Abstract
Proteins encoded by mitochondrial DNA are translated using mitochondrially encoded tRNAs and rRNAs. As with nuclear encoded tRNAs, mitochondrial tRNAs must be processed to become fully functional. The mitochondrial form of ribonuclease P (mt:RNase P) is responsible for 5′-end maturation and is comprised of three proteins; mitochondrial RNase P protein (MRPP) 1 and 2 together with proteinaceous RNase P (PRORP). However, its mechanism and impact on development is not yet known. Using homology searches, we have identified the three proteins composing Drosophila mt:RNase P: Mulder (PRORP), Scully (MRPP2) and Roswell (MRPP1). Here, we show that each protein is essential and localizes with mitochondria. Furthermore, reducing levels of each causes mitochondrial deficits, which appear to be due at least in part to defective mitochondrial tRNA processing. Overexpressing two members of the complex, Mulder and Roswell, is also lethal, and in the case of Mulder, causes abnormal mitochondrial morphology. These data are the first evidence that defective mt:RNase P causes mitochondrial dysfunction, lethality and aberrant mitochondrial tRNA processing in vivo, underscoring its physiological importance. This in vivo mt:RNase P model will advance our understanding of how loss of mitochondrial tRNA processing causes tissue failure, an important aspect of human mitochondrial disease.
INTRODUCTION
Most metazoan mitochondrial genomes encode the same transcripts (1). The 13 proteins encoded by mitochondrial DNA (mtDNA) are required components of the complexes that carry out oxidative phosphorylation. In order for mitochondria to fulfill the ATP demands of the cell, each mitochondrial-encoded protein must be efficiently generated by the mitochondrial machinery. To this end, the mitochondrial ribosomes rely on the large and small ribosomal RNAs and the entire suite of tRNAs encoded by the mtDNA.
Nuclear tRNAs are transcribed as pre-tRNAs that require 5′- and 3′-end cleavage and other modifications in order to mature and become fully functional (2–4). As mitochondria have a bacterial origin, mtDNA is transcribed as polycistrons that must be processed and cleaved to create the final products (5,6). Thus, mtDNA transcription is complex and places unique demands on the mitochondrial RNA processing machinery, differing significantly from the transcription that commonly takes place in the nucleus (7). Mitochondrial tRNAs (mt:tRNAs) punctuate the mRNAs and rRNAs found in mtDNA and thus have the added complication of requiring excision from the polycistron before maturation (8). In the nucleus, the RNAse P ribozyme has long been known to be responsible for the 5′-end pre-tRNA cleavage reaction, while other endonucleases, such as RNase Z and Rex1p carrying out the 3′-end cleavage reaction (9–11). Recently, a new class of protein-only RNase Ps (proteinaceous RNase P (PRORP)) has been identified that can carry out the 5′-end cleavage in human mitochondria (12). In contrast to the RNAse P ribozyme, PRORP (also called Mitochondrial RNase P Protein 3 (MRPP3)) is a metallonuclease without an RNA component. Single protein PRORPs exist in the nucleus and organelles of land plants and unicellular eukaryotes (13,14). In contrast, human PRORP has only been identified in mitochondria and is part of a three-protein complex referred to as mt:RNase P, comprising mitochondrial RNase P Protein 1 (MRPP1/TRMT10C) and mitochondrial RNase P Protein 2 (MRPP2/SDR5C1/HSD10) (12). MRPP1 is required for methylation of the ninth position of certain mitochondrial tRNAs, although this activity is not obligatory for mt:RNAse P function and 5′-end cleavage (15). MRPP2 is a dehydrogenase, however evidence supports that in the context of mt:RNase P it is required for the methyltransferase activity of MRPP1, as well as likely functions as a scaffold protein for the formation of an MRPP1 and 2 sub-complex (15). As such, the MRPP1 and MRPP2 sub-complex catalyzes tRNA methylation and in the context of mt:RNase P supports 5′-end processing by PRORP. The 3′-end cleavage of mt:tRNAs also uses a mitochondrially localized RNase Z, called ELAC2 in humans and RNase ZL in Drosophila. While these two enzyme complexes are distinct, there is evidence that RNase Z/ELAC2 preferentially cleaves products that have already been processed by RNase P (16).
While this newly identified mt:RNAse P complex appears critical for tRNA processing and maturation, it is currently unknown how loss of mt:RNAse P function affects mitochondrial output and health. This is particularly relevant to human health as the majority of mitochondrial diseases caused by mutations in mtDNA are due to point mutations in mt:tRNAs (17). Based on their location, most of the mitochondrial tRNA mutations could affect mt:RNAse P function, leading to unprocessed RNA species and mitochondrial dysfunction. Specific examples include MELAS (mitochondrial encephalopathy, lactic acidosis and stroke-like episodes), mitochondrial myopathy and HSD10 disease (18–23). Highlighting the importance of mt:tRNA processing in human health, mutations in RNase Z/ELAC2 and MRPP2/SDR5C1 are associated with cardiomyopathy and neurodegeneration, and cause mis-regulated mt:tRNA cleavage in cell culture (24,25). For unknown reasons mitochondrial diseases manifest very differently in different organs and tissues and have quite different etiology, making their diagnoses and treatment difficult, therefore it is critical to understand the molecular mechanisms underlying mt:tRNA cleavage in vivo (17,26).
Drosophila is an excellent model system to study the effect of altered mtDNA post-transcriptional processes as Drosophila and human mtDNA genomes are arranged in a similar manner, with mt:tRNAs punctuating the mRNAs and rRNAs (27). In order to determine the developmental and physiological significance of mt:RNase P function, we have used sequence similarities to identify the Drosophila mt:RNase P complex and examined Drosophila lacking the orthologs for PRORP, MRPP2 and MRPP1, called Mulder, Scully and Roswell, respectively. Here we show that Mulder, Scully and Roswell localize to mitochondria as expected and that each can associate with the others. Importantly, reducing any one of the three causes lethality. Furthermore, flies lacking Mulder, Scully or Roswell have associated mitochondrial deficiencies, exhibiting greatly reduced levels of ATP and swollen, damaged-looking mitochondria. In addition, flies with reduced Mulder, Scully or Roswell accumulate unprocessed mt:tRNA indicating that the mitochondrial defects we observe are at least in part direct and specific for the protein complex function. Finally, we find that overexpressing either Mulder or Roswell exerts dominant negative effects on development and can also cause mitochondrial defects. These data are the first to show that loss of mt:RNase P function is detrimental in metazoans and specifically mis-regulates mt:tRNA processing. Understanding the role of the highly conserved mt:RNase P will be useful to dissect a subset of the complex mechanisms underlying mitochondrial diseases.
MATERIALS AND METHODS
Fly stocks
The following fly stocks were obtained from the Bloomington Stock Center: y1 w* mldrB FRT19A/FM7i, ActGFP, y1 w* mldrC FRT19A/FM7i, ActGFP, y1 w* scuA FRT19A/FM7i, ActGFP, y1 w* scuD FRT19A/FM7i, ActGFP, y1 v1; scuTRiP.HMSO2305, y1 sc* v1; scuTRiP.GL01079, y1 sc* v1; rswlTRiP.HMC02423, y1 w*; P{w[+mC] = tubP-GAL4}LL7/ TM3, P{w[+mC] = ActGFP}JMR2, Ser[1], y1 w*; P{w[+mC] = Act5C-GAL4}17bFO1/ TM3, P{w[+mC] = ActGFP}JMR2, Ser, w*; P{w[+mC] = ple-GAL4.F}3, P{w[+mC] = GAL4::VP16-nos.UTR}. The following stocks were obtained from the Vienna Drosophila Research Center: y w1118/ mldrKK108043, w1118; rswl GD12447 (28).
Transgenic flies and constructs
The full-length Open Reading Frames for each respective gene was amplified from the following Drosophila ESTs (Drosophila Genomics Resource Center, Bloomington, IN, USA): rswl (LD44982), scu (IP05285) and mldr (SD17694). The two point mutations present in SD17694 were corrected (R250L and L528M) using the QuikChange method (Stratagene). For transgenic flies, rswl, scu and mldr were sub-cloned into pPWM, pPWF and pPWV plasmids (The Drosophila Gateway Collection, Carnegie Institution of Washington, Baltimore, MD, USA), respectively, then commercially injected (Genetic Services, Inc, Cambridge, MA, USA). For S2R+ tissue culture constructs, rswl, scu and mldr were sub-cloned into pAWM containing a C-terminal myc tag. scu and mldr were also cloned into pAWF containing a C-terminal FLAG tag (The Drosophila Gateway Collection, Carnegie Institution of Washington, Baltimore, MD, USA).
