Abstract
Precise conversion of genetic information into proteins is essential to cellular health. However, a margin of error exists and is at its highest on the stage of translation of mRNA by the ribosome. Here we present three crystal structures of 70S ribosome complexes with messenger RNA and transfer RNAs and show that when a G•U base pair is at the first position of the codon–anticodon helix a conventional wobble pair cannot form because of inescapable steric clash between the guanosine of the A codon and the key nucleotide of decoding center adenosine 1493 of 16S rRNA. In our structure the rigid ribosomal decoding center, which is identically shaped for cognate or near-cognate tRNAs, forces this pair to adopt a geometry close to that of a canonical G•C pair. We further strengthen our hypothesis that spatial mimicry due either to base tautomerism or ionization dominates the translation infidelity mechanism.
INTRODUCTION
The ribosome is the giant macromolecular enzyme that converts genetic information into protein using messenger RNA template and transfer RNA substrates. Accurate protein synthesis depends strongly on the correct selection of the aminoacyl-tRNA complementary to the mRNA codon presented in the ribosomal decoding center. The ribosome occasionally makes mistakes by selecting from the pool of aminoacyl-tRNAs the wrong substrate. Misincorporation of amino acids into proteins may have a dual effect on the organism viability. First, it can result in production of dysfunctional or misfolded proteins, reducing the amount of active molecules and, maybe, producing toxic ones. On the other hand, the effects of single protein mutations on cell fitness are rarely drastic and levels of error tolerance are quite high (1). Moreover, it is shown that certain levels of mistranslation are beneficial for development of drug-resistance or adapting to stress conditions (rev. in (2)), although long-term evolutionary consequences are not yet established.
The frequency of incorporation of a wrong amino acid in the protein was estimated to be in the range of 10−4–10−3. These estimates depend on the selected method of measurement and reporter system (3–5). Overall, the number of proteins and types of substitutions investigated is very limited and does not allow to build a generalized picture. The error rates for transcription and tRNA aminoacylation are one order of magnitude lower (10−5–10−4) (rev in (6) and (7)) and, hence, the decoding of mRNA on the ribosome must be responsible for the majority of mistakes. The structure of genetic code suggests that misreading of the first or second codon position will lead to an almost certain substitution of amino acid, while the third position allows a certain leeway in base-pair geometry or non-Watson–Crick base-pairing. Also, the corresponding anticodon position is frequently heavily modified, adding complexity to interactions. Thus first and second base-pairs of the codon/anticodon helix must be under strict control but the existence and frequency of errors (and ease with which ribosome fidelity can be modulated (8,9)) indicate that there are ways to bypass this control.
Despite the breakthroughs in X-ray crystallography of ribosomes, until now a structural and mechanistic explanation for the discrimination between cognate and near-cognate tRNAs by the ribosome is not completely elucidated. Meanwhile, understanding of the mechanism underlying the incorrect decoding will bring a rationale to such crucial question as accuracy of translation (3,10,11) and will help to predict the possible level of incorrectly translated proteome. A significant number of experimental studies, utilizing a variety of techniques, rate the misincorporations caused by G/A or U/C substitutions and subsequent formation of G•U mismatches, in particular in the first position, as the most frequent ones (5,12–14). For example, mass-spectroscopy investigation of expressed proteins revealed misincorporation frequency of 10−5–10−3 and G•U mismatches to be responsible for 40% of misincorporated amino acids (12).
Recent crystal structures of the rare near-cognate state of the full 70S ribosome complexes with messenger RNA and tRNAs cognate or near-cognate to the A-site codon (with G•U, A•A, U•U, C•A non-complementary pairs) showed that (i) non-canonical pairs at both the first and the second positions of the codon–anticodon mini helix adopt geometry close to canonical Watson–Crick pairs (for example G•C-like geometry instead of expected G•U wobble geometry); (ii) the close van der Waals fit and H-bonding between the key nucleotides of the decoding center (A1493,1492, G530) and the minor groove of codon–anticodon helix is compatible with both canonical Watson–Crick pairing and codon–anticodon duplex carrying a mismatch at the first or second position; (iii) both cognate and near-cognate 70S ribosome structures revealed an identical locked conformation of the shoulder domain of the 30S ribosomal subunit (15–18).
