Abstract
Alternative splicing and adenosine to inosine (A to I) RNA-editing are major factors leading to co- and post-transcriptional modification of genetic information. Both, A to I editing and splicing occur in the nucleus. As editing sites are frequently defined by exon–intron basepairing, mRNA splicing efficiency should affect editing levels. Moreover, splicing rates affect nuclear retention and will therefore also influence the exposure of pre-mRNAs to the editing-competent nuclear environment. Here, we systematically test the influence of splice rates on RNA-editing using reporter genes but also endogenous substrates. We demonstrate for the first time that the extent of editing is controlled by splicing kinetics when editing is guided by intronic elements. In contrast, editing sites that are exclusively defined by exonic structures are almost unaffected by the splicing efficiency of nearby introns. In addition, we show that editing levels in pre- and mature mRNAs do not match. This phenomenon can in part be explained by the editing state of an RNA influencing its splicing rate but also by the binding of the editing enzyme ADAR that interferes with splicing.
INTRODUCTION
Alternative splicing and adenosine (A) to inosine (I) RNA-editing are the two major co-transcriptional processes that greatly expand the diversity of mammalian transcriptomes. Both processes are coordinated with transcription and occur in the nucleus (1–3). Virtually every mammalian protein-coding transcript is subject to RNA-editing and about 95% of multiexon genes undergo alternative splicing (4,5). Hence, both processes show a large overlap in the transcriptome. In particular, both mechanisms expand receptor diversity in the central nervous system (CNS) (6,7). Alternative splicing as well as A to I editing have been implicated in a series of neurological disorders. This includes depression or amyotrophic lateral sclerosis in case of A to I editing defects or spinal muscular atrophy, Duchenne muscular dystrophy, schizophrenia and the Rett syndrome for splicing deficiencies (8–11). Together, this demonstrates the importance of a tight control of alternative splicing and RNA editing.
A to I editing is the most abundant form of RNA-editing in mammals. The reaction is mediated by adenosine deaminases acting on RNA (ADARs). In mammals two catalytically active ADAR enzymes, ADAR1 and ADAR2 (also known as ADARB1) and one inactive enzyme, ADAR3 have been identified in the soma (12). ADARs bind double-stranded and structured RNAs and convert adenosines to inosines by hydrolytic deamination. Inosines are interpreted as guanosines by most cellular machines. Thus, different processes can be affected, ranging from recoding of codons in mRNAs, over the masking of endogenous RNAs to the innate immune system, to changes in mRNA splicing (12–16).The consequences of A to I editing on mRNA splicing have been well documented for the transcript encoding glutamate receptor subunit 2 (Gria2). Editing of a single adenosine in exon 11 of the Gria2 transcript, encoding the so-called Q/R site, is essential for mammalian life (17). The editing competent RNA-stem is formed by basepairing between exon 11 and intron 11 (18). Therefore, the pre-mRNA needs to be edited before removal of intron 11. Interestingly, lack of editing prevents splicing of intron 11 but not of other introns (17). Most likely, splicing is regulated by editing of two intronic hotspots (19,20). Thus, editing at the intronic sites may act as a ‘safe-guard’ to ensure that only edited transcripts are spliced, exported and translated.
Most editing sites in the human transcriptome are found in Alu repeats that are typically located in non-coding parts of genes, like introns or UTRs (4,21). However, a small fraction of editing sites is located in exons and can lead to non-synonymous codon changes or alter splice-sites (12). Interestingly, editing levels are highly variable between different substrate sites, different tissues and under different physiological and developmental conditions (20–24). These different editing levels can not solely be explained by varying ADAR protein levels as these have been shown to be relatively constant (24). Instead, additional factors such as regulatory- and competing proteins, RNA helicases or the local RNA-environment may contribute to the regulation of editing levels (25–28).
An important factor controlling the extent of editing may be the rate and efficiency of splicing. Editing sites are defined via base pairing with editing complementary sites (ECSs). For many exonic editing sites that lead to protein recoding, the ECS is located in an adjacent intron (29–31). Therefore, at these sites editing can only occur prior to intron removal. However, a fraction of protein-recoding editing sites rely on an ECS that is located within the same exon as the editing site (29,32). Also in these latter cases, editing might be affected by pre-mRNA splicing as splicing efficiency is one of the most important factors determining nuclear retention time. In sum, it seems reasonable to assume that splicing efficiency may have a strong impact on A to I editing levels of sites residing in protein-coding exons.
To test this hypothesis we chose a set of exonic editing sites that depend on ECSs that are either located within an adjacent intron or within the same exon. To do so, the editing sites located in the transcripts encoding cytoplasmic FMR1-interacting protein 2 (Cyfip2) and the R/G editing site in glutamate receptor subunit 2 (Gria2) were chosen as examples for substrates with an intron-located ECSs (29,31). Editing sites in the transcripts of the insulin-like growth factor-binding protein 7 (Igfbp7) and the gamma-aminobutyric acid receptor subunit alpha-3 (Gabra3) on the other hand, served as references for substrates with ECSs located in the same exon as the editing site (29,32). The impact of splicing efficiency on editing levels was tested in a minigene approach by fusing the exons harboring the editing site including the downstream exonic or intronic ECS to heterologous intron/exon sequences with progressively weaker branch points. To back up data generated by this mutational approach we also used a splicing inhibitor to test the effect of splicing rates on editing of various substrates in primary cells.
MATERIALS AND METHODS
Cloning of minigene constructs
All minigene constructs are fusion constructs of an exonic sequence harboring one or two editing sites and an ECS fused to the last 61 basepairs of intron 2 plus exon 2 of the adenovirus major late transcript (AdML) (33). In the +/− and −/− versions the polypyrimidine tract sequence was mutated to weaken splicing efficiency. All sequences were introduced into pcDNA3.1(−) using EcoRI and BamHI for the test exon–intron part and BamHI plus KpnI for the AdML intron–exon part. The following exon–intron parts were cloned: Gabra3-IS: Exon 9 plus 161 basepairs of intronic sequence. Gabra3: Exon 9 plus 90 basepairs of intronic sequence. Gria2: Exon 13 plus 224 basepairs of intronic sequence. Igfbp7: Exon 1 plus 150 basepairs on intronic sequence. The polypyrimidine tract sequences have been as follows (the sequence including the terminal AG of the 3′ splice site is given; mutations are underlined):
| (+/+) wild type: | 5′-TCCTGTCCCTTTTTTTTCCACAG-3′ |
| (+/−) for Gabra3, Gabra3-IS, Cyfip2 and Igfbp7: | 5′-TCCTGTCCCTTGGGGTTCCACAG-3′ |
| (−/−) for Gabra3, Gabra3-IS, Cyfip2 and Igfbp7: | 5′-TCCTGTCCCTTGGGGAACCACAG-3′ |
| (+/−) for Gria2: | 5′-TCCTGTCCCTTGTTGTTCCACAG-3′ |
| (−/−) for Gria2: | 5′-TCCTGTCCCTTGGGGTTCCACAG-3′ |
In case of Cyfip2, nucleotide +128 (relative to the 5′ splice site) was changed from a G to an A using a mutational PCR in order to remove a cryptic splice site.