Northern blots
Total RNA was extracted from 5 to 10 mg of wild-type and mutant strains of Drosophila larvae using a Tissue and Insect RNA kit (Zymo Research Corporation, St. Louis, MO, USA). Five micrograms from each sample were mixed with an equal amount of 2× urea loading dye (0.05% Bromo-Phenol-Blue, 0.05% Xylene Cyanol dye, 50% m/v urea, 0.1 M EDTA) and run on 6% urea-polyacrylamide gel. Northern blot analysis was performed according to the DIG Northern Starter Kit (Sigma-Aldrich Corp. St. Louis, MO, USA) with a hybridization temperature of 50°C. The size standards of the RNA fragments were detected using DIG-labeled RNA Molecular Weight Marker I (Sigma-Aldrich Corp. St. Louis, MO, USA). Each blot was performed at least three times.
Immunofluorescence
Larval brains and ovaries from well-fattened females were dissected and fixed as previously described (29). S2R+ culture cells were fixed and mounted as described (29). In short, samples were fixed in Grace's Insect Medium (modified) (BioWhittaker, Lonza, Cologne, Germany) containing 4% paraformaldehyde and 20 mM formic acid (Sigma-Aldrich Corp. St. Louis, MO, USA). Antibody staining was performed in antibody wash buffer (1× PBS:0.1% Triton X-100:1% BSA). The following antibodies were used: rabbit anti-GFP (1:2000, Torrey Pines Biolabs, Secaucus, NJ, USA), mouse anti-myc (1:1000, Sigma-Aldrich Corp. St. Louis, MO, USA), anti-FLAG (1:1000, Sigma-Aldrich Corp. St. Louis, MO, USA), mouse anti-ATP synthase (1:1000, CVA, Mitosciences) and Phalloidin (1:250, Invitrogen). The following secondary antibodies were used: goat anti-mouse IgG2b Alexa 488, goat anti-mouse IgG2b Alexa 568, goat anti-mouse IgG1 Alexa 488 and goat anti-rabbit Alexa 488 (Molecular Probes, Lifetechnologies, Grand Island, NY, USA). Images were collected using Zeiss 710 and Zeiss 700 confocal microscopes and 63× Plan Apo NA 1.4 lens.
Western blotting, immunoprecipitation and cell fractionation
Western blotting, immunoprecipitations (IP) and cell fractionations have been performed as described earlier (29,30). The following antibodies were used: mouse anti-myc (1:5000, Sigma-Aldrich Corp. St. Louis, MO, USA), anti-FLAG (1:10000, Sigma-Aldrich Corp. St. Louis, MO, USA), anti-CVA (1:100000, Mitosciences), mouse anti-alpha tubulin (1:5000, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA) and anti-Mldr (1:2000, this study).
Pupation, eclosion and negative geotaxis measurements
Pupation, eclosion and negative geotaxis measurement were performed as described (29,30). P-values were calculated in Microsoft Excel using a two-tailed Student's t-test.
ATP measurements and aconitase assay
ATP measurements and aconitase assays were performed as described (29). In short, for ATP measurement, larval samples were homogenized in ATP extraction buffer (100 mM Tris-Cl, pH 8.0, 4 mM EDTA, pH 8.0; 6 M guanidine hydrochloride) and protein concentrations were measured using Bradford assay. The ATP concentration was determined using an ATP Determination Kit (Molecular Probes, Invitrogen) according to the manufacturer's directions. One hundred microliter assays were performed in a 96-well plate and the luminescence was measured using a Biotek Synergy H1 luminometer. Each sample was processed in duplicate and read in triplicate. The amount of ATP was normalized against protein concentration. For aconitase assays, larval extracts were electrophoresed on cellulose acetate membrane (Cellogel, Accurate Chemicals, Westbury, NY, USA) in running buffer [20 mM potassium phosphate (pH 7.8), 3.6 mM citrate]. Aconitase activities were detected on the membrane by incubating it in staining solution [100 mM potassium phosphate, (pH 6.5), 1 mM NADP1, 25 mM MgCl2, 2 mM cis-aconitic acid, 0.5 mg/ml 2,3-bis-(2-methoxy-4-nitro-5-sulfenyl)-2H-tetrazolium-5 carboxanilide disodium salt (XTT) or MTT, 0.3 mM phenazine methosulfate, 5 units/ml isocitrate dehydrogenase] for 10 min. From three membranes, the band intensities from mitochondrial aconitase activity of each sample were normalized against the respective cytoplasmic aconitase activity for comparison. Analyses were done using ImageJ (31).
Mulder antibody generation
The Drosophila EST SD17694 (Drosophila Genomics Resource Center, Bloomington, IN, USA) was cloned into pMCSG7 vector (gifted from Midwest Center for Structural Genomics, IL, USA) and transformed into BL21(DE3) expression cells (New England Biolabs, MA, USA). The culture was grown at 37°C and the protein was expressed using 200 μM of IPTG at 18°C in TB media (Sigma-Aldrich Corp. St. Louis, MO, USA). The cells were lysed using lysis buffer (8M urea, 100 mM NaH2PO4, 10 mM Tris and 10 mM imidiazole pH 8.0) and sonication. The lysate was centrifuged and the supernatant was passed through Ni-NTA column and washed with a denaturing wash buffer (8M urea, 100 mM NaH2PO4, 150 mM NaCl, 20 mM imidiazole pH 8.0). The protein samples were eluted from the column using 20 mM MOPS pH 7.8,100 mM NaCl, 1 mM TCEP, 500 mM imidazole and 6 M urea. The purity of the sample was verified by SDS PAGE gel and fractions having a single band were pooled and then commercially injected into rabbits (Covance Research Products, Princeton, NJ, USA). A 1 mg/ml protein was used for the injection and the serum was checked by ELISA test for antibody tightness.
RESULTS
Members of the protein only RNase P complex localize to mitochondria in Drosophila
Pre-tRNAs are processed through cleavage of their 5′ and 3′ ends, as well as by the covalent modification of various bases (2,3). In metazoan mitochondria, the protein in the mt:RNase P complex responsible for catalyzing the 5′-end cleavage is PRORP which contains a metallonuclease domain, as well as a pentatricopeptide repeat (PPR) domain thought to bind RNA (Figure 1A). The Drosophila ortholog contains all of the same recognizable domains, including a mitochondrial targeting signal. The Drosophila PRORP ortholog is the previously undescribed gene CG15896 that we have named mulder (mldr) (Figure 1A). MRPP2 is a member of the short-chain dehydrogenase/reductase (SDR) superfamily and is also known as Hydroxysteroid (17-Beta) Dehydrogenase 10 (HSD10 is the protein, HSD17B10 is the gene) (Figure 1A). The Drosophila ortholog Scully (Scu) is highly homologous to MRPP2, with 69% amino acid identity (Figure 1A) (32). MRPP1 is a methyltransferase and Drosophila contain two potential orthologs. However, only one has a mitochondrial targeting signal, which is encoded by the previously undescribed gene CG5190 that we have named roswell (rswl) (Figure 1A).
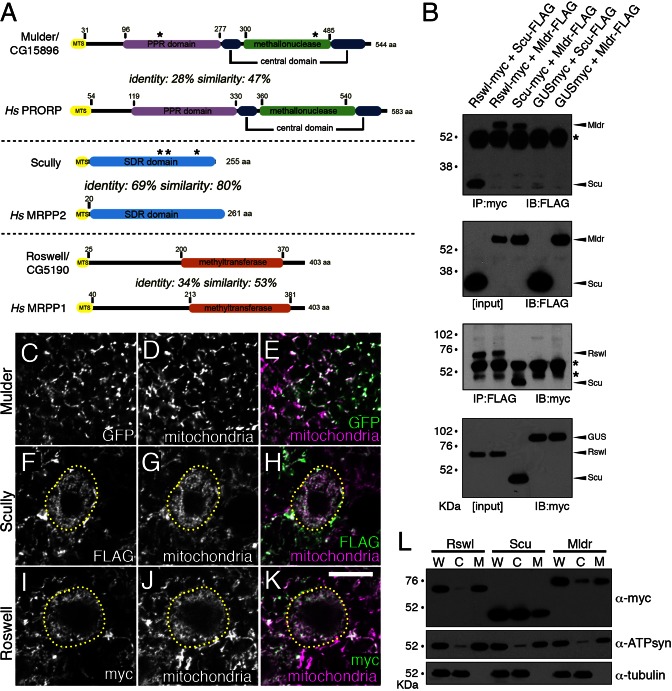
Figure 1.
