for 5 min at 95 °C before electrophoresis and then separated on 10% SDS-polyacrylamide gel electrophoresis 120 V for 2–3 h. Proteins were transferred onto polyvinylidene difluoride membranes by wet electroblotting in a buffer containing 25 mM Tris, 192 mM glycine, and 20% methanol for 1.5 h at 180 mA. Membranes were then blocked for 30 min with specific antibodies. Detection of specific proteins was performed using Chemiluminescence and then exposed to Kodak image station 4000 mm Pro (Carestream Health, Rochester, New York).
Gel retardation
Gel retardation experiments were performed using guinea pig hepatic cytosol according to a previously published standard protocol (Soshilov and Denison, 2014a).
Generation of AhR-deficient YAMC cells
Two AhR CRISPR guide RNAs, in a Cas9 vector which also expresses GFP, were purchased from GenScript (Piscataway, New Jersey). Sequences of the guide RNAs were CGGTCTCTGTGTCGCTTAGA and GAACACAGAGTTAGACCGCC. YAMC cells were cotransfected with both plasmids and 48 h later, cells were FACS sorted to collect the 5% highest GFP expressing cells into individual wells of a 96-well plate. Clonal cells were grown into larger cultures and tested for knock-out of AhR protein.
Statistical analysis
Statistical significance of differences between the treatment groups was determined by an analysis of variance and/or Student’s t test, and levels of probability were noted. At least 3 repeated experiments were determined for each data point, and results are expressed as means ± SD.
Computational homology modeling of AhR
Residues 241 through 400 (sequence HGQNKKGKDG-ALLPPQLALF-AIATPLQPPS-ILEIRTKNFI-FRTKHKLDFT-PIGCDAKGQL-ILGYTEVELC-TRGSGYQFIH-AADMLHCAES-HIRMIKTGES-GMTVFRLFAK-HSRWRWVQSN-ARLIYRNGRP-DYIIATQRPL-TDEEGREHLQ-KRSTSLPFMF) of the mouse AhR were investigated through homology modeling. The homology model of the AhR was derived using I-TASSER (Yang and Zhang, 2015). All binding site residues characterized by mutagenesis studies and known to be critical or influence TCDD binding (Motto et al., 2011; Pandini et al., 2009; Xing et al., 2012), which were also investigated in a previous study (Bisson et al., 2009), are included in our model. The homology model was built using the crystal structure of the hypoxia-inducible factor-2α:AhR nuclear translocator complex (PDB ID: PZP4 [Wu et al., 2015], chain B) as an initial template. The N- and C- terminal ends of the modeled protein were acetylated and amidated to avoid any artifacts which could occur due to the artificial placement of positively and negatively charged groups at the backbone termini of the truncated ends of the modeled systems under investigation.
Generation of docking poses
TCDD and 1,4-DHNA were independently positioned into the binding sites of AhR using AutoDock Vina (Trott and Olson, 2010). The structures for both TCDD and 1,4-DHNA were obtained from the ZINC database (Irwin et al., 2012). The search space used was 20 × 24 × 20 Å so as to include in the binding pocket AhR residues involved in TCDD binding according to mutagenesis studies (Motto et al., 2011; Pandini et al., 2009; Xing et al., 2012). During the initial AutoDock Vina (Trott and Olson, 2010) docking, the side-chains of residues identified as binding pocket residues for TCDD binding to AhR according to mutagenesis studies (Motto et al., 2011; Pandini et al., 2009; Xing et al., 2012) were treated as flexible. The produced complex conformations of TCDD and 1,4-DHNA in complex with AhR with the lowest binding free energy according to AutoDock Vina (Trott and Olson, 2010) were used as initial structures for docking simulation runs performed in CHARMM (Brooks et al., 2009). Six separate docking simulation protocols were introduced independently to investigate the binding of TCDD and 1,4-DHNA to AhR. In summary, during the docking simulations each ligand independently was constrained using harmonic or quartic potential energy functions to the docked binding site through the MMFP module of CHARMM [42]; a quartic potential energy function was used so as to avoid bias toward the initial positioning of the molecules performed by Autodock Vina (Trott and Olson, 2010). In each 20 independent runs comprising 200 of short 2 ps simulations were performed. In each step, prior to the short MD simulation run, the ligands were independently rotated about a randomly generated axis and posterior to the short MD simulation run, the complex conformation was minimized and was saved for evaluation. This procedure resulted in the generation of 4000 binding conformations for each ligand in complex with AhR per protocol. Additional information on the protocols used in the docking simulation runs are provided in the Supplemental Materials. As an initial screening, from each docking simulation protocol, out of the 4000 complex structures produced, we extracted the 3 complex structures with the lowest interaction energy for further analysis. This translated to 18 docking conformations of TCDD in complex with AhR and 18 docking conformations of 1,4-DHNA in complex with AhR were extracted in total.
Molecular dynamics simulations of selected TCDD:AhR and 1,4-DHNA:AhR complexes
In order to refine the ligand:receptor structures, optimize intermolecular interactions, determine the structural stability of the selected binding modes, and assess the most energetically favored binding modes of the TCDD:AhR and 1,4-DHNA:AhR complexes, we performed 36 independent MD simulation runs of which the initial structures corresponded to the 18 selected TCDD:AhR docking conformations, and the 18 selected 1,4-DHNA: AhR docking conformations. All MD simulations of the 36 systems under investigation were performed in explicit solvent using CHARMM (Brooks, et al., 2009) and CHARMM36 topology and parameters (Best et al., 2012) with periodic boundary conditions. Additional information on the MD simulations is provided in the Supplemental Materials.
MM GBSA association free energy calculations
To identify the most energetically favorable conformation of TCDD and 1,4-DHNA in complex with AhR, we calculated the association free energy of the 18 complexes per ligand over the 10 ns production runs using the Molecular Mechanics Generalized Born Surface Area (MM GBSA approximation [Carney et al., 2008; Sambuy et al., 2005; Tamamis et al., 2012]), by extracting snapshots from the simulations every 20 ps. Additional information on the MM GBSA calculations is provided in the Supplemental Materials.
Selection and analysis of the binding modes with lowest association free energy
The simulations of the TCDD:AhR and 1,4-DHNA:AhR binding modes with the most favorable MM GBSA association free energies were selected as the ones representing the most likely naturally occurring binding conformations of the 2 ligands in complex with AhR; the selection was performed similarly to the studies as previously described (Tamamis and Floudas, 2014a,b,c; Tamamis et al., 2014). The selected simulations of both the TCDD:AhR and 1,4-DHNA:AhR binding modes were extended for an additional 20 ns, for a total production run of 30 ns each. To determine the stability of the ligands in the AhR binding pockets, the entire complexes were structurally aligned by the backbone atoms of the pocket residues and the RMSD of the ligand heavy atoms was calculated with respect to the average conformation over the entire 30 ns production run duration using VMD (Eargle et al., 2006; Humphrey et al., 1996). To determine the key interactions occurring in the lowest association free energy binding modes, the average per AhR residue interaction free energies between the AhR protein and each ligand of the structures with the lowest MM GBSA association free energies were calculated for the entire 30 ns production runs (Tamamis et al., 2010, 2011, 2012). Additional information on the per AhR residue interaction free energy calculations is provided in the Supplementary Materials.
