Abstract
In Saccharomyces cerevisiae, ribosomal protein gene (RPG) promoters display binding sites for either Rap1 or Abf1 transcription factors. Unlike Rap1-associated promoters, the small cohort of Abf1-dependent RPGs (Abf1-RPGs) has not been extensively investigated. We show that RPL3, RPL4B, RPP1A, RPS22B and RPS28A/B share a common promoter architecture, with an Abf1 site upstream of a conserved element matching the sequence recognized by Fhl1, a transcription factor which together with Ifh1 orchestrates Rap1-associated RPG regulation. Abf1 and Fhl1 promoter association was confirmed by ChIP and/or gel retardation assays. Mutational analysis revealed a more severe requirement of Abf1 than Fhl1 binding sites for RPG transcription. In the case of RPS22B an unusual Tbf1 binding site promoted both RPS22B and intron-hosted SNR44 expression. Abf1-RPG down-regulation upon TOR pathway inhibition was much attenuated at defective mutant promoters unable to bind Abf1. TORC1 inactivation caused the expected reduction of Ifh1 occupancy at RPS22B and RPL3 promoters, but unexpectedly it entailed largely increased Abf1 association with Abf1-RPG promoters. We present evidence that Abf1 recruitment upon nutritional stress, also observed for representative ribosome biogenesis genes, favours RPG transcriptional rescue upon nutrient replenishment, thus pointing to nutrient-regulated Abf1 dynamics at promoters as a novel mechanism in ribosome biogenesis control.
INTRODUCTION
In Saccharomyces cerevisiae, ribosome biogenesis requires the coordinated expression of more than 750 genes, including 138 genes coding for 79 ribosomal proteins (RPs) and more than 200 genes of the ribosome biogenesis (Ribi) regulon (1). RP gene (RPG) transcription absorbs a large fraction of RNA polymerase II activity in yeast cells growing under optimal nutrient condition, and it is finely regulated in response to different stimuli (2). The target of rapamycin complex I (TORC1) plays a key role in the co-regulation of RP and Ribi genes in response to nutrient availability and other environmental stimuli (3). Transcription of most RPGs requires the general regulatory factor Repressor Activator Protein 1 (Rap1), which binds to one or, more frequently, two specific binding sites located between ∼400 and ∼200 bp upstream of the transcription start site (TSS) (4–6). A small subset (6%) of RPGs, however, appear to be Rap1-independent and to require instead ARS-binding factor 1 (Abf1) (4,6,7). In addition to Rap1, several factors have been found to localize to Rap1-dependent RPGs (Rap1-RPGs) and to participate in their transcriptional regulation. In particular, the forkhead (FH)-like DNA-binding protein Fhl1 localizes almost exclusively at RPGs, ∼100 bp downstream of Rap1 on average, where it might help to recruit Ifh1, a key co-activator of Rap1-RPG transcription. Ifh1 lacks a direct DNA-binding activity and plays a key role in TORC1-dependent control of RPG transcription, getting dissociated from Rap1-RPG promoters upon TORC1 inhibition (8,9). Other factors reported to operate at Rap1-RPG promoters include the HMG-like protein Hmo1 (10–12), the stress- and nutrient-responsive regulator Sfp1 (13,14) and the co-repressor Crf1 (15). Although an Fhl1 binding motif was defined on the basis of protein binding microarray analyses (16,17), such a specific site has been difficult to identify at RPG promoters. Recently, a comprehensive ChIP-seq analysis in S. cerevisiae clearly revealed an enrichment of this sequence element at RPG promoters, but it also demonstrated that Fhl1-binding sites affect the activity of only few promoters, thus suggesting that Fhl1 recruitment to most RPGs does not occur through the interaction with a specific promoter element (6). The same study also highlighted the involvement in RPG transcription of the HMG-like protein Hmo1, that was found associated with about half of RPG promoters (category I promoters) at a position immediately downstream of Fhl1/Ifh1 location, but not with the remaining (category II) promoters. At both these categories of Rap1- RPG promoters, Rap1 thus supports Fhl1/Ifh1 binding and, at category I promoters, Hmo1 would also contribute to Fhl1/Ifh1 recruitment (6). A very recent high-resolution ChIP-exo analysis accurately defined the positional organization at RPG promoters of the regulatory factors cited above and confirmed their involvement in RPG co-regulation through influence on nucleosome and pre-initiation complex positioning (18).
Unlike the other RPG regulatory proteins, the Fhl1/Ifh1 pair appears to be common to all yeasts (19). Such an evolutionary conservation is paralleled by a recognized key role of this protein couple in the signalling pathways controlling RPG transcription in response to growth and stress signals (8,9,15,20). In particular, growth-regulated recruitment at Rap1-RPG promoters was demonstrated for Ifh1 (8,9,15), but not for Fhl1 or Rap1, whose association with promoters is generally considered unaffected by growth conditions. Hmo1 and Sfp1 were also shown to leave RPG promoters upon TOR pathway inactivation (11,14).
While the promoter architecture and transcriptional regulation of Rap1-RPGs has been extensively investigated, much less is known about the small subset of Abf1-associated RPGs (Abf1-RPGs). This small cohort of promoters offers an interesting opportunity to grasp the common logic of RPG promoter architecture and coordinated transcriptional regulation. Abf1-centred promoters are thought to share with the more numerous Rap1-associated promoters principles and mechanisms of transcription activation, a notion reinforced by the functional interchangeability of Rap1 and Abf1 (21,22). Nevertheless, key issues concerning these promoters still await proper investigation. First, the actual involvement of the Fhl1/Ifh1 pair at Abf1-RPG promoters has never been demonstrated experimentally; in fact, recent genome-wide studies noted the general lack of these proteins, as well as of Hmo1, at RPG promoters bound by Abf1 (6,18). Moreover, Abf1 involvement in transcription displays subtle mechanistic differences from Rap1 (e.g. the lack of continuous Abf1 requirement at certain promoters for the maintenance of transcription activation (23,24)). Taken together these observations raise the possibility that transcriptional modulation in response to signalling pathways takes place at Abf1-RPG promoters through different mechanisms from those occurring at Rap1-RPG promoters. Adding further interest to the study of Abf1-RPGs is the possibility of their co-regulation with ribosome biogenesis genes, given previous evidence of a selective deregulation of such genes in an abf1 mutant strain (25).
Here we report a detailed in silico, in vitro and in vivo characterization of the architecture and function of S. cerevisiae Abf1-RPG promoters. We provide evidence for the functional requirement and reciprocal influence of evolutionarily conserved Abf1 and Fhl1 binding sites at these promoters, and for TORC1-dependent modulation of Abf1 association with target RP and Ribi gene promoters.
MATERIALS AND METHODS
Computational analysis of promoter sequences
Information on S. cerevisiae RPGs containing an Abf1 binding site was from (4). The coding sequences were retrieved from the Saccharomyces Genome Database (http://www.yeastgenome.org). Orthologous genes in Saccharomyces paradoxus, Saccharomyces mikatae and Saccharomyces kudriavzevii were identified through BLASTN against the corresponding genomes (http://seq.yeastgenome.org/cgi—bin/blast—fungal.pl). For each set of orthologous genes, the flanking regions (1000 bp upstream of the coding sequence) were aligned using Clustal X (26). The alignments, visualized through GeneDoc, were manually inspected to directly identify conserved sequence blocks. The positions of conserved motifs reported in the text correspond to the distances from the TSS (defined as the 5′ end of the most abundant transcript among those recently mapped to each gene by TIF-seq (27)).
Yeast cell cultures
Yeast cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% dextrose), except when otherwise indicated. For gene expression and ChIP analyses, cell pre-cultures were grown overnight, inoculated at an initial OD600 = 0.15, then cultures were incubated up to an OD600 = 0.6–0.7 before being collected for further analysis. For rapamycin treatment, once reached an OD600 = 0.6 cells were treated for 30 or 60 min with 200 ng/ml rapamycin (LC Laboratories) dissolved in DMSO prior to be harvested for subsequent analysis. For time course of Abf1-RPS28B promoter association and RPS28B expression during entry into and exit from stationary phase, cultures of either wild type (wt) or RPS28B Amut strains were grown into stationary phase (OD600 ∼ 20) and further incubated for 12 h before being diluted into fresh YPD medium at OD600 = 0.6–0.7. Samples were collected for ChIP and gene expression analyses at the following points: during exponential (OD600 = 0.6) and post-diauxic (OD600 = 10) phases, before dilution into fresh YPD medium (i.e. 12 h after reaching OD600 ∼ 20), and 5, 10, 20, 30 and 60 min after dilution.
Strain construction
The genotypes of S. cerevisiae strains used in this study are reported in Supplementary Table S1. Promoter region mutagenesis of native RP loci RPL4B, RPP1A, RPS22B, RPS28A and RPS28B was carried out by a two-step gene replacement strategy, as previously described (28). The Kluyveromyces lactis URA3 mutagenic cassettes for gene disruption were amplified from pUG72 vector (29) by sequential PCRs, in order to add 50–60 bp of target gene-flanking sequences necessary for homologous recombination (30). Gene disruption was carried out in an ABF1-TAP background for all the RPGs (31); RPS22B disruption was also carried out in a 13Myc-FHL1 background (see below). The mutagenic cassettes for gene replacement encompassed the whole chromosome region removed by previous gene disruption, only differing for specific mutations introduced into cis-regulatory elements. These cassettes were generated by mutagenic PCR using an ‘overlap extension’ strategy (32). Disruption and replacement cassettes were synthesized using GoTaq DNA polymerase (Promega) and Phusion DNA polymerase (Thermo Scientific), respectively.
