Abstract
Many acute and chronic lung diseases could benefit from improved regeneration therapy. In development and throughout life, genetically encoded exposure memory systems allow environmental exposures, diet, and infectious agents to direct subsequent phenotypic adaptation and responses. The impact of such memory systems on lung regeneration is currently unknown. This article provides a brief overview of advances in redox biology and medicine as a framework for elucidating exposure memory and delineating spatiotemporal responses in lung regeneration. New imaging and omics methods enable precise definition to advance knowledge of development and the cumulative changes in lung biochemistry, structure, and cell populations occurring from prior and ongoing exposures. Importantly, conditioning steps may be needed to reverse exposure memory and enable effective regeneration. Thus, to complement developmental biology and regenerative medicine, research programs are needed to gain systematic knowledge of how lifelong exposures impact lung biology and support transition of lung regeneration from hypothetical to practical medicine.
Keywords: oxidative stress, redox code, environmental exposure, occupational exposure, aging
Adult lungs exhibit little cell regeneration in the absence of injury and repair but have capacity to rapidly regenerate new cells after injury (1, 2). Failure of these repair and regeneration systems results in chronic lung dysfunction (2), and improved understanding of these systems may result in improved management and treatment strategies. Means to monitor and support lung repair and regeneration in the absence of overt disease are limited, however, and there is an expectation that understanding cumulative lifelong exposures, such as encompassed in the concept of the exposome, will be essential to understand mechanisms and variations among individuals. Thus, opportunity exists to protect against long-term declines in lung function and decrease morbidity and mortality from lung diseases by mapping out responses to common exposures, delineating molecular impediments to efficient repair and regeneration, and developing preconditioning regimens to enhance specific reparative processes.
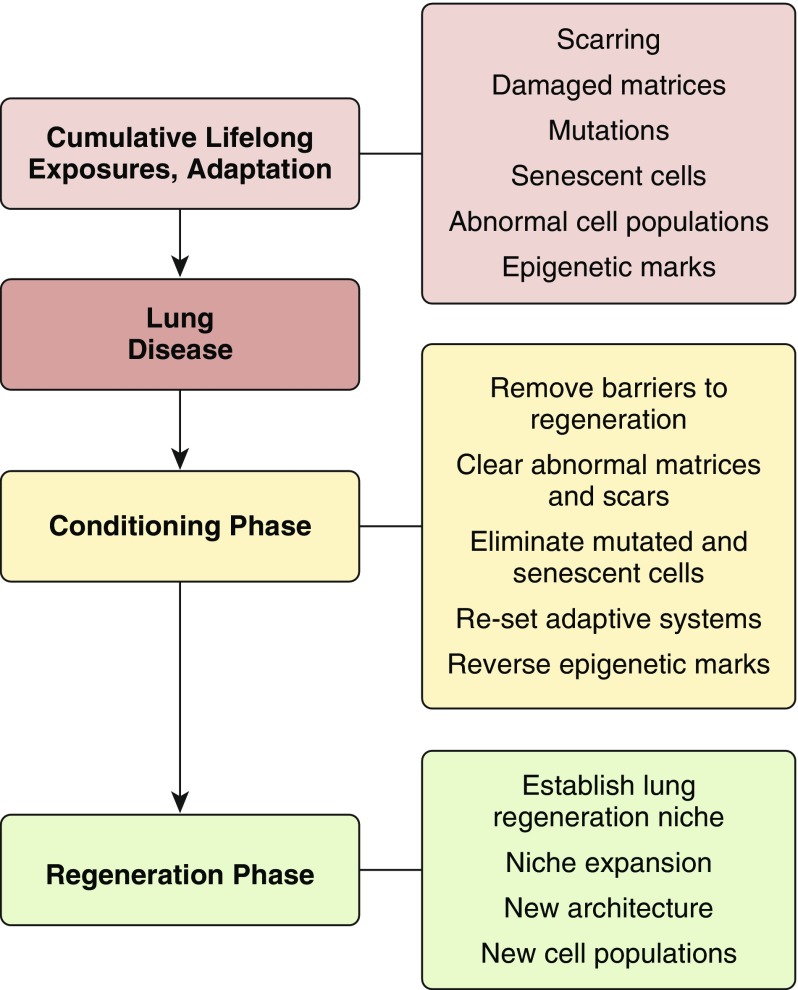
A concept diagram is provided in Figure 1, in which cumulative lifetime exposures (3, 4) and adaptive responses result in lungs with scarring, damaged extracellular matrix, mutations, senescent cells, abnormal cell populations, and epigenetic changes. Some of these contribute to lung disease and/or serve as barriers to regeneration. Regeneration of the diseased lung requires a conditioning phase to remove barriers to regeneration, clear scars and replace dysfunctional extracellular matrix, eliminate mutated and senescent cells, reset adaptive systems, and reverse epigenetic marks. With such conditioning steps, induction of lung regeneration niche and expansion and elaboration of new architecture and cell populations can be pursued to regenerate lung function (5).
Figure 1.
Conceptual diagram for lung regeneration. After accumulation of lifelong exposures and adaptive responses in disease development, a conditioning phase is needed to enable efficient regeneration of lung function.
The complexity of lung anatomy and cell physiology, with approximately 40 cell types and a broad spectrum of lung diseases (6–10), suggests that systematic study will be needed to address the multiple molecular pathways, anatomic diversities, temporal behaviors, and relative intensities of phenotypes in lung regeneration. In addition, the diversity of lung diseases must be addressed. For instance, asthma may be well controlled with infrequent symptoms or uncontrolled and life threatening (11); such differences may reflect underlying differences in exposure history. Respiratory distress syndromes can occur acutely, and prior exposures may determine which repair and regenerative methods are needed to prevent permanent loss of function (12). Similarly, in cystic fibrosis, recurring exacerbations result in long-term lung failure (13), and the lifelong history of exposures may define the need for different conditioning regimens to support regeneration. Improved lung regeneration in this diverse spectrum, as well as in chronic obstructive pulmonary disease, idiopathic pulmonary fibrosis, sarcoidosis, bronchiolitis obliterans, and other chronic lung diseases, will have considerable impact in pulmonary medicine.
