Abstract
During DNA double-strand break and replication fork repair by homologous recombination, the RAD51 recombinase catalyzes the DNA strand exchange reaction via a helical polymer assembled on single-stranded DNA, termed the presynaptic filament. Our published work has demonstrated a dual function of the SWI5–SFR1 complex in RAD51-mediated DNA strand exchange, namely, by stabilizing the presynaptic filament and maintaining the catalytically active ATP-bound state of the filament via enhancement of ADP release. In this study, we have strived to determine the basis for physical and functional interactions between Mus musculus SWI5–SFR1 and RAD51. We found that SWI5–SFR1 preferentially associates with the oligomeric form of RAD51. Specifically, a C-terminal domain within SWI5 contributes to RAD51 interaction. With specific RAD51 interaction defective mutants of SWI5–SFR1 that we have isolated, we show that the physical interaction is indispensable for the stimulation of the recombinase activity of RAD51. Our results thus help establish the functional relevance of the trimeric RAD51–SWI5–SFR1 complex and provide insights into the mechanistic underpinnings of homology-directed DNA repair in mammalian cells.
INTRODUCTION
Homologous recombination (HR) represents an important error-free mechanism for the elimination of DNA double-strand breaks (DSBs) and the restoration of DNA replication forks (RFs) that have stalled or been injured. By its involvement in the repair of chromosome damage, HR is indispensable for maintaining genomic integrity and stability (1,2). Accordingly, HR dysfunction is a major cause of chromosome fragility and cancer susceptibility (3–5).
HR is catalyzed by RAD51 recombinase and its cohort of accessory factors. RAD51 polymerizes on ssDNA generated from the nucleolytic processing of a DSB or a damaged RF, to form a helical protein filament, known as the presynaptic filament. The presynaptic filament catalyzes the search for an intact homologous donor duplex DNA and DNA strand exchange with the donor sequence (6,7). The recombinase activity of RAD51 is regulated by several associated partners, including BRCA2, RAD51AP1 and SWI5–SFR1 (5–10). These and other RAD51 recombinase accessory factors fulfill mechanistically distinct functions in the execution of HR. For example, cytological studies have provided strong evidence that the recruitment of RAD51 to DNA damage as measured by nuclear protein focus formation depends on BRCA2 (5,11,12). Biochemically, purified BRCA2 promotes RAD51 filament assembly on ssDNA over dsDNA and functions as a recombination mediator to assemble the presynaptic filament on ssDNA occupied by the single-strand DNA binding protein RPA. Studies published by several groups have provided mechanistic information regarding the action of BRCA2 as a HR mediator (13–17). On the other hand, RAD51AP1 does not appear to affect presynaptic formation, but, rather, it functions with the presynaptic filament to capture duplex DNA and to facilitate the assembly of the synaptic complex in which the recombining ssDNA and duplex target are homologously aligned and limited base pairing has occurred (18,19).
Cell-based analyses in the fission yeast and mammalian cells have implicated the proteins encoded by the SWI5 and SFR1 genes in RAD51-dependent HR and recombination repair of DNA damage (20–23). In agreement with this, biochemical and biophysical analyses have shown that the fission yeast and mouse SWI5 and SFR1 proteins form a heterodimeric complex that physically interacts with RAD51 and enhances RAD51-mediated DNA strand exchange (24–27). Importantly, working with highly purified mouse proteins, we have furnished evidence that the enhancement of RAD51 activity stems from a dual action of SWI5–SFR1, namely, by stabilizing the presynaptic filament and enhancing the release of ADP from the filament to maintain the presynaptic filament in its active, ATP-bound form (27,28). In this study, we provide evidence that SWI5–SFR1 interacts with the oligomeric, but not the monomeric, form of RAD51. Importantly, in this study, we have found that the C-terminal region of SWI5 makes a major contribution toward complex formation between SWI5–SFR1 and RAD51. Based on this information, we have identified a mutant variant of SWI5 that is proficient in complex formation with SFR1 but is strongly affected for the ability to interact with RAD51. Our biochemical analyses have revealed that the mutant SWI5–SFR1 complex is devoid of any ability to stabilize the RAD51 presynaptic filament, to enhance ATP hydrolysis by RAD51, or to stimulate RAD51-mediated DNA strand exchange. Our results thus provide insights into the nature and functional relevance of the higher order complex consisting of the recombinase RAD51 and SWI5–SFR1 in HR.
MATERIALS AND METHODS
Details on the DNA substrates, construction of expression plasmids, and protein expression and purification are described in the Supplementary Data.
Affinity pulldown
For examining the interaction of RAD51 with SWI5–SFR1, 6 μg of mouse SWI5–SFR1 (or mutant variants) containing a (His)6 tag in SFR1's amino-terminus was incubated with 4 μg strep-tagged mouse RAD51 (or mutant variants) and the indicated amount of mouse BRC4 peptide in 30 μl of buffer A (25 mM Tris–HCl, pH 7.5, 10% glycerol, 0.01% Igepal, 100 mM KCl, 1 mM 2-mercaptoethanol and 5 mM imidazole) for 30 min at 4°C. After being mixed with 10 μl of Talon resin for 45 min at 4°C to capture the (His)6-tagged SWI5–SFR1 and associated RAD51, the resin was washed three times with 30 μl of buffer A and then treated with 25 μl of 2% SDS to elute proteins. The supernatant, last wash, and SDS eluate, 10 μl each, were analyzed by 15% SDS-PAGE and Coomassie Blue staining.
