Abstract
Out of the myriad of potential DNA binding sites of the glucocorticoid receptor (GR) found in the human genome, only a cell-type specific minority is actually bound, indicating that the presence of a recognition sequence alone is insufficient to specify where GR binds. Cooperative interactions with other transcription factors (TFs) are known to contribute to binding specificity. Here, we reasoned that sequence signals preventing GR recruitment to certain loci provide an alternative means to confer specificity. Motif analyses uncovered candidate Negative Regulatory Sequences (NRSs) that interfere with genomic GR binding. Subsequent functional analyses demonstrated that NRSs indeed prevent GR binding to nearby response elements. We show that NRS activity is conserved across species, found in most tissues and that they also interfere with the genomic binding of other TFs. Interestingly, the effects of NRSs appear not to be a simple consequence of changes in chromatin accessibility. Instead, we find that NRSs interact with proteins found at sub-nuclear structures called paraspeckles and that these proteins might mediate the repressive effects of NRSs. Together, our studies suggest that the joint influence of positive and negative sequence signals partition the genome into regions where GR can bind and those where it cannot.
INTRODUCTION
A critical step in the control of gene expression is played by transcriptional regulatory factors (TFs) that bind to genomic DNA binding sites and regulate target genes. In eukaryotes, TFs generally have short (6–20 bp) and degenerate recognition sequences. For example, the glucocorticoid receptor (GR), a hormone-controlled TF, can bind regions matching the consensus sequence at just six positions over the 15 positions of the motif (1). This means that a myriad of potential GR binding sequences (GBSs) are encoded in the genome of which only a cell-type specific minority is bound. Therefore, mechanisms must exist that specify which of the potential GBSs are actually bound. For one, the chromatin context in which GBSs are embedded plays a pivotal role in restricting where in the genome GR can bind with the majority of GR binding occurring at preexisting loci of accessible chromatin as specified by DNase I sensitivity assays (2). Furthermore, nucleosome presence and positioning influence GR binding (3,4) and DNA methylation correlates with TF binding and might directly interfere with the ability of TFs to bind DNA (5). Another level of organization that might influence where TFs bind is provided by the three dimensional organization of the genome in the nucleus. For example, inactive genomic regions are enriched in lamin-associated domains (6) that often accumulate in the nuclear periphery or near nucleoli (7). Similarly, the nucleus contains several distinct nuclear bodies, including paraspeckles, which are linked to regulation of gene expression (8).
The information necessary to specify the genome-wide pattern of TF binding is, at least in part, encoded in the DNA sequence. In theory, such sequence-encoded signals can influence TF binding either positively or negatively. Candidate sequence signals can be identified by computational approaches that search for overrepresented sequences at TF-bound regions. For example, in genomic regions bound by GR, the over-represented motifs correspond to GR binding preferences, in addition to several other sequence motifs of cooperating TFs (9,10). These cooperating factors can play a role in making chromatin accessible to facilitate GR binding to nearby GBSs (9).
Here, we focus instead on sequence motifs that are under-represented at GR bound loci. So far, most of the focus has been on over-represented sequences, which are good candidates to play a role in either directly or indirectly recruiting TFs of interest to defined genomic loci. We reasoned that under-represented motifs might serve the opposite function, i.e. restricting GR binding, and their functional analysis could contribute to understanding which signals (positive and negative) guide GR to the appropriate genomic loci. We tested the activity of such under-represented sequences and found that they can indeed interfere with GR binding to nearby GBSs by mechanisms that appear not to involve changes in chromatin accessibility, but rather implicate proteins associated with a specific sub-nuclear structure called paraspeckles.
MATERIALS AND METHODS
Cell lines, transient transfections and luciferase assays
U2OS and U2OS cells stably expressing GR (11) were grown in DMEM supplemented with 5% fetal bovine serum (FBS). Transient transfections were done essentially as described (12). Luciferase activity was measured using the dual luciferase assay kit (Promega).
ChIP-seq data sets
We identified GR ChIP-seq peaks in U2OS cells previously (13) (ArrayExpress accession:E-MTAB-2731). The other public data sets were downloaded as processed peak BED files from GEO (www.ncbi.nlm.nih.gov/geo/) (MyoD: GSM1197184, HoxD13: GSM1091901). The MyoD and HoxD13 peak coordinates were relative to the mouse assembly mm9 and the chicken assembly galGal3, respectively.
Motif analysis of TF-bound regions
The ChIP-seq peak sequences were first extended (±4 kb around the peak summit) and then scanned with the complete TRANSFAC collection of vertebrate position-specific scoring matrices (PSSMs) (version 2010.1, 907 motifs), using the program RSAT matrix-scan (14,15). Sequence segments were considered as a match if the P-value associated to the weight score was less than 10−3. For each sequence segment, the weight score is a classic log ratio of the likelihood between two models: the motif model (represented by the PSSM) and the background model that should best recapitulate the searched sequences (14). Here, a specific background model was trained for each TF data set on the peak sequences using a Markov chain of order 1, which accounts for the CpG depletion of vertebrate genomes and all other dinucleotide content. To ensure that the results were robust to the chosen background model, we have performed additional tests varying the background model (various training sets or markov orders, including a local sliding window; see Supplementary Notes). The number of matches at each position were then summed and normalized by the number of scanned regions. Scanning a large 8 kb window around the peak summit ensures to visualize the ‘basal’ number of matches for each motif. A P-value of the local under/over-representation of motifs is computed using a chi-square test in 50 bp bins, where the expected number of matches is uniform, by dividing the total number of matches over the 8 kb sequences by the number of bins. We also performed de novo motif discovery as complementary approach to motif scanning (Supplementary Notes).
Comparison of motif depletion between bound and unbound GBSs
To generate the set of bound GBSs, we extracted 300 bp around the peak summits from the above-mentioned 8 kb regions, and retained only the 300 bp fragments having one or more GBS matches (total number of GBSs: 58195). For the control set of unbound GBSs, we extracted GBS-matches (RSAT matrix-scan, P-value threshold 10−3) from all genomic regions that are neither located under a GR ChIP-seq peak assayed in other cell lines available in our lab (U2OS, A549, IMR90, K562, Nalm6), nor in the ENCODE blacklist. Out of all these GBS matches, we randomly selected the same number as in the bound GBSs and extracted 1 kb regions centered on the GBSs. These two data sets (bound and unbound) were then scanned for exact matches of the word TTAATTAA within 10bp bins (RSAT dna-pattern).
Plasmids
Reporter plasmids to test the effect of candidate sequences on nearby GBSs (‘Negative Regulatory Sequences (NRS) reporters’) were constructed by first inserting the GBS sequence of interest (see Supplementary Table S1) by ligating oligonucleotides with overhangs to facilitate direct cloning into the XmaI and BglII sites of pGL3 promoter (Promega). Subsequently, oligonucleotides encoding the candidate or control sequence with overhangs to facilitate direct cloning (see Supplementary Table S1) were ligated into the Asp718 and NotI sites. Constructs with NRS2 or control sequence shifted by 10 bp or 20 bp were cloned using the oligos listed in Supplementary Table S1 to facilitate direct cloning into the BglII and Asp718 sites of PGL3 promoter. Constructs to stably integrate the NRS reporters at the AAVS1 locus were designed as described previously (16). In short, the NRS reporter was amplified using primers JT163 and JT 164 (see Supplementary Table S1), amplicons were digested with Eco31I and cloned into SalI and Asp718 linearized SAA-GFP plasmid (16). NRS reporters for zebrafish were constructed by multisite-gateway cloning (Invitrogen) using p3E-TagRFP, pME-e1b_promoter and p5E-GAB-6GBS as entry clones and pDEST_cry_ECFP (17) as destination vector. Subsequently, two copies of control or candidate NRS sequences were integrated into the AscI site downstream of the 6 GBSs using oligos (see Supplementary Table S1) with overhangs to facilitate direct cloning. To test the effect of NRSs on MyoD-driven transcription, we generated reporters containing 3 copies of the MyoD recognition sequence flanked by a single copy of either control or candidate sequences. This was done by digesting the GRE2Gal42 reporter (18) with PstI and SpeI and subsequent ligations using oligos with overhangs to facilitate direct cloning as listed in Supplementary Table S1. The chicken MyoD expression construct (pVax-MyoD) was a kind gift of the Mundlos laboratory (MPIMG, Berlin). Gal4-DBD fusion proteins of NONO, PSPC1 and SFPQ have been described previously (19). 2×gal4-2×GRE-luciferase (carrying two gal4 binding sites and two GBSs) and matching construct lacking the 2 gal4 binding sites have been described previously (18).
