Abstract
Colonization factor antigen I (CFA/I) is the archetype of eight genetically related fimbriae of enterotoxigenic Escherichia coli (ETEC) designated class 5 fimbriae. Assembled by the alternate chaperone pathway, these organelles comprise a rigid stalk of polymerized major subunits and an apparently tip-localized minor adhesive subunit. We examined the evolutionary relationships of class 5-specific structural proteins and correlated these with functional properties. We sequenced the gene clusters encoding coli surface antigen 4 (CS4), CS14, CS17, CS19, and putative colonization factor antigen O71 (PCFO71) and analyzed the deduced proteins and the published homologs of CFA/I, CS1, and CS2. Multiple alignment and phylogenetic analysis of the proteins encoded by each operon define three subclasses, 5a (CFA/I, CS4, and CS14), 5b (CS1, CS17, CS19, and PCFO71), and 5c (CS2). These share distant evolutionary relatedness to fimbrial systems of three other genera. Subclass divisions generally correlate with distinguishing in vitro adherence phenotypes of strains bearing the ETEC fimbriae. Phylogenetic comparisons of the individual structural proteins demonstrated greater intrasubclass conservation among the minor subunits than the major subunits. To correlate this with functional attributes, we made antibodies against CFA/I and CS17 whole fimbriae and maltose-binding protein fusions with the amino-terminal half of the corresponding minor subunits. Anti-minor subunit Fab preparations showed hemagglutination inhibition (HAI) of ETEC expressing homologous and intrasubclass heterologous colonization factors while anti-fimbrial Fab fractions showed HAI activity limited to colonization factor-homologous ETEC. These results were corroborated with similar results from the Caco-2 cell adherence assay. Our findings suggest that the minor subunits of class 5 fimbriae may be superior to whole fimbriae in inducing antiadhesive immunity.
Enterotoxigenic Escherichia coli (ETEC) is a principal cause of diarrhea in young children in resource-limited countries and also in travelers to these areas (4, 19). ETEC produces disease by adherence to small intestinal epithelial cells and expression of a heat-labile (LT) and/or heat-stable (ST) enterotoxin (31). ETEC typically attaches to host cells via filamentous bacterial surface structures known as colonization factors (CFs). More than 20 different CFs have been described, a minority of which have been unequivocally incriminated in pathogenesis (14).
Firm evidence for a pathogenic role exists for CF antigen I (CFA/I), the first human-specific ETEC CF to be described. CFA/I is the archetype of a family of eight ETEC fimbriae that share genetic and biochemical features (9, 14, 15, 24). This family includes coli surface antigen 1 (CS1), CS2, CS4, CS14, CS17, CS19, and putative CF O71 (PCFO71). The complete DNA sequences of the gene clusters encoding CFA/I, CS1, and CS2 have been published previously (11, 12, 21, 36, 49). The genes for the major subunit of two of the other related fimbriae have been reported previously (13, 15). The four-gene bioassembly operons of CFA/I, CS1, and CS2 are similarly organized, encoding (in order) a periplasmic chaperone, major fimbrial subunit, outer membrane usher protein, and minor fimbrial subunit. CFA/I assembly takes place through the alternate chaperone pathway, distinct from the classic chaperone-usher pathway of type I fimbrial formation and that of other filamentous structures such as type IV pili (37, 52). Based on the primary sequence of the major fimbrial subunit, CFA/I and related fimbriae have been grouped together as class 5 fimbriae (28).
Studies of CS1 have yielded details on the composition and functional features of class 5 fimbriae (46). The CS1 fimbrial stalk consists of repeating CooA major subunits. The CooD minor subunit is apparently localized to the fimbrial tip, comprises an extremely small proportion of the fimbrial mass, and is required for the initiation of fimbrial formation (45). Contrary to earlier evidence suggesting that the major subunit mediates binding (5), recent findings have implicated the minor subunit as the adhesin and identified specific amino acid residues required for in vitro adhesion of CS1 and CFA/I fimbriae (44). The inferred primary amino acid structure of those major subunits that have been sequenced share extensive similarity. Serologic cross-reactivity of native fimbriae is, however, limited, and the pattern of cross-reactivity correlates with phylogenetically defined subtaxons of the major subunits (13).
Implication of the minor subunits of class 5 fimbriae as the actual adhesins entreats scrutiny regarding the degree of their conservation relative to that of the major subunits. We speculated that CooD and its homologs have retained greater similarity due to functional constraints imposed by ligand binding requirements and/or its immunorecessiveness, itself attributable to the extremely large ratio of major to minor subunits in terms of fimbrial composition. The first aim of the present study was to examine the evolutionary relationships of the minor and major subunits of class 5 ETEC fimbriae as well as the two assembly proteins. We then tested whether the defined phylogeny predicted patterns of immunologic cross-reactivity using in vitro binding inhibition as a functional end point. Our findings reveal evolutionary distinctions between the class 5 major and minor fimbrial subunits and provide confirmatory evidence that the minor subunits function as adhesins while having practical implications for vaccine-related research.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The phenotypes of all ETEC strains used in adhesion experiments are shown in Table 1. The type strains that expressed CS1, CS4, CS14, CS17, CS19, and PCFO71 were each isolated from the feces of young children with diarrhea as part of a longitudinal study of childhood diarrhea in Egypt (38). Each of these type strains, except for those that expressed CFA/I, CS1, and CS2, was also the source of DNA for sequence analysis of the corresponding fimbrial operon. E. coli BL21 [F− ompT hsdSB(rB− mB−) gal dcm] was obtained from a commercial source (New England Biolabs, Beverly, Mass.) and used for cloning and expression of maltose-binding protein (MBP) fusions.
TABLE 1.
Identity and phenotype of representative ETEC strains expressing each of the class 5 fimbriae used in adhesion assays and as a source of DNA for fimbrial operon sequence analysis
| Strain | CF type(s) | Fimbrial subclass | Toxin type(s) | Serotype | GenBank accession no. of strain used for DNA sequence analysisa | Source or reference(s) |
|---|---|---|---|---|---|---|
| H10407 | CFA/I | 5a | LT, ST | O78:H11 | Not doneb | 10 |
| WS2560B | CS4 + CS6 | 5a | LT, ST | O25:H− | AY281092c,d | This study |
| WS3294A | CS14 | 5a | ST | O78:H18 | AY283611 | This study |
| WS1974A | CS1 | 5b | ST | O8:H9 | Not done | This study |
| WS2173A | PCFO71 | 5b | LT | O71:H4 | AY513487 | This study |
| WS6788A | CS17 | 5b | LT | O8:H9 | AY515609d | This study |
| WS0115A | CS19 | 5b | LT, ST | O114:H− | AY288101 | This study; 23 |
| C91f | CS2 | 5c | LT, ST | O6:H16 | Published data | 12, 51 |
GenBank accession number for fimbrial operon sequences completed in the course of this study.
Published data on the CFA/I operon sequence are derived from ETEC strain E7473 (21).
Accession number for the CS4 operon sequence.
For routine propagation and protein expression, bacteria were grown in Luria-Bertani medium (47) or in rich medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, and 2 g of glucose per liter). For hemagglutination and tissue culture adherence assays, cultures were grown on CFA agar (8) with or without the addition of 1.5 g of Bacto bile salts no. 3 (Difco, Detroit, Mich.) per liter. Ampicillin (62.5 μg/ml) and kanamycin (50 μg/ml) were added as needed for selection pressure.
DNA preparation and nucleotide sequence analysis.
For DNA sequence analysis, wild-type plasmid DNA was purified by a modified alkaline lysis procedure (plasmid midi kit; QIAGEN, Valencia, Calif.). Using the published sequence for the major subunit sequence of the corresponding CF, primers were designed to read outward from the 5′ and 3′ ends of each gene. The complete DNA sequence of both strands of each fimbrial operon was then generated by primer walking. Wild-type plasmid (1 to 3 μg of DNA) and 400 ng of primer were used for each sequencing reaction. Sequencing reactions were performed with Big Dye terminator enzyme mix (Applied Biosystems, Foster City, Calif.) in a Perkin-Elmer thermal cycler under the following conditions: 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. The reactions were analyzed on an ABI PRISM 3100 genetic analyzer (Applied Biosystems). Analysis of the derived sequences was performed with Sequencher, version 4.1 (Gene Codes Corporation, Ann Arbor, Mich.).
Database searches, multiple alignments, and statistical analyses.
