Abstract
It has been hypothesized that low frequency (1–5% minor allele frequency (MAF)) and rare (<1% MAF) variants with large effect sizes may contribute to the missing heritability in complex traits. Here, we report an association analysis of lipid traits (total cholesterol, LDL-cholesterol, HDL-cholesterol triglycerides) in up to 27 312 individuals with a comprehensive set of low frequency coding variants (ExomeChip), combined with conditional analysis in the known lipid loci. No new locus reached genome-wide significance. However, we found a new lead variant in 26 known lipid association regions of which 16 were >1000-fold more significant than the previous sentinel variant and not in close LD (six had MAF <5%). Furthermore, conditional analysis revealed multiple independent signals (ranging from 1 to 5) in a third of the 98 lipid loci tested, including rare variants. Addition of our novel associations resulted in between 1.5- and 2.5-fold increase in the proportion of heritability explained for the different lipid traits. Our findings suggest that rare coding variants contribute to the genetic architecture of lipid traits.
Introduction
Genome-wide association studies (GWAS) have identified hundreds of mainly common variants that are robustly associated with cardiometabolic traits (1–4). For lipid levels, a series of large-scale meta-analyses (N > 100 000) identified a total of 164 independent single nucleotide polymorphisms (SNPs) in 159 loci contributing to variation in plasma concentrations of total cholesterol (TC), triglycerides (TG), low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) (2,3,5–8). Blood lipid levels have an estimated heritability of 40–70% (9); however, variants reaching genome-wide significance explain only ∼15% of the heritable fraction for these traits (2,3). The clinical relevance of these 164 SNPs of which 71 associate with more than one lipid trait is underscored by an overall excess of significant association signals for coronary artery disease (CAD), fasting glucose, type 2 diabetes, blood pressure traits and body mass index among them (2).
It has been hypothesized that low frequency (1–5% minor allele frequency (MAF)) and rare (<1% MAF) variants with larger effects may account in part for the missing heritability in complex traits (10,11). To test this hypothesis in relation to lipid traits we used the Illumina HumanExome Beadchip (ExomeChip), an array which provides comprehensive coverage of low frequency coding variants (non-synonymous, splice-site and stop altering), to profile 27 312 individuals. The Exome array also includes most of the lead (or a proxy) GWAS variants in the 159 known lipid loci which allowed to assess the independent contribution of additional, mainly low frequency, coding variants in these loci by performing conditional analysis with the GCTA software.
Results
Single-marker analysis
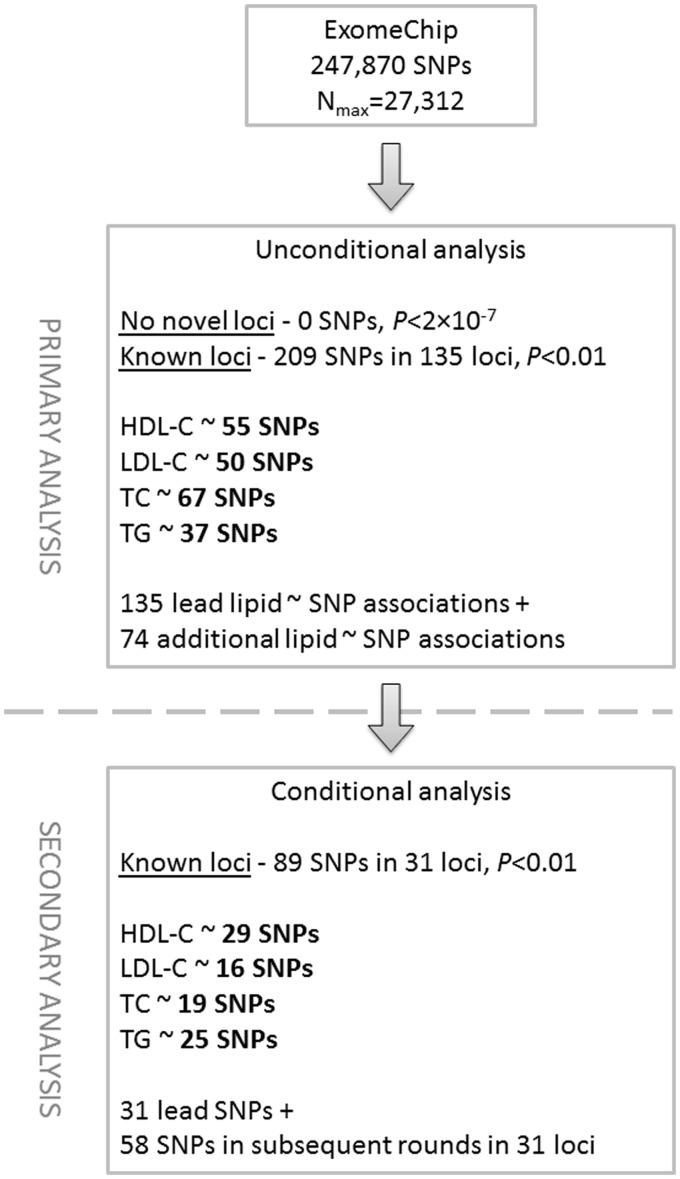
In our single-marker meta-analysis of ExomeChip (Illumina) data in 27 312 individuals (an overview of the study design is given in Fig. 1), we did not find any new variant associated with a lipid trait at either a genome-wide threshold of significance (P < 5 × 10−8) or an array-wide threshold of significance (P < 2 × 10−7) outside the 159 previously reported loci (considering a 1 Mb window centred on the sentinel SNP).
Figure 1.
Flow diagram of the study.
The 159 unique loci known to be associated with one or more lipid traits represent 247 association signals (73 for HDL-C, 58 for LDL-C, 74 for TC and 42 for TG) (3,6–8, 12). The ExomeChip array does not have a good proxy of the reported sentinel SNP (2,3) in 21 of the 159 lipid loci (see Materials and Methods). In our study, we detected 209 association signals with a lipid trait (55 for HDL-C, 50 for LDL-C, 67 for TC and 37 for TG) at P < 0.01 (nominal significance; direction of effect same as published (2)) in 135 of the 159 unique lipid loci (Supplementary Material, Table S1). Of the remaining 24 loci, 9 had no lead SNP or a proxy on the ExomeChip, 4 had the lead SNP or proxy fail QC, and 11 did not show a nominal association in our study.
Further assessing the results of our single-marker analysis, we found that in about half (n = 98) of the 209 association signals (26 for HDL-C, 17 for LDL-C, 35 for TC and 20 for TG), the lead SNP was either the published one or a highly linked proxy (r2 > 0.8; Supplementary Material, Table S1); in two instances, the proxy is a putative functional variant (rs2792751 in GPAM for LDL-C and rs35332062 in MLXIPL for TG) (Supplementary Material, Table S1). In many loci, our top hit was different than the previously published lead SNP (this study) and not in close LD (r2 < 0.8) (Supplementary Material, Table S1). Table 1 lists the 27 most significant of these associations (P < 10−4) of which eight are due to low frequency or rare coding variants. Interestingly, for 16 of these 27 association signals, the new sentinel variant was >1000-fold more significant than the previously published one. These results were also corroborated with one exception by conditional and joint analyses (see below); SNP rs5015480, a downstream variant in CYP26A1 previously reported for T2D (13), was not significant in the joint analysis. For the remaining 11 signals, the new sentinel variant was only marginally more significant than the previously published one (includes two low frequency variants in LPA and KDM2B).
Table 1.