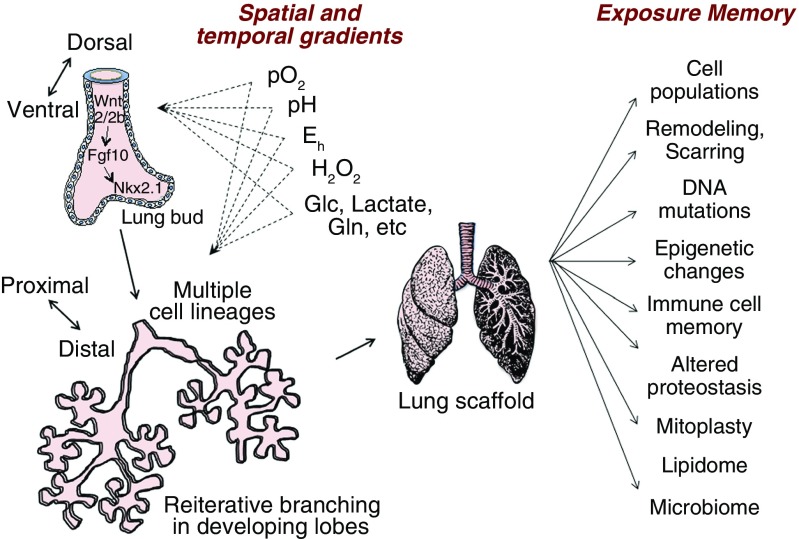
Beers and Morrisey provide an excellent review of lung repair, remodeling, and regeneration, noting a connection between the signaling pathways of lung development and those responding to lung injury (5). They define regeneration as full functional recovery of airway epithelial cell–lined airway and alveolar lined spaces, as opposed to repair, which can include aberrant repopulation, scarring, and fibrosis. They summarize two aspects of developing polarity in organogenesis. Early ventral to dorsal polarity involves Wingless-related MMTV integration site (Wnt) 2/2b → fibroblast growth factor 10 → homeodomain protein gene Nkx2.1 signaling in the mesoderm to delineate the trachea and lungs from the esophagus in the endoderm (Figure 2, left). As discussed below, these processes occur with spatial and temporal redox control, involving O2 delivery, pH control, and H2O2 and redox potential gradients (Figure 2, center). Proximal to distal polarity develops through additional cytokine and growth factor signaling, with extensive and reiterative branching of the endoderm and mulitpotent progenitor cells giving rise to multiple epithelial lineages. Considerable progress has been made in defining the signaling pathways and mapping cell lineages (14).
Figure 2.
Exposure memory and lung regeneration. Signaling processes in development are similar to those in repair and provide strategies for use of chemokines and growth factors in lung regeneration (left). Improved understanding of spatial and temporal redox gradients provides additional strategies to facilitate lung regeneration. These include well-known effects of O2 partial pressure, pH conditions, optimal redox potential, H2O2 signaling, and energy precursor supply. Lung regeneration differs from lung development in that regeneration of lung must accommodate the preexisting lung scaffold (right). Within this scaffold, individual lifelong exposures result in multiple types of exposure memory. The impact of this exposure memory on regeneration is currently unknown. Importantly, exposure memory could impose barriers and require preconditioning for regeneration. Eh = redox potential; Glc = glucose; Gln = glutamine. Based on References 5, 14, 37, and 3. Lung drawing from Pixabay.com.
Although similarities exist between development and post-injury signaling, regeneration after injury differs from lung development in that the latter has a preformed scaffold and occurs within the context of exposure memory (Figure 2, right). The process involves immune cells, proliferation of remaining epithelial cells, and recruitment of different progenitor populations. Prior research shows that fetal alcohol exposure or vitamin C deficiency affects lung cell populations and function in postnatal life (15–17), implying that the character of the scaffold influences regeneration. Systematic knowledge of exposure memory (3, 18) is needed to determine whether conditioning to reverse exposure memory will promote lung regeneration.
This article reviews advances in understanding of oxidative stress and redox biology as a basis to improve strategies for lung regeneration. A brief discussion of newer concepts of oxidants and oxidative stress is followed by consideration of central principles of redox organization as they affect the lung and lung regeneration. We then address emerging research on the exposome and exposure memory systems that enable an organism to adapt during lifetime to environmental exposures. We follow this with more specific consideration of spatial and temporal heterogeneities that occur within aerobic organisms and the implications of such organization for strategies to regenerate lung tissues. Finally, we briefly review progress in integrative omics and the potential to delineate network structures that support normal lung development and regeneration.
Oxidative Stress and Redox Biology
Oxidative stress is an imbalance between oxidants and antioxidants in favor of the oxidants, leading to a disruption of redox signaling and control and/or molecular damage (Box 1) (19). The original definition by Sies (20) encompassed only macromolecular damage, but later discovery of redox signaling mechanisms (21), especially proinflammatory signaling by simple change in the redox potential of the cysteine/cystine couple (22), led to a proposal that oxidative stress should be viewed in terms of disruption of redox signaling and control (21).
Box 1. Terminology for Lung Regeneration
Oxidative stress is defined as an adverse process: An imbalance between oxidants and antioxidants in favor of oxidants, leading to a disruption of redox signaling and control and/or molecular damage. This term is appropriate for conditions causing lung injury but not as a mechanism for regeneration.
Redox signaling is defined as cellular communication involving an oxidant or reductant as a messenger. H2O2 is a common messenger and results in transient, usually reversible oxidation of target proteins.
Redox sensing is defined as biological control processes in which structural organization, trafficking, macromolecular interactions, and complex activities are integrated through redox switches. These are orthogonal to redox signaling and have a sustained time course.
Reactive oxygen species (ROS) is a loosely defined term and should be avoided. Preferred terminology is “oxidant” when uncharacterized and the name of the oxidant when known (e.g., H2O2).
A salient point concerning terminology for regenerative medicine is that oxidative stress is not synonymous with “oxidation.” Oxidation is a chemical term referring to loss of electrons. For every oxidation reaction in biological systems, there is a reduction reaction in which another molecule accepts one or more electrons and becomes reduced. All aerobic life depends on oxidation-reduction reactions for energy supply, so oxidation cannot be equated with oxidative stress. Many biological oxidants, including H2O2, have important biological functions so that increased H2O2 production, such as occurs in redox signaling during development and regeneration, should not be termed “oxidative stress.” A more appropriate description is simply “increased H2O2 production.” Oxidative stress, by definition, always denotes adverse conditions (Box 1).
At nanomolar concentrations, H2O2 regulates redox-dependent cellular signaling mechanisms in development and organ tissue repair. This process is distinct from oxidative stress and is termed “redox signaling,” defined in terms of redox mechanisms of cell communication (Box 1). An overlap of terminologies occurs for wound repair, where oxidative stress can function in redox signaling. Numerous aspects of wound repair, organ adaptation, and regeneration mechanisms (e.g., redox signaling in hypoxia-inducible factor [HIF]-1α regulation of hypoxia [23], redox signaling of growth factor [vascular endothelial growth factor, platelet-derived growth factor, epidermal growth factor, fibroblast growth factor]-dependent cell proliferation) have been studied in detail (24–26). Redox mechanisms also provide an organizational structure for cells through the redox proteome (27, 28). This process has been described in terms of “redox sensing,” defined as mechanisms orthogonal to redox signaling, which control cell organization, trafficking, and functional integration (29). Understanding of oxidative stress, redox sensing, and redox signaling provides a sound foundation to optimize conditions for lung regeneration therapy.
