Abstract
Sexual differentiation of malaria parasites into gametocytes in the vertebrate host and subsequent gamete fertilization in mosquitoes is essential for the spreading of the disease. The molecular processes orchestrating these transitions are far from fully understood. Here, we report the first transcriptome analysis of male and female Plasmodium falciparum gametocytes coupled with a comprehensive proteome analysis. In male gametocytes there is an enrichment of proteins involved in the formation of flagellated gametes; proteins involved in DNA replication, chromatin organization and axoneme formation. On the other hand, female gametocytes are enriched in proteins required for zygote formation and functions after fertilization; protein-, lipid- and energy-metabolism. Integration of transcriptome and proteome data revealed 512 highly expressed maternal transcripts without corresponding protein expression indicating large scale translational repression in P. falciparum female gametocytes for the first time. Despite a high degree of conservation between Plasmodium species, 260 of these ‘repressed transcripts’ have not been previously described. Moreover, for some of these genes, protein expression is only reported in oocysts and sporozoites indicating that repressed transcripts can be partitioned into short- and long-term storage. Finally, these data sets provide an essential resource for identification of vaccine/drug targets and for further mechanistic studies.
INTRODUCTION
The most severe clinical presentation of human malaria is caused by the unicellular protozoan parasite Plasmodium falciparum, which has a complex life cycle, split between a human host and a mosquito vector. Sexual development of Plasmodium parasites is an essential, multistep process involving both hosts (1–4) that is an attractive target for intervention strategies preventing the spread of malaria parasites in the human population (5). Gametocytogenesis, the sexual differentiation of asexual precursors, is the first step that occurs during blood stage development in mammalian hosts. Female and male P. falciparum gametocytes develop in about 10 days through 5 morphologically distinct stages (stage I–V) (6). During their maturation P. falciparum gametocytes sequester in the bone marrow before they re-enter circulation once mature (7). Gametocytes mature inside erythrocytes and gradually elongate the infected cells. This elongated form, which becomes apparent in stage II, is only observed in P. falciparum, while gametocytes of most other mammalian Plasmodium species are round cells (8).
Morphological differences between male and female P. falciparum gametocytes become notable in stage IV, with a relatively small nucleus in female gametocytes (FG) compared to male gametocytes (MG). Stage V fully mature gametocytes are ‘growth-arrested’ cells only further activated once ingested into the midgut of mosquitoes. Within 20 min after activation MG undergo three rounds of genome replication and mitotic cell division releasing up to eight motile microgametes; while ingested FG differentiate into a single immotile macrogamete. The female gamete is fertilized by a flagellated male resulting in the formation of a diploid zygote, which develops into an ookinete within 24 h (4).
The molecular events essential for gametocytes development and transmission to the mosquito vector have so far been studied mainly in the rodent P. berghei malaria model, including the first proteomic study of male and female gametocytes (9). In this species, female gametocytes appear to be supplied with translationally repressed transcripts that are translated into proteins only after gamete formation and fertilization (10,11). Translationally repressed transcripts in P. berghei FG have been shown to be associated with messenger ribonucleoprotein particles (mRNP) which contain the RNA helicase DOZI and the Sm-like protein CITH as translational repressors (12).
However, the substantial differences in developmental timing (13) and morphology (8) of mature gametocytes prevent an a priori extension of the P. berghei findings to P. falciparum. Therefore, it is unknown whether DOZI and CITH-mediated translational repression (TR) is operational in P. falciparum. In addition, certain proteins appear to be expressed in P. falciparum gametocytes while the genes encoding the orthologous proteins in P. berghei have been demonstrated to be translationally repressed (14,15).
Recently, evidence was obtained for translational repression of transcripts in P. falciparum gametocytes by transcriptome profiling of a mutant lacking the RNA binding protein, Puf2 (PF3D7_0417100; (16)). Disruption of the puf2 gene resulted in increased gametocytogenesis, a male-biased sex ratio (17) as well as the deregulation of transcript abundance (16). Interestingly, both up- and down-regulation was observed in the mutant and some of the up-regulated transcripts appeared to be translationally repressed, including orthologs of genes translationally repressed in P. berghei (i.e. Pfs25, Pfs28, plasmepsin VI). Puf2 has so far not been identified to play a similar role in P. berghei gametocytes and it is unknown whether and how the lack of Puf2 expression affects global protein synthesis in P. falciparum gametocytes.
In this study, we provide a comprehensive molecular characterization of P. falciparum MG and FG at both the transcript and protein level. Using sex-specific reporters (9), MG and FG were purified by fluorescence-activated cell sorting (FACS) sorting and subjected to state-of-the art sequencing technologies to generate gender-specific transcriptomes. MG and FG associated proteomes were identified by liquid chromatography tandem mass spectrometry (LC-MS/MS). Functional differences between MG and FG in P. falciparum and P. berghei proteomes were revealed by gene set enrichment analysis (GSEA) and genome-wide translational repression of maternal P. falciparum transcripts was analysed by an integrated analysis of gametocyte transcriptome and proteome. These analyses provide insight into sex-specific molecular processes associated with the formation of male and female gametes. Moreover, our analyses provide evidence for a large set of putatively repressed transcripts in female gametes indicating that translational repression is a major and evolutionarily conserved mechanism underlying post-fertilization Plasmodium development in the mosquito.
MATERIALS AND METHODS
Detailed description of the methods can be found in the Supplementary Methods, including the generation of the gender-specific fluorescent parasite lines PfDynGFP, PfP47GFP and PfDynGFP/PfP47mCherry.
Gametocyte culture and purification
Gametocyte cultures were performed in the semi-automated culture system (18,19) and were started at 5% hematocrit and 0.5% parasitemia. Gametocyte cultures were treated with N-acetyl-glucosamine on day 7 to eliminate asexual parasites. Mature stage IV/V gametocyte production was evaluated in cultures after 13–15 days in Giemsa stained thin blood films (20,21). Male exflagellation capacity was evaluated after stimulation with fetal calf serum at pH 8.0 (22). Gametocytes were concentrated in 37°C culture medium and separated from uninfected erythrocytes and culture debris using a 63% and a 33% Percoll density gradient and subsequently taken up in a 4°C suspended activation (SA) buffer (10 mM Tris, pH 7.3, 170 mM NaCl, 10 mM glucose). Gametocytes were further purified by magnetic separation from uninfected red blood cells using MACS columns (23,24).
Flow cytometry of gametocytes
Male and female gametocytes from the PfDynGFP, PfP47GFP and PfDynGFP/PfP47mCherry lines were sorted using the Coulter Epics Elite flow cytometer (Beckman Coulter) or the BD FACS Aria SORP flow cytometer keeping cells at 4°C in SA buffer. Gametocytes were first separated from uninfected red blood cells using forward and sideward scatters, followed by sorting males and females based on signal intensity of the fluorescent proteins (green fluorescent protein (GFP) or mCherry). An aliquot of sorted cells was reanalysed to determine purity of sorting.
RNA isolation and sequencing
NF54 PfDynGFP P. falciparum parasites carrying PfP47mCherry episomal plasmids were used for RNA-Seq analysis of male or female gametocyte populations. RNA was isolated on RNeasy columns and subsequently enriched for polyA+ mRNA as described (25,26). RNA hydrolysis, cDNA synthesis and strand-specific RNA-Seq was performed as reported earlier (25,26). Libraries were amplified for a total of 16 cycles (4 cycles pre-size-selection PCR, 12 cycles post-size-selection PCR) using a P. falciparum optimized Kapa PCR protocol (KAPA Biosystems). Libraries were sequenced for 92 cycles single-end on a HiSeq2000 system (Illumina).
Single-end RNA-Seq reads were mapped to the PlasmoDB 9.1 spliced transcriptome using BWA (Version 0.6.2-r126, default parameters) and filtered for mapping quality ≥15. Transcript abundance was calculated as RPKM values (reads per kilobase of exon per million mapped reads) for both sense and anti-sense strands. From the distribution of anti-sense expression rates we estimated that with an RPKM cutoff of 4.8 we would achieve a false discovery rate (FDR) of 5%. Details about normalization of the RNA-seq data are described in the Supplementary Methods section. To access experimental variation, RNA-seq libraries from biological replicates have been generated and both the original and replicate samples were sequenced for 75 bp single-read at low depth on NextSeq500 (data available upon request). These replicates show high degree of correlation (R2 = 0.83–0.89, Supplementary Figure S1) and show that our differential gene expression call between MG and FG is reliable with a FDR lower than 5%.
Protein isolation and mass spectrometric analysis
Purified male and female gametocytes from three/four independent parasite cultures were pooled to produce a single male and female gametocyte sample of approximately 2–3 × 107 gender-purified parasites for MS analysis. The parasite samples were lysed by repetitive cycle of freezing in liquid nitrogen and thawing at 37°C, divided into a cytosolic and a membrane fraction and processed by an in gel digestion procedure described in previous studies (9,27,28). Digested samples were acidified to a final concentration of 0.1% trifluoroacitic acid (TFA) and purified by stop and go extraction (STAGE) tips (29). Peptide identification experiments were performed by LC-MS/MS using a 7-Tesla linear ion trap ion cyclotron resonance Fourier transform (LTQ-FT) mass spectrometer (Thermo Fisher, Bremen, Germany) coupled to the Agilent 1100 nano HPLC or to the nano EASY LC chromatographic workstation (Proxeon, Denmark) as described by Silvestrini et al. (28). Mass spectrometry data was analysed with the Andromeda (30) search engine integrated in the proteomics software suite MaxQuant version 1.3.05 (31). For detailed experimental information see Supplementary Methods.