The Drosophila mt:RNase P proteins Mldr, Scu and Rswl localize to mitochondria. (A) Schematics indicating the domains of each protein and their homology to the human homologs. Asterisks indicate the approximate location of the mutations used in this study. The length of the mitochondrial targeting sequence (MTS, yellow) was predicted using the Mitoprot server. Domain boundaries of Drosophila mt:RNase P components were predicted based on alignments with human homologs (Clustal Omega). Sequence identify and similarity were determined by BLAST. (B) Western blots showing each mt:RNase P protein can reciprocally co-immunoprecipitate with the others. GUS = glucuronidase as a negative control. Arrowheads indicate the correct band size. Asterisks indicate the anti-mouse IgG cross-reactive bands. (C–K) GFP-tagged Mldr, FLAG-tagged Scu and myc-tagged Rswl (C, F, I, respectively) co-localize with mitochondria (D, G, J) in larval brains. E, H, K show the merged images. Dashed yellow lines indicate third larval instar neuroblasts. Overexpression of Mldr causes early larval lethality and it is difficult to find NBs in the very young brains, thus there is no NB indicated (C, D). (L) Western blot of fractionated S2R+ cell extract from cells transfected with myc-tagged Mldr, Scu or Rswl shows each protein is located in the mitochondrial fraction (M). Tubulin and ATP synthase are controls for cytoplasmic (C) and mitochondrial (M) fractions, respectively. W = whole cell extract. White = anti-ATP synthase (D, G, J), anti-GFP (C), anti-FLAG (F), anti-myc (I). Green = anti-GFP (E), anti-FLAG (H), anti-myc (K). Magenta = anti-ATP synthase (E, H, K). Scale bar = 10 μm in K for C–K.
For Mldr, Scu and Rswl to exert their function as the Drosophila mt:RNase P, they would have to associate with each other. To test this, we transfected S2R+ cells pairwise with myc- and FLAG-tagged constructs and performed reciprocal immunoprecipitation (IP) (Figure 1B). Rswl and Scu, Rswl and Mldr and Scu and Mldr reciprocally co-IP'd, indicating that the three were able to associate. Myc-tagged glucuronidase (GUS) served as a negative control. Furthermore, the mt:RNase P complex must localize to mitochondria in order to process mt-tRNAs. Mldr, Scu and Rswl contain putative mitochondrial targeting signals (Figure 1A; MTS), however none have been previously shown to localize to mitochondria in vivo. To test the localization of each protein, we developed transgenic flies using the UAS/GAL4 system to conditionally express GFP-tagged Mldr, FLAG-tagged Scu and myc-tagged Rswl (33). As evidenced by immunofluorescence, all three proteins co-localized with mitochondrial ATP synthase in the larval brain (Figure 1C–K) and germ cells (Supplementary Figure S1A–F, Figure 6D–F). S2R+ cells transfected with myc-tagged Mldr, Scu and Rswl also showed strong mitochondrial co-localization with ATP synthase (Supplementary Figure S1G–O). To confirm this localization biochemically, we transfected S2R+ cells with myc-tagged Mldr, Scu and Rswl and separated the extract into cytoplasmic and mitochondrial fractions. By probing the fractions with anti-myc antibody, we saw that each protein is detected in the mitochondrial fraction (Figure 1L). There were minimal amounts of Mldr and Rswl in the cytoplasmic fractions which were likely due to a trace contamination from the mitochondrial fraction judging from the ATP synthase antibody. While immunofluorescence in situ showed predominantly mitochondrial staining, Scu was also found in the cytoplasmic fraction, which could be due to overexpression in S2R+ cells that often led to large cytoplasmic aggregates (Supplementary Figure S2, arrows). ATP synthase and tubulin served as controls for the purity of the mitochondrial and cytoplasmic fractions, respectively. Taken together, these data show that Mldr, Scu and Rswl can associate with each other, and that each is localized to mitochondria in vivo.
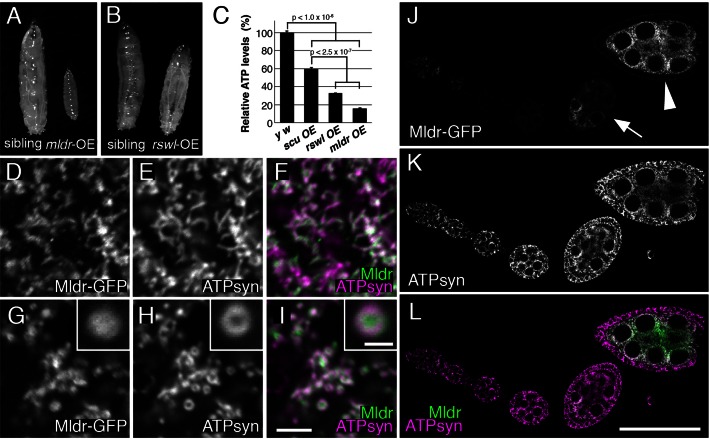
Figure 6.
Overexpressing Mulder causes mitochondrial morphology defects. (A) UAS-mldr-GFP expressing larvae driven by tubGAL4 do not develop past second instar. The sibling control larva is a third instar. (B) UAS-rswl-myc larvae, in contrast, are able to grow to the third instar stage driven by tubGAL4. (C) Graph showing relative ATP levels for UAS-scu, UAS-rswl and UAS-mldr larvae overexpressed using tubGAL4. OE = overexpression. (D–I) Germ cells expressing UAS-mldr-GFP under the control of nosGAL4. (D–F) Germ cells with lower Mldr expression (D) have mitochondria that are of the normal shape and size (E, F is the merge). Germ cells with high expression of UAS-Mldr-GFP (G) have swollen mitochondria (H, I is the merge). The swollen, ring-shaped mitochondria (H, inset) have Mldr-GFP concentrated in the middle (G, inset, I inset is the merge). (J–L) A representative ovariole expressing UAS-mldr-GFP under control of nosGAL4. Weaker expression is seen in younger germ cells (J, arrow) compared to strong Mldr-GFP expression in older ones (J, arrowhead). ATP synthase labeling shows all of the mitochondria throughout the ovariole (K). L is the merge of J and K. White = anti-GFP (D, G, J), anti-ATP synthase (E, H, K). Green = anti-GFP (F, I, L). Magenta = anti-ATP synthase (F, I, L). Scale bars = 5 μm in I for D-I, 1.5 μm in I inset for all insets, 50 μm in L for J–L. Error bars = s.e.m.. P-values were calculated in Microsoft Excel using a two-tailed Student's t-test.
Mulder, Scully and Roswell are orthologous to PRORP, MRPP2 and MRPP1
There are no available structures for Mldr and Scu. We have, however, used known structures of homologous proteins to construct the high confidence models shown in Figure 2. Specifically, Phyre server was used to create a structure model of Mldr using the metallonuclease domain of human PRORP and the full-length structures of A. thaliana PRORP1 and PRORP2 as templates (34–36). The I-Tasser server was used to create a structure model of Scu based on structures of human MRPP2 (37,38). There are two uncharacterized publicly available alleles of mldr that were generated in a large-scale mutagenesis screen: mldrB (Y121D in the PPR domain) and mldrC (W465R in the metallonuclease domain) (Figure 2A, yellow circles) (39). There are three available scu alleles. Two of them were generated in the same genetic screen as was mldr: scuD (S163F) is in a highly conserved residue (Figure 2B, red circle) and scuA (Q159Stop) is predicted to result in a C-terminal truncation (Figure 2B, shown in yellow) (39). scu4058 results from a small X-ray induced deletion which causes a frameshift at E205 leading to a stop codon 60 nucleotides downstream after E236 (32). This results in a smaller truncation than scuA, consisting of the last 19 residues (Figure 2B, shown in green). The putative tetrameric assembly of Scu is presented where the four monomers are differentially color-coded (Figure 2C). It is evident that S163 lies in the interface between two monomers.
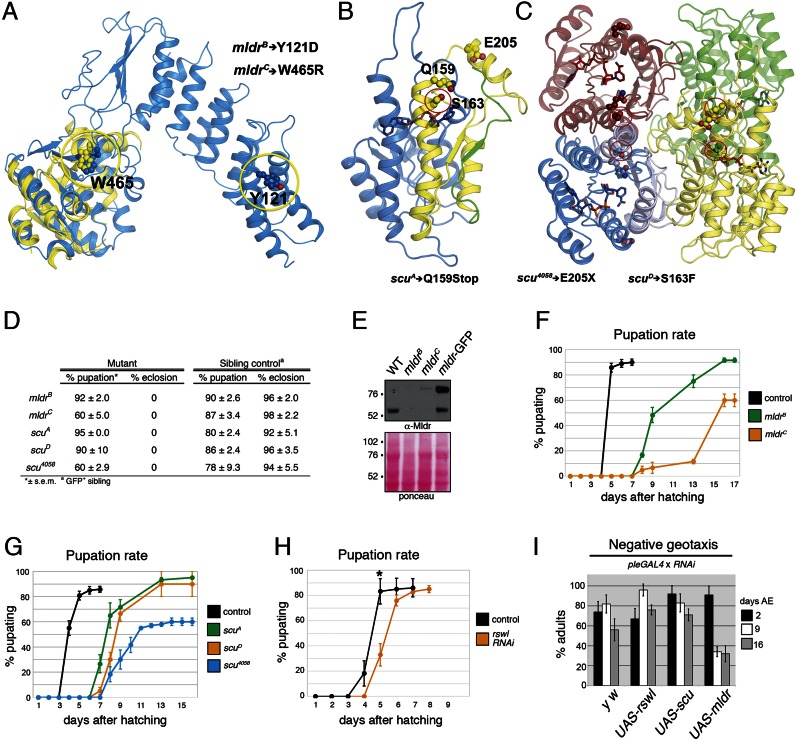
Figure 2.