Both kinetic and thermodynamic parameters are important to describe normal cell function and dysfunction caused by oxidants. Thermodynamics describes the energetics of systems, whereas kinetics describes the rates of processes. Living systems exist as kinetically determined, nonequilibrium states, yet individual components can operate under high-flux, near-equilibrium conditions or under kinetically limited, nonequilibrium conditions. The details of these relationships are beyond the scope of the present discussion, but, importantly, the redox potential, Eh, provides a convenient way to compare the tendency of molecules to donate or accept electrons; in such processes, Eh is expressed in millivolts (21). Redox couples with a more negative Eh are better reductants, whereas those with more positive Eh are better oxidants. The biological range spans nearly 1,000 mV; in healthy systems, steady-state redox potentials of donor/acceptor couples are often maintained within 10- to 30-mV ranges; thus, a large positive shift of Eh that disrupts normal function also denotes oxidative stress. In extracellular spaces, Eh is more positive than in cells (e.g., plasma Eh for glutathione (GSH)/glutathione disulfide (GSSG) is −138 mV [30], median epithelial lung lining fluid Eh is −170 mV [31], and lung Eh is −232 mV [32]). Further variations occur within organelles, with mitochondria and nuclei having more negative Eh (more reducing) than cytosol, and the cisternae of the endoplasmic reticulum having more positive Eh (intermediate between cytoplasm and extracellular pools) (33). These values are important because they point to the need to maintain appropriate redox conditions to support lung regeneration.
Finally, a point of confusion surrounds the term “reactive oxygen species” or ROS. The term was introduced at a time when there was an assumption that hydroxyl radicals generated by reaction of superoxide and a hydroperoxide (Fenton reaction 34]). This is a central mechanism of oxidative stress, human aging, and disease. Although this is correct for ionizing radiation, such as used in cancer therapy, this is quantitatively minor relative to the total range of oxidative processes. Although useful for earlier studies of oxidative stress, this use is now discouraged because it provides little useful information. If evidence is not available to show that an oxidant involves oxygen, then the term can be an overinterpretation or misinterpretation. If there is evidence for increased superoxide anion radical, hydroperoxide, or other oxidizing species, then science is best served by use of the more specific chemical terminology, along with the criteria for interpretation. If an oxidant is uncharacterized, it should be described simply as an “oxidant” (Box 1). In development and regeneration of the lung, O2 is an oxidant that has signaling activity through HIF-1α (35, 36). Many other biologic oxidants have been described, and these oxidants function through specific signaling events as well as less specifically in maintaining networks of redox-sensitive elements (27). More precise use of terminology can be expected to aid in delineating relevant mechanisms for lung regeneration.
The Redox Code
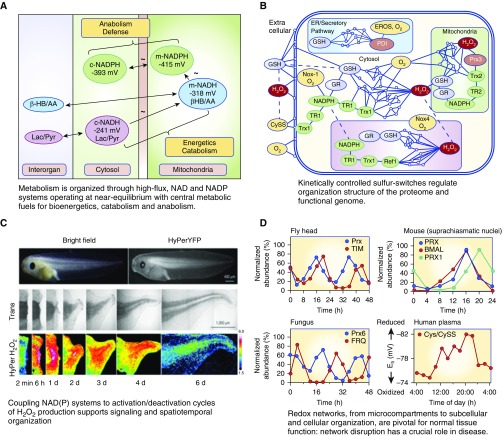
The central principles of redox organization are provided by the redox code (37), where “code” is used to mean a system of principles. These principles help define the molecular logic of redox processes during lung regeneration. Four principles summarize major points of redox theory (i.e., generalizations that describe the redox interactions in bioenergetics, metabolism, and macromolecular structure and function) (Figure 3). The first principle is that metabolism is organized through high-flux, nicotinamide adenine dinucleotide (NAD) and nicotinamide adenine dinucleotide phosphate (NADP) systems operating at near equilibrium with central metabolic fuels for bioenergetics, catabolism, and anabolism (37). This principle provides a foundation for lung regeneration as a critical starting point for understanding the need for specific metabolic fuels to support the energy demands and biosynthetic precursor needs for structural and functional regeneration.
Figure 3.
Principles of the redox code provide rationale for systematic variation of O2, pH, and thiol/disulfide supply systems to optimize regeneration. Reprinted by permission from References 37 (A, B, D) and 49 (C). Cys = cysteine; NAD = nicotinamide adenine dinucleotide; NADH = nicotinamide adenine dinucleotide reduced; NADP = nicotinamide adenine dinucleotide phosphate; NADPH = nicotinamide adenine dinucleotide phosphate reduced; Nox = nicotinamide adenine dinucleotide phosphate reduced oxidase; BMAL = brain and muscle arnt-like; ER = endoplasmic reticulum; EROS = ER oxidase system; FRQ = frequency; GSH = glutathione; GR = glutathione disulfide reductase; HyPer = hydrogen peroxide sensor; HyPerYFP = Hyper tagged with yellow fluorescent protein; PDI = protein disulfide isomerase; Prx = peroxiredoxin; TIM = timeless; TR = thioredoxin reductase; Trx = thioredoxin.
The nicotinamide adenine dinucleotide (NAD) system supports catabolic reactions using dietary fuels (carbohydrates, fats, proteins) and O2 for ATP supply and also accepts electrons from diverse substrates for catabolism and elimination of wastes (38). To achieve these unrelated demands to eliminate wastes while maintaining stable energy supply, the system operates in a near-equilibrium state with major metabolic fuels. This allows elimination processes to proceed at rates dictated by availability of resources for energy demand. Importantly, the cytoplasmic and mitochondrial pools of NADH/NAD+ (reduced/oxidized) are maintained at distinct redox potentials (EhCyt, −240 mV; EhMit, −320 mV) with high-flux transport systems allowing continuous interaction between the compartments (37, 39, 40). Maintenance at proper set points is essential for lung regeneration.
The NADP system is a counterpart to the NAD system, functioning in biosynthetic processes (anabolism), defense, cell signaling, and energy storage. Although the chemical properties of the active niacinamide functional group are identical in NADP and NAD, the specificity of interaction with proteins allows these systems to be poised at different Eh and serve these different biologic functions. To support anabolism and detoxification, the cytoplasmic and mitochondrial NADP systems are maintained at substantially more negative redox potentials (more reducing) than the NAD systems in the same compartments. These values, −390 and −415 mV, respectively, for cytoplasm and mitochondria (37, 40), provide the energy for biosynthesis of nucleotide bases, reduction of ribose to deoxyribose, biosynthesis of fatty acids, heme, and other critical processes of anabolism.