To tag FHL1 with a 13Myc epitope, a tagging cassette was synthesized by PCR (with Phusion DNA polymerase) on the pFA6a-13Myc-kanMX6 vector (33) using primers that allow to fuse the sequence coding for 13xMyc epitope to the C-terminus of FHL1. After transformation with 1 μg of cassette, BY4741 cells were plated on selective medium (YPD containing 0.2 mg/ml G418).
The sequences of all oligonucleotide primers used for promoter mutagenesis and tagging are reported in Supplementary Table S2.
Gene expression analysis
Total RNA was extracted from yeast cells using the RNeasy MiniKit (Promega) with DNase I treatment following manufacturer's suggestions. Five hundred nanogram of total RNA sample was reverse-transcribed using the iScript™ cDNA Synthesis kit (Bio-Rad) with the hexamer random priming protocol. The cDNA samples were used as templates of qPCR reactions conducted with ABI 7300 instrumentation (Applied Biosystems) and SYBR® Green PCR Master Mix (Life Technologies). Primer pairs were designed in order to avoid cross-amplification of paralogous gene cDNAs. For RPL4B and RPS28B, a forward primer within the coding sequence and a reverse primer within the 3′ UTR were used; for RPP1A ad RPS22B, we used a primer annealing within the 5′ UTR. Primer sequences are reported in Supplementary Table S2. The concentration of each primer pair was optimized in order to satisfy the requirements for mRNA level quantification according to the Livak method (34). RPG expression was normalized to the expression of HHT2 gene, chosen as an internal standard. The ΔCT was calculated as CT target gene – CT internal standard; the expression level variations upon promoter mutation were then expressed as 2−ΔΔCT (with ΔΔCT = ΔCT mutant strain - ΔCT wt strain).
Chromatin immunoprecipitation assays
Yeast strains used for direct ChIP measurements were either from the Yeast TAP-Tagged Collection (Open Biosystems; (31)) or obtained by 13Myc tagging of FHL1. ChIP was performed essentially as reported (28,35). For Fhl1-13Myc, immunoprecipitation was carried out using 9E10 mouse monoclonal antibodies (Invitrogen) followed by incubation with Dynabeads M-280 Sheep Anti-Mouse IgG (Life Technologies). For qPCR analysis of ChIP samples, input and immunoprecipitated (IP) DNA samples were analyzed (as described above for expression analysis) using primer pairs producing 100–125 bp amplicons (listed in Supplementary Table S2). ΔCT was first calculated, as the difference between IP and input CT values (after correcting for input sample dilution), for both target gene and HHT2 gene (chosen as an internal standard). The enrichment of target gene in IP DNA was then calculated as ΔΔCT = ΔCT target - ΔCT HHT2.
Protein expression and purification
The sequence coding for the Fhl1 Forkhead domain (FHD; from amino acid 440–580) was PCR-amplified from S. cerevisiae genomic DNA using forward and reverse primers containing recognition sites for NheI and HindIII restriction enzymes, respectively (see Supplementary Table S2). The amplicon was first cloned into pGEM® T-easy (Promega), then a NheI-HindIII fragment from this construct was inserted into pET28b cut with the same enzymes. After transformation of the expression vector into Escherichia coli BL21-Gold(DE3)pLysS competent cells, Fhl1-FHD expression was induced with 1 mM IPTG for 4 h at 30°C. After cell lysis by sonication in lysis buffer (50 mM Tris/HCl pH 7.6, 0.3 M NaCl, 5% (v/v) glycerol, 1 mM β-mercaptoethanol, 1 μM pepstatin, 1 μM leupeptin, 1 mM PMSF) supplemented with 0.2 mg/ml lysozyme, the 6xHis-tagged Fhl1-FHD recombinant polypeptide was purified to apparent homogeneity by affinity chromatography on Ni-NTA agarose (Qiagen).
Electrophoretic mobility shift assays
Radiolabelled DNA fragments for electrophoretic mobility shift assays (EMSA) were prepared by PCR using oligonucleotide primers that had been pre-labelled by T4 polynucleotide kinase in the presence of γ32P-ATP (Perkin-Elmer, 3000 Ci/mmol), using pGEM T-easy plasmids carrying wt or mutated versions of the different RP promoters. The primers and templates used to generate each EMSA substrate are listed in Supplementary Table S3. Radiolabelled amplicons were purified by the GEL/PCR Purification Kit (Fisher Molecular Biology) using the manifacturer's PCR-clean up protocol. Increasing protein amounts were pre-incubated 30 min at 22°C with 25 fmol of non-specific competitor DNA (a PCR linear fragment with no Fhl1 binding sites). Specific radiolabelled DNA probe (25 fmol) was then added, and the incubation continued for 30 min at 22°C in 10 μl (final volume) of 20 mM Tris/HCl pH 7.6, 100 mM KCl, 5% glycerol (v/v), 0.5 mM DTT, 0.1 mg/ml bovine serum albumin, 1 mM EDTA. At the end of incubation, reaction mixtures were loaded onto a 6% native polyacrylamide gel (5.8% acrylamide; 0.2% bisacrylamide) in TBE buffer (45 mM Tris/borate, pH 8.0, 1 mM EDTA). Gels were run 3–4 h at 4°C, vacuum dried and exposed to phosphorimaging plates (Packard Cyclone, Alliance Analytical).
RESULTS
Phylogenetic footprinting analysis of Abf1-demarcated RP gene promoters reveals a conserved Fhl1 binding site
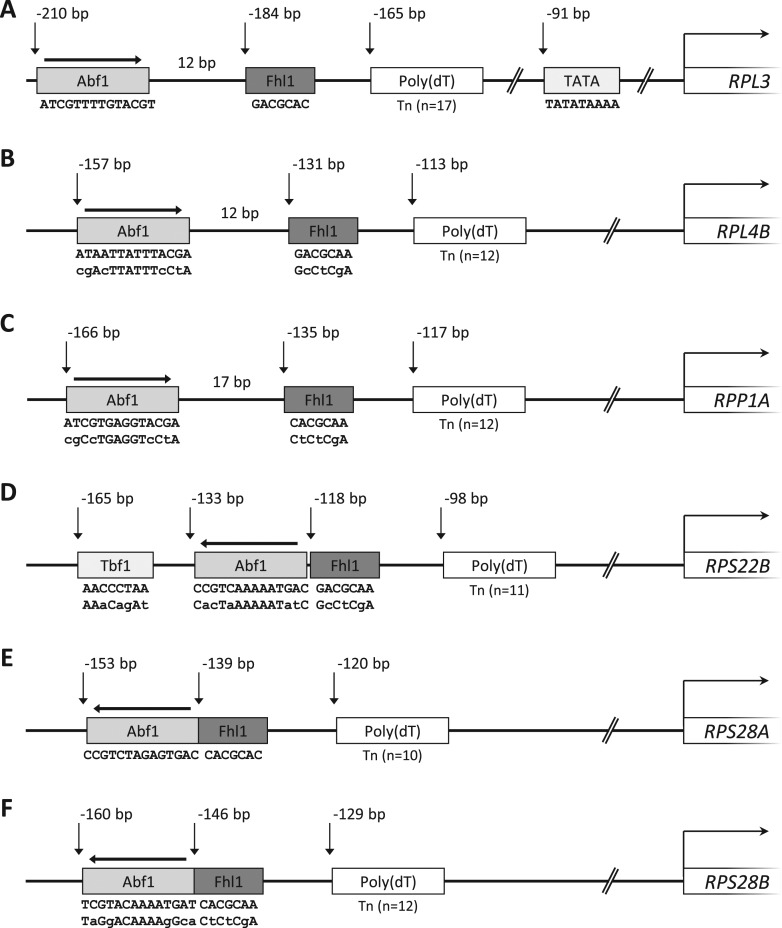
Our study initially focused on eight S. cerevisiae RPGs whose promoters display an Abf1 binding site instead of a Rap1 binding site: RPL3, RPL4A, RPL4B, RPP1A, RPP2B, RPS22B, RPS28A, RPS28B (4). To identify cis-acting control elements and their organization in these promoters, we employed a bioinformatic approach based on the detection of conserved sequence motifs in the aligned orthologous RP promoter sequences of four Saccharomyces species: S. cerevisiae, S. paradoxus, S. mikatae and S. kudriavzevii (28,36). Starting from the S. cerevisiae sequence of each gene, the orthologs in the other three hemiascomycetes were identified. The upstream sequences from the different genomes were then aligned using CLUSTAL X (26). The alignments (Supplementary Figure S1) revealed, for six of these promoters (RPL3, RPL4B, RPP1A, RPS22B, RPS28A and RPS28B), a common architecture in which a single Abf1 binding site is followed downstream by a nearby located, conserved 7-bp sequence element (consensus SACGCAM; S = G or C, M = A or C), which in turn is followed by a 10–17 bp poly(dT) tract at a position ∼120 bp upstream of the TSS. The consensus sequence of the 7-bp element corresponds to the binding site of the transcription factor (TF) Fhl1, as initially shown by studies based on protein binding microarrays (16,17,37) and later confirmed by ChIP-seq (chromatin immunoprecipitation with high-throughput DNA sequencing) and MITOMI (mechanically induced trapping of molecular interactions) (6). As illustrated in Figure 1, the orientation and relative position of Abf1 and Fhl1 binding sites vary according to two architecture sub-types. In the case of RPL3, RPL4B and RPP1A (Figure 1A–C) the Abf1 site (RTCRYnnnnnACGR) has a forward orientation and is separated from the downstream Fhl1 site by 12–17 bp. In RPS22B, RPS28A and RPS28B promoter regions (Figure 1D–F) the Abf1 site has a reversed orientation and is immediately followed by the Fhl1 site. As expected for RPGs, all the promoters are TATA-less, with the notable exception of the essential RPL3 gene, displaying an evolutionarily conserved TATA box 91 bp upstream of the TSS.