Limited proteolysis
Full-length SWI5–SFR1 (3 μg) was digested with proteinase K (2 nM) for the indicated times at 37°C. Digestion was terminated by boiling the samples in SDS-PAGE loading buffer (2% SDS, 50 mM Tris–HCl, pH 6.8, 10% glycerol, 715 mM 2-mercaptoethanol and 0.02% Bromophenol blue). The proteolytic products were resolved by 15% SDS-PAGE and visualized by Coomassie Blue staining. The proteolytic fragments were in-gel digested by trypsin and identified by N-terminal sequencing.
DNA strand exchange
All the reaction steps were carried out at 37°C. The 80-mer Oligo 1 (4.8 μM nucleotides) was incubated with 1.6 μM RAD51 in 10.5 μl of buffer B (35 mM Tris–HCl, pH 7.5, 1 mM DTT, 2.5 mM MgCl2, 35 mM KCl and 100 ng/ml BSA) containing 1 mM ATP for 5 min. The indicated amount of SWI5–SFR1 or mutant variant was then added in 1 μl, followed by a 5-min incubation. The reaction was initiated by adding homologous 32P-labeled 40-mer duplex (2.4 μM base pairs) to 12.5 μl of final volume. After 20 min incubation, a 5 μl aliquot was removed and mixed with an equal volume of 1% SDS containing proteinase K (1 mg/ml) and incubated at 37°C for 10 min. The reaction mixtures were resolved in 10% polyacrylamide gels in TBE buffer (89 mM Tris, 89 mM borate, and 2 mM EDTA, pH 8.0) and the DNA species in dried gels were quantified by phosphorimaging analysis in a Personal FX phosphorimager using the Quantity One software (Bio-Rad).
Exonuclease I protection
RAD51 (1.3 μM) was incubated with 5′-32P-labeled 80-mer Oligo 1 ssDNA (3 μM nucleotides) in 18 μl buffer B containing 0.1 mM ATP at 37°C for 5 min. Following the incorporation of the indicated amount of SWI5–SFR1 or mutant variants and a 5-min incubation, exonuclease I (1.5 units; New England Biolabs) was added to the reaction mixture (20 μl final volume). After 20 min of incubation at 37°C, a 10 μl aliquot was removed and mixed with 2.5 μl of 60 mM EDTA, 0.5% SDS, and proteinase K (1 mg/ml) and incubated at 37°C for 10 min. The samples were subjected to electrophoresis, gel drying, and phosphorimaging analysis.
Analytical ultracentrifugation
In order to determine their molar mass, the sedimentation velocity of SWI5–SFR1 and SWI5 F83A/L85A-SFR1 was examined in a Beckman-Coulter XL-A analytical ultracentrifuge. The proteins were loaded onto standard double sector cells with an Epon charcoal-filled centerpiece and centrifugation was performed at 20°C and 50 000 rpm. The cells were scanned at 280 nm in a continuous mode, and the experimental data were analyzed by the Sedfit program (version 14.1). After the ultracentrifugation experiments, the protein samples were visually checked for clarity, and no indication of precipitation was found.
Circular dichroism analysis
The structures of SWI5–SFR1 and SWI5 F83A/L85A-SFR1 (3.5 μM) were determined by Circular Dichroism (CD) analysis in a Jasco J-815 spectropolarimeter under constant N2 flush. The far UV CD spectra were measured at 195–250 nm with a 1.0 bandwidth and a 0.1 nm resolution at a scan speed of 20 nm/min using a 1 mm path length quartz cuvette. All CD spectra were corrected for their respective buffer blanks and three repetitive scans were averaged at 25°C.
ATPase activity
To examine the effect of SWI5–SFR1 or SWI5 F83A/L85A-SFR1 on ATP hydrolysis by RAD51, RAD51 (1.6 μM) was incubated in 12.5 μl of buffer B containing the final concentration of 0.5 mM ATP, and with or without 80-mer Oligo 1 ssDNA (4.8 μM nucleotides) at 37°C for 5 min, followed by the addition of the indicated amount of SWI5–SFR1 or SWI5 F83A/L85A-SFR1, and then a 5-min incubation. After adding 0.3 μCi [γ-32P] ATP, 1.5 μl aliquots of the reaction mixtures were removed at the indicated times and mixed with an equal volume of 500 mM EDTA to halt the reaction. The level of ATP hydrolysis was determined by thin layer chromatography in polyethyleneimine cellulose sheets (Fluka) with phosphorimaging analysis. The apparent velocity (Vapp.) was calculated before 10% of the radiolabeled ATP had been hydrolyzed.