RESULTS
Induction of Cyp1a1 and Cyp1b1 in Mouse YAMC Cells by TCDD and Naphthalene Compounds
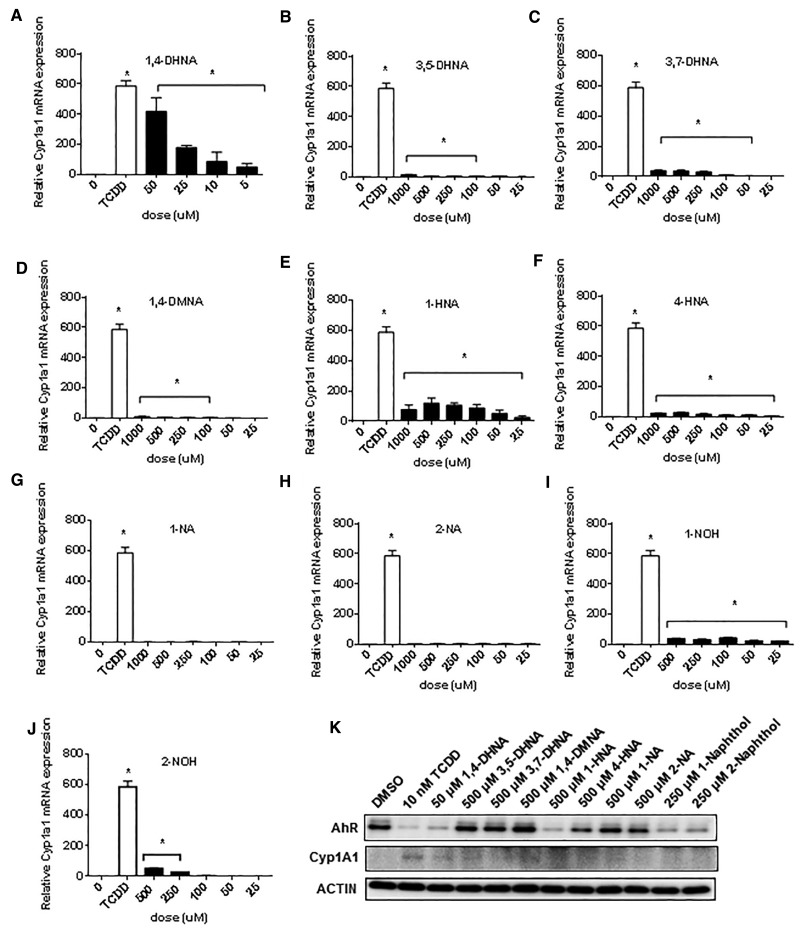
Both TCDD and 1,4-DHNA induce Cyp1a1 and inhibit DSS-induced colitis; however, 1,4-DHNA is approximately 3 orders of magnitude lower in potency (Benson and Shepherd, 2011; Fukumoto et al., 2014; Furumatsu et al., 2011). In contrast to the well-known SARs for dioxin-like compounds, the contributions of the hydroxyl and carboxylic acid substituents and their positions on the naphthalene ring has not previously been reported. In this study, we used mouse YAMC cells as a model for investigating the Ah-responsiveness of naphthalene derivatives on normal colon, and also human Caco2 colon cancer cells which are frequently used as an in vitro model for colonic responses and 1,4-DHNA induced AhR-dependent Cyp1a1 gene expression in this cell line has previously been reported (Fukumoto, et al., 2014). The substituted naphthalene derivatives used in this study included 1,4-DHNA, 3,5- and 3,7-DHNA, 1,4DMNA, 1-HNA, 4-HNA, 2-NA, 1-NA, 2-naphthol (2-NOH), and 1-NOH (Supplementary Figure S1). The effects of these compounds on YAMC and Caco2 cell viability are summarized in Supplementary Figure S2. In YAMC cells, 1,4-DHNA was the most cytotoxic of the substituted naphthalenes, and only 500 and 1000 μM 1- and 2-NOH were more cytotoxic than DHNA. Cyp1a1 induction is widely used as a marker of Ah-responsiveness, and 5-50 μM DHNA induced a concentration-dependent increase in Cyp1a1 mRNA levels (Figure 1A). TCDD (10 nM) induced approximately a 600-fold increase in Cyp1a1 mRNA levels, and the maximal induction by 1,4-DHNA (50 μM) was approximately 450-fold. In contrast, 3,5- and 3,7-DHNA exhibited minimal activity as Cyp1a1 inducers (15- to 40-fold lower induction than TCDD) (Figs. 1B and C), and even lower inducibility was observed for 1,4-DMNA (Figure 1D). Thus, maximal activity was observed for the 1,4-dihydroxy substitution and methylation of these hydroxyl groups resulted in loss of activity. Both 1-HNA and 4-HNA contain a single hydroxyl substituent and these compounds induced Cyp1a1 mRNA (1-HNA > 4-HNA) (Figs. 1E and F), whereas 1- and 2-NA (containing no hydroxyl groups) exhibited low to nondetectable induction (Figs. 1G and H). 1- and 2-NOH maximally induced a 43- and 50-fold enhancement of Cyp1a1 mRNA compared with the approximately 600-fold induction response observed for TCDD (Figs. 1I and J), indicating that loss of the 2-carboxyl substituent resulted in decreased activity. 1,4-Dihydroxynaphthalene is unstable in solution and is oxidized to the 1,4-quinone; however, induction of Cyp1a1 by this compound was also observed (Supplementary Figure S4). Western blot analysis showed that TCDD induced Cyp1a1 protein in YAMC cells with only minimal changes in protein levels by 1,4-DHNA, 1- and 4-HNA; however, TCDD, 1,4-DHNA, 1- and 4-HNA, 1- and 2- NOH decreased expression of the AhR protein (Figure 1K). TCDD, 1,4-DHNA, 1- and 4-HNA, 1- and 2-NOH treatment downregulated AhR expression and minimal effects were observed for the other analogs. Thus, the induction response (Cyp1a1) for 1,4-DHNA and related compounds was maximal for 1,4-DHNA, and the loss of one or both hydroxyl or carboxyl groups decreased potency.
FIG. 1.
Induction of Cyp1a1 in mouse YAMC cells. YAMC cells were treated with different concentrations of 1,4-DHNA (A), 3,5-DHNA (B), 3,7-DHNA (C), 1,4-DMNA (D), 1-HNA (E), 4-HNA (F), 1-NA (G), 2-NA (H), 1-NOH (I), and 2-NOH (J) for 18 h, and Cyp1a1 mRNA levels were determined (in triplicate) by real time PCR as outlined in the Materials and Methods. (K) Western blot analysis. YAMC cells were treated with a single concentration of the naphthalene compounds for 24 h, and whole cell lysates were then analyzed by western blots. TCDD (10 nM) was used a positive control. Significant (P < .05) induction is indicated (*).
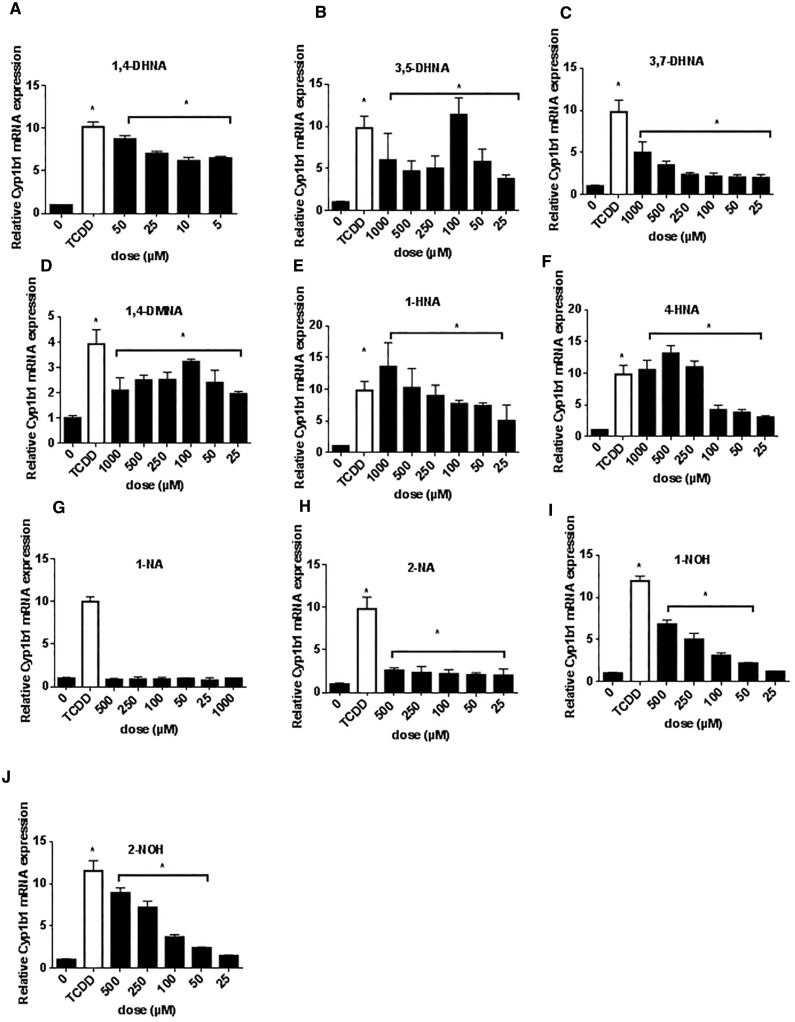
Basal levels of Cyp1b1 mRNA in YAMC cells were higher than observed for Cyp1a1, and 10 nM TCDD induced a 10-fold increase in Cyp1b1 mRNA in this cell line and 50 μM DHNA induced a similar fold induction response (Figure 2A). 3,5- and 3,7-DHNA, 1,4-DMNA, 1- and 4-HNA induced Cyp1b1 mRNA (Figs. 2B–F), and with the exception of 3,7-DHNA, the maximal induction response for the naphthalene derivatives was similar to that observed for TCDD. Both NAs (1-NA and 2-NA) exhibited minimal induction of Cyp1b1 mRNA (Figs. 2G and H), whereas 1- and 2-NOH induced Cyp1b1 levels > 50% of that observed for TCDD (Figs. 2I and 2J). Thus, the fold induction of Cyp1b1 by TCDD was much lower than observed for Cyp1a1 in YAMC cells and although the SARs for the naphthalene compounds were similar for both responses, their fold induction responses compared with TCDD were significantly higher for Cyp1b1 compared with Cyp1a1. The most striking difference in compound-induced gene expression was observed for 1,4-DMNA which did not induce Cyp1a1 but induced levels of Cyp1b1 mRNA >80% of the level observed for 10 nM TCDD (Figure 2D).