Published sequence data for the genes encoding CFA/I (GenBank accession no. [gb] M5561) (21), CS1 Swiss-Prot accession no. [sp] P25731, sp P25730, European Molecular Biology Laboratory accession no. [emb] CAA54229, and emb CAA54230) (11, 20), CS2 (emb Z47800) (12), cable pili (gb AY114293), Tcf fimbriae (NCBI reference sequence accession no. [NC_] 003198) (34), and a putative Yersinia pestis fimbrial system (YPO03797 to YPO3802; NC_003143) (35) were retrieved from searches of the National Center for Biotechnology Information nonredundant databases by using BLAST, version 2.2.5 (2). These sequences along with those generated in this study served as the basis for phylogenetic analyses. The MacVector molecular sequence analysis program, version 7.2 (Accelrys, San Diego, Calif.) was used for translation and other routine analyses of DNA sequence data. The SignalP program (version 1.1) was used to predict the presence and location of signal cleavage sites (33).
Multiple-sequence alignments of sets of homologous proteins were produced by using ClustalX (version 1.81) (54) with the default alignment parameters. An in-house program was used to produce a multiple DNA alignment of the concatenated strings of the aligned amino acid sequences of the four proteins involved in the alternate chaperone assembly pathway. Protein phylogenetic trees were constructed by using the PAUP* program, version 4.0b10 (53), based on neighbor-joining methods (40).
The proportions of polymorphic synonymous (pS) and nonsynonymous (pN) sites and distances (dS and dN) were calculated by using the method of Nei and Gojobori (32). To examine variation in the different fimbrial assembly proteins, these statistics were tabulated in a sliding-window analysis of 30 codons along the gene by the program PSWIN. The ratio of synonymous-to-nonsynonymous differences per site within and between subfamilies was compared by using the MEGA2 program (25). The McDonald-Kreitman test (30), implemented by DnaSP (39), was used to assess the role of natural selection in the divergence within and between subfamilies.
Construction, expression, and purification of MBP fusion proteins.
Recombinant plasmids were designed and constructed by PCR amplification and cloning into the pMAL-p2 vector (New England Biolabs) for expression of the mature form of the CFA/I major subunit (CfaB, residues 24 to 170), the N-terminal half of the mature form of the CFA/I minor subunit (CfaE, residues 23 to 211), the C-terminal half of CfaE (residues 212 to 360), and the N-terminal half of the mature form of the CS17 minor subunit (CsbD, residues 19 to 214). Plasmid preparations of ETEC strains H10407 (CfaB and CfaE) and WS6788A (CsbD) were used as PCR templates. The PCR primers, product sizes, and restriction sites of pMAL-p2 used to make each construct are shown in Table 2. Each PCR product was gel purified, digested with the appropriate restriction enzymes, and cloned into pMAL-p2 to yield the desired protein fusion. The integrity of each construct was confirmed by junctional sequence analysis. All fusion constructs were electrotransformed into E. coli BL21 for further use. Restriction enzymes and ligases were purchased from New England Biolabs and used according to the manufacturer's instructions.
TABLE 2.
Constructs and primers used for making C-terminal fusions with MalE in pMAL-p2
| Plasmid | Fusion proteina | PCR primer (sequence [5′-3′])b | PCR product size (bp) |
|---|---|---|---|
| pAB206 | MBP-CfaB24-170 | CfaBXbaF (GCTCTAGAGTAGAGAAAAATATTACTGTAACAGG) | 467 |
| CfaBHindR (CGCCCCAAGCTTTCATCAGGATCCCAAAGTCATTACAAG) | |||
| pAB202 | MBP-CfaE23-211 | CfaEnEcoF (GGAATTCGCAGATAAAAATCCCGGAAGTGAA) | 585 |
| CfaEnXbaR (GCTCTAGATCAGAACTGTGGTAACCATATCTGAAT) | |||
| pAB201 | MBP-CfaE212-360 | CfaEcEcoF (GGAATTCAAAAGTAACGCTCGTGTCGATCTT) | 468 |
| CfaEcXbaR (GCTCTAGATCACTAGAGTGTTTGACTACTTGGTGT) | |||
| pRA1 | MBP-CsbD19-214 | DMalBamF (GTACTGGATCCGGGCGATACCCGGAAACTACA) | 614 |
| DMalHindR (GTACATAAGCTTCTAAAACCCTGGAAGCCATACCTG) |
The subscript amino acid residue span of the corresponding protein is numbered with respect to the full-length protein.
Primers were used for the generation of pMAL-p2 insertion fragments. Flanking restriction enzyme sites introduced for cloning are underlined.
The expression and purification of MBP fusion proteins was carried out according to protocols recommended by the manufacturer (NEB) with some modifications. Briefly, for BL21 with the recombinant plasmids designed to express MBP-CfaE23-211 (pAB202) and MBP-CfaB24-170 (pAB206), bacteria were grown in Luria-Bertani broth at 37°C to a cell density of 0.5 A600 and fusion protein expression was then induced for 2 h after the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, and periplasmic contents were released by cold osmotic shock. The osmotic shock fluid supernatants were loaded on an amylose affinity column. After washing of unbound proteins, MBP fusion products were eluted with 10 mM maltose. For BL21(pRA1) and BL21(pAB201), the MBP-CsbD19-214 and MBP-CfaE212-360 constructs, respectively, bacteria were grown in rich broth. For BL21(pRA1), induction was carried out at 37°C for 2.5 h. BL21(pAB201) was induced at 16°C for 16 h. Cells were harvested by centrifugation, suspended in column buffer, and frozen at −20°C overnight. After thawing and the addition of 1 mM phenylmethylsulfonyl fluoride (Sigma Chemical), cells were lysed by one cycle of microfluidization (model 1109 apparatus; Microfluidic Corp., Newton, Mass.). The soluble fraction was subjected to amylose affinity chromatography as described above. To ascertain protein mass and relative purity, eluted fusion protein fractions were analyzed by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a Hoefer mighty small II mini-vertical unit. Fusion protein fractions were pooled, and the protein concentration was estimated by the Bio-Rad protein assay (Hercules, Calif.) and adjusted to a final concentration of 1 mg/ml for antibody generation.
Purification of ETEC fimbriae.
CFA/I and CS17 fimbriae were purified as previously described (16, 17). Briefly, bacteria were harvested from CFA plates or CFA plates with bile salt into phosphate-buffered saline (PBS) and heated to 60°C to extract heat-soluble proteins, which were then precipitated with 40% (CFA/I) or 20% (CS17) ammonium sulfate. Purified fimbriae were dialyzed overnight against PBS.
Antibody production.
Rabbit polyclonal antibody preparations were prepared against MBP-CfaB24-170, MBP-CfaE23-211, MBP-CfaE212-360, and MBP-CsbD19-214 and against native CFA/I and CS17 fimbriae. Rabbit immunizations and antiserum collection were performed by Harlan Bioproducts for Science, Inc. (Indianapolis, Ind.). Purified immunoglobulin G (IgG) was derived from each antiserum by using Hi-Trap protein G columns as directed by the manufacturer (Amersham Pharmacia, Piscataway, N.J.). From each of these preparations, Fab fragments were generated by using the ImmunoPure Fab preparation kit (Pierce, Rockford, Ill.). Briefly, 10 mg of IgG at a concentration of 20 mg/ml was digested overnight with papain at 37°C. The samples were washed and loaded onto an immobilized protein A column, and purified Fab fragments were eluted and dialyzed against PBS (pH 7.4). The concentration of purified Fab was determined with the bicinchoninic acid protein assay (Pierce). Each Fab preparation was brought to a final concentration of 2 mg/ml in PBS by ultrafiltration with an Amicon Ultra centrifugal concentrator (Millipore, Bedford, Mass.).
MRHA and inhibition.
ETEC strains were tested for mannose-resistant hemagglutination (MRHA) of human type A, bovine, and chicken erythrocytes based on previously described methods (6). Human erythrocytes were harvested as needed from a single volunteer donor, and bovine and chicken erythrocytes were purchased from Lampire Laboratories (Pipersville, Pa.). Erythrocytes were stored for up to 2 weeks at 4°C in Alsever's solution prior to use. Just before each assay, erythrocytes were washed and suspended in PBS with 0.5% d-mannose to a final concentration of 3%. Bacteria were grown overnight at 37°C and suspended in PBS with 0.5% d-mannose to a final concentration of about 1010 CFU/ml. Equal volumes (25 μl of each) of 3% red cells, bacterial suspension, and PBS with 0.5% d-mannose were added and mixed in wells on a 12-well ceramic tile (CoorsTec, Golden, Colo.), rocked on ice for 20 min, graded by visual inspection, and scored as follows: negative, no MRHA activity; 1+, low, weak reaction; 2+, moderate reaction; 3+, strong reaction; 4+, nearly instantaneous and complete reaction involving all of the erythrocytes.
