Abstract
Genetic engineering projects often require control over when a protein is degraded. To this end, we use a fusion between a degron and an inactivating peptide that can be added to the N-terminus of a protein. When the corresponding protease is expressed, it cleaves the peptide and the protein is degraded. Three protease:cleavage site pairs from Potyvirus are shown to be orthogonal and active in exposing degrons, releasing inhibitory domains and cleaving polyproteins. This toolbox is applied to the design of genetic circuits as a means to control regulator activity and degradation. First, we demonstrate that a gate can be constructed by constitutively expressing an inactivated repressor and having an input promoter drive the expression of the protease. It is also shown that the proteolytic release of an inhibitory domain can improve the dynamic range of a transcriptional gate (200-fold repression). Next, we design polyproteins containing multiple repressors and show that their cleavage can be used to control multiple outputs. Finally, we demonstrate that the dynamic range of an output can be improved (8-fold to 190-fold) with the addition of a protease-cleaved degron. Thus, controllable proteolysis offers a powerful tool for modulating and expanding the function of synthetic gene circuits.
INTRODUCTION
Genetic circuits are built by designing interactions between regulators to perform a computational operation (1–3). When the regulators are proteins, it is desirable to be able to eliminate them quickly from the cell when the input states are changed. However, they are often very stable and their clearance rate is limited by cell division, which varies with growth phase. To overcome this, a set of C-terminal degrons (from ssrA) are often used to tune the degradation half-lives from 40 min to hrs (4,5). These degrons are targeted constitutively by tail-specific proteases (4). The availability of these degrons has aided genetic circuit design, where tagging regulators and fluorescent reporters accelerates switching between states and enables the measurement of circuit dynamics (6,7). However, the proteases are always active and the lower intra-cellular concentration of the target reduces their effectiveness (strength of regulation, the intensity of fluorescence, etc.). To overcome this, Collins and coworkers use orthogonal degradation machinery to induce targeted protein degradation (8). Here, we apply an approach where an N-terminal degron is inaccessible by the constitutive cellular proteases until a recombinant protease is expressed (9). The inducible nature of the system allows it to be integrated into circuits in various ways to tune or change their function.
Our system exploits the N-end rule for protein degradation, which states that the nature of the exposed amino acid at the N-terminus will modify the half-life of a protein; some residues are considered stabilizing and others highly destabilizing (10,11). The endogenous protease machinery of the host carries out the degradation of the protein and the amino acid sequence signal is the ‘degron.’ We use Potyvirus proteases, including tobacco etch virus protease (TEVp), to control the function of a target protein. TEVp belongs to the family of cysteine proteases and is widely used because of the exquisite affinity toward its recognition site (12). Here, we expand this system by utilizing two additional Potyvirus proteases: tobacco vein mottling virus protease (TVMVp), which is known to be orthogonal to TEVp (13,14) and sunflower mild mosaic virus protease (SuMMVp), which has not been characterized to date (15). TEVp and TVMVp have been used in the past to construct genetic circuits (16–20).
These proteases are integrated into simple transcriptional circuits, where the inputs and outputs are defined as promoters. Many such circuits have been built that implement logic operations (21–25). The simplest is a NOT gate, whose output inverts the activity of the input (26). This function can be implemented by arranging the input promoter to drive the expression of a repressor that turns off an output promoter. For example, we recently created a large set of NOT gates based on a library of TetR-family repressors (25). Further, the NOT function can be extended to build 2-input NOR gates, which in turn can be connected in different ways to build more complex circuits (24,25). When building circuits, the repressors underlying the NOT gates are often fused to C-terminal protease tags to increase their degradation rate (7,27,28).
In this manuscript, we demonstrate that the three Potyvirus proteases are highly orthogonal and do not react with each other's cleavage sites. Then, we show that TetR-family repressors can be controlled where the expression of the Potyvirus protease either causes their degradation by exposing a cryptic degron or enables them to bind DNA by releasing a sterically inhibiting bulky domain. These functions are demonstrated in the context of transcriptional logic gates. Then, a polyprotein composed of multiple repressors separated by Potyvirus cleavage sites is built and we show that the domains can be shuffled to modify the response of a circuit, including simultaneous activation and repression of two outputs with a single protease. Finally, we show that the inducible proteases can be used to increase the dynamic range of a circuit output by coordinating the induction of the Potyvirus protease to coincide with the expression of the output gene. This enables ‘actuators’ to impart a greater impact on cellular behavior (3,29. This work introduces new parts to incorporate post-translational control into transcriptional genetic circuits as a means to expand and optimize their behavior.
MATERIALS AND METHODS
Strains and media
Escherichia coli DH10b cells (F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu) 7697 galU galK rpsL nupG λ−) were used in all cloning procedures and experiments. Cells were always grown in LB Miller broth (Difco, 90003–350). Chloramphenicol (34 μg/ml) (Alfa Aesar, AAB20841-14), kanamycin (50 μg/ml) (GoldBio, K-120-10), spectinomycin (100 μg/ml) (GoldBio S-140-5) and ampicillin (75 μg/ml) (GoldBio, A-301-5) were supplemented when necessary. IPTG (GoldBio, I2481C25) and arabinose (Sigma-Aldrich, MO, A3256) were used as inducers.
Liquid assay conditions
For all experiments, inoculants of single colonies grown overnight in LB with the appropriate antibiotics were diluted 1:500 into 800 μl of fresh LB supplemented with antibiotics with and without inducers and grown in 96-deep-well plates (USA Scientific, 1896–2000) covered with air permeable membranes AeraSeal (E&K scientific, CA, T896100) for 6 or 8 h at 37°C in a Multitron Pro incubator shaker (In Vitro Technologies, VIC, Australia) at 900 rpm.