New sentinel SNPs in known lipid loci (P < 10−4) showing stronger association than the previously reported variant
|
New lead SNP |
Published lead SNP |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Locus (gene; amino acid change) | rsID | Trait- raising/ other allele | Publication status | %EAF | N | β | SE | P-value | rsID (r2)a | Trait- raising/ other allele | %EAF | N | β | SE | P-value | |
| New lead SNP >1000-fold more significant than the published lead SNP | |||||||||||||||||
| HDL | LIPG (LIPG;N/S) | rs779603474,5 | G/A | A | 1.29 | 25 373 | 0.289 | 0.040 | 6.42E-13 | rs72419186 (0.006) | T/G | 83.82 | 25 371 | 0.075 | 0.012 | 7.78E-10 | |
| ANGPTL4 (ANGPTL4;E/K) | rs1168430646,5 | A/G | B | 1.88 | 24 406 | 0.254 | 0.034 | 6.79E-14 | rs72554363,2,14(0.02) | A/C | 51.66 | 25 375 | 0.021 | 0.009 | 1.95E-02 | ||
| LIPC | rs18005881,2 | T/C | A | 21.89 | 25 375 | 0.138 | 0.011 | 1.12E-36 | rs15320851,2(0.0004) | A/G | 40.26 | 25 374 | 0.109 | 0.009 | 1.16E-31 | ||
| APOA1 | rs22667881,8 | A/G | A | 91.66 | 25 375 | 0.115 | 0.016 | 3.29E-12 | rs9641841,8(0.5) | C/G | 80.45 | 25 372 | 0.098 | 0.021 | 3.04E-06 | ||
| TRIB1 | rs29540331 | G/A | A | 70.47 | 25 374 | 0.041 | 0.010 | 3.29E-05 | rs29540291,2,10(0.4) | T/A | 44.76 | 25 375 | 0.016 | 0.009 | 6.92E-02 | ||
| APOE | rs7694491,2,10,16 | G/A | B | 88.08 | 25 375 | 0.100 | 0.014 | 7.39E-13 | rs44206381,11(0.6) | A/G | 81.15 | 21 172 | 0.076 | 0.013 | 1.80E-09 | ||
| LDL | APOE (APOE;R/C) | rs74124,5 | C/T | A | 92.79 | 24 361 | 0.561 | 0.029 | 1.43E-80 | rs44206381,11(0.02) | G/A | 18.80 | 20 158 | 0.200 | 0.013 | 3.15E-54 | |
| PCSK9 (PCSK9;(R/L) | rs115911471,4,5 | G/T | A | 98.35 | 24 361 | 0.461 | 0.036 | 1.93E-37 | rs24794091,13(0.009) | G/A | 33.61 | 24 361 | 0.042 | 0.010 | 2.14E-05 | ||
| CILP2 | rs23041301,2,12 | A/G | A | 91.06 | 24 360 | 0.099 | 0.016 | 9.01E-10 | rs104019691,2,14(0.5) | T/C | 91.92 | 22 547 | 0.119 | 0.028 | 1.62E-05 | ||
| APOA1 | rs20752901,2 | C/T | A | 8.26 | 27 312 | 0.120 | 0.016 | 3.52E-14 | rs9641841,8(0.5) | G/C | 19.26 | 27 309 | 0.097 | 0.020 | 2.26E-06 | ||
| TC | FRMD5 (MAP1A;P/L) | rs557071004,5 | T/C | A | 2.60 | 26 483 | 0.186 | 0.028 | 6.87E-11 | rs29292821,2(0.4) | T/A | 4.86 | 26 473 | 0.063 | 0.021 | 2.64E-03 | |
| APOE (APOE;R/C) | rs74124,5 | C/T | B | 92.73 | 27 312 | 0.423 | 0.017 | 7.43E-145 | rs44206381,11(0.02) | G/A | 19.09 | 23 109 | 0.164 | 0.012 | 1.96E-42 | ||
| PCSK9 (PCSK9;R/L) | rs115911471,4,5 | G/T | B | 98.39 | 27 312 | 0.406 | 0.034 | 5.42E-32 | rs24794091,13(0.009) | G/A | 33.96 | 27 312 | 0.039 | 0.009 | 2.57E-05 | ||
| MPP3 (CD300LG;R/C) | rs728365614,5 | T/C | C | 2.69 | 26 484 | 0.133 | 0.027 | 1.21E-06 | rs80778891,11(0.007) | C/A | 20.33 | 26 484 | 0.005 | 0.011 | 6.69E-01 | ||
| APOA1 | rs22667881,8 | G/A | A | 8.34 | 26 484 | 0.265 | 0.016 | 5.97E-61 | rs9641841,8(0.5) | G/C | 19.45 | 26 481 | 0.230 | 0.037 | 7.99E-10 | ||
| WASF5P_HLAB (CDSN) | rs30942162,7,10, 16 | A/G | C | 77.60 | 26 484 | 0.052 | 0.011 | 3.66E-06 | rs22470561,2,10(0.08) | C/T | 75.18 | 26 484 | 0.028 | 0.011 | 1.04E-02 | ||
| New lead SNP nominally more significant than the published lead SNP | |||||||||||||||||
| HDL | ABCA1 | rs39050001,2 | G/A | A | 86.23 | 25 374 | 0.095 | 0.013 | 4.85E-13 | rs18830251,2(0.3) | C/T | 73.92 | 25 374 | 0.074 | 0.010 | 6.85E-13 | |
| MLXIPL | rs11789791,8 | C/T | B | 18.21 | 25 375 | 0.055 | 0.012 | 3.10E-06 | rs171457381,11(0.5) | T/C | 12.03 | 25 375 | 0.056 | 0.014 | 6.07E-05 | ||
| COBLL1 | rs133892191,15 | T/C | B | 38.05 | 25 374 | 0.037 | 0.009 | 9.09E-05 | rs123286751,8(0.2) | C/T | 12.08 | 25 371 | 0.042 | 0.014 | 2.36E-03 | ||
| LDL | BRAP (SH2B3;W/R) | rs31845041,4,5 | C/T | A | 56.67 | 24 361 | 0.040 | 0.010 | 3.28E-05 | rs110659871,6(0.7) | A/G | 61.63 | 24 361 | 0.036 | 0.010 | 2.04E-04 | |
| MYLIP (MYLIP;N/S) | rs93708674,5 | A/G | A | 50.43 | 24 158 | 0.038 | 0.009 | 3.52E-05 | rs37573541,15(0.3) | C/T | 76.91 | 24 359 | 0.035 | 0.011 | 1.18E-03 | ||
| LPA (LPA;I/M) | rs37982201,4,5 | C/T | C | 1.57 | 24 361 | 0.154 | 0.037 | 3.42E-05 | rs15643481,2(0.001) | C/T | 16.55 | 24 361 | 0.032 | 0.012 | 9.31E-03 | ||
| TC | APOB | rs5410411,6 | A/G | A | 83.48 | 27 312 | 0.141 | 0.012 | 2.29E-33 | rs13671174,5(0.06) | A/G | 31.96 | 27 312 | 0.112 | 0.009 | 6.19E-33 | |
| CETP | rs15326241,2, 15 | A/C | A | 44.13 | 27 311 | 0.040 | 0.009 | 5.88E-06 | rs37642611,13(0.6) | A/C | 32.17 | 27 312 | 0.041 | 0.009 | 1.03E-05 | ||
| HNF1A (KDM2B;L) | rs346065627,10,16 | C/A | C | 99.99 | 6885 | 2.810 | 0.708 | 7.20E-05 | rs11692881,4,5,14(0.000009) | C/A | 34.67 | 27 307 | 0.028 | 0.009 | 2.73E-03 | ||
| TG | FTO | rs37518132,9 | T/G | C | 55.18 | 24 410 | 0.037 | 0.009 | 5.77E-05 | rs11219801,2(0.6) | A/G | 43.11 | 26 484 | 0.027 | 0.009 | 2.56E-03 | |
| CYP26A1 | rs50154801,11,17 | C/T | C | 55.75 | 26 480 | 0.038 | 0.009 | 2.62E-05 | rs20688881,11(0.00003) | G/A | 53.36 | 26 483 | 0.037 | 0.009 | 3.97E-05 | ||
Publication status a, previously published variants for the investigated and other lipid traits; publication status b, previously published variants for lipid traits other than the investigated one; publication status c, new variants (not published for any lipids trait).
aPair wise LD estimation between the ExomeChip lead SNP and the published lead SNP was based on the reference panel used for the conditional analysis (n = 11 396 samples).
1NHGRI catalogue,
2intron variant,
3independent secondary hit in the conditional analysis,
4non-synonymous variant,
5missense variant,
6intergenic variant,
7synonymous variant,
83′ prime UTR variant,
9common variant,
10non-coding transcript variant,
11downstream gene variant,
12splice region variant,
13upstream gene variant,
14nonsense-mediated mRNA decay transcript variant,
15regulatory region variant,
16non-coding exon variant,
17did not reach significance threshold in joint analysis (Pj > 0.05; see text).