FIG. 2.
Induction of Cyp1b1 in mouse YAMC cells. YAMC cells were treated with different concentrations of 1,4-DHNA (A), 3,5-DHNA (B), 3,7-DHNA (C), 1,4-DMNA (D), 1-HNA (E), 4-HNA (F), 1-NA (G), 2-NA (H), 1-NOH (I), and 2-NOH (J) for 18 h and Cyp1b1 mRNA levels were determined (in triplicate) by real time PCR as outlined in the “Materials and Methods” section. Significant (P < .05) induction is indicated (*).
Induction of CYP1A1 and CYP1B1 in Human Caco2 Cells by TCDD and Naphthalene Compounds
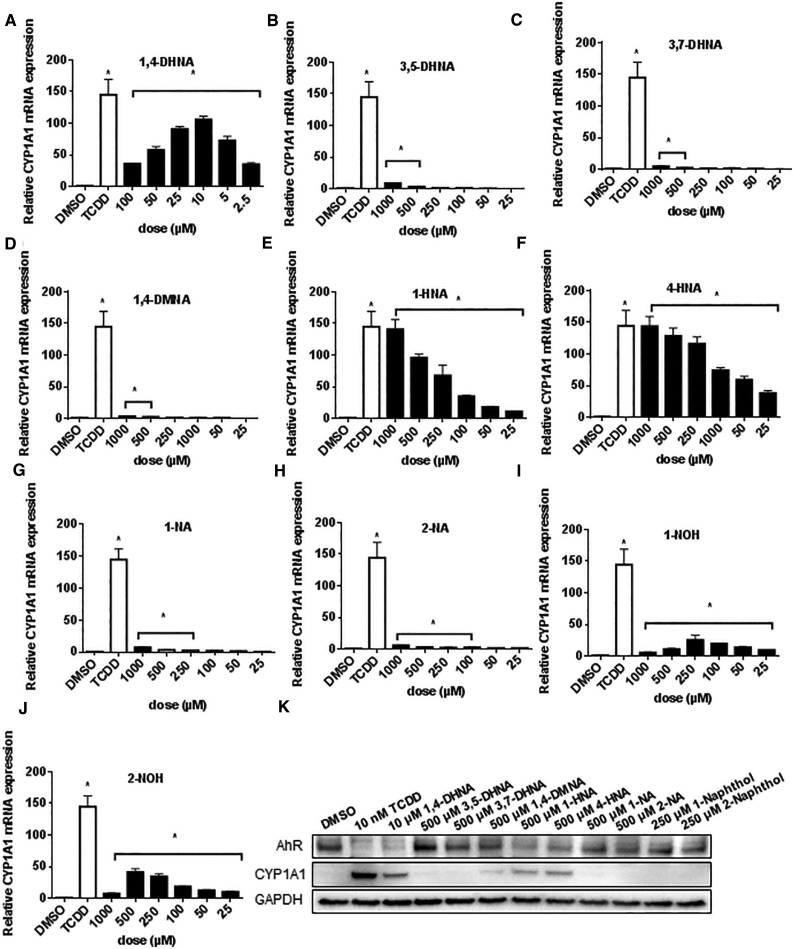
The SARs for 1,4-DHNA and structurally-related analogs were also carried out in human Caco2 cells, a colon cancer cell line used extensively as a model for investigating colonic effects of various drugs and dietary factors (Engle et al., 1998; Sambuy, et al., 2005). 1,4-DHNA (Figure 3A) but not 3,5-DHNA, 3,7-DHNA or 1,4-DMNA (Figs. 3B–D) induced CYP1A1 mRNA levels >50% of that observed for TCDD (140-fold induction). In contrast, both 1-HNA and 4-HNA maximally induced CYP1A1 mRNA (Figs. 3E and F); 1- and 2-NA (Figs. 3G and H) were relatively inactive, and both 1- and 2-NOH (Figs. 3I and J) induced <15% of the maximal response observed for TCDD. 1,4-Dihydroxynaphthalene also induced CYP1A1 in Caco2 cells (Supplementary Figure S4B). Western blot analysis showed that TCDD and 1,4-DHNA induced CYP1A1 protein, and both the 1- and 4-HNA compounds also induced these responses and all 4 compounds decreased AhR protein levels. TCDD, 1,4-DHNA, 1- and 4-HNA induced AhR downregulation and these results were different than their effects on the AhR in YAMC cells (Figure 1).
FIG. 3.
Induction of CYP1A1 in human Caco2 cells. Caco2 cells were treated with different concentrations of 1,4-DHNA (A), 3,5-DHNA (B), 3,7-DHNA (C), 1,4-DMNA (D), 1-HNA (E), 4-HNA (F), 1-NA (G), 2-NA (H), 1-NOH (I), and 2-NOH (J) for 18 h, and CYP1A1 mRNA levels were determined (in triplicate) by real time PCR as outlined in the “Materials and Methods” section. (K) Western blot analysis. Caco2 cells were treated with the various compounds for 24 h, and whole cell lysates were then analyzed by western blots. TCDD (10 nM) was used a positive control. Significant (P < .05) induction is indicated (*).
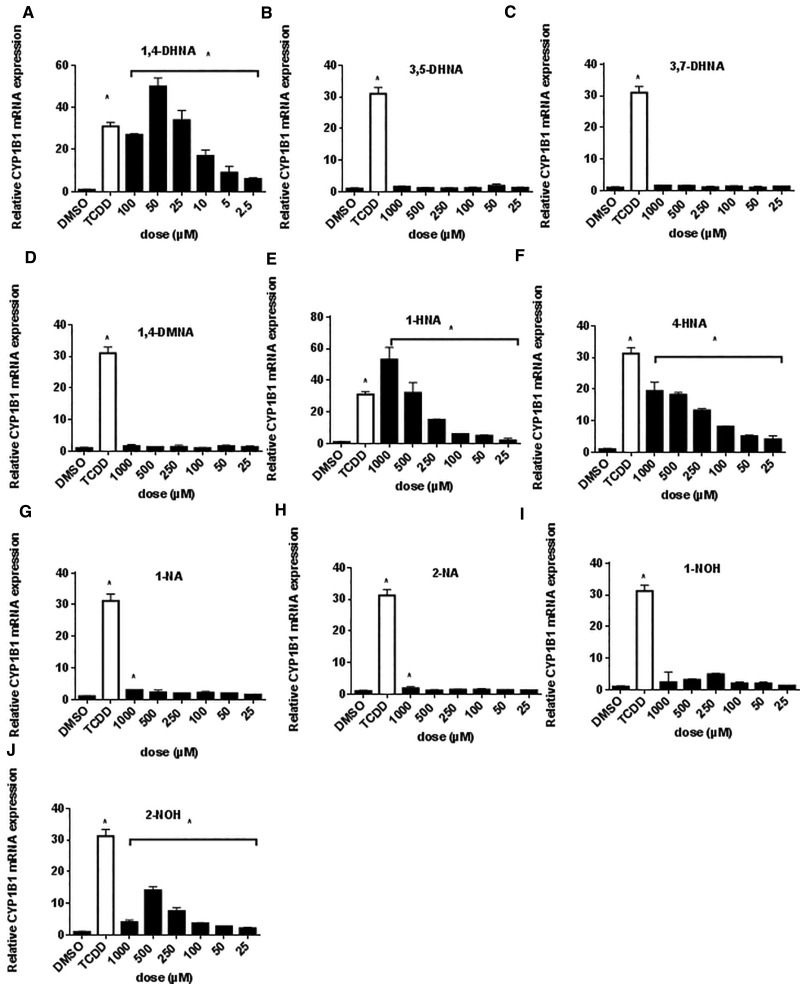
In Caco2 cells, TCDD induced a 31-fold induction of CYP1B1 compared with controls (Figure A), and only minimal to nondetectable induction was observed for 3,5-DHNA, 3,7-DHNA or 1,4-DMNA (Figs. 4B–D), whereas maximal induction responses were observed for 1- and 2- HNA (Figs. 4E and F). Minimal induction was observed for 1- and 2-NA (Figs. 4G and H), whereas both 1-NOH and 2-NOH induced CYP1B1 (50% of TCDD-induced response) (Figs. 4I and J) (2-NOH > 1-NOH). Supplementary Figure S5 compares the induction of CYP1A1/CYP1B1 in YAMC and Caco2 cells after treatment for 6 or 18 h. Induction responses for CYP1A1 (YAMC and Caco2) and CYP1B1 (Caco2) were significantly higher at the latter time point and this is consistent with previous studies on induction of CYP1A1 mRNA in this cell line (de Waard et al., 2008). In contrast, comparable compound-dependent induction of Cyp1b1 mRNA levels was observed YAMC cells after treatment for 6 and 18 h. The SARs for induction of CYP1A1 by 1,4-DHNA and related compounds (compared with TCDD) were similar in Caco2 and YAMC cells; however, both 1- and 4-HNA induction responses were lower in YAMC vs. Caco2 cells, suggestive of species differences in the AhR and/or cell-specific differences in metabolism of these compounds. In contrast, SARs for induction of Cyp1b1/CYP1B1 were highly variable and cell context-dependent for the naphthalene compounds.