Time course analysis
E. coli DH10b cells carrying both the pNus-Tet or pTF-PhlF plus the cognate pTac-TEV and pTac-Su plasmids, respectively, were grown in 5 ml LB plus antibiotics at 37°C for 18 h in an Innova 44 shaker (Eppendorf, CT) at 250 rpm. This culture was diluted 1:100 in 200 μl of fresh LB plus antibiotics and grown at 37°C for 3 h in an ELMI Digital Thermos Microplate shaker (Elmi Ltd, Riga, Latvia) at 1000 rpm. For time courses, this is the t = 0h time point. This culture was further diluted 1:10 in 200 μl final volume of fresh LB with antibiotics in the presence of 2 mM IPTG and samples taken every hour for cytometry analysis. To maintain the cells in exponential growth phase, the culture was diluted 1:5 every 2 h.
Potyvirus proteases orthogonality assay
Cells carrying plasmid constitutively expressing the proteases (pTEV, pTVMV, or pSuMMV, ampicillin resistance) were co-transformed with the reporter plasmids ptevY-GFP, ptvmvY-GFP or psummvY-GFP (chloramphenicol resistance) in all possible combinations. Cells carrying the reporter plasmids only were also grown to calculate the fold reduction in fluorescence levels upon expression of the proteases. Cells were plated on LB agar (1%) supplemented with ampicillin and/or chloramphenicol for 18 h at 37°C. Single colonies were then inoculated into 800 μl liquid media with the appropriate antibiotics and grown overnight in 96-deep-well plates (USA Scientific, 1896–2000) covered with air permeable membranes AeraSeal (E7K Scientific, CA, T896100) at 37°C in a Multitron Pro incubator shaker (In Vitro Technologies, VIC, Australia) at 900 rpm. The cultures were then transferred to 1.5 ml microcentrifuge tubes, spun down at 6000 xg for 5 min in a tabletop microcentrifuge and resuspended in 1X PBS. The GFP fluorescence of 200 μl of these cell suspensions was then measured using a Synergy H1 Hybrid Microplate Reader (Biotek, VM) and the fold reduction in fluorescence calculated as the quotient between the fluorescence levels of strains containing only the reporter plasmids and strains containing both reporter and protease plasmids.
Flow cytometry analysis
2–5 μl of the cultures were diluted into 150 μl 1X phosphate buffered sulphate (PBS) with 2 mg/ml kanamycin. Fluorescence was measured using a MACSQuant VYB (Miltenyi Biotec) with a 488-nm laser for GFP excitation and 561-nm for RFP excitation using 5 × 104 recorded events. FlowJo v10 (TreeStar Inc.) was used to analyze the data. GFP fluorescence values are shown as the geometric average.
RESULTS
Mining Potyvirus proteases and testing orthogonality
Many Potyvirus proteases have been described in the literature (30). Given their high sequence similarity, new protease members can be inferred from genomic data for different Potyvirus species. There are over 170 defined species in the family of Potyviridae and recent studies show that many additional species can be found in wild plants (15,30,31). The mining of new proteases is aided by the fact that their recognition sites can be easily deduced from the flanking sites of the protease gene. From the possibilities, we selected TVMVp (cleavage site ETVRFQ), as it has been shown to not crosstalk with TEVp (14). We also searched uncharacterized putative Potyvirus proteases (15) and identified a candidate from SuMMV (sunflower mild mosaic virus, cleavage site EEIHLQ). We chose this protease because the canonical cleavage site is very different from TEVp and TVMVp.
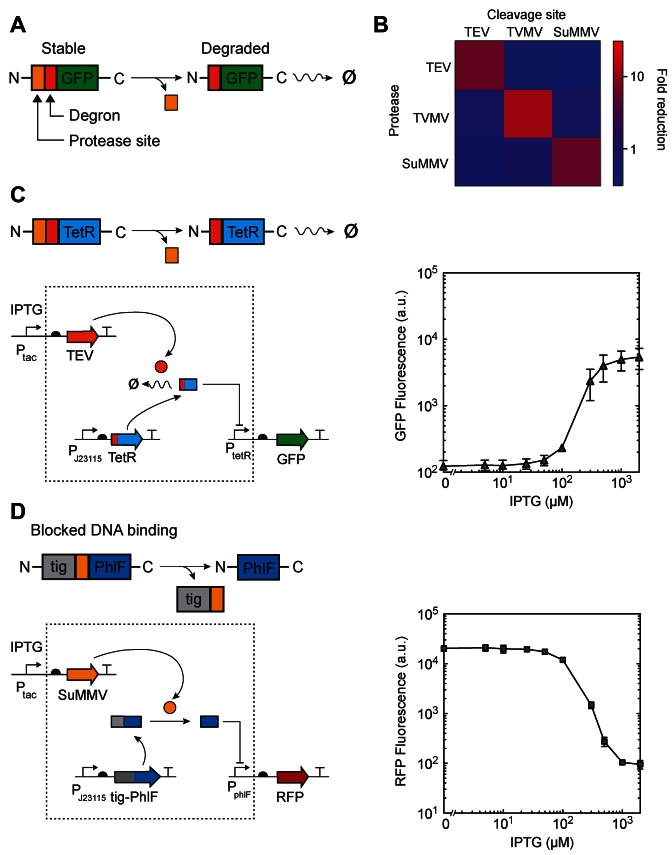
Along with TEVp, the sequences for these proteases were codon optimized to allow for a better expression in E. coli (32) (Supplementary Table S1). We tested for orthogonality by using the cryptic degron approach shown in Figure 1A where GFP is tagged and used as a reporter (Materials and Methods). We cloned the plasmids pTEV, pTVMV and pSuMMV encoding constitutively expressed TEVp, TVMVp or SuMMVp as well as their respective reporter plasmids with a constitutively expressed gfp gene preceded by a cryptic degron and a protease site (ptevY-GFP, ptvmvY-GFP and psummvY-GFP, Supplementary Figure S7). Cells carrying one of the protease plasmids were co-transformed with one of the three different reporters and their GFP levels monitored to generate the orthogonality matrix shown in Figure 1B. A decrease in GFP could only be observed when the correct protease is expressed that corresponds to the reporter with the cognate tag. The results showed that TEVp, TVMVp and SuMMVp are orthogonal and presented no discernible crosstalk (Figure 1B).