RESULTS
SWI5–SFR1 interacts with the oligomeric form of RAD51
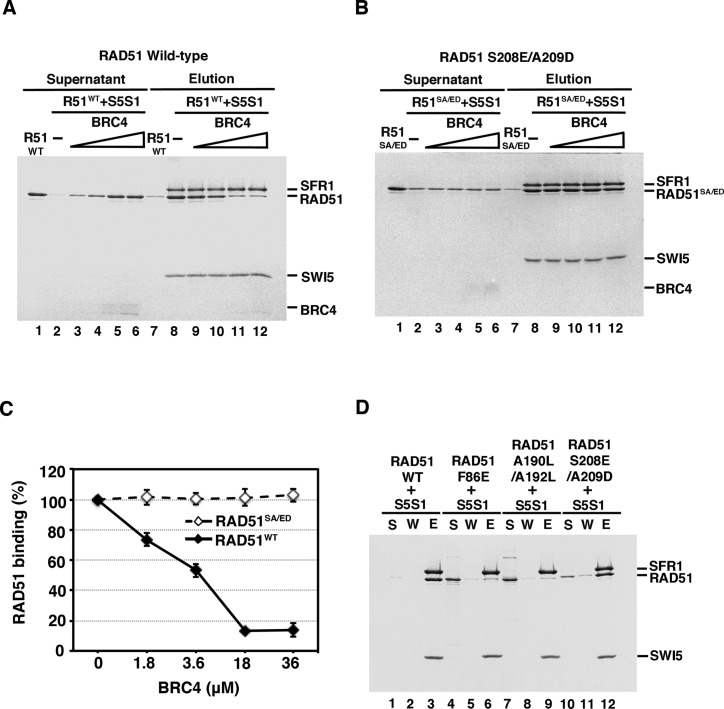
Our previous work has revealed that mouse SWI5–SFR1 physically interacts with mouse RAD51 and stimulates its DNA strand exchange activity partly via the stabilization of the presynaptic filament (27). We wished to delineate how physical interaction between RAD51 and SWI5–SFR1 contributes to the stabilization of the presynaptic filament and to the stimulation of RAD51-mediated DNA strand exchange. RAD51 exists as an oligomeric structure in solution (29–31), and it has been documented that the C-terminal domain and the BRC repeats of BRCA2 interact with the oligomeric form and the monomeric form of RAD51, respectively (30,31). These results prompted us to ask whether the oligomeric state of RAD51 is germane for SWI5–SFR1 interaction. We addressed this question by first obtaining the monomeric form of RAD51 to examine its interaction status with SWI5–SFR1. To do so, we used the BRC4 peptide from the mouse BRCA2 protein to dissociate the oligomeric form of mouse RAD51 into monomers, as verified by gel filtration analysis (Supplementary Figure S1, (32)). Next, the monomerized RAD51 was incubated with SWI5–SFR1 complex and then Talon resin (which recognizes the (His)6 tag on SFR1) was used to pulldown SWI5–SFR1 and any associated RAD51. We found that the interaction between SWI5–SFR1 and RAD51 was significantly decreased by increasing amounts of the BRC4 peptide (Figure 1A and C), suggesting that SWI5–SFR1 has the highest affinity for the RAD51 oligomer. We verified that BRC4 and SWI5–SFR1 do not physically interact (Supplementary Figure S2). To further confirm our results, we generated the RAD51 S208E/A209D mutant that is defective in interaction with BRC4 and verified that it remains oligomeric upon incubation with BRC4 (Supplementary Figure S1, (31,33)). Importantly, as shown in Figure 1B and C, BRC4 had little or no effect on the interaction of RAD51 S208E/A209D with SWI5–SFR1 (Figure 1B and C).
Figure 1.
SWI5–SFR1 interacts with the oligomeric form of RAD51. For affinity pulldown, purified SWI5–SFR1 (S5S1) was incubated with either RAD51 wild-type (A) or RAD51 S208E/A209D (B) in the presence of an increasing concentration of the BRC4 peptide. Reaction mixtures were mixed with Talon resin to capture protein complex through the (His)6 tag on SFR1. The supernatant (lanes 1–6) and SDS eluate (lanes 7–12) from the pulldown reactions were analyzed by 15% SDS-PAGE with Coomassie Blue staining. RAD51 alone was included as a control (lanes 1 & 7). (C) The results from (A) and (B) were quantified and plotted, and error bars represent the standard deviation (±SD) calculated based on at least three independent experiments. (D) SWI5–SFR1 was incubated with RAD51 wild-type (WT), F86E, A190L/A192L, or S208E/A209D for affinity pulldown analyses. The supernatant (S), wash (W), and SDS eluate (E) fractions from the pulldown reactions were analyzed by 15% SDS-PAGE with Coomassie Blue staining. Symbols: R51, RAD51; R51SA/ED, RAD51 S208E/A209D; S5S1, SWI5–SFR1.
It has been well documented that the human RAD51 F86E and A190L/A192L mutant variants are monomeric in nature (30,31,33,34). As expected, both mouse mutant variants also exhibit monomeric status (Supplementary Figure S3, (31)). Then we wished to further address whether these monomeric RAD51 mutant proteins are capable of interaction with SWI5–SFR1. Consistent with results presented earlier, neither RAD51 F86E nor A190L/A192L could interact with SWI5–SFR1 (Figure 1D). The oligomeric RAD51 S208E/A209D mutant was included as a positive interaction control in this (Figure 1D, lanes 10–12). In conclusion, our results provide evidence that oligomeric state of RAD51 facilitates stable association with SWI5–SFR1.