FIG. 4.
Induction of CYP1B1 in human Caco2 cells. Caco2 cells were treated with different concentrations of 1,4-DHNA (A), 3,5-DHNA (B), 3,7-DHNA (C), 1,4-DMNA (D), 1-HNA (E), 4-HNA (F), 1-NA (G), 2-NA (H), 1-NOH (I), and 2-NOH (J) for 18 h, and CYP1B1 mRNA levels were determined (in triplicate) by real time PCR as outlined in the “Materials and Methods” section. Significant (P < .05) induction is indicated (*).
Effects of Naphthalene Compounds as AhR Antagonists and in Transformation of Guinea Pig Cytosol and AhR Dioxin-responsive Element (DRE) Localization in ChIP Assays
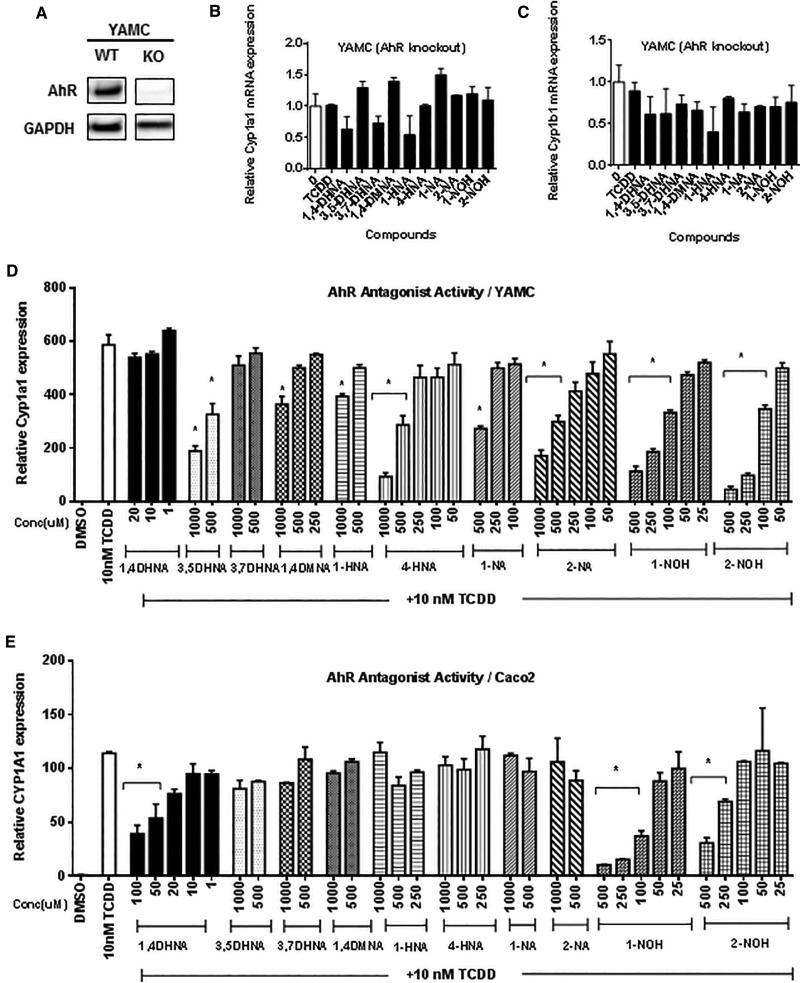
In order to confirm the application of mouse YAMC cells for investigating the Ah-responsiveness of compounds (Cheng et al., 2015), we used the CRISPR/Cas9 technology to generate AhR knockout cells and expression of AhR in one of these cell lines used in this study is illustrated in Figure 5A. Treatment of the knockout cells with TCDD, 1,4-DHNA and related compounds did not induce Cyp1a1 (Figure 5B) or Cyp1b1 (Figure 5C), confirming the AhR-dependence of the induction response in wt YAMC cells (Figs. 1 and 2). We also investigated the potential AhR antagonist activities of 1,4-DHNA and related compounds in mouse YAMC and human Caco2 cells by determining their inhibition of TCDD-induced Cyp1a1 gene expression (Figure 5D). In YAMC cells, all compounds, with the exception of 1,4- and 3,7-DHNA, inhibited TCDD-induced Cyp1a1 mRNA expression at one or more of the higher concentrations. However, AhR antagonist activity for these substituted naphthalenes was observed at concentrations that induced some level of cytotoxicity (Supplementary Figure S2). The only compound that did not act as an AhR antagonist (3,7-DHNA) was not cytotoxic. In contrast, with the exception of 1- and 2-NOH, this series of substituted naphthalenes exhibited minimal cytotoxicity in Caco2 cells (Supplementary Figure S3), suggesting that this cell line may be more suitable for determining AhR antagonist activity. The results (Figure 5E) show that 1,4-DHNA (20–100 μM) significantly inhibited TCDD-induced CYP1A1 gene expression in Caco2 cells, providing an explanation for the reduced reduction response at higher 1,4-DHNA concentrations (Figure 3A). Noncytotoxic concentrations of both 1- and 2-NOH (eg, 250 μM) also exhibited AhR antagonist activity with 1-NOH being highly active (>80% inhibition).
FIG. 5.
1,4-DHNA and related compounds do not activate AhR-deficient YAMC cells and their partial AhR antagonist activity. (A) AhR knockout YAMC cells. The AhR was knocked out (ko) in YAMC cells using CRISPR/Cas9, and expression of the AhR in wt and ko cells was determined by western blots as outlined in the Materials and Methods. ko-YAMC cells were treated with 10 nM TCDD, 10 μM 1,4-DHNA, 500 μM 3,5-DHNA, 3,7-DHNA, 1,4-DMNA, and 1-HNA, 4-HNA, 1-NA, 2-NA, and 250 μM 1-NOH and 2-NOH for 18 h and analyzed for expression of Cyp1a1 (B) and Cyp1b1 (C) mRNA (in triplicate) by real time PCR. YAMC (D) and Caco2 (E) cells were treated with 10 nM TCDD alone and in combination with the hydroxyl/NAs for 18 h, and Cyp1a1/CYP1A1 mRNA levels were determined (in triplicate) by real time PCR. Significant (P < .05) antagonist activity is indicated (*). In the YAMC knockout cells, 3,5-DHNA, 1,4-DMNA and 1-NA slightly induced (<50%) Cyp1a1 mRNA levels.