Loss of mldr and scu results in lethality in Drosophila. (A–C) Predicted structure of Mulder based on the Arabidopsis thaliana PRORP1, PRORP2 and the human PRORP available structures (A) and Scully (B and C) based on the available human MRPP2 structures. The modeled MRPP2 monomer is shown in blue in B whereas the expected tetrameric assembly is shown in C. The point mutations used in this study are indicated with yellow (A) and red (B, C) circles. The scu alleles Q159Stop and E205X result in truncations. The E205X and Q159Stop mutations result in a 19-residue truncation in yellow and an additional 87-residue truncation in green, respectively shown in B. The position of W465, Y121 in Mulder and S163, Q159 and E205 are all shown as ball and sticks. (D) Table summarizing percent pupation and eclosion for mldr and scu alleles. (E) Western blot indicating larvae mutant for mldrB and mldrC have greatly reduced protein levels. (F–H) Pupation rates. Both mldrB and mldrC mutant larvae have delayed pupation with 40% of mldrC mutant larvae failing to pupate (F). Larvae mutant for scu have delayed pupation as well. The scu4058 allele also fails to pupate 40% of the time (G). (H) Expressing UAS-rswl-RNAi using ActGAL4 shows a modest but reproducible pupation delay. Note the different scale in the X-axis. The controls (black lines) for F–H are a representative sibling control for one of the genotypes. The lines stop because the adults eclose. (I) Negative geotaxis assay. Expressing scu or rswl RNAi in dopaminergic neurons using Ple-GAL4 has no effect on adult fly locomotion, whereas expressing mldr-RNAi causes an age-dependent decrease in locomotion. AE = After Eclosion. Error bars = s.e.m. for F, G and s.d. for H. * P = 0.02, determined using a two-tailed Student's t-test in Microsoft Excel.
Reducing Mldr, Scu and Rswl causes lethality in vivo
In order to study the consequences of loss of mt:RNase P in vivo, we examined mutants for mldr and scu, and RNAi knockdown for mldr, scu and rswl. We found the mutant alleles for mldr and scu cause lethality because flies containing mutations in either gene do not make it to the adult stage as we only saw heterozygous sibling adult flies eclose, whereas homozygous mutant adults never eclosed (Figure 2D). While mldrB and mldrC are only point mutations, the larvae contained very low protein levels as judged by western blot (Figure 2E). To determine at which stage of development the lethality occurs, we transferred 20 one-day-old mutant larvae into vials and scored how many pupated (Figure 2F–H). Under these uncrowded conditions, mldrB showed a delay in pupation relative to wild-type sibling controls, with all the larvae finally pupating by day 16 (Figure 2F). mldrC had a stronger phenotype, with a longer pupation delay and only 60% of the larvae pupating compared with sibling controls (Figure 2F). The three available scu alleles gave similar results. scuA and scuD were both lethal and experienced delayed pupation under uncrowded conditions (Figure 2G). scu4058 was the most severe, with a slightly longer pupation delay and only 60% of the larvae pupating (Figure 2G). Pupae mutant for mldr and scu frequently did not develop normally and had reduced spiracle eversion (Supplementary Figure S3). Expressing UAS-rswl-RNAi also gave a modest delay in pupation compared to control (Figure 2H).
In addition to examining both mldr and scu mutants, we ubiquitously expressed available RNAi lines using the highly expressed tubGAL4 and lower expressed ActGAL4 (Table 1 & Supplementary Table S1). Expressing UAS-scu and UAS-mldr RNAi using tubGAL4 allowed larvae to pupate equally well as controls. The scu RNAi knockdown pupae did not develop to adulthood. Approximately 10% of the mldr knockdown pupae eclosed and using ActGAL4 to express the RNAi at lower levels allowed an even larger portion of the adults to eclose (Table 1 and Supplementary Table S1). At present, there are no available alleles for rswl and only one RNAi stock. Expressing rswl-RNAi using ActGAL4 allowed the larvae to pupate, but not develop. While this is only one RNAi line, this result is very similar to our results with mldr and scu. Expressing UAS-rswl RNAi at higher levels using tubGAL4 suppressed even pupation (Table 1).
Table 1. mt:RNase P RNAi knockdown pupation and eclosion rates.
| RNAi | Sibling controlb | |||
|---|---|---|---|---|
| Percent pupationa | Percent eclosion | Percent pupation | Percent eclosion | |
| tubGAL4/ UAS-rswlGD12447 | 0 | 0 | 85 ± 5 | 98 ± 1.8 |
| ActGAL4/ UAS-rswlGD12447 | 92 ± 3.3 | 0 | 98 ± 1.7 | 98 ± 1.7 |
| UAS-scuTRiP.HMS02305 /+; tubGAL4/+ | 86 ± 7.2 | 0 | 98 ± 2.5 | 98 |
| tubGAL4/ UAS-scuTRiP.GL01079 | 92 ± 1.7 | 0 | 95 ± 2.9 | 97 ± 3.3 |
| tubGAL4/ UAS-mldrKK108043 | 83 ± 1.7 | 8 ± 5.1 | 92 ± 1.7 | 97 ± 3.3 |
a± s.e.m.
bGFP+ sibling larvae.
Neurons are particularly sensitive to mitochondrial output. To determine if decreasing Mldr, Scu or Rswl caused neuronal deficits, we expressed UAS-RNAi for each using the tyrosine hydroxylase pleGAL4 specific to dopaminergic neurons and examined the ability of the flies to climb at different ages (Figure 2I). Expressing neither UAS-rswl nor UAS-scu RNAi caused deficits in locomotion. However, knocking down mldr in dopaminergic neurons caused an age-progressive loss of locomotive function (Figure 2I). Taken together, these data support an essential role for mt:RNase P in vivo. Furthermore, ubiquitously knocking down each complex member caused lethality at the same developmental stage and thus gave very similar phenotypes, suggesting they function in the same pathway.
mldr and scu mutants and rswl knockdown larvae have reduced ATP levels but no apparent mitochondrial oxidative stress
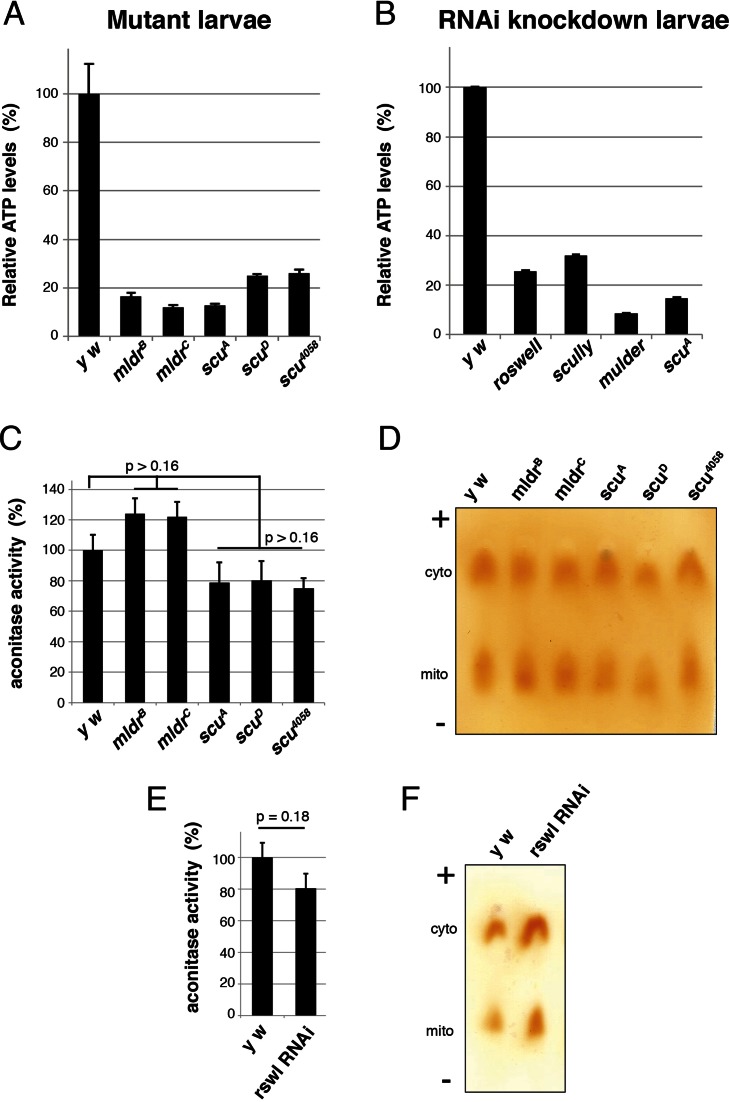
In metazoans, mtDNA encodes the entire suite of tRNAs necessary for translating mtDNA encoded proteins. There are only 13 proteins encoded by mtDNA, all of which are required for the protein complexes used for oxidative phosphorylation. Since human mt:RNase P is involved in mt:tRNA processing in vitro, we tested whether knocking down each component caused ATP levels to decrease in vivo. Larvae mutant for mldr and scu showed greatly reduced ATP levels relative to wild-type control (Figure 3A). Consistent with this result, extract from larvae expressing UAS-RNAi for mldr, scu and rswl also showed low levels of ATP relative to control (Figure 3B). One of the consequences of electron transport during respiration can be the accumulation of reactive oxygen species. If reactive oxygen species accumulate, mitochondria can experience a reduction in ATP due to the build-up of general oxidative damage as a consequence of oxidative stress. For example, Drosophila harboring mutations for various mitochondrial proteins often suffer from mitochondrial oxidative damage and flies lacking mt:RNase Z function were shown to have increased oxidative damage (examples include (29,40–43)). One assay to assess the mitochondrial redox environment is to measure the enzymatic ability of mitochondrial aconitase, an enzyme easily rendered non-functional due to oxidation (44,45). mldr and scu mutants and rswl RNAi knockdown larvae did not show a decrease in mitochondrial aconitase activity relative to cytoplasmic aconitase activity (Figure 3C–F). This suggests that the drop in ATP levels is not due to general mitochondrial oxidative stress.