The amino-terminal half of SFR1 is dispensable for complex formation with SWI5 and interaction with RAD51
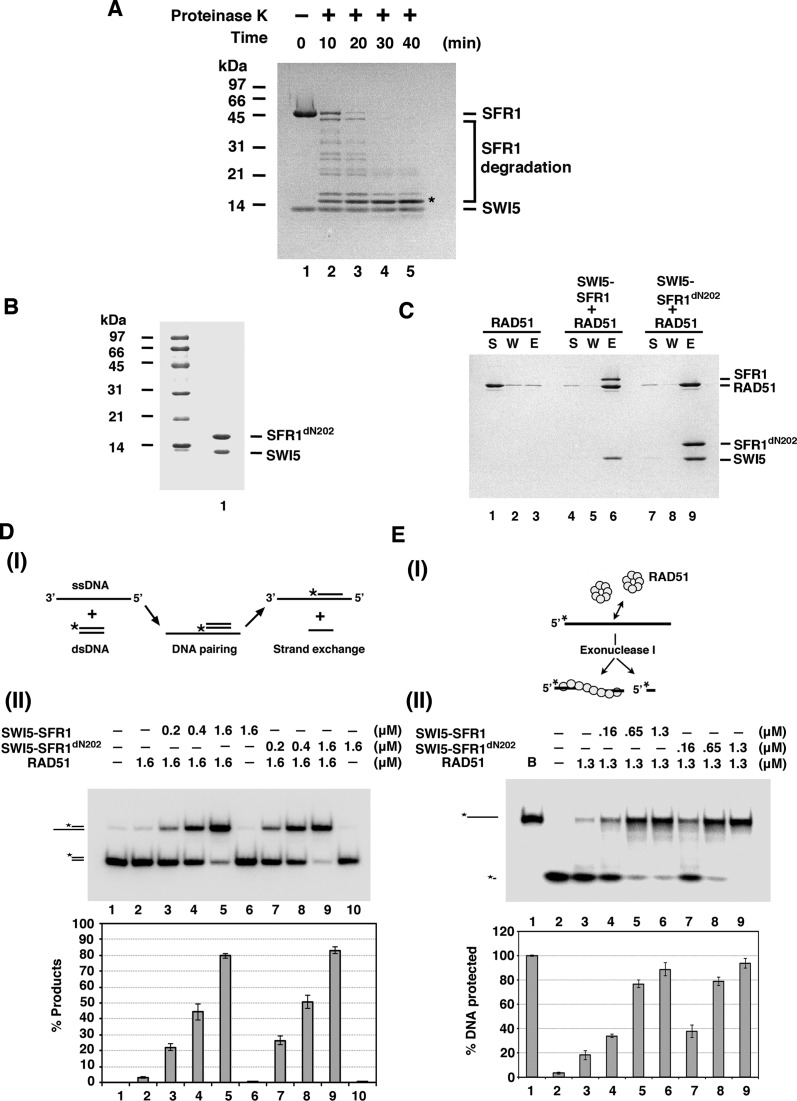
To delineate the functional significance of the complex of SWI5–SFR1 and RAD51, we proceeded to map the region in SWI5–SFR1 needed for RAD51 interaction. Our previous study showed that neither SWI5 nor SFR1 alone interacts with RAD51 (27). Thus, we directed our effort at identifying a mutant variant of the SWI5–SFR1 complex that specifically lacks the ability to interact with RAD51. To do so, we used limited proteolysis approach to isolate the core of the SWI5–SFR1 complex. As shown in Figure 2A, SFR1 was digested into a small fragment around 15 kDa (Figure 2A). Importantly, amino-terminal sequencing of the proteolytic fragment showed that it lacks the first 202 amino acid residues of the SFR1 protein (data not shown). Importantly, the SFR1dN202 mutant protein lacking these residues remains competent for stable complex formation with SWI5 (Figure 2B) and maintains a 1:1 stoichiometric ratio with the latter in the assembled complex as verified by analytical ultracentrifugation (Supplementary Figure S4). Next, we examined the SWI5–SFR1dN202 mutant complex for physical and functional interactions with RAD51. We found that SWI5–SFR1dN202 retains the ability to (1) interact with RAD51 (Figure 2C), (2) stimulate RAD51-mediated DNA strand exchange (Figure 2D) and (3) stabilize the RAD51 presynaptic filament (Figure 2E). Thus, the first 202 amino acid residues of SFR1 are dispensable for complex formation with SWI5 and physical/functional interactions with RAD51 as well.
Figure 2.
Functional characterization of SWI5–SFR1dN202 complex. (A) To map the minimal complex unit of SWI5–SFR1, purified SWI5–SFR1 was incubated with proteinase K for the indicated times. The proteolytic products were resolved by 15% SDS-PAGE and stained by Coomassie Blue staining. The asterisk denotes the SFR1dN202. (B) Purified SWI5–SFR1dN202 complex (3 μg) was subjected to 15% SDS-PAGE and Coomassie Blue staining. (C) SWI5–SFR1dN202 was tested for RAD51 interaction by affinity pulldown as described in Figure 1D. (D) DNA strand exchange assay to monitor the stimulatory effect of SWI5–SFR1 by RAD51. (I) Schematic of the DNA strand exchange assay. The radiolabeled substrate and product are visualized and quantified by phosphorimaging analysis after PAGE. The asterisk denotes the 32P label. (II) DNA strand exchange was conducted with the indicated amounts of SWI5–SFR1 or SWI5–SFR1dN202. The results were graphed. (E) Exonuclease I protection assay to monitor the influence of SWI5–SFR1 on the stability of RAD51 filament. (I) Schematic of the exonuclease I protection assay. The RAD51 filament harboring 5′-32P-labeled DNA is challenged with exonuclease I. The radiolabeled DNA and product are visualized and quantified by phosphorimaging analysis after PAGE. The 32P label is denoted by the asterisk. (II) Treatment of the RAD51 presynaptic filament with exonuclease I in the presence of the indicated concentrations of SWI5–SFR1 or SWI5–SFR1dN202. The results were graphed. (D-E) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments.