Importantly, both the NAD and NADP systems are under rapid equilibration with oxidizable fuels so that catabolism, anabolism, and related functions are coordinated in subcellular compartments and in different organ systems. This has important implications for lung regeneration because it emphasizes that design of regeneration strategies must support anabolic needs for NADPH. The NAD and NADP systems differ in that a large number of oxidizable substrates can reduce NAD+ to NADH, whereas few oxidizable substrates have a sufficiently negative Eh to support reduction of NADP+ to NADPH under the prevailing Eh values in cells. These include substrates undergoing oxidative decarboxylation, especially glucose 6-phosphate going to ribose 5-phosphate catalyzed by glucose 6-phosphate dehydrogenase (G6PDH) and phosphogluconate dehydrogenase. The rate of glucose use to support NADPH via the pentose phosphate cycle has been estimated to be 20% of the rate of glucose use to support ATP production, indicating that in regenerative processes, the capacity to maintain NADPH supply for biosynthesis may become critical (41, 42). Other mechanisms to support NADPH supply, including an NADP-linked isocitrate dehydrogenase, malic enzyme, or the mitochondrial transhydrogenase, may be essential to regeneration.
The second principle of the redox code is that kinetically controlled sulfur switches regulate structural and functional organization of the proteome and functional genome (Figure 3). Accumulating data suggest that 10 to 20% of the more than 200,000 cysteine (Cys) residues encoded in the human genome undergo reversible oxidation (43), a process linking protein folding, protein activation/deactivation, macromolecular interactions, and directional trafficking to the bioenergetic and metabolic regulation of cells (43). The central organization structure is linked to the bioenergetics systems through mitochondrial H2O2 production and H2O2 generation by NADPH oxidases (Nox) (44, 45). Relatively slow, kinetically controlled oxidation of Cys (46, 47) in transcription factors, actin cytoskeleton, mRNA and microRNA (miRNA) processing and trafficking machinery, molecular chaperones and docking proteins, translation and apoptosis machinery, and enzymes for glycolysis, lipid, and protein metabolism, is countered by thioredoxin- or GSH-dependent reduction, each linked to NADPH, to maintain stable steady states (27, 28). The broad impact of oxidative stress on the redox proteome structure has been delineated in lung fibroblasts for low, environmental levels of cadmium exposure (48). Although incompletely characterized for cell differentiation and lung development, lung regeneration will require that this NADPH-dependent oxidant and reductant supply structure is appropriately maintained.
Parallel links between the bioenergetics and metabolic organization and the macromolecular organization and function are also provided by other bioenergetics components. For instance, acetyl-CoA supports protein acetylation, S-adenosyl-methionine supports methylation, and NAD+ supports ADP-ribosylation of proteins. Sirtuins, involved in histone regulation of gene expression, are regulated by the [NADH]/[NAD+] ratio (4), and are likely to be essential for regeneration programming.
The third principle of the redox code is that coupling of NAD(P) systems to activation/deactivation cycles of H2O2 production supports signaling and spatiotemporal organization (37). Incorporation of this principle into regenerative strategies may facilitate proper polarity and timing, factors that are critical for appropriate structural and functional restoration. Tools to monitor spatiotemporal sequence are available to directly visualize tissue-scale gradient of H2O2, as shown for tadpole tail regeneration (Figure 3) (49). Such spatiotemporal sequencing can be expected for lung regeneration, but specific studies will be needed to discover whether hypoxia-oxygenation cycles under control of HIF-1α, peroxide flux driven by mitochondrial activity, Nox-dependent H2O2 activation/deactivation cycles, or another redox system is used.
The fourth principle of the redox code is that redox networks, from microcompartments to subcellular and cellular organization, form an adaptive system to respond to the environment (37). Redox proteomics and redox metabolomics provide a mechanistic interface between the environment and the functional genome (50). This is important for lung regeneration because it emphasizes the need to consider the environmental causes of lung dysfunction; if these are not eliminated, then any regeneration will ultimately fail. In a broader sense, this principle may present the most daunting aspect of lung regeneration because it implies that regeneration will require both elimination of agents causing disease and removal of the molecular memories of prior exposures.
The Exposome and Exposure Memory
Recognition that the redox proteome and redox metabolome serve as an adaptive interface between an organism and its environment (28) laid the foundation to consider redox biology as a critical mechanistic interface between the functional genome and the exposome (50). As indicated above, this is directly relevant to lung regeneration, because at any point in an individual’s life, lung structure and function retain a history of prior exposures. The exposome, originally defined by Wild (4), is the cumulative measure of environmental influences and associated biological responses throughout the lifespan, including exposures from the environment, diet, behavior, and endogenous processes (51). The logical extension of the redox interface concept focuses attention to the molecular logic of metazoan evolution that gave rise to the mammalian respiratory system (3). A key aspect is the evolution of genetically encoded exposure memory systems (3) (i.e., systems that “remembered” thickness of barrier functions, optimal length of bronchioles and alveolar size for O2 delivery, number and location of mitochondria, sensory input, etc.). These memory systems provide mechanisms for an individual to adapt to environment (e.g., food supply, altitude, etc.) during lifespan to provide competitive advantage (3).
The relevance of exposure memory systems (Figure 2) (3) to lung regeneration is readily apparent. For instance, positive and negative selection in the immune system provides memory of prior exposures. In lung regeneration, preservation of beneficial aspects would be desirable, while at the same time, some of these may antagonize regenerative potential. Similarly, epigenetic mechanisms result in long-term changes in control of gene expression. Some of these are likely to function in opposition to regeneration, and others are likely to provide beneficial adaptations. Other exposure memories include epiproteomic changes (e.g., post-translational modifications, proteolytic processing, crosslinking and aggregation in response to environmental exposures) (52–54); lipidomics changes in cell and organellar membranes and other components of the lipidome; glycomic changes from prior dietary compositions (55, 56), infections (57), and other exposures; and heteroplasmic changes of mitochondria to adapt bioenergetics function to match food availability.
An important opportunity exists to map exposure memory in the lung by systematically evaluating environmental exposures, cell populations, mitochondrial populations, and epigenetic and epiproteomic characteristics that accumulate. For instance, methods are available to measure telomere length in different cell types of human lung and gain an understanding of cell senescence and exhaustion of replicative potential. Methods are available to monitor environmental chemical accumulation in the lungs and lung mitochondria (58), and methods are available to measure chemical adducts of long-lived proteins, DNA mutations in nuclear and mitochondrial DNA, and epigenetic changes of specific cell populations. Medical protocols will be needed to remove or correct the molecular memories that are deleterious and establish or optimize regenerative potential.