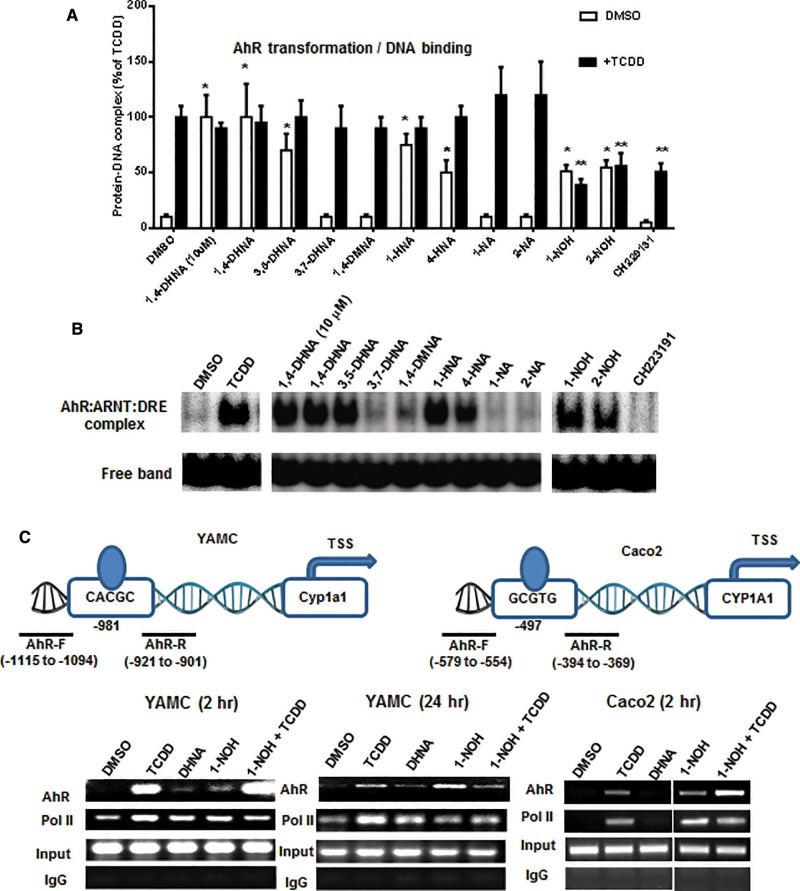
Figures 6A and B summarize the effects of 1,4-DHNA and related compounds on transformation of hepatic cytosol (guinea pig) to its DNA binding form in a gel mobility shift assay. 1,4-DHNA (10 and 100 μM) and 100 μM 3,5-DHNA and 1- and 4-HNA, 1- and 2-NOH alone significantly stimulated AhR transformation/DNA binding of guinea pig hepatic cytosol, whereas AhR transformation/DNA binding was not observed for 3,7-DHNA, 1,4-DMNA, 1- and 2-NA. In combination studies with TCDD, only 1- and 2-NOH inhibited TCDD-induced transformation and this correlated with the AhR antagonist activity observed in the transactivation assays (Figs. 5D and E). As a positive control, we show that the AhR antagonist CH229131 did not stimulate AhR transformation/DNA binding of guinea pig cytosol but inhibited TCDD-induced transformation/DNA binding. These AhR transformation/DNA binding assays confirmed that 1,4-DHNA, 1- and 4-HNA which induced CYP1A1 and CYP1B1 also induced AhR transformation. The results obtained for 3,5-DHNA were somewhat surprising based on the lack of CYP1A1 induction; however, this compound induced Cyp1b1 in YAMC cells (Figure 2). The effects of 1- and 2-NOH (100 μM) alone and in combination with TCDD show that both compounds alone induced transformation/DNA binding but also inhibited TCDD-induced transformation/DNA binding to an extent similar to that observed for the well-characterized AhR antagonist CH229131. These data further confirm the partial AhR agonist/antagonist activities observed for 1- and 2-NA for induction of Cyp1a1 in colon cells.
FIG. 6.
Effects of 1,4-DHNA and related compounds on transformation and DNA binding of guinea pig cytosol and ChIP analysis of the Cyp1a1 promoter. A, Transformation and DNA binding of guinea pig hepatic cytosolis AhR. DMSO (solvent control), 10 μM CH229131, 10 and 100 μM 1,4-DHNA, and 100 μM concentrations of the remaining compounds alone and in combination with TCDD were incubated with guinea pig cytosol and analyzed by gel mobility shift assays as outlined in the Materials and Methods. B, A representative gel showing the ligand-induced transformed AhR-DRE complex is illustrated in this panel. C, ChIP assay. YAMC and Caco2 cells were treated with various compounds and analysis of recruitment of pol II and the AhR to the Cyp1a1/CYP1A1 promoters was determined in a ChIP assay as outlined in the “Materials and Methods” section.
We also examined the effects of TCDD, 1,4-DHNA and 1-NOH alone and in combination with TCDD in a ChIP assay. Treatment with TCDD for 2 h resulted in the recruitment of the AhR and pol II to the DRE region of the CYP1A1 promoter in Caco2 and YAMC cells (Figure 6C). Similar effects were observed for 1-NA and 1,4-DHNA in YAMC and for 1-NOH (but not 1,4-DHNA) in Caco2 cells. Surprisingly, results observed in cells treated with 1-NOH plus TCDD for 2 h appeared to be additive with respect to AhR interactions with the Cyp1a1 promoter. These unexpected ligand-induced effects after treatment for 2 h were further investigated in YAMC cells after treatment for 24 h. TCDD and to a lesser extent 1-NOH recruited the AhR and pol II to the Cyp1a1 promoter, and in the combination treatment (1-NOH plus TCDD), the 1-NOH compound decreased the effects of TCDD on AhR and pol II recruitment. 1-NOH alone which induced minimal expression of Cyp1a1 mRNA (Figure 1I) recruited relatively high levels of the AhR to the Cyp1a1 promoter. Currently, we are further investigating both the time- and compound-dependent recruitment of the AhR, pol II and other nuclear cofactors, including coactivators and corepressors, to the DRE region of the Cyp1a1 and other Ah-responsive genes.
Modeling of TCDD and 1,4-DHNA Interactions with the AhR in Order to Identifying the Most Energetically Favored Binding Conformations
We introduced the MM GBSA approximation and identified the TCDD:AhR and 1,4-DHNA:AhR-binding modes which acquire the lowest MM GBSA association free energy across all 18 simulated TCDD:AhR and 1,4-DHNA:AhR-binding modes, respectively. The average association free energies of the simulated TCDD:AhR and 1,4-DHNA:AhR-binding modes are tabulated in Supplementary Tables S1 and S2, respectively. The simulations encompassing the most energetically favored binding conformations of TCDD:AhR and 1,4-DHNA:AhR according to MM GBSA were both derived from the docking protocols using quartic potential energy functions and are the most likely to correspond to the naturally occurring binding conformations.
Structural Stability of Binding Modes
The stability of lowest association free energy binding modes of TCDD and 1,4-DHNA in complex with AhR was confirmed through RMSD calculations over the 30 ns simulation production runs. The RMSD of the heavy atoms of TCDD in complex with AhR is 0.9 ± 0.5 Å with respect to the average structure of TCDD in complex with AhR, and the RMSD of heavy atoms of 1,4-DHNA in complex with AhR is 0.9 ± 0.5 Å with respect to the average structure of 1,4 in complex with AhR.
Interactions between TCDD and AhR
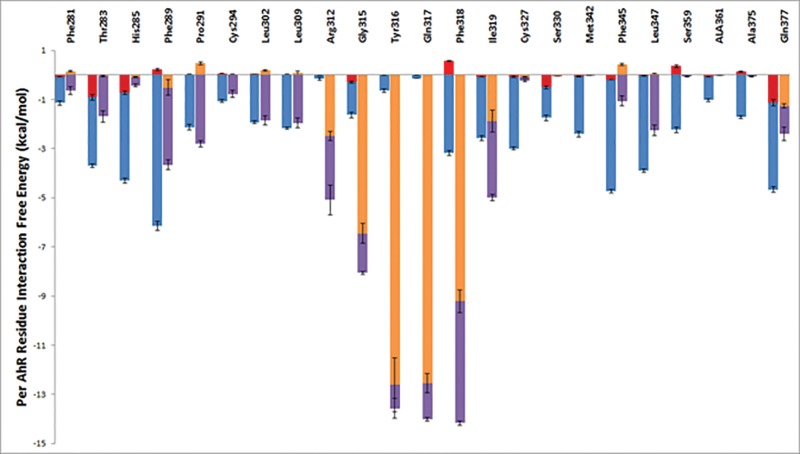
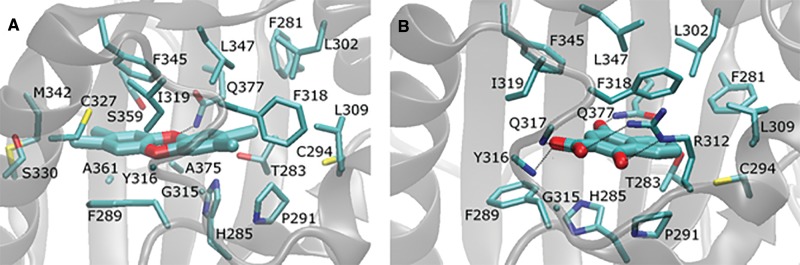
The average per-residue interaction free energy between AhR residues and TCDD are decomposed into polar and nonpolar contributions, and selected interactions are presented in Figure 7. Residues contributing the most interaction free energies are presented in Figure 8A. In the TCDD:AhR-binding mode, the medial oxygen atom of TCDD forms a hydrogen bond with the NE group of Gln377, indicated with a black dotted line in Figure 8A. As predicted by previous studies (Motto et al., 2011; Pandini et al., 2009; Xing et al., 2012), the TCDD-binding pocket of AhR is highly hydrophobic, and the binding of TCDD in AhR is primarily stabilized by nonpolar interactions (Figure 8A). Residues Phe289, Cys327, Met342, Ile319, Phe345, and Leu347 form hydrophobic walls around the left side of the ligand, in the perspective of Figure 8A. The aromatic rings residues Phe289, Phe345, and, less frequently, Phe318 form π-π interactions with the aromatic rings of TCDD. Van der Waals interactions are also formed between TCDD and the side-chain atoms of residues Phe281, Thr283, His285, Pro291, Cys294, Leu302, Leu309, Ile319, Cys327, Ser330, Met342, Leu347, Ser359, Ala361, and Ala375 as well as the backbone atoms of residues Gly315 and Tyr316 due to their close proximity to the bound TCDD molecule.