These findings differ from the results of the earlier studies performed on the crystals of isolated 30S subunits where the nucleotides of decoding center (A1493, A1492, G530) were given a role as monitors and discriminators in the decoding process adopting different conformations depending whether canonical Watson–Crick pairs (U•A) or non-complementary pairs (U•G in wobble geometry) were found in the first and second positions in the codon–anticodon helix (19,20).
The inconsistency between observed geometry of the U4•G36 base pair at the first position of the codon–anticodon mini helix (wobble or Watson–Crick G•C like) can be explained by the differences between the two experimental models, isolated 30S ribosomal subunit and the whole 70S ribosome used in these studies. In the structures of 70S ribosome complexes the messenger RNA forms a kink between the P and A codons, the universal feature of the messenger RNA (21–23), which restricts the movement of the sugar–phosphate backbone of mRNA, so that the first nucleotide of A codon cannot be displaced toward the major groove of the codon–anticodon mini-helix, and thus prevents the formation of U4•G36 wobble geometry (Figure 1A). In contrast, in the 30S model, where A and P codons are not covalently linked, such restraints were absent and the mRNA nucleotide possessed sufficient freedom of movement to form a conventional wobble U4•G36 pair.
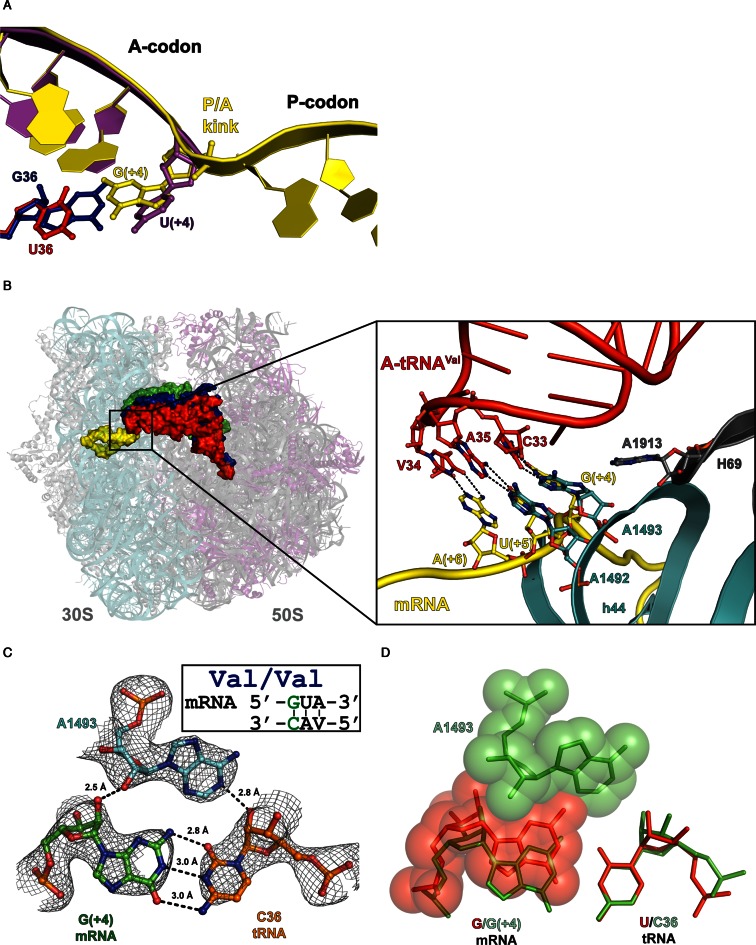
Figure 1.