Figure 3.
Loss of mt:RNase P complex proteins causes decreases in ATP in vivo. (A) Larvae mutant for mldr and scu have greatly reduced levels of ATP compared to wild-type (y w). (B) Larvae ubiquitously expressing RNAi for rswl, scu and mldr using tubGAL4 also have reduced levels of ATP. scuA mutant larvae are included as a control. (C, E) mldr and scu mutant larvae (C) and larvae ubiquitously expressing rswl-RNAi (E) do not have decreased mitochondrial aconitase activity relative to cytoplasmic aconitase activity. (D, F) Representative cellulose acetate membranes for C and E, respectively, showing cytoplasmic (cyto) and mitochondrial (mito) aconitase activity. ‘+’ and ‘−’ indicate the electrophoretic migration. Error bars represent s.e.m.. P - values were calculated using a two-tailed Student's t-test in Microsoft Excel. Information on replicates can be found in the Materials and Methods.
Mitochondria swell in the absence of Mldr and Scu
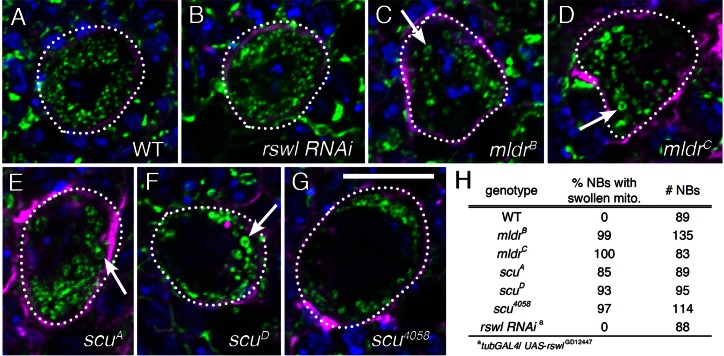
When the inner mitochondrial membrane of a mitochondrion loses its selective permeability due to damage, it osmotically swells and ruptures the outer mitochondrial membrane. This phenomenon gives rise to swollen, ring-like mitochondria when viewed by microscopy. Mldr, Scu and Rswl localized to mitochondria, and there was a decrease in ATP levels when any of the three were knocked down. The lethality we observed with mldr and scu mutants and with UAS-rswl-RNAi knockdown could be due to a variety of non-specific defects. To examine whether there are defects in mitochondrial size and shape with loss of mt:RNase P, we examined mitochondrial morphology using immunofluorescence. Mitochondria in larval brain neuroblasts have a stereotypical, distinctive, small spherical structure (Figure 4A) (29). When we examined neuroblasts in mldr and scu mutant larval brains, we found they were larger and swollen compared to wild-type, and often contained ring-like mitochondria (Figure 4C–G, arrows, versus A). For scu4058, this is consistent with mitochondrial morphological changes seen using transmission electron microscopy (32). Loss of mitochondrial proteins in Drosophila does not always give rise to this phenotype (29). In contrast, the rswl RNAi knockdown using tubGAL4 did not show swollen or ring-like mitochondria, even though these larvae do not grow past second instars (Figure 4B).
Figure 4.
Larvae lacking Mldr and Scu have mitochondrial defects. (A–G) Larval stem cells (dashed circles) from third instar larval brains labeled for mitochondria (green), actin (magenta) and DAPI (blue). Larval stem cells in wild-type brains have small, dispersed mitochondria (A). (C–G) In contrast, mitochondria in the neuroblasts of mldr and scu mutants are grossly swollen with frequent ring-shaped mitochondria (arrows). (B) Ubiquitously expressing rwsl-RNAi using tubGAL4 does not cause defects in mitochondrial morphology. (H) Table indicating the penetrance of neurobasts (NB) with swollen mitochondria and the number of neuroblasts examined. Green = anti-ATP synthase, magenta = phalloidin, blue = DAPI (A–G). Scale bar = 10 μm in G for A–G.
Mitochondrial tRNA processing is altered in the absence of Mldr and Scu
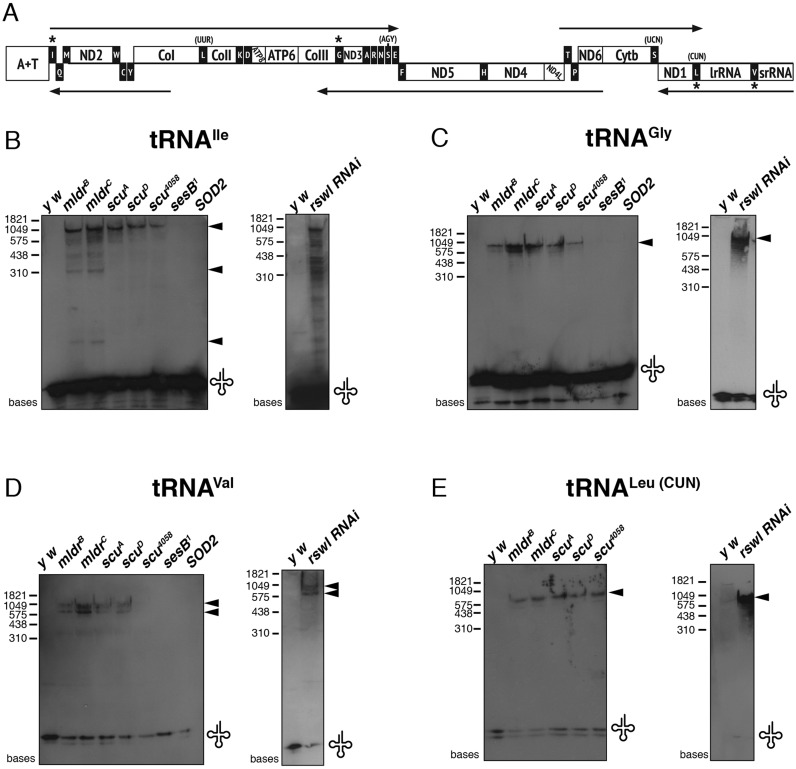
Products encoded by mtDNA do not contain introns and with the exception of the A+T rich region, there are very few nucleotides that do not encode mRNA, rRNA or tRNA (Figure 5A) (27). Drosophila mtDNA is thought to be transcribed as five polycistrons (5,6). tRNAs in these polycistronic units are thought to serve as punctuation marks for 3′ and 5′ end processing of mitochondrial rRNAs and mRNAs (8). As such, most mt:mRNAs and mt:rRNAs are separated by at least one mt:tRNA (Figure 5A). Since the human mt:RNase P complex cleaves the mt:tRNA 5′-ends in vitro, we investigated whether mt:tRNA processing was aberrant in vivo in mldr and scu mutants and with rswl RNAi knockdown. mt:tRNAs are found in different sequence milieux. To test a variety of upstream sequence context, we chose to examine tRNAs that are found downstream of rRNA (mt:tRNALeu(CUN) and mt:tRNAVal), mRNA (Cytochrome c oxidase subunit III (CoIII), mt:tRNAGly) and the A+T rich region (mt:tRNAIle) (Figure 5A, asterisks). mt:tRNA processing by Arabidopsis PRORPs appears to be highly efficient and go to completion at steady-state levels in vitro (46); thus, normally unprocessed tRNAs would not be predicted to be observable by northern analysis. As such, wild-type larvae consistently and completely processed each of the mt:tRNAs we examined, with no evidence of unprocessed higher molecular weight RNA intermediates (Figure 5B–E, y w). In contrast, the mutant alleles of mldr and scu, and the larvae expressing rswl RNAi knockdown accumulated precursor mitochondrial RNAs (Figure 5B–E). mt:tRNAIle and mt:tRNAGly are encoded in the same polycistron. After probing for mt:tRNAIle in mdlr mutants, we observed three distinct bands, in contrast to scu mutants and rswl RNAi knockdown. Probing for mt:tRNAGly, however, showed only a single band (Figure 5C). mt:tRNALeu(CUN) and mt:tRNAVal are in the same polycistron and when probing with each gave bands at approximately the same sizes for mldr and scu mutants and rswl knockdown.