For hemagglutination inhibition (HAI) assays, each bacterial strain was used at a concentration corresponding to two times the minimal hemagglutination titer (MHT). The MHT was determined at the start of each HAI assay day by making serial twofold dilutions of the bacterial suspension (from a starting concentration of 1010 CFU/ml) in PBS. A total of 25 μl of each dilution was added to equal volumes of 3% erythrocyte suspension and PBS with 0.5% d-mannose and rocked on ice. The MHT was defined as the reciprocal of the lowest concentration of bacteria showing at least a 1+ MRHA. To determine the HAI titer of each Fab antibody preparation, a twofold dilution series was made starting with the stock antibody solution (2 mg/ml). A 25-μl volume of each Fab dilution was added to an equal volume of a 2× MHT bacterial suspension in the ceramic tile wells and preincubated at room temperature with rocking for 20 min. An equal volume of erythrocyte suspension (3%) was then added to each well, the tiles were rocked on ice for 20 min, and MRHA was scored as described above. The HAI titer was expressed as the reciprocal of the highest dilution of antiserum that completely inhibited MRHA.
Caco-2 adherence assay and inhibition studies.
Caco-2 cell adherence assays were performed as described previously (7, 23) with minor modifications. Briefly, Caco-2 cells were maintained at 37°C in air supplemented with 5% CO2 in EMEM (Eagle's minimum essential medium in Earle's balanced salt solution) supplemented with 2 mM l-glutamine, 20% fetal bovine serum, 0.1 M nonessential amino acids, 1 mM sodium pyruvate, and 1.5 g of sodium bicarbonate/liter. Cells were seeded in 24-well plates (Costar, Corning, N.Y.), loaded with tissue culture-treated glass coverslips (Fisher Scientific), incubated for 14 days (±1 day) to postconfluence, washed with PBS, and covered with 750 μl of the supplemented EMEM prior to the assay. Bacterial strains were grown on CFA agar with or without bile salts overnight at 37°C and suspended to 109 bacteria/ml in supplemented EMEM with 1% d-mannose. The suspension was added to the tissue culture wells at a final concentration of 2.5 × 108 bacteria/ml. Plates were incubated, washed, fixed, stained, and mounted as described previously (23) and then observed microscopically. The number of bacteria adherent to 100 randomly selected cells was counted to give an average number of cells with at least 1 adherent bacterium (adherence index 1), and the number of bacteria per Caco-2 cell with at least one adherent bacteria was counted (adherence index 2). For each bacterial strain, a minimum of three experiments was done in duplicate to determine the adherence indices, expressed as means ± standard deviations (SD).
For Caco-2 cell adherence inhibition experiments, a 120-μl aliquot of Fab antibody preparation (2 mg/ml starting concentration) was added to 480 μl of the bacterial suspension and preincubated at room temperature for 20 min. The addition of PBS in place of the antibody preparation served as a negative control in each experiment. A 250-μl aliquot of the bacterium-antibody mixture (2.5 × 108 bacteria/ml) was then added to the tissue culture wells. The cells were incubated, processed, and analyzed as described above. The level of inhibition was determined by comparing the primary adherence index with and without the addition of antibody. For each test bacterium-antibody preparation, a minimum of three experiments was performed in duplicate.
Statistical methods.
For Caco-2 cell adherence inhibition studies, adherence in the presence of each antibody preparation was compared to that with the addition of PBS, using a one-tailed Student t test, assuming unequal variance between samples. For HAI experiments, reciprocal titers between experimental groups were compared by using the Wilcoxon signed rank test for paired samples (one-tailed) with XLSTAT data analysis software.
RESULTS
Analysis of fimbrial operons.
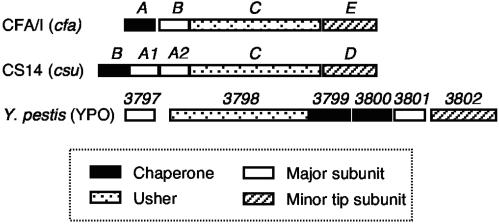
We determined the nucleotide sequence of the gene clusters that encode CS4, CS14, CS17, CS19, and PCFO71 from wild-type diarrhea-associated isolates of ETEC that tested positive for each respective fimbriae by monoclonal antibody-based detection (Table 1). The major subunit alleles of the newly sequenced CS4, CS14, CS17, and CS19 gene clusters each showed 99 to 100% nucleotide sequence identity with corresponding gene sequence(s) previously deposited in GenBank, with no more than four nucleotide differences per allele. Each locus had four open reading frames that encoded proteins with homology to the CFA/I class chaperones, major subunits, ushers, and minor subunits. As previously reported (13), the one exception was for the CS14 gene cluster, which contained two tandem open reading frames downstream of the chaperone gene (Fig. 1). Their predicted protein sequences share 94% amino acid identity with one another and are both homologous to other class 5 fimbriae major subunits.
FIG. 1.
Organizational comparison of class 5 fimbrial system operons. The CFA/I operon that is shown exemplifies the organization of CS1 (cooBACD), CS2 (cotBACD), CS4 (csfBACD), CS17 (csbBACD), CS19 (csdBACD), PCFO71 (cosBACD), the Cbl pili of B. cepacia (cblBACD), and the putative Tcf fimbriae of S. enterica serovar Typhi (tcfABCD) (data not shown). The CS14 gene cluster is distinguished by the presence of two highly similar major subunit genes tandemly arranged in the expected positions (csuA1 and csuA2). The hypothetical fimbrial system operon of Y. pestis features a different gene order and the presence of two chaperone-like genes and two major subunit genes. Y. pestis gene numbers are in reference to the (YPO) genome annotation.
We searched available sequence databases to identify other fimbrial systems with similarity to the class 5 ETEC fimbriae. Homologous chromosomal elements were found in Burkholderia cepacia (strain BC7), encoding the well-characterized cable pilus (29, 41), and in Salmonella enterica serovar Typhi (strain CT18), encoding the putative Tcf fimbriae (34, 55). In the Y. pestis genome, another homologous fimbrial operon was identified (35). It differed from the other fimbrial gene clusters in the order and number of assembly genes (Fig. 1).
Examination of the inferred amino acid sequences of all protein homologs involved in class 5 fimbrial biogenesis reveals many basic similarities. Across genera, each set of homologs generally share similar physicochemical properties in terms of polypeptide length, mass, and theoretical isoelectric point (data not shown). All of the involved proteins contain an amino-terminal signal peptide that facilitates translocation to the periplasm via the type II secretion pathway. None of the major subunit proteins contain any cysteine residues, whereas the number and location of six cysteine residues are conserved for all of the minor subunits except that of the Y. pestis homolog 3802, which contains only four of these six residues.
Phylogenetic analysis of class 5 fimbrial systems.
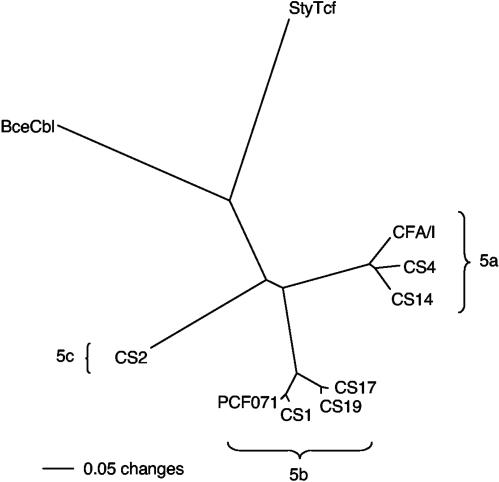
To develop a general representation of the evolutionary relationships among the class 5 fimbrial systems at the protein level, an optimal multialignment of each ordered string of four requisite proteins (chaperone-major subunit-usher-minor subunit) was generated. From this alignment, an unrooted phylogram was constructed from amino acid distances (mean character difference) based on the neighbor-joining algorithm (Fig. 2). The eight ETEC class 5 fimbriae clustered into three subclasses of three (CFA/I, CS4, and CS14), four (CS1, PCFO71, CS17, and CS19), and one (CS2) member(s), which we will refer to as subclasses 5a, 5b, and 5c, respectively. The cable pilus and Tcf fimbriae are more distantly related to one another and to the ETEC fimbrial systems, whereas the Y. pestis fimbrial system is the most evolutionarily distant member of this class.
FIG. 2.
Unrooted phylogram constructed by the neighbor-joining method showing the evolutionary relatedness of a concatenated alignment of the four proteins encoded by the class 5 fimbrial operons. Distances represent the mean character differences among the protein alignments. Abbreviations: BceCbl, B. cepacia Cbl pilus; StyTcf, S. enterica serovar Typhi Tcf fimbriae.