Among the 27 association signals (Table 1), four variants had not been previously associated with a lipid trait. Two of them had a 1000-fold more significant association for TG levels than the previous sentinel SNP: rs72836561 a missense variant in CD300LG (p.R82C) and rs3094216 a synonymous variant (p.C448=) in CDSN. The other two variants were rs3751813 (TG association) an intronic variant in the FTO gene andrs34606562 (TC association) a synonymous change (p.L1174=) in KDM2B both of which overlap a strong peak for H3K27Ac which marks active regulatory elements. In the LPA locus, the missense variant rs3798220 (p.I1891M) previously associated with Lp(a) lipoprotein levels and CAD (14) had the strongest signal for LDL-C.
In 15 of the 135 lipid loci with an association signal in our data (P < 0.01), we lacked the published lead SNP or a proxy (12 not on the ExomeChip and 3 QC failures; Supplementary Material, Table S1, column AJ) and therefore we were unable to undertake a direct comparison of the strength of association between our top hit and the published one. However, in one such locus, ABCA8, which is associated with HDL-C (2,3), we found a missense variant (rs77542162; Cys1319Arg; 1.57% MAF) associated with both LDL-C (P = 6.40 × 10−13) and TG (P = 6.23 × 10−11) but not HDL-C (P = 0.46) located in the ATP-binding cassette, sub-family A (ABC1), member 6 (ABCA6) gene (Supplementary Material, Table S1). ABCA6 encodes a membrane-associated protein and is located together with ABCA8 and three other ABC1 family members on 17q24. ABCA6 may play a role in macrophage lipid homeostasis (15) and in intercellular lipid transport processes in vascular endothelial cells (16).
Conditional analysis
We next undertook conditional analysis which looks for association signals that are independent of the lead SNP from the unconditional analysis, in the 135 unique loci harbouring 209 lipid association signals at a nominal significance level of P < 0.01 (boundaries are listed in Supplementary Material, Table S2) using the GCTA software (17) and meta-analysis summary statistics from all 27 312 samples. We considered a signal from the conditional analysis to be significant if it passed a Bonferroni correction threshold based on the number of SNPs tested across the locus examined (Supplementary Material, Table S2). Therefore only loci which had a lead SNP with Punconditional less than the locus-wide Bonferroni threshold for multiple testing (i.e. based on the number of tested SNPs per locus) were amenable to conditional analysis. Based on the threshold calculated for each locus (Supplementary Material, Table S2), it was possible to examine 98 of the 209 lipid association regions for a secondary signal (see Materials and Methods; Supplementary Material, Table S1). We found 31 (31.6%) of these association regions to have at least one additional independent signal. In total, we identified 89 independent signals (29 for HDL-C, 16 for LDL-C, 19 for TC and 25 for TG) in the 31 association regions (Table 2 and Supplementary Material, Table S3) corresponding to the 31 sentinel SNPs from the unconditional analysis and 58 SNPs from the subsequent rounds of conditional analysis. The largest number of independent signals per locus was 5, in the APOA1 locus for HDL-C as well as in the APOB and APOE loci for LDL-C (the latter illustrated in Supplementary Material, Fig. S1). Approximately 30% of the 89 independent variants were either low frequency (13; 14.6%) or rare variants (14; 15.7%) (Supplementary Material, Table S1).
Table 2.
New variants identified as independent signals in the approximate conditional analysis
|
Single SNP meta-analysis |
Approximate conditional meta-analysis |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chromosome: position | rsID | Trait-raising/ other allele | %EAF | β | SE | P-value | N | β | SE | P-value | Conditioned on | Locus |
| HDL | 4:102816487 | rs1163291291,4 | T/C | 99.97 | 1.835 | 0.448 | 4.23E-05 | 9023 | 1.842 | 0.448 | 3.99E-05 | rs13107325 | SLC39A8 |
| 11:117112526 | rs795541102,3 | T/G | 37.94 | 0.038 | 0.009 | 5.69E-05 | 24 813 | 0.034 | 0.009 | 2.16E-04 | rs2266788, rs35120633, rs186808413 | APOA1 | |
| 11:116707044 | rs1384071551,4 | A/T | 99.88 | 0.589 | 0.143 | 3.98E-05 | 20 910 | 0.505 | 0.143 | 4.09E-04 | rs2266788, rs35120633, rs186808413, rs79554110 | APOA1 | |
| 15:58830639 | rs1400297291,4 | A/G | 99.92 | 1.019 | 0.273 | 1.88E-04 | 8544 | 0.963 | 0.273 | 4.14E-04 | rs1800588, rs10468017 | LIPC | |
| 15:58837989 | rs2006843241,4 | A/G | 0.13 | 0.760 | 0.223 | 6.62E-04 | 7941 | 0.721 | 0.223 | 1.23E-03 | rs1800588, rs10468017, rs140029729 | LIPC | |
| 18:47113165 | rs1176236311,4 | T/C | 0.22 | 0.389 | 0.116 | 8.36E-04 | 17 028 | 0.390 | 0.116 | 8.08E-04 | rs77960347, rs4939883 | LIPG | |
| 19:45448465 | rs51671,4,5 | G/T | 36.02 | 0.042 | 0.009 | 6.19E-06 | 25 337 | 0.046 | 0.009 | 8.70E-07 | rs769449 | APOE | |
| 19:45028231 | rs360532771,4 | G/A | 99.62 | 0.304 | 0.079 | 1.14E-04 | 22 141 | 0.289 | 0.079 | 2.43E-04 | rs769449, rs5167 | APOE | |
| LDL | 6:161018174 | rs77706283,8 | C/T | 45.47 | 0.030 | 0.009 | 1.38E-03 | 23 595 | 0.035 | 0.009 | 2.03E-04 | rs3798220 | LPA |
| 12:121660770 | rs1458147491,4 | G/A | 99.91 | 0.768 | 0.205 | 1.85E-04 | 13 946 | 0.768 | 0.205 | 1.85E-04 | rs34606562 | HNF1A | |
| 19:45448036 | rs11328991,4,5 | T/C | 47.49 | 0.033 | 0.009 | 3.37E-04 | 24 201 | 0.037 | 0.009 | 4.71E-05 | rs7412, rs769449, rs445925, rs3208856 | APOE | |
| TC | 2:21229160 | rs57429041,4 | T/C | 0.05 | 1.676 | 0.260 | 2.53E-11 | 13 764 | 1.679 | 0.260 | 2.38E-11 | rs541041, rs1367117, rs533617 | APOB |
| TG | 2:27550967 | rs104981711 | A/G | 59.83 | 0.053 | 0.009 | 5.42E-09 | 26 031 | −0.030a | 0.007 | 2.54E-05 | rs1260326 | GCKR |
| 11:116701353 | rs763532039,10 | C/T | 99.96 | 1.258 | 0.320 | 8.60E-05 | 11 860 | 1.279 | 0.320 | 6.58E-05 | rs2266788, rs35120633 | APOA1 | |
| 15:42436237 | rs1397889071,4 | G/A | 0.04 | 1.665 | 0.421 | 7.67E-05 | 6731 | 1.668 | 0.421 | 7.49E-05 | rs2412710 | CAPN3 | |
| 16:57015091 | rs58801,4 | C/G | 5.09 | 0.085 | 0.021 | 3.85E-05 | 24 330 | 0.078 | 0.021 | 1.33E-04 | rs3764261 | CETP | |
Non-synonymous variant,
common variant,
intron variant,
missense variant,
nonsense-mediated mRNA decay transcript variant,
non-coding exon variant,
non-coding transcript variant,
NHGRI catalogue,
stop gained variant,
splice region variant,
synonymous rs7770628 has been previously published for LPA eQTL.
Change in the direction of the conditional effect.