Figure 5.
The mt:RNase P complex is required for mt:tRNA processing in Drosophila. (A) Diagram of Drosophila mitochondrial DNA structure (adapted from Xie and Dubrovsky). The five arrows indicate the predicted five transcribed polycistrons. Asterisks denote which mt:tRNAs were assayed via northern blot. ND = NADH dehydrogenase (Complex I), Cytb = ubiquinol: cytochrome c oxidoreductase (Complex III), Co = Cytochrome c oxidase (Complex IV), ATP = ATP synthase (Complex V). (B–E) Northern blots. mldr and scu mutant larvae contain unprocessed mt:tRNAs transcripts for mt:tRNAIle, mt:tRNAGly, mt:tRNAVal and mt:tRNALeu(CUN). Northern blots using extract from rswl RNAi knockdown larvae also show unprocessed, larger molecular weight species. Arrows indicate the major additional bands. sesB1 and SOD2 mutant larvae are used as controls. The mature tRNAs are indicated by the tRNA symbol at the bottom of the blot.
Overexpressing Mldr causes mitochondrial dysfunction
For multiprotein enzyme complexes, a decrease in any one protein often results in changes or loss of enzymatic function. Mutations in mldr and scu, as well as RNAi knockdown of rswl, resulted in pupal death, developmental delays and a decrease in mitochondrial function indicating each is essential in vivo. On the other hand, ectopically increasing enzyme levels can also lead to dominant negative effects through sequestering critical components from their normal function, or through mis-regulated activity. Using tubGAL4 for high, global expression, we found overexpressing UAS-mldr-GFP and UAS-rswl-myc resulted in lethality (Table 2). Larvae overexpressing mldr could develop to second instars, but often did not develop past first instar (Figure 6A). In contrast, larvae overexpressing rswl were able to develop to third instars (Figure 6B). In agreement with the lethality, both genotypes exhibited low levels of ATP production (Figure 6C). Larvae overexpressing scu had increased ATP levels relative to mldr and rswl, albeit lower than wild-type, which did not result in lethality (Figure 6C, Table 2). To determine whether mitochondrial morphology was affected, we examined germ cells. nanosGAL4 (nosGAL4) is expressed exclusively in the germline. The nosGAL4 stock we used does not give uniform expression in all developmental stages of the ovary, with lower expression in early germ cells and very high expression later (Figure 6J). When we examined mitochondrial morphology using immunofluorescence, we observed that in the lower expressing germ cells, mitochondria had the narrow, rod-shaped morphology characteristic of normal germ cell mitochondria (Figure 6D–F). However, mitochondria in germ cells with high expression of UAS-mldr had swollen, ring-shaped mitochondria indicative of a loss of selective permeability (Figure 6G–I). ATP synthase, which is found in the mitochondrial inner-membrane, labeled the outside of the ring-shaped mitochondria (Figure 6H, inset) whereas antibodies against Mldr, which is a mitochondrial matrix protein, labeled the inside of the mitochondrial rings (Figure 6G and I). Although overexpressing UAS-rswl-myc ubiquitously was lethal and decreased ATP levels, in contrast to UAS-mldr, mitochondrial morphology was normal (Figure 1I–K, Supplementary Figure S4). These results indicate that past a threshold expression level, Mldr causes dominant effects.
Table 2. Effect of mt:RNase P overexpression.
| tubGAL4/ TM3 Ser ActGFP x | Number of adults | |
|---|---|---|
| Balancer | non-Balancer | |
| UAS-rswl-myc-4a/Y | 299a | 0 |
| UAS-rswl-myc-5a/TM3 Sb | 163 | 3 |
| UAS-rswl-myc-1b/CyO | 167 | 1 |
| UAS-scu-FLAG-3a | 300 | 327 |
| UAS-scu-FLAG-3a/CyOb | 168 | 70 |
| UAS-scu-FLAG-4a/CyO | 199 | 84 |
| UAS-mldr-GFP-4b | 213 | 0 |
| UAS-mldr-GFP-2a/TM6b Hu Tb | 125 | 0 |
| UAS-mldr-GFP-4a/Y | 108a | 0 |
aincludes all males.
bsame insertion as 3a, maintained as a balanced stock.
DISCUSSION
Each component of mt:RNase P is required in vivo
PRORPs represent a new class of tRNA 5′-end processing enzymes that lack a catalytic RNA component, i.e. a ribozyme. In humans, PRORP requires the obligatory presence of a sub-complex containing MRPP1 and MRPP2 to catalyze the cleavage reaction. The complex was initially identified from mitochondrial extracts and while it has been localized to mitochondria in cell culture, the physiological significance of these proteins during development has not been previously addressed in vivo (47). Here, we have shown that Drosophila contains orthologs of each member of the mt:RNase P complex: Mulder (PRORP), Scully (MRPP2) and Roswell (MRPP1). Each member localizes to mitochondria, and its loss causes lethality. In addition, decreasing the amount of Mldr, Scu or Rswl causes an accumulation of unprocessed mt:tRNAs, supporting that they function in the mt:tRNA processing pathway and that the mitochondrial defects seen in the mutants (loss of ATP, mitochondrial morphology defects) may be due to a decrease in mature mt:tRNAs.
Mldr levels are the most critical in vivo
Overall, the lethality we observed in mldr and scu mutants and rswl RNAi knockdown was very similar and consistent. All were pupal lethal, and mldr and scu mutants showed delayed pupation. Even though mldrB and mldrC are point mutations, there are low levels of protein present (Figure 2E). While both alleles cause a lack of protein, mldrC (W465R) has a more severe developmental defect since 40% of the larvae do not pupate, suggesting that there is a difference in how any potential residual protein functions between the two alleles. The scu mutants we examined also showed differences in lethality, with scu4058 being the most severe which is surprising given that scuA is a larger truncation. One potential explanation is that the additional amino acids left in scu4058 allows it to sequester other proteins, acting as a dominant negative. The human MRPP2 ortholog must form a tetramer in order to function (48,49). The truncations for scu4058 and scuA, shown in only the blue monomer in Figure 2C, and color-coded with two lighter shades of blue, form the majority of the dimerization interface. It seems that even if these truncations were to result in viable coded proteins they would be unable to form a tetramer. Thus, it is likely these scu mutations could lead to disruption of tetramer formation with subsequent detrimental effects to the mt:RNase P function. In addition, MRPP2 (also known as HSD10 (HSD17B10 is the gene)) is known to function in the isoleucine biosynthetic pathway. However, patients suffering from HSD10 disease have symptoms that do not correlate with an increase in toxic metabolites due to misregulated isoleucine biosynthesis. Rather, the symptoms are related to mitochondrial dysfunction and apoptotic cell death (50). Thus, while MRPP2/HSD10 has dehydrogenase activity, it is likely the loss of mitochondrial function associated with MRPP2/HSD17B10 mutations is primarily due to its non-enzymatic function in the mt:RNase P.
In addition to the common phase of lethality, knockdown of mldr, scu and rswl exhibited greatly reduced ATP levels and accumulation of unprocessed mt:RNAs (Figures 3 and 5). However, our more detailed analyses of mitochondrial function revealed some critical differences between the three proteins. For example, only mldr RNAi caused an age-related decrease in locomotion when expressed in dopaminergic neurons (Figure 2I). In addition, mldr and scu mutants had swollen and ring-like mitochondria in neuroblasts, whereas rswl-RNAi did not (Figure 4). In general, when mitochondria have this morphology, it is indicative of a loss of selective permeability. Once this is lost, the mitochondrial outer membrane fragments, leading to a lack of ATP production. However, normal morphology does not necessarily mean ATP production is at appropriate levels. These data suggest that while all three proteins are essential, cells are most sensitive to reductions in Mldr.
Interestingly, while decreasing Mldr, Scu and Rswl caused lethality, overexpressing Mldr and Rswl had the same effect (Table 2). In contrast, overexpressing UAS-scu-FLAG in a wild-type background resulted in the expected Mendelian ratio of adults, suggesting that too much of Scu does not appear to alter the function of the complex, cause a deleterious increase in dehydrogenase activity or sequester any critical components in a dominant negative fashion. In contrast, overexpressing Mldr was lethal, led to decreased ATP levels and also caused defects in mitochondrial morphology (Figure 6). Overexpressing Rswl was lethal, but unlike Mldr, did not appear to change mitochondrial morphology as judged by immunofluorescence (Supplementary Figure S4). One possible explanation is that too much Mldr causes indiscriminate nucleotide cleavage in the mitochondrion resulting in a more severe effect. However, while the lethal outcome of Rswl overexpression may be more specific to mt:tRNA processing, we cannot rule out there is a concomitant deleterious increase in nonselective methyltransferase activity. It could be that an increase in indiscriminate methyltransferase activity is simply not as deleterious as nucleotide cleavage while still being important. Thus, decreasing and increasing Rswl expression causes a decrease in ATP and lethality, but does not give rise to any mitochondrial morphological changes. Looking at all of the phenotypes caused by lack and overexpression of the three mt:RNase P complex members, it is apparent based on our data that Drosophila is most sensitive to alterations in the levels of Mldr.