Transfection, RNA isolation, RT-PCR and Sanger sequencing
Twenty-four hours prior to transfection HEK293 cells were seeded in 35 mm dishes (2 × 105 cells). The respective minigene plasmid was co-transfected with Flag-rADAR2 using jetPEI (Polyplus, France) following the manufacturer's instructions. To generate biological replicates, at least three independent transfections were made. For all constructs (except Gabra3 constructs) 2 μg of minigene plasmid plus 2 μg Flag-rADAR2 were transfected. For Gabra3 constructs we observed very high expression levels as compared to other constructs when we transfected 2 μg of plasmid and only moderate editing levels as compared to an earlier report (34). Thus, we reduced the amount of transfected minigene plasmid to 0.5 μg for Gabra3, to compensate for the elevated expression, and raised the amount of transfected Flag-rADAR2 to 3.5 μg. Forty-eight hours after transfection, RNA was isolated using TriFast (Peqlab, Germany) according to the manufacturer's guidelines. To remove traces of genomic DNA and plasmid DNA, 8 μg of isolated RNA were incubated with 3 μl of FastDigest BamHI (Thermo Scientific) at 37°C for 45 min (targeting the minigene constructs) followed by a digestion with 27 units of DNase I (Thermo Scientific) for 45 minutes at 37°C. Subsequently, RNA was phenol extracted. 1.7 μg of total RNA were reverse transcribed using random hexamer priming and RevertAid H Minus reverse transcriptase (Thermo Scientific) following manufacturer's instructions. A control without reverse transcriptase was included for every sample. To amplify products for Sanger sequencing, Taq-polymerase (Thermo Scientific) was used (except for Igfbp7 constructs) with the following standard PCR protocol: 3 min at 95°C followed by 33 cycles 95°C (30 s), 58°C (30 s), 72°C (30 s). For Igfbp7 constructs Phusion polymerase (Thermo Scientific) and GC buffer was used: 30 s 98°C, followed by 30 cycles of 98°C (10 s), 60°C (30 s), 72°C (20 s) and a final elongation of 3 min at 72°C. A semi-nested PCR was done for Igfbp7 pre-mRNA and mRNA, Gabra3 pre-mRNA, Cyfip2 pre-mRNA and mRNA and Gria2 pre-mRNA. The PCR products representing pre-mRNAs and mRNAs were gel purified. Sanger sequencing was done using the eluted PCR products and the reverse primer of the first or the semi-nested PCR. Only in case of Gabra3, Gabra3-IS, CyFip2 and Gria2 PCR products the respective forward primer was used for sequencing. The percentage of editing is defined as the height of the G peak divided by the sum of the A + G peaks (In case of the reverse primer: the height of the T peak divided by the sum of the T + C peaks). In order to estimate splicing efficiency of the minigene constructs PCR cycle numbers were reduced (Gabra3 constructs: 25 or 22 cycles; Igfbp7 and Cyfip2 constructs: 30 cycles; Gria2 constructs: 25 cycles). If not stated otherwise, a unique forward primer specific for the respective construct was combined with the universal reverse primer binding in AdML exon 2. Primer sequences are as follows: Universal AdML exon 2 primer: Rev 5′-GAAAGACCGCGAAGAGTTTG-3′, Gabra3 constructs: Fwd 5′-TGCTTACCATGACCACCTTG-3′, pre-mRNA-nested Rev 5′-CTATAGGCTGCCCACTCCTG-3′, Cyfip2 constructs: Fwd 5′-GCCACTGTGCTGGATATCTG-3′, pre-mRNA-nested Rev 5′-CGGGAGCCGAGAGACATTAC-3′, mRNA-nested Fwd 5′-CAGCTGCAGGTGGTGC-3′, Gria2 constructs: Fwd 5′-CAAAGGAAGCCTTGCGACAC-3′, pre-mRNA-nested Rev 5′-CATCAGGGTAGGTGGGATACT-3′, Igfbp7-constructs: mRNA Fwd 5′-TCTTCCTCCTCTTCGGACAC-3′, mRNA-nested Rev 5′-GGGTAGCGGCTCTTGCAC-3′, pre-mRNA Fwd 5′-GGCTGCTGCCCTATGTGC-3′, pre-mRNA Rev 5′-CCAACTCTTTCCCTCCCATC-3′, pre-mRNA-nested Rev 5′-AGGGTTGGAGAGGGAAGC-3′.
Actinomycin D treatment and nuclear versus cytoplasmic separation
For actinomycin D (AMD) (Sigma Aldrich, MO) treatment, HEK293 cells were co-transfected as described above and treatment with AMD was done 48 h after transfection for 0, 2 and 6 h at a concentration of 5 μg/ml. For nuclear versus cytoplasmic separation, cells were seeded into 10 cm dishes (1 × 106 cells) 24 h prior to transfection. Transfection was done as described above, but using 4 μg of Flag-rADAR2 and 4 μg of the least efficiently spliced version of the Igfbp7 minigenes. Forty-eight hours after transfection nuclear versus cytoplasmic separation was done as described previously (35). Antibodies against histone H3 or GAPDH were used to probe for the nuclear and cytoplasmic fractions, respectively. RNA isolation, RT and PCR was done as described above.