Out of the 58 additional signals identified from the conditional analysis, 42 have previously been associated with a lipid trait (34 reported for the investigated lipid trait and 8 for a lipid trait other than the investigated one; Supplementary Material, Table S3) and 16 have not been previously associated with any lipid trait (Table 2). Among the 34 lipid signals previously reported, two variants, rs439401 in APOE (TC) and rs35120633 in APOA1 (TG and HDL-C), showed an increase in their effect size after conditioning for the top hit in the corresponding region. In both instances, the very strong association signal of the lead SNP in the region (e.g. rs7412 P = 7.43 × 10−145 in the APOE locus) appears to partially mask the weaker secondary signal. In the unconditional analysis, SNP rs439401 had an effect size (β) of −0.051 per T allele (P = 1.77 × 10−8) whereas after conditioning on rs7412 the β doubled to −0.103 (P = 3.70 × 10−31); this variant has been associated with HDL-C and TG as a bivariate phenotype (18). Similarly, the association of rs35120633 with TG and HDL-C (unconditional P = 2.67 × 10−40 and P = 2.02 × 10−9, respectively) became stronger after conditioning on rs2266788 (P = 5.21 × 10−46 and P = 1.38 × 10−10, respectively).
Of the 16 conditional signals not previously associated with a lipid trait, 10 are rare variants (Table 2). These variants are missense except rs76353203 (MAF 0.04%; β = −1.258 per T allele) which introduces a stop codon in APOC3 (Arg19TER) and is known to cause hyperalphalipoproteinaemia 2. At several loci, conditional analysis identified variants with much larger effect sizes than the sentinel SNP, e.g. missense variant rs116329129 (rs116329129:T > C, p.V280A; MAF 0.03%) in BANK1 which was associated with HDL-C, had a β of −1.835 per C allele compared with −0.104 for the lead variant rs13107325 (MAF 6.02%) which is located in SLC39A8. The B-cell scaffold protein with ankyrin repeats 1 (BANK1) gene encodes a B-cell-specific scaffold protein involved in B-cell receptor-induced calcium mobilization from intracellular stores. Variants in BANK1 have been associated with susceptibility to systemic lupus erythematosus (19). Similarly, the missense variant rs139788907 (MAF 0.03%) in the phospholipase A2, group IVF (PLA2G4F) gene (rs139788907:A > G, p.L326P; deleterious change per SIFT) which was associated with TG levels, had a β = 1.665; ∼1.98 mmol/l per G allele) 10-fold higher than that of the intronic lead variant rs2412710 (MAF 1.8%; β = 0.165; 0.20 mmol/l per G allele) which is located in CAPN3. PLA2G4F encodes a calcium-dependent phospholipase A2 that selectively hydrolyzes glycerophospholipids in the sn-2 position.
Joint analysis
Regional analyses can determine the specific contribution that each locus makes to the trait heritability. The iterative rounds of conditional analyses within GCTA described above enabled the identification of independently associated variants at each locus. Joint analyses can simultaneously estimate the effects of each of these significant variants adjusted for all other effects.
The joint analyses were performed in both the discovery studies with appropriate ethical approval for sharing individual level data (16 of the 19 cohorts; N = 24 894) and the replication studies (four cohorts; N = 9029), in order to (i) validate the original conditional analyses based on GCTA regarding the handling of rare and low-frequency variants as well as check for any impact of sample size difference and (ii) confirm consistency between the discovery and replication studies, to ensure that the replication data are sufficiently concordant to be used for a risk score analyses (see below).
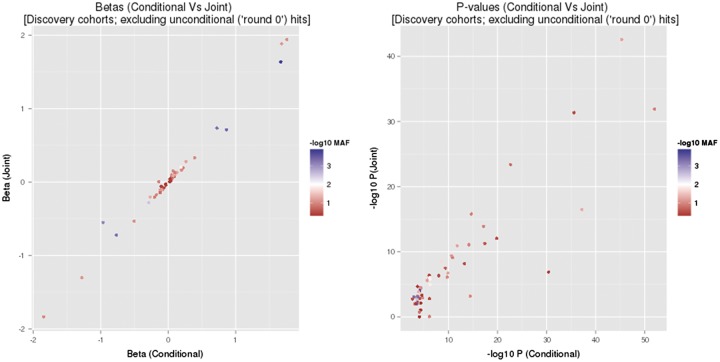
Over 95% of the variants that were significant in the conditional analyses also had P < 0.05 in the joint analyses (29/29 for HDL-C; 17/18 for LDL-C; 18/19 for TC; and 18/20 for TG) (Supplementary Material, Table S4). A comparison of the βs between conditional and joint analyses (Fig. 2) revealed close agreement between the two analyses, with almost perfect directional consistency (55 of 56 variants). Comparison of the P-values from each analysis (Fig. 2) also showed close agreement for most variants despite a difference in sample size between the two analyses and the more conservative nature of the joint tests. Importantly, the relationship between P-values did not appear to depend on MAF, and none of the variants with noticeably discordant P-values was rare or of low frequency. Of the four variants with P > 0.05 we excluded rs5015480 and rs2068888 (TG signals in CYP26A1) from further analyses but retained rs3208856 (APOE-LDL) and rs920915 (LIPC-TC) given that they are established associations. In summary, we found good concordance between the joint and conditional analyses within the discovery studies.
Figure 2.
Comparison of results between conditional and joint analyses. Note that these figures do not include the lead SNP in each region (i.e. round > 0 means that variants from the conditional analysis are shown only), as the conditional analyses do not produce adjusted estimates of their effects in contrary to the joint analyses.
Joint analysis in the replication studies detected an effect in the same direction for 91.8% of the variants (Supplementary Material, Table S4), showing good concordance between the discovery and replication data sets.
Locus-specific genetic score analysis
Next we calculated an overall genetic risk score association for each region except CYP26A1 (see joint analysis above), assessing the combined effects of all independent variants within a locus. In such analyses, the score is weighted by the effect sizes of each included variant. We used the beta estimates from the conditional analyses as risk score weights and performed the analyses in the replication set which comprised four independent studies (N = 9029). It is paramount to use an independent data set in order to minimize any bias.
The genetic score analyses identified several strong effects (Table 3; effect estimates are from the unweighted model and are expressed as per one-allele increment); for example, in the CETP locus each trait-increasing allele associated with 12.4% of an SD (∼0.06 mmol/l) increase in HDL-C accounting for 3.1% of the overall trait variation. We also note the PCSK9 locus in which each trait-increasing allele was associated with 19.2% of an SD (∼0.18 mmol/l) increase in LDL-C, but this region accounted for only 0.4% of the variation. PCSK9 was also associated with a large effect on TC (16.6% of an SD; 0.18 mmol/l per trait-increasing allele), and explained 0.3% of the variation. For TG, the strongest effect was found at the APOA1 locus (17.2% of an SD; ∼0.20 mmol/l per trait-increasing allele accounting for 1.7% of the variation). Cumulatively per trait, all regions tested accounted for 6.3% (HDL-C), 2.9% (LDL-C), 2% (TC), and 3.8% (TG) of the variation (Table 3).
Table 3.