Complex of the Thermus thermophilus 70S ribosome with mRNA and cognate tRNAVal in the A-site. (A) Differences between the position of the first A codon nucleotide in the 70S structure (yellow/red) and in the 30S model (PDB ID: 1N32) (magenta/blue). (B) Right, side view of the 70S ribosome complex with three tRNAs bound at the A—(red), P—(blue) and E—(green) sites and mRNA (yellow). Left, the close up view on the codon–anticodon duplex and essential nucleotides of the decoding center (G530 of 16S rRNA is omitted). (C) A-minor groove interactions of the first A-site base pair with 16S A1493 in the cognate complex with possible hydrogen bonds (distances ≤3 Å) and interatomic distances indicated. 2Fo–Fc electron density map is contoured at 1.2 σ. (D) Hypothetical overlay of a wobble base pair (red) on the first codon–anticodon base pair (green), demonstrating a possible clash with the 16S A1493 nucleotide.
In contrast to the canonical Watson–Crick pairs, which are isosteric upon their reversal, the U•G wobble pair is not isosteric with the wobble pair G•U. Hence, here we address the question what will be the geometry of a mismatch with guanosine being the first nucleotide of the mRNA A codon? As it was mentioned above the P/A kink imposes certain restraints on the sugar-phosphate moiety of this first A codon nucleotide against the movement toward the major groove. However, the same kink cannot prevent the movement of the first codon nucleotide toward the minor groove that would potentially result in conventional wobble geometry of G4•U36 pair.
In this study we present three crystal structures of Thermus thermophilus 70S ribosome complexes programmed with a long mRNA, carrying guanosine at the first position of the A-site codon. We solved the structure of 70S ribosome in cognate state with tRNAValVAC bound to the cognate valine codon GUA in the A site. We also succeeded to solve structures of the 70S ribosome complexes in the rare near-cognate state when tRNALysSUU was bound to the near-cognate glutamic acid GAA codon in the A site (forming a G4•U36 pair) in the presence and in the absence of the aminoglycoside antibiotic paromomycin.
The structures we are describing deepen the understanding of the tRNA discrimination mechanism on the ribosome, and give structural and mechanistic explanation to the occurrences of rare missense errors caused by the incorporation of near-cognate tRNAs.
MATERIALS AND METHODS
Materials
The 70S ribosomes from T. thermophilus strain HB8 were purified according to the published protocol (16). Uncharged native individual tRNAVal, tRNALys and tRNAfMet from Escherichia coli were ordered from Chemical Block (Russia). Two mRNA constructs whose sequences are specified below were purchased in Thermo Scientific (USA) and deprotected following the supplier procedure. Aminoglycoside antibiotic paromomycin was purchased from Sigma-Aldrich.
Complex formation and crystallization
The ribosomal complexes were formed in 10 mM Tris-acetate, 40 mM KCl, 7.5 mM Mg(CH3COO)2, 0.5 mM DTT at pH7.0 at 37°C. For all complexes the 70S ribosomes (3 μM) were incubated with 5-fold stoichiometric excess of mRNA and three to 5-fold excess of tRNAs for 15 min. For both the cognate and near-cognate 70S ribosome complexes the mRNA sequences contained GGC.AAG.GAG.GCA.AAA (Z) at the 5′-end (21). The cognate complex mRNA sequence contained valine A codon and was as follows: mRNA-1 = ZAUGGAAA8. The near-cognate complex modeled the G4•U36 mismatch at the first codon–anticodon positions contained glutamine A codon and was as follow: mRNA-2 = ZAUGGAAA7 (the start codon and the Shine-Dalgarno sequence are underlined).
The 70S ribosomes (3 μM) were pre-incubated with mRNA-1 or mRNA-2 and tRNAfMet for 15 min to fill the P-site. The complexes modeling cognate or near-cognate (G4•U36) states were obtained by followed incubation with tRNAVal and tRNALys respectively. Complex with paromomycin was obtained by including the antibiotic (60 μM) into the incubation mixture containing 70S/tRNAfMet/mRNA-2/tRNALys.