Stable integration of NRS reporters using ZFNs
Stable integration of reporter genes at the AAVS1 (‘safe harbor’) locus of U2OS cells stably expressing GR was performed as described previously (16). In short, cells were transfected with 10 μg of donor reporter construct and 0.5 μg of an expression construct encoding zinc-finger-nucleases against the AAVS1 locus using the Amaxa nucleofector Kit V (Lonza). Three weeks post transfection, GFP-positive cell pools were isolated by fluorescence-activated cell sorting (FACS). From this GFP-positive population of cells, we isolated single-cell derived clones that were genotyped for correct integration of the reporter at the AAVS1 locus using one primer targeting the donor construct (Luc-fw: tcaaagaggcgaactgtgtg) and another primer targeting the AAVS1 locus directly flanking the site of integration (R5 ctgggataccccgaagagtg).
Chromatin immunoprecipitation (ChIP)
ChIP experiments targeting GR were essentially done as described (1) using protein A/G beads (Santa Cruz) except for the washing step which consisted of 5 washes with 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 500 mM NaCl, 5% Glycerol, 0.1% Sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 0.5 mg/l BSA, followed by 2 washes with 20 mM Tris, pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate. GR binding at the integrated GBS locus and at the endogenous FKBP5 locus (positive control) and RPL19 locus (negative control) was analyzed by qPCR.
Stable integration and analysis of NRS-reporters in zebrafish
Stable integration of NRS reporters in zebrafish was done using Tol2-transgenesis using the Tol2Kit (20). Briefly, 10 μg plasmid pCS2FA-transposase from the kit was linearized with NotI, treated with proteinase K and subsequently purified by phenol-chloroform extraction. The DNA was precipitated and washed once with 70% EtOH. Linearized DNA was recovered in 20 μl DEPC water and used for in vitro transcription using the kit mMessage mMachine by Ambion according to the manufacturer's protocol. For injection, 10–20 ng/μl NRS reporter plasmid was mixed with 25 ng/μl transposase mRNA and injected into embryos at the one-cell stage (∼30 pg DNA per injection) as described (20). Forty eight hours post injection, fish were treated with dexamethasone or DMSO as vehicle control for 8 h. TagRFP expression in transgenic fish, which were identified by CFP expression in the eye lens, was analyzed using a confocal laser scanning microscope (Zeiss LSM 700). For quantification of mRNA levels (primers listed in Supplementary Table S2), RNA was purified, reverse transcribed using random primers and cDNA was analyzed by quantitative real-time PCR (qRT-PCR). Data were analyzed using the ΔΔCt method using efla, a gene not affected by dexamethasone treatment, for normalization. To control for differences in the number of integrations, we used ECFP mRNA concentration as a reference.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed as described (1) using either an oligo encoding a GBS (in bold) flanked by the control sequence (underlined) or by the NRS2 sequence (underlined).
Control: TAGGTACGAGGTAGGCTTGCTAGCCCGGAGAACAAAATGTTCTGATC
NRS2: TAGGTATTAATTCAATTAACTAGCCCGGAGAACAAAATGTTCTGATC
DNase I assays
DNase I assays were done as described (21) by growing U2OS cells in 6 well tissue culture plates to confluency, washing them with phosphate buffered saline (PBS) and scraping them into 1 ml DNase I buffer (20 mM HEPES pH 7.4, 0.5 mM CaCl2, 5 % glycerol, 3 mM MgCl2, 0.2 mM spermine, 0.2 mM spermidine) plus 0.2% NP40 alternative. Next, cells were homogenized by vortexing, incubated on ice for 5 min and centrifuged for 5 min at 500 g, 4°C to pellet nuclei. Nuclei were resuspended in 200 μl DNase I buffer and 50 μl aliquots were DNase I treated (or mock treated to normalize for the amount of chromatin in input) by the addition of 25 μl of DNase I buffer containing 1.5 μl DNase I (Qiagen, 2.7 u/μl) and then incubated at 37°C for 8 min. The reaction was stopped by addition of an equal volume of 2x stop buffer (50 mM Tris pH 7.4; 200 mM NaCl; 100 mM EDTA; 2% SDS, 200 μg/ml proteinase K) and samples were incubated at 65°C for 4 h to remove proteins. Finally, DNA was purified using the PCR purification kit (Qiagen) and regions of interest were analyzed by qPCR (primers listed in Supplementary Table S2).
MNase I assays
Nucleosome occupancy was analyzed essentially as described (22), with slight modifications to enable analysis by qPCR. Cells were grown to confluence in 10 cm dishes and treated with 1 μM dexamethasone or 0.1% ethanol respectively for 60 min. Prior to harvesting, cells were washed once with PBS, resuspended in 11.3 μl ice-cold lysis buffer (10 mM Tris pH 7.4; 10 mM NaCl; 3 mM MgCl2; 0.5 % IGEPAL) per cm2 and transferred to 1.5 ml Eppendorf tubes. Chromatin was collected by centrifugation at 1600 RPM for 15 min at 4°C. Resulting pellets were resuspended in 200 μl storage buffer (50 mM Tris pH 7.4; 40 % glycerol; 5 mM MgCl2; 0.1 mM EDTA) per 107 cells. Chromatin samples were flash-frozen in liquid nitrogen as aliquots, and then stored at −80°C. Prior to measuring the concentration of total DNA, aliquots were diluted 1:10 in storage buffer and 0.4 volumes of 5 M NaCl was added to disrupt protein/DNA interactions other than those between histones and DNA. Subsequently, each sample was split in two of which one half was treated with MNase whereas the other half was untreated to normalize for the amount of input. For each sample containing 0.8 μg genomic DNA in 50 μl storage buffer, an equal volume of MNase reaction buffer (50 mM Tris pH 7.4; 25 mM KCl; 2.5 mM CaCl2; 5 mM MgCl2; 12.5 % glycerol) was added containing 1 μl MNase (NEB; ∼200 kunitz) for MNase-treated samples. Samples were incubated for 10 min at 30°C, and the reaction stopped by adding 100 μl stopping buffer (50 mM Tris pH 7.4; 200 mM NaCl; 100 mM EDTA; 2% SDS) supplemented with 15 μl Proteinase K. Samples were incubated for 120 min at 60°C, phenol-chloroform extracted and purified mono-nucleosomal DNA from the test conditions was purified by cutting out the gel-slice corresponding to a DNA size of approximately (100–220 bp) from an 1.5% agarose gel using the NucleoSpin Gel and PCR clean-up kit (Macherey and Nagel). qPCR was used to analyze the samples using primers targeting either the GAPDH promoter (region with low predicted nucleosome occupancy), the +1 nucleosome of GAPDH (region of high predicted nucleosome occupancy) or the genomically integrated NRS reporters (primers listed in Supplementary Table S2).
DNA pull-down and mass spectrometry
Biotin-labeled oligos encoding either control or candidate sequences as indicated below were used for DNA pull-down assays performed essentially as described previously (23).