The carboxyl-terminal region of SWI5 is critically important for RAD51 interaction
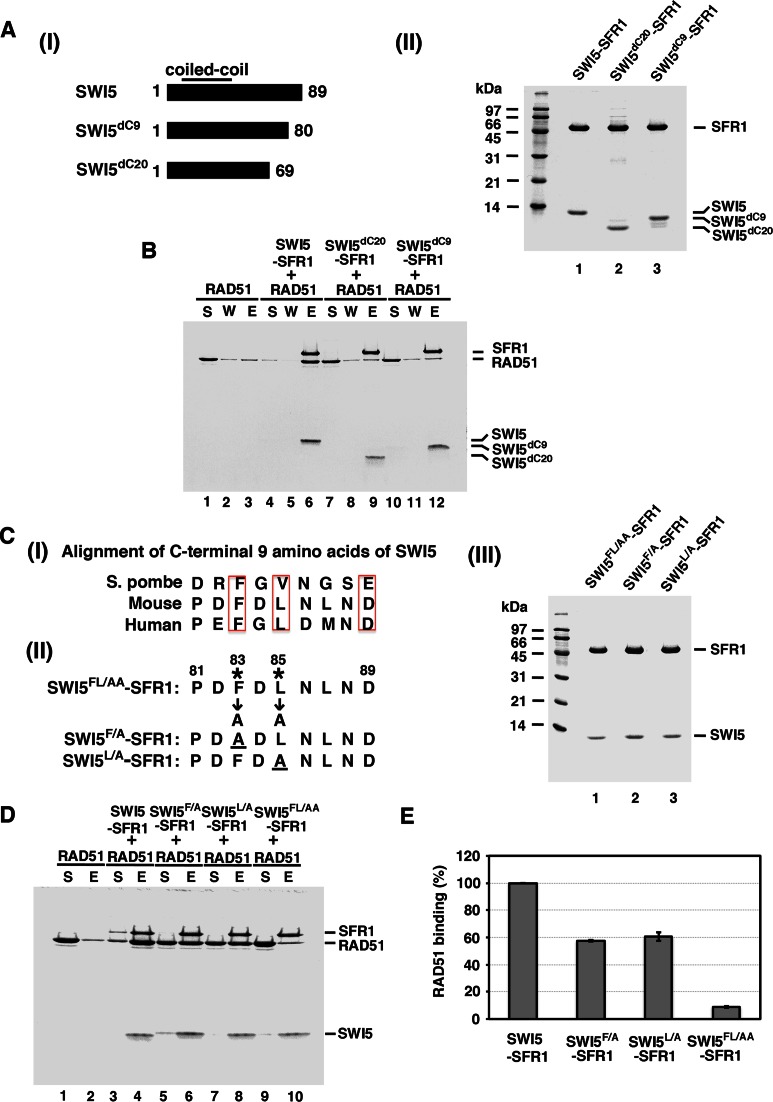
Complex formation between SWI5 and SFR1 is mediated by coiled-coil domains located within the N-terminal region of SWI5 and the C-terminal region of SFR1 (22,26). Since much of the N-terminal region of SFR1 is dispensable for RAD51 interaction, we turned our attention to the C-terminal part of SWI5 in our attempt to isolate a RAD51 interaction defective variant of SWI5–SFR1. To this end, we examined the effect of deleting the last 9 or 20 amino acid residues from SWI5 on RAD51 interaction (Figure 3A, panel I). We could obtain highly purified complexes of SWI5–SFR1 that harbor these mutant SWI5 (SWI5dC9 and SWI5dC20) proteins (Figure 3A, panel II). Importantly, we found that the mutant SWI5–SFR1 complexes are significantly defective in RAD51 interaction (Figure 3A and B). The results thus indicate that the last nine residues of SWI5 are indispensable for the interaction between SWI5–SFR1 and RAD51. An alignment of the nine residues from SWI5 orthologs from yeast, mouse to human led the identification of three highly conserved residues, namely, F83, L85 and D89 in the mouse protein (Figure 3C, panel I). We mutated these three amino acids to alanine to generate the mutant variants with either a single or a double substitution (Figure 3C, panel II). We first verified that these SWI5 mutants are proficient in complex formation with SFR1 and then tested the mutant complexes for RAD51 interactions. The results showed that (1) substitution of F83 or L85 to alanine (the F83A and L85A mutations) in SWI5 does not affect complex formation with SFR1 but attenuates the affinity of the mutant complexes for RAD51, and (2) the F83A/L85A double mutation again has no effect on complex formation with SFR1 but it greatly diminishes the affinity of the mutant complex for RAD51 (Figure 3C–E). The results therefore demonstrated that the conserved SWI5 residues F83 and L85 are indispensable for RAD51 interaction. We note that we have not been able to verify whether D89 in SWI5 is also important for RAD51 interaction, as the SWI5D89A–SFR1 mutant complex that we constructed is highly susceptible to proteolysis during purification (data not shown).
Figure 3.
SWI5dC20–SFR1, SWI5dC9–SFR1 and SWI5FL/AA–SFR1 are defective in RAD51 interaction. (A) (I) Schematic of C-terminal truncation of SWI5 mutant variants made in this study. (II) Purified SWI5–SFR1, SWI5dC20–SFR1 and SWI5dC9–SFR1 were resolved by 15% SDS-PAGE and stained with Coomassie Blue. (B) Purified SWI5dC20–SFR1 and SWI5dC9–SFR1 mutant complexes were examined for RAD51 interaction by affinity pulldown through the (His)6 tag on SFR1. Wild-type SWI5–SFR1 was included as positive control. (C) (I) Alignment of the C-terminal 9 amino acid residues of SWI5 from S. pombe, mouse, and human. (II) The SWI5–SFR1 mutants made in this study. (III) Purified SWI5 F83A/L85A–SFR1 (SWI5FL/AA–SFR1), SWI5 F83A–SFR1 (SWI5F/A–SFR1) and SWI5 L85A–SFR1 (SWI5L/A–SFR1) were resolved by 15% SDS-PAGE and stained with Coomassie Blue. (D) Purified SWI5FL/AA–SFR1, SWI5F/A–SFR1 and SWI5L/A–SFR1 mutants were examined for RAD51 interaction by affinity pulldown. (E) The results from (D) were graphed. Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments.