Bioinformatic analysis
Relative LFQ expression profiles of proteins detected by at least two ‘razor and unique’ peptides in one of gametocyte genders were clustered by the self-organizing tree algorithm (SOTA) using The Institute of Genomic Research (TIGR) MultiExperiment Viewer (MeV4.2) (http://www.tm4.org/mev.html) software package.
GSEA with GSEA v2.1.04 (http://www.broad.mit.edu/gsea/index.jsp) was applied to test whether expression of a predefined gene set correlates with a ranked expression list of proteins in male and female gametocytes without applying a threshold on expression ratio's prior to statistical analysis (32).
Gene ontology (GO) enrichment analysis of translationally repressed P. falciparum transcripts was performed with the software package Ontologizer (33), with GO terms taken from GO consortium (http://geneontology.org/page/download-annotations), with predicted GO terms downloaded from PlasmoDB and with terms collected by manual assembling. GO term enrichment relative to the background of all.
Detailed information about bioinformatics analysis is provided in Supplementary Methods.
Data deposition
The Gene Expression Omnibus accession number for the strand-specific RNA-seq data reported in this paper is GSE75795. The RNA-seq and proteomic data have been submitted to PlasmoDB (www.plasmoDB.org).
RESULTS AND DISCUSSION
Generation of sex-specific reporter lines and isolation of male and female P. falciparum gametocytes
In order to enable separation of gametocyte genders, we generated transgenic P. falciparum lines expressing fluorescent proteins in a sex-specific manner. For the male-specific reporter line, the pDynGFP construct containing the gfp gene under control of the promoter region of dynein heavy chain protein gene (PF3D7_1023100; the ortholog of a male-specific P. berghei protein (9)) was stably integrated into the genome of NF54 parasites (Supplementary Figure S2A). For generation of a GFP expressing female-specific reporter line, NF54 parasites were transfected with a construct containing the gfp gene under control of the promoter region of PfP47 (PF3D7_1346800; a protein exclusively expressed at the surface of female gametocytes and gametes (21)) (Supplementary Figure S2B). Furthermore, we generated a double-transgenic reporter line by transfection of the construct pPFCENv2-PfP47mCherry (Supplementary Figure S2C) into the male-specific PfDynGFP line (see above). This construct contains mCherry under control of the promoter region of PfP47 and a centromere for stable maintenance of the episomal construct (34).
Diagnostic polymerase chain reaction (PCR) confirmed correct integration of the pDynGFP construct into the non-essential Pf52 locus (PF3D7_0404500 (21)) (Supplementary Figure S2A) and detected the presence of episomal constructs in the PfP47GFP and PfDynGFP/P47mCherry lines (Supplementary Figure S2B and C) using primers against the bsd resistance marker (Supplementary Table S1).
Sex-specific reporter gene expression was first analysed by live fluorescence during the egress of motile males from the infected RBC in a so-called exflagellation assay. All exflagellation centers in PfDynGFP were positive for GFP fluorescence while no GFP signal was observed in the exflagellating centers of PfP47GFP parasites (data not shown). Sex-specific GFP expression in PfP47GFP and PfDynGFP gametocytes was further determined by immunofluorescence analysis (IFA) using antibodies recognizing sex-specific marker proteins α-Tubulin-II (male) and P47 (female) of fixed parasite samples (Figure 1A). Notably, in parasites of the PfDynGFP/PfP47mCherry line we observed some gametocytes that were both GFP and mCherry positive. Morphologically these double fluorescent parasites appeared to be stage IV male gametocytes (data not shown).
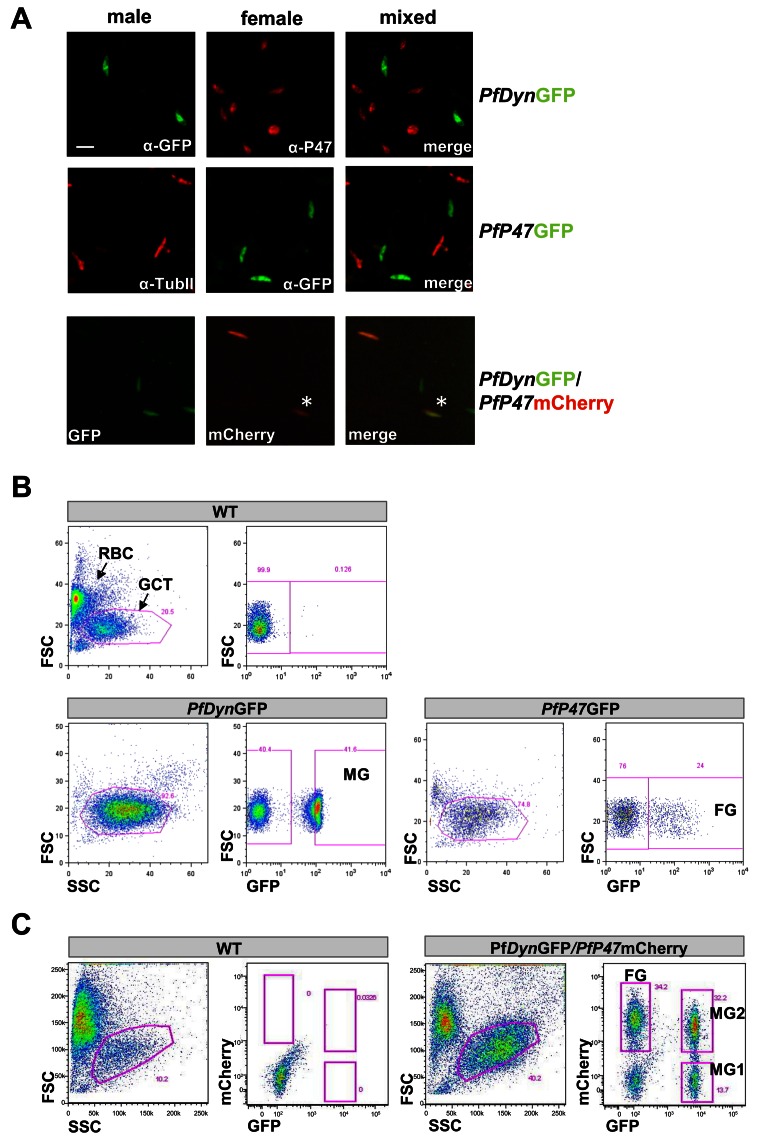
Figure 1.
Fluorescent marker expression in male- and female-specific reporter parasites and FACS sorting of male (MG) and female (FG) gametocytes. (A) Top panel: MG-specific expression of GFP in the PfDynGFP line, visualized by staining of gametocytes with anti-GFP antibodies (green) in the absence of staining with antibodies against the female-specific marker P47 (red). Middle panel: FG-specific expression of GFP in the PfP47GFP line, visualized by staining of gametocytes with anti-GFP antibodies (green) in the absence of staining with antibodies against the male-specific marker Tubulin-II (anti-TubII; red). Note that in this line not all female parasites express GFP and the percentage of GFP positive parasites declines over time due to blasticidin resistance and loss of the episome. Bottom panel: In the PfDynGFP/PfP47mCherry line GFP-positive (MG) and mCherry-positive (FG) gametocytes are present and in addition a population of GFP/mCherry double-positive cells (yellow colour in the overlay) as visualized by live fluorescence analysis. (B) For proteomic analysis MG were selected on basis of GFP-fluorescence intensities from PfDynGFP (gate MG) and FG from PfP47GFP (gate FG). Gametocyte-infected erythrocytes (GCT) were selected by gating on forward and side scatter (FS/SS). WT-infected erythrocytes (upper panel) show low GFP fluorescence intensity. RBC: red blood cells. (C) For transcriptomic analysis MG were selected on basis of GFP expression (gate MG1) and FM on basis of mCherry expression (gate FG). In addition a population of cells were selected that were both GFP and mCherry positive (gate MG2). GCT were selected by gating on FS/SS.
GFP-positive PfDynGFP gametocytes were obtained by FACS purification (Figure 1B), resulting in a population of >99% male gametocytes as determined by IFA using anti-P47 antibody (data not shown). Similarly, FACS purification of GFP-positive PfP47GFP gametocytes (Figure 1B) resulted in >95% female gametocytes as determined by anti-α-Tubulin-II antibody (data not shown). Subsequently, purified PfDynGFP male and PfP47GFP female gametocytes were subjected to proteome analyses. Additional populations of male or female gametocytes were obtained from the PfDynGFP/PfP47mCherry line by FACS sorting into either GFP-positive (MG1) or mCherry-positive (FG) parasites, respectively. In addition we FACS sorted the cells that were both GFP- and mCherry positive (MG2). These were subjected to transcriptome analyses.