Figure 1.
Controlled degradation and release of transcriptional repressors. (A) A schematic of controlled degradation of GFP mediated by a protease is shown. Orange square, protease recognition site; red square, N-terminal degron. (B) Orthogonality matrix for the proteases used in this study depicted as the fluorescence fold reduction in cultures of cells containing combinations of pTEV, pTVMV and pSuMMV plasmids and GFP reporter plasmids (Supplementary Figure S7). (C) Left panels, a schematic of controlled degradation of TetR mediated by a protease (TEV) is shown. Right panel, the response function for the TEV-mediated controlled degradation of TetR is shown. Cells containing ptevF-TetR and pTac-TEV were grown at different concentrations of IPTG for 6 h and the resulting GFP fluorescence was measured by flow cytometry. (D) Left panels, a schematic of controlled release of PhlF mediated by SuMMV is shown. Grey square, bulky fusion partner fused to the N-terminus of PhlF (tig, trigger-factor). Right panel, the response function for the SuMMV-mediated controlled release of PhlF is shown. Cells containing pTig-PhlF and pTac-Su were grown at different concentrations of IPTG for 6 h and the resulting RFP fluorescence was measured by flow cytometry. In all panels, error bars correspond to the standard deviation of three experiments performed on different days.
Controlled degradation of a target protein using cryptic degrons
Cryptic N-terminal degrons (N-degrons) can be constructed by fusing an additional cleavable sequence at their N-terminus, such as a TEVp cleavage site. This method has been successfully applied in yeast (9,33). Since the TEVp cleavage sequence does not contain any destabilizing residues, the tagged protein is stable until TEVp is expressed and exposes the internal cryptic N-degron (Figure 1A). In the case of E. coli, the N-degron will be recognized by the adaptor protein ClpS and degraded by the endogenous ClpAP machinery (34).
We tested if this method could be applied in E. coli. To do this, we first modified a aTc-inducible system based on TetR that has green fluorescent protein (GFP) as the output (25). The N-terminus of TetR was tagged with a TEVp site (ENLYFQ) followed by the ‘F-degron’ (FLFVQ) (35). Thus, the degradation of TetR is controlled by induction of TEVp. However, because TEVp aggregates when overexpressed in E. coli (36), we also included a self-cleavable maltose-binding protein domain (MalE-TEVp) and used a truncated version of TEVp, both of which are known to improve solubility and stability (37,38). The protease gene is placed under IPTG control (Ptac) on a pSC101 plasmid (Supplementary Figure S6). Finally, TEVp is a very active enzyme and even small amounts of protease can cleave its substrate (39). For this reason, we designed an RBS library of MalE-TEVp and selected for one that yielded low basal activity and a large dynamic range of protease activity.
Cells carrying the F-degron-fused TetR and IPTG-inducible TEVp were exposed to varying amounts of inducer. The activity of the protease is assessed by its ability to derepress the PtetR promoter (25) by degrading TetR. The promoter is transcriptionally fused to gfp and its activity is measured by flow cytometry (Materials and Methods). The response curve after 6 h of induction is shown in Figure 1C. The output is induced 44-fold by the protease. This is similar to the level of induction that can be achieved by the addition of aTc to induce TetR (Supplementary Figure S1B). The same experiment was performed with an F-degron tagged PhlF circuit with RFP as an output, yielding 50-fold activation (Supplementary Figure S1C).
The degree to which a protein is degraded can also be controlled by modifying the first residue of the cryptic degron (35), which affects both the rate of cleavage by TEVp (39) and the rate of degradation in the cell. We tested the same TetR circuit tagged with the alternate N-degron YLFVQ (Y-degron), known to be degraded at a lower rate than the F-degron (35). Indeed, this circuit showed only 12-fold activation (Supplementary Figure S1A).
We performed time course experiments in circuits containing tevF-TetR, tevY-TetR and tevF-PhlF in the presence of pTac-TEV (Supplementary Figure S2). Our results show that protease-controlled circuits still respond at similar timescales as transcription/translation-based circuits, with times needed to reach half-maximal activity of 2 h (for tev-F-TetR) and 2.6 h (for tev-F-PhlF).
Controlled release of a repressor
Another application of the Potyvirus proteases is to release an inhibitory domain, thus activating the target protein. Here, we tested whether the ability of a transcriptional repressor to bind to DNA could be sterically blocked in a way that binding could be recovered when the domain is proteolytically cleaved. TetR family repressors form active dimers that are able to bind their cognate DNA operator through their N-terminal HTH domains (40). We hypothesized that fusing a bulky domain to the N-terminus of a repressor would generate binding-impaired variants, as the presence of this large moiety would sterically hinder the recognition of the operator. By using an amino acid linker corresponding to a protease recognition site, the repressor will be released and bind to its operator upon expression of the cognate protease, hence repressing transcription (Figure 1D).
To test this, we constructed a constitutively expressed fusion protein composed of the bulky tig gene (trigger factor) (41) and the PhlF repressor linked with a SuMMVp cleavage site. A PphlF promoter, which is repressed by PhlF (25), reports repressor activity via the expression of RFP (Supplementary Figure S6). Trigger factor was chosen as a partner gene because of its large size (432 residues, 42 kDa) and because it is known to act as a chaperone and has been used to increase solubility of proteins (42). As with TEVp, we constructed a self-cleavable MalE-SuMMVp fusion under the control of IPTG-inducible Ptac and built an RBS library to find a variant with low leakiness and high induction fold change.