Control: CAAAAGATCGCTGCAGACTTGAACCGAGGTAGGCTTGCTAGCCCGG
NRS1: CAAAAGATCGCTGCAGACTTGAACAGGTTAATTAACACTAGCCCGG
NRS2: CAAAAGATCGCTGCAGACTTGAACTTAATTCAATTAACTAGCCCGG
For each pull-down, ∼0.5 mg each of pre-cleared nuclear proteins from HeLa cells and dsDNA-loaded Dynal MyOne C1 streptavidin magnetic beads (Invitrogen) were mixed and incubated for 3 h at 4°C. Next, beads were washed 4 times with 20 mM Tris-HCl pH 7.3, 10% glycerol, 0.1 M KCl, 0.2 mM EDTA, 10 mM K glutamate, 0.04% NP40. DNA-bound proteins were either released by direct trypsin digestion on beads. Alternatively, proteins were released by restriction digestion of the DNA, precipitated with ice-cold acetone (−20°C, overnight) and subsequently trypsin digested. Digests were desalted by C18 StageTips and dissolved in 5% acetonitrile, 2% formic acid for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. LC-MS/MS was carried out by nanoflow reverse phase liquid chromatography (RPLC) (Agilent, Santa Clara, CA, USA) coupled online to a Linear Ion Trap (LTQ)-Orbitrap XL mass spectrometer (Thermo-Electron Corp). Briefly, the LC separation was performed using a PicoFrit analytical column (75 μm ID × 150 mm long, 15 μm Tip ID (New Objectives, Woburn, MA, USA)) in-house packed with 3 μm C18 resin (Reprosil-AQ Pur, Dr Maisch, Germany). Peptides were eluted using a nonlinear gradient from 2 to 40% solvent B over 160 min at a flow rate of 200 nl/min (solvent A: 97.9% H2O, 2% acetonitrile, 0.1% formic acid; solvent B: 97.9% acetonitrile, 2% H2O, 0.1% formic acid). A total of 1.8 kV were applied for nanoelectrospray generation. A cycle of one full FT scan mass spectrum (300–2000 m/z, resolution of 60 000 at m/z 400) was followed by 10 data-dependent MS/MS scans acquired in the linear ion trap with normalized collision energy (setting at 35%). Target ions already selected for MS/MS were dynamically excluded for 60 s.
Raw MS data were processed with MaxQuant software (version 1.5.0.0) (24), applying the label-free quantification algorithm (25) and searched against the human proteome database UniProtKB with 88.717 entries, released in 11/2014. A false discovery rate (FDR) of 0.01 for proteins and peptides and a minimum peptide length of 7 amino acids were required. A maximum of two missed cleavages was allowed for the tryptic digest. Cysteine carbamidomethylation was set as fixed modification, while N-terminal acetylation and methionine oxidation were set as variable modifications. MaxQuant-processed output files can be found in Supplementary Table S3.
esiRNA knockdown
For esiRNA knockdown experiments, 10.000 transgenic cells were seeded per well of a 48-well plate. The next day, these cells were transfected with a mix of 75 ng of each esiRNAs (Sigma-Aldrich) against SFPQ and NONO (EHU 158661, EHU 071361) or a non-target esiRNA control (RL) using lipofectamine 2000 (Invitrogen). Thirty two hours post transfection, cells were treated for 16 h with 1 μM dexamethasone, or 0.1% ethanol as vehicle control. Cells were lysed and luciferase activity was measured.
RESULTS
Identification of under-represented sequence motifs at GR-bound loci
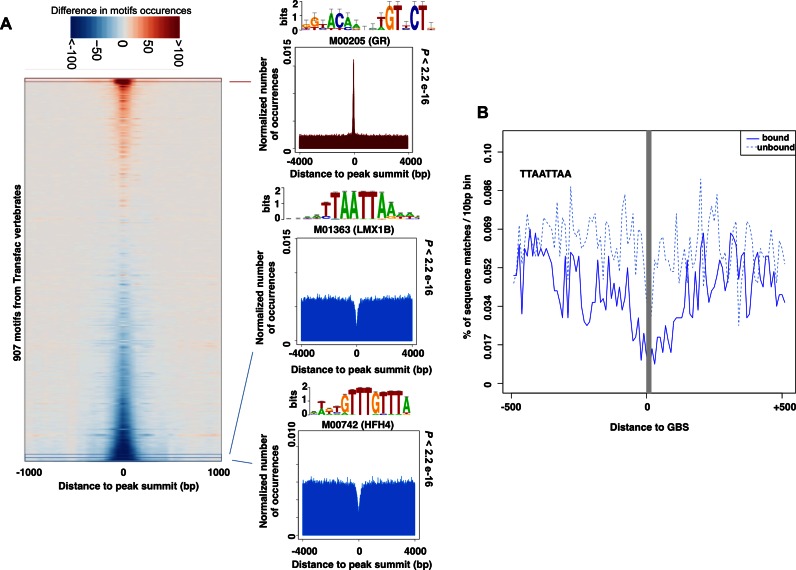
We performed motif analysis in GR bound regions, by scanning a window of 8 kb centered on the peak summit of GR binding peaks (based on ChIP combined with next generation sequencing (ChIP-seq)) with the complete collection of motifs from the TRANSFAC database. As expected, this analysis showed a local enrichment at the peak summit of the canonical GBS for all cell lines examined, and of several other motifs (e.g. AP1) (Figure 1A and data not shown). Interestingly, we also identified many sequence motifs that were under-represented at GR-bound regions across all cell lines tested (Figure 1A and data not shown). As many of these under-represented sequences have a high AT content (examples of depleted sequences: TTTGTTT, TTAATTAA), we first assumed an artifact of the ChIP procedure as AT-rich sequences tend to be depleted during the ChIP procedure (26). Nevertheless, a study of a large panel of ENCODE ChIP-seq data sets also concluded that a GC-rich environment is a general feature of TF binding (27). The observed local depletion of AT-rich sequences thus concurs with this study and, rather than being an experimental artifact, might also reflect the fact that AT-depletion could be beneficial for TF binding.
Figure 1.
Analysis of motif occurrence around GR-bound regions. (A) DNA sequences from GR ChIP-Seq peaks in U2OS cells stably expressing GR were aligned at the peak summit, and flanking genomic DNA ±4000 bp was extracted. The entire collection of TRANFAC vertebrate motifs was used to scan these 8 kb regions. The normalized number of hits for each motif is shown as a heatmap focusing on the most central 2 kb region, where red and blue denote motif enrichment and under-representation, respectively. Representative examples of motifs that are either enriched or under-represented around the peak summit of GR-bound regions are displayed. The motif for GR (M00205) is made from very few binding sites but nonetheless is very similar to GR motifs constructed from ChIP-seq data sets. The detailed list of matrices and ranking is available as supplementary file 2. (B) Percentage of 10 bp bins with a sequence match to the word TTAATTAA in 1 kb regions centered on 58195 GBSs, extracted from bound (regions located within GR ChIP-seq peaks in U2OS, plain line) or unbound (regions located outside of any ChIP-seq peaks in five cell lines, dotted line). The central grey area indicates the localization of GBSs (regions incompatible with the word TTAATTAA).
We thus hypothesized that the observed under-representation could reflect a role in modulating GR binding. If the under-represented motifs indeed play a role in preventing GR binding to nearby GBSs, the depletion should be more pronounced for bound GBSs when compared to other GBS-like sequences in the genome not bound by GR. To test this, we generated two sets of 1 kb sequences centered on a GBS. The first set consisted of sequences from GR-bound regions, whereas the second set contains GBS matches in genomic regions that are not bound by GR in any of the five cell lines we examined (U2OS, A549, IMR90, K562, Nalm6). Next, we compared the occurrence profiles of the word TTAATTAA (candidate sequence used later in the experimental validations) and found that the depletion was more pronounced for the bound set of GBSs (Figure 1B). Together, our analyses uncovered several sequence motifs that are under-represented at GR-bound regions.