Association results from the genetic score analyses estimating the combined effect of all statistically significant SNPs within a region by regressing a genetic score against each lipid trait
|
Replication studies |
|||||||
|---|---|---|---|---|---|---|---|
| Trait | Locus | rsIDs | N | β | SE | P-value | R2 |
| HDL | |||||||
| LPL | rs328, rs268, rs13702, rs1801177 | 9025 | 0.098 | 0.010 | 1.07E-21 | 0.010 | |
| ABCA1 | rs3905000, rs1883025 | 9025 | 0.050 | 0.011 | 1.53E-05 | 0.003 | |
| APOA1 | rs2266788, rs35120633, rs186808413, rs79554110, rs138407155 | 9025 | 0.060 | 0.013 | 8.90E-14 | 0.003 | |
| LIPC | rs1800588, rs10468017, rs140029729, rs200684324 | 7634 | 0.116 | 0.013 | 6.19E-18 | 0.010 | |
| CETP | rs3764261, rs5880, rs9939224, rs5882 | 9025 | 0.124 | 0.007 | 1.12E-75 | 0.031 | |
| LCAT | rs2271293, rs4986970 | 9025 | 0.064 | 0.020 | 1.15E-03 | 0.001 | |
| APOE | rs769449, rs5167, rs36053277 | 9025 | 0.055 | 0.013 | 1.51E-04 | 0.003 | |
| LDL | ABCG5/8 | rs6756629, rs4245791 | 8697 | 0.057 | 0.014 | 3.87E-05 | 0.002 |
| LPA | rs7770628, rs3798220 | 8697 | 0.037 | 0.016 | 4.31E-05 | 0.001 | |
| PCSK9 | rs505151, rs11591147 | 8697 | 0.192 | 0.034 | 3.73E-14 | 0.004 | |
| APOE | rs1132899, rs3208856, rs445925, rs769449, rs7412 | 8697 | 0.116 | 0.013 | 5.13E-110 | 0.011 | |
| APOB | rs41288783, rs5742904, rs533617, rs541041, rs1367117 | 8697 | 0.099 | 0.011 | 1.57E-19 | 0.010 | |
| TC | ABCG5/8 | rs4245791, rs6756629 | 9029 | 0.057 | 0.013 | 2.09E-05 | 0.003 |
| PCSK9 | rs11591147, rs505151 | 9029 | 0.166 | 0.033 | 2.37E-11 | 0.003 | |
| APOA1 | rs2075290, rs35120633 | 9029 | 0.091 | 0.022 | 5.66E-05 | 0.002 | |
| LIPC | rs1532085, rs1800588, rs920915 | 9029 | 0.017 | 0.014 | 1.73E-02 | 0.000 | |
| LIPG | rs4939883, rs77960347 | 9029 | 0.063 | 0.019 | 4.72E-04 | 0.002 | |
| APOE | rs7412, rs439401, rs769449, rs445925 | 9029 | 0.046 | 0.011 | 4.05E-57 | 0.002 | |
| APOB | rs541041, rs1367117, rs533617, rs5742904 | 9029 | 0.086 | 0.011 | 2.30E-18 | 0.008 | |
| TG | LPL | rs15285, rs268, rs1801177, rs328 | 8729 | 0.111 | 0.011 | 3.26E-26 | 0.013 |
| TRIB1 | rs2954033, rs2954029 | 8729 | 0.049 | 0.009 | 7.72E-08 | 0.004 | |
| APOA1 | rs76353203, rs35120633, rs7350481, rs2266788 | 8729 | 0.172 | 0.015 | 2.70E-37 | 0.017 | |
| LIPC | rs1800588, rs1532085 | 8729 | 0.030 | 0.012 | 9.63E-03 | 0.001 | |
| CETP | rs5880, rs3764261 | 8729 | 0.035 | 0.015 | 2.87E-02 | 0.001 | |
| GCKR | rs1049817, rs1260326 | 8729 | 0.077 | 0.018 | 3.32E-17 | 0.002 | |
Betas, standard errors (SEs) and R2 estimates taken from the non-weighted models; betas are expressed as per one-allele increment in the risk score, P-values estimated from the weighted models; only regions with a P-value <0.05 are presented. Note that R2 estimates were synthesized by taking a weighted average over contributing studies (with weights based on sample size).
MAF - Minor Allele Frequency,
SNP - Single Nucleotide Polymorphism,
GWAS - Genome-wide association studies,
TC - Total Cholesterol,
TG - Triglycerides,
LDL-C - low-density lipoprotein cholesterol,
HDL-C - high-density lipoprotein cholesterol,
CAD - Coronary Artery Disease,
CVD - Cardiovascular Disease,
QC - Quality Control,
LD - Linkage Disequilibrium
Heritability
First, we assessed heritability in the 135 unique known lipid loci which reached P < 0.01 in our study, considering only the published lead SNP (or proxy) and estimated a 7.12% heritability for HDL-C, 6.52% for LDL-C, 7.03% for TC and 6.31% for TG. When we considered for the same loci all independent sentinel SNPs from our study (lead and secondary signals as per Supplementary Material, Table S1) we observed a between 1.5- and 2.5-fold increase in the heritability estimates (14.73% for HDL-C, 15.06% for LDL-C, 13.49% for TC and 9.62% for TG).
Finally, after exclusion of the CYP26A1 locus (two variants) we assessed the incremental contribution of the multiple independent signals we detected by conditional analysis in the remaining 30 loci (87 in total) to heritability. Accounting for all signals per locus increased their contribution to heritability estimates for all lipid traits; 4.78% versus 11.07% (HDL-C), 1.26% versus 8.89% (LDL-C), 2.29% versus 7.00% (TC) and 5.70% versus 6.55% (TG) when comparing heritability estimates based on the known sentinel SNPs alone.
Discussions
We undertook an association study in 27 312 individuals to test the hypothesis that low-frequency and rare coding variants contribute to the genetic architecture of the four main lipid traits, TC, TG, HDL-C and LDL-C explaining some of the missing heritability in large-scale genetic studies of common variants (2,3). Of the 203 350 non-synonymous (missense, nonsense, splice-site and frameshift) variants present on the ExomeChip, ∼64 000 had an MAF above 0.1% to allow for single variant association testing. We did not find any new loci to be significant at the genome-wide level of significance in addition to the 159 loci known to be robustly associated (P < 5 × 10−8) with plasma concentrations of these lipid traits. Our findings are in agreement with other recent studies that have used exome sequencing, exome arrays or 1000 Genome Project imputed GWAS studies to investigate circulating blood lipid levels or related traits (20–22) that have also not found new loci harbouring low frequency/rare coding variants with large effect sizes (21–23).
To extend our assessment of the impact of low frequency/rare coding variation on lipid levels, we also examined the 159 known lipid (2,3) by assessing the results of both the single-marker and conditional analyses at these loci; for the latter, we took advantage of the presence of previously reported index lipid-associated variant (or a good proxy) at 135 of these loci on the ExomeChip. We note that a recent study by the ENGAGE consortium (24) has identified an additional 10 unique loci associated with lipid traits but the ExomeChip does not harbour the sentinel SNP or a good proxy to allow conditional analysis (seven loci have a variant on the array reaching nominal significance; Supplementary Material, Table S5). Interestingly, in 16 of the loci tested in our study we detected lead variants having an index signal at least 1000-fold more significant than the previously reported sentinel SNP (Table 1); these included variants previously reported in the literature for either the investigated and other lipid traits (10) or lipid traits other than the investigated one (4) as well as two variants not previously associated with a lipid trait (rs3094216 and rs72836561). SNP rs3094216 (MAF 77.6%) is located in the CDSN gene, corneodesmosin, which encodes a protein found in human epidermis and other cornified squamous epithelia. Furthermore, rs3094216 is in strong LD with rs3095318 a missense variant in CDSN (p. M18L). Mutations in CDSN are known to cause peeling skin syndrome type B disease, a rare recessive genodermatosis, whereas a common synonymous SNP (rs1062470) has been associated with psoriasis (25). The other variant, rs72836561, is a low-frequency missense variant in CD300LG (p.R82C; MAF 2.69%). CD300LG encodes the CD300 molecule-like family member G protein; a type I cell surface glycoprotein that contains a single immunoglobulin V-like domain and has a role in lymphocyte binding and transmigration.
In addition to rs72836561 (CD300LG) described above, two more low frequency or rare coding variants had not been previously associated with a lipid trait: the missense variant rs3798220 (p. I1891M) in LPA which was associated with LDL-C levels and rs34606562 a synonymous change (p.L1174) in KDM2B associated with TC levels. KDM2B encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. KDM2B gene has been recently associated with methylation in adipose tissue and our lead variant (rs34606562), which overlaps a strong peak for H3K27Ac, is located 95.6 kb away of the methylation probe (cg13708645) used to detect it (26). In total, after taking into consideration the results of the conditional analyses, we found 27 (13 low frequency and 14 rare) variants to be associated with lipid levels. Interestingly, we observed higher effect sizes for variants with MAF below 3% compared with more common SNPs (Supplementary Material, Fig. S2; power calculations based on TC showed 80% power to detect a minimum effect size of 0.125 at 1% MAF). However, this may reflect the fact that our study only had power to detect rare variants with higher effect sizes.
Overall, our study identified 14 missense variants not previously reported to be associated with a lipid trait. Two of them, rs3798220 and rs72836561, were new sentinel SNPs (Table 1) and the remaining 12 were identified as distinct additional signals through conditional analysis (rs116329129, rs138407155, rs140029729, rs200684324, rs117623631, rs5167, rs36053277, rs145814749, rs1132899, rs5742904, rs139788907, rs5880; Table 2). In total, 21 unique lipid loci (28 regions of association; all lipid traits) where we had the known sentinel SNP on the ExomeChip array had a missense variant as lead SNP (P < 10−4) (Supplementary Material, Table S1).