Isolation and culturing of primary cells and splicing inhibition
Bone marrow was isolated as described previously (36). Briefly, bone marrow was isolated from the femurs and tibiae of 8–10 week old mice. Cultures were established in RPMI medium (supplemented with 10% FBS, penicillin streptomycin, gentamycin and L-glutamine). Sixteen hours after establishing the cultures, splicing inhibition was performed using meayamycin at a concentration of 5 nM for 6 h. DMSO was used as a vehicle control. Meayamycin is a potent inhibitor of splicing which interferes with the U2 snRNP associated protein SF3b and is active already at low nanomolar concentrations (37). Primary neuronal cultures were established from mouse embryos at embryonic day 11.5. Each embryo isolate was split into two wells of a 6-well dish coated with poly-D-lysine. Neuronal cells were subsequently cultured in neurobasal A medium (supplemented with B27, penicillin, streptomycin, gentamycin and L-glutamine) for 2 weeks. To prevent outgrowth of dividing cells, Cytosine β-D-arabinofuranoside (Ara-C) (Sigma Aldrich, MO) was added to 1 μM after 48 h. After 96 h the concentration was raised to 5 μM. For splicing inhibition, cells were treated with meayamycin at a concentration of 15 nM or DMSO as control. RNA was isolated using TriFast (PeqLab) and DNaseI treated (Thermo Fisher Scientific) following the manufacturer's instructions. RT and PCR was done as described above. The following primer sequences were used: Cog3 Fwd 5′-TCAGATTGATGGACAACTTTTCTT-3′, Cog3 Rev 5′-AAGGCATTGTTGCTATTCAGC-3′, Cyfip2 Fwd 5′-TGCAGGTGGTACCCCTTT-3′, Cyfip2 Rev 5′-ATCCCGGATCTGAACCATCTG-3′, Exoc8 Fwd 5′-CAAGGGTTTCTCTGTATAGC-3′, Exoc8 Rev 5′-GGTTTCAGCACCCACATTCT-3′, Gabra3 Fwd 5′-TCTCACCATGACCACCTTGA-3′, Gabra3 Rev 5′-GTTGGAGCTGCTGGTGTTTT-3′, Gria2 Fwd 5′-GTGAGGACTACGGCAGAAGG-3′, Gria2 Rev 5′-GTCCAACAGGCCTTGTTCAT-3′, Igfbp7 Fwd 5′-CCCCTCTCCTCTTCCTCCTC-3′, Igfbp7 Rev 5′-GGTAGCGGCTCTTGCACA-3′, Pum2 Fwd 5′-AAACTGGCTTGCAGTCTGGT-3′, Pum2 Rev 5′-TGTCAGGGACATTATAGGGCAG-3′, Sfi1 Fwd 5′-CGTCTCCAGAACTGGTTTCAG-3′, Sfi1 Rev 5′-GAGTCTCTGTGCCAGGAGTTG-3′, Vcp Fwd 5′-CGATGAGCTTGATGCCATT-3′, Vcp Rev 5′-CTCTGCTTTAGGCCATCCAT-3′.
RESULTS
Editing sites guided by intronic ECSs are affected by splicing rates
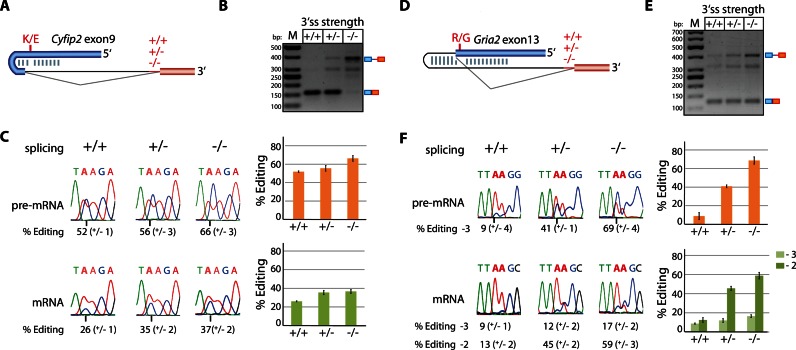
To test whether splicing efficiency controls the rate of A to I editing we chose a set of model substrates. The transcript of Cyfip2 contains one editing site (K/E-site) in exon 9 at position -35 relative to the next downstream 5′ splice site (5′ ss). The ECS is situated in the adjacent intron, approximately 10 to 20 nucleotides downstream of the 5′ ss. To manipulate splicing efficiency, we used a minigene approach. The Cyfip2 exon 9 including parts of the downstream intron (spanning the ECS) was fused to three variants of a heterologous intron–exon sequence that differed exclusively in their polypyrimidine tracts giving rise to gradually reduced splice efficiency (Figure 1A). The minigene constructs were co-transfected with a plasmid expressing rat ADAR2 into HEK293 cells. As expected and confirmed by RT-PCR the polypyrimidine tract mutations led to reduced splicing efficiency (Figure 1B). When editing levels of corresponding pre-mRNAs and mRNAs were determined by Sanger sequencing, strong changes were observed (Figure 1C). With decreasing splicing efficiency, editing increased from 26 to 37% in the mRNA and from 52 to 66% in the pre-mRNA. Interestingly, editing levels are generally higher in pre-mRNAs compared to the corresponding mRNAs. Also, editing levels in the pre-mRNA increase continuously with a reduction in splicing while the corresponding mRNA-editing levels reach a plateau at intermediate splicing levels (Figure 1C). The generally higher editing levels in nuclear pre-mRNAs together with a plateau in mRNA editing suggests that mRNA-maturation steps such as splicing and export may control the level of edited cytoplasmic mRNA.
Figure 1.
Reduced splicing efficiency enhances exonic editing at sites depending on intronic ECSs (A) Model for the K/E editing site in Cyfip2 exon 9 that is defined between an exon and intron (exon in blue, downstream exon in red, intron is depicted as a thin line; +/+, +/−, −/− indicates a strong, intermediate or weak polypyrimidine tract). (B) RT-PCR with exon-specific primers to estimate splicing efficiency of Cyfip2 transcripts with a strong, intermediate or weak polypyrimidine tract (+/+, +/−, −/−). Pre-mRNA and mRNA are indicated at the right side of the panel. M = size standard. bp = basepairs. (C) Sanger sequencing of the RT-PCR-products derived from Cyfip2 pre-mRNA or mRNA. Shown is a representative sequencing chromatogram. The average plus standard deviation of three biological replicates is depicted next to the chromatograms as a bar chart (orange = pre-mRNA, green = mRNA) and in numbers below the chromatograms. The edited adenosine is marked by a vertical line underneath the chromatogram. Panels (D–F): Model, splicing efficiency and sequencing chromatograms for the Gria2 R/G-site. For the editing targets Cyfip2 and Gria2, editing frequency increases when splicing efficiency is impaired. Both editing sites are coordinated with an ECS in the downstream intron. Interestingly, pre-mRNA editing is higher than mRNA editing for both targets.
The Gria2 R/G site was used as another model substrate where editing is guided by intronic elements. The R/G site is located in exon 13 at position -2 relative to the 5′ ss. Again three fusion minigenes were constructed, each encompassing exon 13 of the Gria2 transcript including a part of the adjacent downstream intron 13 (including the ECS) followed by heterologous downstream intron–exon sequences with altered branch points (Figure 1D). As for the Cyfip2 minigenes, reduced splicing efficiencies were observed with weaker polypyrimidine tracts (Figure 1E). Subsequently, editing levels were determined by direct sequencing of PCR products obtained for pre-mRNAs and spliced mRNAs. Strikingly, the editing frequencies at the R/G site (position - 2) gradually increased from 13 over 45% and even 59% with reduced splicing efficiency (Figure 1F). Editing was also detected at position -3 where editing levels similarly increased when splicing efficiency was reduced. Editing in the Gria2 pre-mRNA was not precisely quantifiable at position -2 as the G-peak was fused with the neighboring G-peak in most chromatograms (Figure 1F). Still, while not exactly quantifiable, it was obvious that editing levels also increased in the Gria2 pre-mRNA with reduced splicing efficiency. Pre-mRNA-editing at position -3 dramatically increased upon reduced splicing efficiency from about 10% for the most efficiently spliced reporter construct (+/+) to almost 70% for the most inefficiently spliced reporter (−/−). Again, as seen for the Cyfip2 minigenes higher editing levels were observed in pre-mRNAs compared to mRNAs. Thus, for both substrates in which editing depends on intronic elements a strong correlation between reduced splicing efficiency and increased editing levels can be observed.