Under-represented sequences interfere with GR binding
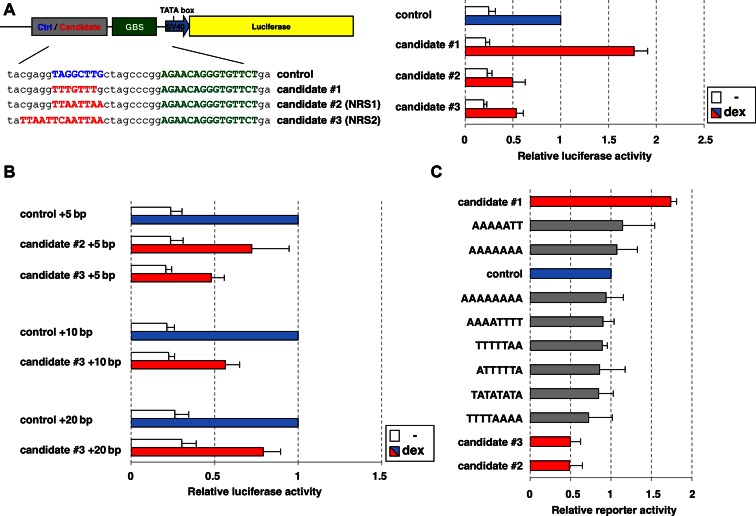
To test if the under-represented sequences are not simply an experimental artifact, we set out to test if their presence interferes with GR binding to GBSs nearby. As a proxy for reduced GR binding, we first determined if the presence of an under-represented sequence influences the ability of GR to activate transcription from a nearby GBS. We therefore constructed reporters that encode a GBS sequence upstream of a minimal promoter driving the expression of a luciferase reporter gene. In addition, the GBSs were flanked by either a control sequence (random) or by one of three under-represented candidate sequences (Figure 2A). The first candidate sequence was chosen because its depletion is cell-type specific with a marked depletion at GR-bound regions for U2OS and Nalm-6 cells, whereas no obvious depletion was observed for IMR90 or A549 cells. The second and third candidate sequence are derived from motifs that are depleted across all cell types and resemble a large collection of highly similar depleted motifs that are recognized by members of the homeobox family of transcription factors.
Figure 2.
Effects of depleted sequences on GR-dependent gene-activation. (A) U2OS cells were transfected with ‘NRS-reporters’ that contain a GR binding sequence (GBS) flanked by either a control or one of 3 depleted sequences from GR-bound genomic regions (candidates) and a luciferase reporter gene. Normalized luciferase activity for cells either treated with vehicle control (−) or dexamethasone (dex) ± standard error of mean (SEM) (n = 3) is shown. (B) Same as for (A), except that spacing between GBS and control/depleted sequence was increased by 5, 10 or 20 base pairs, respectively. (C) Effect of presence of sequences as indicated in NRS-reporters was tested for a broad panel of AT-rich sequences. Fold activation upon dex treatment ±SEM (n = 3) is shown. Activities relative to control reporter (set at 1) are shown.
As expected, GR activated transcription of reporters with GBSs flanked by the control sequence (Figure 2A). Next, we examined the effect of flanking the GBS by one of the candidate sequences. For candidate #1, GR-dependent activation was not compromised, in fact stronger than that seen for the control sequence indicating that this sequence does not influence GR's ability to bind to GBSs nearby. In contrast, when the GBSs were flanked by candidate sequences #2 or #3, a marked decrease in activation was observed for all GBSs examined (Figure 2A, other GBS sequences tested showed the same results: data not shown). Notably, the effect was specific for the hormone-dependent activation of the reporters, as the basal activity was not affected by the presence of either candidate sequence (Figure 2A).
One possible explanation for the effects observed for candidates #2 and #3 is that they are bound by proteins, which prevent GR from interacting with the nearby GBSs by sterical hindrance. To test this, we increased the spacing between GBS and test sequences by 5 bp, the equivalent of approximately half a DNA helical turn and by 10 bp and 20 bp to increase the spacing. Similar to the observation with the initial reporters, GR-dependent activation was reduced (Figure 2B). This indicates that these sequences do not require an exact positioning relative to the GBS to exert their effect although the effect appears to become weaker with increased spacing (Figure 2B). A common feature of many under-represented candidate sequences was their high AT-content. To test if the effects observed for candidates #2 and #3 were sequence-specific or a general feature of AT-rich sequences, we tested the effect of several additional AT-rich sequences in our reporters (Figure 2C). This analysis showed that although some other AT-rich sequences also had a small negative effect on GR-dependent activation, their effects were markedly smaller than those observed for candidates #2 and #3 (Figure 2C) indicating that their effect is sequence-specific rather than a simple consequence of being AT-rich.
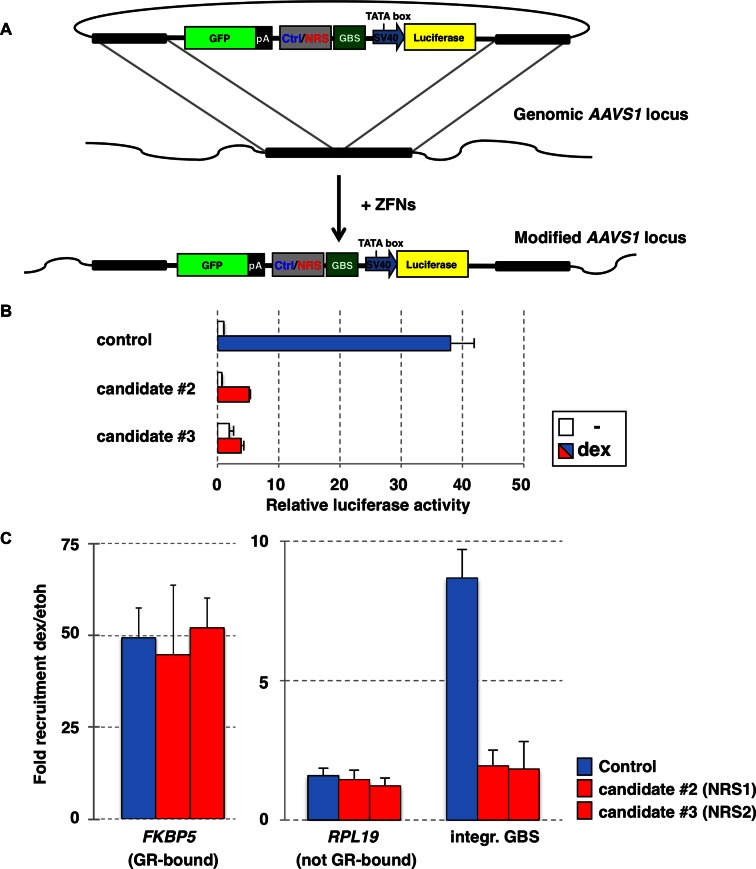
The negative effect of the candidate sequences on GR-dependent activation suggests that these sequences interfere with GR's ability to bind to GBSs nearby. To test this hypothesis directly, we set out to generate cell lines with stably integrated reporters with GBSs flanked by candidate sequences using zinc finger nucleases targeting the AAVS1 ‘safe harbor’ locus (1) (Figure 3A). Mirroring what we saw for the transiently transfected reporters, the integrated reporters showed a robust activation of the luciferase reporter gene upon hormone treatment when the GBS was flanked by the control sequence (Figure 3B). In contrast, the cell-lines encoding candidate sequences #2 or #3 showed a reduced GR-dependent activation compared to the control cell line (Figure 3B: ∼75% reduction for candidate #2 and ∼95% for candidate #3 when compared to control).
Figure 3.
Depleted sequences attenuate GR-dependent activation and binding in the genomic context. (A) Schematic of donor NRS-reporter construct and of the AAVS1 locus after targeted integration of the reporter. (B) Populations of reporter cell lines were treated overnight with 1 μM dexamethasone (dex) or 0.1% ethanol as vehicle control. Luciferase activity was measured and average activity, relative to ethanol-treated control cell line, ±SEM (n = 3) is show. (C) Effect of depleted sequences on genomic GR binding. Populations of cell lines with stably integrated reporters as indicated, were treated for 90 min with 1 μM dexamethasone (dex) or 0.1% ethanol as vehicle control. Fold increase in genomic GR binding upon dex treatment ±SEM (n = 3) as determined by chromatin immunoprecipitation for each line was determined by qPCR for bound (FKBP5) and unbound (RPL19) control regions and at the GBS locus of the integrated NRS reporters (integr. GBS).
Next, we tested the effect of the candidate sequences on GR binding at the integrated reporter using ChIP assays. To compare the efficiency and specificity of the ChIP assay between our cell lines with stably integrated reporters, we first examined GR binding at the endogenous GR target site FKBP5 and found a comparable enrichment for all cell lines examined (Figure 3C). As expected, no binding was observed for any of the clonal lines at the RPL19 control locus where GR does not bind. In contrast, at the integrated GBS, a robust GR recruitment was observed when the GBS was flanked by the control sequence (∼8-fold increase in binding upon dexamethasone-treatment) but not when the GBS was flanked by either candidate #2 or #3, (<2-fold increase upon dexamethasone treatment, Figure 3C). Taken together, these results show that candidate sequences #2 and #3 interfere with the binding of GR to nearby GBSs and we, therefore, refer to these sequences from now on as negative regulatory sequences 1 and 2 (NRS1 and NRS2).