Figure 1.
Architecture of Abf1-dependent RP gene promoters. Graphical representation of promoter region architecture for RPL3 (A), RPL4B (B), RPP1A (C), RPS22B (D), RPS28A (E), RPS28B (F). The indicated positions of conserved sequence elements refer to the distance from the TSS, corresponding to the 5′ end of the most abundant transcript among those recently mapped by TIF-seq (27). The wt sequences of the cis-regulatory elements and the corresponding mutated versions considered in this study are reported below the corresponding boxes (mutated bases in lowercase).
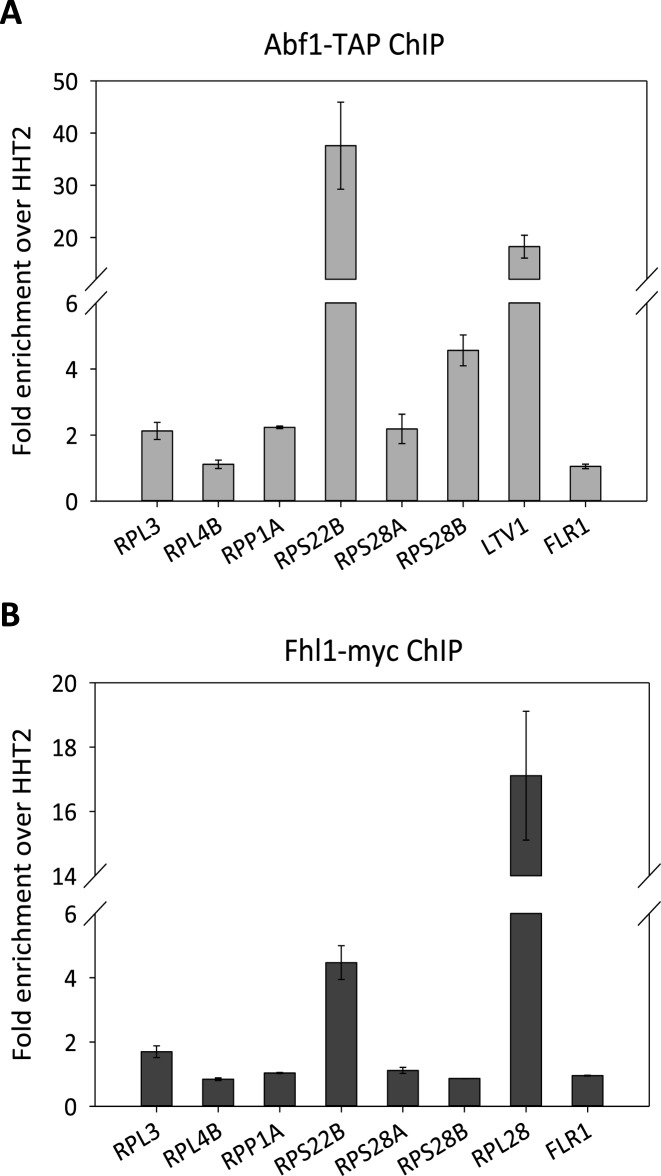
Variable extents of Abf1 and Fhl1 association with RP gene promoters
Actual binding of Abf1 and Fhl1 to their cognate sites within RPG promoter regions was addressed by chromatin immunoprecipitation followed by quantitative PCR analysis (ChIP-qPCR) using a TAP-tagged ABF1 strain (31) and an FHL1-13MYC strain. Abf1 was found enriched at least 2-fold (with respect to HHT2 used as a standard) at RPL3, RPP1A, RPS22B, RPS28A and RPS28B promoters, but not at RPL4B promoter. RPS22B promoter region displayed by far the strongest Abf1 enrichment (Figure 2A). In contrast, Fhl1 was not found to be enriched at the tested RP promoters, with the notable exception of RPS22B and, to a much lesser extent, of RPL3 (1.7-fold enrichment) (Figure 2B). The general lack of ChIP-detectable association of Fhl1 with these promoters was confirmed using other tags (TAP-tag, HA-tag; data not shown), and is in agreement with the results of previous ChIP analyses, which also reported only a modest enrichment of Fhl1 at RPL3 promoter (20,38) and an ∼5-fold enrichment of the same protein at RPS22B promoter (6,8).
Figure 2.
ChIP-qPCR analysis of Abf1 and Fhl1 association to RP gene promoters. (A) Abf1 ChIP analysis was conducted using a strain carrying a TAP-tagged ABF1 allele. Fold-enrichment of the indicated RP gene promoters was calculated relative to HHT2 used as an internal standard. Positive (LTV1) and negative (FLR1) controls for Abf1 association were analyzed in the same experiment. (B) Fhl1 ChIP analysis was performed using a strain carrying a 13myc-tagged allele of FHL1. Positive (RPL28) and negative (FLR1) controls for Fhl1 association were analyzed in the same experiment. For both panels, data are the average of two independent replicates, with bars indicating the standard error.
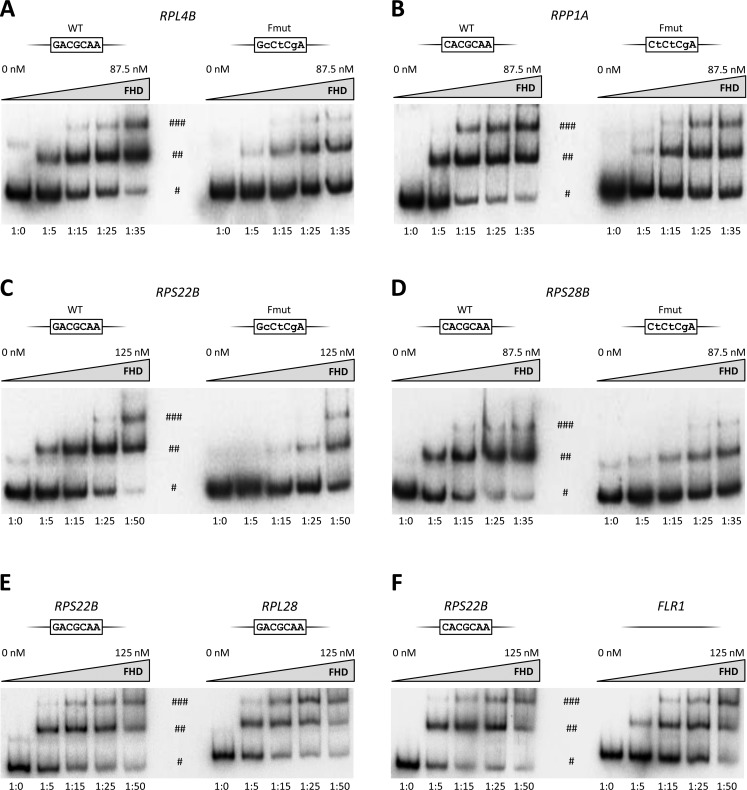
In vitro evidence of Fhl1 binding to Abf1-associated RP gene promoters
Given the evolutionary conservation of Fhl1 binding sites and their conformity to consensus sequence, the lack of detection by ChIP of the interaction between Fhl1 and its cognate sites in most of the tested RPG promoters is puzzling. As a possible explanation, the N-terminal tag in Fhl1 might be poorly accessible to the ChIP antibody or the protein could be inefficiently cross-linked, specifically at these promoters (but not at the RPL28 promoter used as positive control). Alternatively, the putative Fhl1 binding sites in the particular sequence context of RPG promoters might represent largely sub-optimal Fhl1 targets in the genome (39). To gain insight into this issue, we expressed in Escherichia coli and purified the DNA binding domain of Fhl1 (FHD; see Supplementary Figure S2), and tested its interaction with RPG promoters by EMSA. In particular we investigated the interactions of Fhl1 with the promoter regions of RPL4B, RPP1A, RPS22B and RPS28B. As in ChIP analysis, the promoter regions of RPL28 and FLR1 were used as positive and negative controls, respectively. As shown in Figure 3, FHD was found to associate with each of the four Abf1-RPG promoters, as well as with RPL28 promoter, with an appreciably higher affinity than it did with promoters whose Fhl1-binding site had been disrupted. Residual FHD binding to mutated promoters was observed in all cases, together with the appearance (occurring with all used probes) of a super-shifted band at the highest FHD concentrations, suggesting that more than one monomer might be accommodated onto the fragment, possibly by virtue of the high isoelectric point of the polypeptide (estimated pI = 9.8). Such a basic character of FHD is probably responsible also for low-affinity binding to promoter DNA displaying no Fhl1-binding motif (Figure 3F) and to mutated RPG promoter regions. Overall, these results suggest that the Fhl1 binding sites within the Abf1-RPG promoter regions can be specifically recognized by the FHD of Fhl1. Albeit not conclusive, this observation prompted us to address with further analysis the contribution of both Fhl1 and Abf1 binding sites to promoter activity.
Figure 3.
Sequence-specific binding of Fhl1 FH domain to RPG promoters. EMSA analyses performed to evaluate the association of purified Fhl1 FH domain (FHD) with the promoter region of RPL4B (A), RPP1A (B), RPS22B (C) and RPS28B (D). In each of these cases, wt and mutant (Fmut, altered in the Fhl1 binding site) promoter region fragments were compared for their ability to associate with increasing concentrations of FHD (indicated above the gel images). The wt and mutated sequences of Fhl1 binding site for each probe are indicated on the top of each panel. The results of EMSA evaluating the association of FHD with RPL28 (positive control) and FLR1 (non-specific DNA, negative control) promoter regions are shown in panels E and F, respectively, in comparison with binding to RPS22B promoter. The promoter DNA:FHD molar ratio is reported below each lane. (#) Free radiolabelled probe; (##) FHD-probe shifted complex; (###) supershifted complex likely corresponding to DNA fragments bound by two FHD molecules.