This circuit represents an alternative architecture for a NOT gate. Here, the input promoter drives the expression of a protease, which activates a constitutively expressed inactive repressor. The response function of the gate was characterized by varying the induction of the input promoter and measuring fluorescence of the output at steady-state (6 h) (Figure 1D). In the absence of IPTG, the output is high, which confirms that the Tig-PhlF fusion is not able to bind its operator. The expression of the protease leads to a 200-fold dynamic range, which is an improvement over an equivalent NOT gate where Ptac directly drives the expression of PhlF (85-fold, Supplementary Figure S3) (25). While the transition is cooperative, intermediate levels of RFP fluorescence between the ON and OFF states are observed, which suggests that the expression of SuMMVp protease can be tuned to release only a fraction of the PhlF molecules present in the cell. We also tested a similar construct composed of a NusA moiety (495 residues, 54.9 kDa) fused to TetR and separated by a TEVp site (Supplementary Figure S3). As with the trigger factor, NusA has been successfully used to increase solubility of proteins (43). This confirms that the approach works with different hindering domains and repressors.
Finally, we explored the impact of the residue immediately after the cleavage site (P1′ position) on the efficiency of repressor release (Supplementary Figure S4). As for TEV, SuMMV also seems to present variability in its cleaving efficiency depending on the P1′ position (39). Thus, via the modification of a single amino acid a wide range of response functions with different OFF levels can be generated (200-fold for glycine; 2-fold for tyrosine).
Polyproteins as modular units in genetic circuit design
Many viral genomes, including Potyvirus, are expressed as a single protein that is then cleaved into subunits by genomically encoded proteases (44). Polyproteins have been used as a strategy to express more than one protein from a single transcriptional unit in organisms where genetic manipulation is tedious, such as plants (45). The first approaches involved the use of self-cleavable peptides, a technique that has also been effectively applied in diverse organisms with different objectives, such as plant metabolic engineering, co-production of proteins in yeast or reprogramming mammalian somatic cells (46–49). The second approach, based on the expression of a polyprotein that is cleaved by a site-specific protease, has also been successfully applied to co-express proteins in plants, IgG antibodies and even as a MALDI-MS calibration technique (50–52). One of the advantages of using the cleavable polyprotein approach is that the subunits will be generated in equimolar amounts (53), a feature that has been successfully applied in yeast metabolic engineering (54).
We explored the possibility of exploiting polyproteins as modular units in the design of genetic circuits: all the components, including their regulatory parts, are encoded in a single cistron, translated as a single protein and then cleaved into functional subunits. This principle was applied to design a ‘polyrepressor’, in which bulky fusion partners, protease cleavage sequences, cryptic degrons and transcriptional repressors can be constitutively expressed as a polyprotein and controlled by a protease. This has the advantage of splitting the outputs of a gate with a minimal number of parts and genes or when stoichiometric amounts of proteins are needed. The approach offers many levers of control while maintaining a single gene in the circuit.
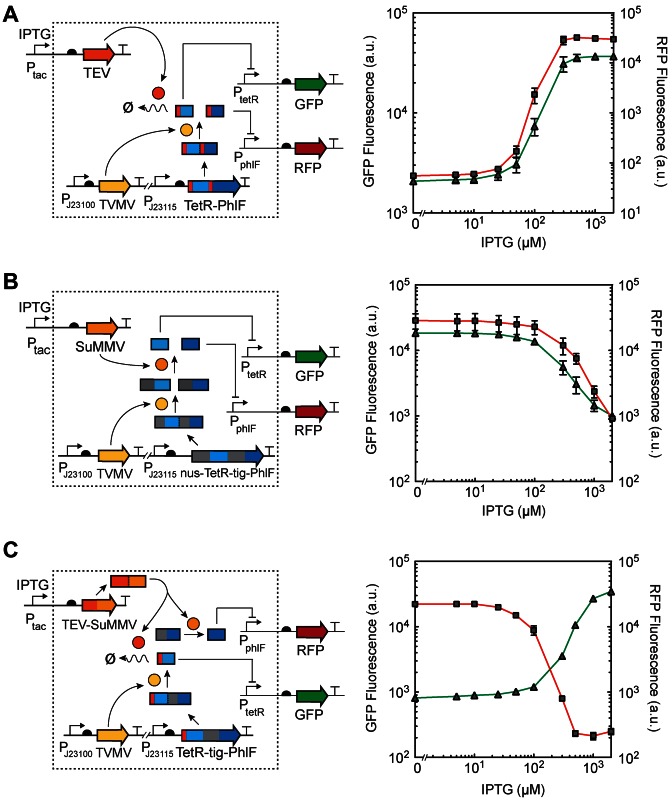
First, we tested the simultaneous control of the degradation of TetR and PhlF with a single protease (TEVp) (Figure 2A). This splits the output of the gate to the control of PtetR and PphlF, which is reported by GFP and RFP, respectively. The polyrepressor comprises two repressor subunits with a cryptic degron each (as in Figure 1C), but separated by a linker corresponding to the cleavage sequence of TVMVp. TVMV protease is constitutively expressed and cleaves the polyrepressor into two functional subunits (F-degron-TetR and F-degron-PhlF) upon which TEVp can act (Supplementary Figure S8). Hence, TVMVp does not have any direct functional role in the control of the circuit output. The RBS controlling the expression of the polyrepressor was tuned as before. The response functions for the gate were then measured (Figure 2A). This demonstrates that TEVp is able to control both TetR and PhlF responses simultaneously and the same threshold of inducer is required to activate both.
Figure 2.
Polyproteins as modular units to control genetic circuits. For all figures, the left panel shows a schematic diagram of the system and the right panel shows the corresponding response functions for the two outputs. Constitutively expressed TVMVp cleaves the polyprotein in two functional subunits in all cases. Cells containing a plasmid encoding a given polyprotein design expressed from a PJ23115 constitutive promoter and a Ptac-inducible protease in a separate plasmid were exposed to different concentrations of IPTG for 6 h. The resulting GFP and RFP fluorescences were measured using flow cytometry. For all of the panels, red lines-squares indicate RFP fluorescence and green lines-triangles indicate GFP fluorescence. (A) Simultaneous controlled degradation of TetR and PhlF by TEV protease in cells containing pPoly-Deg and pTac-TEV. (B) Simultaneous release of TetR and PhlF by SuMMV protease in cells containing pPoly-Rel and pTac-Su. (C) Simultaneous degradation and release of TetR and PhlF by TEV and SuMMV proteases, respectively, in cells containing pPoly-Switch and pTac-TEV-Su. The colored squares represent the same parts as in Figure 1C and D. In all figures, error bars correspond to the standard deviation of three experiments performed on different days.