NRSs interference is conserved across vertebrate species
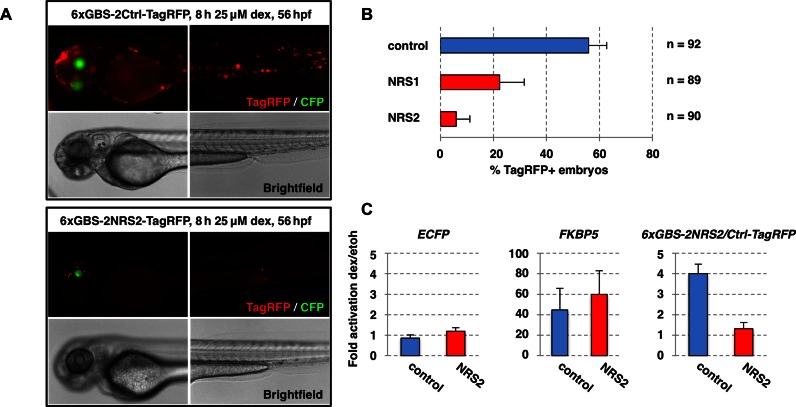
To investigate if the effects of NRSs on GR binding are conserved across species and to test for potential cell-type specificity, we set out to study their effects in Danio rerio (zebrafish). Zebrafish have functional glucocorticoid receptor signaling (28) and have diverged from humans some 450 million years ago (29). Similar to the experiments described above in mammalian cells, we tested the effects of NRSs in zebrafish using reporter constructs for which GBS sequences were flanked by NRS or control sequences upstream of a e1b promoter driving expression of a TagRFP reporter gene (Figure 4A). The reporter constructs contained a cassette that drives expression of ECFP in the eye lens to score transgenic fish and were randomly integrated using the Tol2 transposase (20). For all reporter constructs, little to no RFP expression was observed when the transgenic fish were treated with DMSO as vehicle control (data not shown). Upon treatment with dexamethasone to activate GR, visual inspection of transgenic fish showed expression of the RFP reporter for 56% of transgenic fish when the GBS was flanked by the control sequence (Figure 4B). The fraction of RFP-positive fish was markedly smaller when the GBS was flanked by either NRS1 or NRS2, (22% for NRS1, 6% for NRS2). Furthermore, the absence of RFP expression was not restricted to specific cells or tissues, but observed throughout the animal, arguing that the effects of NRSs occur throughout the body. Because visual quantification is subjective, we also quantified hormone-induced activation of the TagRFP mRNA. Therefore, we compared TagRFP mRNA levels between DMSO (vehicle) and dexamethasone-treated fish. In line with our visual inspection, we found that hormone-dependent activation of TagRFP was lower when the GBSs were flanked by NRS2 (1.3-fold) when compared to GBSs flanked by control (4.0-fold) whereas regulation of the endogenous GR-responsive gene FKBP5 gene was comparable for both populations of transgenic fish and no obvious regulation was seen for the ECFP gene, which was used to score transgenic fish (Figure 4C). Together, these experiments show that the activity of NRSs is conserved across species and active regardless of tissue examined.
Figure 4.
Effect of depleted sequences on GR is conserved across species and active throughout the body. (A) NRS reporter constructs with 6 GBSs flanked by either 2 control or depleted candidate sequences were randomly integrated into the zebrafish genome and dexamethasone-induced expression was quantified by counting RFP+ transgenic fish. (B) Average % of transgenic embryos with RFP expression for each reporter tested ± standard deviation are shown. n = total number of fish analyzed from 3 experiments. (C) Change in mRNA levels upon hormone treatment was determined by qPCR for pools of transgenic fish with reporter construct as indicated (control or NRS2). ECFP: negative control, reporter gene to score transgenic fish; FKBP5: endogenous GR target-gene; TagRFP: integrated NRS reporter. Average fold induction ±SEM (n = 3) is shown.
NRSs do not majorly affect DNA conformation and chromatin accessibility
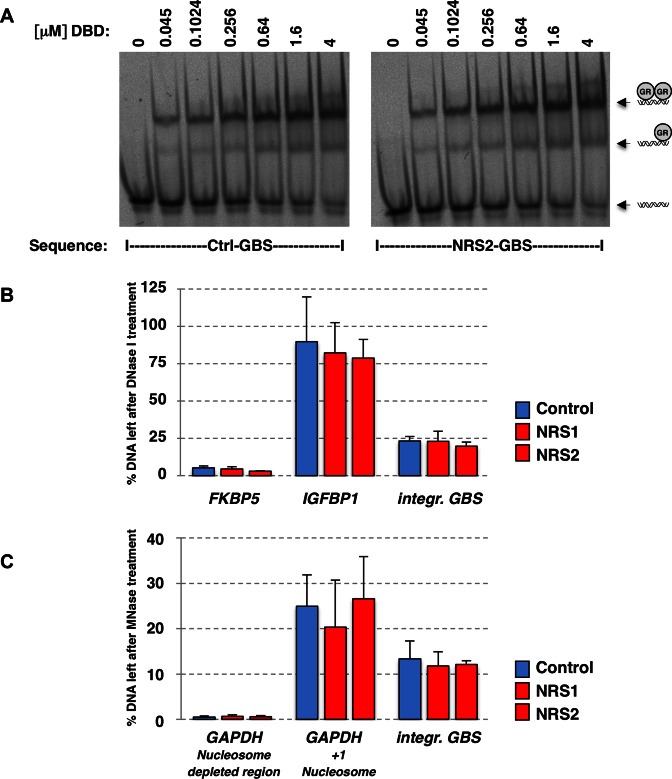
To understand how NRSs interfere with GR binding, we tested several possible mechanisms by which they might exert their effects. First, we tested a possible role of NRS-induced changes in DNA conformation. We therefore compared the binding of GR between a GBS flanked by the control sequence with that seen when the GBS is flanked by NRS2 using EMSAs. These experiments showed that the binding was indistinguishable between the two sequences (Figure 5A), indicating that the ability of NRSs to interfere with GR binding requires additional cellular components that are not present when analyzing naked DNA.
Figure 5.
Characterization of the NRS sequences. (A) EMSA comparing binding of the DNA binding domain of GR to a GBS sequence flanked by either the control sequence (left) or by NRS2 (right). (B) DNase I accessibility assay was performed with populations of transgenic cells with stably integrated reporters as indicated. Regions of interest were analyzed by qPCR (FKBP5: control accessible region; IGFBP1: control inaccessible region; integr. GBS: integrated reporter region). Results are shown as % of input remaining after DNAse I digestion ±SEM (n = 3). (C) Nucleosome occupancy was analyzed using micrococcal nuclease (MNase) assays for populations of transgenic cells with stably integrated reporters as indicated. Regions of interest were analyzed by qPCR. Integr. GBS: integrated reporter region. Results are shown as % of input remaining after MNase digestion ±SEM (n = 3).
Genomic GR binding is strongly correlated with accessible chromatin, with the majority of binding (>90%) occurring at DNase I hypersensitive sites (2). Thus, one explanation for the effect of NRSs could be that they render nearby genomic regions inaccessible. To test this, we assayed the influence of NRSs on chromatin accessibility using DNase I assays. We first established assay conditions at which the majority of a control ‘open’ region, FKBP5, was degraded after DNase I treatment, whereas the majority of DNA was resistant to DNase I treatment for a control ‘closed’ region, IGFBP1 (Figure 5B). These assays showed that we can indeed discriminate between open and closed regions and that comparable results are obtained for these control regions for all transgenic cell lines examined. For the GBS locus of the integrated reporter, we found that the locus was relatively sensitive to DNase I digestion (∼80% degraded) indicative of an ‘open’ chromatin context. Surprisingly however, the presence of an NRS sequence did not result in a change in the sensitivity to DNase I, suggesting that NRSs do not prevent GR binding by changing chromatin accessibility (Figure 5B).