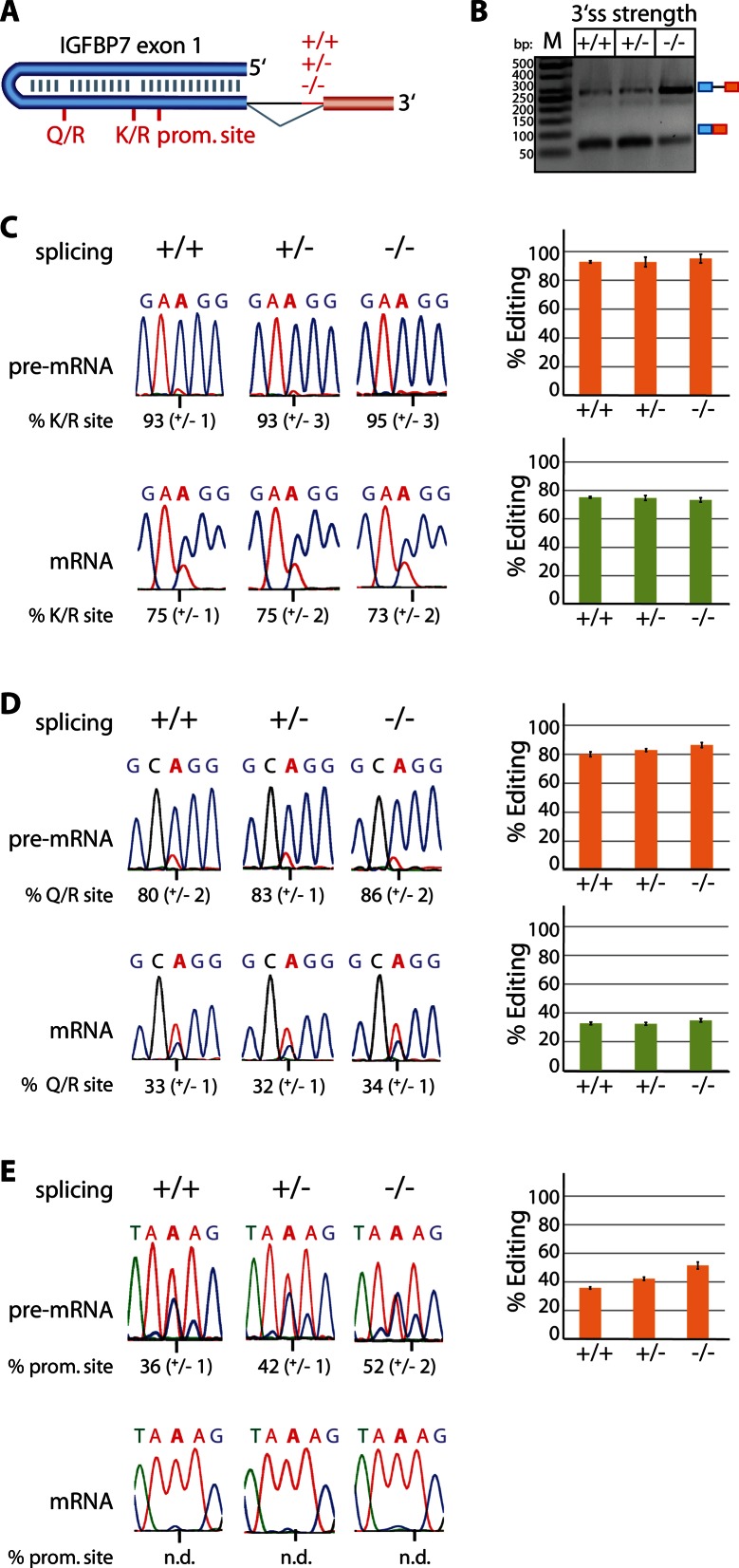
Next, we tested whether splicing can also control editing if the double-stranded RNA (dsRNA) structure required for editing is formed within the exon itself. In this case, splicing efficiency might indirectly control editing by influencing nuclear retention times. To address this question, we chose Igfbp7 as a model substrate. The Igfbp7 transcript contains two editing sites, a Q/R site and a K/R site. Both sites are located in exon 1 and are distant to a splice site (Figure 2A). Again the sequence for exon 1 and a part of the downstream intron 1 was fused to intron–exon sequences with progressively weaker 3′ splice sites. The impact on splicing was confirmed using RT-PCR (Figure 2B). In contrast to constructs where the editing sites were defined by basepairing of exonic and intronic sequences, editing levels were unaffected by splicing efficiency in mRNAs with strictly exon-dependent editing sites. Here, editing levels stayed at about 33 and 74% for the Q/R and K/R site, respectively (Figure 2C and D). Interestingly, editing levels were again higher in pre-mRNAs than in the mature mRNAs. For the Q/R site a moderate increase in editing from 80 to 86% was observed in the pre-mRNA upon reduction of splicing efficiency while only negligible changes were observed at the K/R site (93 to 95%). We also detected promiscuous editing at several sites in the Igfbp7 pre-mRNA that increased with reduced splicing efficiency. One of these sites we quantified and observed an increase in editing from 36 to 52% (compare chromatograms +/+ and −/− in Figure 2E). Most interestingly, no detectable promiscuous editing could be seen in the mature mRNA (Figure 2E), again indicating selective splicing, export or turnover of edited and unedited transcripts. Taken together, the findings seen for the Igfbp7 editing sites suggest that splicing does not control the percentage of editing for the strictly exonically defined Q/R and K/R editing sites but may have an effect on the specificity of editing sites.
Figure 2.