Spatial and Temporal Heterogeneities
Advances in understanding growth factor signaling in lung development portend considerable advances in lung regeneration (14). Elucidation of the specific signaling events in the epithelial differentiation, vascularization, mesenchyme, and immune development will enable targeted activation and switching in cell and molecular phenotypes. Yet all of these processes occur naturally within well-controlled microenvironments. Thus, one can expect that effective management of physical and chemical/metabolic environments will be essential for regeneration.
The study by Hwang and Sinskey (59) showed that mammalian cells in culture have Po2, pH, and Eh requirements for optimal growth (60). Tissue and intracellular O2 gradients were shown for mammalian cells in the 1970s and 1980s (61–63). The importance of HIF-1α signaling microvascular development (64) and lung cell phenotypic switching (65, 66) suggests that specific control of Po2 will be critical for lung regeneration. Similarly, intracellular pH control and directional pH gradients could be essential for appropriate growth and differentiation responses. Intracellular pH gradients have been detected (67) but not characterized for lung cells during development. Fluorescent O2 and pH indicators are available to facilitate mapping of these gradients in lung regeneration models, and application of these strategies is likely to aid in understanding mechanistic details to improve regenerative potential.
Although gradients of Po2 and pH have been known for decades, experiments to test whether modulation of gradients affects cell or tissue regeneration appear to be unavailable. In principle, agents that stimulate mitochondrial biogenesis (68) or alter mitochondrial homeostasis (69, 70) can be used to manipulate O2 gradients. Similarly, inhibitors of polarized transporters, such as bicarbonate transporters and H+-coupled cation transporters, can be used to control transcellular pH gradients (67). Evidence for tissue O2 gradients occurs in different organ systems (71), but cellular studies are mostly restricted to differentiated cells. There is an important need for basic research to explore O2 and pH gradient effects in stem cells and organogenesis.
Thiol/disulfide redox gradients are more complex and difficult to map but will also be critical to strategies for lung regeneration. Protein abundance varies over nine orders of magnitude (72), and reactivities of thiols with oxidants such as H2O2 vary over six orders of magnitude (73). If polarity in redox organization is critical for regenerative signaling, then it will be necessary to apply advanced proteomics approaches to characterize regenerative models. Transient expression of targeted reductant proteins, such as nuclear localization signal–thioredoxin-1 (74), similar peroxiredoxin fusion proteins (75), or targeted oxidant proteins, such as nuclear localization signal–D-amino acid oxidase (76), could be used to induce redox gradients during organogenesis or regeneration. This represents a frontier for regenerative medicine, but rapid development of imaging methods suggests that future strategies can take advantage of the new technologies to substantially advance progress.
Integrative Omics
Perhaps the most promising advances for lung regeneration lie in the explosive development of omics technologies. Recent advances in miRNA research are beginning to show the miRNAs that direct and block functional responses in the lung (77). Transcription can be measured on a single-cell basis to enable reconstruction of transcriptional programs (78). Mass spectrometry–based proteomics methods have not yet achieved sensitivity to measure changes in a single cell, but fluorescent fusion proteins, epitope-tagged proteins, and fluorescent enzyme products can be used to map changes in protein abundance, distribution, and catalytic activity at a single-cell level.
Redox proteomics now allows measurement of oxidation of hundreds of specific protein Cys (28, 79, 80). In principle, this can be increased to thousands using existing technologies (81–83). By combining redox proteomics with high-resolution metabolomics, mechanistic links can be obtained from changes in specific proteins with changes in products of associated enzymatic activities. Similarly, transcriptome–metabolome interactions have been used to identify critical hubs in toxicologic mechanisms (84) and show that, in principle, such approaches can be used to define central organizational hubs for tissue and organ regeneration.
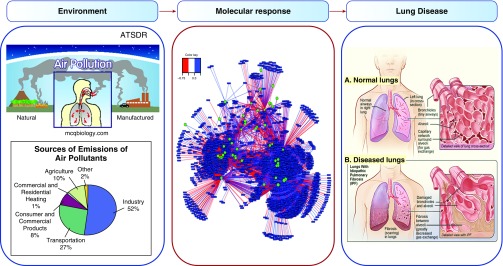
Efforts to sequence the exposome (18) are expected to further facilitate protocol development by providing understanding of molecular and cellular responses to the environment (Figure 4). For instance, components of the microbiome may be activating or inhibitory factors for specific lung cells and/or structures (Figure 4). Recent research on microbiome–metabolome interactions (85) shows that microbial groups associate with metabolic pathways, indicating that microbial populations could affect regeneration potential (Figure 4). Elucidation of miRNA-directed and/or exposure-related epigenetic changes may provide further means to direct lung regeneration. Studies of environmental chemical burden associated with disease may help identify residual chemicals that interfere with lung regeneration. Characterization of long-lived macromolecular structures and barrier remodeling in the lung may lead to new methods to eliminate scarring and diffusion barriers and enhance restoration of function. Similarly, improved understanding of immune cell populations can further be expected to guide elimination of dysfunctional components while retaining beneficial immunologic memory. Importantly, integrative omics tools are now available to directly link these information-rich data toward applications in lung regeneration.
Figure 4.
The cumulative lifelong perturbations in metabolic and cellular functions contribute to lung disease. The lungs are exposed to airborne particulates, environmental chemicals, and infectious agents. Within the lungs, the body burden of these environmental agents causes many types of molecular responses. Central panel reprinted by permission from Reference 85; right panel is from wikipdedia.org for idiopathic pulmonary fibrosis (https://en.wikipedia.org/wiki/Idiopathic_pulmonary_fibrosis). ATSDR = Agency for Toxic Substances and Disease Registry.
Especially during the last decade, increased numbers of omics studies, including genomics, epigenomics, metabolomics, and proteomics, have been used in studies for lung development, remodeling, and regeneration (86–88). In many cases, discovery was limited and partially understood due to single-molecule approaches at the level of cellular and molecular signaling. Therefore, to improve our understanding of the complicated paradigm of lung development and regeneration, increased emphasis on systems biology analyses combining multiple omics studies, including genomics, proteomics, redox proteomics, and metabolomics with bioinformatics analysis will be needed. The results of this powerful approach will provide the critical pathways and associated molecules to establish a systems-level understanding of lung regeneration.