The P. falciparum gametocyte transcriptome
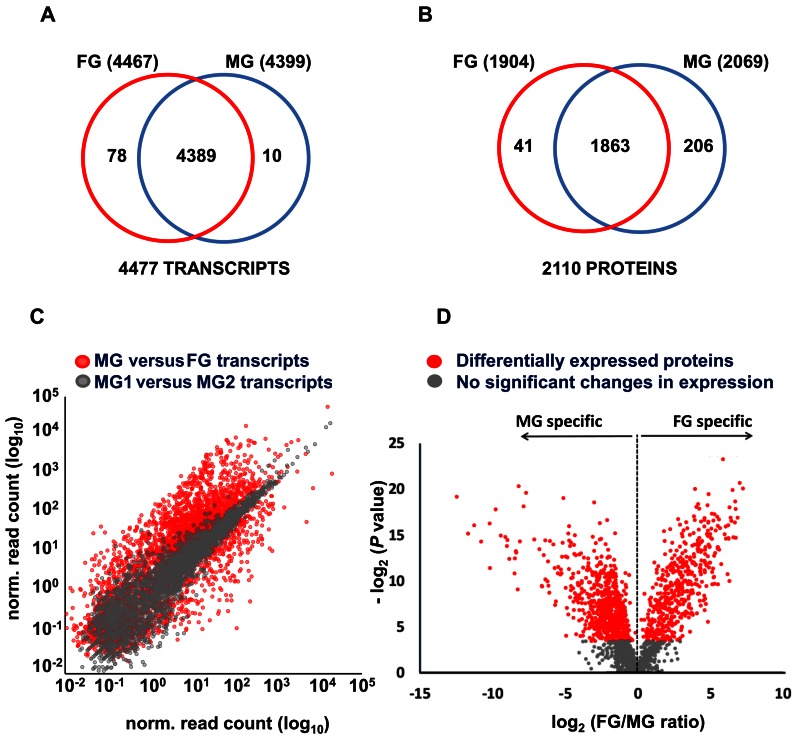
Messenger RNA abundance was analysed in purified MG1, MG2 and FG populations of PfDynGFP/PfP47mCherry with strand-specific RNA sequencing technology by calculating normalized read count per kilobase coding sequence. With a false discovery rate of 5% calculated based on the antisense read count (see Materials and Methods for details) 4477 different sense transcripts were identified, 98% of which were shared between MG and FG (Figure 2A, Supplementary Table S2). This indicates that nearly all genes are transcribed to some extent in gametocytes similarly to what has been observed for asexual blood stage parasites (35,36).
Figure 2.
Transcriptome and proteome of purified P. falciparum MG and FG. (A) Venn diagram depicting the overlap between the transcriptomes of MG and FG. (B) Venn diagram depicting the overlap between the proteomes of MG and FG. (C) Scatter plot of transcript abundance (normalised read counts) of genes in FG and MG (in red). In grey, normalized read count values for genes in the two MG populations, MG1 and MG2 are shown. (D) Volcano plot of protein abundance in FG and MG, showing differential expression of proteins in the two genders. Depicted in the plot is the comparison of FG/MG ratios versus P–values (Student's t-test between replicate measurements). Red dots: Differentially Expressed Proteins (DEP) with significant P-value. Grey dots: Proteins not significantly differentially expressed between MG and FG.
A very high level of correlation (Pearson r = 0.93) was observed between the expression values of the MG1 and MG2 samples (Figure 2C and Supplementary Figure S3B) corroborating the above-mentioned light microscopic analyses. Notably, this correlation was even higher than the correlation between biological replicates of MG1 and MG2 (r2 = 0.83 and 0.85), respectively (see Supplementary Figure S1). This confirms that both MG1 (GFP+) and MG2 (GFP +/ mCherry+) populations mainly consist of male gametocytes. Therefore, we have used the average mRNA abundance value for all genes from these two populations to represent the MG transcriptome for all follow-up analyses.
A scatter plot of MG and FG expression values revealed significant quantitative differences between the male and female transcriptomes (Figure 2C). Accordingly, SOTA clustering identified 1893 transcripts upregulated in FG and 2078 transcripts upregulated in MG, respectively of which 2918 transcripts were significantly differentially transcribed (Supplementary Table S2). The combined data show that a large proportion of the gametocyte transcriptome (66.4%) is differentially transcribed in MG and FG indicating for the first time that global, sex-specific transcriptional regulation plays a major role in gametocyte differentiation into MG or FG.
Interestingly, 20 out of the 27 ApiAP2-type transcription factors also show differential mRNA abundance (Supplementary Table S2) with PfAP2-G identified to be key for the asexual to sexual developmental switch (37). Some of these transcription factors might play a role in MG and FG differentiation, while others are involved in transcriptional regulation in the stages after gametocyte activation, i.e. in gametes or zygotes.
The P. falciparum gametocyte proteome
The protein abundance levels in MG (PfDynGFP) and FG (PfP47GFP) were determined by tandem mass spectrometry. Parasite lysates were separated into a cytosolic and a pellet fraction and subsequently fractionated by one-dimensional SDS-PAGE into 10 gel slices. Tryptic digests were measured in triplicate by nano-LC-MS/MS (180 runs) resulting in the identification of a total of 2110 P. falciparum proteins in MG and FG. After combining these data sets with the proteome of mixed-sex stage V gametocytes we identified 2241 proteins with a false discovery rate of 1% (Figure 2B, Supplementary Table S3), comprising altogether 34 316 unique peptide sequences (Supplementary Table S4). Proteins that were detected multiple times in either MG or FG (1926 out of the 2110) were subjected to a label-free quantitative analysis (38–41). Strong correlations (0.903 < r < 0.993) between technical replicates of mass spectrometry runs demonstrated a high reproducibility (Supplementary Figure S3A). Significant quantitative differences between MG and FG were assessed by Student's t-tests and visualized in a Volcano plot (Figure 2D). About half (47.6%) of the quantifiable proteome (1926 proteins) was significantly differentially expressed between MG and FG with 469 proteins up-regulated in FG, and 446 in MG (Supplementary Table S3).
Validation of gametocyte transcriptome and proteome
The transcriptome and proteome expression patterns in MG and FG were verified with a set of marker genes (9,42–50). All 16 known male-specific markers were up-regulated in both the MG proteome and transcriptome (Table 1), except for ACTII that was upregulated in the FG transcriptome. Similarly, 12 known female-specific markers were nearly all detected at a higher abundance in the FG transcriptome and proteome (9–11,15,50–54) (Table 1). Based on protein and transcript expression of these markers the purities of isolated MG and FG were estimated to be >95%, which is supported by the IFA and exflagellation assays (see above).
Table 1. Transcript and protein levels, shown as female/male ratio, of known marker-proteins of Plasmodium female (FG) or male (MG) gametocytes.
| gene ID | Product | P. falciparumA | P. bergheiB | Marker | |
|---|---|---|---|---|---|
| FG/MG protein ratio | FG/MG transcript ratio | FG/MG protein ratio | |||
| PF3D7_1250100 | Pfg377 (51) | 5.9E+03 | 3.8E+01 | 4.9E+01 | FG |
| PF3D7_1346800 | 6-cysteine protein (P47) (54) | 2.2E+03 | 2.2E+01 | 1.7E+01 | FG |
| PF3D7_1475500 | LCCL protein (CCp1) (15) | 2.1E+03 | 2.3E+01 | 6.1E+00 | FG |
| PF3D7_1426500 | ABC transporter, (EPP family)(53) | 1.5E+03 | 4.4E+01 | N.D | FG |
| PF3D7_1407000 | LCCL protein (CCp3) (9,15) | 8.1E+02 | 7.4E+00 | 6.8E+00 | FG |
| PF3D7_0719200 | NIMA related kinase 4 (NEK4)(9) | 9.7E+01 | 2.8E+01 | 2.0E+01 | FG |
| PF3D7_1474900 | trailer hitch homolog (CITH) (11) | 9.7E+01 | 1.6E+00 | 1.1E+01 | FG |
| PF3D7_0320800 | ATP-dep.RNA helicase (DOZI) (10) | 5.3E+01 | 3.1E+00 | 1.1E+01 | FG |
| PF3D7_0729900 | dynein heavy chain, putative (9) | 2.3E+01 | 5.9E-01 | 2.3E+00 | FG |
| PF3D7_0415600 | adenylate kinase (GAK) (50) | 7.0E+00 | 1.2E+01 | N.D | FG |
| PF3D7_0525900 | NIMA related kinase 2 (NEK2) (52) | 4.6E+00 | 2.7E+01 | N.D | FG |
| PF3D7_1031000 | Pfs25 (15) | 2.3E+00 | 3.6E+01 | N.D | FG |
| PF3D7_0422300 | alpha tubulin 2 (9) | 7.6E-01 | 2.5E-02 | 2.5E-01 | MG |
| PF3D7_1441400 | FACT-S (48) | 4.0E-01 | 2.0E-01 | 6.1E-02 | MG |
| PF3D7_0517400 | FACT-L (9,48) | 1.7E-01 | 1.8E-01 | 1.4E-02 | MG |
| PF3D7_1412500 | actin II (ACT2) (42) | 1.5E-01 | 1.6E+00 | 1.1E-01 | MG |
| PF3D7_1113900 | MAP2 protein kinase (9,49,50) | 9.1E-02 | 8.6E-02 | 1.1E-01 | MG |
| PF3D7_1014200 | HAP2 (9,47) | 8.3E-02 | 1.5E-02 | N.D | MG |
| PF3D7_0717500 | CDPK4 protein kinase (50) | 7.9E-02 | 3.8E-02 | 1.4E-01 | MG |
| PF3D7_0302100 | SRPK1 protein kinase (50) | 6.6E-02 | 1.7E-01 | N.D | MG |
| PF3D7_1216700 | perforin like protein 2 (PPLP2) (43) | 6.0E-02 | 2.2E-01 | N.D. | MG |
| PF3D7_1122900 | dynein heavy chain, putative (9) | 5.6E-02 | 1.1E-02 | 1.9E-01 | MG |
| PF3D7_0905300 | dynein heavy chain, putative (9) | 5.5E-02 | 2.8E-03 | 1.3E-01 | MG |
| PF3D7_1023100 | dynein heavy chain, putative (9) | 5.1E-02 | 2.6E-02 | 1.6E-01 | MG |
| PF3D7_1228300 | NEK1 protein kinase (44) | 4.1E-02 | 1.4E-01 | 4.7E-01 | MG |
| PF3D7_1465800 | dynein beta chain, putative (9) | 3.3E-02 | 8.4E-02 | 5.7E-01 | MG |
| PF3D7_0111000 | kinesin, putative (9) | 2.0E-02 | 9.1E-02 | 1.6E-01 | MG |
| PF3D7_0208900 | 6-cysteine protein (P230p) (45) | 1.9E-02 | 1.9E-02 | 6.3E-01 | MG |
A: Determined in this study; B: Proteome evidence of the P. berghei orthologs; based on reprocessed proteome data from Khan et al. Cell, 121, 675-687 (2005). Quantification of P. berghei protein levels was performed in this study; N.D.: Not Detected ; Gene ID underlined: reporter markers for constructs purified by FACS.