Decreasing Drosophila mt:RNase P complex proteins does not cause oxidative stress
We saw a consistent loss of ATP for mldr and scu mutants, as well as RNAi knockdown of mldr, scu and rswl. The ATP loss is very striking, and is not necessarily observed in other larvae mutant for proteins involved in various mitochondrial functions (29,43). However, we observed no accompanying oxidative stress by assaying mitochondrial aconitase function. Mitochondrial aconitase is well established as being exceedingly sensitive to oxidation, and thus loss of its function is often used as an indirect assay for an increased mitochondrial oxidative environment (45,51). Mutating Drosophila proteins involved in mitochondrial function does not always lead to oxidative stress in larvae (29). One possible reason is that since mitochondria fulfill a variety of biochemical functions, lowering levels of different pathways would not necessarily cause oxidative stress. Another possible reason is that there is evidence that larvae primarily use glycolysis, and not respiration, to maintain ATP levels (52). Respiration is primarily responsible for generating reactive oxygen species in mitochondria since the electron transport chain can be ‘leaky’.
RNAse Z is the enzyme responsible for 3′-end tRNA cleavage. Drosophila lacking mitochondrial RNAse Z have been reported to have cell cycle defects, as well as oxidative damage (43). In contrast, we did not observe any particularly small imaginal disks (indicative of cell cycle defects), and while larvae development showed a delay, the larval brains were of relatively normal size, as were the pupae that formed. It is still possible that mutant clones would be smaller relative to their wild-type siblings. Thus, it appears that while flies lacking either mt:RNAse Z or mt:RNase P accumulate unprocessed mitochondrial RNA species, they have different effects on ATP levels and oxidative stress (43).
Reducing Mldr, Scu and Rswl results in accumulation of unprocessed mitochondrial RNAs
Larvae mutant for mldr and scu, as well as rswl RNAi knockdown, accumulate unprocessed mt:RNAs (Figure 5). There are several conclusions that can be gleaned from these experiments. Wild-type larvae consistently and completely processed the four mt:tRNAs we examined, as did the sesB1 and SOD2 mutants – two genes mutated in unrelated processes (53–55). Thus, accumulation of mt:RNAs was specific for Mldr, Scu and Rswl and not due to general mitochondrial dysfunction. For mt:tRNAIle and mt:tRNAGly, which are encoded in the polycistron downstream of the A + T rich region, the amount of processed mt:tRNA was larger compared to mt:tRNAVal and mt:tRNALeu(CUN), both encoded in the polycistron that also encodes the large and small mt:rRNAs. There are several possible explanations for this observation. First, both ribozyme and protein-only RNase P enzymes have differential affinities and cleavage rates for some RNA substrates in vitro (46,56–58). Thus the differences in the amount of processed product could be due to different cleavage efficiencies of mt:RNase P for the two polycistrons. However, given that there is more processed product for mt:tRNAIle and mt:tRNAGly, the two polycistrons would also need to be differentially transcribed, as happens in humans (59). Since the same amount of total RNA was added for all the blots, it is possible the two polycistrons are differentially transcribed. However, counterintuitively, the polycistron encoding the mt:rRNAs appears to be at lower levels than the one downstream of the A + T rich region, in contrast to what is thought to happen for human mtDNA (59). The difference in transcript levels does not appear to be due to non-specific binding of the probes to other tRNAs, as performing BLAST analysis with each full-length mt:tRNA probe (Supplementary Table S2) did not recognize any other tRNAs. Alternatively, this could be explained by differences in probe binding, however, the unprocessed mt:mRNAs were of similar intensity for all the blots.
In addition, mldrC mutant larvae had the largest accumulation of unprocessed mt:tRNAs. This is consistent with it being the more severe mldr allele, as well as with our observations that Drosophila are most sensitive to perturbation in Mldr levels. For mt:tRNAIle processing, mldr mutants showed several band sizes of unprocessed mt:tRNAs, which may reflect the potential influence of mldr mutations on the specificity of the 5′ end cleavage (Figure 5B). In contrast, the scu mutations affected mt:tRNAIle 5′-end processing of the same cleavage sites equally. rswl RNAi also showed a variety of band sizes for mt:tRNAIle cleavage. By size, these bands cannot be clearly associated with putative RNA transcripts including tRNAIle. It is possible that these detected bands are products of miscleavage by Rswl deficient mt:RNase P, suggesting a potential role of Rswl in restricting substrate recognition.
mt:RNase P and disease
Many mitochondrial diseases are associated with point mutations in mtDNA (60). While mt:tRNAs encode for a relatively small proportion of mtDNA, over 50% of the disease-causing mutations found in mtDNA are found in the mt:tRNAs (61). However, most of these mt:tRNA mutations do not occur in the anticodon or discriminator bases that are necessary for codon recognition and aminoacylation. Therefore, these disease-causing mt:tRNA mutations likely impact such processes as post-transcriptional modifications, interaction with the translation machinery or mt:tRNA processing. There is evidence that normal mt:tRNA processing is critical as mutations in precursor mt:tRNAs that impair mt:tRNA 5′-end maturation, as well as in human PRORP, can give rise to disease (19–20,23,62). Studying how loss of Mldr, Scu or Rswl function affects cellular function is therefore important for promoting the understanding of the mechanisms of these mitochondrial diseases.
Supplementary Material
Acknowledgments
We would like to thank Drs Frank Shewmaker and Xin Xiang for critically reading the manuscript. We would also like to thank the USU Biomedical Instrumentation Core. The Drosophila Genomics Resource Center, supported by NIH grant 2P40OD010949-10A1, provided reagents. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and Vienna Drosophila Resource Center were used in this study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [1R01GM117141 to M.K.]; Uniformed Services University [USUHS BIO-71-3019 to R.T.C.]; National Institutes of Health/Department of Defense [CHIRP HU0001-14-2-0041 to R.T.C.]. Funding for open access charge: NIH R01 GM117141.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gissi C., Iannelli F., Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb.) 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- 2.Jackman J.E., Alfonzo J.D. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paris Z., Fleming I.M., Alfonzo J.D. Determinants of tRNA editing and modification: avoiding conundrums, affecting function. Semin. Cell Dev. Biol. 2012;23:269–274. doi: 10.1016/j.semcdb.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthier F., Renaud M., Alziari S., Durand R. RNA mapping on Drosophila mitochondrial DNA: precursors and template strands. Nucleic Acids Res. 1986;14:4519–4533. doi: 10.1093/nar/14.11.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres T.T., Dolezal M., Schlotterer C., Ottenwalder B. Expression profiling of Drosophila mitochondrial genes via deep mRNA sequencing. Nucleic Acids Res. 2009;37:7509–7518. doi: 10.1093/nar/gkp856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.M., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A., et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann R.K., Gossringer M., Spath B., Fischer S., Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Progress Mol. Biol. Translat. Sci. 2009;85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 10.Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochim. et Biophys. Acta. 2012;1819:1017–1026. doi: 10.1016/j.bbagrm.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 12.Holzmann J., Frank P., Löffler E., Bennett K.L., Gerner C., Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–536. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Gobert A., Gutmann B., Taschner A., Gossringer M., Holzmann J., Hartmann R.K., Rossmanith W., Giege P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 14.Taschner A., Weber C., Buzet A., Hartmann R.K., Hartig A., Rossmanith W. Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multicomponent RNA-protein complex. Cell Reports. 2012;2:19–25. doi: 10.1016/j.celrep.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzezniak L.K., Bijata M., Szczesny R.J., Stepien P.P. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–626. doi: 10.4161/rna.8.4.15393. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Nagao A., Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 18.Bindoff L.A., Howell N., Poulton J., McCullough D.A., Morten K.J., Lightowlers R.N., Turnbull D.M., Weber K. Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. A potential disease mechanism. J. Biological Chem. 1993;268:19559–19564. [PubMed] [Google Scholar]