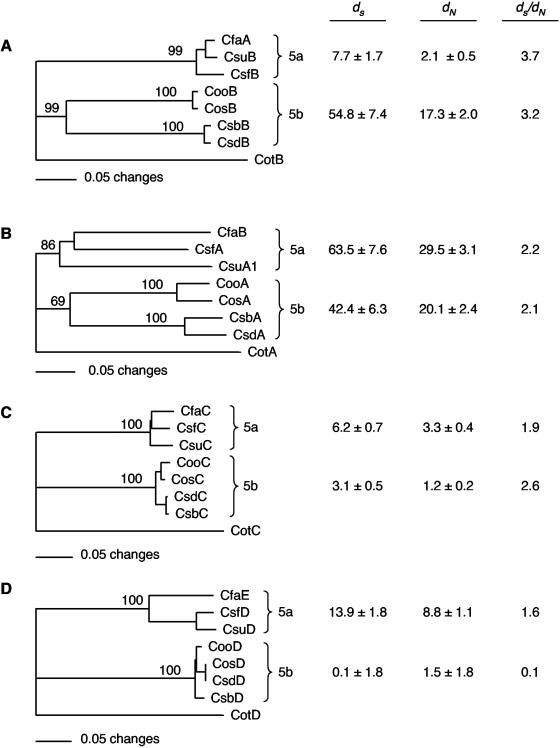
To test the hypothesis that the minor subunits are more highly conserved than the major subunits of class 5 fimbriae, we constructed distance-based phylogenetic trees of each of these two structural homolog groupings as well as of the chaperone and usher protein homologs. Since here we were primarily interested in ETEC, we confined these analyses to the eight ETEC class 5 fimbriae. The topology of the trees for each of the fimbrial structural proteins and of the accessory proteins is similar, with bootstrap values supporting the grouping of the homologs into the 5a and 5b subclasses (Fig. 3). The exception is the phylogeny for the major subunits (Fig. 3B), where the longer internal branches have bootstrap confidence values of 86 and 69% for the 5a and 5b subclasses. A comparison of the major and minor subunit trees (Fig. 3B and D) indicates a similar level of divergence between the subclasses. For example, the divergences between the 5a and 5b major subunit subclasses are 98.6 ± 9.6 and 41.9 ± 4.3 at synonymous (dS) and nonsynonymous (dN) sites, respectively, and 105.1 ± 10.3 and 47.0 ± 3.6 for the 5a and 5b minor subunit subclasses, respectively. The comparative distance separating protein taxa within each subclass, however, is much less among the minor than the major subunits, as measured by the branch lengths at the amino acid level and by both dS and dN at the nucleotide level. This comparison indicates that the major subunits within each of these subclasses have diverged at a faster rate (or for a much longer time) than their minor subunit counterparts. It is also notable that, for the outer membrane usher proteins (Fig. 3C), the distance between the internal nodes separating each subclass is less than that for any of the other proteins and that intertaxon distances within subclasses of ushers are on the same order of magnitude as those for the minor subunits. This suggests that both the ushers and the minor subunits have evolved at similarly slow rates compared to the major subunits.
FIG. 3.
Phylogenetic trees showing evolutionary relationships of each of the proteins required for biosynthesis of class 5 ETEC fimbriae. Using the neighbor-joining method, distances represent mean character differences among the aligned proteins. Comparisons of chaperone (A), major subunit (B), outer membrane usher (C), and minor subunit (D) homologs are shown. In each case, the CS2 (Cot) protein homolog was defined as the out-group to root the tree. Bootstrap values shown in each tree were calculated by using MEGA2 with synonymous sites and the minimum evolution algorithm. The numbers at the nodes are the percentage of 1000 replicate trees that included each clade. Columns to the right of each tree show the synonymous (dS) and nonsynonymous (dN) differences per 100 sites that are variable among alleles within the ETEC 5a and 5b clades. Also shown are the ratios of synonymous and nonsynonymous differences (dS/dN).
The accelerated evolutionary rates of the major subunit compared with the two downstream genes encoding the ushers and minor subunits, respectively, may be attributable to marked differences in the intensity of selection on the different sites (e.g., due to stronger functional constraints on the latter two alleles) or to intraspecies horizontal gene transfers in which the recombination event did not affect the major subunit alleles. An overall measure of the degree of functional constraint is the ratio of the synonymous to nonsynonymous change which is expected to be >1 for purifying (negative) selection against amino acid replacements and <1 for diversifying (positive) selection favoring amino acid change (18). The dS/dN ratio, measuring divergence within subclasses, reveals that overall the subunits encoded by the ETEC class 5 fimbrial gene clusters are under purifying selection (dS/dN > 1), with the exception of the 5b subclass of the minor subunits. The greatest level of constraint is seen for the chaperone subunit (Fig. 3A) (dS/dN > 3), and the lowest levels are seen for the minor subunits (Fig. 3D).
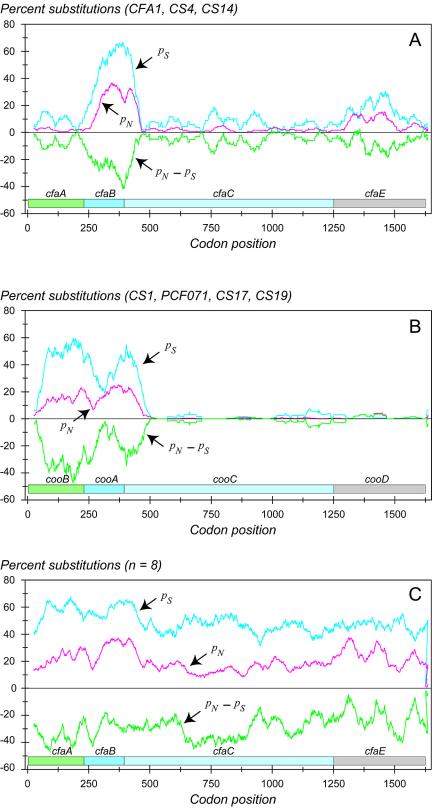
Although overall evidence indicates negative selection, certain regions of the molecules could be subjected to positive selection for amino acid change. To ascertain the changes in selective constraint for different regions of the molecules, we calculated the percentage of sites that were polymorphic for nonsynonymous (pN) and synonymous (pS) for subsets of 30 codons in a sliding window for the length of the fimbrial operon (Fig. 4). We used the difference, pN − pS, as a measure of the degree of functional constraint, whereby the more negative the value, the less the contribution of replacement substitutions and the greater the contribution of synonymous substitutions. The zero difference line indicates selectively neutral variation, where the per site rates of synonymous and nonsynonymous substitutions are equal. A positive difference, where amino acid replacements exceed the silent substitutions, suggests the action of positive selection. The sliding window analysis for the 5a subclass reveals that the rate of silent changes varies three- or fourfold through the gene cluster with a maximum level in the major subunit (CfaB). There is a secondary peak in pN and pS in the minor subunit. This is consistent with the longer branches of the 5a subclass in the gene tree for the major and minor subunits in Fig. 3. The 5b subclass shows a similar peak of divergence in the major subunit and, in contrast, an additional strong peak in the chaperone subunits, reflecting the long internal branches for these subunits in the gene trees (Fig. 3). Over all eight sequences, pN − pS was <0, except for a few positions where it approaches zero.
FIG. 4.
Sliding-window plot of the percentage of substitutions per 100 sites for synonymous (pS) and nonsynonymous (pN) sites between subclasses 5a (A) and 5b (B) and all class 5 ETEC fimbrial operons (C). The difference (pN − pS) is a measure of the level of selective constraint on the different proteins and domains within each protein. The gene names of the first fimbriae shown in each cluster are shown above the corresponding coding segment.
The sliding-window analysis suggests that the level of functional constraint on the fimbrial components is generally strong, as reflected by a pN − pS of <0, and that it varies both across the gene clusters and between subclasses. The fact that the synonymous rate varies substantially across the gene clusters, even for the same subunit between subclasses, supports the idea that recombination has contributed to sequence divergence. This is particularly striking, for example, in the chaperone subunit of the 5b subclass where the divergence between CooB and CosB and CsbB and CsdB varies dramatically along the length of the sequence. The genes of these two groups of chaperones are mosaics, as detected by Maxchi analysis (50), with a sequence divergence ranging from 4% (bp 1 to 162) to 34% (bp 163 to 612) and 54% (bp 613 to 702) (data not shown).