We then compared our proteome data with putative sets of MG and FG enriched P. falciparum proteins, that were identified by a comparison of proteomes of parasites from a MG and FG producing line with that of parasites from a line producing only FG (55). In this study, 258 genes were found to be upregulated in FG compared to MG of which 223 were shared with our male and female gametocyte proteome and 171 of these genes (77%) were also upregulated in our FG proteome. Of the 174 genes that were found to be upregulated in MG (55), 156 genes were also identified in our data set, including 136 (87%) that were also upregulated in our MG proteome. The large overlap in both FG and MG upregulated genes between the two studies supports our purification strategy in obtaining male and female gametocytes.
We next examined the degree of similarity in protein abundance between gametocytes of P. falciparum and gametocytes of P. berghei. We first quantified protein abundance in P. berghei gametocytes by analysing the published P. berghei MG and FG proteomes (9) (879 proteins; Supplementary Table S5) using the peptide counting method emPAI (56). Clustering analysis revealed that most of these proteins exhibited similar sex-specificity in the two different species (Supplementary Figure S4A). Pearson correlation analyses of FG/MG protein ratios between both species (Supplementary Figure S4B) revealed a moderate concordance (r = 0.62). In P. berghei MG and FG differential protein expression was observed for a number of protein kinases and phosphatases with comparable expression profiles for most of these proteins in P. falciparum MG and FG. In the proteomes of P. falciparum gametocytes 5 additional protein kinases and 10 phosphatases were identified that were upregulated in FG and 10 additional kinases and 3 phosphatases that were upregulated in MG (Supplementary Table S6) likely due to increased proteomic coverage compared to the P. berghei gametocyte proteome.
Taken together the expression patterns of established markers in purified male and female gametocytes and the similarity in expression patterns with large scale FG and MG proteomes confirm the validity of our data set.
Omics data underline gender-specific biology of gametocytes
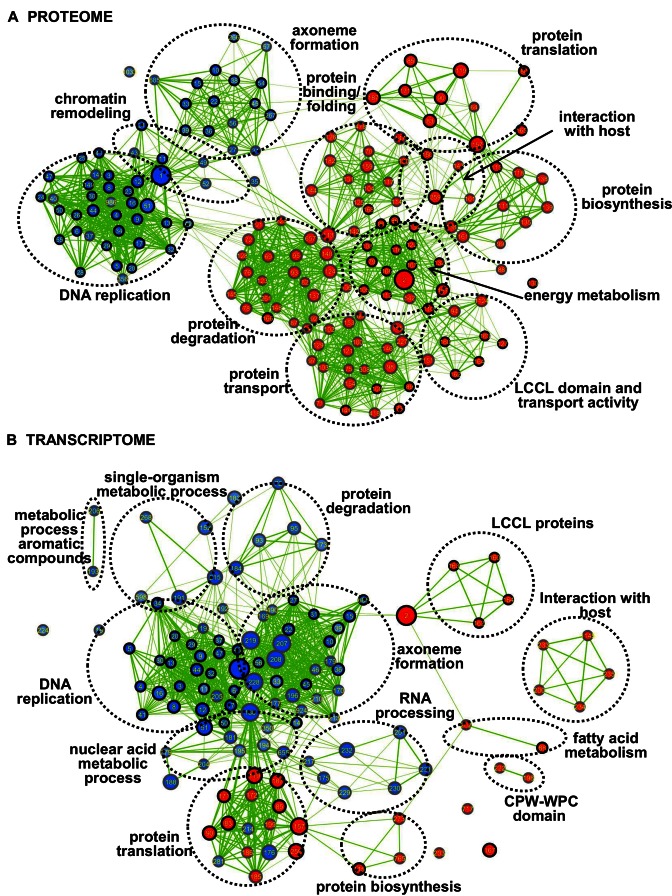
To discover gender-specific biological processes and functions we performed GSEA of quantified P. falciparum proteome and transcriptome. We first compiled gene sets from various sources including metabolic pathways, GO terms, protein domains, manually-curated annotations and protein–protein interaction (PPI) subnetworks. We included 23 subnetworks that were identified by the MCODE clustering algorithm from the putative gametocyte protein interaction network composed of 1167 nodes (proteins) and 11 223 edges (interactions) functionally annotated by GO enrichment analyses. The MG and FG enrichment maps of the proteome and transcriptome are shown in Figure 3 for gene sets passing moderate conservative statistical significance thresholds (p-value < 0.005, FDR < 0.075). In the proteome we identified 109 FG enriched gene sets (red nodes) and 57 gene sets (blue nodes) upregulated in MG (Figure 3A, Supplementary Table S7A) while in the transcriptome there was enrichment for 32 FG gene sets and 87 MG sets (Figure 3B). Sex-specific functionally related gene sets were found as clusters in the enrichment maps representing subnetworks of overlapping gene sets (Figure 3).
Figure 3.
GSEA enrichment maps of protein and transcripts in purified P. falciparum MG and FG identifying gender-enriched biological networks. Enrichment maps from GSEA showing gene sets in an interaction network with nodes of (A) proteins or (B) transcripts in either MG (blue) or FG (red) with moderately conservative statistical significance (p < 0.005, FDR < 0.075 and overlap coefficient = 0.2). Clustered sub networks of nodes (depicted as dotted circles) reflect generic functional networks. Nodes represent enriched gene sets, where node size corresponds to the number of genes and colour intensity corresponds to statistical significance (P-value). Edges represent overlap between gene sets with line thickness correlating to the degree of overlap. Gene sets identified in P. falciparum and P. berghei proteome are shown in (A) with node border colour in black. Gene sets identified in P. falciparum proteome and transcriptome are depicted in (B) with node border colour in black. Supplementary Table S7 provides the details of the GESA analysis and the identification of the different nodes and edges.
These clusters highlight the divergent roles that MG and FG require for life cycle progression in the mosquito host. MG show up-regulation of proteins involved in DNA replication and axoneme formation linked to the rapid formation of eight haploid flagellar and motile male gametes after activation of the MG in the mosquito midgut. Furthermore, GSEA of MG transcriptomes identified 48 gene sets that were (in part) related to RNA processing, nuclear acid metabolic processes as well as DNA replication and axoneme formation. In contrast, FG show up-regulation of transcriptome and proteome sub-networks of genes involved in protein synthesis, metabolic activity and protein translation, proteins that may play a role for the development of the zygote after fertilisation of the female gamete. Despite the clear morphological differences between gametocytes of P. berghei and P. falciparum the majority (85%) of the gene sets identified by GSEA in the proteomes of P. berghei gametocytes (Supplementary Table S7B and C) were also identified by GSEA in the P. falciparum gametocytes. This could indicate a conservation of differentiation pathways in gametocytes of both species.
Importantly, this analysis appears to be powerful in allocating putative functions to so far uncharacterized proteins. For example, we detected two subnetworks (MCODE clusters) in male gametocytes annotated to DNA replication that contained three conserved Plasmodium proteins (PF3D7_0503200, PF3D7_1319400 and PF3D7_1334100) without annotation. Their location in the ‘DNA replication subnetwork’ suggests that these proteins are involved in DNA replication. In line with this, delta-BlastP identified homology of PF3D7_0503200 and PF3D7_1334100 with the ORC1/CDC6 family of archaeal proteins (TIGR02928), and eukaryotic CDC6 proteins (COG1474) (Supplementary Figure S5) with functions in replication, recombination and repair. This together with the recent identification of Alba-domain proteins in Plasmodium (57), key organizer of the archaean Sulfolobus genome together with Sir2 (58) suggest that the malaria parasite has retained aspects of ancient regulatory mechanisms.