Another possible explanation for the effect of NRSs could be that they change the positioning or presence of nucleosomes. To analyze the effect of NRSs on the presence of nucleosomes, we performed MNase assays. In this assay, the presence of a nucleosome protects DNA from digestion by MNase, whereas regions lacking nucleosomes are more efficiently digested. Consistent with expectation, the MNase assay showed that the +1 nucleosome region of GAPDH was resistant to MNase digestion, whereas the nucleosome-depleted region upstream of the TSS of GAPDH was very sensitive to MNase digestion (Figure 5C). We next examined the nucleosome occupancy at the GBS locus of the integrated luciferase reporter genes for each of the cell lines and found that this locus was relatively resistant to MNase digestion but that the presence of an NRS sequence did not result in a marked change in MNase sensitivity (Figure 5C).
Together, these experiments indicate that the effects of NRSs do not appear to be a consequence of changes in either chromatin accessibility or nucleosome density. However, given the resolution of the data, we cannot rule out that NRSs induce subtle changes in the positioning of nucleosomes that might influence whether the GBS is available for GR binding or not.
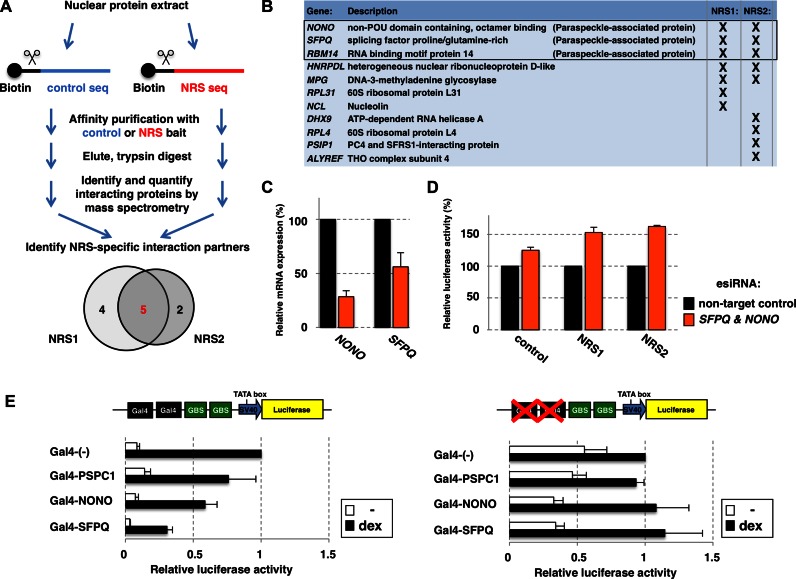
NRSs interact with paraspeckle-associated proteins
Since the NRS sequences appeared to neither exert their effect by changing the conformation of the DNA nor by influencing the local chromatin accessibility, we set out to identify NRS-interacting proteins reasoning that this might provide mechanistic insights. Therefore, we used the NRS sequences as baits for DNA pull-down assays and compared bound nuclear proteins between NRS and control sequences by mass spectrometry (Figure 6A). These experiments resulted in the identification of 11 proteins that were at least 2-fold enriched for NRS sequences relative to control in at least two out of three experiments (Figure 6B). Five of these 11 proteins are enriched for both NRS sequences, which is not that surprising given that they are similar in sequence (both contain TTAATT) and might thus interfere with GR binding by interacting with the same protein(s). Of these five shared NRS-binding proteins, three are known paraspeckle-associated proteins (Figure 6B). Arguing for a possible role of these proteins in mediating the effects of NRSs, published studies have shown that SFPQ and NONO act as co-repressors and interfere with DNA binding of the androgen and progesterone receptor, two close homologs of GR (30,31). Given their established role in repressing the activity of steroid hormone receptors and the fact that we identified multiple paraspeckle-associated proteins, we decided to focus our attention on these proteins. To test if paraspeckle proteins play a role in mediating the repressive effects of the NRS sequences, we tested the effect of reducing SFPQ and NONO levels using esiRNAs. Because SFPQ and NONO have been reported to play redundant roles (19), we knocked-down their expression simultaneously (Figure 6C). Indicative of a role of paraspeckle proteins in mediating the effects of NRSs, we found that reducing SFPQ and NONO levels resulted in an increase in reporter activity when the GBS was flanked by either NRS1 or 2 (Figure 6D: 53% for NRS1; 63% for NRS2;). Arguing that this effect is at least somewhat specific, a smaller (∼25%) effect was observed when the GBS was flanked by the control sequence (Figure 6D). To further test if paraspeckle-associated protein can influence GR binding to GBSs, we tested the effect of specifically recruiting candidate proteins to GBS-driven reporters. The targeted recruitment was accomplished by using a reporter for which the GBS is flanked by gal4 DNA binding sites (18) and by expressing the candidate proteins as fusion proteins with the Gal4 DNA binding domain (Figure 6E). Consistent with the ability of paraspeckle-associated proteins to negatively influence GR binding to GBSs nearby, we found that the expression of Gal4 fusion proteins of SFPQ and NONO resulted in a reduced ability of GR to activate expression of the luciferase reporter gene (Figure 6E). Furthermore, another paraspeckle-associated protein, PSPC1, also resulted in small decrease in reporter activity. To test if these effects were a consequence of targeted recruitment and not simply an effect of over-expression, we also assayed their effect on a control reporter where the GBS sequences were not flanked by Gal4 DNA binding sites. For this reporter however, co-expression of none of the paraspeckle-associated proteins showed a marked effect (Figure 6E), indicating that their effect requires the targeted recruitment of these proteins to the GBS locus. Together, these data argue that paraspeckle-associated proteins binding to the NRS sequences may play a role in preventing GR from binding to GBSs nearby.
Figure 6.
Identification and characterization of NRS-associated proteins. (A) Workflow for the label-free identification of NRS-specific interaction partners was as follows: DNA pull-downs were performed to identify and quantify proteins from a nuclear extract using biotin-labeled oligos encoding either NRS1, NRS2 or control sequence. Shown is the overlap of NRS interacting proteins (>2-fold enrichment over control bait in at least 2 out of 3 experiments). (B) Identity and description of the 11 NRS-interacting proteins that were identified. (C) Efficacy of esiRNA knockdown. RNA levels for genes as indicated were quantified by qPCR, two days after transfection with esiRNAs targeting SFPQ and NONO. Percentage relative to non-target control ±SEM (n = 4) is shown. (D) Effect of esiRNA knockdown of paraspeckle genes SFPQ and NONO on genomically-integrated NRS-reporters. Thirty six hours after populations of cell lines, with stably integrated reporters as indicated, were transfected with esiRNAs, cells were treated overnight with EtOH as vehicle control or 1 μM dex. Percentage activity relative to dex-treated non-target control ±SEM (n = 3) is shown. (E) Targeted recruitment of paraspeckle-associated proteins interfered with GR-dependent transcriptional activation. Expression constructs for Gal4 DNA binding domain fusion proteins as indicated were co-transfected with luciferase reporters encoding two GBSs next to two Gal4 binding sequences (left) or with reporters lacking Gal4 binding sequences as control (right). Reporter activity relative to dex-treated cells expressing the Gal4-DBD only ± SEM (n = 3) is shown.
NRSs affect other transcription factors
To test if NRS can also influence other TFs, we examined the motif occurrence around genomic regions bound by MyoD, a muscle-specific basic helix-turn-helix transcription factor (Supplementary Figure S1A). We found that, similar to GR-bound regions, certain motifs were over-represented around MyoD-bound regions (e.g. the MyoD consensus motif) whereas other motifs were under-represented, including two motifs from which the NRS sequences were derived (Supplementary Figure S1A). To test if these under-represented sequence could negatively influence DNA-binding and transcriptional regulation by MyoD, we flanked three MyoD binding sites by a single control or NRS sequence upstream of a luciferase reporter gene. Since little to no MyoD expression is observed in U2OS cells (32), activation of this reporter could be achieved by co-transfecting a MyoD-expression construct. Similar to our results for GR, NRS2 resulted in a marked decrease in MyoD-dependent transcriptional activation when compared to control sequence (Supplementary Figure S1B). In contrast to GR however, the presence of NRS1 did not have a marked effect on MyoD-dependent activation indicating that the activity of this NRS might be TF-specific. Collectively, these experiments show that the activity of NRSs is not restricted to GR and indicate that NRSs might play a more global role in controlling where in the genome TFs can bind.