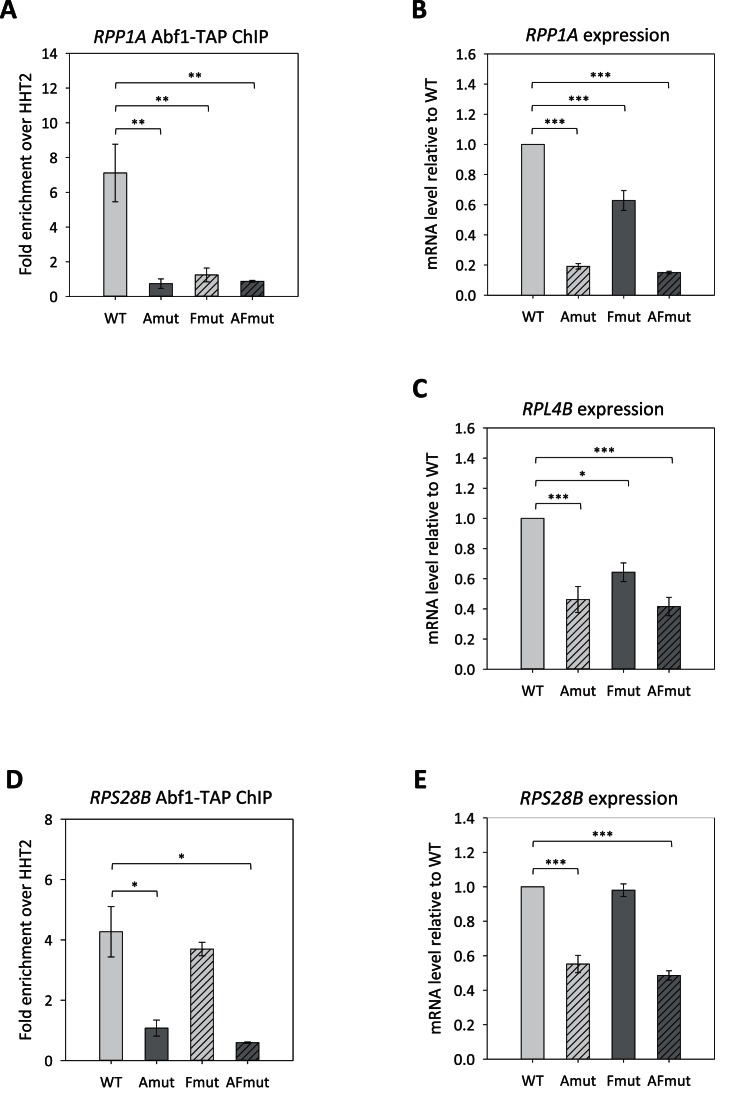
In vivo mutational analysis of RPL4B, RPP1A and RPS28B promoters
We chose to mutagenize the four promoters (RPL4B, RPP1A, RPS28B and RPS22B) whose binding by FHD had been verified by EMSA. Disrupting base substitutions were introduced into Abf1 and Fhl1 binding sites, alone (generating the mutants indicated as ‘Amut’ and ‘Fmut’, respectively) and in combination (producing the mutants named ‘AFmut’), at the RPL4B, RPP1A and RPS28B genomic loci by a two-step gene replacement strategy, as previously described (28). Due to its complex architecture, the RPS22B promoter region required more extensive mutagenesis and analysis, as detailed in the dedicated section below.
For each wt and promoter mutant strain, mRNA levels were measured by RT-qPCR. Abf1 enrichment was also evaluated by ChIP-qPCR, but not for RPL4B whose interaction with Abf1 was not easily detectable by ChIP (see above, Figure 2A). The results for RPL4B, RPP1A and RPS28B promoters are shown in Figure 4. Abf1 site mutation produced a significant decrease in RPG expression, most evident in the case of RPP1A (∼5-fold reduction). For RPP1A and RPS28B, expression reduction was concomitant with the loss of Abf1 association with the promoter region. Considering RPP1A, it is worth noting that disruption of the Fhl1 recognition site strongly reduced Abf1 binding (Figure 4A), thus suggesting a role of Fhl1 in favouring the association of Abf1 with its binding site located ∼20 bp upstream. For both RPP1A and RPL4B Fhl1 site disruption resulted in a moderate, yet significant decrease of RPG expression (Figure 4B and C). By contrast, Fhl1 site mutation within the RPS28B promoter context did not produce any significant reduction in gene expression (Figure 4E). Double mutants, in which both Abf1 and Fhl1 recognition motifs were mutationally altered, displayed RPG expression levels only slightly lower than those observed upon disruption of Abf1 site alone. Residual expression levels generally observed in mutants of both TF binding sites are likely due to the positive contribution of poly(dT) elements to RPG transcription (7,40).
Figure 4.
Abf1 and Fhl1 role and interactions at three RP gene promoters. The Abf1 and Fhl1 binding sites were mutated, alone (Amut, Fmut) or in combination (AFmut), in the promoter regions of RPP1A (A, B), RPL4B (C) and RPS28B (D, E), in a TAP-tagged ABF1 strain. For each promoter mutant, Abf1-promoter association (A, D) was then assessed by ChIP-qPCR and expression levels (B, C, E) were measured by RT-qPCR. Data are represented as mean ± SEM of three independent experiments. A one-way ANOVA test was used to compare the means of measurements for the set of strains in each experiment. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 using a Tukey post-hoc test.
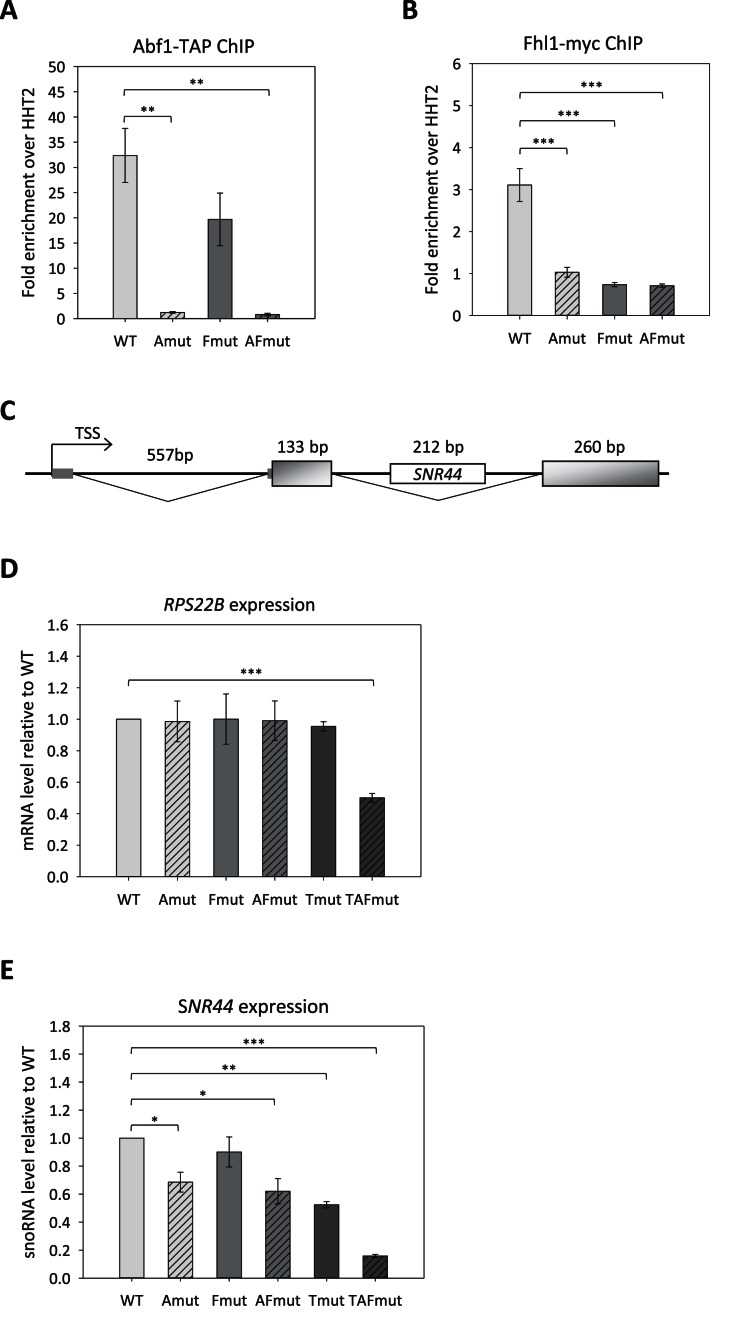
In vivo mutational analysis of RPS22B promoter
In the promoter region of RPS22B, the orientation and relative position of Abf1 and Fhl1 sites is very similar to the one of RPS28B; in particular the Abf1 site is immediately followed by an Fhl1 motif which, as revealed by ChIP, is bound in vivo by this TF (Figure 2B). RPS22B is characterized by a complex transcription unit organization (Figure 5C) with two introns of ∼500 bp, the first one located into the 5′ UTR sequence and the second one interrupting the coding region. Moreover, the snoRNA gene SNR44 is hosted within the downstream RPS22B intron. Interestingly, promoter alignment revealed the presence of an additional, evolutionarily conserved sequence block, located ∼20 bp upstream of the Abf1 site, that corresponds to the recognition site for Tbf1, known to be involved in snoRNA gene expression (Supplementary Figure S1 and Figure 1D). Accordingly, Tbf1 association with this promoter was previously revealed by ChIP (28). As shown in Figure 5, when the Abf1 and Fhl1 sites were mutated, individually or in combination, even though both Abf1 and Fhl1 associations were compromised (panels A and B), no effect on RPS22B expression was observed (Figure 5D), while a moderate but significant reduction could be detected for SNR44 expression (panel E). Remarkably, the association of Fhl1 with this promoter region was severely affected both by mutation of its cognate site and by Abf1 site disruption, thus indicating that efficient Fhl1 recruitment needs not only a specific recognition element on DNA but also the simultaneous presence of closely bound Abf1.