Integrated analysis of gametocyte transcriptome and proteome identifies a putative set of translationally repressed transcripts in FG
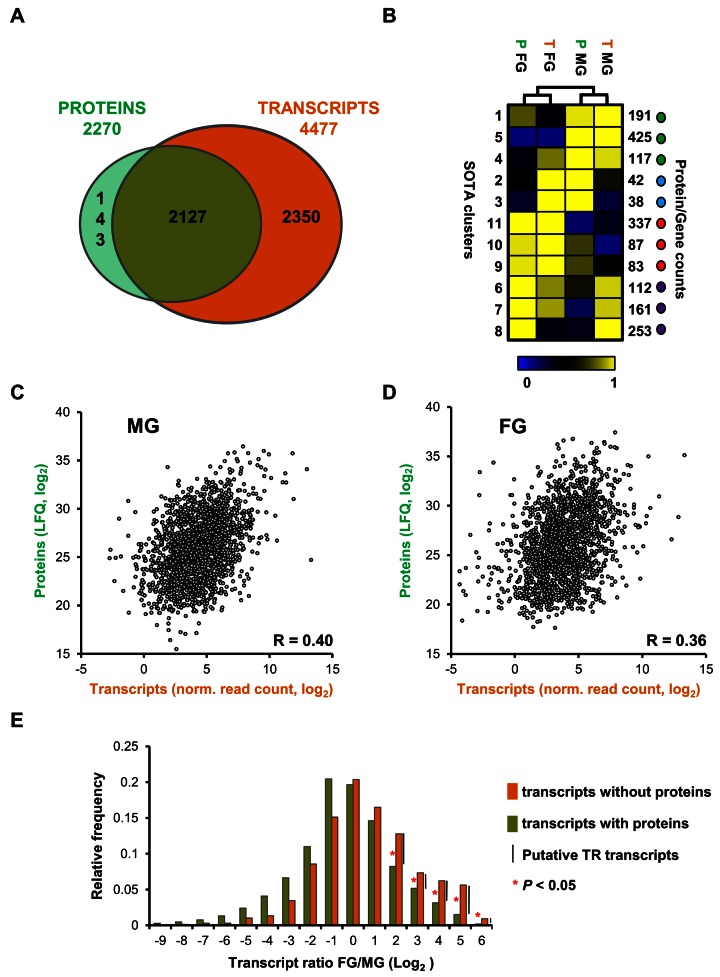
To investigate the correlation between transcriptome and proteome, we compared global transcript and protein levels for all genes for which we had obtained quantitative data in both the proteome and transcriptome data sets (Figure 4A). Comparison through regression analysis showed rather moderate correlation between transcript and protein levels in both MG (Pearson r = 0.40 Figure 4C) and FG (Pearson r = 0.36, Figure 4D), consistent with previously reported correlation coefficients between transcript and protein levels identified in populations of mixed MG and FG of P. falciparum (Spearman r = 0.37) (59). SOTA clustering analysis using relative expression of transcripts and proteins (Figure 4B), however, suggested that for most genes (67%) the transcript and the protein show similar sex-specificity. We observe better correlation between FG/MG ratios in the proteome and transcriptome (Pearson r = 0.60, Supplementary Figure S6). In conclusion, while there is a rather low global correlation between mRNA and protein abundance, sex-specificity of the transcript is a good indicator for the sex-specificity of the protein and hence sex-specific function.
Figure 4.
Comparison between transcriptome and proteome of purified P. falciparum MG and FG. (A) Venn diagram showing the overlap of genes identified in proteomes and transcriptomes (MG and FG combined). (B) Self-organising tree algorithm (SOTA) clustering of genes based on relative abundance of their transcripts and proteins identifies clusters of genes that are co-expressed (green and red dots) and clusters of genes that show opposite expression when transcript and protein levels are compared (blue and purple dots). (C) Scatter plot depicting protein abundance (log2LFQ) and transcript abundance (log2 (normalized read count)) for genes identified in purified MG. (D) Scatter plot depicting protein abundance (log2LFQ) and transcript abundance (normalized read count) for genes identified in purified FG. Pearson correlation values are shown in the corner of the plots. (E) FG/MG ratio distribution of transcripts with (green) or without (orange) protein evidence, showing that for upregulated genes in female gametocytes a significant higher percentage of genes has transcripts without proteins compared to genes with both transcript and protein.
In addition to the genes that were expressed as both transcripts and proteins in gametocytes (Figure 4A), we detected 143 genes that were expressed only as protein and a much larger set of 2350 genes that were identified only at the transcript level. The identification of a large set of genes that had no (quantifiable) proteome evidence but were present as transcripts was not unexpected since we found nearly all genes expressed to some extent, similar to asexual blood parasites (36).
However, this set also includes genes, which are among the most highly transcribed. Moreover, there is a clear FG bias for these genes, which is apparent from Figure 4E where we assessed the FG/MG gene ratio distribution of these genes. The presence of highly abundant transcripts without corresponding protein expression in P. falciparum FG is reminiscent of a large set of TR transcripts in FG of the rodent parasite, P. berghei. Such transcripts are stored in cytoplasmic mRNP for translation during post-fertilization development (12,16).
From the genes expressed only as transcripts we generated a list of putative TR genes in P. falciparum FG (Supplementary Table S8). We first identified 810 upregulated FG transcripts with significant difference in FG/MG gene ratio distribution (p < 0.05, 2-sided Fisher's exact test) and without quantitative proteomics data. To prevent including genes in this list for which lack of proteomic evidence could potentially be explained by low level transcription, we implemented a high expression level cut-off value of 15 normalised tag count in FG and excluded 298 transcripts. This resulted in a set of 512 putative TR transcripts in P. falciparum FG.
This set of TR transcripts included 18 out of 33 P. falciparum orthologs of P. berghei genes (Supplementary Table S9) for which experimental evidence of translational repression in FG has been reported including the genes pfs28, the ApiAP2 transcription factor ap2-0 and plasmepsin IV (10,12,60,61).
These TR transcripts also shows a substantial overlap with a putative set of 731 TR transcripts that have been identified in P. berghei FG, based on the association of transcripts with translational repressors DOZI or CITH (12). From the 687 corresponding P. falciparum orthologs, we found 427 transcripts without quantified proteins in female gametocytes of which 185 are present in the set of P. falciparum TR transcripts. Accordingly, there is a clear overrepresentation of DOZI/CITH transcripts in this set relative to non-TR genes corresponding to a 5.9-fold enrichment (p = 1.7 E-54, 2-sided Fisher's exact test). Notably, in the P. falciparum FG proteome we found upregulation of all 16 protein homologues to the protein components of the P. berghei CITH/DOZI complex (11) (Supplementary Table S10), suggesting that similar mRNPs exist in rodent and human parasites regulating translation in female gametocytes during transmission.
In a P. falciparum mutant lacking the RNA binding protein Puf2, 326 genes were identified that showed increased transcripts levels total gametocytes (stage III and V) compared to wild type, and hence been suggested to be translationally repressed (16). For 153 of these genes we did not find quantifiable proteins including 86 transcripts that are present in our set of TR transcripts. Thus, our list of putative TR transcripts shows a 9.9-fold enrichment (p = 1.0 E-38) for transcripts whose stability is regulated by Puf2.
Combining all TR studies, our set of P. falciparum TR transcripts in FG consist of 252 genes which have been associated previously with TR in gametocytes and we report 260 novel putative TR transcripts in P. falciparum FG that had not been found to be associated with CITH/DOZI or Puf2. The large number of novel TR transcripts could indicate that P. falciparum FG express additional proteins and/or mRNP involved in TR. This is also suggested by the analysis of the P. falciparum mutant lacking expression of Puf2 (16). The DOZI mRNP complex in P. berghei does not contain Puf proteins (10,11), suggesting DOZI and Puf2 are components of different protein complexes involved in TR. In addition, there was only limited overlap between deregulated transcripts in the P. falciparum puf2 gene-deletion mutant and DOZI-associated transcripts in P. berghei, indicating that DOZI and Puf2 function in different pathways, even though they may act upon a subset of similar TR transcripts (16,62). In P.berghei, Puf2 also regulates translation in sporozoites during stage transition to the rodent host (16,62). A sub set of 26 Puf2-mediated TR transcripts in sporozoites (63) is found in our list of translationally repressed transcripts in female gametocytes (Supplementary Table S11). Further research aiming to explore interactions between TR transcripts and different protein complexes is required to reveal whether fundamental differences exist between rodent and human parasites with respect to control of gene expression during sexual differentiation and further development in the mosquito.