DISCUSSION
Sequences matching the GR motif are ubiquitously found in the genome, yet only a cell-type-specific minority of these potential binding sites appears to be actually occupied. This raises the question: what discriminates the subset that is GR-bound from the subset of potential binding sites that is not? For one, regions bound by GR are typically enriched for other sequence motifs that are bound by other factors that cooperate with GR to facilitate selective recruitment to a subset of potential recognition sequences (10). Such factors can play a role in the establishment or maintenance of accessible chromatin, which is a major determinant of GR binding. However, even within the universe of accessible chromatin, the presence of a GBS is a poor predictor of GR binding, as many GBSs with a high motif score are not occupied by GR ((2), unpublished data from the Meijsing lab). We reasoned that other sequence signals may prevent GR binding to GBSs nearby. If such sequences existed, they should be depleted at GR-bound sites and scanning of GR-bound regions with collections of known motifs identified several such candidate sequences. Similarly, de novo motif discovery approaches focusing on under-represented motifs could be used to uncover such sequences. When we compare bound regions with unbound regions with a GBS match (300 bp window centered on GBS), we find an exact match to the TTAATTAA NRS1 sequence for 0.8% for bound regions whereas this number is 1.6% for unbound regions. This indicates that NRS1 could explain the lack of binding for a small fraction of unbound GBSs. Notably, we identify several additional depleted sequences and other studies have also described sequences that, when present near GBSs, negatively influence GR's ability to regulate transcription (33). These sequences were identified using naturally occurring deletion mutants of the mouse mammary tumor virus (MMTV), a model system to study GR signaling. The effects of these sequences were attributed to cut-like homeobox 1 (Cux1) and special AT-rich sequence binding protein 1 (SATB1) (34,35). Interestingly, our genome-wide analysis uncovered that the recognition sequences of Cux1 (Transfac M00102) and SATB1 (Transfac M01232) are depleted near GR binding sites (Supplementary Figure S2), indicating that their effect is not restricted to the MMTV but may play a more general role in controlling where in the genome GR binds.
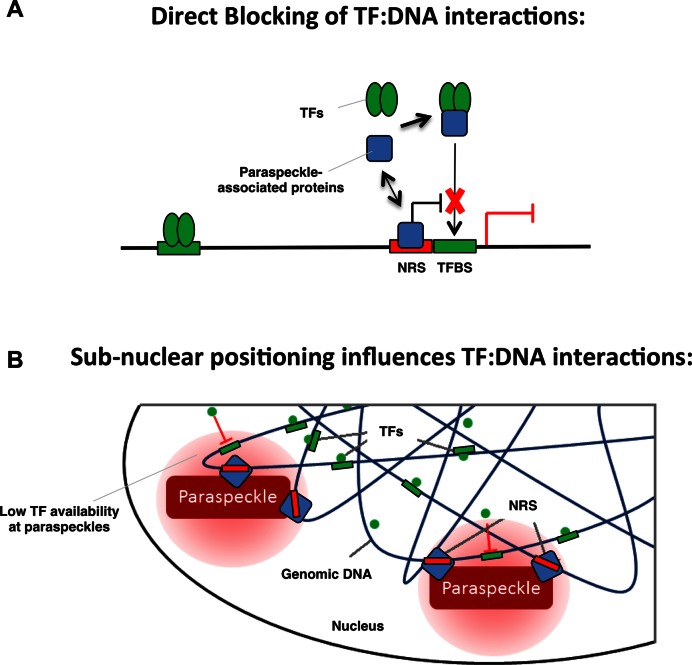
Our mechanistic studies indicated that neither changes in DNA shape, nor in chromatin accessibility or nucleosome density appear to be responsible for mediating the effects of NRSs. To our surprise, this suggests that a mechanism other than chromatin accessibility is responsible for mediating the effect of NRSs. One possible mechanism could be that proteins binding to NRSs might interfere with GR binding by sterical hindrance. However, we do not think this is the mechanism responsible, for several reasons. First, when we shifted the NRS by 5 bases, half a DNA helical turn, NRSs still exerted their effect, indicating that their activity does not require a precise positioning relative to the GBS. Similarly, the NRSs and TF binding sites had a different relative positioning in the reporters used to study the effect of NRSs in zebrafish and on MyoD, yet NRSs exerted their effect. Furthermore, when we used baits in our DNA pull-down assays where we flanked the GBS by an NRS, we found no effect of having a flanking NRS on GR binding to the nearby GBS (data not shown) indicating that the protein(s) binding to the NRS sequence need a cellular and or chromatinized context to exert their effect. Similarly, our in vitro binding studies (EMSAs) using naked DNA, showed that NRSs require cellular components to exert their effect. To identify proteins involved in mediating the effects of NRSs, our mass-spectrometry analysis uncovered 3 paraspeckle-associated proteins that were reproducibly identified as NRS-binders. Functional assays (Figure 6D and E) indicated that these proteins may indeed play a role in mediating the effects of NRSs. We envision several mechanisms by which paraspeckle-associated proteins might interfere with GR binding (Figure 7). First, SFPQ interacts with the corepressor Sin3A which in turn interacts with histone deacetylases (36) which are linked to gene repression possibly by influencing TF:DNA interactions. Alternatively, paraspeckle-associated proteins might directly bind to GR and thereby prevent it from interacting with GBSs (Figure 7A). In this scenario, NRSs would dynamically interact with the paraspeckle-associated proteins resulting in an increased local concentration. Arguing for a direct role of paraspeckle-associated proteins, GR directly interacts with SFPQ (37). Furthermore, studies have show that SFPQ interferes directly with DNA binding of the androgen receptor (30) and the progesterone receptor (31), which have DNA binding domains virtually indistinguishable from GR. Another explanation could be that NRSs might serve as anchoring sites that direct the NRS-encoding genomic loci to subnuclear regions, like paraspeckles, which may be less permissive to TF binding (Figure 7B). Of note, genomic regions associated with nuclear lamina often have a high AT content, similar to the NRSs, and are lowly expressed (38). Furthermore, Cux1 and SATB1, that bind the negative regulatory sequences identified in a previous study, can bind to matrix attachment regions that play a role in chromatin looping and nuclear architecture (39). Although our analysis indicated that paraspeckle-associated proteins may play a role in mediating the effect of NRS, their knockdown only resulted in a partial reduction of NRS activity. Our attempts to ChIP SFPQ at NRSs failed to show binding arguing that perhaps other or additional, yet unknown, mechanisms are in place that mediate the effects of NRSs. Alternatively, the partial reversion could reflect the fact that the knock-down of paraspeckle components, which are expressed at high levels in the cell, was incomplete (Figure 6C). Moreover, compensatory effects of other (unknown) factors might have resulted in only partial reduction of NRS activity.
Figure 7.
Cartoon depicting how NRS sequences and paraspeckle-associated proteins might cooperate to influence TF binding to nearby response elements. (A) By blocking TF:DNA interactions or (B) by influencing sub-nuclear positioning.
NRSs can influence genomic binding by GR and possibly other TFs, and our experiments in zebrafish indicate that they influence GR binding throughout the body. This prompts the question about the biological significance of NRS-mediated restriction of genomic binding by GR and other TFs might be. One possibility could be that NRSs constitutively restrict where TFs can bind in the genome to reduce the search-space for TFs to find regulatory sequences and to assure that the limited pool of TFs expressed in a cell localizes to the appropriate binding sites. NRSs resemble the recognition sequence of TFs from the homeobox family including Hox proteins, a family of TFs that define different cellular identities along the anterior-posterior axis (40). Thus, a possible function of these sequences might be that they play a role in preventing the binding of certain TFs, like GR which is expressed throughout the body, to binding sites near promoters of Hox-controlled target genes to prevent spurious activation of these genes at either the wrong place or the wrong time. Importantly, our studies indicated that the NRS sequences restrict binding by mechanisms other than simply restricting chromatin accessibility. This would provide the possibility for some TFs, that are resistant to the actions of NRSs, to bind and regulate transcription while at the same time preventing binding of other TFs that might interfere with proper regulation of those genes. Accordingly, when we examined genomic regions bound by HoxD13 (Supplementary Figure S3A), we find an enrichment of the HoxD13 consensus motif and for the motifs with marked depletions at GR-bound regions (Supplementary Figure S3B: M00742, M1363 and M01294). Conversely, the GR consensus motif is depleted at HoxD13-bound regions (Supplementary Figure S3C) perhaps as a consequence of the fact that GR cannot bind and therefore GR recognition sequences are lost over time in these regions.