Our joint analyses validated the results from the conditional analyses showing that the GCTA software is also suitable for handling low-frequency variants, despite being designed for common variant analysis. We removed from further analyses one locus for TG, CYP26A1, based on the results of the joint analysis as both variants had Pj > 0.05. This was partly due to using a random effect model for rs2068888 which showed significant heterogeneity. In the genetic score analyses, estimating the combined effect of genetic variants within each locus, we observed substantial overall effects on lipid traits in several loci (CETP, PCSK1, APOA1); for example, in the CETP locus each trait-increasing allele associated with 12.4% of an SD (∼0.06 mmol/l) increase in HDL-C accounting for 3.1% of the overall trait variation.
As shown by others (24,27–30), we found a substantial increase in the explained variance when we assessed heritability estimates based on the 209 variants (all traits) identified by both the unconditional and conditional analyses compared to the published lead SNPs in the corresponding 135 loci. In some loci, the inclusion of new independent secondary signals contributes only marginally to heritability estimates. For example, in the APOE locus, 96.26% of the locus-specific heritability for TC was explained by rs7412 and rs769449 (72.41% and 23.84%, respectively) which capture the APOE 2/3/4 alleles. SNP rs7412 was the most significant variant for TC and LDL-C (P = 7.43 × 10−145 and P = 1.43 × 10−80, respectively) in our study whereas rs769449, a proxy of rs429358 (r2 0.82), was the lead variant for HDL-C (P = 7.29 × 10−13) and a secondary independent signal for TC and LDL-C. For LDL-C, these two variants explain 91.51% of the locus-specific heritability (70.95% and 20.56%, respectively). Among the three additional secondary signals in the APOE locus for LDL-C, the low-frequency missense variant rs3208856 explained most of the remaining variance (7.55%). Overall the inclusion of low frequency/rare variants appears to significantly impact heritability estimates, for example, we observed a 7-fold increase in LDL-C variance explained cumulative when comparing only the loci that harboured secondary signals. But rare coding variants with large effect sizes are not likely to explain the overall missing fraction of the genetic component of lipid traits.
Some important limitations of our study merit to be highlighted. First, the list of tested coding variants is by no means exhaustive especially at the rare end of the frequency spectrum. Hamond et al. estimated the Exome array to capture 72.5% and 66.2% of loss-of-function and missense variation with MAF 0.5% and 0.1%, respectively (31). Second, our study does not have sufficiently high power to detect very low-frequency and/or rare variants with small effect sizes. Power calculations for our study (based on TC) showed that we had 80% power to detect a minimum effect size of 0.07 at a 3% MAF, 0.125 at 1% MAF, 0.4 at 0.1% and 1.25 at 0.01% MAF. Therefore, even larger sample sizes will be required to identify new rare variants with small effect sizes.
In conclusion, we demonstrate that low frequency/rare coding variants contribute to the genetic architecture and heritability of lipid traits despite a paucity of low-frequency coding variants with large effect sizes.
Materials and Methods
Samples and phenotypes
We collected summary statistics for ExomeChip SNPs from 19 studies (N ∼ 26 000). Among these, 17 studies consisted primarily of individuals of European ancestry, and two studies consisted of individuals of South Asian descent (see Supplementary Material, Note and Table S6 for details). Both population-based studies and case–control studies were included; for case–control studies, cases and control samples were analysed separately. Results for blood lipid levels were provided in mmol/l units and trait residuals within each cohort were adjusted for age, age2 and sex, and then inverse-rank normalized. Individuals known to be on lipid-lowering medication were excluded from the analysis (Supplementary Material, Table S6).
Genotyping
A total of 247 870 genetic variants were genotyped using the Illumina ExomeChip array. The ExomeChip variants comprise 203 350 non-synonymous, 10 690 splice and 5641 stop variants as well as 4761 SNPs from the GWAS NHGRI catalogue. Genotypes were called with GenCall, subjected to QC (Supplementary Material, Table S7) to remove poor quality samples and finally recalled using zCall, an algorithm optimized for rare variant detection (32). Average standard errors for association statistics from each study were plotted against study sample size to identify outlier studies. Allele frequencies were inspected to ensure all analyses used the same strand assignment.
Primary linear regression analysis
Analyses were performed for each trait (HDL-C, LDL-C, TC and TG) using the assumption of an additive genetic model. Individual SNP association tests were performed using linear regression with the inverse normal transformed trait values as the dependent variable and the expected allele count for each individual as the independent variable. Explicit adjustments for population sub-structure using principal components (33) were carried out. These analyses were performed using a range of analytical software (Supplementary Material, Table S7).
Meta-analysis
An inverse-variance weighted meta-analysis using a fixed effect model was performed, using both GWAMA (34) and METAL (35) and results were compared and checked for consistency. SNPs were excluded from the meta-analysis if they had MAF >5% and were absent in >90% of the samples or had MAF <5% and were absent in >25% of the samples and present in at least two studies and/or failed cluster plot evaluation. Heterogeneity was evaluated using Cochran’s Q- and I2-statistic. For SNPs with non-significant heterogeneity (P for Q > 0.01), we report the results from the fixed effect model whereas in the presence of significant heterogeneity (P for Q < 0.01) we used a random effect model. Signals were considered to be novel if they reached a genome-wide significance (P < 5 × 10 − 8) in the meta-analysis and were > ±500 kB away from the nearest previously described lipid locus. For the previously published lipid loci, we considered replication at nominal significance level of P < 0.01. We note that for 21 loci the published lead SNP was not present on the ExomeChip and for a further 7 loci it was removed during QC (Supplementary Material, Table S8).
Approximate conditional analysis
Conditional analysis was implemented in GCTA (17) using meta-analysis summary statistics from all 27 312 samples. A subset of 11 396 samples (part of the contributing studies: BC1958, BRIGHT, FIA3, EPIC and GoDARTS) of European origin was used as a reference panel for LD calculations. We considered in total 159 published lipid loci (2,3) and 247 lipid association signals (73 for HDL-C, 58 for LDL-C, 74 for TC and 42 for TG). SNPs failing the cluster plot inspection were replaced by the next most significant SNP in the locus. Subsequent rounds of stepwise conditional analysis were performed in each locus until no significant SNP could be identified. The level of significance for each round of the conditional analysis was defined as 0.05/(locus SNP content − conditional SNPs) to account for multiple testing (Supplementary Material, Table S2).
Joint analyses
Joint analyses were performed for any loci identified in the conditional analyses as containing more than one statistically significant SNP. The joint tests estimated the associations between the phenotype and all statistically significant independent SNPs within a region simultaneously (by fitting one linear regression model per region).
Locus-specific genetic score analyses
The genetic score analyses estimated the combined effect of all statistically significant SNPs within a region (Supplementary Material, Table S2) by regressing a genetic score against the phenotype. Genetic scores were derived in two ways: (i) by summing the number of trait-increasing alleles (as defined by the estimated directions of the SNP effects in the conditional analyses) carried by each individual; and (ii) by producing a weighted sum of the number of trait-increasing alleles against the phenotype (36). In this latter scenario, the genetic scores were weighted by multiplying genotypes by the corresponding estimated SNP effect (i.e. the ‘β’) from the conditional analysis. Joint tests and genetic score analyses were performed on the inverse-rank normalized trait values, which had been adjusted for age, age2 and sex. Adjustments for principal components were also made, where applicable, to control for any potential population stratification within each study.
The joint analyses were run in a total of 20 cohorts (Nmax = 33 923). Of these, 16 (N = 24 894) contributed to the individual SNP meta- and conditional analyses and were considered ‘discovery’ cohorts, whereas a further four cohorts (N = 9029) that did not contribute to the preceding analyses were also included as ‘replication’ cohorts. The genetic score analyses were only run in the replication cohorts in order to minimize bias, due to using weights estimated from the discovery meta-analyses. Only studies with unrelated individuals were included in these analyses. Studies with any missing data (i.e. where an SNP had been dropped during QC) within a particular region did not contribute to the overall result for that region.
Linear regression tests and genetic score analyses were conducted separately by each study. Meta-analyses were performed using the metafor package in R (37). Overall estimates of the proportion of variation explained by each region (R2) were derived by taking a weighted average over contributing studies (with weights based on sample size).
Heritability
Heritability estimates were calculated using the multifactorial liability threshold model (38). The calculations are performed using the inverse normal transformed traits meta-analysis results, based on a population SD of 1 and under the additive genetic model assumption. All variants included in the heritability calculations per trait were not in LD (r2 < 0.3).