Functional annotation analysis of the putative TR transcripts in P. falciparum FG
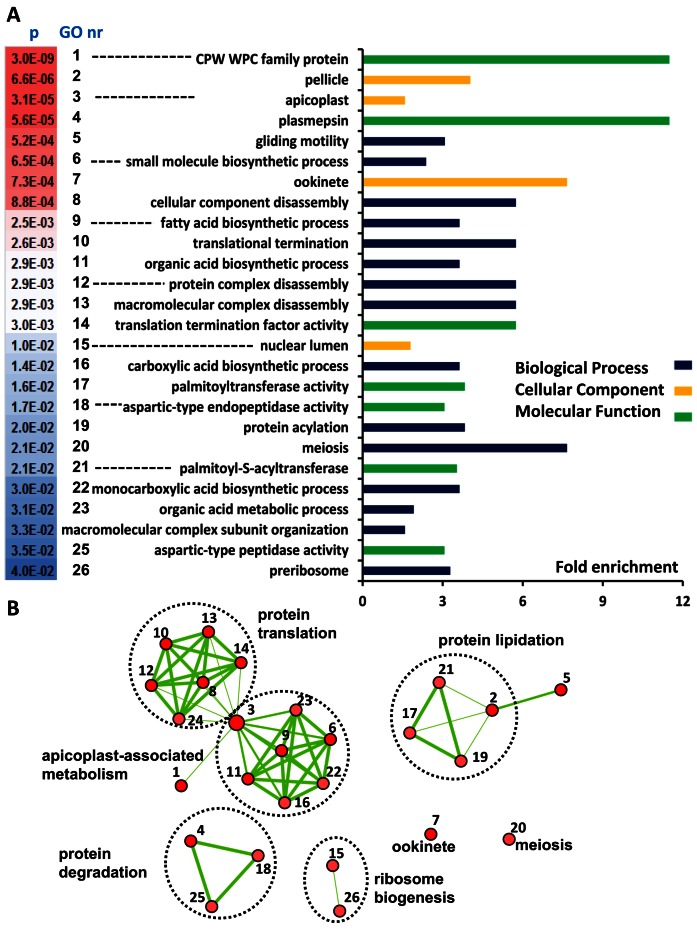
The functions of the set of putative TR transcripts were identified by GO enrichment analysis against a background of all P. falciparum genes (Supplementary Table S12). The degree of over representation for statistical significant GO terms represented by fold enrichment is depicted in Figure 5A with overlap between GO terms visualized by edges in a network (Figure 5B), where overlapping GO terms group together in clusters.
Figure 5.
Gene Ontology Enrichment analysis of the putative set of TR transcripts in P. falciparum FG. (A) Fold enrichment of Gene Ontology (GO) terms of the putative set of TR transcripts relative to the background of all P. falciparum genes. Gene Ontologies representing Biological Processes are presented in dark blue lines, Cellular Component in orange and Molecular Function in green. (B) Enrichment map of GO terms in an interaction network. Nodes represent enriched GO terms with colour intensities reflecting statistical significance and numbers referring to descriptions provided in (A). Edges represent overlap between GO terms with line thickness correlating to the degree of overlap. Supplementary Table S12 provides the details of the GO enrichment analysis and the genes involved.
The statistically most significant and most enriched term in the set of TR transcripts in FG is the CPW-WPC protein family with all 8 members present. This includes the ortholog of PyCPW-WPC-1 that was found to be translationally repressed in gametocytes of the rodent parasite P. yoelii where it is only translated after fertilization and trafficked to the surface of developing ookinetes (59).
Five clusters of GO terms are found in the enrichment network representing a variety of biological roles for the translationally repressed transcripts in apicoplast-associated metabolism, protein translation, -degradation, -lipidation and ribosome biogenesis. The largest cluster, apicoplast-associated metabolism, is comprised of 7 nodes containing metabolic GO terms in the apicoplast including type II fatty acid synthesis with 4 repressed transcripts (PF3D7_0420200 (Holo-ACP synthase); PF3D7_0615100 (Fabl); PF3D7_1020800 (Dihydrolipoamide S-acetyltraferase) and PF3D7_1124500 (Pyruvate dehyrdogenase E1 subunit) in the pathway (Supplementary Figure S7). We previously found an important distinction for this pathway between rodent and human Plasmodium species (64). Whereas type II fatty acid synthesis is not essential for development of P. berghei blood- nor mosquito-stages, this pathway was essential for complete development of P. falciparum oocysts in the mosquito.
The significance of TR genes in the fatty acid biosynthesis pathway is supported by the presence of the ‘protein lipidation’ cluster including 4 members of the palmitoyl-S-acyltransferases (PATs) protein family (DHHC3-4, DHHC 9–10). Notably, the PATs DHHC2, DHHC3 and DHHC10 are also present in the set of TR transcripts in P. berghei (65). In our P. falciparum data DHHC3 and DHHC10 are present, but not DHHC2 that was found to be upregulated in the male gametocyte proteome. Recently, protein palmitoylation has been shown to be essential for developmental progression of P. berghei zygotes (65). The presence of PATs in the P. falciparum repressome corroborates the important role for protein palmitoylation in development during further progression in the mosquito.
The enrichment map of TR transcripts (Figure 5B) shows two unconnected nodes that do not share genes with other nodes. Meiosis is implicated in translation repression with the transcripts DMC1 (PF3D7_0816800; meiotic recombination protein) and MND1 (PF3D7_1461500; meiotic nuclear division protein 1) and several transcripts expressed in ookinetes are repressed, involving genes encoding the ookinete adhesin P28 and multiple putatively secreted ookinete proteins (PSOP6, 13, 20). These findings are supported by previous studies reporting the involvement of surface adhesins and micronemal proteins in midgut recognition and cell traversal of ookinetes (12,66,67).
Finally the enrichment map (Figure 5B) contains several nodes associated with translation and ribosome biogenesis. These contain numerous moderately enriched molecular functions relevant, e.g. for preribosome, protein complex disassembly and translational termination. This is not unexpected given that FG after fertilization will develop further from zygotes into ookinetes, and during this development protein biosynthesis will primarily rely on regulated processing and translation of transcripts stored in mRNP particles and where transcription is minimal (12).
Function of translationally repressed genes extends beyond the zygote and ookinete stage
To investigate at which developmental stage TR transcripts in FG are functional we compared our list of TR genes with the phenotype of published P. berghei mutants that lack those genes (available in the database www.pberghei.eu). For 10 TR genes (Supplementary Table S8), gene disruption affects development of the zygote/ookinete development in P. berghei. These include the genes encoding P28, two Inner Membrane Complex proteins—also known as alveolins—DMC1 (meiotic recombination protein 1), the protein kinase NEK2, the phosphatase PPKL, the transcription factor AP2-O and Gamer. Disruption of the P. berghei gamer gene affected not only zygote development but also fertilization since it is also expressed as a protein in male gametes of P. berghei where it plays a role in gamete release (68). Next to the 10 genes with a role in zygote/ookinete development our set of TR transcripts contains 12 genes (Supplementary Table S8) where gene disruption results in a phenotypic effect only after ookinete formation, i.e. during development of oocyst and/or sporozoites. Gene disruption of plasmepsin VI and rhomboid protease rom3 affects both oocyst and sporozoite development while gene knockouts of trap, trap-like (tlp) and fabI exclusively impairs sporozoite development. This delayed phenotypic effect suggests that translational repression in FG is not only in preparation of rapid protein synthesis in zygotes just after fertilization as has been proposed previously (10–12,68), but that TR transcripts can be stored for prolonged periods in order to produce proteins that are required much later during parasite development in the mosquito.
Indeed, in P. berghei prolonged storage of translational repressed transcripts has been shown for several members of the LCCL protein family, PbLAP4-6 (PbCCP2,PbPFNA and Pb CCP4, respectively) (14). The transcripts are translated after fertilisation and their protein products are associated with crystalloid bodies in maturing ookinetes (14). However, mutants lacking expression of all protein family members only show a phenotype during further development of the oocyst, i.e. in the formation of sporozoites. This may indicate that not only transcripts but also proteins are stored for later use. Also in P. falciparum, several LCCL members are implicated in oocyst/sporozoite development in the mosquito (69). Interestingly, translational repression is presumably absent for all members of the P. falciparum LCCL family members (15) and it has been suggested that these proteins in P. falciparum play also a role in fertilization (70). We also found protein evidence for five LCCL members in female gametocytes. Combined, these observations on expression of LCCL proteins and their roles during development of oocysts and sporozoites indicate that in addition to transcripts, Plasmodium species also store proteins for later use.
Evidence for longer term storage of TR transcripts for later development was also obtained through comparison with Plasmodium proteome data of different mosquito stages, such as gametes (27,68), zygotes (71), ookinetes (65,71), oocysts and oocyst-derived sporozoites (72) and salivary gland sporozoites (72,73). We found protein evidence in these stages for 196 TR genes and in all stages we find proteins that were not detected in the preceding life cycle stage. This observation is indeed in support of prolonged storage of the TR transcripts in P. falciparum and translation in multiple life cycle stages throughout complete development within the mosquito.
CONCLUSIONS
Sexual reproduction is an essential process in the lifecycle of malaria parasites and key to P. falciparum parasite transmission via the mosquito vector. In this study we report the first genome-wide transcriptome analysis of separated male and female gametocytes of P. falciparum together with the most comprehensive Plasmodium proteome of both sexes to characterize the molecular events underlying sexual differentiation of gametocytes. We observed that sex-specific gametocyte morphologies are driven by clear diversification of the gene expression program as 66% of the transcriptome and 47% of the proteome show differential abundance between the sexes. By an integrated analysis of the proteome and transcriptome we identified for the first time a putative set of translationally repressed P. falciparum transcripts in female gametocytes comprising 512 genes, including 260 novel putative translational repressed transcripts. Many of these transcripts lack protein evidence and/or essential function directly after gamete formation and fertilization suggesting prolonged storage of these transcripts for translation by yet unidentified proteins.