NRS activity might also be context-specific to facilitate the condition-specific binding and activation of transcriptional programs by GR. In this regard, it is known that the paraspeckle component SFPQ binds to the promoter of anti-viral genes like IL8 and represses its expression (41). Upon viral infection, expression of the long non-coding RNA NEAT1 is induced, which in turn sequesters SFPQ from the IL8 promoter, thereby releasing its repressive effect and activating expression of the IL8 gene. This raises the intriguing possibility that viral infection might change the repertoire of available GR binding sites by releasing the repressive effect of paraspeckle-associated proteins on GR-binding to GBSs near NRSs.
In summary, our study shows that the analysis of TF-bound genomic loci can not only identify sequences that play a positive role in TF binding but also has the potential to uncover sequences that restrict TF binding to certain parts of the genome. In this study, we uncovered two sequences that showed the potential to restrict TF binding. Our analysis moreover uncovered additional under-represented sequences, indicating that the spectrum of sequences that play a role in restricting where in the genome TFs bind might be larger. Our simple approach to identify sequences that restrict TF binding to certain parts of the genome can be applied to any ChIP data and can thus help uncover how the combined action of positive and negative sequence signals specify where in the genome TFs bind.
ACCESSION NUMBERS
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (42) with the data set identifier PXD002252.
Supplementary Material
Acknowledgments
We thank Edda Einfeldt and Katja Borzym for excellent technical support, Samuel Collombet for technical help with R scripts and the German Ministry for Education and Research (BMBF, grant numbers 0315082, 01EA1303), the European Commission (FP7/2007–2013 under grant agreement 262055 ESGI), and the Max-Planck Society for financial support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Ministry for Education and Research [BMBF, 0315082 and 01EA1303]; European Commission [FP7/2007–2013 under grant agreement 262055 ESGI]; Max-Planck Society for financial support. Funding for open access charge: Institutional funding, Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Starick S.R., Ibn-Salem J., Jurk M., Hernandez C., Love M.I., Chung H.R., Vingron M., Thomas-Chollier M., Meijsing S.H. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 2015;25:825–835. doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Wrange O. Translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev. 1993;7:2471–2482. doi: 10.1101/gad.7.12a.2471. [DOI] [PubMed] [Google Scholar]
- 4.Perlmann T. Glucocorticoid receptor DNA-binding specificity is increased by the organization of DNA in nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 1992;89:3884–3888. doi: 10.1073/pnas.89.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiench M., John S., Baek S., Johnson T.A., Sung M.H., Escobar T., Simmons C.A., Pearce K.H., Biddie S.C., Sabo P.J., et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F., van Lohuizen M., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Bickmore W.A., van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Mao Y.S., Zhang B., Spector D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biddie S.C., John S., Sabo P.J., Thurman R.E., Johnson T.A., Schiltz R.L., Miranda T.B., Sung M.H., Trump S., Lightman S.L., et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siersbaek R., Nielsen R., John S., Sung M.H., Baek S., Loft A., Hager G.L., Mandrup S. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogatsky I., Trowbridge J.M., Garabedian M.J. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol. 1997;17:3181–3193. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas-Chollier M., Watson L.C., Cooper S.B., Pufall M.A., Liu J.S., Borzym K., Vingron M., Yamamoto K.R., Meijsing S.H. A naturally occurring insertion of a single amino acid rewires transcriptional regulation by glucocorticoid receptor isoforms. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17826–17831. doi: 10.1073/pnas.1316235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turatsinze J.V., Thomas-Chollier M., Defrance M., van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 15.Thomas-Chollier M., Defrance M., Medina-Rivera A., Sand O., Herrmann C., Thieffry D., van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeKelver R.C., Choi V.M., Moehle E.A., Paschon D.E., Hockemeyer D., Meijsing S.H., Sancak Y., Cui X., Steine E.J., Miller J.C., et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dona E., Barry J.D., Valentin G., Quirin C., Khmelinskii A., Kunze A., Durdu S., Newton L.R., Fernandez-Minan A., Huber W., et al. Directional tissue migration through a self-generated chemokine gradient. Nature. 2013;503:285–289. doi: 10.1038/nature12635. [DOI] [PubMed] [Google Scholar]
- 18.Freeman B.C., Yamamoto K.R. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 19.Kowalska E., Ripperger J.A., Muheim C., Maier B., Kurihara Y., Fox A.H., Kramer A., Brown S.A. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol. Cell. Biol. 2012;32:4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 21.Shipp L.E., Lee J.V., Yu C.Y., Pufall M., Zhang P., Scott D.K., Wang J.C. Transcriptional regulation of human dual specificity protein phosphatase 1 (DUSP1) gene by glucocorticoids. PloS One. 2010;5:e13754. doi: 10.1371/journal.pone.0013754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham C.D., Sims H.I., Archer T.K., Schnitzler G.R. Multiple distinct stimuli increase measured nucleosome occupancy around human promoters. PloS One. 2011;6:e23490. doi: 10.1371/journal.pone.0023490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittler G., Butter F., Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillier L.W., Marth G.T., Quinlan A.R., Dooling D., Fewell G., Barnett D., Fox P., Glasscock J.I., Hickenbotham M., Huang W., et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y., et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prykhozhij S.V., Marsico A., Meijsing S.H. Zebrafish Expression Ontology of Gene Sets (ZEOGS): a tool to analyze enrichment of zebrafish anatomical terms in large gene sets. Zebrafish. 2013;10:303–315. doi: 10.1089/zeb.2012.0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T.X., Zhou Y., Kanki J.P., Deng M., Rhodes J., Yang H.W., Sheng X.M., Zon L.I., Look A.T. Evolutionary conservation of zebrafish linkage group 14 with frequently deleted regions of human chromosome 5 in myeloid malignancies. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6136–6141. doi: 10.1073/pnas.072560099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X., Sweet J., Challis J.R., Brown T., Lye S.J. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol. Cell. Biol. 2007;27:4863–4875. doi: 10.1128/MCB.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong X., Yu C., Shynlova O., Challis J.R., Rennie P.S., Lye S.J. p54nrb is a transcriptional corepressor of the progesterone receptor that modulates transcription of the labor-associated gene, connexin 43 (Gja1) Mol. Endocrinol. 2009;23:1147–1160. doi: 10.1210/me.2008-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLellan W.R., Xiao G., Abdellatif M., Schneider M.D. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bramblett D., Hsu C.L., Lozano M., Earnest K., Fabritius C., Dudley J. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J. Virol. 1995;69:7868–7876. doi: 10.1128/jvi.69.12.7868-7876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Q., Dudley J.P. CDP binding to multiple sites in the mouse mammary tumor virus long terminal repeat suppresses basal and glucocorticoid-induced transcription. J. Virol. 2002;76:2168–2179. doi: 10.1128/jvi.76.5.2168-2179.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J., Bramblett D., Zhu Q., Lozano M., Kobayashi R., Ross S.R., Dudley J.P. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathur M., Tucker P.W., Samuels H.H. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyson-Capper A.J., Shiells E.A., Robson S.C. Interplay between polypyrimidine tract binding protein-associated splicing factor and human myometrial progesterone receptors. J. Mol. Endocrinol. 2009;43:29–41. doi: 10.1677/JME-09-0001. [DOI] [PubMed] [Google Scholar]
- 38.Meuleman W., Peric-Hupkes D., Kind J., Beaudry J.B., Pagie L., Kellis M., Reinders M., Wessels L., van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galande S., Purbey P.K., Notani D., Kumar P.P. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Rezsohazy R., Saurin A.J., Maurel-Zaffran C., Graba Y. Cellular and molecular insights into Hox protein action. Development. 2015;142:1212–1227. doi: 10.1242/dev.109785. [DOI] [PubMed] [Google Scholar]
- 41.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Vizcaino J.A., Cote R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J., et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.