Crystals were grown at 24°C via vapor diffusion in sitting-drop plates (CrysChem, Hampton Research) as described before (17).
Data collection, processing and structure determination
Data for all complexes were collected at the PXI beamline of Swiss Light Source, Switzerland, at 100K. A very low dose mode was used and huge redundancy data were collected (24). The data were indexed, integrated and scaled using XDS (25), using CC(1/2) as data quality indicator (26). All crystals belong to space group P212121 and contain two ribosomes per asymmetric unit. One of the previously published structures (16), with tRNA, mRNA and metal ions removed, was used for refinement with Phenix (27). The initial model was placed within each data set by rigid body refinement with each biopolymer chain as a rigid body. The resulting electron density maps were inspected in Coot (28) and the tRNA and mRNA chains were built in. During several cycles of manual rebuilding followed by coordinate and isotropic B-factor refinement, magnesium ions were added and the final refinement round took place. The data collection and refinement as well as model geometry statistics are presented in the Table 1.
Table 1. Data collection and refinement statistics.
| Cognatea | UG mismatchb | UG mismatch + paroc | |
|---|---|---|---|
| PDB ID | 5IBB | 5IB8 | 5IB7 |
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 209.5 448.9 620.8 | 209.15 448.16 617.8 | 209.3 448.8 620.2 |
| α, β, γ (°) | 90.0 90.0 90.0 | 90.0 90.0 90.0 | 90.0 90.0 90.0 |
| Resolution (Å) | 170–2.96 (3.04–2.96)* | 170–3.13 (3.21–3.13) | 170–2.99 (3.07–2.99) |
| Rmeas | 44.1 (509.4) | 42.1 (435.9) | 34.6 (471) |
| I/σI | 15.26 (1.00) | 15.91 (0.97) | 14.73 (1.00) |
| CC(1/2) (26) | 99.9 (60.1) | 99.9 (47.6) | 99.8 (52.0) |
| Completeness (%) | 99.8 (99.5) | 100 (100) | 100 (100) |
| Redundancy | 112.23 (113.56) | 133.67 (57.82) | 69.52 (64.06) |
| Refinement | |||
| Resolution (Å) | 146.681–2.96 | 154.45–3.13 | 147.082–2.99 |
| No. reflections | 1 198 197 | 1 008 227 | 1 162 271 |
| Rwork/Rfree | 20.06/24.26 | 19.20/24.37 | 20.15/24.11 |
| No. atoms | |||
| RNA | 201 656 | 201 873 | 202 442 |
| Protein | 90 546 | 89 783 | 90 174 |
| Ligand/ion/water | 4609 | 5344 | 4868 |
| B-factors | |||
| RNA | 96.22 | 108.41 | 102.54 |
| Protein | 102.17 | 113.76 | 109.08 |
| Ligand/ion/water | 79.17 | 88.24 | 79.81 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.009 | 0.009 |
| Bond angles (°) | 1.558 | 1.491 | 1.581 |
*Values in parentheses are for highest-resolution shell.
Number of crystals used for data collection: a1, b4, c3.
RESULTS
The guanosine at the first position of cognate A-site codon
We have determined the structure of the T. thermophilus 70S ribosome programmed by 30 nucleotides long messenger RNA, with tRNAfMet bound to the AUG codon in the peptidyl-tRNA binding site (P-site) and with tRNAValVAC bound to cognate valine codon GUA in the aminoacyl-tRNA binding site (A-site) (Figure 1B, Table 1). As expected A1493 formed type I minor interactions with the first codon–anticodon pair G4•C36 (19) (Figure 1C). Overall analysis of the 70S structure showed as was previously observed that upon binding of A-site tRNA the formation of the rigid decoding center is ensured by conformational rearrangement of the ‘shoulder’ domain of the 30S subunit resulting in ‘shoulder locking’ state (15–18,29).