Figure 5.
Abf1and Fhl1 role and interactions at the RPS22B promoter. Abf1 and Fhl1 binding sites were mutated alone (Amut, Fmut) or in combination (AFmut) within the promoter region of RPS22B both in a TAP-tagged ABF1 and in a 13myc-tagged FHL1 strain. For each promoter mutant, (A) Abf1- and (B) Fhl1-promoter association was assessed by ChIP-qPCR. (C) Schematic representation of RPS22B transcription unit. (D) RPS22B and (E) SNR44 expression levels measured in Abf1-TAP wt and derivative strains carrying mutated Abf1, Fhl1 and Tbf1 binding sites, alone (Amut, Fmut, Tmut) or in combination (AFmut, TAFmut). Experiments in panels A, B, D and E were performed in triplicate. Data are represented as mean ± SEM. A one-way ANOVA test was used to compare the means of measurements for the set of strains in each experiment. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 using a Tukey post-hoc test.
The lack of RPS22B expression impairment by Abf1/Fhl1 site disruption might be due to the presence of the Tbf1 site in the promoter region. Indeed, its disruption in combination with Abf1 and Fhl1 site mutations produced a 2-fold, statistically significant reduction in RPS22B expression. Remarkably the expression levels of SNR44, nested within the second intron of RPS22B, were much more sensitive to promoter mutations than host gene expression. The combination of Abf1 and Fhl1 site mutations produced an almost 2-fold decrease in snR44 RNA levels, that dropped to 16% of the wt in the triple (Abf1, Fhl1, Tbf1 site) promoter mutant (Figure 5E).
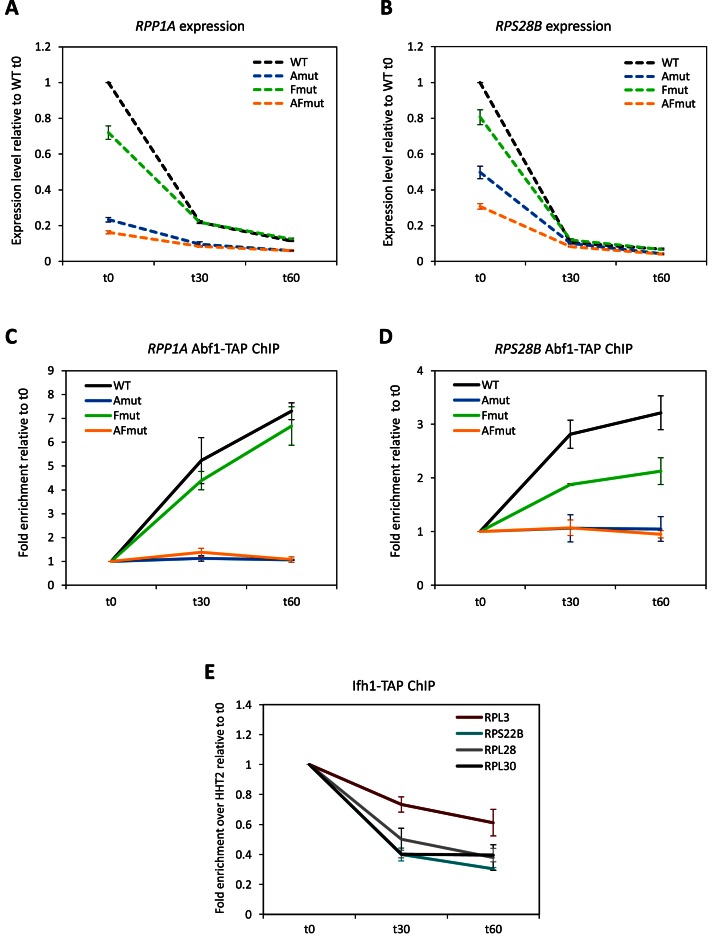
Involvement of Abf1 in TOR-dependent RP gene regulation
RPG expression is known to be down-regulated upon inhibition of TORC1-dependent signalling by rapamycin (41,42). To investigate the occurrence of such a response at Abf1-dependent RP promoters, and to assess its Abf1 binding requirement, we subjected to rapamycin treatment, followed by Abf1 promoter association and expression analyses, the wt, rpp1a and rps28b promoter mutant strains (see above, Figure 4), all constructed within an ABF1-TAP background. As shown in Figure 6A and B, intact RPP1A and RPS28B promoters supported a typical RPG response to rapamycin, with expression decreasing by 5- to 10-fold after 30 min of treatment. The Fhl1 binding site was not required for this response. Mutating the Abf1 binding site, either alone or in combination with Fhl1 motif, resulted as expected in a decreased starting level of RPG expression, more severe in the case of RPP1A. Rapamycin treatment still caused a decrease of transcription from ‘Amut’ and ‘AFmut’ RPP1A mutants. Such a decrease, however, was much more modest than the one observed with wt promoter, not unexpectedly given the much lower starting level of transcription (Figure 6A). In the case of RPS28B, where the lack of Abf1 binding only caused a ∼2-fold decrease in expression, rapamycin-dependent down-regulation was rather strong even with an Abf1 site-less promoter (Figure 6B). Therefore, Abf1 is required at these promoters to support expression levels susceptible to full down-regulation in response to rapamycin. Nevertheless, appreciable repression can still occur at promoters unable to bind Abf1.
Figure 6.
Response of Abf1-dependent RPG promoters to TOR pathway inactivation. (A–D) Exponentially growing, ABF1 TAP-tagged yeast strains, carrying either wt or promoter-mutated RP genes were treated with rapamycin (200 ng/ml). After 30 or 60 min of treatment, cells were collected for expression and Abf1 ChIP analyses of RPP1A (A, C) and RPS28B (B, D) genes. Expression levels are reported as relative to untreated wt cells; Abf1 enrichment for each strain is reported as relative to untreated cells of the same strain (t0). Data were collected from three independent replicates and are presented as mean ± SEM. (E) An exponentially growing, IFH1 TAP-tagged strain was treated with rapamycin (200 ng/ml). After 30 or 60 min of treatment, cells were collected for Ifh1 ChIP analysis of the indicated RPGs. Ifh1 enrichment for each gene is reported relative to the association measured in untreated cells (t0). Data were collected from three independent replicates and are presented as mean ± SEM. Data relative to RPL4B, RPP1A, RPS28A and RPS28B (see text) are not reported in the figure, as no significant enrichment over HHT2 was observed even at t0.
Since previous studies reported that Rap1 association to its target RPG promoters does not change in response to rapamycin treatment (8,9), we asked whether Abf1 association is similarly unaffected by TORC1 inhibition. Surprisingly, at RPP1A and RPS28B promoters rapamycin-dependent down-regulation was accompanied by a large increase in Abf1 association, which was strictly dependent on the presence of Abf1 binding sites (and also appeared to be influenced by the Fhl1 site in the case of RPS28B) (Figure 6C and D). RPL4B and RPS22B promoters behaved similarly, with rapamycin-induced transcriptional down-regulation accompanied by largely increased recruitment of Abf1 but not of Fhl1 (Supplementary Figure S3). To ascertain whether this Abf1 behaviour is specific to RPGs, we investigated in the same rapamycin-treated cell samples the possible changes in Abf1 association with five additional Abf1-bound genes: the ribosome-unrelated IPP1, PIK1 and YKT6 genes, and the Ribi genes LTV1 and NOP12. As shown in Supplementary Figure S4, a 2- to 4-fold increase in Abf1 association was observed at each of the three ribosome-unrelated promoters 30 min after rapamycin treatment, followed by a decrease at 60 min post-rapamycin (Supplementary Figure S4, panels C–E). These changes at promoters were accompanied by small fluctuations in gene expression levels. Such molecular changes clearly differed from the ones observed at RPG promoters, where Abf1 association increased progressively after rapamycin treatment and expression decreased dramatically. Remarkably at the two Ribi genes, LTV1 and NOP12, which both carry an Abf1 binding site in the promoter region, the rapamycin-dependent changes in Abf1 association occurred with a time course very similar to the RPG one. Here, however, the expression response was not perfectly overlapping with RPG response: Ribi gene expression quickly dropped at 30 min post-rapamycin but tended to recover at 60 min post-treatment (Supplementary Figure S4A and B). This observation is in agreement with previous studies showing that Ribi gene expression reacts to environmental/nutritional stress conditions more promptly than RPGs (43,44). These results together reveal a widespread, yet differentiated behaviour of Abf1 at its target promoters. Such a behaviour can not be accounted for by a non-specific phenomenon such as increased abundance of Abf1 upon rapamycin treatment (which was also excluded by Western blot analysis shown in Supplementary Figure S5), and is rather suggestive of a complex role of Abf1 in yeast genomic response to nutritional cues.