Summary and Perspective
Improved use of terminology for oxidative stress will facilitate communication and critical thinking concerning challenges of lung regeneration. Simple concepts of an imbalance of prooxidants and antioxidants have been replaced by more realistic descriptions in terms of redox biology. Central principles of redox biology are now enumerated and provide a complement to molecular signaling needed for strategies leading to successful recovery of lung function. These principles provide a central logic for regeneration model development. Bioenergetic and metabolic organization is linked to macromolecular structure and controlled through activation–deactivation cycles of oxidant generation. Characterization of the redox switches and spatiotemporal organization in the developing lung with ultrahigh-resolution omics methods will provide the knowledge base to support systematic improvement of lung regeneration methods and outcomes. A relatively unexplored barrier to successful recovery of lung function lies in the complex and poorly defined exposure memories of prior environmental, dietary, and infectious agents. Like many great challenges, lung regeneration must be viewed realistically, one step at a time. Emerging knowledge and new tool development support optimism that systematic use of combined molecular and redox control procedures will enable precise and effective restoration of diseased lungs.
Footnotes
Supported by National Institutes of Health grants ES023485, ES025632, HL113451, and AG038746.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Demoly P, Simony-Lafontaine J, Chanez P, Pujol JL, Lequeux N, Michel FB, Bousquet J. Cell proliferation in the bronchial mucosa of asthmatics and chronic bronchitics. Am J Respir Crit Care Med. 1994;150:214–217. doi: 10.1164/ajrccm.150.1.7912988. [DOI] [PubMed] [Google Scholar]
- 2.Akram KM, Patel N, Spiteri MA, Forsyth NR. Lung regeneration: endogenous and exogenous stem cell mediated therapeutic approaches. Int J Mol Sci. 2016;17:E128. doi: 10.3390/ijms17010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones DP. Redox theory of aging. Redox Biol. 2015;5:71–79. doi: 10.1016/j.redox.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 5.Beers MF, Morrisey EE. The three R’s of lung health and disease: repair, remodeling, and regeneration. J Clin Invest. 2011;121:2065–2073. doi: 10.1172/JCI45961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, He J, Wei J, Cho WC, Liu X. Diversity of epithelial stem cell types in adult lung. Stem Cells Int. 2015;2015:728307. doi: 10.1155/2015/728307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolt MW, Racz WJ, Brien JF, Bray TM, Massey TE. Differential susceptibilities of isolated hamster lung cell types to amiodarone toxicity. Can J Physiol Pharmacol. 1998;76:721–727. doi: 10.1139/cjpp-76-7-8-721. [DOI] [PubMed] [Google Scholar]
- 8.Kaminsky DA, Irvin CG, Sterk PJ. Complex systems in pulmonary medicine: a systems biology approach to lung disease. J Appl Physiol (1985) 2011;110:1716–1722. doi: 10.1152/japplphysiol.01310.2010. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso WV, Whitsett JA. Resident cellular components of the lung: developmental aspects. Proc Am Thorac Soc. 2008;5:767–771. doi: 10.1513/pats.200803-026HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters SP, Ferguson G, Deniz Y, Reisner C. Uncontrolled asthma: a review of the prevalence, disease burden and options for treatment. Respir Med. 2006;100:1139–1151. doi: 10.1016/j.rmed.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 12.Del Sorbo L, Slutsky AS. Acute respiratory distress syndrome and multiple organ failure. Curr Opin Crit Care. 2011;17:1–6. doi: 10.1097/MCC.0b013e3283427295. [DOI] [PubMed] [Google Scholar]
- 13.Amadori A, Antonelli A, Balteri I, Schreiber A, Bugiani M, De Rose V. Recurrent exacerbations affect FEV(1) decline in adult patients with cystic fibrosis. Respir Med. 2009;103:407–413. doi: 10.1016/j.rmed.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 14.Kotton DN, Morrisey EE. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Probyn ME, Cuffe JS, Zanini S, Moritz KM. The effects of low-moderate dose prenatal ethanol exposure on the fetal and postnatal rat lung. J Dev Orig Health Dis. 2013;4:358–367. doi: 10.1017/S2040174413000305. [DOI] [PubMed] [Google Scholar]
- 16.Gauthier TW, Ping XD, Harris FL, Wong M, Elbahesh H, Brown LA. Fetal alcohol exposure impairs alveolar macrophage function via decreased glutathione availability. Pediatr Res. 2005;57:76–81. doi: 10.1203/01.PDR.0000149108.44152.D3. [DOI] [PubMed] [Google Scholar]
- 17.Brown LA, Harris FL, Jones DP. Ascorbate deficiency and oxidative stress in the alveolar type II cell. Am J Physiol. 1997;273:L782–L788. doi: 10.1152/ajplung.1997.273.4.L782. [DOI] [PubMed] [Google Scholar]
- 18.Jones DP. Sequencing the exposome: a call to action. Toxicol Rep. 2016;3:29–45. doi: 10.1016/j.toxrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sies H, Jones DP. San Diego: Academic Press; 2007. pp. 45–47. Oxidative stress. In: Fink G, editor. Encyclopedia of stress, 2nd ed. [Google Scholar]
- 20.Sies H. editor. Oxidative stress. London: Academic Press; 1985. p. 1e8. [Google Scholar]
- 21.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 22.Go YM, Jones DP. Intracellular proatherogenic events and cell adhesion modulated by extracellular thiol/disulfide redox state. Circulation. 2005;111:2973–2980. doi: 10.1161/CIRCULATIONAHA.104.515155. [DOI] [PubMed] [Google Scholar]
- 23.Semenza GL, Prabhakar NR. HIF-1-dependent respiratory, cardiovascular, and redox responses to chronic intermittent hypoxia. Antioxid Redox Signal. 2007;9:1391–1396. doi: 10.1089/ars.2007.1691. [DOI] [PubMed] [Google Scholar]
- 24.Ushio-Fukai M. VEGF signaling through NADPH oxidase-derived ROS. Antioxid Redox Signal. 2007;9:731–739. doi: 10.1089/ars.2007.1556. [DOI] [PubMed] [Google Scholar]
- 25.Simon AR, Takahashi S, Severgnini M, Fanburg BL, Cochran BH. Role of the JAK-STAT pathway in PDGF-stimulated proliferation of human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1296–L1304. doi: 10.1152/ajplung.00315.2001. [DOI] [PubMed] [Google Scholar]
- 26.Goldkorn T, Balaban N, Matsukuma K, Chea V, Gould R, Last J, Chan C, Chavez C. EGF-receptor phosphorylation and signaling are targeted by H2O2 redox stress. Am J Respir Cell Mol Biol. 1998;19:786–798. doi: 10.1165/ajrcmb.19.5.3249. [DOI] [PubMed] [Google Scholar]