Supplementary Material
Acknowledgments
The authors thank the Nijmegen Proteomics Facility for usage of the LC-MS/MS instrumentation to carry out this study. The authors gratefully acknowledge Prof. Hagai Ginsburg for providing MPMP data included in the GSEA analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Netherlands Organization for Scientific Research [NWO-Vidi 864.11.007 to R.B.]; The National Institutes of Health (NIH) [EuPathDB-Driving Biological Project sub-award # 553539 to R.B.]; VIDI fellowship from the Netherlands Organization for Scientific Research [NWO; project number 016.158.306 to S.R. and T.B.]; Network of Excellence EviMalaR [Health-2009-2.3.2-1-242095 to C.J.J, E.L, R.B and R.W.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Baker D.A. Malaria gametocytogenesis. Mol. Biochem. Parasitol. 2010;172:57–65. doi: 10.1016/j.molbiopara.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bousema T., Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 2011;24:377–410. doi: 10.1128/CMR.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kooij T.W., Matuschewski K. Triggers and tricks of Plasmodium sexual development. Curr. Opin. Microbiol. 2007;10:547–553. doi: 10.1016/j.mib.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Kuehn A., Pradel G. The coming-out of malaria gametocytes. J. Biomed. Biotechnol. 2010:976827. doi: 10.1155/2010/976827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stone W., Goncalves B.P., Bousema T., Drakeley C. Assessing the infectious reservoir of falciparum malaria: past and future. Trends Parasitol. 2015;31:287–296. doi: 10.1016/j.pt.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Hawking F., Wilson M.E., Gammage K. Evidence for cyclic development and short-lived maturity in the gametocytes of Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 1971;65:549–559. doi: 10.1016/0035-9203(71)90036-8. [DOI] [PubMed] [Google Scholar]
- 7.Rogers N.J., Hall B.S., Obiero J., Targett G.A., Sutherland C.J. A model for sequestration of the transmission stages of Plasmodium falciparum: adhesion of gametocyte-infected erythrocytes to human bone marrow cells. Infect. Immun. 2000;68:3455–3462. doi: 10.1128/iai.68.6.3455-3462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon M.W., Dearnley M.K., Hanssen E., Gilberger T., Tilley L. Shape-shifting gametocytes: how and why does P. falciparum go banana-shaped? Trends Parasitol. 2012;28:471–478. doi: 10.1016/j.pt.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Khan S.M., Franke-Fayard B., Mair G.R., Lasonder E., Janse C.J., Mann M., Waters A.P. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell. 2005;121:675–687. doi: 10.1016/j.cell.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Mair G.R., Braks J.A., Garver L.S., Wiegant J.C., Hall N., Dirks R.W., Khan S.M., Dimopoulos G., Janse C.J., Waters A.P. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mair G.R., Lasonder E., Garver L.S., Franke-Fayard B.M., Carret C.K., Wiegant J.C., Dirks R.W., Dimopoulos G., Janse C.J., Waters A.P. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerreiro A., Deligianni E., Santos J.M., Silva P.A., Louis C., Pain A., Janse C.J., Franke-Fayard B., Carret C.K., Siden-Kiamos I., et al. Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte. Genome Biol. 2014;15:493–508. doi: 10.1186/s13059-014-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon M.W., Thompson J., Gardiner D.L., Trenholme K.R. Sex in Plasmodium: a sign of commitment. Trends Parasitol. 2008;24:168–175. doi: 10.1016/j.pt.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Saeed S., Carter V., Tremp A.Z., Dessens J.T. Translational repression controls temporal expression of the Plasmodium berghei LCCL protein complex. Mol. Biochem. Parasitol. 2013;189:38–42. doi: 10.1016/j.molbiopara.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholz S.M., Simon N., Lavazec C., Dude M.A., Templeton T.J., Pradel G. PfCCp proteins of Plasmodium falciparum: gametocyte-specific expression and role in complement-mediated inhibition of exflagellation. Int. J. Parasitol. 2008;38:327–340. doi: 10.1016/j.ijpara.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Miao J., Fan Q., Parker D., Li X., Li J., Cui L. Puf mediates translation repression of transmission-blocking vaccine candidates in malaria parasites. PLoS Pathog. 2013;9:e1003268. doi: 10.1371/journal.ppat.1003268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao J., Li J., Fan Q., Li X., Li X., Cui L. The Puf-family RNA-binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite Plasmodium falciparum. J. Cell Sci. 2010;123:1039–1049. doi: 10.1242/jcs.059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ifediba T., Vanderberg J.P. Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature. 1981;294:364–366. doi: 10.1038/294364a0. [DOI] [PubMed] [Google Scholar]
- 19.Ponnudurai T., Lensen A.H., Leeuwenberg A.D., Meuwissen J.H. Cultivation of fertile Plasmodium falciparum gametocytes in semi-automated systems. 1. Static cultures. Trans. R. Soc. Trop. Med. Hyg. 1982;76:812–818. doi: 10.1016/0035-9203(82)90116-x. [DOI] [PubMed] [Google Scholar]
- 20.Ponnudurai T., Lensen A.H., Meis J.F., Meuwissen J.H. Synchronization of Plasmodium falciparum gametocytes using an automated suspension culture system. Parasitology. 1986;93:263–274. doi: 10.1017/s003118200005143x. [DOI] [PubMed] [Google Scholar]
- 21.van Schaijk B.C., Janse C.J., van Gemert G.J., van Dijk M.R., Gego A., Franetich J.F., van de Vegte-Bolmer M., Yalaoui S., Silvie O., Hoffman S.L., et al. Gene disruption of Plasmodium falciparum p52 results in attenuation of malaria liver stage development in cultured primary human hepatocytes. PLoS One. 2008;3:e3549. doi: 10.1371/journal.pone.0003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponnudurai T., Lensen A.H., Van Gemert G.J., Bensink M.P., Bolmer M., Meuwissen J.H. Infectivity of cultured Plasmodium falciparum gametocytes to mosquitoes. Parasitology. 1989;98:165–173. doi: 10.1017/s0031182000062065. [DOI] [PubMed] [Google Scholar]
- 23.Fivelman Q.L., McRobert L., Sharp S., Taylor C.J., Saeed M., Swales C.A., Sutherland C.J., Baker D.A. Improved synchronous production of Plasmodium falciparum gametocytes in vitro. Mol. Biochem. Parasitol. 2007;154:119–123. doi: 10.1016/j.molbiopara.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Trang D.T., Huy N.T., Kariu T., Tajima K., Kamei K. One-step concentration of malarial parasite-infected red blood cells and removal of contaminating white blood cells. Malar. J. 2004;3:7–13. doi: 10.1186/1475-2875-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kensche P.R., Hoeijmakers W.A., Toenhake C.G., Bras M., Chappell L., Berriman M., Bartfai R. The nucleosome landscape of Plasmodium falciparum reveals chromatin architecture and dynamics of regulatory sequences. Nucleic Acids Res. 2015;44:2110–2124. doi: 10.1093/nar/gkv1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoeijmakers W.A., Bartfai R., Stunnenberg H.G. Transcriptome analysis using RNA-Seq. Methods Mol. Biol. 2013;923:221–239. doi: 10.1007/978-1-62703-026-7_15. [DOI] [PubMed] [Google Scholar]
- 27.Lasonder E., Ishihama Y., Andersen J.S., Vermunt A.M., Pain A., Sauerwein R.W., Eling W.M., Hall N., Waters A.P., Stunnenberg H.G., et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 28.Silvestrini F., Lasonder E., Olivieri A., Camarda G., van Schaijk B., Sanchez M., Younis Younis S., Sauerwein R., Alano P. Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics. 2010;9:1437–1448. doi: 10.1074/mcp.M900479-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 30.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 31.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer S., Grossmann S., Vingron M., Robinson P.N. Ontologizer 2.0–a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 34.Iwanaga S., Kato T., Kaneko I., Yuda M. Centromere plasmid: a new genetic tool for the study of Plasmodium falciparum. PLoS One. 2012;7:e33326. doi: 10.1371/journal.pone.0033326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartfai R., Hoeijmakers W.A., Salcedo-Amaya A.M., Smits A.H., Janssen-Megens E., Kaan A., Treeck M., Gilberger T.W., Francoijs K.J., Stunnenberg H.G. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 2010;6:e1001223. doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto T.D., Wilinski D., Assefa S., Keane T.M., Sarry L.R., Bohme U., Lemieux J., Barrell B., Pain A., Berriman M., et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kafsack B.F., Rovira-Graells N., Clark T.G., Bancells C., Crowley V.M., Campino S.G., Williams A.E., Drought L.G., Kwiatkowski D.P., Baker D.A., et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol. Cell. Proteomics. 2014;3:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Hubner N.C., Bird A.W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisser H., Nahnsen S., Grossmann J., Nilse L., Quandt A., Brauer H., Sturm M., Kenar E., Kohlbacher O., Aebersold R., et al. An automated pipeline for high-throughput label-free quantitative proteomics. J. Proteome Res. 2013;12:1628–1644. doi: 10.1021/pr300992u. [DOI] [PubMed] [Google Scholar]