The McDonald-Kreitman test is a method for detecting selection at the molecular level based on a comparison of synonymous and nonsynonymous (amino acid replacement) variation within and between species (lineages). Under the neutral mutation hypothesis, the ratio of synonymous (S) to replacement (R) polymorphisms within species should be the same as the ratio of synonymous to replacement differences fixed between species. In application, we tested whether the S/R ratio was statistically the same for substitutions between alleles within a subclass versus differences between subclasses. There is no evidence for positive selection fixing amino acid replacements between subclasses relative to the variation between alleles within a subclass in three of the four comparisons, where the null hypothesis for equal ratios cannot be rejected and the neutrality index is near unity (Table 3). For the minor subunits, however, there is a significant difference in the ratio, primarily as a result of the small number of synonymous substitutions. The small number of synonymous polymorphisms, particularly for the 5b subclass of minor subunits, is consistent with the idea of a selective sweep reducing the variation of this subclass.
TABLE 3.
Results of the McDonald-Kreitman testa
| ETEC fimbrial system component (protein)b | No. of polymorphic sites
|
No. of fixed sites
|
NIc | ||
|---|---|---|---|---|---|
| S | R | S | R | ||
| Chaperone (CfaA) | 121 | 110 | 83 | 85 | 0.89 |
| Major subunit (CfaB) | 120 | 111 | 27 | 31 | 0.81 |
| Outer membrane usher (CfaC) | 98 | 97 | 395 | 345 | 1.13 |
| Minor subunit (CfaE) | 53 | 91 | 170 | 195 | 1.50d |
Numbers of synonymous (S) and replacement (R) sites that are polymorphic within alleles of a subclass or that represent fixed differences between subclasses for each fimbrial component are shown. Under the neutral mutation theory, the S/R ratio should be equivalent for polymorphic and fixed sites under strict neutrality.
Protein function CFA/I homolog.
NI, neutrality index.
ΔG = 4.03; df = 1; P = 0.04 (two-tailed Fisher's exact test).
Characterization of in vitro adherence phenotypes.
Type strains that individually express each of the class 5 ETEC fimbriae were characterized with respect to erythrocyte adherence by MRHA with type A human, bovine, and chicken erythrocytes. The results (Table 4) confirmed previously reported findings, with the exception that our results with chicken erythrocyte MRHA differ from those of a previous report with respect to CS17 and CS19 ETEC (23).
TABLE 4.
In vitro adherence phenotypes of ETEC type strains bearing CFA/I and related class 5 fimbriae
| Strain | CF type | MRHA score for erythrocyte type
|
Result for Caco-2 cell adherence indexa:
|
|||
|---|---|---|---|---|---|---|
| Human type A | Bovine | Chicken | 1b | 2c | ||
| H10407 | CFA/I | 4+ | 4+ | 3+ | 54.3 ± 15.4 | 14.2 ± 2.7 |
| WS2560B | CS4 | 2+ | 2+ | 1+ | 26.7 ± 7.0 | 2.9 ± 1.6 |
| WS3294A | CS14 | 2+ | 3+ | 3+ | 63.3 ± 5.8 | 8.2 ± 2.4 |
| WS1974A | CS1 | −d | 3+ | − | 12.7 ± 8.6 | 2.1 ± 1.1 |
| WS2173A | PCFO71 | − | 4+ | 2+ | 12.7 ± 6.2 | 1.8 ± 0.6 |
| WS6788A | CS17 | − | 4+ | − | 10.0 ± 2.6 | 1.1 ± 0.2 |
| WS0115A | CS19 | − | 4+ | 2+ | 19.3 ± 6.0 | 1.8 ± 0.8 |
| C91f | CS2 | − | 3+ | 3+ | 69.3 ± 4.7 | 15.1 ± 4.7 |
Represents the mean of the results from at least three experiments, each done in duplicate.
Mean proportion of Caco-2 cells with at least one adherent bacterium (±SD).
Mean number of adherent bacteria per Caco-2 cell with at least one adherent bacterium (±SD).
−, negative.
It has previously been reported that ETEC bearing CFA/I, CS2, CS4, CS14, and CS19 manifest adherence to cultured Caco-2 cells (15, 58). We performed Caco-2 cell adherence assays on each of the ETEC type strains bearing the class 5 fimbriae to confirm these findings and quantify the level of adherence for each strain. The results (Table 4) indicated that, indeed, the strains bearing CFA/I, CS4, CS14, and CS2 each showed a moderate to high level of Caco-2 cell adherence while a lower level of adherence was observed for the CS19-bearing strain. In contrast, the strains expressing CS1, CS17, and PCFO71 manifest marginal levels of adherence. Transformation of the strains bearing subclass 5b fimbriae with a plasmid containing the CFA/I positive regulator cfaD was associated with an increase in Caco-2 cell adherence only for the CS19-ETEC strain WS0115A (data not shown).
Considering the evolutionary relationships of the class 5 ETEC fimbriae, it can be seen that there are some distinguishing functional characteristics that correlate with their phylogeny. Subclass 5a fimbriae are distinct from the others by virtue of their ability to cause MRHA of human type A erythrocytes (Table 4). With the exception of the CS19 ETEC, subclass 5b fimbriae show weak, if any, adherence to cultured Caco-2 cells, differentiating them from the other two subclasses.
Localization of the binding domain of CFA/I fimbriae.
Conflicting data have been published regarding which of the component subunits of CFA/I and CS1 mediate adherence (5, 44). We approached this question indirectly by assessing the adherence inhibition activity of antibodies to intact CFA/I fimbriae, CfaB (major subunit), and the nonoverlapping amino-terminal (residues 23 to 211) and carboxy-terminal (residues 212 to 360) halves of CfaE (minor subunit) in two different in vitro adherence models. Rabbit polyclonal antiserum was generated to each of the subunit constituents in the form of MBP fusions from which purified IgG Fab preparations were produced. Fab fractions were selected for evaluation to avoid a potential nonspecific agglutinating effect of bivalent IgG preparations on bacterial suspensions.
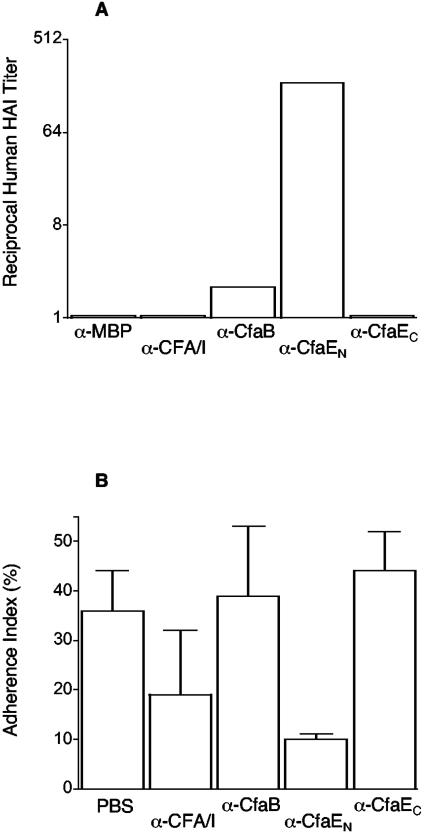
Each of these four antibody preparations was assessed for the ability to inhibit the adherence of strain H10407 (CFA/I) in MRHA and Caco-2 cell adherence assays. The highest human A erythrocyte HAI activity was observed with Fab specific for CfaE23-211 while CfaB antibodies manifested a much lower level of HAI activity (Fig. 5A). No HAI activity was detectable with Fab antibodies against CFA/I or CfaE212-360. Consistent findings were observed in Caco-2 cell adherence inhibition assays, in that the highest inhibitory activity was attributable to anti-CfaE23-211 Fab fractions (Fig. 5B). In this assay, anti-CFA/I Fab antibodies showed a lower level of inhibition and preparations specific for CfaB and CfaE212-360 showed no detectable effect. Taken together, these findings suggest that the most important domain for CFA/I adherence resides in the amino-terminal half of CfaE.
FIG. 5.
Inhibitory effects of different Fab antibody preparations on adherence of strain H10407 (CFA/I) in two in vitro adherence models. (A) Median reciprocal HAI titers of Fab antibodies specific for MBP, CFA/I, CfaB, CfaE23-211 (denoted as CfaEN), and CfaE212-360 (denoted as CfaEC) plotted on a log2 scale. Values below a reciprocal of 2 (limit of detection) were arbitrarily plotted as 1.05 for graphing purposes. (B) Mean Caco-2 cell adherence index (percentage of Caco-2 cells with at least 1 adherent bacterium ± SD) of H10407 after preincubation of bacteria with Fab antibodies with the same specificities. All preparations were tested in at least three experiments, each done in duplicate (panels A and B). α, anti.