- 27.Go YM, Chandler JD, Jones DP. The cysteine proteome. Free Radic Biol Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Go YM, Jones DP. The redox proteome. J Biol Chem. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DP. Redox sensing: orthogonal control in cell cycle and apoptosis signalling. J Intern Med. 2010;268:432–448. doi: 10.1111/j.1365-2796.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DP, Carlson JL, Mody VC, Cai J, Lynn MJ, Sternberg P. Redox state of glutathione in human plasma. Free Radic Biol Med. 2000;28:625–635. doi: 10.1016/s0891-5849(99)00275-0. [DOI] [PubMed] [Google Scholar]
- 31.Yeh MY, Burnham EL, Moss M, Brown LA. Chronic alcoholism alters systemic and pulmonary glutathione redox status. Am J Respir Crit Care Med. 2007;176:270–276. doi: 10.1164/rccm.200611-1722OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Go YM, Kang SM, Roede JR, Orr M, Jones DP. Increased inflammatory signaling and lethality of influenza H1N1 by nuclear thioredoxin-1. Plos One. 2011;6:e18918. doi: 10.1371/journal.pone.0018918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsou TC, Yang JL. Formation of reactive oxygen species and DNA strand breakage during interaction of chromium (III) and hydrogen peroxide in vitro: evidence for a chromium (III)-mediated Fenton-like reaction. Chem Biol Interact. 1996;102:133–153. doi: 10.1016/s0009-2797(96)03740-4. [DOI] [PubMed] [Google Scholar]
- 35.Whyte MK, Walmsley SR. The regulation of pulmonary inflammation by the hypoxia-inducible factor-hydroxylase oxygen-sensing pathway. Ann Am Thorac Soc. 2014;11:S271–S276. doi: 10.1513/AnnalsATS.201403-108AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karuppagounder SS, Alim I, Khim SJ, Bourassa MW, Sleiman SF, John R, Thinnes CC, Yeh TL, Demetriades M, Neitemeier S, et al. Therapeutic targeting of oxygen-sensing prolyl hydroxylases abrogates ATF4-dependent neuronal death and improves outcomes after brain hemorrhage in several rodent models. Sci Transl Med. 2016;8:328ra29. doi: 10.1126/scitranslmed.aac6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DP, Sies H. The redox code. Antioxid Redox Signal. 2015;23:734–746. doi: 10.1089/ars.2015.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sies H, Stahl W, Sundquist AR. Antioxidant functions of vitamins: vitamins E and C, beta-carotene, and other carotenoids. Ann N Y Acad Sci. 1992;669:7–20. doi: 10.1111/j.1749-6632.1992.tb17085.x. [DOI] [PubMed] [Google Scholar]
- 39.Bücher T, Brauser B, Conze A, Klein F, Langguth O, Sies H. State of oxidation-reduction and state of binding in the cytosolic NADH-system as disclosed by equilibration with extracellular lactate-pyruvate in hemoglobin-free perfused rat liver. Eur J Biochem. 1972;27:301–317. doi: 10.1111/j.1432-1033.1972.tb01840.x. [DOI] [PubMed] [Google Scholar]
- 40.Sies H.editor. Nicotinamide nucleotide compartmentation London: Academic Press; 1982205–231. [Google Scholar]
- 41.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gumaa KA, McLean P. The pentose phosphate pathway of glucose metabolism: enzyme profiles and transient and steady-state content of intermediates of alternative pathways of glucose metabolism in Krebs ascites cells. Biochem J. 1969;115:1009–1029. doi: 10.1042/bj1151009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 45.Lassègue B, Sorescu D, Szöcs K, Yin Q, Akers M, Zhang Y, Grant SL, Lambeth JD, Griendling KK. Novel gp91(phox) homologues in vascular smooth muscle cells : nox1 mediates angiotensin II-induced superoxide formation and redox-sensitive signaling pathways. Circ Res. 2001;88:888–894. doi: 10.1161/hh0901.090299. [DOI] [PubMed] [Google Scholar]
- 46.Lim JB, Huang BK, Deen WM, Sikes HD. Analysis of the lifetime and spatial localization of hydrogen peroxide generated in the cytosol using a reduced kinetic model. Free Radic Biol Med. 2015;89:47–53. doi: 10.1016/j.freeradbiomed.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic Biol Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Go YM, Orr M, Jones DP. Actin cytoskeleton redox proteome oxidation by cadmium. Am J Physiol Lung Cell Mol Physiol. 2013;305:L831–L843. doi: 10.1152/ajplung.00203.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Go YM, Jones DP. Redox biology: interface of the exposome with the proteome, epigenome and genome. Redox Biol. 2014;2:358–360. doi: 10.1016/j.redox.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller GW, Jones DP. The nature of nurture: refining the definition of the exposome. Toxicol Sci. 2014;137:1–2. doi: 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basha MR, Murali M, Siddiqi HK, Ghosal K, Siddiqi OK, Lashuel HA, Ge YW, Lahiri DK, Zawia NH. Lead (Pb) exposure and its effect on APP proteolysis and Abeta aggregation. FASEB J. 2005;19:2083–2084. doi: 10.1096/fj.05-4375fje. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Dong Z, Nomura M, Zhong S, Chen N, Bode AM, Dong Z. Signal transduction pathways involved in phosphorylation and activation of p70S6K following exposure to UVA irradiation. J Biol Chem. 2001;276:20913–20923. doi: 10.1074/jbc.M009047200. [DOI] [PubMed] [Google Scholar]
- 54.Xue P, Hou Y, Zhang Q, Woods CG, Yarborough K, Liu H, Sun G, Andersen ME, Pi J. Prolonged inorganic arsenite exposure suppresses insulin-stimulated AKT S473 phosphorylation and glucose uptake in 3T3-L1 adipocytes: involvement of the adaptive antioxidant response. Biochem Biophys Res Commun. 2011;407:360–365. doi: 10.1016/j.bbrc.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An HJ, Kronewitter SR, de Leoz ML, Lebrilla CB. Glycomics and disease markers. Curr Opin Chem Biol. 2009;13:601–607. doi: 10.1016/j.cbpa.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sierpina VS, Murray RK. Glyconutrients: the state of the science and the impact of glycomics. Explore (NY) 2006;2:488–494; quiz 495–497. doi: 10.1016/j.explore.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 57.Gui HL, Gao CF, Wang H, Liu XE, Xie Q, Dewaele S, Wang L, Zhuang H, Contreras R, Libert C, et al. Altered serum N-glycomics in chronic hepatitis B patients. Liver Int. 2010;30:259–267. doi: 10.1111/j.1478-3231.2009.02170.x. [DOI] [PubMed] [Google Scholar]