A well-characterized consequence of TORC1 inhibition at Rap1-RPG promoters is a reduction in Ifh1 association accompanied by transcriptional down-regulation (8,9,15). We thus examined Ifh1 binding at Abf1-RPG promoters before and after rapamycin treatment to ascertain whether the same mechanism applies to these genes as to Rap1-RPGs. In particular, we considered the Rap1-regulated RPL28 and RPL30 genes and the Abf1-associated RPL3, RPL4B, RPP1A, RPS22B, RPS28A and RPS28B genes (Figure 6E). As expected Ifh1 was found associated with RPL28 and RPL30 promoters during exponential growth, and its enrichment levels dropped after rapamycin treatment. As to Abf1-RPGs, the only promoter that displayed a clearly detectable enrichment of Ifh1 during exponential growth was RPS22B. Ifh1 was also weakly enriched at the RPL3 promoter, in agreement with the very low, yet detectable enrichment of Fhl1 at the same promoter (see Figure 2B). The general failure to detect by ChIP any clear association of Ifh1 with the other Abf1-RPGs confirms what recently observed in other studies (6,18) (but see Cai et al. (45), where weak Ifh1 binding to RPP1A, RPL3 and RPS28B was reported). As shown in Figure 6E, changes in Ifh1 enrichment at the RPS22B promoter upon TOR pathway inactivation closely matched those observed for the Rap1-RPGs. Ifh1 enrichment at RPL3, albeit initially very low, was similarly noted to further decrease upon rapamycin treatment, thus suggesting that reduced Ifh1 association is a general feature of RPG regulatory response to TOR pathway inactivation, common to Rap1- and Abf1-RPGs.
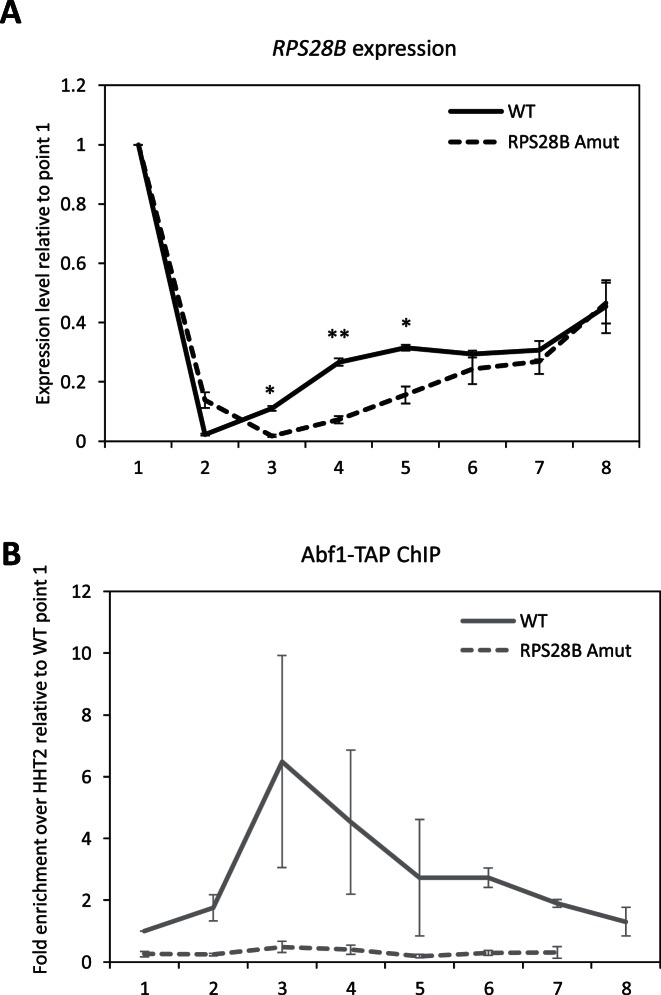
Since Abf1 was observed to play, albeit to different extents, an activator role for all tested Abf1-RPGs (Figures 4 and 5), it seemed unlikely that its increased association with RPG promoters upon TORC1 inhibition might reflect a repressor role in RPG down-regulation. Instead, it seemed more plausible that the increase in Abf1 occupancy may aid in reactivation of Abf1-RPGs once favourable growth conditions and signals are re-established. This hypothesis was experimentally addressed by exploiting the unique properties of RPS28B, whose transcriptional response to TORC1 inactivation is partially maintained and well detectable even when its promoter lacks an Abf1 binding site (see Figure 6B), and by using cell exit from stationary phase as a condition entailing TORC1-dependent RPG reactivation (9,46). Strains carrying wt or Abf1 site-mutated RPS28B were grown to stationary phase in YPD, then diluted into fresh medium and subjected to time-course analysis of Abf1 chromatin association and mRNA levels. As shown in Figure 7, RPS28B expression dropped during stationary phase in both strains. In the wt strain (but obviously not in the Abf1 site mutant) reduced transcription was accompanied by increased Abf1 association. Remarkably, at short times (5 and 10 min) after dilution of stationary phase cultures into fresh medium, the rate of RPS28B transcriptional recovery was, respectively, 3.7- and 2-fold higher in the wt than in the promoter mutant strain. The time dependence of transcription recovery appears to be roughly biphasic in the wt (Figure 7A), with an initial phase of higher recovery rate resulting in a clear anticipation of response to environmental shift in the wt with respect to mutant strain. At longer times (30–60 min) post-dilution into fresh medium, the two strains had attained a comparable extent of transcriptional recovery.
Figure 7.
Time course of Abf1-RPS28B promoter association and RPS28B expression during entry into and exit from stationary phase. Cultures of either wt or RPS28B Amut strains were grown into stationary phase, then diluted into fresh YPD medium. Samples of the two cultures were collected during exponential (1), post-diauxic (2) and stationary phase (3), and 5 min (4), 10 min (5), 20 min (6), 30 min (7) and 60 min (8) after dilution. At each time point the association of Abf1 with RPS28B promoter region was assessed by ChIP-qPCR and RPS28B mRNA levels were measured by RT-qPCR. RPS28B expression levels (A) were normalized to the geometric averaging of TAF10 and ALG9 (chosen as internal standards) expression levels, and are represented as relative to exponentially growing cells (point 1). Abf1 enrichments (B) are reported as relative to exponentially growing (point 1) wt cells. Data were collected from two independent replicates and are presented as mean value ±SEM. An unpaired t test was used to compare the means of measurements for the two strains at each time point. (*) P < 0.05, (**) P < 0.01.
DISCUSSION
In this study we addressed the transcriptional regulation of a small, yet significant subset of S. cerevisiae RPGs whose promoters, unlike most of the 138 RPGs in the budding yeast genome, have long been known to be characterized by the presence of Abf1, instead of Rap1, binding sites (4). Taken together, our data delineate a scenario in which Abf1-dependent RPGs constitute a third, specific promoter type, distinct from the two recently described categories of Rap1-dependent RPG promoters (6). This difference not only merely concerns the presence of a different DNA binding protein demarcating the upstream promoter border, but also the presence and the strictly determined location of an Fhl1 binding site and, more importantly, the regulated recruitment of Abf1 in response to stimuli, which does not occur in the case of Rap1.
In their recent identification of different types of RPG promoter architectures, Knight et al. grouped together (in category III) the promoters of a few RPGs sharing the property of being bound in vivo by Abf1 instead of Rap1, but not by any of the other RPG-associated TFs (Fhl1, Ifh1 and Hmo1) (6). Our study extends the information on these promoters, by including RPS22B promoter and by providing evidence that, together with Abf1, Fhl1 is also involved in transcription of at least some of them.
As revealed by in silico analysis, an Abf1 binding site, in either of its two possible orientations, was invariably present at a position ∼160 bp upstream of the TSS. This site was found to be followed by an Fhl1 binding motif occurring either immediately or ∼15 bp downstream of the Abf1 binding site, depending on the orientation of the latter. Mutations in Abf1 and/or Fhl1 binding motifs affected expression to different extents at the different promoters. At RPP1A, RPL4B and RPS28B Abf1 site mutation produces a sharp decrease in gene expression, particularly evident for RPP1A. The milder effect observed in the case of RPL4B and RPS28B might be a consequence of an RPG autoregulation mechanism acting at the mRNA decay level, previously demonstrated for both of these genes (47,48). Considering Fhl1 site mutation effects, it is notable that at RPS28B and RPS22B promoters, where the Fhl1 binding site is located immediately downstream of the Abf1 binding site, disruption of the Fhl1 site had no significant effect on RP gene expression. In contrast, at promoters where the Fhl1 binding site is located more downstream, like RPL4B and RPP1A, disruption of the Fhl1 site caused a modest (∼35%) yet significant decrease in RPG expression. Suggestive, even though far from being conclusive, is the hypothesis that in the presence of a spatial separation between Abf1 and Fhl1 binding sites the disruption of the latter could affect gene expression, since at these genes Fhl1 could directly interact with promoter region, while in the presence of adjacent Abf1 and Fhl1 binding sites transcription might be unaffected by Fhl1 binding site disruption because of an indirect, Abf1-mediated recruitment of Fhl1. In the particular case of RPP1A, Fhl1 site disruption also impaired Abf1 interaction with its site, an indication that Abf1 binding, at least to this promoter, likely involves the concomitant presence of another TF. TF cooperation in RPG promoter binding was also evident in the case of RPS22B, where Abf1 site disruption also severely affected Fhl1 recruitment (Figure 5B).