Physical interaction is prerequisite for RAD51 activity regulated by SWI5–SFR1
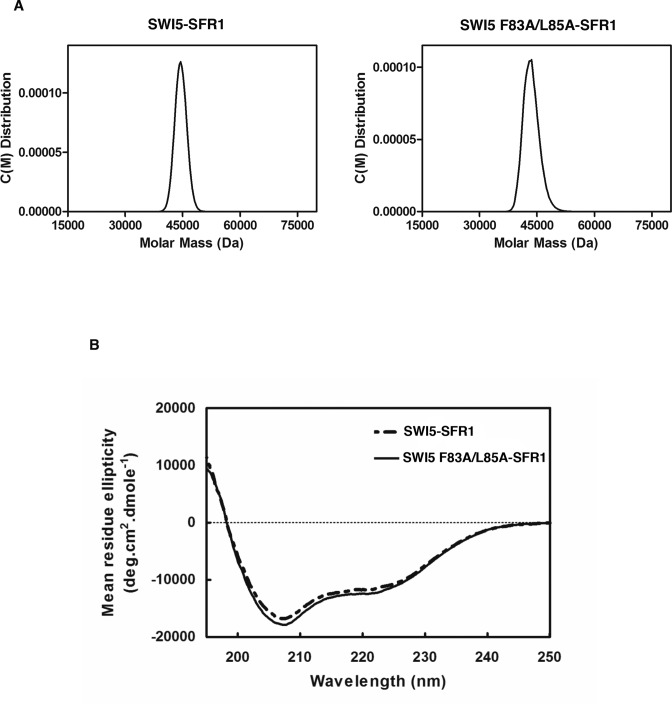
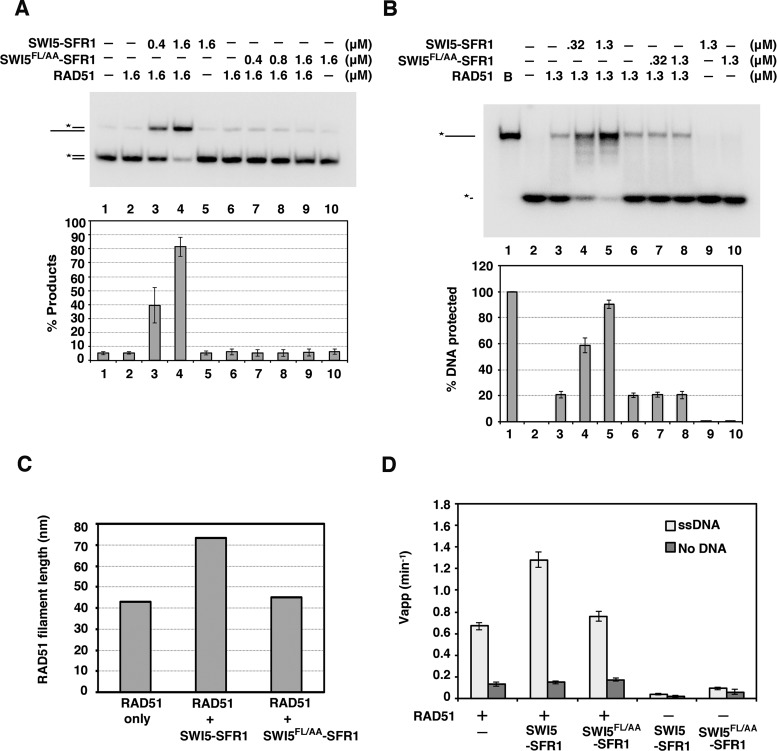
The isolation of the SWI5 F83A/L85A (SWI5FL/AA) mutant provided a valuable tool to address whether RAD51 interaction is important for the function of SWI5–SFR1 in RAD51-mediated DNA strand exchange. First, by analytical ultracentrifugation, we verified that SWI5FL/AA–SFR1 retains the heterodimeric nature of the wild-type complex (Figure 4A). Using circular dichrorism, we also confirmed that the mutant complex has the same global protein structure as its wild-type counterpart (Figure 4B). Then, we asked whether SWI5FL/AA–SFR1 is capable of stimulating RAD51-catalyzed DNA strand exchange. Importantly, the results revealed that SWI5FL/AA–SFR1 is devoid of any stimulatory effect on the strand exchange reaction (Figure 5A). Our previous studies provided evidence that SWI5–SFR1 enhances DNA strand exchange by stabilizing the presynaptic filament and facilitating the release of ADP to maintain the ATP-bound, active state of the presynaptic filament (27,28). To assess whether the SWI5FL/AA–SFR1 is able to stabilize the presynaptic filament, protection of ssDNA bound within the presynaptic filament against digestion by E. coli Exo I was examined, as in our published work (27). While, as expected (27), the wild-type SWI5–SFR1 complex provided protection against Exo I action, SWI5FL/AA–SFR1 was unable to do so (Figure 5B). To corroborate the above findings, we performed electron microscopy (EM) to compare the stability of the presynaptic filament without or with wild-type SWI5–SFR1 or SWI5FL/AA–SFR1. We collected 225 filaments for each group from EM images to obtain the average length of RAD51 filaments (Supplementary Figure S5). As shown in Figure 5C, the average length of the presynaptic filament generated in the presence of SWI5FL/AA-SFR1 is shorter than wild-type SWI5–SFR1 was used, again indicating that SWI5FL/AA–SFR1 lacks the presynaptic filament stabilization attribute of the wild-type complex (Figure 5C). Next, we compared the wild-type and mutant SWI5–SFR1 complexes for the ability to enhance ATP hydrolysis by the presynaptic filament. Again, the SWI5–SFR1 mutant variant lacks any stimulatory effect on the RAD51 ATPase activity (Figure 5D). Taken together, our results demonstrated that the physical interaction between SWI5–SFR1 and RAD51 is a prerequisite for the regulation of presynaptic filament dynamics and enhancement of DNA strand exchange.