Examination of the A-site codon conformation in the present 70S ribosome structure suggested that the guanosine in the first position of valine (GUA) codon could not pair with uracil in conventional G•U wobble geometry (Figure 1D). Comparison of the 70S ribosome structures bearing various mismatches in the codon–anticodon duplex in the decoding center shows that the nucleotides 35 and 36 of the tRNAs anticodons are tightly fixed in place (15–18,29). Therefore, we presumed to model the G•U pair in wobble geometry by the shift of the G nucleotide of the A codon toward to minor groove of the hypothetical pair. This shift is potentially possible because the sugar phosphate backbone of the first nucleotide of A codon is pre-positioned by the P/A kink that there is no constrains for its displacement toward the minor groove of the codon–anticodon helix, contrary to its prohibited movement toward to major groove.
However, analysis of our structure suggest that the shift of G4 nucleotide toward to minor groove, which is necessary for the formation of the wobble geometry of G4•U36 pair, will lead to the clear steric clash of the guanosine nucleobase with the key nucleotide of the decoding center A1493 (Figure 1D).
The G•U mismatch in the 70S decoding center
We succeeded in the determination of the structure of the T. thermophilus 70S ribosome complex in the near-cognate state with G•U mismatch in the decoding center. 70S ribosome was programmed by 30 nucleotides long messenger RNA, with tRNAfMet bound to the AUG codon in the P-site and with tRNALysSUU bound to near-cognate glutamic acid GAA codon in the A site (Figure 2A, Table 1).
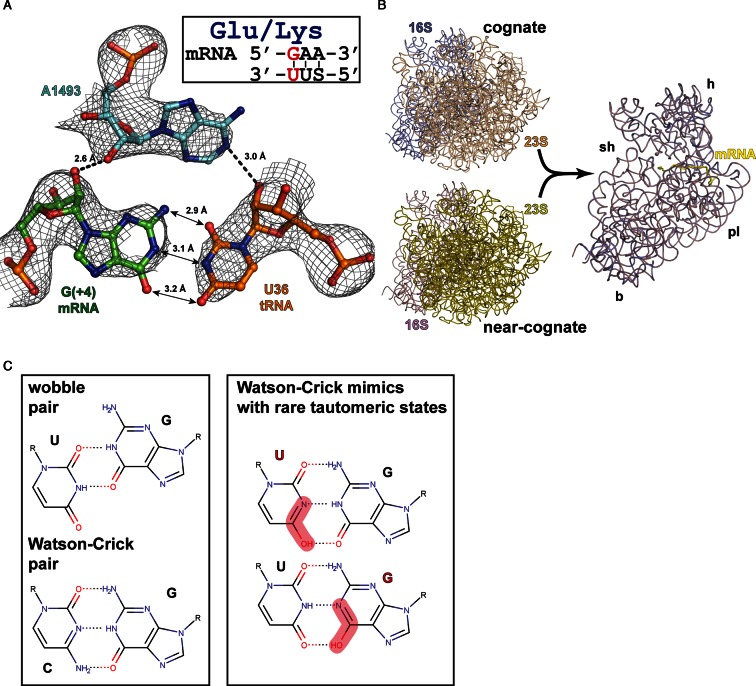
Figure 2.
Complex of the Thermus thermophilus 70S ribosome with mRNA and near-cognate tRNALys in the A-site. (A) A-minor groove interactions of the mismatched A-site base pair with 16S A1493 in the near-cognate complex. Possible hydrogen bonds (distances ≤3 Å) and interatomic distances are indicated. 2Fo–Fc electron density map is contoured at 1.2 σ. (B) Superposition of 23S rRNA from the near-cognate and cognate structures exemplifies identical conformations of 16S rRNA in both states including the conformation of the shoulder domain. For clarity of the representation ribosomal proteins are omitted; mRNA position and the main domains of the small subunit are indicated (h—head, sh—shoulder, pl—platform, b—body). (C) Geometry of non-canonical wobble pair and canonical Watson-Crick pair (left). Watson–Crick-like pairs formed by rare tautomeric states of uracil or guanosine (right).