The architecture of RPS22B transcription unit turned out to be more composite and complicated by the presence of SNR44 gene in the second intron of this RPG. In this case Abf1 association was lost without any effect on RPG expression, but with a significant reduction of SNR44 expression. Only by disrupting an additional Tbf1 binding site, located ∼20 bp upstream of the Abf1 site, together with Abf1 and Fhl1 sites, it was possible to produce a significant RPS22B expression decrease (again accompanied by a much more marked reduction of snR44 levels). As already suggested by Knight et al. (6), the presence of Tbf1 at this promoter might represent a snapshot in evolution, showing the transition (perhaps favoured by an intermediate stage centred on sequence-specific binding of Fhl1 to target promoters) from a Tbf1-based to a Rap1-based mode of RPG transcription, the former discovered in Candida albicans and being present in most ascomycete yeasts, the latter dominating the RPG regulon in S. cerevisiae and closer ascomycete species (19,49). We further note, however, that the presence of a Tbf1 binding site could also be related to the regulation of the intron-hosted SNR44 gene. Indeed, Tbf1 site mutation, which did not produce any effect on RPS22B expression, caused a significant, 2-fold decrease of SNR44 expression, as observed for other Tbf1-controlled snoRNA genes (28). Among the possible explanations for the generally more severe effect of promoter mutations on SNR44 than on RPS22B expression, we tend to exclude the co-existence at this locus of two independent transcription units controlled by the same promoter region. Such hypothesis contrasts with the lack of evidence for the presence of distinct transcription preinitiation complexes for RPS22B and SNR44 (50), and with the general tenet that intron-located snoRNAs are generated from host pre-mRNA processing (51). Alternative explanations are that Abf1 could be required both for RPS22B transcription and for RPS22B mRNA decay, as previously shown for Rap1 at the RPL30 promoter (52), or the existence of an autoregulation mechanism (47,48) for RPS22B, in which a decrease in RPG transcription and thus in the cytosolic levels of the encoded r-protein would result in reduced mRNA (but not snoRNA) destabilization by the corresponding r-protein.
The evolutionary and/or physiological reasons for the maintenance in the budding yeast genome of a small subset of Abf1-dependent RPGs seem to be elusive. These genes do not appear to code for RPs sharing some particular features or roles within the ribosome. Their gene products comprise both universally conserved and eukaryote-specific RPs (like Rps22 and Rps28, respectively), that play roles either in core ribosome activity (e.g. Rpl4) or in mediating regulatory interactions of the ribosome (e.g. P1α). Moreover, these genes are regulated in coordination with the other RPGs in response to environmental changes (44), thus excluding that their peculiar promoter organization reflects a sharp regulatory uncoupling from other RPGs. It remains plausible, however, that the Abf1-centred promoter architecture is related to more subtle regulatory needs. Intriguing appears the possibility that this subset of RPGs, perhaps because of an involvement of their products in ribosome assembly, have to be strictly co-regulated with the large set of genes required for ribosome biogenesis (Ribi genes), whose promoters, known to be characterized by repressive regulatory sites absent from RPG promoters (PAC and RRPE), are also particularly enriched in Abf1 binding sites ((25); M.C. Bosio, in preparation). Several lines of evidence suggest that Ribi and RP genes respond similarly to perturbations, being both repressed (albeit with slightly different time courses) in the global genome expression program previously referred to as Environmental Stress Response (44). These two groups of co-regulated genes also appear to share key mechanisms of TORC1-dependent modulation, involving Sch 9-mediated phosphorylation of common transcription proteins that control the recruitment of RPDL3 histone deacetylase (53). It would thus not be too surprising if a subset of RPGs would display an even more tight sharing of regulatory strategies with Ribi genes. In support to this hypothesis we found that Abf1-RPGs share with Ribi genes, but not with other Abf1-bound genes, a similar response to TORC1 inactivation in terms of Abf1 association changes (see below).
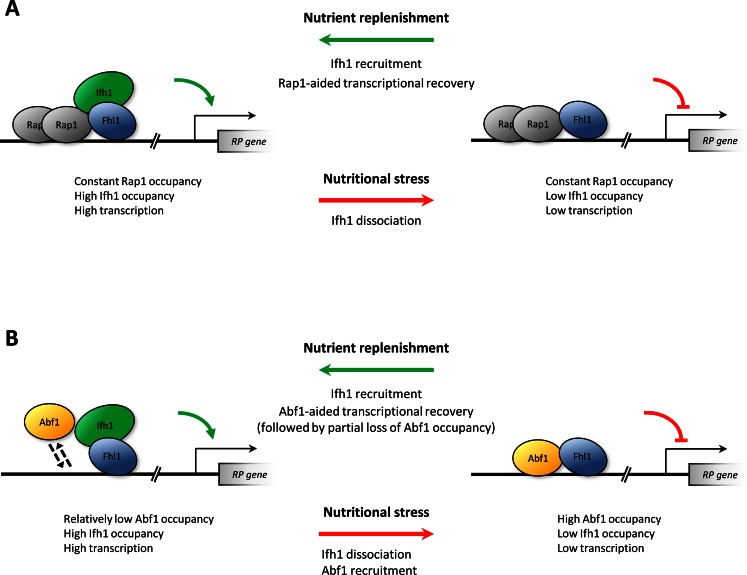
The observation that rapamycin-induced RPG down-regulation is accompanied by increased recruitment of Abf1 came largely unexpected. Abf1, like Rap1, is required for full RPG expression. Based on our data and on previous works (7,40), RPG promoter mutations preventing the binding of this TF generally result in a significant reduction of RPG transcription, as expected for a transcriptional activator. Moreover, the results of previous studies support the common notion that Rap1 association to its cognate sites in RP promoters is not affected by environmental stimuli, such as amino acid starvation, heat shock and TOR pathway inactivation (8,9,54,55), which instead influence the association of other factors, like Ifh1, Hmo1 and, at least in some strains, the corepressor Crf1 (11,15,56). A similar, perturbation-refractory behaviour has been reported for Abf1 association with RPGs, in contrast with our observations (54). We point out, however, that the two RP promoters analysed in the latter study to assess changes in Abf1 occupancy under different growth conditions, RPL9A and RPS11B, are not among those whose promoters rely exclusively on an Abf1 binding site (4,6). It is thus likely that our findings reveal an important difference between Rap1 and Abf1 regulatory interactions with RP (and perhaps other) gene promoters. While Rap1 would be permanently associated with its target promoters, where it would act as a ‘placeholder’ preventing nucleosome deposition and favouring the reversible assembly of other TFs (57), Abf1 association with target promoters would be more dynamic and responsive to environmental stimuli. Which could be the functional significance of Abf1 recruitment upon TOR pathway inactivation? Abf1 has been previously involved in silencing at mating type loci (58) and it could participate in repression mechanisms at some promoters by helping in the recruitment of a co-repressor complex. Therefore, a first possibility is that stable promoter binding by Abf1 is more strictly required for transcriptional repression under starvation conditions than for full gene activation during exponential growth on rich media, which could only require transient Abf1-promoter interaction. Indeed, according to its known properties, Abf1 activity in gene activation does not necessarily require its continuous binding to promoter sites but, at least at some promoters, it may involve a ‘memory effect’ whereby the activated state is maintained even after dissociation of Abf1 from its binding site (23–25). One could thus speculate that low levels of associated Abf1 are sufficient for maintaining the activated state of RPG promoters, and that Abf1 occupancy must increase when starvation-dependent down-regulation is to be supported. Not completely consistent with this view, however, is the observation that rapamycin-induced down-regulation also occurs for RPS28B and RPP1A even after disruption of their Abf1 binding sites (Figure 6A and B). As an alternative, more intriguing hypothesis, we considered that Abf1 recruitment upon TOR inactivation, far from being mechanistically involved in repression, could be part of a regulatory strategy in which promoter priming by Abf1 would warrant rapid transcription reactivation once more favourable growth conditions are re-established. In such a suggestive scenario, Abf1 would act as a pioneer TF anticipating gene expression rescue without interfering with the repression induced during starvation. In support to this hypothesis, we have found that during re-growth from stationary phase the time course of transcriptional rescue of RPS28B, chosen as representative of Abf1-RPGs, is influenced by Abf1, being slower when the Abf1 binding site is disrupted. More precisely, the Abf1-bound promoter was found to support an initial phase of higher recovery rate than the Abf1-less promoter, accounting for a clear anticipation of response to environmental shift which could in principle result in faster adaptation and increased fitness.
Based on these observations and on the results with rapamycin-treated cells, we propose that the reduction of TORC1 signalling under nutritional stress conditions results in similarly decreased transcription of both Rap1-RPGs and Abf1-RPGs, probably involving Ifh1 dissociation. When growth-favouring conditions are re-established, at Rap1-RPGs transcription can be rapidly rescued because Rap1, independently from growth conditions, is stably bound to the promoter region where it can directly recruit the transcription machinery (59). In contrast, Abf1 interaction with target promoters is generally more dynamic, and its presence at some promoters is not continuously required to support transcription. In the case of Abf1-RPGs, but also of at least some Ribi genes, its association with promoters under pro-growth conditions might be relatively low without affecting ongoing transcription rate. Upon nutritional stress Abf1 association with RPG/Ribi promoters would increase, in order to allow them a rapid rescue following stress cessation. These possibilities are illustrated by the model in Figure 8, that also speculate about Ifh1 dynamics based on observations that could only be made for RPS22B and RPL3 promoters. The temporary increase of Abf1 occupancy at other (perhaps many) ribosome-unrelated target promoters in response to TORC1 inactivation (suggested by data in Supplementary Figure S4) could represent an intriguing new facet of yeast global genome response to stress that deserves further investigation. It seems likely that changes in Abf1 phosphorylation state play a role in these phenomena, as Abf1 expression levels were not found by previous genome-wide studies to undergo, in response to rapamycin treatment, an increase which could justify the observed 3- to 7-fold increase in promoter enrichment (60,61), nor could we detect any significant change in its abundance upon TORC1 inhibition. Abf1 is known to be a phosphoprotein (62) and a substrate of casein kinase II (63). Phosphorylated Abf1 forms were found to predominate under nitrogen starvation conditions, while dephosphorylated forms were shown to be enriched in cells grown on a fermentable carbon source (64). Even though no differential phosphorylation states of Abf1 was put in light in previous analyses of the rapamycin-sensitive yeast phosphoproteome (60,65,66), a recent dynamic phosphoproteomics study included Abf1 among candidate proximal targets of TORC1 (67). It is also worth noting that, as recently suggested, TORC1 may regulate via its effector kinase Kns1 the association of CK2 with some of its substrates, by phosphorylating the Ckb1 subunit (68). Abf1 could be one of these substrates, and changes in its phosphorylation state might be part of the pathway through which Kns1 and Mck1 kinases mediate RPG response to TOR pathway inhibition (69).