FIG. 7.
Average interaction free energies (kcal/mol) decomposed into polar (red or orange) and nonpolar (blue or purple) contributions for AhR interacting residues in complex with TCDD (first bar per residue) and in complex with DHNA (second bar per residue). The sum of polar and nonpolar contributions corresponds to the total average per AhR residue interaction free energy. Only residues with at < −0.5 kcal/mol average interaction free energy are presented. Results were calculated using the ensemble of snapshots extracted from simulation trajectories of the most energetically favored binding conformations. The average and standard deviation values for the polar and nonpolar components of the interaction free energies were calculated over 4 “measurements”, where the first, second, third, and fourth measurement corresponds to the individual average interaction free energy components of the first, second, third, and fourth 7.5 ns segment of the 30 ns MD simulation production run.
FIG. 8.
Molecular graphics images of TCDD (A) and 1,4-DHNA (B) in complex with AhR, which correspond to snapshots extracted from the most energetically favored binding conformations. The ligand molecules are shown in licorice representation in both panels. Interacting AhR protein residues are shown in thin licorice representation, and the entire AhR protein is shown in transparent, gray new cartoon representation. Hydrogen bonds are indicated using black dotted lines. Q377 forms hydrogen bonds with both TCDD and 1,4-DHNA.
Interactions between 1,4-DHNA and AhR
As with the TCDD:AhR complex, the average per residue interaction free energy between AhR residues and 1,4-DHNA are decomposed into polar and nonpolar contributions, and selected interactions are presented in Figure 7. Residues with the largest associated interaction free energy contributions are presented in Figure 8B. The hydroxyl group of 1,4-DHNA farthest from its carboxylic acid group forms a hydrogen bond with the NE group of Gln377; the hydroxyl group of 1,4-DHNA closest to its carboxylic acid group forms a hydrogen bond with the NE group of Arg312, and the oxygen atoms of the carboxylic acid group of 1,4-DHNA forms hydrogen bonds with the backbone amino groups of Tyr316 and Gln317 as well as a low interacting salt-bridge with the NH group of Arg312. These hydrogen bonds are indicated using black dotted lines in Figure 8B. Hydrophobic residues Phe281, Phe289, Pro291, Cys294, Leu302, Leu309, Phe318, Ile319, Phe345, and Leu347 predominantly form the 1,4-DHNA binding pocket of AhR (Figure 8B). The aromatic rings of residues Phe318 and, occasionally, Phe289 participate in π–π interactions with the aromatic rings of 1,4-DHNA. Due to their close proximity, strong van der Waals interactions are formed between 1,4-DHNA and the side-chains of residues Phe281, Thr283, Pro291, Cys294, Leu302, Leu309, Ile319, Phe345, and Leu347 as well as the backbone atoms of Gly315 and Tyr316.
DISCUSSION
TCDD and structurally-related halogenated aromatic compounds have been characterized as widespread and persistent environmental contaminants, and risk assessment of dioxin-like compounds have been developed and are based on well-known SARs (Denison et al., 2011; Van den Berg et al., 2006). In contrast, SARs for other structural classes of AhR ligands including pharmaceuticals, microbiota-derived compounds such as 1,4-DHNA, endogenous AhR ligands, and food-derived compounds have not been determined and despite some insights on their intake and levels of exposure, it is difficult to predict their potencies and also their interactions with “dioxin-like” compounds.
Among PAHs, naphthalene is not an AhR ligand (Till et al., 1999) and therefore the 1- and 4-hydroxyl and carboxylic acid groups are responsible for the activity of 1,4-DHNA which induces near maximal expression (compared with 10 nM TCDD) of Cyp1a1 and Cyp1b1 in YAMC and Caco2 cells. The loss of both hydroxyl groups to give 2-NA (or 1-NA) or replacement with methoxyl substituent (1,4-DMNA) resulted in significant loss of Cyp1a1/Cyp1b1 inducibility in both cell lines. Both 3,5- and 3,7-DHNA exhibited minimal activity as Cyp1a1 inducers in both cell lines; however, in YAMC but not Caco2 cells, these compounds induced Cyp1b1 expression, indicating that the requirement of the dihydroxy groups was gene-dependent and specific to the cell context of YAMC and Caco2 cell lines. Differences in SARs for induction of CYP1B1 may also be species-dependent and due not only to differences between the human and mouse AhR but also in expression of cofactors. Hydroxy NAs are bacterial metabolites of PAHs, and previous studies on their developmental toxicity in medaka fish embryos show that 1-HNA was not only the most toxic compound but also induced Cyp1a1 expression in medaka and mouse Hepa1c1c7 liver cancer cell lines (Carney et al., 2008). We also observed that both 1- and 4-HNA induced Cyp1a1/Cyp1b1 in YAMC and Caco2 cells and with the exception of relative low induction of Cyp1a1 in YAMC cells, their maximal induction responses were comparable to 1,4-DHNA and TCDD but 5-20 fold less potent than the former compounds. We further confirmed the role of the AhR in mediating compound-induced Cyp1a1 and Cyp1b1 using AhR knockout YAMC cells obtained using the CRISPR/Cas9 technology (Figure 5). It was interesting to note that compounds such as 3,5-DHNA, 3,7-DHNA and 1,4-DMNA that exhibited minimal to nondetectable induction of Cyp1a1 were effective inducers of Cyp1b1 (>50% of the response induced by 10 nM TCDD). The differential induction of 2 AhR-responsive genes in the same cell line is typical of selective AhR modulators and due, in part, to gene/promoter histone and chromatin differences and is currently being investigated.
1,4-Dihydroxynaphthalene is readily oxidized to the quinone; however, our results show that although this dihydroxy/quinone mixture is cytotoxic, we also observed induction of CYP1A1 in Caco2 and YAMC cells (Supplementary Figure S4). Thus, the loss of the carboxyl group from 1,4-DHNA to give 1,4-dihydroxynaphthalene decreases but does not abrogate AhR activity of this compound. 1- and 2-NOH are biomarkers of human exposure to PAH and smoking (Jain, 2016; Nethery et al., 2012; Sudakin et al., 2013; Sul et al., 2012; Wilhelm et al., 2008), and both compounds exhibited weak induction of Cyp1a1/CYP1A1 (YAMC and Caco2) and CYP1B1 (Caco2) mRNA; however, Cyp1b1 was induced to >50% of the maximal TCDD-induced response in YAMC cells. Both 1- and 2-NOH inhibited TCDD-induced Cyp1a1 gene expression in YAMC and Caco2 cells (Figs. 5D and E), and their AhR antagonist activity was also observed in the gel mobility shift assay (Figure 6A), suggesting that 1- and 2-NOH represent a new class of partial AhR antagonists. We also observed some inconsistencies in the effects of 1-NOH in ChIP assays in YAMC and Caco2 cells (Figure 6C) with respect to recruitment of the AhR to the DRE region of the Cyp1a1 promoter, and this is currently being investigated.