- 19.Chatfield K.C., Coughlin C.R. 2nd, Friederich M.W., Gallagher R.C., Hesselberth J.R., Lovell M.A., Ofman R., Swanson M.A., Thomas J.A., Wanders R.J., et al. Mitochondrial energy failure in HSD10 disease is due to defective mtDNA transcript processing. Mitochondrion. 2015;21:1–10. doi: 10.1016/j.mito.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutschmann A.J., Amberger A., Zavadil C., Steinbeisser H., Mayr J.A., Feichtinger R.G., Oerum S., Yue W.W., Zschocke J. Mutation or knock-down of 17beta-hydroxysteroid dehydrogenase type 10 cause loss of MRPP1 and impaired processing of mitochondrial heavy strand transcripts. Hum. Mol. Genet. 2014;23:3618–3628. doi: 10.1093/hmg/ddu072. [DOI] [PubMed] [Google Scholar]
- 21.Li R., Guan M.X. Human mitochondrial leucyl-tRNA synthetase corrects mitochondrial dysfunctions due to the tRNALeu(UUR) A3243G mutation, associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms and diabetes. Mol. Cell. Biol. 2010;30:2147–2154. doi: 10.1128/MCB.01614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossmanith W., Karwan R.M. Characterization of human mitochondrial RNase P: novel aspects in tRNA processing. Biochem. Biophys. Res. Commun. 1998;247:234–241. doi: 10.1006/bbrc.1998.8766. [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Li R., Fettermann A., Li Z., Qian Y., Liu Y., Wang X., Zhou A., Mo J.Q., Yang L., et al. Maternally inherited essential hypertension is associated with the novel 4263A>G mutation in the mitochondrial tRNAIle gene in a large Han Chinese family. Circ. Res. 2011;108:862–870. doi: 10.1161/CIRCRESAHA.110.231811. [DOI] [PubMed] [Google Scholar]
- 24.Haack T.B., Kopajtich R., Freisinger P., Wieland T., Rorbach J., Nicholls T.J., Baruffini E., Walther A., Danhauser K., Zimmermann F.A., et al. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2013;93:211–223. doi: 10.1016/j.ajhg.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilardo E., Rossmanith W. Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 2015;43:5112–5119. doi: 10.1093/nar/gkv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarham J.W., Elson J.L., Blakely E.L., McFarland R., Taylor R.W. Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. RNA. 2010;1:304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 27.Lewis D.L., Farr C.L., Kaguni L.S. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect. Mol. Biol. 1995;4:263–278. doi: 10.1111/j.1365-2583.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 28.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 29.Sen A., Damm V.T., Cox R.T. Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage. PLoS One. 2013;8:e54283. doi: 10.1371/journal.pone.0054283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen A., Kalvakuri S., Bodmer R., Cox R.T. Clueless, a protein required for mitochondrial function, interacts with the PINK1-Parkin complex in Drosophila. Dis. Model Mech. 2015;8:577–589. doi: 10.1242/dmm.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torroja L., Ortuno-Sahagun D., Ferrus A., Hammerle B., Barbas J.A. scully, an essential gene of Drosophila, is homologous to mammalian mitochondrial type II L-3-hydroxyacyl-CoA dehydrogenase/amyloid-beta peptide-binding protein. J. Cell Biol. 1998;141:1009–1017. doi: 10.1083/jcb.141.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 34.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F., Liu X., Zhou W., Yang X., Shen Y. Auto-inhibitory mechanism of the human mitochondrial RNase P protein complex. Sci. Rep. 2015;5:9878. doi: 10.1038/srep09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinhard L., Sridhara S., Hallberg B.M. Structure of the nuclease subunit of human mitochondrial RNase P. Nucleic Acids Res. 2015;43:5664–5672. doi: 10.1093/nar/gkv481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lustbader J.W., Cirilli M., Lin C., Xu H.W., Takuma K., Wang N., Caspersen C., Chen X., Pollak S., Chaney M., et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 38.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haelterman N.A., Jiang L., Li Y., Bayat V., Sandoval H., Ugur B., Tan K.L., Zhang K., Bei D., Xiong B., et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 2014;24:1707–1718. doi: 10.1101/gr.174615.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirby K., Hu J., Hilliker A.J., Phillips J.P. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. U S A. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macchi M., El Fissi N., Tufi R., Bentobji M., Lievens J.C., Martins L.M., Royet J., Rival T. The Drosophila inner-membrane protein PMI controls crista biogenesis and mitochondrial diameter. J. Cell. Sci. 2013;126:814–824. doi: 10.1242/jcs.115675. [DOI] [PubMed] [Google Scholar]
- 42.Sandoval H., Yao C.K., Chen K., Jaiswal M., Donti T., Lin Y.Q., Bayat V., Xiong B., Zhang K., David G., et al. Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. Elife. 2014;3:e03558. doi: 10.7554/eLife.03558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X., Dubrovsky E.B. Knockout of Drosophila RNase ZL impairs mitochondrial transcript processing, respiration and cell cycle progression. Nucleic Acids Res. 2015;43:10364–10375. doi: 10.1093/nar/gkv1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flint D.H., Tuminello J.F., Emptage M.H. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 45.Gardner P.R., Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 46.Howard M.J., Karasik A., Klemm B.P., Mei C., Shanmuganathan A., Fierke C.A., Koutmos M. Differential substrate recognition by isozymes of plant protein-only Ribonuclease P. RNA. 2016;22:782–792. doi: 10.1261/rna.055541.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez M.I., Mercer T.R., Davies S.M., Shearwood A.M., Nygard K.K., Richman T.R., Mattick J.S., Rackham O., Filipovska A. RNA processing in human mitochondria. Cell Cycle. 2011;10:2904–2916. doi: 10.4161/cc.10.17.17060. [DOI] [PubMed] [Google Scholar]
- 48.Kissinger C.R., Rejto P.A., Pelletier L.A., Thomson J.A., Showalter R.E., Abreo M.A., Agree C.S., Margosiak S., Meng J.J., Aust R.M., et al. Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics. J. Mol. Biol. 2004;342:943–952. doi: 10.1016/j.jmb.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 49.Powell A.J., Read J.A., Banfield M.J., Gunn-Moore F., Yan S.D., Lustbader J., Stern A.R., Stern D.M., Brady R.L. Recognition of structurally diverse substrates by type II 3-hydroxyacyl-CoA dehydrogenase (HADH II)/amyloid-beta binding alcohol dehydrogenase (ABAD) J. Mol. Biol. 2000;303:311–327. doi: 10.1006/jmbi.2000.4139. [DOI] [PubMed] [Google Scholar]
- 50.Rauschenberger K., Scholer K., Sass J.O., Sauer S., Djuric Z., Rumig C., Wolf N.I., Okun J.G., Kolker S., Schwarz H., et al. A non-enzymatic function of 17beta-hydroxysteroid dehydrogenase type 10 is required for mitochondrial integrity and cell survival. EMBO Mol. Med. 2010;2:51–62. doi: 10.1002/emmm.200900055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner P.R., Nguyen D.D., White C.W. Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and in rat lungs. Proc. Natl Acad. Sci. U.S.A. 1994;91:12248–12252. doi: 10.1073/pnas.91.25.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tennessen J.M., Baker K.D., Lam G., Evans J., Thummel C.S. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metabol. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celotto A.M., Frank A.C., McGrath S.W., Fergestad T., Van Voorhies W.A., Buttle K.F., Mannella C.A., Palladino M.J. Mitochondrial encephalomyopathy in Drosophila. J. Neurosci. 2006;26:810–820. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fergestad T., Bostwick B., Ganetzky B. Metabolic disruption in Drosophila bang-sensitive seizure mutants. Genetics. 2006;173:1357–1364. doi: 10.1534/genetics.106.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y.Q., Roote J., Brogna S., Davis A.W., Barbash D.A., Nash D., Ashburner M. stress sensitive B encodes an adenine nucleotide translocase in Drosophila melanogaster. Genetics. 1999;153:891–903. doi: 10.1093/genetics/153.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu F., Altman S. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell. 1994;77:1093–1100. doi: 10.1016/0092-8674(94)90448-0. [DOI] [PubMed] [Google Scholar]
- 57.Sugita C., Komura Y., Tanaka K., Kometani K., Satoh H., Sugita M. Molecular characterization of three PRORP proteins in the moss Physcomitrella patens: nuclear PRORP protein is not essential for moss viability. PLoS One. 2014;9:e108962. doi: 10.1371/journal.pone.0108962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koutmou K.S., Zahler N.H., Kurz J.C., Campbell F.E., Harris M.E., Fierke C.A. Protein-precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial ribonuclease P. J. Mol. Biol. 2010;396:195–208. doi: 10.1016/j.jmb.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terzioglu M., Ruzzenente B., Harmel J., Mourier A., Jemt E., Lopez M.D., Kukat C., Stewart J.B., Wibom R., Meharg C., et al. MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metabol. 2013;17:618–626. doi: 10.1016/j.cmet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 60.Wallace D.C. Diseases of the mitochondrial DNA. Annual Rev. Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rossmanith W., Karwan R.M. Impairment of tRNA processing by point mutations in mitochondrial tRNA(Leu)(UUR) associated with mitochondrial diseases. FEBS Lett. 1998;433:269–274. doi: 10.1016/s0014-5793(98)00928-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.