Editing in Igfbp7 mRNA containing an exonic ECS is not affected by splicing while pre-mRNA editing is increased. (A) Model of the editing sites in Igfbp7 exon1. Two canonical editing sites (Q/R and K/R) and a promiscuous editing site (prom. site) are given (the exon is depicted in blue, downstream exon in red, intron is depicted as a thin line; +/+, +/−, −/− indicates a strong, intermediate or weak polypyrimidine tract). (B) RT-PCR with exon-specific primers to estimate splicing efficiency of Igfbp7 transcripts with a strong, intermediate or weak polypyrimidine tract (+/+, +/−, −/−). Pre-mRNA and mRNA are indicated at the right side of the panel. M = size standard. bp = basepairs. (C–E) Sanger sequencing of the RT-PCR-products representing Igfbp7 pre-mRNA or mRNA. Shown is a representative sequencing chromatogram and the average plus standard deviation of three biological replicates is given below in numbers and next to the chromatograms as bar charts (orange = pre-mRNA, green = mRNA). The edited adenosine is marked by a vertical line underneath the chromatogram. (C and D) The editing frequencies for pre-mRNA and mRNAs in Ifgbp7 do not change when splicing efficiency decreases. Note that the editing-competent duplex is formed within the exon itself. Interestingly, the editing levels for pre-mRNAs are higher than for mRNAs. E) A promiscuous editing site was detected in Igfbp7 pre-mRNA, which could not be seen in mRNA at all. Thus, this represents an extreme case where edited adenosines in pre-mRNA are completely unedited in the respective mRNA.
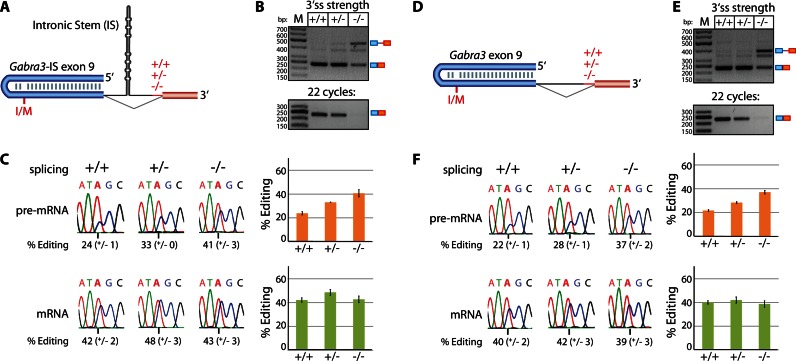
Finally, we choose the Gabra3 transcript as an additional model substrate for a strictly exonically defined editing site. Gabra3 contains the so-called I/M editing site in exon 9. It was also shown that an intronic stem structure in intron 9 can enhance editing at the I/M site (34). Thus, we reasoned that this enhancing effect might also be affected by splicing rates. To test this, two sets of Gabra3 reporter constructs were generated; one encompassing the entire intronic stem structure (Gabra3-IS) and one with the stem structure deleted (Gabra3; Figure 3A and D). Again, editing levels of the pre-mRNA were increasing upon reduced splicing efficiency for both, the Gabra3–IS and the Gabra3 reporter constructs (Figure 3B and E). However, editing levels in the mature mRNA remained constant (Figure 3C and F). Again a particular editing state might be preferentially spliced, exported or stabilized and thus compensate for the differences seen between pre-mRNA and mRNA. The Gabra3–IS and Gabra3 reporter constructs differ only slightly in the extent of editing. Editing levels were only moderately higher for the Gabra3–IS constructs than the Gabra3 constructs, both, at the pre-mRNA and mRNA levels. Thus, at least in the context of our transiently transfected reporter constructs the intronically located stem structure has little effect on editing levels (34).
Figure 3.
I/M site editing in Gabra3 transcripts with or without an intronic stem. (A) The I/M editing site is defined by an ECS in exon 9. (The exon is depicted in blue, downstream exon in red, intron is depicted as a thin line; +/+, +/−, −/− indicates a strong, intermediate or weak polypyrimidine tract). Intron 9 contains a long intronic stem (intronic stem, IS). (B) RT-PCR using exon-specific primers shows a reduction of splice efficiency for Gabra3-IS transcripts with a strong, intermediate or weak polypyrimidine tract (+/+, +/−, −/−). Pre-mRNA and mRNA are indicated at the right side of the panel. M = size standard. bp = basepairs. The PCR has been done also with 22 cycles (lower panel) (C) Sanger sequencing of the RT-PCR-products representing Gabra3-IS pre-mRNA or mRNA. Shown is a representative sequencing chromatogram and the average plus standard deviation of three biological replicates is given below and as a bar chart next to the chromatograms (orange = pre-mRNA, green = mRNA). The edited adenosine is marked by a vertical line underneath the chromatogram. While no differences in editing levels can be seen in the mature mRNAs, a reduction of splicing efficiency leads to a clear increase in editing in the pre-mRNA. (D) A model of the Gabra3 editing site in exon 9 but lacking the intronic stem. (E and F) Compare with panels (B and C) for description. The lack of the intronic stem only modestly affects editing levels and exhibits the same response to reduced splicing efficiency as observed for Gabra3-IS.
Editing and ADAR2-binding of splice-proximal regions affect splicing rates
Our data have shown a surprising difference between editing levels in pre-mRNA and mRNA. This discrepancy could be the result of differential nuclear export, stability or selective processing of edited versus unedited transcripts.
We therefore separated nuclei from cytoplasm of transfected cells and determined editing levels in spliced transcripts of the least efficiently spliced Igfbp7 reporter in both fractions (Supplementary Figure S1). This Igfbp7 reporter had shown the strongest difference in editing levels of pre-mRNA and mRNA. However, no difference in editing levels was observed between spliced nuclear and cytoplasmic transcripts, indicating that selective nuclear export is probably not responsible for the different editing levels observed in pre-mRNAs and mRNAs. Next, we tested whether the stability of mRNAs would be affected by their editing levels. We therefore inhibited transcription of cells transfected with reporter constructs using actinomycin D (AMD). Subsequently cells were harvested and RNAs extracted after 0, 2 and 6 h of AMD treatment. Editing levels were determined in those RNAs. However, no differences in editing levels were observed over time, indicating that edited and unedited mRNAs had the same stability (data not shown).
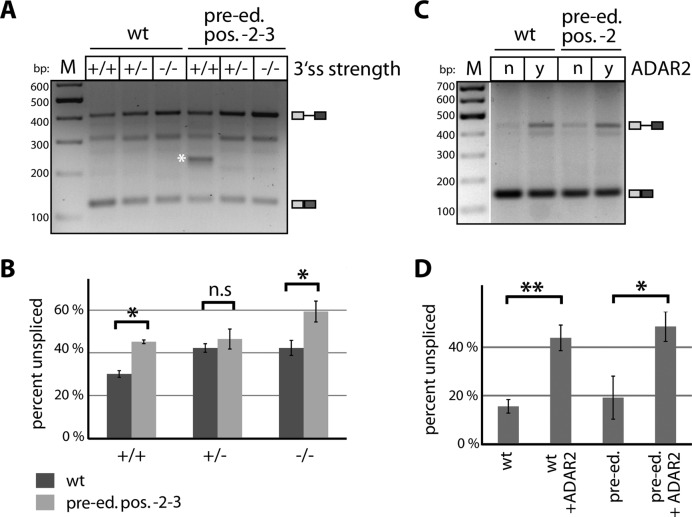
We therefore tested whether edited and unedited pre-mRNAs might be selectively spliced. To do so we mutated the Gria2 minigenes that contain a canonical editing site at position -2 and a facultative editing site at -3, relative to the 5′ splice site. To mimic editing, the A at position -2 was replaced by a ‘pre-edited’ G. Similarly, we made constructs with guanosines at positions -2 and -3. The constructs termed ‘pre-edited -2’ and ‘pre-edited -2-3′ were again co-transfected with ADAR2, with the ‘wildtype’ constructs serving as controls. The ratios of pre-mRNA to mRNA were determined by PCR using a reduced number of cycles. A significant increase of pre-mRNA was observed for the pre-edited -2 versions for one of the Gria2 minigenes (+/−; Supplementary Figure S2). An even stronger accumulation of pre-mRNA was observed for the pre-edited -2-3 constructs in context with the strongest and weakest branch points (Figure 4A and B). This demonstrates that guanosines and therefore most likely also inosines introduced by editing can interfere with splicing of the Gria2 minigene.