Heterologous inhibitory effects of antibodies to the CFA/I and CS17 minor subunit N-terminal domain.
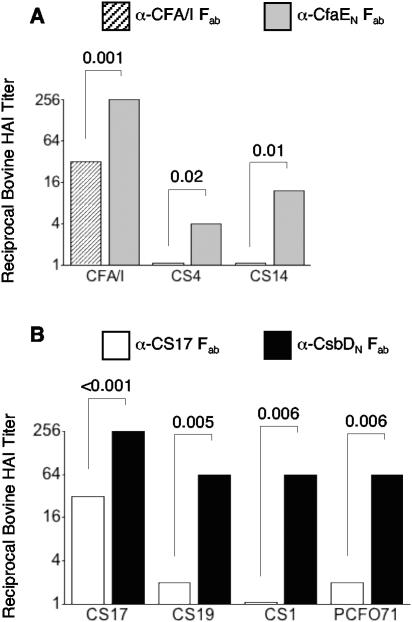
The experiments described above provide corroborative evidence that the minor subunits of CFA/I and other class 5 fimbriae are the receptor binding moiety (44). Since we also observed low levels of sequence divergence of the minor subunits within fimbrial subclasses 5a and 5b, we speculated that these evolutionary relationships would correlate with cross-reactivity of antibodies against the amino-terminal half of minor subunits representing each of these two subclasses. To test this hypothesis, we assessed the inhibitory effect of anti-CfaE23-211 Fab on the adherence of wild-type strains expressing heterologous class 5 fimbriae. Consistent with our predictions, anti-CfaE23-211 inhibited bovine MRHA of CS4-ETEC and CS14-ETEC (Fig. 6A). In comparison, anti-CFA/I Fab antibodies inhibited bovine MRHA of CFA/I-ETEC to a lesser degree than the anti-CfaE23-211 while failing to inhibit MRHA of ETEC bearing CS4 or CS14. Identical results were obtained with human erythrocytes, except that anti-CFA/I Fab failed to display CFA/I-ETEC HAI (data not shown). Neither antibody preparation inhibited bovine MRHA of ETEC bearing heterologous CFs of the other two subclasses (data not shown).
FIG. 6.
Median reciprocal bovine HAI titers (plotted on a log2 scale) of Fab antibody preparations against whole fimbriae or the amino-terminal domain of the minor fimbrial subunit of CFA/I (A) and CS17 (B) for ETEC type strains expressing the colonization factor indicated along the x axis. Results represent the median of the results from at least four experiments, each performed in duplicate. P values are for the differences in HAI titers between the whole fimbriae and minor subunit antibody preparations. α, anti.
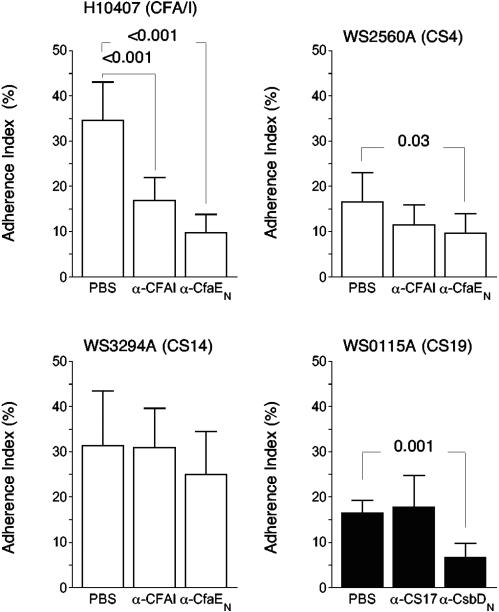
These findings were corroborated by measuring the inhibitory effects of each Fab preparation in the Caco-2 cell adherence assay. Anti-CfaE23-211 antibodies inhibited the adherence of CS4-ETEC and CS14-ETEC when compared to the adherence level when bacteria were preincubated with PBS (Fig. 7) or with anti-MBP antibodies (data not shown). The diminished adherence of CS14-ETEC did not, however, achieve statistical significance. At the same concentration, anti-CFA/I antibodies inhibited the Caco-2 cell adherence of H10407 (CFA/I), though to a significantly lesser degree than did anti-CfaE23-211 Fab. Anti-CFA/I Fab did not, however, inhibit the binding of ETEC bearing heterologous CFs of the same (Fig. 7) or different subclasses (data not shown).
FIG. 7.
Inhibitory effects of Fab antibodies against intact fimbriae and the N-terminal half of the minor subunit of CFA/I (white bar graphs) and CS17 (black bar graph) in Caco-2 cell adherence assays with ETEC bearing homologous (CFA/I only, upper left panel) and heterologous fimbriae. The strain used in the experiments is shown above each graph. The y axes indicate the Caco-2 cell adherence index (percentage of Caco-2 cells with at least one adherent bacteria). Results represent the means (±SD) of the results from at least three experiments, each performed in duplicate. P values are for the differences between the negative control (PBS) and the indicated antibody preparation. α, anti.
To strengthen these findings further, we produced antibodies to the amino-terminal half of the CS17 (subclass 5b) minor subunit CsbD and assessed its inhibitory activity along with that of anti-CS17 fimbrial antibodies in the MRHA and Caco-2 tissue culture cell model systems. Both anti-CS17 and anti-CsbD19-214 Fab antibodies exhibited bovine erythrocyte HAI activity for ETEC bearing CS17, with the HAI titer of anti-CsbD19-214 being significantly higher (Fig. 6B). Distinct from the anti-CS17 Fab antibodies, the anti-CsbD19-214 Fab fraction also manifested significant HAI activity for ETEC bearing each of the other subclass 5b fimbriae. Notably, the intrasubclass CF heterologous HAI activity of anti-CsbD19-214 antibodies was closer in magnitude to its CS17-ETEC HAI activity than for the comparable effects of anti-CfaE23-211 antibodies. This finding was anticipated, given the higher degree of identity of the minor subunits within subclass 5b. Neither preparation inhibited bovine MRHA of ETEC bearing CFs of the other two subclasses (data not shown).
In the Caco-2 cell adherence assay, we assessed the inhibitory effects of the same antibody preparations for CS19-ETEC, the only subclass 5b fimbriae that appears to specifically adhere to Caco-2 cells. Here too we found that anti-CsbD19-214, but not anti-CS17, antibodies showed significant inhibition of CS19-ETEC adherence (Fig. 7). Neither preparation inhibited Caco-2 cell adherence of ETEC expressing representative subclass 5a or 5c fimbriae (data not shown).
DISCUSSION
Since its discovery nearly 30 years ago (9), CFA/I has come to be recognized as the archetype of the largest class of ETEC fimbriae (13, 14). Though all are serologically distinct, each of these so-called class 5 fimbriae is assembled by a similar pathway and shares the functional attribute of mediating human intestinal adhesion. In the present study, we have identified fimbrial systems in two other gram-negative pathogens, which join the cable pilus system of B. cepacia as class 5 fimbriae distantly related to these ETEC fimbriae. In a detailed examination of the evolutionary relationships of the eight ETEC class 5 fimbriae, greater conservation was observed for the minor fimbrial subunits than for the major fimbrial subunits. Our analyses suggest that horizontal gene transfer has contributed to this pattern of conservation. Of practical importance, we found that antibodies to the amino-terminal half of two different minor subunits inhibit the in vitro adherence of bacteria that express homologous fimbriae as well as those expressing closely related heterologous fimbriae. The latter findings corroborate the report of Sakellaris et al., implicating the minor subunit of this class as the adhesive fimbrial component (44), and have potential ramifications for ETEC vaccine development.
Though our main focus was on the class 5 ETEC fimbriae, it is noteworthy that the genetic determinants of similar fimbrial systems are found not only in B. cepacia but also in S. enterica serovar Typhi and Y. pestis. The latter two fimbrial systems have been revealed by genome sequence analysis (34, 35, 55), and their expression and function have yet to be evaluated. Cable pili of B. cepacia are thought to play an auxiliary role in adherence to matrix proteins of the respiratory epithelium (42, 43), a role akin to that of its more distant ETEC relatives.
The eight class 5 fimbriae of ETEC are distinguishable by serologic methods, and monoclonal antibodies generated against unique epitopes of most of these have been exploited for diagnostic purposes (26, 27, 57). The major fimbrial subunit comprises the primary antigenic determinant of these fimbriae, and our finding that this correlates with the pattern of evolutionary divergence of their primary amino acid sequence builds upon previously published observations (13). Despite considerable divergence of their exposed surfaces, each has apparently preserved the ability to adhere to intestinal epithelial cells, thereby maintaining its fitness for attachment, survival, and replication upon entry into the human small intestine. This feature, along with published data indicating that the minor subunits are the actual adhesin, prompted a detailed phylogenetic and functional analysis centered on this component.