- 58.Go YM, Uppal K, Walker DI, Tran V, Dury L, Strobel FH, Baubichon-Cortay H, Pennell KD, Roede JR, Jones DP. Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods Mol Biol. 2014;1198:43–73. doi: 10.1007/978-1-4939-1258-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang C, Sinskey AJ. The role of oxidation-reduction potential in monitoring growth of cultured mammalian cells. In: Spier RE, Griffiths JB, Meignier B, editors. Production of biologicals from animal cells in culture. City; Halley Court publisher; Oxford. 1991. pp. 548–568. [Google Scholar]
- 60.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 61.Jones DP. Effect of mitochondrial clustering on O2 supply in hepatocytes. Am J Physiol. 1984;247:C83–C89. doi: 10.1152/ajpcell.1984.247.1.C83. [DOI] [PubMed] [Google Scholar]
- 62.Jones DP, Mason HS. Gradients of O2 concentration in hepatocytes. J Biol Chem. 1978;253:4874–4880. [PubMed] [Google Scholar]
- 63.Sies H. Oxygen gradients during hypoxic steady states in liver: urate oxidase and cytochrome oxidase as intracellular O2 indicators. Hoppe Seylers Z Physiol Chem. 1977;358:1021–1032. doi: 10.1515/bchm2.1977.358.2.1021. [DOI] [PubMed] [Google Scholar]
- 64.Jiang X, Malkovskiy AV, Tian W, Sung YK, Sun W, Hsu JL, Manickam S, Wagh D, Joubert LM, Semenza GL, et al. Promotion of airway anastomotic microvascular regeneration and alleviation of airway ischemia by deferoxamine nanoparticles. Biomaterials. 2014;35:803–813. doi: 10.1016/j.biomaterials.2013.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agustí A. Phenotypes and disease characterization in chronic obstructive pulmonary disease: toward the extinction of phenotypes? Ann Am Thorac Soc. 2013;10:S125–S130. doi: 10.1513/AnnalsATS.201303-055AW. [DOI] [PubMed] [Google Scholar]
- 66.Agusti A, Sobradillo P, Celli B. Addressing the complexity of chronic obstructive pulmonary disease: from phenotypes and biomarkers to scale-free networks, systems biology, and P4 medicine. Am J Respir Crit Care Med. 2011;183:1129–1137. doi: 10.1164/rccm.201009-1414PP. [DOI] [PubMed] [Google Scholar]
- 67.Aw TY, Jones DP. Heterogeneity of pH in the aqueous cytoplasm of renal proximal tubule cells. FASEB J. 1989;3:52–58. doi: 10.1096/fasebj.3.1.2910737. [DOI] [PubMed] [Google Scholar]
- 68.Rasbach KA, Schnellmann RG. Isoflavones promote mitochondrial biogenesis. J Pharmacol Exp Ther. 2008;325:536–543. doi: 10.1124/jpet.107.134882. [DOI] [PubMed] [Google Scholar]
- 69.Corona JC, Duchen MR. Impaired mitochondrial homeostasis and neurodegeneration: towards new therapeutic targets? J Bioenerg Biomembr. 2015;47:89–99. doi: 10.1007/s10863-014-9576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Busanello EN, Amaral AU, Tonin AM, Zanatta A, Viegas CM, Vargas CR, Wajner M. Disruption of mitochondrial homeostasis by phytanic acid in cerebellum of young rats. Cerebellum. 2013;12:362–369. doi: 10.1007/s12311-012-0426-y. [DOI] [PubMed] [Google Scholar]
- 71.Sies H. Cytochrome oxidase and urate oxidase as intracellular O2 indicators in studies of O2 gradients during hypoxia in liver. Adv Exp Med Biol. 1977;94:561–566. doi: 10.1007/978-1-4684-8890-6_75. [DOI] [PubMed] [Google Scholar]
- 72.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 73.Nagy P, Winterbourn CC. Redox chemistry of biological thiols. San Diego: Elsevier: 2010. [Google Scholar]
- 74.Go YM, Orr M, Jones DP. Increased nuclear thioredoxin-1 potentiates cadmium-induced cytotoxicity. Toxicol Sci. 2013;131:84–94. doi: 10.1093/toxsci/kfs271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hansen JM, Moriarty-Craige S, Jones DP. Nuclear and cytoplasmic peroxiredoxin-1 differentially regulate NF-kappaB activities. Free Radic Biol Med. 2007;43:282–288. doi: 10.1016/j.freeradbiomed.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Halvey PJ, Hansen JM, Johnson JM, Go YM, Samali A, Jones DP. Selective oxidative stress in cell nuclei by nuclear-targeted D-amino acid oxidase. Antioxid Redox Signal. 2007;9:807–816. doi: 10.1089/ars.2007.1526. [DOI] [PubMed] [Google Scholar]
- 77.Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ. 2013;3:315–328. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Go YM, Roede JR, Orr M, Liang Y, Jones DP. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of cd toxicity. Toxicol Sci. 2014;139:59–73. doi: 10.1093/toxsci/kfu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Go YM, Roede JR, Walker DI, Duong DM, Seyfried NT, Orr M, Liang Y, Pennell KD, Jones DP. Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems. Mol Cell Proteomics. 2013;12:3285–3296. doi: 10.1074/mcp.M113.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 82.Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M.System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap Mol Cell Proteomics 201211M111.013722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR., III Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roede JR, Uppal K, Park Y, Tran V, Jones DP. Transcriptome–metabolome wide association study (TMWAS) of maneb and paraquat neurotoxicity reveals network level interactions in toxicologic mechanism. Toxicol Rep. 2014;1:435–444. doi: 10.1016/j.toxrep.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cribbs SK, Uppal K, Li S, Jones DP, Huang L, Tipton L, Fitch A, Greenblatt RM, Kingsley L, Guidot DM, et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4:3. doi: 10.1186/s40168-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonvillain RW, Danchuk S, Sullivan DE, Betancourt AM, Semon JA, Eagle ME, Mayeux JP, Gregory AN, Wang G, Townley IK, et al. A nonhuman primate model of lung regeneration: detergent-mediated decellularization and initial in vitro recellularization with mesenchymal stem cells. Tissue Eng Part A. 2012;18:2437–2452. doi: 10.1089/ten.tea.2011.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hagood JS, Ambalavanan N. Systems biology of lung development and regeneration: current knowledge and recommendations for future research. Wiley Interdiscip Rev Syst Biol Med. 2013;5:125–133. doi: 10.1002/wsbm.1205. [DOI] [PubMed] [Google Scholar]
- 88.Yang G, Biswasa C, Lin QS, La P, Namba F, Zhuang T, Muthu M, Dennery PA. Heme oxygenase-1 regulates postnatal lung repair after hyperoxia: role of β-catenin/hnRNPK signaling. Redox Biol. 2013;1:234–243. doi: 10.1016/j.redox.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]