A similar approach was pursued to design a polyrepressor that releases two repressors controlled by SuMMVp (Figure 2B). In this case, the individual modules NusA-TetR and Tig-PhlF were separated by a TVMVp cleavage site and their release simultaneously controlled by pTac-SuMMVp (Supplementary Figure S8). The RBS was tuned to find the optimal strength. The response functions were measured and the two outputs follow NOT logic with identical thresholds (Figure 2B).
Regulatory polyproteins offer the possibility of using an input to differentially control multiple outputs. To demonstrate this, a hybrid polyprotein was designed in which one of the repressors was tagged with a cryptic degron (tevF-TetR) while the other contained a bulky fusion partner (Tig-PhlF) (Figure 2C). The TVMV protease is constitutively expressed and cleaves the polyprotein to release the (active) TetR and (inactive) PhlF. A protease polyprotein was then constructed where TEVp and SuMMVp are fused separated by a SuMMVp cleavage site. Both TEVp and SuMMVp moieties can cleave themselves from this fusion protein as it is being produced to generate two active proteases (14,37). The protease polyprotein was placed under the control of Ptac and the response function measured. In the absence of inducer, the TetR output was OFF and the PhlF output was ON. Upon induction of the TEV-SuMMV protease fusion, we could observe that the output of TetR switched on, as TEVp degraded TetR, while the PhlF output switched off, as it was released by SuMMVp. This demonstrates that the single polyrepressor can be designed to switch between two outputs in opposite ways. This could be applied to turn one cellular process off as the other is turned on, and vice versa.
Amplification of an output
It is difficult to control the dynamic range of the output of a genetic circuit. This is important for controlling cellular processes where the circuit must have sufficient dynamic range to trigger the response. The basal level of the OFF state is particularly challenging to control. To this end, we use Potyvirus proteases to increase the dynamic range by reducing the background of the OFF state. The approach is based on degradation rescue, which is conceptually the opposite of controlled degradation. The target protein is tagged with a constitutive degron that is cleaved away only when a protease is present, removing the tag and stabilizing the protein (55).
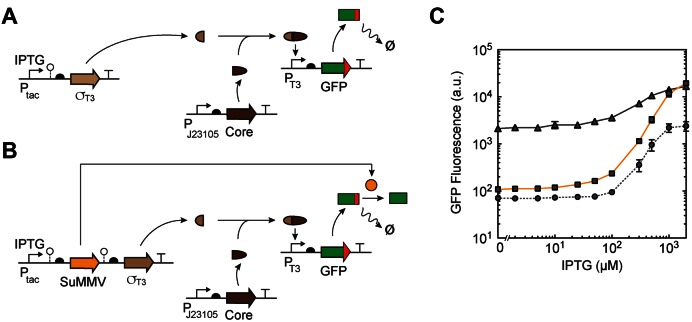
This approach was applied to amplify the connection between a genetic circuit and an actuator (cellular response, metabolic pathway, etc). The linkage is often carried out using phage RNA polymerases, where the output promoter of the circuit drives the expression of the polymerase that in turn controls the actuator (56,57). This organization enables the separation of the controller (sensors and circuitry) from the actuators. Previously, we built a set of orthogonal ‘σ factors’ by splitting T7 RNAP into multiple genes (29) so that the DNA-binding domain and the remainder of the core RNAP are encoded separately. This approach reduces the size of the gene that has to be connected to the circuit's output promoter and enables resource sharing between multiple outputs (Figure 3A).
Figure 3.
Increasing dynamic range with degradation rescue. (A) Schematic of the original split-polymerase system (29) showing the constitutively expressed resource allocator (Core), IPTG-inducible controller (σT3) and constitutively degraded actuator (GFP). o– symbol, RiboJ ribozyme (85). (B) A schematic of degradation rescue with SuMMV protease transcribed as a polycistronic mRNA with σT3 from a single Ptac promoter encoded in pTac-T3-Su. Red squares, C-terminal degradation tag. o– symbol, BydvJ ribozyme (SuMMV) and RiboJ ribozyme (T3) (85,86). (C) Response functions of the circuits shown in (A) and (B). Cells carrying pCore, pTac-T3-Su and pT3-GFP-LVA were grown at different concentrations of IPTG for 8 h and the fluorescence tracked by flow cytometry. Solid grey line, original split-polymerase system without SuMMV protease and untagged GFP. Dashed grey line, split-polymerase system without SuMMV protease and with degron-tagged GFP output. Orange line, split-polymerase system with SuMMV protease expressed from a polycistronic mRNA with σT3. Error bars correspond to the standard deviation of three experiments performed on different days.
One problem with this approach is that leaky σ expression produces sufficient quantities to trigger the response. This can be seen in Figure 3C (continuous black line), where the σT3 promoter is used to drive the expression of GFP. The IPTG-inducible Ptac is the surrogate for the output of a circuit and the core RNAP is expressed from a constitutive promoter (Supplementary Figure S9). While this yields an 8-fold response, there is a high basal level of expression in the absence of inducer (Ptac is in the OFF state).
Next, we tagged GFP with a C-terminal ssrA degradation tag (LVA variant) (5) preceded by a SuMMVp site (Figure 3A). This generates an unstable GFP variant, reducing the background fluorescence in the absence of IPTG (dashed line in Figure 3C). This maintains a 40-fold response upon induction, but there is a significant knockdown of the level of the fully induced ON state. This is characteristic of the addition of a tag that directs proteins to constitutive cellular proteases.