We found that as a result of binding of near-cognate tRNA the rigid decoding center forms and the 30S subunit undergoes an identical conformational rearrangement of the ‘shoulder’ domain (Figure 2B). In agreement with our previous results we observed that the interaction pattern of universally conserved nucleotides of the ribosomal decoding center (16S rRNA G530, A1492, A1493) with the minor groove of the codon–anticodon duplex with G4•U36 base pair remains the same as for cognate case 15–18).
The G4•U36 base pair does not adopt the anticipated wobble geometry, forming instead a base pair similar to Watson–Crick canonical G•C pair (Figure 2A). As we proposed earlier this geometry can be rationalized by a rare tautomeric state of one of the nucleobases (Figure 2C). It is likely that spatial and conformational restraints imposed by the decoding center and conserved structure of the tRNA ASL force preferential selection of the more rare isomers upon interaction between tRNA and mRNA in the A site. It appears plausible that the mimicry of mismatches to canonical base pairs is the mechanism by which these mismatches escape discrimination by the ribosome and result in translational errors.
Additional structure of the near-cognate state with tRNALysSUU determined in the presence of the miscoding aminoglycoside antibiotic paromomycin (Table 1) confirmed that binding of the antibiotic induces the same slight movement of the tip Helix 69 of 23SrRNA toward the near-cognate tRNA and result in rearrangement of the inter subunit bridge B2a between h44 and H69 as we described previously (18).
DISCUSSION
To discriminate between near-cognate (when there is one mismatch in the first or second position of a codon–anticodon duplex) and cognate tRNAs, a structural dialog between the decoding center of the small ribosomal subunit, including the A codon of mRNA and anticodon of tRNA obviously must be established.
In this structural study we challenged the question of tRNA discrimination on the ribosome and showed that when 70S ribosome is primed with covalently intact mRNA where the first nucleotide of A codon is a guanosine, the formation of G4•U36 pair in conventional wobble geometry in the codon–anticodon helix is stringently forbidden. Despite the fact that the universal feature of the P/A kink in the mRNA allows for the shift of guanosine (G4) of the A codon toward the minor groove, necessary for the formation of the standard wobble geometry G4•U36 pair, our structure demonstrates that this movement of G4 will lead to a steric clash with the universally conserved nucleotide A1493 in the ‘out’ conformation. Here it is important to stress that during elongation, at the moment when the A site of 70S ribosome is vacant, this universally conserved nucleotide A1493 stays in the ‘protruded’ conformation from h44 helix (‘out’ from h44) (23,30).
Analysis of the decoding center in the present structures together with our previous library of 70S ribosome complexes showed that when near-cognate tRNAs with different non-complementary pairs in the A-site (G•U, U•G, A•A, A•C, C•A, U•U) are bound to the ribosome, the key nucleotides A1493, A1492 and G530 of 16S rRNA interact with these pairs identically to the way they interact with canonical Watson–Crick base pairs (C•G/G•C and U•A/A•U) (15–19) (Figures 1C and 2A). The resulting formation of identical type I and type II A-minor groove interactions between the key nucleotides of decoding center and the first two codon–anticodon pairs, demonstrates that the ribosome recognizes first of all the shapes of the base pairs. In other words, G530, A1492 and A1493 upon binding of a cognate or near-cognate tRNAs form part of the grip of the decoding center, defining its spatial and stereochemical properties.
These findings differ from that of earlier studies performed on 30S subunits where the nucleotides at the decoding center (A1493, A1492, G530) were given a role as monitors and discriminators during the decoding process that would adopt different conformations depending on whether canonical Watson–Crick pairs or non-canonical (wobble geometry of G•U pair for example) were found in the first and second positions in the codon–anticodon helix (19,20). As it was already discussed previously (18,30) and in the introduction of the present article, the main origin of the observed discrepancy between the observed geometries of G•U base pairs resides in the implementation of two different experimental models used in these studies, on one side the isolated small 30S ribosomal subunit and on the other side the whole 70S ribosome.