Figure 4.
Editing and ADAR2-binding reduces splicing efficiency at the Gria2 R/G site. (A) RT-PCR shows a gradual reduction of splicing in the presence of a strong (+/+), intermediate (+/−) or weak (−/−) polypyrimidine tract in wildtype (wt) or pre-edited (pre-ed) versions of a Gria2 exon 13 minigene. Pre-mRNA and mRNA are indicated at the right side of the panel. M = size standard. bp = basepairs. A white asterisk marks an aberrant splicing product. (B) The relative splicing efficiency—measured from the gel in panel A—is given as % unspliced product. Pre-editing reduces splicing efficiency. Quantification was done from three independent biological replicates. (* P-value < 0.05; n.s. not significant). Dark gray = wt, light gray = pre-edited at position -2-3. (C) RT-PCR of Gria2 reporter transcripts in the presence (y) or absence (n) of ADAR2 indicate a strong reduction in splicing in the presence of ADAR2. Pre-mRNA and mRNA are indicated at the right side of the panel. M = size standard. bp = basepairs. (D) The splicing efficiency—deduced from the gel in panel (C)—in percent unspliced transcript is given. The presence of ADAR2 reduces splicing efficiency. The quantification was done from three independent biological replicates (n.s. not significant, * P-value < 0.05; ** P-value < 0.01).
Surprisingly, when we transfected the Gria2 minigene (+/+) without the addition of the ADAR2 plasmid we observed a strong reduction in pre-mRNA (Figure 4C and D). Conversely, an increase in pre-mRNA accumulation was observed when the pre-edited -2 version of Gria2 was co-transfected with ADAR2. It thus appears that ADAR2 binding alone impairs splicing, even when editing cannot take place. Possibly, binding of ADAR2 interferes with spliceosome assembly at the stage of U1 snRNP binding. Taken together, our data show that editing at positions -2 and -3 as well as ADAR2 binding reduce splicing efficiency of Gria2 with ADAR2 binding having the more pronounced effect. Jointly, both effects can explain the observed differences in pre-mRNA and mRNA editing levels since unedited pre-mRNAs not bound by ADAR2 will be preferentially spliced.
Inhibition of splicing increases editing in endogenous exon–intron substrates
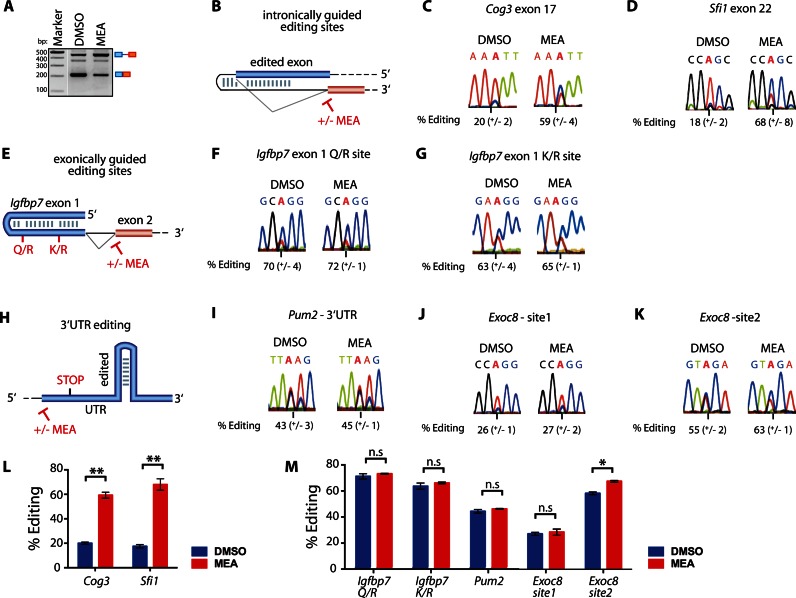
The minigene-based experiments have demonstrated that editing levels increase with a decrease in splicing efficiency when editing sites are defined by intron–exon basepairing. Strictly exon-dependent editing sites are much less affected. To test if these findings can be reproduced in a chromosomal environment, we employed the splicing inhibitor meayamycin that interferes with early spliceosomal assembly (37). For this we needed to identify cells that express and edit endogenous substrates at constant and reproducible levels. Screening of several tissues identified bone marrow cells isolated from 8 to 10 weeks old mice and primary neurons isolated from mouse embryos at embryonic day 11.5 as a suitable system. Meayamycin at a concentration of 5 nM was sufficient to efficiently inhibit splicing in bone marrow (Figure 5A). A target where editing is guided by an intronic element is Cog3 containing an I/V recoding editing site in exon 17 at position -28 relative to the 5′ ss (38) (Figure 5B). Meayamycin treatment led to a 3-fold increase in editing (Figure 5C). Sfi1 contains several editing sites (39). A synonymous editing site is located in exon 22, which is again guided by intronic elements. Editing levels increased 4-fold upon meayamycin treatment (Figure 5D). Next, we tested editing in endogenous Igfbp7 where the editing sites contain an ECS in the same exon (Figure 5E). As in the reporter assays editing efficiencies did not respond to splicing inhibition neither for the Q/R nor the K/R site (Figure 5F and G). In order to expand the panel of tested substrates even more, we picked two additional substrates where the tested editing site is located in the 3′ UTR (Figure 5H). First, we monitored editing in the Pum2 transcript. Editing levels remained unchanged upon meayamycin treatment (Figure 5I). For the transcript coding for the protein Exocyst Complex Component 8 (EXOC8) we observed a small increase in editing of one out of two tested sites. The Exoc8 transcript contains only one exon and should not be directly affected by splicing inhibition. As expected, editing does not change for editing site 1 (Figure 5J). Surprisingly, however, a modest increase in editing was observed at site 2 (Figure 5K), which we cannot explain at present. Still, taken together these experiments show that inhibition of splicing strongly affects editing of endogenous substrates when the exonic editing event is guided by intronic elements (Figure 5L). As expected, editing sites that do not depend on intronic structures are almost unaffected by inhibition of splicing (Figure 5M).