A second architecture was then tested where SuMMVp is placed under the control of Ptac as an operon with σT3 (Figure 3B). The RBS controlling the protease was modified to tune the expression level. The expression of SuMMVp causes the cleavage of the LVA degron from GFP, thus stabilizing it. In the absence of inducer, insufficient protease is produced to stabilize any GFP that results from leaky expression. However, when Ptac is fully induced, this has the dual effect of increasing expression from the T3 promoter and stabilizing the GFP product. Thus, the response to the absence of inducer moves toward that observed when the GFP is constitutively degraded, but the response in the presence of inducer moves toward the level when GFP is stable (orange line, Figure 3C). This has a large impact on the dynamic range, increasing it to 190-fold.
To circumvent the leakiness of our system due to Ptac, we generated a PBAD-inducible SuMMVp variant to decouple the expression of SuMMV from the main circuit (Supplementary Figure S5). For this circuit, both the σT3 fragment and the SuMMV protease can be independently induced with IPTG and arabinose, respectively. This approach allowed for OFF levels indistinguishable from the uninduced system while reaching the same high ON levels.
DISCUSSION
Degron-mediated protein knockdown is common to all kingdoms of life (10,58). Here, we harness this approach using orthogonal proteases gleaned from Potyvirus. By combining them with bulky domains and degrons, protein activity and degradation can be controlled in an inducible manner. This is applied to genetic circuit design, where a transcriptional NOT gate can be constructed by having the input promoter drive the expression of the proteases, which then activates a constitutively expressed repressor. The incorporation of proteases into transcriptional genetic circuits introduces new levers of control that theoretical models predict could improve the performance of many types of dynamic circuits (59). Proteases also allow multiple transcription factors to be continuously expressed, but only become active when the single protease gene is expressed. We demonstrated this by building a repressor-based polyprotein, where the repressors are released upon proteolysis. This offers the possibility of coordinating the activation of many processes simultaneously. For example, natural regulatory proteins could be combined with synthetic ones to induce multiple cellular responses at a defined layer of a genetic circuit.
We show that degradation rescue is an effective tool for amplifying the dynamic range of an output. Proteases can serve as signal amplifiers because they have catalytic activity, and so a single molecule of protease can act upon many molecules of substrate as opposed to a transcription factor that can bind to one site at a time (20). There are several alternative approaches to amplifying a signal from a genetic circuit, including signal inversion using a NOT gate (60), feedback loops (61–64), recombinases (65–67), phosphorylation (68,69), non-coding RNAs (70), upstream activating sequences (71,72), the use of sequestration at the protein level (73–76) or by incorporating decoy operators (69,77). The advantage of using our protease system as shown in Figure 3 is that it does not require changing the circuit itself or the means by which it is connected to an actuator. Rather the actuator gene(s) are simply fused to the degron in order to achieve the amplifying effect. Because it is itself inactive, the tag does not impact activity until after the Potyvirus protease is expressed.
Potyvirus proteases have been shown to function across a wide range of organisms (78–80). TEV in particular is a workhorse and is used extensively in biotechnology. TEV has been engineered to function as a sensor by splitting the protease and using an external signal to induce dimerization which has been applied to the detection of protein–protein interactions and small molecules and has also been engineered to sense light (19,79,81,82). The two additional Potyviruses provide orthogonal means for the cell to respond to additional signals. Here, we demonstrate that these proteases can be used to build simple genetic logic gates. These could form the basis of converting the signal-responsive proteases to a transcriptional output, which is required to connect sensors to transcriptional circuits. They also could form the basis of incorporating protease cascades—common in natural signaling networks—into synthetic circuit design. More broadly, this reflects a need to expand the toolbox for the application of post-translational control into genetic circuit design, including methods of regulating the concentration and activity of a protein without the necessity of triggering a genetic event: proteins can be chemically modified, form part of complexes, sequestered, targeted to different compartments and degraded (83,84).
ACCESSION NUMBERS
Accession numbers are available in Supplementary Table S2.
Supplementary Material
Acknowledgments
C.A.V and J.F.R. conceived of the study and designed the experiments. J.F.R. performed the experiments and analyzed the data. C.A.V. and J.F.R. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Science Foundation Synthetic Biology Engineering Research Center [SynBERC EEC0540879]; Office of Naval Research Multidisciplinary University Research Initiative [N00014-11-1-0725 and N00014-13-1-0074]. Funding for open access charge: US National Science Foundation Synthetic Biology Engineering Research Center [SynBERC EEC0540879]; Office of Naval Research Multidisciplinary University Research Initiative [N00014-11-1-0725 and N00014-13-1-0074].
Conflict of interest statement. None declared.