URLs
http://genome.sph.umich.edu/wiki/Exome_Chip_Design
http://www.metafor-project.org/http://www.wvbauer.com
The results of the meta-analysis are available upon request and will be made available at http://www.qmul.ac.uk/ExomeChip.Lipids.SummaryStatistics.zip
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
1958BC: We are grateful for being able to use the British 1958 Birth Cohort DNA collection.
ASCOT: We thank all ASCOT trial participants, physicians, nurses and practices in the participating countries for their important contribution to the study. The study was investigator-led and was conducted, analysed and reported independently of Pfizer who funded the trial.
BRIGHT: The BRIGHT study is extremely grateful to all the patients who participated in the study and the BRIGHT nursing team.
DIABNORD: We are grateful to the study participants who dedicated their time and samples to these studies. We also thank the VHS, the Swedish Diabetes Registry and Umeå Medical Biobank staff for biomedical data and DNA extraction. We also thank M. Sterner, M. Juhas and P. Storm for their expert technical assistance with genotyping and genotype data preparation.
EFSOCH: We are extremely grateful to the study participants and the study team. The opinions given in this article do not necessarily represent those of NIHR, the NHS or the Department of Health.
EPIC: The authors would like to acknowledge the contribution of the staff and participants of the EPIC Study.
EPIC_incidentT2D: The authors would like to acknowledge the contribution of the staff and participants of the EPIC-Norfolk Study.
Fenland: We are grateful to all the volunteers for their time and help, and to the General Practitioners and practice staff for assistance with recruitment. We thank the Fenland Study Investigators, Fenland Study Co-ordination team and the Epidemiology Field, Data and Laboratory teams. Biochemical assays were performed by the National Institute for Health Research, Cambridge Biomedical Research Centre, Core Biochemistry Assay Laboratory, and the Cambridge University Hospitals NHS Foundation Trust, Department of Clinical Biochemistry.
FIA3: We are indebted to the study participants who dedicated their time and samples to these studies. We also thank J. Hutiainen and Å. Ågren (Umeå Medical Biobank) for data organization and K. Enquist and T. Johansson (Västerbottens County Council) for technical assistance with DNA extraction.
GLACIER: We are indebted to the study participants who dedicated their time and samples to these studies. We thank J. Hutiainen and Å. Ågren (Umeå Medical Biobank) for data organization and K. Enquist and T. Johansson (Västerbottens County Council) for technical assistance with DNA extraction. We also thank M. Sterner, M. Juhas and P. Storm for their expert technical assistance with genotyping and genotype data preparation.
GoDARTS: We are grateful to all the participants who took part in this study, to the general practitioners, to the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We acknowledge the support of the Health Informatics Centre, University of Dundee for managing and supplying the anonymized data and NHS Tayside, the original data owner.
GRAPHIC: We are grateful to the participants of the GRAPHIC Study and to the nurses who recruited the patients.
HELICMANOLIS: The MANOLIS cohort is named in honour of Manolis Giannakakis, 1978-2010. We thank the residents of Anogia and surrounding Mylopotamos villages for taking part. The HELIC study has been supported by many individuals who have contributed to sample collection (including Olina Balafouti, Christina Batzaki, Georgios Daskalakis, Eleni Emmanouil, Chrisoula Giannakaki, Margarita Giannakopoulou, Anastasia Kaparou, Vasiliki Kariakli, Stella Koinaki, Dimitra Kokori, Maria Konidari, Hara Koundouraki, Dimitris Koutoukidis, Eirini Mamalaki, Eirini Mpamiaki, Maria Tsoukana, Dimitra Tzakou, Katerina Vosdogianni, Niovi Xenaki), data entry (Thanos Antonos, Dimitra Papagrigoriou, Betty Spiliopoulou), sample logistics (Sarah Edkins, Emma Gray), genotyping (Robert Andrews, Hannah Blackburn, Doug Simpkin, Siobhan Whitehead), research administration (Anja Kolb-Kokocinski, Carol Smee) and informatics (Martin Pollard, Josh Randall).
HELICPomak: We thank the residents of the Pomak villages for taking part. The HELIC study has been supported by many individuals who have contributed to sample collection (including Olina Balafouti, Christina Batzaki, Georgios Daskalakis, Eleni Emmanouil, Chrisoula Giannakaki, Margarita Giannakopoulou, Anastasia Kaparou, Vasiliki Kariakli, Stella Koinaki, Dimitra Kokori, Maria Konidari, Hara Koundouraki, Dimitris Koutoukidis, Eirini Mamalaki, Eirini Mpamiaki, Maria Tsoukana, Dimitra Tzakou, Katerina Vosdogianni, Niovi Xenaki), data entry (Thanos Antonos, Dimitra Papagrigoriou, Betty Spiliopoulou), sample logistics (Sarah Edkins, Emma Gray), genotyping (Robert Andrews, Hannah Blackburn, Doug Simpkin, Siobhan Whitehead), research administration (Anja Kolb-Kokocinski, Carol Smee) and informatics (Martin Pollard, Josh Randall).
KORA: The authors thank Nadine Lindemann, Viola Maag and Franziska Scharl for excellent technical support. The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München—German Research Center for Environmental Health.
LOLIPOP: We thank the participants and research staff who made the study possible.
NFBC1986: The authors are grateful to the NFBC1986 participants and their families and to the NFBC research staff for data collection. We also thank late Professor Paula Rantakallio (launch of NFBC1966 and 1986), Ms Outi Tornwall and Ms Minttu Jussila (DNA biobanking). The DNA extractions, sample quality controls, biobank up-keeping and aliquotting were performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki.
SCARF: The authors would like to thank all participants in this study.
Conflict of Interest statement. None declared.
Funding
1958BC was funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. This work forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Unit which is supported and funded by the National Institute for Health Research. PD is supported by British Heart Foundation grant RG/14/5/30893. ASCOT was supported by Pfizer, New York, NY, USA, Servier Research Group, Paris, France and by Leo Laboratories, Copenhagen, Denmark. BRIGHT was supported by the Medical Research Council of Great Britain (grant number G9521010D), the British Heart Foundation (grant number PG/02/128) and the Wellcome Trust Strategic Awards 083948A and 085475. AFD was supported by the British Heart Foundation (grant numbers RG/07/005/23633, SP/08/005/25115); and by the European Union Ingenious HyperCare Consortium: Integrated Genomics, Clinical Research, and Care in Hypertension (grant number LSHM-C7-2006-037093). DIABNORD was funded by Novo Nordisk, the Swedish Research Council, Påhlssons Foundation, the Swedish Heart Lung Foundation, and the Skåne Regional Health Authority (all to PWF). The EFSOCH study was supported by South West NHS Research and Development, Exeter NHS Research and Development, the Darlington Trust, and the Peninsula NIHR Clinical Research Facility at the University of Exeter. Timothy Frayling is supported by the European Research Council grant: SZ-245 50371-GLUCOSEGENES-FP7-IDEAS-ERC. EPIC and EPIC-Norfolk is supported by the Medical Research Council programme grants (G0401527, G1000143) and Cancer Research UK programme grant (C864/A8257). The Fenland Study is funded by the Medical Research Council (MC_U106179471). FIA3 was supported in part by a grant from the Swedish Heart-Lung Foundation (grant no. 2020389 to PW Franks). GLACIER was funded by Novo Nordisk, the Swedish Research Council, Påhlssons Foundation, the Swedish Heart Lung Foundation, and the Skåne Regional Health Authority (all to PWF). GoDARTS was supported by the Wellcome Trust (Wellcome Trust UK Type 2 Diabetes Case Control Collection) and the Scottish Health Informatics Programme. The Chief Scientist Office supported informatics. Project funded by the UK Medical Research Council (G0601261). GRAPHIC exome array genotyping was funded by the NIHR and the Wellcome Trust. NJS holds a Chair funded by the British Heart Foundation and is a NIHR Senior Investigator. NM is funded by the NIHR Leicester Cardiovascular Biomedical Research Unit. The GRAPHIC Study is part of the portfolio of studies supported by the NIHR Leicester Cardiovascular BRU. HELICMANOLIS and HELICPomak were funded by the Wellcome Trust (098051) and the European Research Council (ERC-2011-StG 280559-SEPI). KORA was funded by the German Federal Ministry of Education and Research and by the State of Bavaria, DZHK (German Centre for Cardiovascular Research) and the BMBF (German Ministry of Education and Research). LOLIPOP is supported by the National Institute for Health Research (NIHR), the British Heart Foundation (SP/04/002), the Medical Research Council (G0601966,G0700931), the Wellcome Trust (084723/Z/08/Z) the NIHR (RP-PG-0407-10371), European Union FP7 (EpiMigrant, 279143) and Action on Hearing Loss (G51). NFBC1986: was supported by the Academy of Finland (project grants 104781, 120315, 129269, 1114194, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland (75617), the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NIH/NIMH (5R01MH63706:02), the Medical Research Council, UK (G0500539, G0600705, PrevMetSyn/SALVE) and the Wellcome Trust (project grant GR069224), UK, ENGAGE project and grant agreement HEALTH-F4-2007-201413, and the EU Framework Programme 7 small-scale focused research collaborative project EurHEALTHAgeing 277849. SCARF was funded by the Foundation for Strategic Research, the Swedish Heart-Lung Foundation, the Swedish Research Council (8691, 12660, 20653), the European Commission (LSHM-CT-2007-037273), the Knut and Alice Wallenberg Foundation, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular and Diabetes Programmes of Karolinska Institutet and the Stockholm County Council, and the Stockholm County Council (560183). BS acknowledge funding from the Magnus Bergvall Foundation and the Foundation for Old Servants. MF acknowledge funding from the Swedish e-Science Research Center (SeRC).