Our current studies offers the structural basis for understanding how incorrect decoding can occur during the least accurate process of polymerization in the cell, translation. The present observation of Watson–Crick-like geometry for G4•U36 and the previous results on G•U mismatches with Watson–Crick G•C-like geometry (18) resonate with the hypothesis of spontaneous mutagenesis, suggested by Watson–Crick, which states that mutations arise from the naturally occurring rare nucleobase tautomers (31). In 1970s, Topal and Fresco widened the concept of complementarity and analyzed the consequence of base tautomerization in the replication and translation (32,33). They discussed the base pairing schemes, some of which involve tautomerism, that possess the dimensions and shapes close to the complementary Watson–Crick pairs. These base pairs can be accommodated by or pass through the sieve formed by the steric and geometric constrains imposed by the ribosome (34,35). This long-standing concept has been difficult to demonstrate directly until recently, when first structural evidence for errors in replication via a G•T and C•A mismatches with Watson–Crick geometry was procured (36–38). Other non-structural experimental findings also demonstrate that steric or shape complementarity rather than H-bonding, is paramount in replication and translation (39–42).
The hypothesis of base tautomerism as one of the foundations for translational errors still requires experimental proof. For structural evidence, data at a resolution of ∼1Å would be necessary, unattainable for ribosomal complexes at the moment. In general, detection of rare anionic or tautomeric states in polynucleotides or nucleoprotein complexes is a non-trivial task due to low abundance and short life times of such energetically unfavorable states. A recent nuclear magnetic resonance relaxation dispersion study showed that wobble dG•dT and rG•rU mispairs in DNA and RNA duplexes exist in dynamic equilibrium with short-lived, low-populated Watson–Crick-like mispairs (about 0.01%) that are stabilized by rare enolic or anionic states of nucleobases (43), correlating with measured mismatch frequencies (also ∼10−4) for individual codons (12).
Based on our structural studies we can conclude that the rare translational mistakes caused by the incorporation of near-cognate tRNAs can be rationalized mostly by the probability of formation of Watson–Crick-like base pairs in some codon/anticodon combinations due to either deprotonated or rare tautomeric states of nucleobases, or in some cases by a mismatch randomly escaping discrimination by preserving base pair geometry close to the Watson–Crick pair. The present mechanism further establishes that discrimination between tRNAs is primarily founded on spatial fit rather than on the number of hydrogen bonds between the ‘closed’ decoding center and the codon–anticodon duplex.
ACCESSION NUMBERS
The atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 5IBB, 5IB8 and 5IB7.
Supplementary Material
Acknowledgments
We are grateful to the staff of the PXI beamline at the Swiss Light Source (Switzerland) and especially Takashi Tomizaki and Meitian Wang for assistance during synchrotron data collection. We thank Natalia Demeshkina (National Heart, Blood and Lung Institute/NIH, BD, USA) for the help at the early stage of the work.