REFERENCES
- 1.Brophy J.A.N., Voigt C.A. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purcell O., Lu T.K. Synthetic analog and digital circuits for cellular computation and memory. Curr. Opin. Biotechnol. 2014;29:146–155. doi: 10.1016/j.copbio.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen A.A.K., Segall-Shapiro T.H., Voigt C.A. Advances in genetic circuit design: novel biochemistries, deep part mining, and precision gene expression. Curr. Opin. Chem. Biol. 2013;17:878–892. doi: 10.1016/j.cbpa.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman S., Roche E., Zhou Y., Sauer R.T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen J.B., Sternberg C., Poulsen L.K., Bjørn S.P., Givskov M., Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elowitz M.B., Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 7.Stricker J., Cookson S., Bennett M.R., Mather W.H., Tsimring L.S., Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron D.E., Collins J.J. Tunable protein degradation in bacteria. Nat. Biotechnol. 2014;32:1276–1281. doi: 10.1038/nbt.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taxis C., Stier G., Spadaccini R., Knop M. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol. Syst. Biol. 2009;5:267. doi: 10.1038/msb.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasaki T., Sriram S.M., Park K.S., Kwon Y.T. The N-end rule pathway. Annu. Rev. Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 12.Waugh D.S. An overview of enzymatic reagents for the removal of affinity tags. Protein Expr. Purif. 2011;80:283–293. doi: 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nallamsetty S., Kapust R.B., Tözsér J., Cherry S., Tropea J.E., Copeland T.D., Waugh D.S. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 2004;38:108–115. doi: 10.1016/j.pep.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Sun P., Austin B.P., Tözsér J., Waugh D.S. Structural determinants of tobacco vein mottling virus protease substrate specificity. Protein Sci. Publ. Protein Soc. 2010;19:2240–2251. doi: 10.1002/pro.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams M.J., Antoniw J.F. DPVweb: a comprehensive database of plant and fungal virus genes and genomes. Nucleic Acids Res. 2006;34:D382–D385. doi: 10.1093/nar/gkj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daringer N.M., Dudek R.M., Schwarz K.A., Leonard J.N. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth. Biol. 2014;3:892–902. doi: 10.1021/sb400128g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonzarić J., Lebar T., Majerle A., Manček-Keber M., Jerala R. Locked and proteolysis-based transcription activator-like effector (TALE) regulation. Nucleic Acids Res. 2016;44:1471–1481. doi: 10.1093/nar/gkv1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnea G., Strapps W., Herrada G., Berman Y., Ong J., Kloss B., Axel R., Lee K.J. The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray D.C., Mahrus S., Wells J.A. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein V., Alexandrov K. Protease-based synthetic sensing and signal amplification. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15934–15939. doi: 10.1073/pnas.1405220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Rodriguez J., Yang L., Gorochowski T.E., Gordon D.B., Voigt C.A. Memory and combinatorial logic based on DNA inversions: Dynamics and evolutionary stability. ACS Synth. Biol. 2015;4:1361–1372. doi: 10.1021/acssynbio.5b00170. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen A.A.K., Voigt C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamsir A., Tabor J.J., Voigt C.A. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanton B.C., Nielsen A.A.K., Tamsir A., Clancy K., Peterson T., Voigt C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokobayashi Y., Weiss R., Arnold F.H. Directed evolution of a genetic circuit. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu S., Mehreja R., Thiberge S., Chen M.-T., Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guet C.C., Elowitz M.B., Hsing W., Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 29.Segall-Shapiro T.H., Meyer A.J., Ellington A.D., Sontag E.D., Voigt C.A. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 2014;10:742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revers F., García J.A. Chapter Three - Molecular Biology of Potyviruses. Adv. Virus Res. 2015;92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Roossinck M.J. Plant Virus Metagenomics: Biodiversity and Ecology. Annu. Rev. Genet. 2012;46:359–369. doi: 10.1146/annurev-genet-110711-155600. [DOI] [PubMed] [Google Scholar]
- 32.Puigbò P., Guzmán E., Romeu A., Garcia-Vallvé S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jungbluth M., Renicke C., Taxis C. Targeted protein depletion in Saccharomyces cerevisiae by activation of a bidirectional degron. BMC Syst. Biol. 2010;4:176. doi: 10.1186/1752-0509-4-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D.A., Bukau B. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 35.Wang K.H., Oakes E.S.C., Sauer R.T., Baker T.A. Tuning the strength of a bacterial N-end rule degradation signal. J. Biol. Chem. 2008;283:24600–24607. doi: 10.1074/jbc.M802213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucast L.J., Batey R.T., Doudna J.A. Large-scale purification of a stable form of recombinant tobacco etch virus protease. BioTechniques. 2001;30:544–546. doi: 10.2144/01303st06. [DOI] [PubMed] [Google Scholar]
- 37.Kapust R.B., Waugh D.S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. Publ. Protein Soc. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blommel P.G., Fox B.G. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapust R.B., Tözsér J., Copeland T.D., Waugh D.S. The P1’ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 2002;294:949–955. doi: 10.1016/S0006-291X(02)00574-0. [DOI] [PubMed] [Google Scholar]
- 40.Ramos J.L., Martínez-Bueno M., Molina-Henares A.J., Terán W., Watanabe K., Zhang X., Gallegos M.T., Brennan R., Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooke E., Wickner W. Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5216–5220. doi: 10.1073/pnas.84.15.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishihara K., Kanemori M., Yanagi H., Yura T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 2000;66:884–889. doi: 10.1128/aem.66.3.884-889.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis G.D., Elisee C., Newham D.M., Harrison R.G. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 1999;65:382–388. [PubMed] [Google Scholar]
- 44.Adams M.J., Antoniw J.F., Beaudoin F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 2005;6:471–487. doi: 10.1111/j.1364-3703.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- 45.Halpin C., Cooke S.E., Barakate A., Amrani A.E., Ryan M.D. Self-processing 2A-polyproteins – a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J. 1999;17:453–459. doi: 10.1046/j.1365-313x.1999.00394.x. [DOI] [PubMed] [Google Scholar]
- 46.Ryan M.D., Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ralley L., Enfissi E.M.A., Misawa N., Schuch W., Bramley P.M., Fraser P.D. Metabolic engineering of ketocarotenoid formation in higher plants. Plant J. Cell Mol. Biol. 2004;39:477–486. doi: 10.1111/j.1365-313X.2004.02151.x. [DOI] [PubMed] [Google Scholar]