References
- 1.Consortium C.A.D., Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J. et al. (2013) Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet., 45, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Lipids Genetics C., Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L. et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chasman D.I., Pare G., Mora S., Hopewell J.C., Peloso G., Clarke R., Cupples L.A., Hamsten A., Kathiresan S., Malarstig A. et al. (2009) Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet., 5, e1000730.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S., Willer C.J., Peloso G.M., Demissie S., Musunuru K., Schadt E.E., Kaplan L., Bennett D., Li Y., Tanaka T. et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet., 41, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandhu M.S., Waterworth D.M., Debenham S.L., Wheeler E., Papadakis K., Zhao J.H., Song K., Yuan X., Johnson T., Ashford S. et al. (2008) LDL-cholesterol concentrations: a genome-wide association study. Lancet, 371, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M. et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet., 40, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaess B., Fischer M., Baessler A., Stark K., Huber F., Kremer W., Kalbitzer H.R., Schunkert H., Riegger G., Hengstenberg C. (2008) The lipoprotein subfraction profile: heritability and identification of quantitative trait loci. J. Lipid Res., 49, 715–723. [DOI] [PubMed] [Google Scholar]
- 10.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. (2011) Clan genomics and the complex architecture of human disease. Cell, 147, 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schork N.J., Murray S.S., Frazer K.A., Topol E.J. (2009) Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev., 19, 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S. et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G. et al. (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet., 42, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke R., Peden J.F., Hopewell J.C., Kyriakou T., Goel A., Heath S.C., Parish S., Barlera S., Franzosi M.G., Rust S. et al. (2009) Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med., 361, 2518–2528. [DOI] [PubMed] [Google Scholar]
- 15.Kaminski W.E., Wenzel J.J., Piehler A., Langmann T., Schmitz G. (2001) ABCA6, a novel a subclass ABC transporter. Biochem. Biophys. Res. Commun., 285, 1295–1301. [DOI] [PubMed] [Google Scholar]
- 16.Gai J., Ji M., Shi C., Li W., Chen S., Wang Y., Li H. (2013) FoxO regulates expression of ABCA6, an intracellular ATP-binding-cassette transporter responsive to cholesterol. Int. J. Biochem. Cell Biol., 45, 2651–2659. [DOI] [PubMed] [Google Scholar]
- 17.Yang J., Ferreira T., Morris A.P., Medland S.E., Genetic Investigation of A.T.C., Replication D.I.G., Meta-analysis C., Madden P.A., Heath A.C., Martin N.G. et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraja A.T., Vaidya D., Pankow J.S., Goodarzi M.O., Assimes T.L., Kullo I.J., Sovio U., Mathias R.A., Sun Y.V., Franceschini N. et al. (2011) A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes, 60, 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AS J., C A., P S.G., S C. (2014) Systemic lupus erythematosus: old and new susceptibility genes versus clinical manifestations. Curr. Genomics, 15, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. (2012) Five years of GWAS discovery. Am. J. Hum. Genet., 90, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange L.A., Hu Y., Zhang H., Xue C., Schmidt E.M., Tang Z.Z., Bizon C., Lange E.M., Smith J.D., Turner E.H. et al. (2014) Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet., 94, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peloso G.M., Auer P.L., Bis J.C., Voorman A., Morrison A.C., Stitziel N.O., Brody J.A., Khetarpal S.A., Crosby J.R., Fornage M. et al. (2014) Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet., 94, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenthal E.A., Ranchalis J., Crosslin D.R., Burt A., Brunzell J.D., Motulsky A.G., Nickerson D.A., Project N.G.E.S., Wijsman E.M., Jarvik G.P. (2013) Joint linkage and association analysis with exome sequence data implicates SLC25A40 in hypertriglyceridemia. Am. J. Hum. Genet., 93, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surakka I., Horikoshi M., Mägi R., Sarin A.P., Mahajan A., Lagou V., Marullo L., Ferreira T., Miraglio B., Timonen S. et al. (2015) The impact of low-frequency and rare variants on lipid levels. Nat. Genet., 47, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capon F., Allen M.H., Ameen M., Burden A.D., Tillman D., Barker J.N., Trembath R.C. (2004) A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum. Mol. Genet., 13, 2361–2368. [DOI] [PubMed] [Google Scholar]
- 26.Demerath E.W., Guan W., Grove M.L., Aslibekyan S., Mendelson M., Zhou Y.H., Hedman Å.K., Sandling J.K., Li L.A., Irvin M.R. et al. (2015) Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet., 24, 4464–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asselbergs F.W., Guo Y., van Iperen E.P., Sivapalaratnam S., Tragante V., Lanktree M.B., Lange L.A., Almoguera B., Appelman Y.E., Barnard J. et al. (2012) Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet., 91, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanna S., Li B., Mulas A., Sidore C., Kang H.M., Jackson A.U., Piras M.G., Usala G., Maninchedda G., Sassu A. et al. (2011) Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet., 7, e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tada H., Won H.H., Melander O., Yang J., Peloso G.M., Kathiresan S. (2014) Multiple associated variants increase the heritability explained for plasma lipids and coronary artery disease. Circ. Cardiovasc. Genet., 7, 583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y., Waite L.L., Jackson A.U., Sheu W.H., Buyske S., Absher D., Arnett D.K., Boerwinkle E., Bonnycastle L.L., Carty C.L. et al. (2013) Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet., 9, e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmen O.L., Zhang H., Zhou W., Schmidt E., Hovelson D.H., Langhammer A., Løchen M.L., Ganesh S.K., Mathiesen E.B., Vatten L. et al. (2014) No large-effect low-frequency coding variation found for myocardial infarction. Hum. Mol. Genet., 23, 4721–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstein J.I., Crenshaw A., Carey J., Grant G.B., Maguire J., Fromer M., O'Dushlaine C., Moran J.L., Chambert K., Stevens C. et al. (2012) zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics, 28, 2543–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdi H., Williams L.J. (2010) Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat., 2, 433–459. [Google Scholar]
- 34.Magi R., Morris A.P. (2010) GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics, 11, 288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren H., Casas J.P., Hingorani A., Dudbridge F., Whittaker J. (2014) Genetic prediction of quantitative lipid traits: comparing shrinkage models to gene scores. Genet. Epidemiol., 38, 72–83., [DOI] [PubMed] [Google Scholar]
- 37.Viechtbauer W. (2010) Conducting meta-analyses in R with the metaphor package. J. Stat. Softw., 36, 1–48. [Google Scholar]
- 38.Falconer D.S. (1967) The inheritance of liability to diseases with variable age of onset, with particular reference to diabetes mellitus. Ann. Hum. Genet., 31, 1–20. [DOI] [PubMed] [Google Scholar]