The most energetically favored binding mode of TCDD in complex with AhR according to our computational studies is in strong accordance with previous experimental and computational studies examining TCDD binding to AhR. Recent experiments suggest that residues Phe318, Ile319, and Ala375 are key to ligand selectivity (Soshilov and Denison, 2014b). Other mutagenesis studies revealed that substitutions in residues Thr283, His285, Phe289, Pro291, Leu302, Leu309, Cys327, Phe345, and Leu347 result in a reduction in TCDD binding (Motto et al., 2011; Pandini et al., 2009; Soshilov and Denison, 2011). Finally, both computational methods and further mutagenesis studies performed on Gln377 support the contention that the polar side-chain atoms of Gln377 form hydrogen bonds with the medial oxygen of TCDD (Xing et al., 2012). The excellent agreement of our work in comparison to previous studies support the validity of the computational protocol introduced here, and suggest that the in silico identified binding modes most likely represent the naturally occurring binding modes of TCDD and 1,4-DHNA with residues in the AhR ligand binding pocket
The presence of more polar groups which include the negatively charged carboxylic group in 1,4-DHNA compared with TCDD contributes in general to stronger polar interactions between 1,4-DHNA and AhR compared with TCDD and AhR. Although in the TCDD:AhR complex, only one hydrogen bond is formed with the NE group of Gln377 and the medial oxygen of TCDD, 1,4-DHNA forms a hydrogen bond with the NE group of Gln377, the NE group of Arg312, and the backbone amino groups of Tyr316 and Gln317. Additionally, the negatively charged group of 1,4-DHNA forms a low interacting salt-bridge with the NH group of Arg312. The binding of both TCDD and 1,4-DHNA in AhR is also stabilized by nonpolar interactions. Both the aromatic rings of TCDD and 1,4-DHNA form π-π interactions with the aromatic rings of Phe289 and Phe318. Both TCDD and 1,4-DHNA also form strong van der Waals interactions with the side-chains of residues Phe281, Thr283, Pro291, Cys294, Leu302, Leu309, Ile319, Phe345, and Leu347 as well as the backbone atoms of Gly315 and Tyr316. Alanine mutagenesis has shown that Thr283, Phe289, Leu302, Leu309 Phe318, Ile319, Phe345, and Leu347 are critical for TCDD binding (Motto et al., 2011; Soshilov and Denison, 2011). These similarities are largely due to TCDD and 1,4-DHNA sharing the same binding site and both ligands containing aromatic rings. Owing to the smaller size of DHNA, and thus a lesser amount of possible van der Waals interaction and π–π interaction sites, interactions between 1,4-DHNA and the side-chains of residues His285, Cys327, Ser330, Met342, Ser359, Ala361, and Ala375 are weaker compared with the TCDD:AhR complex. Thus, the modeling studies show that both 1,4-DHNA and TCDD interact within the same binding pocket of the AhR but 1,4-DHNA binds with lower affinity. This observation is consistent with similar efficacies of 1,4-DHNA and TCDD for induction of Cyp1a1 but differences in their potencies. To further understand the differences between both 1,4-DHNA and TCDD in comparison to a minimally active compound, 3,7-DHNA, we performed a preliminary study of the latter in complex with AhR using a similar strategy to the one used in the study of TCDD and 1,4-DHNA, with the only difference that the simulation entailing the binding mode with the lowest association free energy was not extended for an additional 20 ns. In comparison with both 1,4-DHNA and TCDD, 3,7-DHNA forms weaker interactions with Thr283, Pro291, Cys294, Leu302, Leu309, and Phe318. Alanine mutagenesis experiments suggest that Thr283, Leu302, Leu309 and Phe318 are critical to TCDD binding (Motto et al., 2011; Soshilov and Denison, 2011), and Phe318 has been shown to be an “agonist/antagonist switch” (Soshilov and Denison, 2014b).
In summary, this study confirms that 1,4-DHNA is a relatively potent AhR agonist in both YAMC and Caco2 cells; however, the potential impact of endogenous 1,4-DHNA alone or in combination with other AhR agonists/antagonists on gut health is unknown and in the future, we hope to more accurately quantitate these compounds and determine their combined effects. Structure activity studies show that both the hydroxyl and carboxyl groups and their positions on the naphthalene ring are important for AhR activity. Both TCDD and 1,4-DHNA are ligands for the AhR and predominantly form strong interactions with the same AhR residues. More polar interactions occur in the AhR:1,4-DHNA complex in comparison to the AhR:TCDD complex due, in part, to differences in overall charge and substituent interactions with various amino acid side-chains. Our results demonstrate that for some of the hydroxyl NA analogs, there are differences in their activation of Cyp1a1 versus Cyp1b1 in the same cell line and also differences in the mouse (YAMC) versus human (Caco2) colon-derived cells. However, 1,4-DHNA induced >70% of the response observed for 10 nM TCDD for Cyp1a1 and Cyp1b1 in both the mouse and human cell lines. This suggests possible efficacy for 1,4-DHNA in humans as an AhR agonist that is protective in the gut; however, this will require more definitive proof from dietary studies and manipulation of gut microorganisms.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This work was supported by National Institutes of Health (R01-ES025713, R01-ES007685, P30-ES023512, R35-CA197707, and R01-CA202697); Cancer Prevention Research Institute of Texas; Texas AgriLife Research; the Office of Graduate and Professional Studies and the Artie McFerrin Department of Chemical Engineering at Texas A&M University; and the Sid Kyle Endowment.
Supplementary Material
REFERENCES
- Benson J. M., Shepherd D. M. (2011). Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn's disease. Toxicol. Sci. 120, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R., Meganathan R. (1982). Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 46, 241–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., Mackerell A. D., Jr. (2012). Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson W. H., Koch D. C., O'Donnell E. F., Khalil S. M., Kerkvliet N. I., Tanguay R. L., Abagyan R., Kolluri S. K. (2009). Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 52, 5635–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. (2009). CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614. 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D., Delzenne N. M. (2011). The gut microbiome as therapeutic target. Pharmacol. Ther. 130, 202–212. 10.1016/j.pharmthera.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Carney M. W., Erwin K., Hardman R., Yuen B., Volz D. C., Hinton D. E., Kullman S. W. (2008). Differential developmental toxicity of naphthoic acid isomers in medaka (Oryzias latipes) embryos. Mar. Pollut. Bull. 57, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Jin U. H., Allred C. D., Jayaraman A., Chapkin R. S., Safe S. (2015). Aryl hydrocarbon receptor activity of tryptophan metabolites in young adult mouse colonocytes. Drug Metab. Dispos. 43, 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waard P. W., de Kok T. M., Maas L. M., Peijnenburg A. A., Hoogenboom R. L., Aarts J. M., van Schooten F. J. (2008). Influence of TCDD and natural Ah receptor agonists on benzo[a]pyrene-DNA adduct formation in the Caco-2 human colon cell line. Mutagenesis 23, 67–73. [DOI] [PubMed] [Google Scholar]
- Denison M. S., Soshilov A. A., He G., DeGroot D. E., Zhao B. (2011). Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eargle J., Wright D., Luthey-Schulten Z. (2006). Multiple Alignment of protein structures and sequences for VMD. Bioinformatics 22, 504–506. [DOI] [PubMed] [Google Scholar]
- Engle M. J., Goetz G. S., Alpers D. H. (1998). Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell Physiol. 174, 362–369. [DOI] [PubMed] [Google Scholar]
- Eom J. E., Kwon S. C., Moon G. S. (2012). Detection of 1,4-dihydroxy-2-naphthoic acid from commercial Makgeolli products. Prev. Nutr. Food Sci. 17, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S., Toshimitsu T., Matsuoka S., Maruyama A., Oh-Oka K., Takamura T., Nakamura Y., Ishimaru K., Fujii-Kuriyama Y., Ikegami S., et al. (2014). Identification of a probiotic bacteria-derived activator of the aryl hydrocarbon receptor that inhibits colitis. Immunol. Cell Biol. 92, 460–465. [DOI] [PubMed] [Google Scholar]
- Furrie E., Macfarlane S., Kennedy A., Cummings J. H., Walsh S. V., O'Neil D. A., Macfarlane G. T. (2005). Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: A randomised controlled pilot trial. Gut 54, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumatsu K., Nishiumi S., Kawano Y., Ooi M., Yoshie T., Shiomi Y., Kutsumi H., Ashida H., Fujii-Kuriyama Y., Azuma T., et al. (2011). A role of the aryl hydrocarbon receptor in attenuation of colitis. Dig. Dis. Sci. 56, 2532–2544. [DOI] [PubMed] [Google Scholar]
- Hubbard T. D., Murray I. A., Perdew G. H. (2015). Indole and tryptophan metabolism: Endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 43, 1522–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). VMD: Visual molecular dynamics. J. Mol. Graph 14, 33–38. 27-8. [DOI] [PubMed] [Google Scholar]
- Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S., Coleman R. G. (2012). ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model 52, 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa K., Hojo K., Yoda N., Kamiyama T., Makino S., Saito M., Sugano H., Mizoguchi C., Kurama S., Shibasaki M., et al. (2002). Isolation and identification of a new bifidogenic growth stimulator produced by Propionibacterium freudenreichii ET-3. Biosci. Biotechnol. Biochem. 66, 679–681. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Matsumoto S., Ohashi Y., Imaoka A., Setoyama H., Umesaki Y., Tanaka R., Otani T. (2011). Beneficial effects of probiotic bifidobacterium and galacto-oligosaccharide in patients with ulcerative colitis: A randomized controlled study. Digestion 84, 128–133. [DOI] [PubMed] [Google Scholar]