Our finding that the minor subunits of class 5 ETEC fimbriae, at least within each of two defined subclasses, are much less divergent than their counterpart major subunits may in part reflect the imposition of constraints that promote the preservation of epithelial binding capacity or the recent origin and spread of new alleles. The latter condition is most likely the case for the 5b subclass, in which the sequences are very similar, with mutations of only ca. 0.1% of the synonymous sites. We know little about the details of the binding epitope of any one of the class 5 ETEC fimbriae other than data presented here and elsewhere that indicate localization to the amino-terminal half of the minor subunit (44). One may expect this constraint to impose the greatest penalties for maladaptive mutations in the amino acid residues either directly or indirectly involved in receptor binding, but we found that the pattern of conservation involves much of the entire minor subunit sequence. This appears to be explained by horizontal gene transfer events within the two subclasses studied, with a crossover between the major subunit and usher genes, the result of which is the sharing of more recent progenitors of the adhesin gene between fimbrial operons than for the corresponding major subunits. Thus, our working hypothesis is that the near identity of sequences of the 5b subclass is a consequence of the recent spread and recombination of an adaptive variant of the minor adhesive subunit. The spread of this adaptive variant resulted in a selective sweep and loss of existing variation in this part of the gene cluster.
The findings of greatest practical importance derive from adherence inhibition studies with antibodies against native fimbriae and specific fimbrial component protein domains. A few issues deserve mention that bear on interpretation of the relative effects of anti-fimbrial and anti-minor subunit antibodies. First, we have simply presented the data on the inhibitory activity of Fab antibody fractions, eliminating the impact of nonspecific effects that may be attributable to the bulkiness or bivalence of intact IgG molecules. In most experiments, however, the counterpart IgG fraction of each antiserum preparation was run side-by-side, with the results of which consistently indicating the superiority of anti-minor subunit antibodies with respect to adherence inhibition (data not shown). Second, the anti-subunit antibodies were generated against MBP fusions with each domain of interest. Since these fusions tended to form high molecular aggregates (data not shown) and the carrier itself is highly immunogenic, the antibody response to each passenger protein may have been negatively affected. This would, however, have tended to bias our results to the null in terms of inhibitory effects of antibodies to the passenger subunit components. More specifically, the relative enzyme-linked immunosorbent assay titers in each preparation fit best with the interpretation that anti-CfaE23-211 antibodies inhibit in vitro adhesion. Anti-CFA/I (largely composed of polymeric CfaB) titers were 50-fold higher in the CFA/I antiserum than in the MBP-CfaE23-211 antiserum, whereas anti-CfaE titers were 10-fold higher in the MBP-CfaE23-211 antiserum than in the CFA/I-specific antiserum (data not shown).
The results of in vitro adherence inhibition experiments indirectly support a role for the minor subunits of CFA/I and CS17, more specifically the amino-terminal halves of these proteins, as the actual adhesins. The consistency of findings between HAI and Caco-2 cell adherence inhibition adds to the credibility of this conclusion. The demonstration that antibodies against CfaE23-211 and CsbD19-214 also inhibit the adherence of bacteria expressing heterologous fimbriae of the same subclass provides a functional correlate of their evolutionary linkage. CsbD19-214 antibodies showed greater heterologous inhibitory effects within subclass 5b than do CfaE23-211 antibodies within subclass 5a, reflecting the relative degrees of conservation of the subclass member minor subunits. The fimbrial subclass 5b minor subunits, in fact, share no less than 97% amino acid identity between any two members, compared with ≥80% for subclass 5a minor subunits. On the other hand, it seems somewhat incongruous that there is less variability of in vitro adherence phenotypes of subclass 5a and 5b fimbriae. CFA/I, CS4, and CS14 (subclass 5a) all have the same in vitro adherence phenotypes, whereas only two (CS19 and PCFO71) of four subclass 5b fimbriae hemagglutinate chicken erythrocytes and only one (CS19) exhibits appreciable adherence to Caco-2 cells. This suggests that the small number of variant residues of the subclass 5b minor subunits includes one or more localized to the binding domain that account for these minor phenotypic differences.
Besides providing an evolutionary framework for understanding the conserved functional characteristics of class 5 ETEC fimbriae, our results suggest a refinement of current strategies for the development of an ETEC vaccine. There are several approaches being pursued by various groups, including a whole-cell killed ETEC vaccine (48), live vaccines consisting of attenuated ETEC or Shigella that express ETEC antigens (3, 56), and purified subunit vaccines (22). Common to each of these strategies is the incorporation of a mixture of intact fimbriae as primary antigenic components. Our data suggest that immunization with the adhesive subunits of class 5 fimbriae and perhaps other ETEC colonization factors may provide superior antiadhesive immunity. An additional advantage suggested by the functional cross-reactivity of minor subunit antibodies is that fewer antigens may be required to elicit immunity to ETEC expressing a wider array of related fimbriae. The large number of antigenically distinct adhesive fimbriae implicated in the pathogenesis of ETEC diarrhea warrants further investigation of such an approach.
Acknowledgments
This work was supported by the Navy Medical In-House Laboratory Independent Research (ILIR) Program (award no. A0120) from the Office of Naval Research and the Henry M. Jackson Foundation for the Advancement of Military Medicine.
The opinions expressed in this paper are those of the authors and do not reflect the official policy of the Department of the Navy, Department of Defense, or the U.S. Government.
We thank Neil Agravante and Norma Diaz for excellent technical assistance and David Tribble and Eric Hall for helpful review and discussions of the manuscript.
Editor: J. D. Clements
REFERENCES
- 1.Altboum, Z., M. M. Levine, J. E. Galen, and E. M. Barry. 2003. Genetic characterization and immunogenicity of coli surface antigen 4 from enterotoxigenic Escherichia coli when it is expressed in a Shigella live-vector strain. Infect. Immun. 71:1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, E. M., Z. Altboum, G. Losonsky, and M. M. Levine. 2003. Immune responses elicited against multiple enterotoxigenic Escherichia coli fimbriae and mutant LT expressed in attenuated Shigella vaccine strains. Vaccine 17:333-340. [DOI] [PubMed] [Google Scholar]
- 4.Black, R. E. 1990. Epidemiology of travelers' diarrhea and relative importance of various pathogens. Rev. Infect. Dis. 12(Suppl. 1):S73-S79. [DOI] [PubMed] [Google Scholar]
- 5.Buhler, T., H. Hoschutzky, and K. Jann. 1991. Analysis of colonization factor antigen I, an adhesin of enterotoxigenic Escherichia coli O78:H11: fimbrial morphology and location of the receptor-binding site. Infect. Immun. 59:3876-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cravioto, A., S. M. Scotland, and B. Rowe. 1982. Hemagglutination activity and colonization factor antigens I and II in enterotoxigenic and non-enterotoxigenic Escherichia coli isolated from humans. Infect. Immun. 36:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darfeuille-Michaud, A., D. Aubel, G. Chauviere, C. Rich, M. Bourges, A. Servin, and B. Joly. 1990. Adhesion of enterotoxigenic Escherichia coli to the human colon carcinoma cell line Caco-2 in culture. Infect. Immun. 58:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, D. G., D. J. Evans, Jr., W. S. Tjoa, and H. L. DuPont. 1978. Detection and characterization of colonization factor of enterotoxigenic Escherichia coli isolated from adults with diarrhea. Infect. Immun. 19:727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D. J., Jr., and D. G. Evans. 1973. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect. Immun. 8:322-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froehlich, B. J., A. Karakashian, L. R. Melsen, J. C. Wakefield, and J. R. Scott. 1994. CooC and CooD are required for assembly of CS1 pili. Mol. Microbiol. 12:387-401. [DOI] [PubMed] [Google Scholar]
- 12.Froehlich, B. J., A. Karakashian, H. Sakellaris, and J. R. Scott. 1995. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect. Immun. 63:4849-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaastra, W., H. Sommerfelt, L. van Dijk, J. G. Kusters, A. M. Svennerholm, and H. M. Grewal. 2002. Antigenic variation within the subunit protein of members of the colonization factor antigen I group of fimbrial proteins in human enterotoxigenic Escherichia coli. Int J. Med. Microbiol. 292:43-50. [DOI] [PubMed] [Google Scholar]
- 14.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 15.Grewal, H. M., H. Valvatne, M. K. Bhan, L. van Dijk, W. Gaastra, and H. Sommerfelt. 1997. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect. Immun. 65:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, R. H., D. R. Maneval, Jr., J. H. Collins, J. L. Theibert, and M. M. Levine. 1989. Purification and analysis of colonization factor antigen I, coli surface antigen 1, and coli surface antigen 3 fimbriae from enterotoxigenic Escherichia coli. J. Bacteriol. 171:6372-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess, S., F. J. Cassels, and L. K. Pannell. 2002. Identification and characterization of hydrophobic Escherichia coli virulence proteins by liquid chromatography-electrospray ionization mass spectrometry. Anal. Biochem. 302:123-130. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, A. L. 1999. Adaptive evolution of genes and genomes. Oxford University Press, New York, N.Y.