- 48.de Amorim Araújo J., Ferreira T.C., Rubini M.R., Duran A.G.G., De Marco J.L., de Moraes L.M.P., Torres F.A.G. Coexpression of cellulases in Pichia pastoris as a self-processing protein fusion. AMB Express. 2015;5:84. doi: 10.1186/s13568-015-0170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. U.S.A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelloniemi J., Mäkinen K., Valkonen J.P.T. Three heterologous proteins simultaneously expressed from a chimeric potyvirus: infectivity, stability and the correlation of genome and virion lengths. Virus Res. 2008;135:282–291. doi: 10.1016/j.virusres.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Giritch A., Marillonnet S., Engler C., van Eldik G., Botterman J., Klimyuk V., Gleba Y. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14701–14706. doi: 10.1073/pnas.0606631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidmann S., Barylyuk K., Nespovitaya N., Mädler S., Zenobi R. A new, modular mass calibrant for high-mass MALDI-MS. Anal. Chem. 2013;85:3425–3432. doi: 10.1021/ac400129h. [DOI] [PubMed] [Google Scholar]
- 53.Chen X., Pham E., Truong K. TEV protease-facilitated stoichiometric delivery of multiple genes using a single expression vector. Protein Sci. Publ. Protein Soc. 2010;19:2379–2388. doi: 10.1002/pro.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghiaci P., Norbeck J., Larsson C. 2-Butanol and butanone production in Saccharomyces cerevisiae through combination of a B12 dependent dehydratase and a secondary alcohol dehydrogenase using a TEV-based expression system. PloS One. 2014;9:e102774. doi: 10.1371/journal.pone.0102774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kostallas G., Löfdahl P.-Å., Samuelson P. Substrate profiling of tobacco etch virus protease using a novel fluorescence-assisted whole-cell assay. PLoS One. 2011;6:e16136. doi: 10.1371/journal.pone.0016136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temme K., Hill R., Segall-Shapiro T.H., Moser F., Voigt C.A. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012;40:8773–8781. doi: 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Temme K., Zhao D., Voigt C.A. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7085–7090. doi: 10.1073/pnas.1120788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. Publ. Protein Soc. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchler N.E., Gerland U., Hwa T. Nonlinear protein degradation and the function of genetic circuits. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9559–9564. doi: 10.1073/pnas.0409553102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karig D., Weiss R. Signal-amplifying genetic circuit enables in vivo observation of weak promoter activation in the Rhl quorum sensing system. Biotechnol. Bioeng. 2005;89:709–718. doi: 10.1002/bit.20371. [DOI] [PubMed] [Google Scholar]
- 61.Becskei A., Séraphin B., Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sayut D.J., Niu Y., Sun L. Construction and engineering of positive feedback loops. ACS Chem. Biol. 2006;1:692–696. doi: 10.1021/cb6004245. [DOI] [PubMed] [Google Scholar]
- 63.To T.-L., Maheshri N. Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science. 2010;327:1142–1145. doi: 10.1126/science.1178962. [DOI] [PubMed] [Google Scholar]
- 64.Zhao W., Bonem M., McWhite C., Silberg J.J., Segatori L. Sensitive detection of proteasomal activation using the Deg-On mammalian synthetic gene circuit. Nat. Commun. 2014;5:3612. doi: 10.1038/ncomms4612. [DOI] [PubMed] [Google Scholar]
- 65.Bonnet J., Subsoontorn P., Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8884–8889. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonnet J., Yin P., Ortiz M.E., Subsoontorn P., Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 67.Yang L., Nielsen A.A.K., Fernandez-Rodriguez J., McClune C.J., Laub M.T., Lu T.K., Voigt C.A. Permanent genetic memory with >1-byte capacity. Nat. Methods. 2014;11:1261–1266. doi: 10.1038/nmeth.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilgiriwala K.S., Jiménez J., Rivera P.M., Del Vecchio D. Synthetic tunable amplifying buffer circuit in E. coli. ACS Synth. Biol. 2015;4:577–584. doi: 10.1021/sb5002533. [DOI] [PubMed] [Google Scholar]
- 69.Lee T.H., Maheshri N. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol. Syst. Biol. 2012;8:576. doi: 10.1038/msb.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine E., Zhang Z., Kuhlman T., Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Curran K.A., Leavitt J.M., Karim A.S., Alper H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 2013;15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 72.Redden H., Alper H.S. The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 2015;6:7810. doi: 10.1038/ncomms8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchler N.E., Louis M. Molecular titration and ultrasensitivity in regulatory networks. J. Mol. Biol. 2008;384:1106–1119. doi: 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 74.Buchler N.E., Cross F.R. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol. Syst. Biol. 2009;5:272. doi: 10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen D., Arkin A.P. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 2012;8:620. doi: 10.1038/msb.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rhodius V.A., Segall-Shapiro T.H., Sharon B.D., Ghodasara A., Orlova E., Tabakh H., Burkhardt D.H., Clancy K., Peterson T.C., Gross C.A., et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013;9:702. doi: 10.1038/msb.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X., Wu B., Szary J., Kofoed E.M., Schaufele F. Functional sequestration of transcription factor activity by repetitive DNA. J. Biol. Chem. 2007;282:20868–20876. doi: 10.1074/jbc.M702547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathieu J., Warthmann N., Küttner F., Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in arabidopsis. Curr. Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 79.Wehr M.C., Laage R., Bolz U., Fischer T.M., Grünewald S., Scheek S., Bach A., Nave K.-A., Rossner M.J. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 80.Pauli A., Althoff F., Oliveira R.A., Heidmann S., Schuldiner O., Lehner C.F., Dickson B.J., Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams D.J., Puhl H.L., Ikeda S.R. Rapid modification of proteins using a rapamycin-inducible tobacco etch virus protease system. PloS One. 2009;4:e7474. doi: 10.1371/journal.pone.0007474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen D.P., Mahesh M., Elsässer S.J., Hancock S.M., Uttamapinant C., Chin J.W. Genetic Encoding of Photocaged Cysteine Allows Photoactivation of TEV Protease in Live Mammalian Cells. J. Am. Chem. Soc. 2014;136:2240–2243. doi: 10.1021/ja412191m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rakhit R., Navarro R., Wandless T.J. Chemical Biology Strategies for post-translational control of protein function. Chem. Biol. 2014;21:1238–1252. doi: 10.1016/j.chembiol.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olson E.J., Tabor J.J. Post-translational tools expand the scope of synthetic biology. Curr. Opin. Chem. Biol. 2012;16:300–306. doi: 10.1016/j.cbpa.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 85.Lou C., Stanton B., Chen Y.-J., Munsky B., Voigt C.A. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.