- 42.Deligianni E., Morgan R.N., Bertuccini L., Kooij T.W., Laforge A., Nahar C., Poulakakis N., Schuler H., Louis C., Matuschewski K., et al. Critical role for a stage-specific actin in male exflagellation of the malaria parasite. Cell. Microbiol. 2011;13:1714–1730. doi: 10.1111/j.1462-5822.2011.01652.x. [DOI] [PubMed] [Google Scholar]
- 43.Deligianni E., Morgan R.N., Bertuccini L., Wirth C.C., Silmon de Monerri N.C., Spanos L., Blackman M.J., Louis C., Pradel G., Siden-Kiamos I. A perforin-like protein mediates disruption of the erythrocyte membrane during egress of Plasmodium berghei male gametocytes. Cell. Microbiol. 2013;15:1438–1455. doi: 10.1111/cmi.12131. [DOI] [PubMed] [Google Scholar]
- 44.Dorin-Semblat D., Schmitt S., Semblat J.P., Sicard A., Reininger L., Goldring D., Patterson S., Quashie N., Chakrabarti D., Meijer L., et al. Plasmodium falciparum NIMA-related kinase Pfnek-1: sex specificity and assessment of essentiality for the erythrocytic asexual cycle. Microbiology. 2011;157:2785–2794. doi: 10.1099/mic.0.049023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eksi S., Suri A., Williamson K.C. Sex- and stage-specific reporter gene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 2008;160:148–151. doi: 10.1016/j.molbiopara.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eksi S., Williamson K.C. Male-specific expression of the paralog of malaria transmission-blocking target antigen Pfs230, PfB0400w. Mol. Biochem. Parasitol. 2002;122:127–130. doi: 10.1016/s0166-6851(02)00091-9. [DOI] [PubMed] [Google Scholar]
- 47.Hirai M., Arai M., Mori T., Miyagishima S.Y., Kawai S., Kita K., Kuroiwa T., Terenius O., Matsuoka H. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr. Biol. 2008;18:607–613. doi: 10.1016/j.cub.2008.03.045. [DOI] [PubMed] [Google Scholar]
- 48.Laurentino E.C., Taylor S., Mair G.R., Lasonder E., Bartfai R., Stunnenberg H.G., Kroeze H., Ramesar J., Franke-Fayard B., Khan S.M., et al. Experimentally controlled downregulation of the histone chaperone FACT in Plasmodium berghei reveals that it is critical to male gamete fertility. Cell. Microbiol. 2011;13:1956–1974. doi: 10.1111/j.1462-5822.2011.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rangarajan R., Bei A.K., Jethwaney D., Maldonado P., Dorin D., Sultan A.A., Doerig C. A mitogen-activated protein kinase regulates male gametogenesis and transmission of the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:464–469. doi: 10.1038/sj.embor.7400404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tewari R., Straschil U., Bateman A., Bohme U., Cherevach I., Gong P., Pain A., Billker O. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Koning-Ward T.F., Olivieri A., Bertuccini L., Hood A., Silvestrini F., Charvalias K., Berzosa Diaz P., Camarda G., McElwain T.F., Papenfuss T., et al. The role of osmiophilic bodies and Pfg377 expression in female gametocyte emergence and mosquito infectivity in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2008;67:278–290. doi: 10.1111/j.1365-2958.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- 52.Reininger L., Tewari R., Fennell C., Holland Z., Goldring D., Ranford-Cartwright L., Billker O., Doerig C. An essential role for the Plasmodium Nek-2 Nima-related protein kinase in the sexual development of malaria parasites. J. Biol. Chem. 2009;284:20858–20868. doi: 10.1074/jbc.M109.017988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran P.N., Brown S.H., Mitchell T.W., Matuschewski K., McMillan P.J., Kirk K., Dixon M.W., Maier A.G. A female gametocyte-specific ABC transporter plays a role in lipid metabolism in the malaria parasite. Nat. Commun. 2014;5:4773–4785. doi: 10.1038/ncomms5773. [DOI] [PubMed] [Google Scholar]
- 54.van Schaijk B.C., van Dijk M.R., van de Vegte-Bolmer M., van Gemert G.J., van Dooren M.W., Eksi S., Roeffen W.F., Janse C.J., Waters A.P., Sauerwein R.W. Pfs47, paralog of the male fertility factor Pfs48/45, is a female specific surface protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 2006;149:216–222. doi: 10.1016/j.molbiopara.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 55.Tao D., Ubaida-Mohien C., Mathias D.K., King J.G., Pastrana-Mena R., Tripathi A., Goldowitz I., Graham D.R., Moss E., Marti M., et al. Sex-partitioning of the Plasmodium falciparum stage V gametocyte proteome provides insight into falciparum-specific cell biology. Mol. Cell. Proteomics. 2014;13:2705–2724. doi: 10.1074/mcp.M114.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Reddy B.P., Shrestha S., Hart K.J., Liang X., Kemirembe K., Cui L., Lindner S.E. A bioinformatic survey of RNA-binding proteins in Plasmodium. BMC Genomics. 2015;16:890–915. doi: 10.1186/s12864-015-2092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bell S.D., Botting C.H., Wardleworth B.N., Jackson S.P., White M.F. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 59.Kangwanrangsan N., Tachibana M., Jenwithisuk R., Tsuboi T., Riengrojpitak S., Torii M., Ishino T. A member of the CPW-WPC protein family is expressed in and localized to the surface of developing ookinetes. Malar. J. 2013;12:129–138. doi: 10.1186/1475-2875-12-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braks J.A., Mair G.R., Franke-Fayard B., Janse C.J., Waters A.P. A conserved U-rich RNA region implicated in regulation of translation in Plasmodium female gametocytes. Nucleic Acids Res. 2008;36:1176–1186. doi: 10.1093/nar/gkm1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall N., Karras M., Raine J.D., Carlton J.M., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 62.Cui L., Lindner S., Miao J. Translational regulation during stage transitions in malaria parasites. Ann. N.Y. Acad. Sci. 2015;1342:1–9. doi: 10.1111/nyas.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomes-Santos C.S., Braks J., Prudencio M., Carret C., Gomes A.R., Pain A., Feltwell T., Khan S., Waters A., Janse C., et al. Transition of Plasmodium sporozoites into liver stage-like forms is regulated by the RNA binding protein Pumilio. PLoS Pathog. 2011;7:e1002046. doi: 10.1371/journal.ppat.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Schaijk B.C., Kumar T.R., Vos M.W., Richman A., van Gemert G.J., Li T., Eappen A.G., Williamson K.C., Morahan B.J., Fishbaugher M., et al. Type II fatty acid biosynthesis is essential for Plasmodium falciparum sporozoite development in the midgut of Anopheles mosquitoes. Eukaryot Cell. 2014;13:550–559. doi: 10.1128/EC.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santos J.M., Kehrer J., Franke-Fayard B., Frischknecht F., Janse C.J., Mair G.R. The Plasmodium palmitoyl-S-acyl-transferase DHHC2 is essential for ookinete morphogenesis and malaria transmission. Sci. Rep. 2015;5:16034–16043. doi: 10.1038/srep16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Angrisano F., Tan Y.H., Sturm A., McFadden G.I., Baum J. Malaria parasite colonisation of the mosquito midgut–placing the Plasmodium ookinete centre stage. Int. J. Parasitol. 2012;42:519–527. doi: 10.1016/j.ijpara.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 67.Lal K., Prieto J.H., Bromley E., Sanderson S.J., Yates J.R., 3rd, Wastling J.M., Tomley F.M., Sinden R.E. Characterisation of Plasmodium invasive organelles; an ookinete microneme proteome. Proteomics. 2009;9:1142–1151. doi: 10.1002/pmic.200800404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Talman A.M., Prieto J.H., Marques S., Ubaida-Mohien C., Lawniczak M., Wass M.N., Xu T., Frank R., Ecker A., Stanway R.S., et al. Proteomic analysis of the Plasmodium male gamete reveals the key role for glycolysis in flagellar motility. Malar. J. 2014;13:315–326. doi: 10.1186/1475-2875-13-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pradel G., Hayton K., Aravind L., Iyer L.M., Abrahamsen M.S., Bonawitz A., Mejia C., Templeton T.J. A multidomain adhesion protein family expressed in Plasmodium falciparum is essential for transmission to the mosquito. J. Exp. Med. 2004;199:1533–1544. doi: 10.1084/jem.20031274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon N., Scholz S.M., Moreira C.K., Templeton T.J., Kuehn A., Dude M.A., Pradel G. Sexual stage adhesion proteins form multi-protein complexes in the malaria parasite Plasmodium falciparum. J. Biol. Chem. 2009;284:14537–14546. doi: 10.1074/jbc.M808472200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patra K.P., Johnson J.R., Cantin G.T., Yates J.R., 3rd, Vinetz J.M. Proteomic analysis of zygote and ookinete stages of the avian malaria parasite Plasmodium gallinaceum delineates the homologous proteomes of the lethal human malaria parasite Plasmodium falciparum. Proteomics. 2008;8:2492–2499. doi: 10.1002/pmic.200700727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lasonder E., Janse C.J., van Gemert G.J., Mair G.R., Vermunt A.M., Douradinha B.G., van Noort V., Huynen M.A., Luty A.J., Kroeze H., et al. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 2008;4:e1000195. doi: 10.1371/journal.ppat.1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindner S.E., Swearingen K.E., Harupa A., Vaughan A.M., Sinnis P., Moritz R.L., Kappe S.H. Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol. Cell. Proteomics. 2013;12:1127–1143. doi: 10.1074/mcp.M112.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.