Figure 5.
Splicing inhibition increases editing rates at intron-dependent sites in vivo. (A) RT-PCR shows a reduction of splicing efficiency of Sfi1 exon 22 upon addition of meayamycin. Total RNA was prepared from bone marrow cells MOCK-treated (DMSO) or treated with the splicing inhibitor meayamycin (MEA). Pre-mRNA and mRNA are indicated at the right side of the panel. M = size standard. bp = basepairs. (B) Scheme of an exonic editing site defined by an ECS in the intron. +/− MEA indicates splicing inhibition or control. (C and D) Sanger sequencing of the Cog3 or Sfi1 mRNAs RT-PCR-products shows a strong increase in editing upon addition of the splicing inhibitor meayamycin (MEA). (E) Scheme of a generic exonic editing site defined by an ECS in the exon. (F and G) Sanger sequencing of the mRNA of the Igfbp7 Q/R or K/R editing site shows no change in editing patterns upon splicing inhibition. The average and standard error of the mean of three biological replicates is given below each chromatogram. (H) Scheme of a generic 3′ UTR editing site defined by an ECS in the exon. (I, J andK) Sanger sequencing of the mRNA of an editing site in the 3′ UTR of Pum2 or two sites in the 3′ UTR of Exoc8 shows significant effects on editing upon inhibition of splicing only for Exoc8-site2. (L) Graphs of editing frequencies of all intronically guided transcripts or (M) exonically defined editing sites in Figure 5. The significance level is given on top (n.s. not significant, * P < 0.05, ** P < 0.01). Blue = DMSO, red = meayamycin.
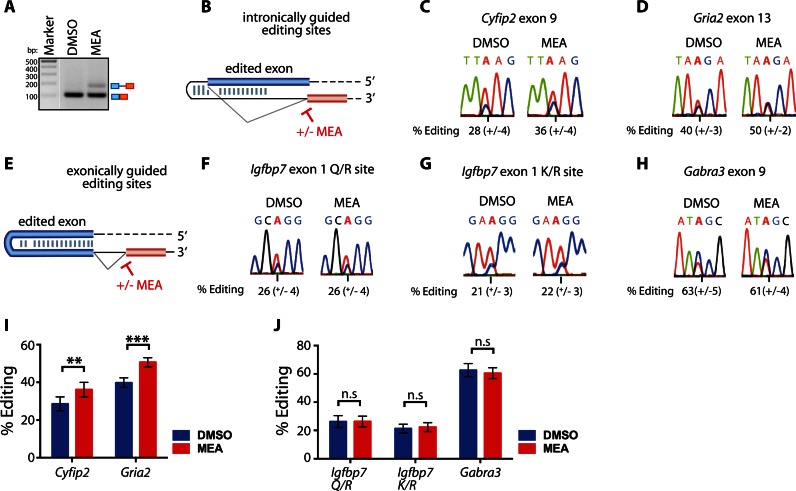
Finally, we used primary neurons to follow editing of the endogenously expressed transcripts Cyfip2, Gria2, Gabra3 and Igfbp7 (Figure 6). Primary neuronal cultures were established from embryos at embryonic day 11.5. In contrast to bone marrow meayamycin was used at a concentration of 15 nM to affect splicing (Figure 6A). We observed similar trends as we have seen for the minigene constructs and for the bone marrow setting. Editing for intronically guided editing sites increases (Figure 6B–D and I), whereas editing for Gabra3 and Igfbp7—both containing editing sites coordinated within the exon—remain the same (Figure 6 E–H and J).
Figure 6.
Splicing inhibition in primary neurons leads to an increase in editing rates of intron-dependent editing. (A) RT-PCR to estimate splicing efficiency upon treatment with meayamycin using the short intron 8 in the Vcp transcript coding for the valosin containing protein. As editing targets shown in the figure contain large introns, the Vcp transcript was used as an independent control to validate splicing inhibition. Primers bind to exons 8 and 9. M = size standard. bp = basepairs. (B) Scheme of a generic exonic editing site defined by an ECS in the intron. +/− MEA indicates splicing inhibition or control. Sanger sequencing shows an increase in editing rates for intron-dependent editing sites (B, C and D) but not for exon-dependent editing sites (E, F, G and H) upon meayamycin treatment (MEA). The average and standard error of the mean of six biological replicates is given below each chromatogram. (I) Graphs depicting editing frequencies of all intronically and exonically (J) defined editing sites in the figure. The significance level is given on top (n.s.: not significant, ** P < 0.01, *** P < 0.001). Blue = DMSO, red = meayamycin.
DISCUSSION
In this study, we have addressed the impact of splicing efficiency on editing frequencies both in pre-mRNAs and in mRNAs. Using two different approaches we find that a reduction in splicing efficiency leads to a clear increase in editing levels when the editing competent stem is formed between an exon and an intron. Thus, splicing efficiency can have a dramatic impact on the level of editing. However, when the editing competent duplex is formed within the edited exon itself, editing levels do not change in the mature mRNA. Extrapolating from the eight substrates tested, this principle most likely will apply to most if not all exonic editing sites. Interestingly, we observe different Igfbp7 editing levels for the Q/R and K/R site in bone marrow and primary neuronal cultures, respectively (compare Figures 5 and 6). While editing of the Q/R and K/R site reaches 70 and 60%, respectively, in bone marrow cells, the levels are at 26 and 22% in the primary neuronal cultures. This is most likely resulting from developmental differences in editing levels which are known to increase with age. As the neuronal cultures were derived from early embryos while the bone marrow cells were derived from adult mice, differences in editing levels can be expected (24,40).
A striking difference between editing levels in pre-mRNA and mRNA was observed, in particular for the Gria2 substrate where editing is higher in the pre-mRNA than in the mRNA (Figure 1). Our experiments indicate that this can be explained by selective splicing and ADAR2-binding to the pre-mRNA. Apparently ADAR-binding but also the presence of guanosines (and most likely also inosines) at a position -2 relative to a 5′ splice site interferes with binding of the U1 snRNP. These findings are in agreement with earlier data that show selective splicing of Gria2 exon 13 containing the R/G editing site (19). Feed-back of the editing state on pre-mRNA splicing was also reported in other contexts (20,41). The finding that ADAR2 binding reduces splicing efficiency also fits very well with the assumption that editing needs to happen before splicing as exonic editing sites are frequently coordinated by an intronic ECS (31). This notion is also supported by analysis of nascent RNA by RNA-seq that shows that editing occurs co-transcriptionally before most of the introns are removed (42). ADAR2-binding may also explain the higher editing levels in the pre-mRNAs of other constructs. Moreover, ADAR-binding might generally interfere with spliceosomal assembly or the binding of accessory splicing factors, thereby assuring that editing can occur before the mRNA is matured and exported from the editing-competent nucleus.