- 19.Huilan, S., L. G. Zhen, M. M. Mathan, M. M. Mathew, J. Olarte, R. Espejo, U. Khin Maung, M. A. Ghafoor, M. A. Khan, Z. Sami, et al. 1991. Etiology of acute diarrhoea among children in developing countries: a multicentre study in five countries. Bull. W. H. O. 69:549-555. [PMC free article] [PubMed] [Google Scholar]
- 20.Jordi, B. J., A. H. van Vliet, G. A. Willshaw, B. A. van der Zeijst, and W. Gaastra. 1991. Analysis of the first two genes of the CS1 fimbrial operon in human enterotoxigenic Escherichia coli of serotype 0139:H28. FEMS Microbiol. Lett. 64:265-270. [DOI] [PubMed] [Google Scholar]
- 21.Jordi, B. J. A. M., G. A. Willshaw, A. M. van der Zeijst, and W. Gaastra. 1992. The complete nucleotide sequence of region 1 of the CFA/I fimbrial operon of human enterotoxigenic Escherichia coli. DNA Sequence 2:257-263. [DOI] [PubMed] [Google Scholar]
- 22.Katz, D. E., A. J. DeLorimier, M. K. Wolf, E. R. Hall, F. J. Cassels, J. E. van Hamont, R. L. Newcomer, M. A. Davachi, D. N. Taylor, and C. E. McQueen. 2003. Oral immunization of adult volunteers with microencapsulated enterotoxigenic Escherichia coli (ETEC) CS6 antigen. Vaccine 21:341-346. [DOI] [PubMed] [Google Scholar]
- 23.Khalil, S. B., F. J. Cassels, H. I. Shaheen, L. K. Pannell, N. El-Ghorab, K. Kamal, M. Mansour, S. J. Savarino, and L. F. Peruski, Jr. 1999. Characterization of an enterotoxigenic Escherichia coli strain from Africa expressing a putative colonization factor. Infect. Immun. 67:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil, S. B., F. J. Cassels, H. I. Shaheen, L. K. Pannell, K. A. Kamal, B. T. Pittner, M. Mansour, R. Frenck, S. J. Savarino, and L. F. Peruski. 2000. PCFO71: a putative colonization factor identified on an enterotoxigenic Escherichia coli strain isolated from an Egyptian child with diarrhea, abstr. B-79. In Abstr. 100th Gen. Meet. Am. Soc. Microbiol., Los Angeles, Calif., 21 to 25 May 2000. ASM Press, Washington, D.C.
- 25.Kumar, S., K. Tamura, I. Jakobsen, and M. Nei. 2000. MEGA 2: molecular evolutionary genetics analysis program, version 2.0. Pennsylvania State University, University Park.
- 26.Lopez-Vidal, Y., P. Klemm, and A. M. Svennerholm. 1988. Monoclonal antibodies against different epitopes on colonization factor antigen I of enterotoxin-producing Escherichia coli. J. Clin. Microbiol. 26:1967-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Vidal, Y., and A. M. Svennerholm. 1990. Monoclonal antibodies against the different subcomponents of colonization factor antigen II of enterotoxigenic Escherichia coli. J. Clin. Microbiol. 28:1906-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low, D., B. Braaten, and M. Van der Woude. 1996. Fimbriae, p. 146-157. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 29.Mahenthiralingham, E., J. Govan, C. A. Hart, and P. Vandamme. 2003. Burkholderia cepacia genome sequence. http://www.sanger.ac.uk/Projects/B_cepacia/. Sanger Institute, London, United Kingdom.
- 30.McDonald, J. H., and M. Kreitman. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652-654. [DOI] [PubMed] [Google Scholar]
- 31.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 35.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Casal, J., J. S. Swartley, and J. R. Scott. 1990. Gene encoding the major subunit of CS1 pili of human enterotoxigenic Escherichia coli. Infect. Immun. 58:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramer, S. W., G. K. Schoolnik, C. Y. Wu, J. Hwang, S. A. Schmidt, and D. Bieber. 2002. The type IV pilus assembly complex: biogenic interactions among the bundle-forming pilus proteins of enteropathogenic Escherichia coli. J. Bacteriol. 184:3457-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, M. R., R. Abu-Elyazeed, S. J. Savarino, A. B. Naficy, T. F. Wierzba, I. Abdel-Messih, H. Shaheen, R. W. Frenck, A. M. Svennerholm, and J. D. Clemens. 2003. High disease burden due to enterotoxigenic Escherichia coli diarrhea in early life among rural Egyptian children. J. Clin. Microbiol. 41:4862-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 40.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 41.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sajjan, U. S., F. A. Sylvester, and J. F. Forstner. 2000. Cable-piliated Burkholderia cepacia binds to cytokeratin 13 of epithelial cells. Infect. Immun. 68:1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sajjan, U. S., H. Xie, M. D. Lefebre, M. A. Valvano, and J. F. Forstner. 2003. Identification and molecular analysis of cable pilus biosynthesis genes in Burkholderia cepacia. Microbiology 149:961-971. [DOI] [PubMed] [Google Scholar]
- 44.Sakellaris, H., G. P. Munson, and J. R. Scott. 1999. A conserved residue in the tip proteins of CS1 and CFA/I pili of enterotoxigenic Escherichia coli that is essential for adherence. Proc. Natl. Acad. Sci. USA 96:12828-12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakellaris, H., V. R. Penumalli, and J. R. Scott. 1999. The level of expression of the minor pilin subunit, CooD, determines the number of CS1 pili assembled on the cell surface of Escherichia coli. J. Bacteriol. 181:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakellaris, H., and J. R. Scott. 1998. New tools in an old trade: CS1 pilus morphogenesis. Mol. Microbiol. 30:681-687. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Savarino, S. J., R. Abu-Elyazeed, M. R. Rao, R. W. Frenck, I. Abdel-Messih, E. R. Hall, S. Putnam, H. El-Mohamady, T. Wierzba, B. Pittner, K. Kamal, P. Moyer, B. Z. Mersg, A. M. Svennerholm, Y. J. Lee, and J. D. Clemens. 2003. 6th Annu. Conf. Vaccine Res., abstr. 02-A-43. National Foundation of Infectious Diseases, Bethesda, Md.
- 49.Scott, J. R., J. C. Wakefield, P. W. Russell, P. E. Orndorff, and B. J. Froehlich. 1992. CooB is required for assembly but not transport of CS1 pilin. Mol. Microbiol. 6:293-300. [DOI] [PubMed] [Google Scholar]
- 50.Smith, J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 51.Smyth, C. J. 1982. Two mannose-resistant haemagglutinins on enterotoxigenic E. coli of serotype O6:K15:H16 or H-isolated from travellers' and infantile diarrhoea. J. Gen. Microbiol. 128:2081-2096. [DOI] [PubMed] [Google Scholar]
- 52.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0 beta. Sinauer Associates, Inc., Sunderland, Mass.
- 54.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Baumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner, A. K., T. D. Terry, D. A. Sack, P. Londono-Arcila, and M. J. Darsley. 2001. Construction and characterization of genetically defined aro omp mutants of enterotoxigenic Escherichia coli and preliminary studies of safety and immunogenicity in humans. Infect. Immun. 69:4969-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viboud, G. I., N. Binsztein, and A. M. Svennerholm. 1993. Characterization of monoclonal antibodies against putative colonization antigens of enterotoxigenic Escherichia coli and their use in an epidemiological study. J. Clin. Microbiol. 31:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viboud, G. I., M. M. McConnell, A. Helander, and A. M. Svennerholm. 1996. Binding of enterotoxigenic Escherichia coli expressing different colonization factors to tissue-cultured Caco-2 cells and to isolated human enterocytes. Microb. Pathog. 21:139-147. [DOI] [PubMed] [Google Scholar]