- Jain R. B. (2016). Association between polycyclic aromatic hydrocarbons and thyroid function among males and females: Data from NHANES 2007-2008. Int. J. Environ. Health Res. 26, 405–419. [DOI] [PubMed] [Google Scholar]
- Jin U. H., Lee S. O., Sridharan G., Lee K., Davidson L. A., Jayaraman A., Chapkin R. S., Alaniz R., Safe S. (2014). Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol. Pharmacol. 85, 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. E., Kim T. J., Moon G. S. (2015). A novel Lactobacillus casei LP1 producing 1,4-dihydroxy-2-naphthoic acid, a bifidogenic growth stimulator. Prev. Nutr. Food Sci. 20, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Mizuno S., Umesaki Y., Ishii Y., Sugitani M., Imaoka A., Otsuka M., Hasunuma O., Kurihara R., Iwasaki A., et al. (2004). Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol. Ther. 20, 1133–1141. [DOI] [PubMed] [Google Scholar]
- Mok C. F., Xie C. M., Sham K. W., Lin Z. X., Cheng C. H. (2013). 1,4-Dihydroxy-2-naphthoic acid Induces apoptosis in human keratinocyte: Potential application for psoriasis treatment. Evid. Based Complement Alternat. Med. 2013, 792840.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone I., Rizzo A., Sarra M., Sica G., Sileri P., Biancone L., MacDonald T. T., Pallone F., Monteleone G. (2011). Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248. [DOI] [PubMed] [Google Scholar]
- Mori H., Sato Y., Taketomo N., Kamiyama T., Yoshiyama Y., Meguro S., Sato H., Kaneko T. (1997). Isolation and structural identification of bifidogenic growth stimulator produced by Propionibacterium freudenreichii. J. Dairy Sci. 80, 1959–1964. [DOI] [PubMed] [Google Scholar]
- Motto I., Bordogna A., Soshilov A. A., Denison M. S., Bonati L. (2011). New aryl hydrocarbon receptor homology model targeted to improve docking reliability. J. Chem. Inf. Model 51, 2868–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Inatsu S., Tanaka M., Sato H., Kouya T., Taniguchi M., Fukuda Y. (2010). The bifidogenic growth stimulator inhibits the growth and respiration of Helicobacter pylori. Helicobacter 15, 422–429. [DOI] [PubMed] [Google Scholar]
- Nethery E., Wheeler A. J., Fisher M., Sjodin A., Li Z., Romanoff L. C., Foster W., Arbuckle T. E. (2012). Urinary polycyclic aromatic hydrocarbons as a biomarker of exposure to PAHs in air: A pilot study among pregnant women. J. Expo. Sci. Environ. Epidemiol. 22, 70–81. [DOI] [PubMed] [Google Scholar]
- Okada Y., Tsuzuki Y., Miyazaki J., Matsuzaki K., Hokari R., Komoto S., Kato S., Kawaguchi A., Nagao S., Itoh K., et al. (2006). Propionibacterium freudenreichii component 1,4-dihydroxy-2-naphthoic acid (DHNA) attenuates dextran sodium sulphate induced colitis by modulation of bacterial flora and lymphocyte homing. Gut 55, 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Tsuzuki Y., Narimatsu K., Sato H., Ueda T., Hozumi H., Sato S., Hokari R., Kurihara C., Komoto S., et al. (2013). 1,4-Dihydroxy-2-naphthoic acid from Propionibacterium freudenreichii reduces inflammation in interleukin-10-deficient mice with colitis by suppressing macrophage-derived proinflammatory cytokines. J. Leukoc. Biol. 94, 473–480. [DOI] [PubMed] [Google Scholar]
- Pandini A., Soshilov A. A., Song Y., Zhao J., Bonati L., Denison M. S. (2009). Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry 48, 5972–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Fioramonti J., Francois A., Robinson T., Neant F., Matuchansky C. (2005). Review article: Bifidobacteria as probiotic agents – physiological effects and clinical benefits. Aliment Pharmacol. Ther. 22, 495–512. [DOI] [PubMed] [Google Scholar]
- Sambuy Y., De Angelis I., Ranaldi G., Scarino M. L., Stammati A., Zucco F. (2005). The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 21, 1–26. [DOI] [PubMed] [Google Scholar]
- Singh N. P., Singh U. P., Singh B., Price R. L., Nagarkatti M., Nagarkatti P. S. (2011). Activation of aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of experimental colitis. PLoS One 6, e23522–e23510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshilov A., Denison M. S. (2011). Ligand displaces heat shock protein 90 from overlapping binding sites within the aryl hydrocarbon receptor ligand-binding domain. J. Biol. Chem. 286, 35275–35282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshilov A. A., Denison M. (2014a). DNA binding (Gel Retardation Assay) analysis for identification of aryl hydrocarbon (Ah) receptor agonists and antagonists In Optimization in Drug Discovery: In Vitro Methods (Caldwell G. W., Yan Z., Eds.), pp. 207–219. Springer Science, New York. [Google Scholar]
- Soshilov A. A., Denison M. S. (2014b). Ligand promiscuity of aryl hydrocarbon receptor agonists and antagonists revealed by site-directed mutagenesis. Mol. Cell Biol. 34, 1707–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin D. L., Smit E., Cardenas A., Harding A. (2013). Naphthalene biomarkers and relationship with hemoglobin and hematocrit in White, Black, and Hispanic adults: Results from the 2003-2004 National Health and Nutrition Examination Survey. J. Med. Toxicol. 9, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul D., Ahn R., Im H., Oh E., Kim J. H., Kim J. G., Kim P., Kim H. A., Park W. Y., Son B. S., et al. (2012). Korea National Survey for Environmental Pollutants in the human body 2008: 1-hydroxypyrene, 2-naphthol, and cotinine in urine of the Korean population. Environ. Res. 118, 25–30. [DOI] [PubMed] [Google Scholar]
- Tamamis P., Floudas C. A. (2014a). Elucidating a key anti-HIV-1 and cancer-associated axis: The structure of CCL5 (Rantes) in complex with CCR5. Sci. Rep. 4, 5447.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Floudas C. A. (2014b). Elucidating a key component of cancer metastasis: CXCL12 (SDF-1alpha) binding to CXCR4. J Chem. Inf. Model 54, 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Floudas C. A. (2014c). Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One 9, e95767.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Kieslich C. A., Nikiforovich G. V., Woodruff T. M., Morikis D., Archontis G. (2014). Insights into the mechanism of C5aR inhibition by PMX53 via implicit solvent molecular dynamics simulations and docking. BMC Biophys. 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Lopez de Victoria A., Gorham R. D., Jr., Bellows-Peterson M. L., Pierou P., Floudas C. A., Morikis D., Archontis G. (2012). Molecular dynamics in drug design: New generations of compstatin analogs. Chem. Biol. Drug Des. 79, 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Morikis D., Floudas C. A., Archontis G. (2010). Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins 78, 2655–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Pierou P., Mytidou C., Floudas C. A., Morikis D., Archontis G. (2011). Design of a modified mouse protein with ligand binding properties of its human analog by molecular dynamics simulations: The case of C3 inhibition by compstatin. Proteins 79, 3166–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till M., Riebniger D., Schmitz H. J., Schrenk D. (1999). Potency of various polycyclic aromatic hydrocarbons as inducers of CYP1A1 in rat hepatocyte cultures. Chem. Biol. Interact. 117, 135–150. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk H. F., Kolar S. S., Fan Y. Y., Cozby C. A., Lupton J. R., Chapkin R. S. (2011). Linoleic acid and butyrate synergize to increase Bcl-2 levels in colonocytes. Int. J. Cancer 128, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg M., Birnbaum L. S., Denison M., De Vito M., Farland W., Feeley M., Fiedler H., Hakansson H., Hanberg A., Haws L., et al. (2006). The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. 93, 223–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weige C. C., Allred K. F., Allred C. D. (2009). Estradiol alters cell growth in nonmalignant colonocytes and reduces the formation of preneoplastic lesions in the colon. Cancer Res. 69, 9118–9124. [DOI] [PubMed] [Google Scholar]
- Whitehead R. H., VanEeden P. E., Noble M. D., Ataliotis P., Jat P. S. (1993). Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 90, 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt S., Nordgaard I., Hansen U., Brockmann E., Rumessen J. J. (2011). A randomised double-blind placebo-controlled trial with Lactobacillus acidophilus La-5 and Bifidobacterium animalis subsp. lactis BB-12 for maintenance of remission in ulcerative colitis. J. Crohns. Colitis 5, 115–121. [DOI] [PubMed] [Google Scholar]
- Wilhelm M., Hardt J., Schulz C., Angerer J., Human Biomonitoring Commission of the German Federal Environment A. (2008). New reference value and the background exposure for the PAH metabolites 1-hydroxypyrene and 1- and 2-naphthol in urine of the general population in Germany: Basis for validation of human biomonitoring data in environmental medicine. Int. J. Hyg. Environ. Health 211, 447–453. [DOI] [PubMed] [Google Scholar]
- Wu D., Potluri N., Lu J., Kim Y., Rastinejad F. (2015). Structural integration in hypoxia-inducible factors. Nature 524, 303–308. [DOI] [PubMed] [Google Scholar]
- Xing Y., Nukaya M., Satyshur K. A., Jiang L., Stanevich V., Korkmaz E. N., Burdette L., Kennedy G. D., Cui Q., Bradfield C. A. (2012). Identification of the Ah-receptor structural determinants for ligand preferences. Toxicol. Sci. 129, 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang Y. (2015). I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.