Figure 4.
Biophysical properties of SWI5 F83A/L85A-SFR1 mutant complex. (A) Analytical ultracentrifugation with sedimentation velocity analysis of SWI5–SFR1 and SWI5 F83A/L85A–SFR1 complexes. The experimental data were analyzed by the Sedfit program (version 14.1), which yielded an estimated molecular mass of 45.8 kDa for SWI5–SFR1, and 44.7 kDa for SWI5 F83A/L85A–SFR1. (B) Circular dichroism spectra of SWI5–SFR1 and SWI5 F83A/L85A–SFR1 complexes. The spectra showed no significant difference in secondary structure between SWI5–SFR1 and SWI5 F83A/L85A–SFR1.
Figure 5.
SWI5 F83A/L85A-SFR1 is functionally impaired. (A) The effect of SWI5–SFR1 or SWI5FL/AA–SFR1 on RAD51-mediated DNA strand exchange was examined. The results were graphed. (B) Exonuclease I protection assay was conducted with the indicated concentrations of SWI5–SFR1 and SWI5FL/AA–SFR1. The results were graphed. (C) The average length of RAD51 filaments with negative staining was determined by electron microscopy (see Supplementary Figure S5). RAD51 was examined alone or with SWI5–SFR1 or SWI5FL/AA–SFR1. The total 225 filaments were counted in each reaction. We note that the average length of the presynaptic filament in the presence of SWI5–SFR1 is much longer than expected. This could be due to the end-to-end association between two DNA molecules by RAD51 as described by Baumann et al. (35). (D) Thin-layer chromatography to monitor the hydrolysis of [γ-32P] ATP by RAD51 in the absence or presence of indicated concentrations of SWI5–SFR1 or SWI5FL/AA–SFR1. The results were graphed. (A, B and D) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments.
DISCUSSION
The physical interaction is prerequisite for the enhancement of SWI5–SFR1 by RAD51
Our previous work has shown that SWI5–SFR1 stabilizes the RAD51 presynaptic filament and also facilitates the release of ADP from the filament (27,28). In this study, the functional interaction between SWI5–SFR1 and RAD51 is dependent on physical interaction between them. We have provided evidence that SWI5–SFR1 interacts with the oligomeric form of RAD51 and have identified F83 and L85 of SWI5 to be indispensable for the interaction between SWI5–SFR1 and RAD51. Importantly, interaction-defective SWI5–SFR1 mutants are compromised for the ability to enhance RAD51-mediated DNA strand exchange and to accelerate ADP release from the presynaptic filament. Altogether, our results establish the functional significance of the RAD51–SWI5–SFR1 complex in DNA strand exchange. Since complex formation between SWI5 and SFR1 is needed for RAD51 interaction (27), it is possible that SFR1 induces a conformation in SWI5 competent for RAD51 binding. Alternatively, SFR1, within the context of the SWI5–SFR1 complex, may also contribute a RAD51 binding surface.
Differential interaction modes of RAD51 and SWI5–SFR1 from yeast to mammal
The S. pombe SWI5–SFR1 complex also physically interacts with Rad51 (24,26). Two distinct Rad51 interaction surfaces within S. pombe SWI5–SFR1 complex have been found, with one residing within the amino-terminal region of Sfr1 and the other within the carboxyl-terminus of this protein (26). Moreover, these N-terminal and C-terminal domains of Sfr1 are needed for functional cooperation with Rad51 in DNA strand exchange (26). In contrast, we have provided evidence for an important role of SWI5 in conferring the SWI5–SFR1 complex with the ability to physically interact with RAD51, and our analysis of the SWI5–SFR1dN202 mutant lacking the N-terminal 202 residues of SFR1 has shown that the N-terminal half of this protein is dispensable for RAD51 interaction. In line with those observations, we suggest that mammalian SWI5–SFR1 interacts with RAD51 via a protein domain distinct from those in the S. pombe ortholog.
The implication of SWI5–SFR1 interacts with oligomeric form of RAD51
The C-terminal region of BRCA2 interacts with oligomeric, but not monomeric, RAD51, suggesting that this region contacts the ATPase core domain of RAD51 spanning two RAD51 protomers (30,31). It is tempting to speculate that SWI5–SFR1 also recognizes an interface located between the ATPase core of two RAD51 protomers. Such a contact between SWI5–SFR1 and RAD51 may then lead to an enhanced stability of the presynaptic filament and the release of ADP from the filament as we have documented previously (27,28). Finally, it will be of considerable interest to determine whether the S. pombe SWI5–SFR1 complex also interacts with the oligomeric form of Rad51.