Figure 8.
Model of Rap1- versus Abf1-based RPG transcriptional regulation. Simplified representation of TF association/dissociation in response to nutritional changes at Rap1-RPG (A) and Abf1-RPG (B) promoters. The behaviour of Ifh1 at Abf1-RPGs is illustrated as suggested by the data reported in Figure 6E.
Supplementary Material
Acknowledgments
We thank Enrico Baruffini (University of Parma), Olivier Lefebvre (CEA-Saclay, France) and Sebastien Fribourg (IECB, Pessac, France) for plasmids and E. coli expression strains, and Rodolfo Negri (Sapienza Università di Roma) and Nicola Doniselli (University of Parma) for discussions.
Footnotes
Present address: Maria Cristina Bosio, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Celoria 26, 20133 Milano, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Education, University and Research (MIUR, PRIN 2009); Italian Association for Cancer Research (AIRC IG 16877). Funding for open access charge: Italian Association for Cancer Research (AIRC IG2015-16877).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bosio M.C., Negri R., Dieci G. Promoter architectures in the yeast ribosomal expression program. Transcription. 2011;2:71–77. doi: 10.4161/trns.2.2.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 3.Loewith R., Hall M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lascaris R.F., Mager W.H., Planta R.J. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. doi: 10.1093/bioinformatics/15.4.267. [DOI] [PubMed] [Google Scholar]
- 5.Lieb J.D., Liu X., Botstein D., Brown P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 6.Knight B., Kubik S., Ghosh B., Bruzzone M.J., Geertz M., Martin V., Denervaud N., Jacquet P., Ozkan B., Rougemont J., et al. Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev. 2014;28:1695–1709. doi: 10.1101/gad.244434.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lascaris R.F., Groot E., Hoen P.B., Mager W.H., Planta R.J. Different roles for abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene. Nucleic Acids Res. 2000;28:1390–1396. doi: 10.1093/nar/28.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wade J.T., Hall D.B., Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- 9.Schawalder S.B., Kabani M., Howald I., Choudhury U., Werner M., Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 10.Hall D.B., Wade J.T., Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger A.B., Decourty L., Badis G., Nehrbass U., Jacquier A., Gadal O. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell Biol. 2007;27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasahara K., Ohtsuki K., Ki S., Aoyama K., Takahashi H., Kobayashi T., Shirahige K., Kokubo T. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marion R.M., Regev A., Segal E., Barash Y., Koller D., Friedman N., O'Shea E.K. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl Acad. Sci. U.S.A. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin D.E., Soulard A., Hall M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 16.Zhu C., Byers K.J., McCord R.P., Shi Z., Berger M.F., Newburger D.E., Saulrieta K., Smith Z., Shah M.V., Radhakrishnan M., et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badis G., Chan E.T., van Bakel H., Pena-Castillo L., Tillo D., Tsui K., Carlson C.D., Gossett A.J., Hasinoff M.J., Warren C.L., et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reja R., Vinayachandran V., Ghosh S., Pugh B.F. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 2015;29:1942–1954. doi: 10.1101/gad.268896.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavoie H., Hogues H., Mallick J., Sellam A., Nantel A., Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudra D., Zhao Y., Warner J.R. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarragudi A., Miyake T., Li R., Morse R.H. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell Biol. 2004;24:9152–9164. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goncalves P.M., Maurer K., van Nieuw Amerongen G., Bergkamp-Steffens K., Mager W.H., Planta R.J. C-terminal domains of general regulatory factors Abf1p and Rap1p in Saccharomyces cerevisiae display functional similarity. Mol. Microbiol. 1996;19:535–543. doi: 10.1046/j.1365-2958.1996.404939.x. [DOI] [PubMed] [Google Scholar]
- 23.Schroeder S.C., Weil P.A. Genetic tests of the role of Abf1p in driving transcription of the yeast TATA box bindng protein-encoding gene, SPT15. J. Biol. Chem. 1998;273:19884–19891. doi: 10.1074/jbc.273.31.19884. [DOI] [PubMed] [Google Scholar]
- 24.Paul E., Tirosh I., Lai W., Buck M.J., Palumbo M.J., Morse R.H. Chromatin mediation of a transcriptional memory effect in yeast. G3 (Bethesda) 2015;5:829–838. doi: 10.1534/g3.115.017418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarragudi A., Parfrey L.W., Morse R.H. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:193–202. doi: 10.1093/nar/gkl1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelechano V., Wei W., Steinmetz L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preti M., Ribeyre C., Pascali C., Bosio M.C., Cortelazzi B., Rougemont J., Guarnera E., Naef F., Shore D., Dieci G. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell. 2010;38:614–620. doi: 10.1016/j.molcel.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston M., Riles L., Hegemann J.H. Gene disruption. Methods Enzymol. 2002;350:290–315. doi: 10.1016/s0076-6879(02)50970-8. [DOI] [PubMed] [Google Scholar]
- 31.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 32.Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 33.Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Braglia P., Dugas S.L., Donze D., Dieci G. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:1545–1557. doi: 10.1128/MCB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa S., Gisselbrecht S.S., Rogers J.M., Hartl D.L., Bulyk M.L. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl Acad. Sci. U.S.A. 2013;110:12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levo M., Zalckvar E., Sharon E., Dantas Machado A.C., Kalma Y., Lotam-Pompan M., Weinberger A., Yakhini Z., Rohs R., Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015;25:1018–1029. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goncalves P.M., Griffioen G., Minnee R., Bosma M., Kraakman L.S., Mager W.H., Planta R.J. Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Res. 1995;23:1475–1480. doi: 10.1093/nar/23.9.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powers T., Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardenas M.E., Cutler N.S., Lorenz M.C., Di Como C.J., Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade C., Shea K.A., Jensen R.V., McAlear M.A. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell Biol. 2001;21:8638–8650. doi: 10.1128/MCB.21.24.8638-8650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai L., McCormick M.A., Kennedy B.K., Tu B.P. Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep. 2013;4:1063–1071. doi: 10.1016/j.celrep.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galdieri L., Mehrotra S., Yu S., Vancura A. Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS. 2010;14:629–638. doi: 10.1089/omi.2010.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badis G., Saveanu C., Fromont-Racine M., Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 48.Presutti C., Ciafre S.A., Bozzoni I. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 1991;10:2215–2221. doi: 10.1002/j.1460-2075.1991.tb07757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogues H., Lavoie H., Sellam A., Mangos M., Roemer T., Purisima E., Nantel A., Whiteway M. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell. 2008;29:552–562. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer A., Lidschreiber M., Siebert M., Leike K., Soding J., Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 51.Dieci G., Preti M., Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Bregman A., Avraham-Kelbert M., Barkai O., Duek L., Guterman A., Choder M. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–1483. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P., Tyers M., Rougemont J., Beyer A.L., Loewith R. Sch 9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30:3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohde J.R., Cardenas M.E. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell Biol. 2003;23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reid J.L., Iyer V.R., Brown P.O., Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y., McIntosh K.B., Rudra D., Schawalder S., Shore D., Warner J.R. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell Biol. 2006;26:4853–4862. doi: 10.1128/MCB.02367-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharya A., Warner J.R. Tbf1 or not Tbf1? Mol. Cell. 2008;29:537–538. doi: 10.1016/j.molcel.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Loo S., Laurenson P., Foss M., Dillin A., Rine J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papai G., Tripathi M.K., Ruhlmann C., Layer J.H., Weil P.A., Schultz P. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature. 2010;465:956–960. doi: 10.1038/nature09080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iesmantavicius V., Weinert B.T., Choudhary C. Convergence of ubiquitylation and phosphorylation signaling in rapamycin-treated yeast cells. Mol. Cell Proteomics. 2014;13:1979–1992. doi: 10.1074/mcp.O113.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hardwick J.S., Kuruvilla F.G., Tong J.K., Shamji A.F., Schreiber S.L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl Acad. Sci. U.S.A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Francesconi S.C., Eisenberg S. The multifunctional protein OBF1 is phosphorylated at serine and threonine residues in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. U.S.A. 1991;88:4089–4093. doi: 10.1073/pnas.88.10.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Upton T., Wiltshire S., Francesconi S., Eisenberg S. ABF1 Ser-720 is a predominant phosphorylation site for casein kinase II of Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:16153–16159. doi: 10.1074/jbc.270.27.16153. [DOI] [PubMed] [Google Scholar]
- 64.Silve S., Rhode P.R., Coll B., Campbell J., Poyton R.O. ABF1 is a phosphoprotein and plays a role in carbon source control of COX6 transcription in Saccharomyces cerevisiae. Mol. Cell Biol. 1992;12:4197–4208. doi: 10.1128/mcb.12.9.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soulard A., Cremonesi A., Moes S., Schutz F., Jeno P., Hall M.N. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch 9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliveira A.P., Ludwig C., Zampieri M., Weisser H., Aebersold R., Sauer U. Dynamic phosphoproteomics reveals TORC1-dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci. Signal. 2015;8:rs4. doi: 10.1126/scisignal.2005768. [DOI] [PubMed] [Google Scholar]
- 68.Sanchez-Casalongue M.E., Lee J., Diamond A., Shuldiner S., Moir R.D., Willis I.M. Differential phosphorylation of a regulatory subunit of protein kinase CK2 by target of rapamycin complex 1 signaling and the Cdc-like kinase Kns1. J. Biol. Chem. 2015;290:7221–7233. doi: 10.1074/jbc.M114.626523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J., Moir R.D., McIntosh K.B., Willis I.M. TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell. 2012;45:836–843. doi: 10.1016/j.molcel.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.