Author Contributions: A.R. and G.Y. conducted experiments and performed analysis. All authors discussed the results and commented on the manuscript. The authors declare no competing interests.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Research Agency [ANR-15-CE11-0021-01 to G.Y., E.W]; European Research Council advanced grant [294312 to M.Y.]. Funding for open access charge: French National Research Agency [ANR-15-CE11-0021-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ruan B., Palioura S., Sabina J., Marvin-Guy L., Kochhar S., Larossa R.A., Soll D. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribas de Pouplana L., Santos M.A., Zhu J.H., Farabaugh P.J., Javid B. Protein mistranslation: friend or foe? Trends Biochem. Sci. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Ieong K.W., Johansson M., Ehrenberg M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9602–9607. doi: 10.1073/pnas.1506823112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manickam N., Nag N., Abbasi A., Patel K., Farabaugh P.J. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014;20:9–15. doi: 10.1261/rna.039792.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libby R.T., Gallant J.A. The role of RNA polymerase in transcriptional fidelity. Mol. Microbiol. 1991;5:999–1004. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 7.Francklyn C.S. DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression. Biochemistry. 2008;47:11695–11703. doi: 10.1021/bi801500z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pape T., Wintermeyer W., Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurland C.G., Ehrenberg M. Optimization of translation accuracy. Prog. Nucleic Acid Res. Mol. Biol. 1984;31:191–219. doi: 10.1016/s0079-6603(08)60378-5. [DOI] [PubMed] [Google Scholar]
- 10.Drummond D.A., Wilke C.O. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer E.B., Farabaugh P.J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Shah B., Bondarenko P.V. G/U and certain wobble position mismatches as possible main causes of amino acid misincorporations. Biochemistry. 2013;52:8165–8176. doi: 10.1021/bi401002c. [DOI] [PubMed] [Google Scholar]
- 13.Blanchet S., Cornu D., Argentini M., Namy O. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:10061–10072. doi: 10.1093/nar/gku663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy B., Leszyk J.D., Mangus D.A., Jacobson A. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc. Natl. Acad. Sci. U.S.A. 2015;112:3038–3043. doi: 10.1073/pnas.1424127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenner L., Demeshkina N., Yusupova G., Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 16.Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G. Structural insights into the translational infidelity mechanism. Nat. Commun. 2015;6:7251. doi: 10.1038/ncomms8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozov A., Demeshkina N., Khusainov I., Westhof E., Yusupov M., Yusupova G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016;7:10457. doi: 10.1038/ncomms10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 19.Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 20.Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 21.Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 22.Selmer M., Dunham C.M., Murphy F.V.T., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 23.Jenner L.B., Demeshkina N., Yusupova G., Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 24.Mueller M., Wang M., Schulze-Briese C. Optimal fine phi-slicing for single-photon-counting pixel detectors. Acta Crystallogr. D Biol. Crystallogr. 2012;68:42–56. doi: 10.1107/S0907444911049833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karplus P.A., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demeshkina N., Jenner L., Yusupova G., Yusupov M. Interactions of the ribosome with mRNA and tRNA. Curr. Opin. Struct. Biol. 2010;20:325–332. doi: 10.1016/j.sbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G. New structural insights into the decoding mechanism: translation infidelity via a G.U pair with Watson-Crick geometry. FEBS Lett. 2013;587:1848–1857. doi: 10.1016/j.febslet.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Watson J.D., Crick F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 32.Topal M.D., Fresco J.R. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 33.Topal M.D., Fresco J.R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976;263:289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- 34.Westhof E. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett. 2014;588:2464–2469. doi: 10.1016/j.febslet.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 35.Westhof E., Yusupov M., Yusupova G. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 2014;6:19. doi: 10.12703/P6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W., Hellinga H.W., Beese L.S. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bebenek K., Pedersen L.C., Kunkel T.A. Replication infidelity via a mismatch with Watson-Crick geometry. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1862–1867. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S., Konigsberg W.H. Mispairs with Watson-Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant. Protein Sci. 2014;23:508–513. doi: 10.1002/pro.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales J.C., Kool E.T. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 40.Kool E.T., Sintim H.O. The difluorotoluene debate–a decade later. Chem. Commun. (Camb) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 41.Khade P.K., Shi X., Joseph S. Steric complementarity in the decoding center is important for tRNA selection by the ribosome. J. Mol. Biol. 2013;425:3778–3789. doi: 10.1016/j.jmb.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noller H.F. How does the ribosome sense a cognate tRNA? J. Mol. Biol. 2013;425:3776–3777. doi: 10.1016/j.jmb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Kimsey I.J., Petzold K., Sathyamoorthy B., Stein Z.W., Al-Hashimi H.M. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015;519:315–320. doi: 10.1038/nature14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.