Supplementary Material
Footnotes
Present address: Chan-I Chung, Chemical Resources Laboratory, Tokyo Institute of Technology, 4259-R1-18, Nagatsuta-cho, Midori-ku, Yohohama, Kanagawa, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academia Sinica, National Taiwan University [NTU-EPR-105R8955-3]; Ministry of Science and Technology [MOST 102-2628-B-002-044-MY3]. Funding for open access charge: Ministry of Science and Technology [MOST 102-2628-B-002-044-MY3].
Conflict of interest statement. None declared.
REFERENCES
- 1.Mehta A., Haber J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heyer W.D. Regulation of recombination and genomic maintenance. Cold Spring Harb. Perspect. Biol. 2015;7:a016501. doi: 10.1101/cshperspect.a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 4.Venkitaraman A.R. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- 5.Prakash R., Zhang Y., Feng W., Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Ehmsen K.T., Heyer W.D., Morrical S.W. Presynaptic filament dynamics in homologous recombination and DNA repair. Crit. Rev. Biochem. Mol. Biol. 2011;46:240–270. doi: 10.3109/10409238.2011.576007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung P., Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell. Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 9.Zelensky A., Kanaar R., Wyman C. Mediators of homologous DNA pairing. Cold Spring Harb. Perspect. Biol. 2014;6:a016451. doi: 10.1101/cshperspect.a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowalczykowski S.C. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2015;7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan S.S., Lee S.Y., Chen G., Song M., Tomlinson G.E., Lee E.Y. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 12.Godthelp B.C., Artwert F., Joenje H., Zdzienicka M.Z. Impaired DNA damage-induced nuclear Rad51 foci formation uniquely characterizes Fanconi anemia group D1. Oncogene. 2002;21:5002–5005. doi: 10.1038/sj.onc.1205656. [DOI] [PubMed] [Google Scholar]
- 13.San Filippo J., Chi P., Sehorn M.G., Etchin J., Krejci L., Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Doty T., Gibson B., Heyer W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao W., Vaithiyalingam S., San Filippo J., Maranon D.G., Jimenez-Sainz J., Fontenay G.V., Kwon Y., Leung S.G., Lu L., Jensen R.B., et al. Promotion of BRCA2-Dependent Homologous Recombination by DSS1 via RPA Targeting and DNA Mimicry. Mol. Cell. 2015;59:176–187. doi: 10.1016/j.molcel.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiese C., Dray E., Groesser T., San Filippo J., Shi I., Collins D.W., Tsai M.S., Williams G.J., Rydberg B., Sung P., et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modesti M., Budzowska M., Baldeyron C., Demmers J.A., Ghirlando R., Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 20.Akamatsu Y., Dziadkowiec D., Ikeguchi M., Shinagawa H., Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akamatsu Y., Tsutsui Y., Morishita T., Siddique M.S., Kurokawa Y., Ikeguchi M., Yamao F., Arcangioli B., Iwasaki H. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J. 2007;26:1352–1362. doi: 10.1038/sj.emboj.7601582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akamatsu Y., Jasin M. Role for the mammalian SWI5–SFR1 complex in DNA strand break repair through homologous recombination. PLoS Genet. 2010;6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan J., Chen J. The role of the human SWI5-MEI5 complex in homologous recombination repair. J. Biol. Chem. 2011;286:9888–9893. doi: 10.1074/jbc.M110.207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haruta N., Kurokawa Y., Murayama Y., Akamatsu Y., Unzai S., Tsutsui Y., Iwasaki H. The SWI5–SFR1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- 25.Kokabu Y., Murayama Y., Kuwabara N., Oroguchi T., Hashimoto H., Tsutsui Y., Nozaki N., Akashi S., Unzai S., Shimizu T., et al. Fission yeast SWI5–SFR1 protein complex, an activator of Rad51 recombinase, forms an extremely elongated dogleg-shaped structure. J. Biol. Chem. 2011;286:43569–43576. doi: 10.1074/jbc.M111.303339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwabara N., Murayama Y., Hashimoto H., Kokabu Y., Ikeguchi M., Sato M., Mayanagi K., Tsutsui Y., Iwasaki H., Shimizu T. Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the SWI5–SFR1 complex. Structure. 2012;20:440–449. doi: 10.1016/j.str.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Tsai S.P., Su G.C., Lin S.W., Chung C.I., Xue X., Dunlop M.H., Akamatsu Y., Jasin M., Sung P., Chi P. Rad51 presynaptic filament stabilization function of the mouse SWI5–SFR1 heterodimeric complex. Nucleic Acids Res. 2012;40:6558–6569. doi: 10.1093/nar/gks305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su G.C., Chung C.I., Liao C.Y., Lin S.W., Tsai C.T., Huang T., Li H.W., Chi P. Enhancement of ADP release from the RAD51 presynaptic filament by the SWI5–SFR1 complex. Nucleic Acids Res. 2014;42:349–358. doi: 10.1093/nar/gkt879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin D.S., Pellegrini L., Daniels D.S., Yelent B., Craig L., Bates D., Yu D.S., Shivji M.K., Hitomi C., Arvai A.S., et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies O.R., Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esashi F., Galkin V.E., Yu X., Egelman E.H., West S.C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 32.Davies A.A., Masson J.Y., McIlwraith M.J., Stasiak A.Z., Stasiak A., Venkitaraman A.R., West S.C. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 33.Yu D.S., Sonoda E., Takeda S., Huang C.L., Pellegrini L., Blundell T.L., Venkitaraman A.R. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol. Cell. 2003;12:1029–1041. doi: 10.1016/s1097-2765(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 35.Baumann P., Benson F.E., West S.C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.