Surprisingly, Gabra3 mRNAs showed higher editing levels than the corresponding pre-mRNAs. This may indicate selective maturation of the edited transcript. Alternatively, as Gabra3 editing does not require intronic sequences, editing may still continue after splicing as long as the mRNA has not left the editing-competent nucleus. For Gabra3 we have also tested two minigene versions. One version contains an intronic stem while the other is lacking the intronic stem. Previous work has shown that the intronic stem in Gabra3 increases editing at the nearby exonic site (34). Using our constructs we only observe a minor increase in editing upon addition of the intronic stem which is in contrast to the strong increase reported earlier (34). A likely explanation in the observed differences may lie in the different design of the constructs used. While we used an exon–intron–exon context, the most dramatic effect of the intronic stem was observed for an exon–intron construct only. Addition of a second exon did in fact also diminish the effect exerted by the intronic stem (34).
Supplementary Material
Acknowledgments
We thank the following students for their help with cloning and testing constructs: Richard Wallner, Christoph Geier, Mariya Licheva and Anna Siegert. For technical assistance we thank Peter Burg and Celine Brunner. The rat ADAR2 clone was a kind gift of Ron Emeson (Vanderbilt University). Meayamycin was a kind gift of Prof. Kazunori Koide, University of Pittsburgh.
Author contribution: K.L. and M.J. have written the manuscript. K.L. and M.J. have conceived the experiments. K.L. has conducted most of the experiments. U.K. has generated the endogenous editing data in Figures 5 and 6. E.M. has contributed to Figure 1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds zur Förderung der wissenschaftlichen Forschung [P26882, P26845, SFB 43-13, W1207]; Deutsche Forschungsgemeinschaft [LI 2431 2-1]. Funding for open access charge: Fonds zur Förderung der wissenschaftlichen Forschung [P26845] .
Conflict of interest statement. None declared.
REFERENCES
- 1.Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Licatalosi D.D., Darnell R.B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurencikiene J., Kallman A.M., Fong N., Bentley D.L., Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E., et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q., Shai O., Lee L.J., Frey J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Raj B., Blencowe B.J. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Pullirsch D., Jantsch M.F. Proteome diversification by adenosine to inosine RNA editing. RNA Biol. 2010;7:205–212. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 8.Licatalosi D.D., Darnell R.B. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich I., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 10.Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 12.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellaker C., Vesely C., Ponting C.P., McLaughlin P.J., et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon O., Oren S., Safran M., Deshet-Unger N., Akiva P., Jacob-Hirsch J., Cesarkas K., Kabesa R., Amariglio N., Unger R., et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tajaddod M., Jantsch M.F., Licht K. The dynamic epitranscriptome: A to I editing modulates genetic information. Chromosoma. 2016;125:51–63. doi: 10.1007/s00412-015-0526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 18.Maas S., Melcher T., Herb A., Seeburg P.H., Keller W., Krause S., Higuchi M., O'Connell M.A. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 19.Schoft V.K., Schopoff S., Jantsch M.F. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res. 2007;35:3723–3732. doi: 10.1093/nar/gkm314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penn A.C., Balik A., Greger I.H. Steric antisense inhibition of AMPA receptor Q/R editing reveals tight coupling to intronic editing sites and splicing. Nucleic Acids Res. 2013;41:1113–1123. doi: 10.1093/nar/gks1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Z., Cheng Y., Tan B.C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X., et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 22.Sanjana N.E., Levanon E.Y., Hueske E.A., Ambrose J.M., Li J.B. Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics. 2012;192:281–287. doi: 10.1534/genetics.112.141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stulic M., Jantsch M.F. Spatio-temporal profiling of Filamin A RNA-editing reveals ADAR preferences and high editing levels outside neuronal tissues. RNA Biol. 2013;10:1611–1617. doi: 10.4161/rna.26216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahlstedt H., Daniel C., Enstero M., Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tariq A., Garncarz W., Handl C., Balik A., Pusch O., Jantsch M.F. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 2013;41:2581–2593. doi: 10.1093/nar/gks1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garncarz W., Tariq A., Handl C., Pusch O., Jantsch M.F. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jepson J.E., Savva Y.A., Jay K.A., Reenan R.A. Visualizing adenosine-to-inosine RNA editing in the Drosophila nervous system. Nat. Methods. 2011;9:189–194. doi: 10.1038/nmeth.1827. [DOI] [PubMed] [Google Scholar]
- 28.Marcucci R., Brindle J., Paro S., Casadio A., Hempel S., Morrice N., Bisso A., Keegan L.P., Del Sal G., O'Connell M.A. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levanon E.Y., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., SandersBush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 31.Higuchi M., Single F.N., Kohler M., Sommer B., Sprengel R., Seeburg P.H. Rna editing of ampa receptor subunit Glur-B—a base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 32.Ohlson J., Pedersen J.S., Haussler D., Ohman M. Editing modifies the GABA(A) receptor subunit alpha 3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konarska M.M., Sharp P.A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 34.Daniel C., Veno M.T., Ekdahl Y., Kjems J., Ohman M. A distant cis acting intronic element induces site-selective RNA editing. Nucleic Acids Res. 2012;40:9876–9886. doi: 10.1093/nar/gks691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibley C.R., Emmett W., Blazquez L., Faro A., Haberman N., Briese M., Trabzuni D., Ryten M., Weale M.E., Hardy J., et al. Recursive splicing in long vertebrate genes. Nature. 2015;521:371–375. doi: 10.1038/nature14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroeger K., Collins M., Ugozzoli L. The preparation of primary hematopoietic cell cultures from murine bone marrow for electroporation. J. Vis. Exp. 2009;23 doi: 10.3791/1026. doi:10.3791/1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y., Vogt A., Forsyth C.J., Koide K. Comparison of splicing factor 3b inhibitors in human cells. Chembiochem. 2013;14:49–52. doi: 10.1002/cbic.201200558. [DOI] [PubMed] [Google Scholar]
- 38.Holmes A.P., Wood S.H., Merry B.J., de Magalhaes J.P. A-to-I RNA editing does not change with age in the healthy male rat brain. Biogerontology. 2013;14:395–400. doi: 10.1007/s10522-013-9433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillman A.A., Cookson M.R., Galter D. ADAR2 affects mRNA coding sequence edits with only modest effects on gene expression or splicing in vivo. RNA Biol. 2016;13:15–24. doi: 10.1080/15476286.2015.1110675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J.B., Levanon E.Y., Yoon J.K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 41.Ryman K., Fong N., Bratt E., Bentley D.L., Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez J., Menet J.S., Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol. Cell. 2012;47:27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.